Abstract
Testing of the DNA of TT virus (TTV) was done with serum samples obtained from 191 persons working in a public hospital of the city of Rio de Janeiro, Brazil. TTV DNA was detected by PCR in the sera of 125 (65.4%) individuals. PCR products were cloned, and sequences with a length of 159 bases surrounding the TATA signal region were determined for 100 clones derived from 31 individuals. One clone from each of 23 subjects was sequenced, while 7 to 19 clones from eight individuals were sequenced. None of the sera contained a viral sequence identical to that of any other individual. Phylogenetic analysis revealed the existence of a divergent TTV genotype possessing a single-base deletion at position 140. Among the eight persons for whom various sequences were analyzed, six were coinfected with between two and seven TTV strains belonging to different genotypes. The results suggest that coinfection with multiple TTV strains belonging to different genotypes is a common event in healthy Brazilian adults.
TT virus (TTV) is a newly discovered human virus that was first detected in the serum of a Japanese patient (initials, T.T.) with posttransfusion hepatitis (12, 13). The TTV genome is constituted by a single-stranded, circular DNA of negative polarity (9, 13). The TTV nucleotide sequence (3,818 to 3,853 nucleotides) does not show a significantly high homology to the sequence of any other virus. Several TTV isolates have been entirely sequenced (4, 6, 8, 9, 14), revealing a high degree of divergence among strains and the existence of at least 16 genotypes, which are separated by an evolutionary distance greater than 0.30 (15).
Although TTV DNA titers closely correlated with aminotransferase levels in the sera of some patients during posttransfusion hepatitis (12), no clear association between TTV infection and human liver disease has been established at this time. Very high prevalences (62 to 96%) of TTV infection have been found in healthy populations of Japan (15, 20) as well as in developing Asian, African, and South American countries (1, 11, 17).
As was initially demonstrated, TTV transmission occurs through the parenteral route (12). However, very high prevalences in healthy populations indicate the existence of other routes of transmission.
Coinfection with multiple TTV strains has been described for people exposed to blood and blood products (3, 5, 21), as well as for patients with liver disease (2, 7, 22). Here we show that such a coinfection is a common event in Brazilian health care workers and that a healthy person can be coinfected by at least seven strains.
MATERIALS AND METHODS
Population studied.
In 1999, all persons working in a public hospital of the city of Rio de Janeiro, Brazil, were invited to receive immunization against hepatitis B. At the occasion of the first-dose injection, blood samples from 1,104 people were collected. Testing of the DNA of TTV was done with serum samples obtained from 191 subjects, selected among persons aged 19 to 50 years who declared not having received blood transfusion.
Extraction and amplification of DNA.
Two hundred and fifty microliters of serum was treated with 0.5 mg of proteinase K per ml in the presence of 0.2 M NaCl–0.25% sodium dodecyl sulfate for 4 h at 37°C. DNA was then extracted by phenol-chloroform and precipitated by ethanol. The pellet was dried and resuspended in 50 μl of distilled water. DNA was amplified by a single round of PCR performed with 5 μl of DNA in a 50-μl reaction mixture. AmpliTaq Gold polymerase (Perkin-Elmer Applied Systems, Foster City, Calif.) and primers T801 and T935 were used according to the protocol described by Takahashi et al. (20). Ten microliters of PCR products was loaded onto a 2% agarose gel, electrophoresed, and stained with ethidium bromide to visualize bands with an expected length of 199 bp.
Molecular cloning and nucleotide sequencing.
PCR products were cloned into the pCR2.1-TOPO vector (TOPO TA cloning kit; Invitrogen, San Diego, Calif.). Recombinant plasmids were purified and insertion DNAs were sequenced by using Cy5-labelled forward and reverse M13 primers and the reagents provided in the Autoread Sequencing Kit (Amersham Pharmacia Biotech, Uppsala, Sweden). Sequencing products were analyzed on an ALFexpress automated sequencer (Amersham Pharmacia).
Computer analysis of TTV sequences.
Alignment of multiple nucleic acid sequences was performed with the University of Wisconsin Genetic Computer Group program PILEUP. Phylogenetic analysis was performed with the program GROWTREE from the same package. Genetic distances between TTV strains were calculated by using the program Dnadist from the package PHYLIP, version 3.5c.
RESULTS
Seroprevalence of TTV DNA.
One hundred ninety-one health care workers (47 men, 144 women) of a public hospital of the city of Rio de Janeiro, Brazil, were enrolled in this study. Serum samples were analyzed for the presence of TTV DNA. One hundred twenty-five samples were positive, corresponding to a prevalence of 65.4%.
Phylogenetic analysis.
PCR products were cloned, and sequences of 159 bases (nucleotides 26 to 184) surrounding the TATA signal region localized upstream of open reading frame 2 (ORF2) were determined for 100 clones derived from 31 individuals. One clone from each of 23 subjects was sequenced, while 7 to 19 clones were sequenced from eight individuals. A total of 59 different sequences was thus obtained.
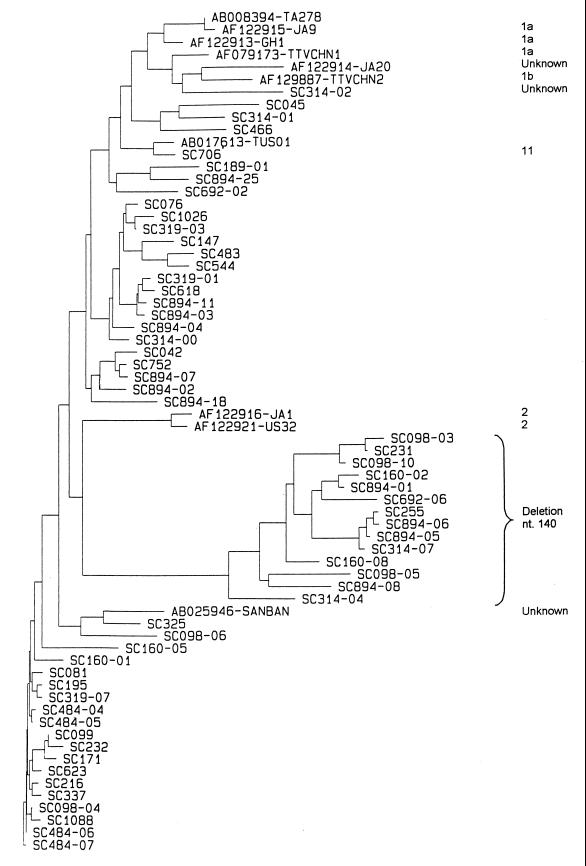
Figure 1 shows a phylogenetic tree which includes these 59 sequences along with 10 sequences available in databases and belonging to different TTV genotypes. A large genetic diversity was observed among the isolates from this study. No two sequences were identical when derived from different persons. Evolutionary distances between our sequences were up to 0.49. A group of 14 sequences constituted a separate branch (Fig. 1). These had in common a 1-nucleotide deletion at position 140. The genetic distances between strains of this cluster were less than 0.30.
FIG. 1.
Phylogenetic tree of 69 TTV isolates. The Jukes-Cantor algorithm was used for distance determination, and the tree was constructed with the neighbor-joining method. Ten sequences available in the databases are identified by their GenBank accession numbers, followed by the names of the isolates. The genotypes of seven of them (AB008394, AF122915, AF122913, AF122914, AF122916, AF122921, AB017613) are known and indicated at the right. Fifty-nine sequences, whose names begin by the letters SC, are from this work. Different clones from a single patient are distinguished by a number after the hyphen. Fourteen sequences contain a single-base deletion at position 140.
Despite the large genetic diversity observed here, some stretches of the genome were perfectly conserved in all sequences. This was notably the case for the TATAA motif at nucleotides 86 to 90 and an ATG codon (position 107) which had been initially proposed to be the translation initiation codon of ORF2 (13).
Coinfection with multiple TTV isolates.
Several clones derived from the same serum were sequenced to determine if coinfection (or superinfection) with multiple TTV strains occurred. This was performed for eight individuals whose demographic, professional, serological, and clinical data are shown in Table 1.
TABLE 1.
Demographic, professional, serological, and clinical data of the subjects for whom several TTV clones were sequenced
| Subject | Sex | Age | Profession | Presence of protein in seruma
|
Clinical history
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anti-HAV | HBsAg | Anti-HBs | Anti-HBc | Anti-HCV | ALT (IU/liter) | AST (IU/liter) | Blood transfusion | Surgery | ||||
| SC098 | F | 44 | Assistant nurse | Pos | Neg | Neg | Neg | Neg | 23 | 80 | No | No |
| SC160 | F | 40 | Assistant nurse | Pos | Neg | Neg | Neg | Neg | 15 | 34 | No | Yes |
| SC189 | F | 22 | Medical student | Neg | Neg | Neg | Neg | Neg | 16 | 16 | No | Yes |
| SC314 | F | 43 | Assistant nurse | Pos | Neg | Pos | Pos | Posb | 11 | 11 | No | Yes |
| SC319 | M | 50 | Assistant nurse | Pos | Neg | Neg | Neg | Neg | 28 | 20 | No | Yes |
| SC484 | M | 42 | Attendant | Pos | Neg | Neg | Neg | Neg | 36 | 34 | No | Yes |
| SC692 | F | 45 | Attendant | Pos | Neg | Neg | Neg | Neg | 32 | 43 | No | No |
| SC894 | F | 50 | Assistant nurse | Pos | Neg | Neg | Neg | Neg | 24 | 28 | No | No |
HAV, hepatitis A virus; HCV, hepatitis C virus; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
Hepatitis C virus RNA negative.
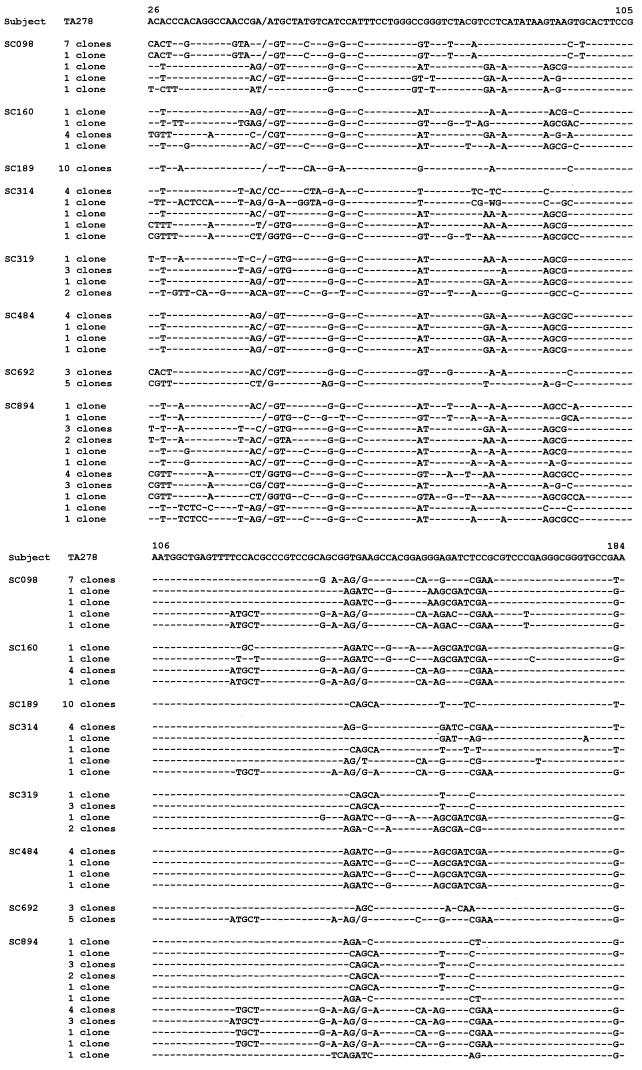
Altogether, 77 clones (7 to 19 from each person) were sequenced. Figure 2 shows the alignment of the nucleotide sequences, and Table 2 summarizes the characteristics of the sequences derived from each person. Surprisingly, in only one case (a young medical student, subject SC189), all the clones showed identical sequences. In another individual (attendant SC484), four closely related sequences (evolutionary distances less than or equal to 0.02) were found (Table 2). In the other subjects (five assistant nurses and one attendant), between two and eleven distinct sequences were found. The different TTV sequences deriving from single individuals could be genetically very close or very separated (evolutionary distances from 0.006 to 0.45). By using sequences of TTV strains of known genotypes and subtypes, genetic distances between two genotypes and between subtypes of the same genotype were calculated. All distances (calculated for the genome segment covering nucleotides 26 to 184) were greater than or equal to 0.11. On this basis, it could be concluded that six of the eight persons under study were coinfected with between two and seven different TTV genotypes or subtypes (Table 2, last column).
FIG. 2.
Alignment of partial nucleotide sequences of 36 TTV variants found in eight health care workers. The number of clones showing identical sequences is given. The sequence of prototype isolate TA278 of genotype 1a (13) is indicated at the top. Dashes represent the same nucleotides as in the TA278 isolate. In two clones from subject SC319, an insertion of an A nucleotide occurs at position 45; slashes indicate the absence of this nucleotide in the other sequences.
TABLE 2.
Distribution of the different sequences from the subjects for whom several TTV clones were sequenceda
| Subject | No. of clones sequenced | No. of clones with deletion at nt 140 | No. of clones with insertion at nt 45 | No. of distinct sequences | No. of clones of each sequence | Distance between sequences | No. of genotypes and/or subtypes (d > 0.10) |
|---|---|---|---|---|---|---|---|
| SC098 | 11 | 9 | 0 | 5 | 7, 1, 1, 1, 1 | 0.05—0.39 | 4 |
| SC160 | 7 | 5 | 0 | 4 | 4, 1, 1, 1 | 0.11—0.31 | 4 |
| SC189 | 10 | 0 | 0 | 1 | 10 | 1 | |
| SC314 | 8 | 2 | 0 | 5 | 4, 1, 1, 1, 1 | 0.14—0.45 | 5 |
| SC319 | 7 | 0 | 2 | 4 | 3, 2, 1, 1 | 0.04—0.27 | 3 |
| SC484 | 7 | 0 | 0 | 4 | 4, 1, 1, 1 | 0.006—0.02 | 1 |
| SC692 | 8 | 5 | 0 | 2 | 5, 3 | 0.27 | 2 |
| SC894 | 19 | 9 | 0 | 11 | 4, 3, 3, 2, 1, 1, 1, 1, 1, 1, 1 | 0.02—0.32 | 7 |
nt, nucleotide; d, genetic distance.
DISCUSSION
TTV is a virus with a wide nucleotide sequence divergence (6, 15, 18, 23). The PCR assays developed soon after the discovery of the virus were not able to amplify DNA of all genotypes, and the TTV prevalences in the populations of different countries have therefore been dramatically underestimated. Recently, improved PCR protocols and new sets of primers have led to increased rates of TTV DNA detection. The PCR method employed in this study has thus allowed the detection of TTV in 92% of the serum samples of a group of Japanese blood donors (20), a prevalence much higher than the 23% obtained with the same samples by using the previously designed primers NG059, NG061, and NG063 (13). Another independent study has shown that the method allows the detection of five- to sixfold more TTV-positive samples than two other protocols (7). Using PCR primers whose design was based on NG059, NG061, and NG063, but which were degenerated to allow the detection of a higher number of TTV variants, we recently reported a TTV seroprevalence of 62% in blood donors living in the city of Rio de Janeiro, Brazil (11). The prevalence (65.4%) obtained now in a group of 191 health care workers living in the same city was not significantly different.
Although the DNA segment analyzed here is relatively short and is more conserved than other regions of the TTV genome (4, 14), no two sequences were identical when derived from different persons. Eighty-nine percent of the genetic distances between two of our strains were higher than 0.10 (17% higher than 0.30), with a maximum value at 0.49. These findings demonstrate the large genetic diversity of TTV strains circulating in Brazil. At the moment, few TTV genotypes have been sequenced in the genome region under study. This made it difficult to ascribe a genotype, from the 16 previously described (15), to each of our samples. However, it is interesting that 14 sequences, with a single-nucleotide deletion at position 140, constituted a separate genotype (Fig. 1). Examination of the nucleotide sequences deposited in Genbank by using the BLAST program revealed that two sequences (AF109811 and AF109812), from Chinese TTV isolates, presented the 1-nucleotide deletion at position 140. These two sequences presented an overlapping of only 72 bp with ours. On this small genome segment, a very high homology (93 to 98%) was observed between the Chinese sequences and the 14 Brazilian sequences belonging to the separate genotype.
The ORF2 translation initiation codon was initially localized to position 107 (9, 13). However, further studies suggested that another ATG codon, at position 263, is preferentially recognized as the start codon (4, 6, 14). The existence of a single-nucleotide deletion at position 140 in 14 sequences, which would introduce a frameshift mutation, reinforced this hypothesis. However, it is noteworthy that there was no case in which all TTV clones derived from the same individual contained that deletion. Therefore, a complementation between wild-type and deletion mutant strains cannot be excluded.
Mixed infections of TTV have been reported in individuals at high risk for infection with parenterally transmitted viruses, such as intravenous drug users (9), hemophiliacs (9, 21), and hemodialysis patients (3, 5), as well as in patients with liver disease (2, 7, 9, 22). Recently, coinfections with two or three TTV strains have been reported to be in some healthy Japanese individuals (15, 16). Here we show that such a mixed infection is a common event in healthy Brazilian people, at least in health care workers. Our results confirm and extend previous observations showing that infection by a given genotype is not protective against the superinfection by another type (5, 15). Furthermore, we show that the number of TTV isolates infecting an individual can be high. For example, for subject SC894, the nucleotide sequences of 19 clones were determined and 11 distinct sequences were obtained (Table 2). Genetic distances between two TTV sequences present in the same serum could be very close or very divergent (up to 0.45 for subject SC314). Although the mutation rate of the TTV genome as well as the duration of infection in the persons under study are unknown, it is likely that very close sequences (genetic distance < 0.02) derived from a single source of infection (quasispecies). On the contrary, there is no doubt that subject SC314 was infected from multiple sources. Using a cutoff value of 0.10, it was demonstrable that TTV strains belonging to at least seven genotypes or subtypes may infect the same healthy person (Table 2).
A recent study has demonstrated similar prevalences of TTV infection in medical workers and in age-matched controls (10). It remains to be determined if the phenomenon of multiple infection occurs at a comparable frequency in both groups. In a previous paper, we showed that the TTV seroprevalence increased continuously with age (19). Here, the only individual (SC189) for whom all TTV clones were identical was the youngest of a group of eight persons. She was 22 years old, whereas all others were 40 to 50 years old. Further studies are necessary to determine if an accumulation of TTV strains occurs during the life of healthy individuals or if such a tendency is restricted to health care workers.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to the GenBank database under accession no. AF216433 through AF216491.
ACKNOWLEDGMENTS
We are grateful to M. dos Reis Arantes and T. E. Palmer for providing us the blood samples and to R. R. S. da Silva for technical assistance. We also thank K. Rispeter and R. Hallett for the critical reading of the manuscript.
This work has been supported by CNPq.
REFERENCES
- 1.Abe K, Inami T, Asano K, Miyoshi C, Masaki N, Hayashi S, Ishikawa K-I, Takebe Y, Win K M, El-Zayadi A R, Han K H, Zhang D Y. TT virus infection is widespread in the general populations from different geographic regions. J Clin Microbiol. 1999;37:2703–2705. doi: 10.1128/jcm.37.8.2703-2705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J K, Curran R, Berridge S, Grabowska A M, Jameson C L, Thomson B J, Irving W L, Sharp P M. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J Gen Virol. 1999;80:1759–1768. doi: 10.1099/0022-1317-80-7-1759. [DOI] [PubMed] [Google Scholar]
- 3.Biagini P, Gallian P, Attoui H, Cantaloube J F, de Micco P, de Lamballerie X. Determination and phylogenetic analysis of partial sequences from TT virus isolates. J Gen Virol. 1999;80:419–424. doi: 10.1099/0022-1317-80-2-419. [DOI] [PubMed] [Google Scholar]
- 4.Erker J C, Leary T P, Desai S M, Chalmers M L, Mushahwar I K. Analyses of TT virus full-length genomic sequences. J Gen Virol. 1999;80:1743–1750. doi: 10.1099/0022-1317-80-7-1743. [DOI] [PubMed] [Google Scholar]
- 5.Gallian P, Berland Y, Olmer M, Raccah D, de Micco P, Biagini P, Simon S, Bouchouareb D, Mourey C, Roubicek C, Touinssi M, Cantaloube J F, Dussol B, de Lamballerie X. TT virus infection in French hemodialysis patients: study of prevalence and risk factors. J Clin Microbiol. 1999;37:2538–2542. doi: 10.1128/jcm.37.8.2538-2542.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hijikata M, Takahashi K, Mishiro S. Complete circular DNA genome of a TT virus variant (isolate name SANBAN) and 44 partial ORF2 sequences implicating a great degree of diversity beyond genotypes. Virology. 1999;260:17–22. doi: 10.1006/viro.1999.9797. [DOI] [PubMed] [Google Scholar]
- 7.Irving W L, Ball J K, Berridge S, Curran R, Grabowska A M, Jameson C L, Neal K R, Ryder S D, Thomson B J. TT virus infection in patients with hepatitis C: frequency, persistence, and sequence heterogeneity. J Infect Dis. 1999;180:27–34. doi: 10.1086/314825. [DOI] [PubMed] [Google Scholar]
- 8.Miyata H, Tsunoda H, Kazi A, Yamada A, Khan M A, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582–3586. doi: 10.1128/jvi.73.5.3582-3586.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushahwar I K, Erker J C, Muerhoff A S, Leary T P, Simons J N, Birkenmeyer L G, Chalmers M L, Pilot-Matias T J, Desai S M. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177–3182. doi: 10.1073/pnas.96.6.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagano K, Fukuda Y, Yokozaki S, Okada K, Tanaka K, Funahashi K, Hayakawa T. Low risk of TT virus (TTV) infection in medical workers. J Hosp Infect. 1999;42:243–246. doi: 10.1053/jhin.1999.0589. [DOI] [PubMed] [Google Scholar]
- 11.Niel C, de Oliveira J M, Ross R S, Gomes S A, Roggendorf M, Viazov S. High prevalence of TT virus infection in Brazilian blood donors. J Med Virol. 1999;57:259–263. [PubMed] [Google Scholar]
- 12.Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–97. doi: 10.1006/bbrc.1997.7765. [DOI] [PubMed] [Google Scholar]
- 13.Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1–16. [Google Scholar]
- 14.Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437–448. doi: 10.1006/viro.1999.9769. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428–436. doi: 10.1006/viro.1999.9770. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, Kato N, Iizuka H, Tsuda F, Miyakawa Y, Mayumi M. Distinct genotypes of a nonenveloped DNA virus associated with posttransfusion non-A to G hepatitis (TT virus) in plasma and peripheral blood mononuclear cells. J Med Virol. 1999;57:252–258. doi: 10.1002/(sici)1096-9071(199903)57:3<252::aid-jmv7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 17.Prescott L E, Simmonds P. Global distribution of transmission-transmitted virus. N Engl J Med. 1998;339:776–777. doi: 10.1056/NEJM199809103391118. [DOI] [PubMed] [Google Scholar]
- 18.Prescott L E, MacDonald D M, Davidson F, Mokili J, Pritchard D I, Arnot D E, Riley E M, Greenwood B M, Hamid S, Saeed A A, McClure M O, Smith D B, Simmonds P. Sequence diversity of TT virus in geographically dispersed human populations. J Gen Virol. 1999;80:1751–1758. doi: 10.1099/0022-1317-80-7-1751. [DOI] [PubMed] [Google Scholar]
- 19.Saback F L, Gomes S A, de Paula V S, da Silva R R S, Lewis-Ximenez L L, Niel C. Age-specific prevalence and transmission of TT virus. J Med Virol. 1999;59:318–322. doi: 10.1002/(sici)1096-9071(199911)59:3<318::aid-jmv10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Hoshino H, Ohta Y, Yoshida N, Mishiro S. Very high prevalence of TT virus (TTV) infection in general population of Japan revealed by a new set of PCR primers. Hepatol Res. 1998;12:233–239. [Google Scholar]
- 21.Takayama S, Yamazaki S, Matsuo S, Sugii S. Multiple infection of TT virus (TTV) with different genotypes in Japanese hemophiliacs. Biochem Biophys Res Commun. 1999;256:208–211. doi: 10.1006/bbrc.1999.0270. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka H, Okamoto H, Luengrojanakul P, Chainuvati T, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis patients and healthy blood donors in Thailand. J Med Virol. 1998;56:234–238. doi: 10.1002/(sici)1096-9071(199811)56:3<234::aid-jmv10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 23.Viazov S, Ross R S, Niel C, de Oliveira J M, Varenholz C, Roggendorf M. Sequence variability in the putative coding region of TT virus: evidence for two rather than several major types. J Gen Virol. 1998;79:3085–3089. doi: 10.1099/0022-1317-79-12-3085. [DOI] [PubMed] [Google Scholar]