Highlights
-
•
LC targeted multiplexed SRM/MS system is very sensitive for the identification of many Pls species.
-
•
This system could identify low abundance Pls species.
-
•
This system could identify true peak of Pls species from multiple peaks in the transition used for quantification.
-
•
This system could quantify the concentration of different Pls species.
Abbreviations: CS, commercial standard; IS, internal standard; LC, liquid chromatography; MS/MS, tandem mass spectrometry; MTBE, methyl tert-butyl ether; PLs, glycerophospholipids; Pls, plasmalogens; Pls-PC, plasmalogens choline; Pls-PE, plasmalogens ethanolamine; PUFAs, polyunsaturated fatty acids; RT, retention time; SRM, selected reaction monitoring
Keywords: LC-MS/MS, Targeted multiplexed SRM/MS‘, Plasmalogens, Phospholipids, Identification, Quantification
Abstract
Plasmalogens (Pls) levels are reported to be altered in several neurological and metabolic diseases. Identification of sn-1 fatty alcohols and sn-2 fatty acids of different Pls species is necessary to determine the roles and mechanisms of action of Pls in different diseases. Previously, full-scan tandem mass spectrometry (MS/MS) was used for this purpose but is not effective for low-abundance Pls species. Recently, multiplexed selected reaction monitoring MS (SRM/MS) was found to be more selective and sensitive than conventional full-scan MS/MS for the identification of low-abundance compounds. In the present study, we developed a liquid chromatography (LC)-targeted multiplexed SRM/MS system for the identification and quantification of different Pls choline (Pls-PC) and Pls ethanolamine (Pls-PE) species. We determined five precursor-product ion transitions to identify sn-1 and sn-2 fragments of each Pls species. Consequently, sn-1 and sn-2 fatty acyl chains of 22 Pls-PC and 55 Pls-PE species were identified in mouse brain samples. Among them, some species had C20:0 and C20:1 fatty alcohols at the sn-1 position. For quantification of Pls species in mouse brain samples, a single SRM transition was employed. Thus, our results suggest that the LC-targeted multiplexed SRM/MS system is very sensitive for the identification and quantification of low-abundance lipids such as Pls, and is thus expected to make a significant contribution to basic and clinical research in this field in the future.
1. Introduction
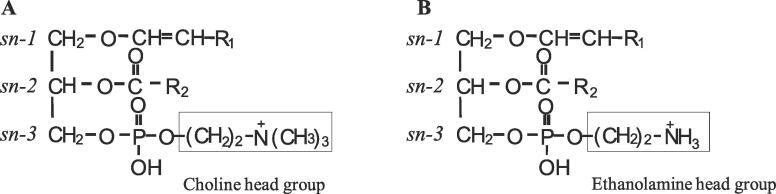
Plasmalogens (Pls) are important glycerophospholipids (PLs) with 1-O-vinyl ether bonds at their sn-1 position (Fig. 1). The fatty alcohols of Pls at the sn-1 position typically consist of C16:0 (palmitoyl), C18:0 (stearoyl), or C18:1 (oleoyl) carbon chains. The sn-2 position is occupied by various fatty acids, including polyunsaturated fatty acids (PUFAs) [[1], [2]]. In mammalian cells, most of the Pls head groups are either ethanolamine (Pls-PE) (Fig. 1B) or choline (Pls-PC) (Fig. 1A). Pls-PE levels are higher than those of Pls-PC in most tissues except the heart [3]. Recent studies have revealed that Pls are involved in many biological functions, such as membrane formation and fusion [[4], [5], [6]], antioxidation [[7], [8]], phagocytosis [9], neuronal and lymphatic cell survivability [[10], [11]], and organization and stability of lipid rafts [12]. Patients with peroxisomal diseases [[13], [14]], neurodegenerative diseases [[15], [16], [17]], and cancer [[18], [19]] were shown to have altered levels of Pls. Although the importance of Pls in biological systems is becoming apparent, corresponding analysis at the molecular species level remains difficult.
Fig. 1.
Chemical structure of target plasmalogen (Pls). Fatty alcohols are present in the sn-1 position, whereas the sn-2 position is occupied by fatty acids. The structure of Pls-PC with a choline head group at the sn-3 position (A). The structure of Pls-PE with an ethanolamine head group at the sn-3 position (B).
In the past, Pls analysis was performed using gas chromatography (GC) [20] or liquid chromatography (LC) [[21], [22]]. However, these methods do not offer the capability to distinguish individual molecular species from other PLs or to identify the compositions of fatty acyl chains at the sn-1 and sn-2 positions. Since 2004, full-scan tandem mass spectrometry (MS/MS) [[16], [23], [24]] has been used as an alternative to previous methods. Although full-scan MS/MS analysis has high resolution and mass precision, it cannot characterize and quantify low-abundance Pls species. For example, the amount of Pls-PC is 20–30 times less than that of diacyl-PC in plasma [25], which makes corresponding analysis difficult. Subsequently, selected reaction monitoring (SRM) analysis has become popular for the quantification of Pls [[26], [27]]. The low noise level of this SRM analysis results in considerably higher sensitivity than that of conventional full-scan MS/MS analysis [28]. However, this single SRM analysis also presents some limitations. For example, the coexistence of lipids with the same m/z in biological samples makes their identification very complicated. Additionally, this analysis is unable to distinguish diacyl-, alkyl-acyl-, and Pls-type species. To determine the target identity, confirmation by full-scan MS/MS analysis [27] and/or pretreatment with acid hydrolysis [24] are required. Therefore, it is necessary to develop a simple method that can simultaneously identify and quantify Pls species with multiple SRM/MS analyses.
Recently, SRM/MS analysis with multiple transitions, namely, LC-targeted multiplexed SRM/MS systems, has received much attention for both identification and quantification in MS-based targeted proteomics [[29], [30], [31]]. This analysis targets multiple preselected precursor-product ion transitions for each analyte. Multiple filtering stages with several structurally specific product ions of the target compound improve the selectivity and reliability by reducing the probability of false positive identification. Because compounds containing the same m/z produce different structural ions due to differences in molecular structure, only structure-specific ions are detected after multiple transitions analysis [32]. However, a previous study reported [28] that the selection of multiple precursor-product ion transitions is the major challenge for the development of this system. For this purpose, laboratory-based discovery experiments are conducted to determine the target analyte. However, this experiment-based method is not effective for low-abundance compounds. The complex backgrounds of biological samples compromise the reliability of MS/MS spectra of low-abundance analytes. An alternative way to overcome this problem is through the use of information collected in a database. Here, we selected five precursor-product ion transitions for each Pls species based on the information in previous full-scan MS/MS-based studies [[23], [24]] or in the LipidBlast in-silico MS/MS database (MSMS-prediction-distribute-v49) [2].
Many Pls molecules have the same molecular weight and composition as other PLs. The LC-targeted multiplexed SRM/MS system is expected to be useful for the identification and analysis of these low-abundance lipids. In this study, we established and validated a method for both the identification and quantification of Pls species using this system.
2. Material and methods
2.1. Chemicals and reagents
The commercial standards (CSs) 1-(1Z-octadecenyl)-2-oleoyl-sn-glycero-3-phosphocholine (PC (P-36:1) (P-18:0/18:1) or C18(plasm-18:1 PC) and the internal standards (ISs) 1-pentadecanoyl-2-oleoyl(d7)-sn-glycero-3-phosphocholine (PC (d7–33:1)) and 1-pentadecanoyl-2-oleoyl(d7)-sn-glycero-3-phosphoethanolamine (PE (d7–33:1)) were purchased from Avanti Polar Lipids (Alabaster, AL). LC-MS grade ammonium formate and ammonium acetate were purchased from Sigma-Aldrich (St. Louis, USA). LC-MS grade methanol and methyl tert-butyl ether (MTBE) were obtained from Wako Pure Chemical Industries (Osaka, Japan).
2.2. Animal experiments
Male wild-type mice of AD model mouse, J20 [33] (3 months old) were used in this study. These mice were kept in fasting for 6 h before sample collection. The study was approved by the Ethical Committee of Shimane University School of Medicine (approval number: IZ29–28). All animal experimental procedures were performed following the guidelines and the regulations for experimentation at Shimane University.
2.3. Sample preparation
PLs were extracted according to a previously reported lipid extraction method with some modifications [34]. Briefly, 10 mg of weighed white matter tissue from the mouse brain was homogenized in 462 μL ice-cold methanol containing ISs (133 pmol PC (d7–33:1) and 282 pmol PE (d7–33:1)). Each homogenized sample was centrifuged at 6000 rpm for 2 min at 4 °C, and the supernatant was collected. Then, 1540 μL MTBE was added and incubated overnight with shaking at room temperature. Afterward, 128 μL of 0.15 M ammonium acetate solvent was added to 1/3 of the supernatant and centrifuged at 3000 rpm for 15 min at 4 °C. The upper organic layer was collected, and the bottom layer was re-extracted with 411 μL solvent mixture (10:3:2.5 MTBE:methanol:0.15 M ammonium acetate, by volume). Finally, the collected lipid extract was dried under a vacuum evaporator and stored at −80 °C until LC-MS analysis. The dried lipid extract was dissolved in 667.3 μL of methanol/10 mM ammonium formate (9:1, by volume) before LC-MS analysis.
2.4. LC-MS/MS conditions
A Shimadzu HPLC system coupled to a triple quadrupole mass spectrometer equipped with an electrospray ionization (ESI) source (Nexera X2, LCMS-8030, Shimadzu Co., Kyoto, Japan) was used. The chromatograph consisted of a DGU-14 AM degasser, an LC-10AD pump, a SIL-HTC autosampler (held at 4 °C), and a thermostatted column compartment. The ESI source settings were as follows: heat-block and dissolved-line temperatures, 400 °C and 250 °C, respectively, and nebulizer and drying gas flow rates, 2 and 15 L/min, respectively. Nitrogen gas was used as the collision and nebulizer gas. The mobile phase was 10 mM ammonium formate (solvent A) and 100% methanol (solvent B). A sample was loaded onto a YMC-Triart C18 analytical column (3 μm, 2.0 × 100 mm, YMC Co., Kyoto, Japan) with a guard cartridge held at 40 °C. Separation was performed using a binary gradient system at a flow rate of 0.2 mL/min. The gradient program was as follows: 0 min, 80% B; 8 min, 100% B; 15 min, 100% B; 15.1 min, 80% B; and 25 min, 80% B.
Data acquisition and analyses were performed using LabSolutions software (version 5.60SP2 software; Shimadzu). The quantification of Pls-PC and Pls-PE was performed with the calibration curves of PC (d7–33:1) (753.6 > 184) and PE (d7–33:1) (711.6 > 570.5), respectively. To eliminate the matrix effect, the area of each target was corrected by the IS area ratio of sample to standard. The quantitative values were determined using the corrected area with IS calibration curves. The measurement point of PC (d7–33:1) was 6, from 0.00014 to 0.4256 pmol/2 µL injection, and that of PE (d7–33:1) was 5, from 0.00072 to 0.45120 pmol/2 µL injection. The peak areas of all analyte Pls species were within the range of the calibration curve. Finally, amounts were expressed as nmol/mg of tissue sample.
2.5. Statistical analysis
The collected data were analyzed using Microsoft Excel. All quantitative data are presented as the mean ± SD (standard deviation). Analyses and figure preparation were performed using Microsoft Excel.
3. Results
3.1. Identification of Pls-PC by targeted multiplexed SRM/MS analysis
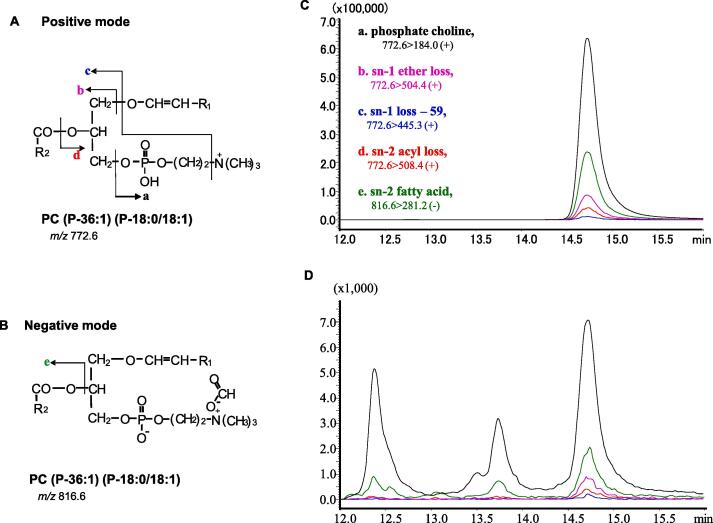
First, we identified and characterized the commercial standard (CS) PC (P-36:1) (P-18:0/18:1) (Fig. 2). We selected the precursor ion m/z 772.6 [M + H]+ in the positive mode (Fig. 2A) or m/z 816.6 [M + 45]− in the negative mode (Fig. 2B), as previously reported [35]. Four product ions of m/z 184.0, 504.4, 445.3, and 508.4 (Fig. 2Aa-d) in the positive mode [23] and one product ion at m/z 281.2 (Fig. 2Be) in the negative mode were chosen. These product ions were identified as phosphate-choline, sn-1 ether loss, sn-1 loss −59, sn-2 acyl loss, and sn-2 fatty acid carboxylate ions in the LipidBlast in-silico MS/MS database (MSMS-prediction-distribute-v49) (Fig. 2A and B) [2]. Next, the parameters for each transition, such as collision energy (CE), were optimized (Table 1). Analysis of the CS with five SRM transitions showed that all peaks were detected at the same retention time (RT) (Fig. 2C).
Fig. 2.
Identification of PC (P-36:1) (P-18:0/18:1) by LC-targeted multiplexed SRM/MS. The structure of PC (P-36:1) and the fragmentation pattern in the positive (A) and negative (B) modes. Identification of the PC (P-36:1) peak by matching the RT of five SRM transitions from the commercial standard (C) and brain samples (D). Note, R1: C16H33 (m/z 225.2582); R2: C17H33 (m/z 237.2582). Here, the mentioned intensity of the 772.6 > 184.0 transition was 1/20th of the original intensity.
Table 1.
SRM transitions of PC (P-36:1) (P-18:0/18:1) and PE (P-36:1) (P-18:0/18:1).
| Target | Mode | SRM transitions |
CE | |
|---|---|---|---|---|
| Precursor ion | Product ion | |||
| PC (P-36:1) (P-18:0/18:1) |
+ | 772.6 | 184.0 | −32 |
| 504.4 | −33 | |||
| 445.3 | −37 | |||
| 508.4 | −33 | |||
| − | 816.6 | 281.2 | +42 | |
| PE (P-36:1) (P-18:0/18:1) |
+ | 730.6 | 589.6 | −20 |
| 462.3 | −22 | |||
| 392.3 | −22 | |||
| 339.3 | −22 | |||
| − | 728.6 | 281.2 | +30 | |
Note: +, positive; −, negative; CE, collision energy.
In mouse brain samples, the transition 772.6 > 184.0 produced three peaks (Fig. 2D). Among those three peaks, only the RT of the third peak matched that of the other four SRM transitions. Furthermore, the RT matched that of the standard (Fig. 2C). Thus, the 3rd peak was confirmed as PC (P-36:1) in the mouse brain sample.
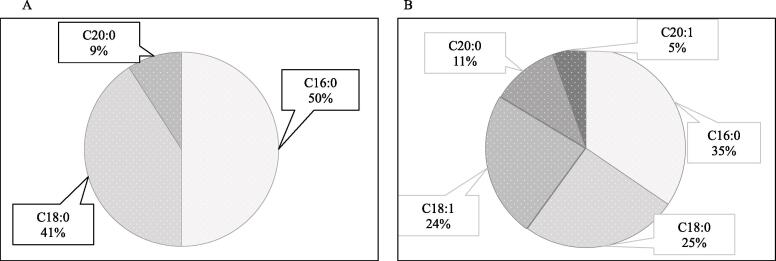
To identify different Pls-PC species from the mouse brain sample, first, a precursor ion scan between m/z 400.0 and m/z 900.0 for the product of m/z 184.0 was performed. Most of the expected precursor ions from this scan were not found (data not shown here). Consequently, the precursor ions of Pls-PC molecular species containing sn-1 C16:0, C18:0, and C20:0 were selected from the database, and transitions with respect to their common product ion 184.0 were determined. The precursor ions for target candidates of identification were selected from the transitions [M + H]+ >184.0, which produced good peak intensity. For these precursor ions, respective product ions were chosen from the database following PC (P-36:1). After the same analysis as that for PC (P-36:1), the correct peak for each Pls-PC species from mouse brain samples was identified by matching the RTs of at least two of the five SRM transitions, and the corresponding peak was quantified. This identification method allowed us to identify 22 Pls-PC species from mouse brain samples (Table S1). Among these species, the percentages of sn-1 C16:0, C18:0 and C20:0 Pls-PC species were 50%, 41%, and 9%, respectively (Fig. 3A).
Fig. 3.
Summary of identified Pls-PC and Pls-PE species. The percentages of sn-1 C16:0, C18:0, and C20:0 Pls-PC species (A) and sn-1 C16:0, C18:0, C18:1, C20:0, and C20:1 Pls-PE species (B) are shown.
3.2. Identification of Pls-PE by targeted multiplexed SRM/MS analysis
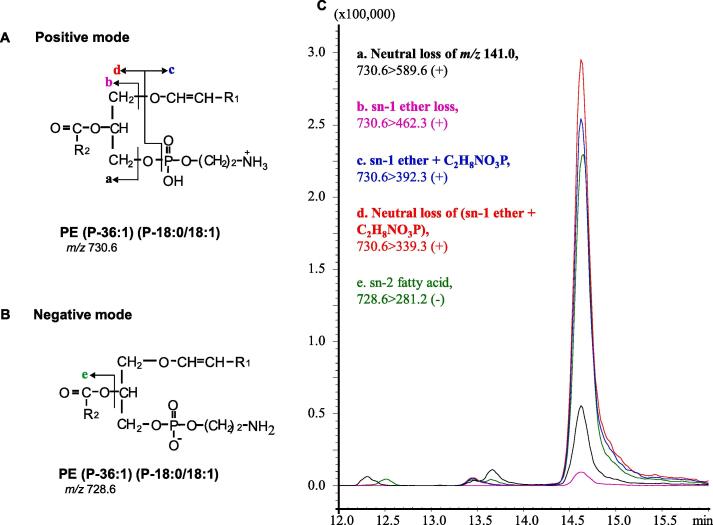
Since Pls-PE concentrations are higher than those of Pls-PC in brain tissue, we did not use the CS to develop a method for Pls-PE detection, but instead utilized five transitions for PE (P-36:1) (P-18:0/18:1) using brain tissue in the same way as for PC (P-36:1). Identification of PE (P-36:1) is shown in Fig. 4. First, we selected the precursor ion, m/z 730.6 [M + H]+ in the positive mode (Fig. 4A) or m/z 728.6 [M−H]− in the negative mode (Fig. 4B), as previously reported [35]. Then, the four product ions m/z 589.6, 462.3, 392.3, and 339.3 in the positive mode (Fig. 4Aa-d) [23] and one product ion m/z 281.2 in the negative mode (Fig. 4Be) were selected. These product ions were identified as neutral loss of m/z 141.0, sn-1 ether loss, sn-1 ether + C2H8NO3P, neutral loss of sn-1 ether + C2H8NO3P and the sn-2 fatty acid carboxylate ion in the database (Fig. 4A and B) [2]. Next, we optimized the parameters of transitions directly using the brain sample (Table 1). During brain sample analysis, three peaks appeared at the 730.6 > 589.6 transition, but the RT of only the 3rd peak was matched with the peaks of four other SRM transitions (Fig. 4C), which allowed for the correct identification of PE (P-36:1).
Fig. 4.
Identification of PE (P-36:1) (P-18:0/18:1) by targeted multiplexed SRM/MS. The structure of PE (P-36:1) and the fragmentation pattern in the positive (A) and negative (B) modes. Identification of the PE (P-36:1) peak by matching the RTs of five SRM transitions from brain samples (C). Note, R1: C16H33 (m/z 225.2582); R2: C17H33 (m/z 237.2582).
Next, to identify Pls-PE molecular species other than PE (P-36:1) from the sample, a precursor ion scan between m/z 400.0 and m/z 900.0 for the product neutral loss of m/z 141.0 was performed. However, most of the expected precursor ions were not found like Pls-PC. Next, five SRM transitions of Pls-PE species containing sn-1 C16:0, C18:0, and C20:0 were obtained using the database in the same manner as for PE (P-36:1). For the Pls-PE species containing sn-1 C18:1 and C20:1, product ions m/z 390.0 and m/z 418.0, respectively [24], were selected. Other product ions of these species were theoretically calculated. The target Pls-PE species were identified by matching the RTs of these five SRM transitions. This analysis allowed us to identify 55 Pls-PE species from mouse brain samples (Table S2). The percentages of sn-1 C16:0, C18:0, C18:1, C20:0 and C20:1 Pls-PE species were 35%, 25%, 24%, 11%, and 5%, respectively (Fig. 3B).
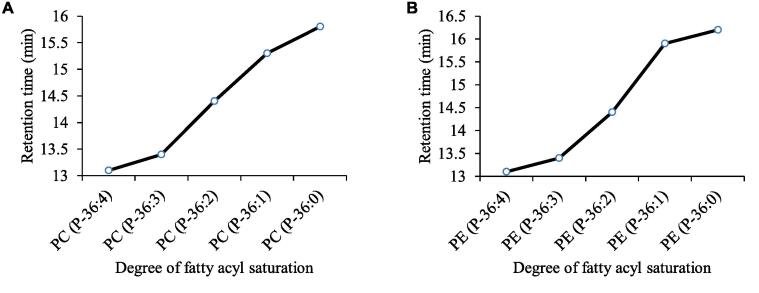
3.3. Comparison of RTs for Pls with the same carbon number according to the difference in the degree of fatty acyl chain unsaturation
We evaluated the effect of double bonds on the elution of the same carbon-containing Pls species. Fig. 5 shows the RT trend for five molecular species with the same carbon number and different degrees of saturation in Pls-PC (Fig. 5A) and Pls-PE (Fig. 5B). In the cases of both Pls-PC and Pls-PE, the RTs were shorter with a higher degree of unsaturation (Tables S1 and S2).
Fig. 5.
The RTs of Pls with the same carbon number depend on the degree of unsaturation of the fatty acyl chain. The RT tendencies of the five species of Pls-PC (A) and Pls-PE (B) are shown.
3.4. Quantification of Pls-PE and Pls-PC in mouse brain samples
After identification of the Pls, we quantified them by a single SRM/MS method. Pls-PC species were quantified using the transition [M + H]+ >184.0, which had the highest intensity among the five transitions. This product ion was derived from phosphate-choline, a common structure among PC group PLs [36]. On the other hand, Pls-PE species were quantified using the transition [M + H]+ > neutral loss of m/z 141.0. This product ion was derived from ethanolamine phosphate, a common structure of PE-type PLs [36]. Through these standard transitions, Pls having the same molecular weight can simultaneously be detected independently of their fatty acyl chain composition.
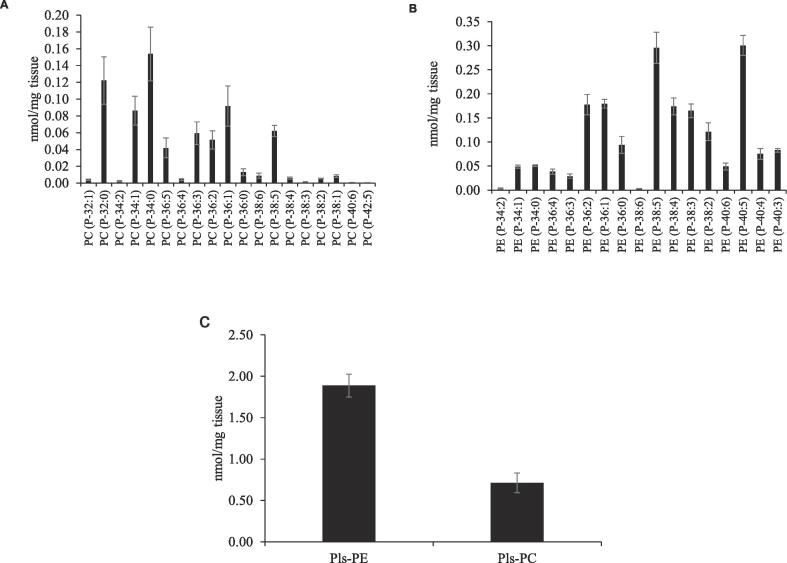
The major Pls-PC species in the mouse brains were PC (P-32:0), PC (P-34:1), PC (P-34:0), PC (P-36:3), PC (P-36:1) and PC (P-38:5) (Fig. 6A). On the other hand, the major Pls-PE species were PE (P-36:2), PE (P-36:1), PE (P-38:5), PE (P-38:4), PE (P-38:3) and PE (P-40:5) (Fig. 6B). Many Pls-PE species have positional isomer species, and the relative abundance of these isomer species was determined (data not shown) [37]. Most species were PUFAs containing Pls (Fig. 6B and Table S2). The concentration of total Pls-PE (1.89 ± 0.14 nmol/mg tissue) was much higher than that of Pls-PC (0.71 ± 0.12) (Fig. 6C).
Fig. 6.
Quantification of Pls species from mouse brain samples. Concentrations of individual molecular species of Pls-PC (A) and Pls-PE (B). The total concentrations of Pls-PC and Pls-PE were also calculated (C). The data represent the means ± SD (standard deviation) obtained from 4 mice.
4. Discussion
In the present study, we developed an LC-targeted multiplexed SRM/MS system for both the identification and quantification of different Pls species. It was reported that Pls identification, especially that of Pls-PC, is difficult in the absence of alkali metal adducts due to the little fragmentation pattern in full-scan MS/MS analysis. Some studies have used alkali metal adducts to solve this problem, but high alkali metal adduct concentrations can damage MS instrumentation, and low concentrations may cause poor Pls peak separation [23]. However, here, we applied an LC-targeted multiplexed SRM/MS system and successfully identified sn-1 and sn-2 fatty acyl chains of 22 Pls-PC and 55 Pls-PE species using ammonium formate as the elution buffer in one run. Thus, targeted identification in multiplexed SRM/MS analysis does not require full-scan MS/MS analysis, pretreatment with acid hydrolysis or the use of an alkali metal. This system allows for relatively simple, specific, and sensitive analysis.
To develop a multiplexed SRM/MS system, five product ions were selected from a database. The regulatory body provides a guideline [38] for LC-MS/MS analysis which states that the RTs of at least two of multiple transitions should be matched for identification of a compound. A previous study [30] also reported that the same RT of peaks from multiple transitions for the same precursor ion ensured correct identification of a compound. We identified individual species by matching the RTs of at least two of five SRM transitions for Pls-PC and all five SRM transitions for Pls-PE.
Importantly, among the identified Pls species, we detected C20:0 fatty alcohols at the sn-1 position in Pls species such as PC (P-36:0) (P-20:0/16:0), PE (P-36:1) (P-20:0/16:1), PE (P-36:0) (P-20:0/16:0), PE (P-38:2) P-(20:0/18:2), PE (P-40:4) (P-20:0/20:4), and PE (P-40:3) (P-20:0/20:3) (Tables S1 and S2). These species were reported in the database [2], not in Pls analysis-related studies [26]. We also detected some low-abundance species containing C20:1 fatty alcohols at the sn-1 position, such as PE (P-40:5) (P-20:1/20:4), PE (P-40:4) (P-20:1/20:3), and PE (P-40:3) (P-20:1/20:2) (Table S2). The low abundance of these species might be the reason for their not being reported in full-scan MS/MS analysis. However, this system allowed us to detect such low-abundance Pls species from mouse brain tissue. This result suggests that the LC-targeted multiplexed SRM/MS system can be used for the characterization of low-abundance lipid molecules in lipidomic studies.
Previously, quantification of Pls by a single SRM/MS analysis was reported [[26], [27]]. Although several peaks were detected by this analysis, identification of correct peaks of individual species is necessary. For example, our results showed that the transition used for quantification with product ions at m/z 184.0 for Pls-PC and the neutral loss of m/z 141.0 for Pls-PE produced multiple peaks. However, after analysis with five transitions, only one peak was identified for individual Pls species. Thus, this system selectively and specifically identified Pls from complex biological samples. The use of multiple transitions for the same precursor ion increased the selectivity and reliability of SRM/MS analysis and reduced the possibility of false positive identification. In addition, this identification system provides a simple means of validating many Pls species that can be false positively identified by a single SRM/MS analysis.
We developed a single SRM/MS method for quantification using ISs. Validation guidelines [38] recommend independent methods to confirm the identity of an analyte in LC-MS analysis. Previous studies as well as regulatory guidelines recommend IS addition-based matrix effect correction [39]. In this study, protonated Pls-PC and Pls-PE were quantified with transitions using common product ions at m/z 184.0 and neutral loss of m/z 141.0, respectively. Although the transition using the neutral loss of m/z 141.0 did not show the highest peak intensity among the five transitions, it was chosen to be the same transition as that of the IS PE (d7–33:1). Thus, we could quantify positional isomer species having the same molecular weight by one transition. For example, PE (P-36:1) (m/z 730.6) has four positional isomer species: P-16:0/20:1, P-18:0/18:1, P-18:1/18:0 and P-20:0/16:1. We quantified the total concentration of PE (P-36:1) (0.18 ± 0.009 nmol/mg tissue) through this system. In addition, the purpose of this study was to check the changes of Pls species levels in clinical diseases, not to quantify the absolute concentration of Pls molecular species. Thus, we followed a semi-quantitative method using a few internal standards previously described [40]. In our future study, we are planning to quantify the individual concentrations of these positional isomer species by monitoring unique transitions. Our result that the concentrations of Pls-PE were higher than those of Pls-PC in the mouse brain was supported by previous studies [3]. Our calculated Pls concentrations were almost the same as those in previous reports [[40], [41]].
It was found that the more saturated fatty acyl chains there were with the same carbon number, the greater the RT [[42], [43]]. In this study, we tried to confirm that the respective RTs of the same carbon-containing Pls species will depend on the number of double bonds of the fatty acyl chains [44]. We found that the RTs of Pls-PC and Pls-PE species with the same carbon number depended on the number of double bonds present in the fatty acyl chain. Thus, determination of the elution order for Pls species with the same carbon number aided in their identification.
We identified and quantified C16:0, C18:0, and C20:0 fatty alcohols at the sn-1 position in Pls species as targeted products and found them in the database (MSMS-prediction-distribute-v49). However, information about C18:1 and C20:1 fatty alcohols at the sn-1 position in Pls species was not found in the database. A previous full scan MS/MS study reported product ions of Pls-PE species having C18:1 and C20:1 fatty alcohols at the sn-1 position were 390.0 and 418.0, respectively [24]. Based on this information, we identified Pls-PE positional isomer species containing C18:1 and C20:1 fatty alcohols at the sn-1 position (Table S2). Further studies are warranted to determine the targeted product ions and to identify C18:1 and C20:1 fatty alcohols at the sn-1 position in Pls-PC species. In addition, alkyl-acyl-PC and alkyl-acyl-PE are intermediate products of the Pls biosynthesis pathway. The difference in the alkyl-acyl-type structure with respect to the Pls-type structure is the presence of only the ether (instead of vinyl ether) linkage at the sn-1 fatty acyl chain. Product ions, such as those used for Pls-PC and Pls-PE identification, are not found in the database (MSMS-prediction-distribute v49). In this study, the precursor ions of alkyl-acyl-PC and alkyl-acyl-PE were selected from the database, and their targeted product ions were theoretically calculated. After analysis, five alkyl-acyl-PCs and two alkyl-acyl-PEs were identified (data not shown here). More detailed studies are warranted to differentiate the characteristics such as elution order and the effects of different linkages at the sn-1 fatty acyl chain on the RT among diacyl-, alkyl-acyl- and Pls-type lipids. Nevertheless, this study will be of importance to identify and quantify additional Pls species as well as low-abundance PLs species using lipidomic database information.
In conclusion, our results suggest that this system is a very sensitive identification and quantification method for Pls. The developed method can be applied not only to Pls analysis but also to other lipid analyses, and is expected to fuel research in the lipidomics field. Moreover, this method will help to build understanding of the physiological roles of Pls in different clinical diseases.
Author contributions
AKA, HK, AS, HS, HO and AN contributed to the conception and design of this study. AKA, HK, AS, HS, HO, AH and AN contributed to the data acquisition, analysis, and interpretation of the results. AKA, HK, HO and AN contributed to the writing of the manuscript. HK, AS, SY and AN contributed to critical revision of the article for important intellectual content. AKA and HO contributed to statistical analysis. AN contributed to the provision of the reagents and other study materials. All authors contributed to drafting of the manuscript, and all authors have approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We would like to acknowledge the technical expertise of the Interdisciplinary Center for Science Research, Organization for Research and Academic Information, Shimane University. We would like to thank the members of the ‘Metabolizumo project’ for their support in this study. We would also like to thank Ms. Misa Tanaka (Department of Pediatrics, Shimane University Faculty of Medicine) for the helpful suggestions.
Funding
No external funding was used for this study.
Role of the funding source
Not applicable.
Data availability
All data are available upon request to the corresponding author of this manuscript
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2021.09.004.
Contributor Information
Abul Kalam Azad, Email: akazad@med.shimane-u.ac.jp.
Hironori Kobayashi, Email: bakki@med.shimane-u.ac.jp.
Abdullah Md. Sheikh, Email: abdullah@med.shimane-u.ac.jp.
Harumi Osago, Email: biochem1@med.shimane-u.ac.jp.
Hiromichi Sakai, Email: hisakai@med.shimane-u.ac.jp.
Md. Ahsanul Haque, Email: ahsanul@uap-bd.edu.
Shozo Yano, Email: syano@med.shimane-u.ac.jp.
Atsushi Nagai, Email: anagai@med.shimane-u.ac.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fahy E., Subramaniam S., Brown H.A., Glass C.K., Merrill A.H., Murphy R.C., Raetz C.R.H., Russell D.W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M.S., White S.H., Witztum J.L., Dennis E.A. A comprehensive classification system for lipids. J. Lipid Res. 2005;107(5):337–364. doi: 10.1002/(ISSN)1438-931210.1002/ejlt.v107:510.1002/ejlt.200405001. [DOI] [PubMed] [Google Scholar]
- 2.Kind T., Liu K.-H., Lee D.Y., DeFelice B., Meissen J.K., Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods. 2013;10(8):755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul S., Lancaster G.I., Meikle P.J. Plasmalogens: a potential therapeutic target for neurodegenerative and cardiometabolic disease. Prog. Lipid Res. 2019;74:186–195. doi: 10.1016/j.plipres.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Deng Y., Almsherqi Z.A. Evolution of cubic membranes as antioxidant defence system. Interface Focus. 2015;5(4):20150012. doi: 10.1098/rsfs.2015.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glaser P.E., Gross R.W. Plasmenylethanolamine facilitates rapid membrane fusion: a stopped-flow kinetic investigation correlating the propensity of a major plasma membrane constituent to adopt an HII phase with its ability to promote membrane fusion. Biochemistry. 1994;33(19):5805–5812. doi: 10.1021/bi00185a019. [DOI] [PubMed] [Google Scholar]
- 6.Rog T., Koivuniemi A. The biophysical properties of ethanolamine plasmalogens revealed by atomistic molecular dynamics simulations. Biochim. Biophys. Acta, Lipids Lipid Metab. 2016;1858(1):97–103. doi: 10.1016/j.bbamem.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maeba R., Ueta N. A novel antioxidant action of ethanolamine plasmalogens in lowering the oxidizability of membranes. Biochem. Soc. Trans. 2004;32:141–143. doi: 10.1042/BST0320141. [DOI] [PubMed] [Google Scholar]
- 8.Reiss D., Beyer K., Engelmann B. Delayed oxidative degradation of polyunsaturated diacyl phospholipids in the presence of plasmalogen phospholipids in vitro. Biochem. J. 1997;323:807–814. doi: 10.1042/bj3230807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubio J.M., Astudillo A.M., Casas J., Balboa M.A., Balsinde J. Regulation of phagocytosis in macrophages by membrane ethanolamine plasmalogens. Front. Immunol. 2018;9:1723. doi: 10.3389/fimmu.2018.01723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahabieh M.S., Ha ZongYi, Di Pietro E., Nichol J.N., Bolt A.M., Goncalves C., Dupéré-Richer D., Pettersson F., Mann K.K., Braverman N.E., del Rincón S.V., Miller W.H. Peroxisomes protect lymphoma cells from HDAC inhibitor-mediated apoptosis. Cell Death Differ. 2017;24(11):1912–1924. doi: 10.1038/cdd.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hossain M.S., Ifuku M., Take S., Kawamura J., Miake K., Katafuchi T., Amin A.R.M.R. Plasmalogens rescue neuronal cell death through an activation of AKT and ERK survival signaling. PLoS One. 2013;8(12):e83508. doi: 10.1371/journal.pone.0083508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pike L.J., Han X., Chung K.N., Gross R.W. Lipid rafts are enriched in arachidonic acid and plasmenylethanolamine and their composition is independent of caveolin-1 expression: a quantitative electrospray ionization/mass spectrometric analysis. Biochemistry. 2002;41:2075–2088. doi: 10.1021/bi0156557. [DOI] [PubMed] [Google Scholar]
- 13.Fujiki Y., Yagita Y., Matsuzaki T. Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease. Biochim. Biophys. Acta, Lipids Lipid Metab. 2012;1822(9):1337–1342. doi: 10.1016/j.bbadis.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Messias M.C.F., Mecatti G.C., Priolli D.G., De Oliveira Carvalho P. Plasmalogen lipids: functional mechanism and their involvement in gastrointestinal cancer. Lipids Health Dis. 2018;17:41. doi: 10.1186/s12944-018-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabelo N., Martín V., Santpere G., Marín R., Torrent L., Ferrer I., Díaz M. Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson's disease and incidental Parkinson's disease. Mol. Med. 2011;17(9-10):1107–1118. doi: 10.2119/molmed.2011.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodenowe D.B., Cook L.L., Liu J., Lu Y., Jayasinghe D.A., Ahiahonu P.W.K., Heath D., Yamazaki Y., Flax J., Krenitsky K.F., Sparks D.L., Lerner A., Friedland R.P., Kudo T., Kamino K., Morihara T., Takeda M., Wood P.L. Peripheral ethanolamine plasmalogen deficiency: a logical causative factor in Alzheimer's disease and dementia. J. Lipid Res. 2007;48(11):2485–2498. doi: 10.1194/jlr.P700023-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Grimm M.O., Kuchenbecker J., Rothhaar T.L., Grösgen S., Hundsdörfer B., Burg V.K., Friess P., Müller U., Grimm H.S., Riemenschneider M., Hartmann T. Plasmalogen synthesis is regulated via alkyl-dihydroxyacetonephosphate-synthase by amyloid precursor protein processing and is affected in Alzheimer's disease. J. Neurochem. 2011;116:916–925. doi: 10.1111/j.1471-4159.2010.07070.x. [DOI] [PubMed] [Google Scholar]
- 18.Dean J.M., Lodhi I.J. Structural and functional roles of ether lipids. Protein Cell. 2018;9(2):196–206. doi: 10.1007/s13238-017-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv J., Lv C.Q., Xu L., Yang H. Plasma content variation and correlation of plasmalogen and GIS, TC, and TPL in gastric carcinoma patients: a comparative study. Med. Sci. Monit. Basic Res. 2015;21:157–160. doi: 10.12659/MSMBR.893908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dacremont G., Vincent G. Assay of plasmalogens and polyunsaturated fatty acids (PUFA) in erythrocytes and fibroblasts. J. Inherit. Metab. Dis. 1995;18(Suppl 1):84–89. doi: 10.1007/BF00711431. [DOI] [PubMed] [Google Scholar]
- 21.Mawatari S., Okuma Y., Fujino T. Separation of intact plasmalogens and all other phospholipids by a single run of high-performance liquid chromatography. Anal. Biochem. 2007;370(1):54–59. doi: 10.1016/j.ab.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Murphy E.J., Stephens R., Jurkowitz-Alexander M., Horrocks L.A. Acidic hydrolysis of plasmalogens followed by high-performance liquid chromatography. Lipids. 1993;28(6):565–568. doi: 10.1007/BF02536090. [DOI] [PubMed] [Google Scholar]
- 23.Otoki Y., Nakagawa K., Kato S., Miyazawa T. MS/MS and LC-MS/MS analysis of choline/ethanolamine plasmalogens via promotion of alkali metal adduct formation. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015;1004:85–92. doi: 10.1016/j.jchromb.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Berry K.A.Z., Murphy R.C. Electrospray ionization tandem mass spectrometry of glycerophosphoethanolamine plasmalogen phospholipids. J. Am. Soc. Mass Spectrom. 2004;15(10):1499–1508. doi: 10.1016/j.jasms.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Bräutigam C., Engelmann B., Reiss D., Reinhardt U., Thiery J., Richter W.O., Brosche T. Plasmalogen phospholipids in plasma lipoproteins of normolipidemic donors and patients with hypercholesterolemia treated by LDL apheresis. Atherosclerosis. 1996;119(1):77–88. doi: 10.1016/0021-9150(95)05632-7. [DOI] [PubMed] [Google Scholar]
- 26.Ikuta A., Sakurai T., Nishimukai M., Takahashi Y., Nagasaka A., Hui S.P., Hara H., Chiba H. Composition of plasmalogens in serum lipoproteins from patients with non-alcoholic steatohepatitis and their susceptibility to oxidation. Clin. Chim. Acta. 2019;493:1–7. doi: 10.1016/j.cca.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Nishimukai M., Yamashita M., Watanabe Y., Yamazaki Y., Nezu T., Maeba R., Hara H. Lymphatic absorption of choline plasmalogen is much higher than that of ethanolamine plasmalogen in rats. Eur. J. Nutr. 2011;50(6):427–436. doi: 10.1007/s00394-010-0149-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y., Brasier A.R. Applications of selected reaction monitoring (SRM)-mass spectrometry (MS) for quantitative measurement of signaling pathways. Methods. 2013;61(3):313–322. doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charnot A., Gouveia D., Armengaud J., Almunia C., Chaumot A., Lemoine J., Geffard O., Salvador A. Multiplexed assay for protein quantitation in the invertebrate Gammarus fossarum by liquid chromatography coupled to tandem mass spectrometry. Anal. Bioanal. Chem. 2017;409(16):3969–3991. doi: 10.1007/s00216-017-0348-0. [DOI] [PubMed] [Google Scholar]
- 30.Gianazza E., Banfi C. Post-translational quantitation by SRM/MRM: applications in cardiology. Expert Rev. Proteomics. 2018;15(6):477–502. doi: 10.1080/14789450.2018.1484283. [DOI] [PubMed] [Google Scholar]
- 31.Wesseling H., Gottschalk M.G., Bahn S. Targeted multiplexed selected reaction monitoring analysis evaluates protein expression changes of molecular risk factors for major psychiatric disorders. Int. J. Neuropsychopharmacol. 2015;18:pyu015. doi: 10.1093/ijnp/pyu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruve A., Rebane R., Kipper K., Oldekop M.L., Evard H., Herodes K., Ravio P., Leito I. Tutorial review on validation of liquid chromatography-mass spectrometry methods: part I. Anal. Chim. Acta. 2015;870:29–44. doi: 10.1016/j.aca.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Galvan V., Gorostiza O.F., Banwait S., Ataie M., Logvinova A.V., Sitaraman S., Carlson E., Sagi S.A., Chevallier N., Jin K., Greenberg D.A., Bredesen D.E. Reversal of Alzheimer's-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc. Natl. Acad. Sci. U.S.A. 2006;103(18):7130–7135. doi: 10.1073/pnas.0509695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott S.K., Jenner A.M., Mitchell T.W., Brown S.H.J., Halliday G.M., Garner B. An improved high-throughput lipid extraction method for the analysis of human brain lipids. Lipids. 2013;48(3):307–318. doi: 10.1007/s11745-013-3760-z. [DOI] [PubMed] [Google Scholar]
- 35.Xia Y.-Q., Jemal M. Phospholipids in liquid chromatography/mass spectrometry bioanalysis: comparison of three tandem mass spectrometric techniques for monitoring plasma phospholipids, the effect of mobile phase composition on phospholipids elution and the association of phospholipids with matrix effects. Rapid Commun. Mass Spectrom. 2009;23(14):2125–2138. doi: 10.1002/rcm.v23:1410.1002/rcm.4121. [DOI] [PubMed] [Google Scholar]
- 36.Pulfer M., Murphy R.C. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22(5):332–364. doi: 10.1002/(ISSN)1098-278710.1002/mas.v22:510.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi H., Iida Y., Shimizu T., Taguchi R. Separation and quantification of sn-1 and sn-2 fatty acid positional isomers in phosphatidylcholine by RPLC-ESIMS/MS. J. Biochem. 2010;147(2):245–256. doi: 10.1093/jb/mvp171. [DOI] [PubMed] [Google Scholar]
- 38.European Commission, 2020. Analytical Quality Control and Method Validation Procedures for Pesticide Residues Analysis in Food and Feed. Document No. SANTE/2015/11945. European Commission, Health and Consumer Protection Directorate of General, Brussel, Belgium. https://www.eurl-pesticides.eu/docs/public/tmplt_article.asp?CntID=727.
- 39.Wang M., Wang C., Han X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom. Rev. 2017;36(6):693–714. doi: 10.1002/mas.v36.610.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han X., Holtzman D.M., McKeel D.W., Jr. Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 41.Palavicini J.P., Wang C., Chen L., Hosang K., Wang J., Tomiyama T., Mori H., Han X. Oligomeric amyloid-beta induces MAPK-mediated activation of brain cytosolic and calcium independent phospholipase A2 in a spatial-specific manner. Acta Neuropathol. Commun. 2017;5(1):56. doi: 10.1186/s40478-017-0460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salazar C., Jones M.D., Sturtevant D., Horn P.J., Crossley J., Zaman K., Chapman K.D., Wrona M., Isaac G., Smith N.W., Shulaev V. Development and application of sub-2-μm particle CO2 -based chromatography coupled to mass spectrometry for comprehensive analysis of lipids in cottonseed extracts. Rapid Commun. Mass Spectrom. 2017;31(7):591–605. doi: 10.1002/rcm.7825. [DOI] [PubMed] [Google Scholar]
- 43.Takashima S., Toyoshi K., Shimozawa N. Analyses of the fatty acid separation principle using liquid chromatography-mass spectrometry. Med. Mass Spectrom. 2018;2:21–33. doi: 10.24508/mms.2018.06.002. [DOI] [Google Scholar]
- 44.Aicheler F., Li J., Hoene M., Lehmann R., Xu G., Kohlbacher O. Retention Time Prediction Improves Identification in Nontargeted Lipidomics Approaches. Anal. Chem. 2015;87(15):7698–7704. doi: 10.1021/acs.analchem.5b01139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request to the corresponding author of this manuscript