Abstract
Rhodopsin comprises an opsin attached to a retinal chromophore and is the only visual pigment conferring dim-light vision in vertebrates. On activation by photons, the retinal group becomes detached from the opsin, which is then inactive until it is recharged. Of all vertebrate species, those that dive face unique visual challenges, experiencing rapid decreases in light level and hunting in near darkness. Here, we combine sequence analyses with functional assays to show that the rhodopsin pigments of four divergent lineages of deep-diving vertebrates have undergone convergent increases in their retinal release rate. We compare gene sequences and detect parallel amino acids between penguins and diving mammals and perform mutagenesis to show that a single critical residue fully explains the observed increases in retinal release rate in both the emperor penguin and beaked whale. At the same time, we find that other shared sites have no significant effect on retinal release, implying that convergence does not always signify adaptive significance. We propose that accelerated retinal release confers rapid rhodopsin recharging, enabling the visual systems of diving species to adjust quickly to changing light levels as they descend through the water column. This contrasts with nocturnal species, where adaptation to darkness has been attributed to slower retinal release rates.
Keywords: scotopic vision, visual pigment, retinal release rate, functional convergence, amniotes
Introduction
The independent transition to an aquatic niche across divergent groups of amniote vertebrates has been accompanied by widespread changes in the visual system for coping with dim-light conditions (Reuter and Peichl 2008). Of all aquatic/semiaquatic species, those that have evolved deep diving arguably encounter the most extreme conditions, with many taxa reaching depths of >1,000 m (aphotic zone), where sunlight is absent (Doyle et al. 2008; Robinson et al. 2012; Berrow et al. 2018). Since the origin of terrestrial amniotes, diving to depths of >150 m into the so-called mesopelagic zone (Warrant and Locket 2004) has evolved convergently in birds, reptiles and two groups of mammals (pinnipeds and cetaceans) (Levenson 2004), with the most extreme cases observed in whales (Schorr et al. 2014; Berrow et al. 2018). Molecular studies indicate that deep-diving mammals have partially or fully lost cone-mediated color vision, resulting from pseudogenization of the short-wavelength-sensitive opsin gene (Levenson et al. 2006; Emerling et al. 2015) and, in many cetaceans, also the middle/long-wavelength-sensitive opsin gene (Meredith et al. 2013).
Unlike cone opsins, the rhodopsin (RH1) pigment, encoded by the RHO gene, has been retained by almost all species (but see Niemiller et al. [2013]), highlighting its critical importance in low light conditions (Davies et al. 2012). Rhodopsin also shows broad conservation in spectral tuning (∼500 nm) across vertebrates, with the exception of some marine mammals and deep-sea fishes, in which spectral shifts to <490 nm are considered as adaptations to blue light environments of the deep ocean (Lythgoe and Dartnall 1970; McFarland 1971; Crescitelli 1990; Fasick and Robinson 2000; Yokoyama et al. 2008). Yet despite its high level of evolutionary conservation, there is growing evidence that rhodopsin kinetics varies across taxa.
When activated by light, rhodopsin is converted to its light-active state metarhodopsin II (Meta II), after which the retinal group starts to detach from the opsin (Farrens and Khorana 1995). The rate at which the retinal group is released (Meta II decay), measured as a half-life (Farrens and Khorana 1995), may affect light sensitivity, and variation across taxa appears to relate to visual adaptation. For example, in nocturnal mammals as well as some birds and fishes, slow rates appear to confer increased sensitivity to low light (Van Hazel et al. 2016; Hauser et al. 2017; Gutierrez et al. 2018), and this relationship has also been used to propose well-developed scotopic vision in early amniotes (Liu et al. 2019). Although physiological evidence is lacking, experimental work suggests that the slow release rate reflects the persistence of Meta II (Chen et al. 2012; Gutierrez et al. 2018). Alternatively, the slow release of the chromophore might have evolved as a mechanism to reduce the build-up of toxic all-trans-retinal in rod photoreceptors (Gutierrez et al. 2018).
More recently, observations from some fishes have uncovered cases of accelerated release rates (Van Nynatten et al. 2021). Although the adaptive significance of this result is not fully understood, rapid release may promote fast recharging of the rhodopsin pigment, thereby conferring adaptation to changing light conditions (Van Nynatten et al. 2021). If this explanation is correct, then diving species—which experience decreasing light levels during foraging bouts—might also benefit from rapid rhodopsin kinetics. Dungan and Chang (2017) compared the retinal release rates of in vitro-expressed rhodopsin pigments based on the killer whale and hippopotamus, and found similar values, although both were slower than the cow pigment (Dungan and Chang 2017). Despite this, it was suggested that deeper diving cetacean species might benefit from faster release rates, either for sensing bioluminescence or for adjusting to rapidly changing light conditions during dives (Dungan and Chang 2017).
To test whether the evolution of rhodopsin in diving taxa has indeed involved a reduction in light sensitivity, here we perform functional assays of retinal release in several divergent vertebrate lineages, spanning penguins, turtles, pinnipeds, and both baleen and toothed whales. Given that members of these former three groups can enter the water from land, and also do not echolocate, their reliance on rapidly recharging rhodopsin might be even greater than that of fully aquatic cetaceans.
Results and Discussion
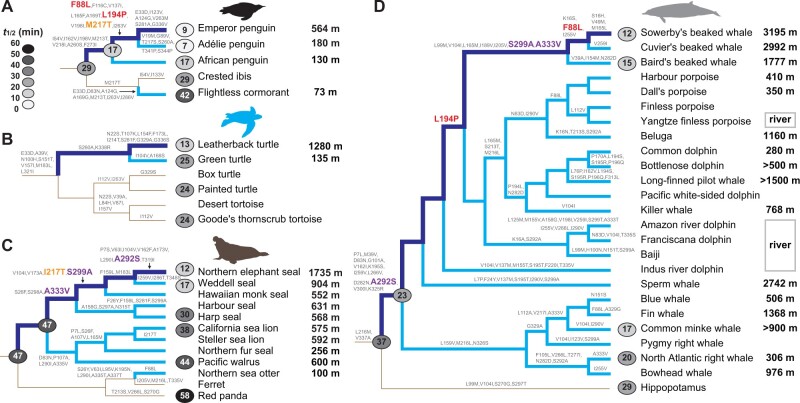
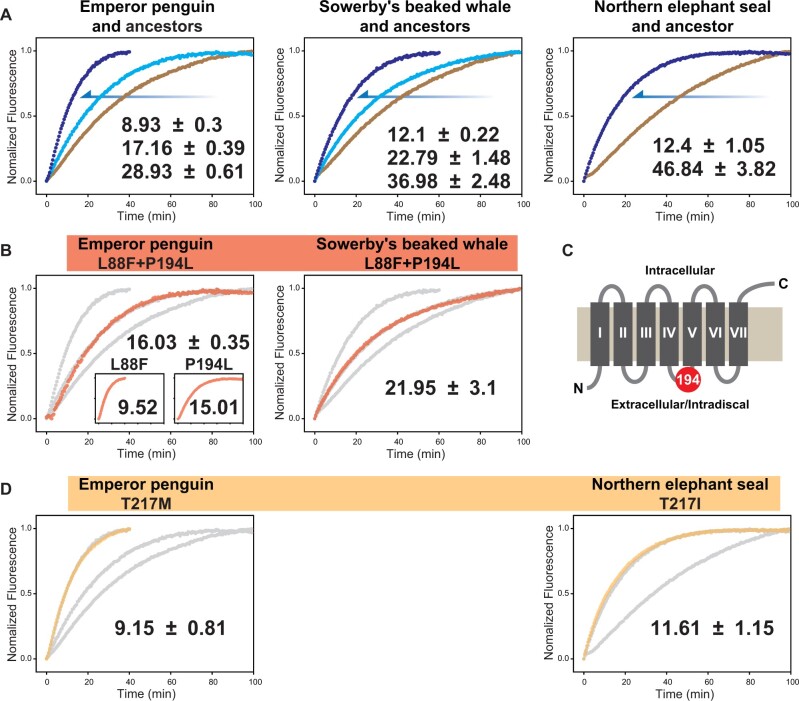
To determine whether the independent evolution of deep diving in birds, reptiles, and mammals was accompanied by molecular adaptations for dim-light vision, we expressed and measured the retinal release rates and spectral tuning of 21 extant pigments and five ancestral pigments. First, to assess how retinal release rates have evolved with the origin of deep diving in birds, we resurrected the ancestral pigment of the core waterbirds (Aequornithes), a monophyletic clade containing both diving and nondiving species. We recorded a retinal release half-life (t1/2) of 28.93 ± 0.61 min (fig. 1A and supplementary fig. S1A, Supplementary Material online, also see supplementary data 1, Supplementary Material online) that was similar to that of our published value for the ancestral bird (30.51 min) (Liu et al. 2019), indicative of little phenotypic change in the rhodopsin since the origin of birds. We also obtained and compared retinal release rates of several members of this clade and found that crested ibis (Nipponia nippon) retained a similar rate to the ancestral form (29.46 ± 2.96 min), but that the flightless cormorant (Nannopterum harrisi) showed a slower rate (42.14 ± 2.64 min) (two-tailed t-test, P = 0.002). In contrast, we recorded a much more rapid retinal release rate for the inferred rhodopsin protein of the ancestral penguin (17.16 ± 0.39 min), with further significant increases in the emperor penguin (Aptenodytes forsteri) and Adélie penguin (Pygoscelis adeliae) (t1/2 = 8.93 ± 0.3 and 7.25 ± 0.54 min, respectively) (P < 0.001), but not in the African penguin (Spheniscus demersus), which showed no amino acid differences. Such variation within this clade appears to correspond to differences in diving depths, implying that the observed changes are adaptive. Notably, the emperor penguin is known to dive up to 564 m (Wienecke et al. 2007), Adélie penguin up to 180 m (Watanuki et al. 1997), and the African penguin up to 130 m (Wilson 1985). In contrast to the dramatic shifts in retinal release rates in penguins, we detected slight changes in the absorption wavelength (λmax) (supplementary fig. S2A and see supplementary data 1, Supplementary Material online).
Fig. 1.
Convergent increases in rhodopsin retinal release rate associated with independent evolution of deep diving in birds, reptiles, and mammals. Molecular and phenotypic evolution of rhodopsin in penguins (A), turtles (B), pinnipeds (C), and cetaceans (D). Diving lineages are shown in pale blue, and the evolutionary path to the deepest diving species (emperor penguin, leatherback turtle, elephant seal, and Sowerby’s beaked whale) from its respective terrestrial ancestor is shown in dark blue. Amino acid substitutions on the four paths are orange if convergent between the emperor penguin and elephant seal, red if convergent between emperor penguin and Sowerby’s beaked whale, and purple between elephant seal and Sowerby’s beaked whale. For each phylogeny, retinal release rates for extant and ancestral pigments are shown on the tips and nodes, respectively, scaled by shade from pale (fast) to dark (slow). Maximum diving depths are listed on the right (also see supplementary table S1, Supplementary Material online).
Given that the phylogenetic relationship among the three main clades of penguins remains somewhat equivocal based on recent studies (Gavryushkina et al. 2017; Pan et al. 2019), we also reconstructed the ancestral protein sequence under an alternative topology using an expanded data set of newly available genomes (Pan et al. 2019). We found that although this second ancestral rhodopsin differed slightly in its retinal release rate (15.75 ± 0.63 min) compared with the first one, it nonetheless still supports an increase in retinal release at the origin of penguins (P < 0.001) (supplementary fig. S3, Supplementary Material online).
We also studied rhodopsin evolution in marine turtles and their terrestrial relatives (order Testudines), and found that the pigment in the leatherback turtle (Dermochelys coriacea) was characterized by a retinal release rate of t1/2 = 12.65 ± 0.44 min, approximately twice as fast as that of the other three testudine species examined (∼24–25 min) (P < 0.001): green turtle (Chelonia mydas), painted turtle (Chrysemys picta) and Goode’s thornscrub tortoise (Gopherus evgoodei) (fig. 1B and supplementary fig. S1B, Supplementary Material online). Although the green turtle is also a marine species, its maximum diving depth (135 m) (Rice and Balazs 2008) is much shallower than that of leatherback turtle (1,280 m) (Doyle et al. 2008), which can dive into the bathypelagic (aphotic) zone (>1,000 m) where the only light is from bioluminescence (Warrant and Locket 2004). In spite of this dramatic retinal release rate change, the leatherback showed a very slight downward shift in spectral tuning of 1–2 nm over the other turtle and tortoise species (supplementary fig. S2B, Supplementary Material online), strongly suggesting that adaptive evolution of rhodopsin has led to improved temporal acuity without altering spectral sensitivity.
Among mammals, deep diving has evolved in two main groups: the pinnipeds (seals and allies) and the cetaceans (whales, dolphins, and porpoises). We determined the rhodopsin phenotypes of nine species of pinniped and found further evidence of adaptive changes for light sensing linked to diving depth. Although the ancestral pinniped rhodopsin showed no amino acid replacements, implying no phenotypic change during the origin of this group (t1/2 = 46.84 ± 3.82 min), the early diversification of pinnipeds appears to have been accompanied by marked functional changes for altered light sensitivity (fig. 1C and supplementary fig. S1C, Supplementary Material online). Specifically, we recorded a significant increase in the retinal release rate of the California sea lion (Zalophus californianus) (37.72 ± 3.11 min) (P = 0.02), and independent, more dramatic changes in the eared seals (Phocidae). The two fastest pigments were recorded for the northern elephant seal (Mirounga angustirostris) and Weddell seal (Leptonychotes weddellii) (12.4 ± 1.05 and 16.73 ± 1.14 min, respectively) (P < 0.001), which are known to dive deep up to 1,735 m (Robinson et al. 2012) and 904 m (Heerah et al. 2013), respectively. These values support the results of behavioral experiments that revealed that the northern elephant seal is better adapted to dim-light environments compared with other pinnipeds and the human (Levenson and Schusterman 1999). In contrast, we found that the harp seal (Phoca groenlandica)—a more shallow diver that can reach ∼500 m—showed a less extreme shift in retinal release rate of 29.51 ± 4.76 min (P = 0.003), and that the walrus (Odobenus rosmarus) exhibited no significant shift (43.56 ± 3.83 min). Further support for a link between rhodopsin evolution and diving comes from distantly related terrestrial carnivores, with the dog (Canis familiaris) and red panda (Ailurus fulgens) exhibiting slower rates of retinal release (42.65 ± 2.86 and 57.62 ± 1.48 min, respectively) than most pinnipeds. Finally, we also recorded the spectral tuning of rhodopsins from pinnipeds and their relatives and found similar values to those already published (Lythgoe and Dartnall 1970; Southall et al. 2002), with the only major λmax-shift (−14 nm) in the northern elephant seal (supplementary fig. S2C, Supplementary Material online).
Cetaceans have evolved the most extreme diving depths of all the amniote vertebrates (Berrow et al. 2018). We determined the phenotypes of representative toothed whales (Odontoceti) and baleen whales (Mysticeti), and their sister taxon, the hippopotamus. We detected a marked shift in rhodopsin retinal release rate at the origin of modern cetaceans compared with their common ancestor with the hippopotamus (t1/2 = 22.79 ± 1.48 vs. 36.98 ± 2.48 min, respectively) (P = 0.001) (fig. 1D and supplementary fig. S1D, Supplementary Material online). This finding implies that the ancestor of extant whales was likely a deep diver, also supporting a previous study’s findings based on cetacean rhodopsins (Dungan 2018). Indeed, as expected, the rhodopsin retinal release rate of the semiaquatic hippopotamus (Hippopotamus amphibious) (28.95 ± 2.89 min) was found to be slower than that of its fully aquatic relatives.
Comparisons of rhodopsin phenotypes within cetaceans revealed further evolutionary shifts, with retinal release rates ranging from 20.14 ± 0.38 min in the right whale (Eubalaena glacialis) to 12.1 ± 0.22 min in Sowerby’s beaked whale (Mesoplodon bidens). Beaked whales dive into the aphotic zone, and Sowerby’s beaked whale has been recorded to dive up to 3,195 m, exceeding all other cetaceans (Minamikawa et al. 2007; Berrow et al. 2018). Thus, our functional assays of rhodopsin retinal release—the first to be conducted for beaked whales—provides compelling evidence that rhodopsin has undergone molecular adaptations related to deep diving. These results are particularly interesting given that, unlike our other focal groups (i.e., penguins, sea turtles, and pinnipeds), beaked whales are also able to echolocate. As such, our results suggest these taxa rely on both sensory modalities while diving (also see recent findings based on genomic analyses [McGowen et al. 2020]). In addition to the detected changes in retinal release rate, we also recorded previously reported shifts in spectral sensitivity at the origin of extant whales and during the subsequent diversification of this group (McFarland 1971; Fasick and Robinson 2000) (supplementary fig. S2D, Supplementary Material online).
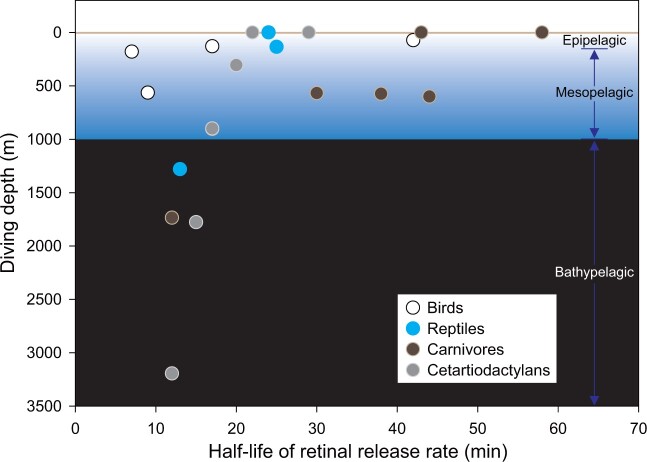
Taken together, our findings from vertebrates show that four independent origins of diving behavior have involved convergent evolution of the rhodopsin protein to increase the retinal release rate, with the shortest half-life values typically observed in species that dive to the greatest depths (fig. 2). To assess whether observed functional convergence has arisen through identical molecular changes, for each group we identified all amino acid substitutions occurring on the branch between the deepest diving taxon and its terrestrial ancestor. We detected a single convergent substitution between the emperor penguin and northern elephant seal (M217T and I217T, respectively), two parallel substitutions between the emperor penguin and Sowerby’s beaked whale (F88L and L194P), and three parallel changes between the northern elephant seal and Sowerby’s beaked whale (A292S, S299A, and A333V, fig. 1). All six of these replacements showed high posterior probabilities of being convergent (P > 0.95, PCOC), providing additional support that rhodopsin phenotypic convergence is driven at least partially by the same molecular mechanisms. Previously, the replacements A292S and S299A were reported in some cetartiodactyls, where they were suggested to modulate retinal release via epistatic effects (Dungan and Chang 2017), with the first substitution also showing a major effect on spectral tuning (Yokoyama et al. 2008). At the same time, however, it is intriguing that we detected no shared derived residues between the leatherback turtle and any of the other three deepest diving taxa, implying that functional convergence in this lineage has evolved via a separate as yet unknown mechanism, also see Chi et al. (2020).
Fig. 2.
Retinal release of rhodopsin versus diving depth. Retinal release rates and diving depths show for species from four vertebrate clades. The diving depths of terrestrial relatives are set at zero. The ocean is divided into three zones: epipelagic (<∼150 m), mesopelagic (∼150–1,000 m), and bathypelagic (>1,000 m, also called aphotic) (Warrant and Locket 2004).
To determine whether the observed functional convergence between the emperor penguin and each of the beaked whale and elephant seal has been driven by the same molecular mechanisms (fig. 3A), we examined retinal release rates in wild-type pigments in which we replaced either derived single residues or pairs of residues with their respective ancestral states. We first showed that the double mutants (L88F and P194L) constructed for both the emperor penguin and beaked whale rhodopsins each almost recovered the phenotypes of their respective ancestors. Next, by producing single mutants for the emperor penguin based on these respective substitutions, we confirmed that P194L was the key substitution underlying the retinal release rate shift (15.01 ± 1.37 min) (P = 0.014) (fig. 3B and see supplementary data 1, Supplementary Material online). This site is located in an intradiscal domain, which is considered critical for rhodopsin folding and Meta II kinetics (Sakai et al. 2010) (fig. 3C). In contrast, L88F had no significant effect on retinal release (fig. 3B), and single mutants for penguin and elephant seal rhodopsins based on T217M and T217I also revealed no functional impacts (fig. 3D). Thus our comparisons of diving vertebrates highlight how phenotypic convergence can result from both convergent and divergent molecular adaptations, and also demonstrates cases whereby such convergent molecular changes appear to have no clear bearing on the phenotype. In this respect, it is also noteworthy that none of these mutations appeared to alter the spectral sensitivity of rhodopsin (supplementary fig. S4, Supplementary Material online), including the previously reported tuning site 194 (Yokoyama et al. 2008; Borges et al. 2015).
Fig. 3.
Impact of convergent/parallel amino acid substitutions on retinal release. (A) Convergence in rhodopsin retinal release during the evolution of the emperor penguin, Sowerby’s beaked whale, and the northern elephant seal. In each case, the retinal release rate undergoes a downward shift from the terrestrial ancestor (brown) to the first aquatic ancestor (pale blue) to the extant taxon (dark blue). (B) For the two parallel substitutions between emperor penguin and Sowerby’s beaked whale (red), double mutants of each wildtype were constructed in which the shared residues were replaced by the ancestral forms (L88F and P194L). For the emperor penguin, these backward substitutions were also tested separately using single mutants (see inset). Gray lines indicate the ancestral and wild-type values. (C) The position of site 194 (red circle) mapped onto rhodopsin structure, with transmembrane helices I–VII shown as boxes. (D) For the convergent substitution between emperor penguin and elephant seal (orange), two single mutants of the wild-type were constructed, in which the shared residue was replaced by alternate ancestral forms (T217M or T217I). For each plot, the mean t1/2 value and standard deviation are given. Gray lines indicate the ancestral and wild-type values.
We observed clear differences in retinal release rate and spectral tuning between diving and nondiving taxa across four groups of vertebrates. To assess whether these patterns were significant after accounting for phylogenetic nonindependence in the data, we ran separate analyses of phylogenetically independent contrasts between diving depth and each of these parameters and found that both were significant (supplementary fig. S5, Supplementary Material online). However, given that these two phenotypic traits also co-vary, and that molecular adaptations for spectral tuning along the rhodopsin molecule could theoretically influence retinal release rate, and vice versa, we also ran generalized linear mixed models (GLMMs) to assess whether diving could explain variance in retinal release after accounting for the effects of spectral tuning and phylogeny. Although spectral tuning was a significant predictor of retinal release (deviance information criteria [DIC] = 30.411, PMCMC = 0.012, mean posterior = 1.09), we found that adding log-diving depth led to a substantial improvement in model fit (DIC = 18.195; log diving: PMCMC = 0.094, mean posterior = −0.017; spectral: PMCMC = 0.751 and mean posterior = 0.2). Finally, to confirm the importance of log-diving depth as a predictor of retinal release, we also fitted a univariate model and found further improvement in model fit (DIC = 7.193, PMCMC = 0.002, mean posterior = −0.019). These results thus imply that the evolution of rapid rhodopsin kinetics in diving species is not solely an artefact of shifts to blue light. Certainly, these two aspects of dim-light vision are likely to be driven by different ecological factors, also see Fasick and Robinson (2000). Indeed, it is noteworthy that although all four focal lineages of deep divers showed faster rates of retinal release, the penguins and sea turtles both showed little change in spectral tuning (supplementary fig. S2A and B, Supplementary Material online). From a plot of retinal release versus spectral tuning, the largest residual values correspond to the two species of beaked whale, a baleen whale, and the elephant seal, all of which show more extreme shifts to blue light than other divers with similar retinal release rates (supplementary fig. S6, Supplementary Material online). The reason for this is unclear; however, one possibility is that these taxa specialize on blue light-emitting bioluminescent prey, a scenario that is also supported by light sensor data collected for the elephant seal (Vacquie-Garcia et al. 2012).
Despite the strong signature that rhodopsin has undergone functional convergence for more rapid retinal release across diving vertebrates, the adaptive significance of these changes in kinetics is not fully clear. One potential explanation for the association between retinal release rate and diving depth is the nonlinear attenuation of visible light in water (Kirk 1977). Indeed, light intensity varies widely across the mesopelagic zone (∼150–1,000 m) (Warrant and Locket 2004), and deep-diving species will experience more rapidly depleting light levels than those that dive to more shallow depths. Therefore, for such diving species to be able to detect prey while also descending rapidly (Levenson and Schusterman 1997, 1999), they may benefit from the rapid recharging of rhodopsin driven by fast retinal release (Ala-Laurila et al. 2006; Dungan and Chang 2017). Such enhanced so-called “dark adaptation” has also been recorded in some fishes (Van Nynatten et al. 2021). This evolutionary pressure is expected to scale with diving depth although it is noteworthy that the retinal release half-life showed no further decrease in species that dive beyond 1000 m, suggesting that this represents a lower limit (half-life) for conferring light sensitivity under the sea. Finally, it is also plausible that by conferring the ability to detect changes in light levels, the rapid recharging of rhodopsin mediated by retinal release may also allow diving species to monitor depth, information that is critical in determining when to resurface for oxygen. This explanation could account for why rhodopsin-mediated light sensing might remain important in toothed whales, which can also use echolocation for hunting.
Materials and Methods
RH1 Sequences from Deep-Sea Diving Vertebrates
To determine whether the evolution of deep diving in vertebrates has involved phenotypic adaptation in the rhodopsin pigment, we focused on four divergent groups: penguins (Aves: Sphenisciformes), turtles (Reptilia: Testudines), pinnipeds (Mammalia: Carnivora), and cetaceans (Mammalia: Cetartiodactyla). For each group, we obtained published RH1 sequences from NCBI (https://www.ncbi.nlm.nih.gov, last accessed October 2019) of representative taxa spanning the full range of diving depths, and combined these with data from their respective nondiving relatives.
For penguins, we retrieved complete RH1 sequences for two penguins, two other nondeep-diving members of the same clade of core waterbirds, and 12 outgroups (see supplementary table S1, Supplementary Material online). We also generated one new gene sequence, for the African penguin (Spheniscus demersus), using muscle tissue from a dead individual obtained from Shanghai Zoo. For this, genomic DNA was extracted using the TIANamp kit (Tiangen) and amplified as three segments using custom-designed primer sets (supplementary table S2, Supplementary Material online). Amplicons were purified, ligated with pGEM-T Easy vector (Promega), and transformed into Trans5α competent cells (TransGen Biotech). Positive clones from each of the three reactions were sequenced by ABI 3730 (Applied Biosystems). We also obtained a newly available set of RH1 sequences from 18 penguin species (Pan et al. 2019) from the China National GeneBank DataBase (https://db.cngb.org/cnsa). For turtles, we obtained RH1 sequences for two diving marine species and five nonmarine species. For pinnipeds, we obtained RH1 sequences for five eared seals (Phocidae), three earless seals (Otariidae), the walrus (Odobenidae), and 13 carnivoran relatives. Finally, for whales, we obtained RH1 sequences for 24 species, covering 12 cetacean families and both Odontoceti and Mysticeti. All RH1 sequences analyzed in this study with corresponding database accession numbers are listed in supplementary table S1, Supplementary Material online.
Molecular Evolution of Vertebrate Rhodopsin
Amino acid sequences of rhodopsin were aligned separately for each group using MEGA X (Kumar et al. 2018). We estimated the best-fitting amino acid substitution model based on the corrected Akaike information criterion in ProtTest 3 (Darriba et al. 2011). The best-fitting models for birds was MtMam+I+Г+F, and, for reptile, carnivore and Cetartiodactyla, it was JTT+Г+F. Data sets were used to infer rhodopsin ancestral sequences using Codeml in PAML 4 (Yang 2007), based on published species tree topologies (see supplementary data 2, Supplementary Material online). For the penguins, a further ancestral reconstruction of rhodopsin was conducted using an expanded data set and also a different species phylogeny (Pan et al. 2019). For each of the four focal groups of diving species, we inferred all amino acid substitutions between the deepest diving species (emperor penguin, Aptenodytes forsteri; leatherback turtle, Dermochelys coriacea; northern elephant seal, Mirounga angustirostris; and Sowerby’s beaked whale, Mesoplodon bidens) and its respective terrestrial ancestor. We compared the four sets of substitutions and identified shared replacements, which we confirmed were convergent by running the software PCOC (Rey et al. 2018) on the relevant pairs of branches under the species tree topology.
In Vitro Functional Assays for Rhodopsins
To quantify the phenotypic consequences of amino acid replacements that occurred in the evolution of deep diving, we performed in vitro expression of wild type and mutant rhodopsin pigments. For each of our four focal groups, we first determined the phenotypes of rhodopsin pigments in several deep-diving species and their nondiving relatives. From the birds, we focused on the emperor penguin (Aptenodytes forsteri) and Adélie penguin (Pygoscelis adeliae), African penguin (Spheniscus demersus), ibis (Nipponia nippon), and cormorant (Nannopterum harrisi); from the reptiles the leatherback turtle (Dermochelys coriacea) (family Dermochelyidae), green turtle (Chelonia mydas) (Cheloniidae), and two nonmarine species (Chrysemys picta and Gopherus evgoodei); from pinnipeds, three eared-seals (harp seal, Phoca groenlandica; northern elephant seal, Mirounga angustirostris; Weddell seal, Leptonychotes weddellii), one earless-seal (California sea lion, Zalophus californianus), and the walrus (Odobenus rosmarus), and two terrestrial relatives (Ailurus fulgens and Canis familiaris); and finally, from four cetaceans, Mesoplodon bidens, Berardius bairdii, Balaenoptera acutorostrata, Eubalaena glacialis, and also the hippopotamus (Hippopotamus amphibious).
To express rhodopsin, we added a 5′-Kozak sequence (CCACC) and a 3′-purification tag (ACA GAG ACC AGC CAA GTG GCG CCT GCC) into RH1 coding sequence. Genes were cut (HindIII and XhoI sites) and ligated into a pcDNA3.1 (+) vector (Invitrogen), and then transfected (Xfect reagent, Clontech) into HEK293T cells. Cells that showed surface expression of rhodopsin were harvested after 48 h, to which we added a chromophore 11-cis-retinal group to regenerate visual pigments. Rhodopsin pigments were solubilized with detergent n-dodecyl β-d-maltoside (Macklin) and then purified (Rho 1D4 monoclonal antibody, University of British Columbia). Finally, rhodopsins were eluted in a HEPES buffer (50 mM, pH = 6.6) containing 140 mM NaCl, 3 mM MgCl2, 0.1% maltoside, 20% glycerol, and 40 μM rhodopsin epitope (GenScript) (also see Liu et al. [2018)]).
Retinal release rates were recorded for each rhodopsin using a Cary Eclipse fluorescence spectrophotometer (Agilent). After photobleaching, measurements were carried out at 20°C and with a 30-s interval. The excitation wavelength was set at 295 nm with a 2.5-nm slit, and the emission wavelength at 330 nm with a 10-nm slit. The rate of retinal release was fitted following the equation y = y0 + a(1 − e−bx) (all r2 > 0.99), and the half-life of the release rate was calculated by t1/2 = ln2/b following published procedures (Farrens and Khorana 1995; Liu et al. 2019). For each rhodopsin pigment examined, we repeated the experimental expression 3–4 times and took independent replicate measurements of retinal release rate from which we obtained the respective means and standard deviations. Half-life values between pairs of rhodopsin pigments were compared (two-tailed t-test). We also measured spectral tuning (λmax) by using a U-3900 spectrophotometer (Hitachi) for each pigment.
To assess whether variance in spectral tuning and retinal release could be explained by diving depth, we ran phylogenetically independent contrasts using PDAP in Mesquite 3 (Maddison WP and Maddison DR 2019). For this, we used species divergence times based on literatures (see supplementary fig. S7, Supplementary Material online). We also built more complex generalized linear mixed effects models using the MCMCglmm package (Hadfield 2010) in which we assessed the impact of diving depth (log-transformed) on retinal release after accounting for spectral tuning. For these models, we used the same ultrametric tree to generate a random effect of phylogenetic distance. We compared models containing each trait in isolation (as fixed effects) with an additive model containing both traits. Models were run for 5,000,000 iterations, with a burn-in phase of 50,000 iterations, and a thinning interval of 500. Uninformative priors were used in all models, with an inverse-Gamma distribution for both the residual variance structure of the covariates, and the random factor (V = 1, nu = 0.02). Models were run three times, and the model with the median DIC was selected. Model fit was compared using DIC, with significance ascertained using PMCMC, which can be interpreted as Bayesian equivalents of a P value (Hadfield et al. 2013).
To assess whether parallel increases in retinal release between some deep-diving lineages were driven by specific convergent substitutions, we conducted site-directed mutagenesis. We focused on positions 88 and 194 (parallel substitutions between emperor penguin and Sowerby’s beaked whale) and also for 217 (convergent substitution between emperor penguin and northern elephant seal). We introduced backward mutations into the wild-type rhodopsins by using TransStart FastPfu DNA polymerase (TransGen Biotech) with the target site changed in the primers. After amplification, DpnI enzyme (New England Biolabs) was added to digest the original plasmid templates. PCR products were transformed into DH5α competent cells (Tiangen Biotech) and verified by sequencing. The phenotypes (t1/2 and λmax) of mutant rhodopsin were also measured as described above.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Y. Koutalos for helpful comments on the manuscript, and R. Crouch (Medical University of South Carolina) and L. Neuhold (National Eye Institute, National Institutes of Health) for providing 11-cis-retinal. This work was supported by the Fundamental Research Funds for the Central Universities (GK202102006), the Natural Science Basic Research Program of Shaanxi (2021JM-197), and the National Natural Science Foundation of China (31601855) to Y.L., and also funded by the European Research Council Starting grant (310482) to S.J.R.
Author Contributions
Y.L. designed the project. Y.L. and A.W. contributed experimental materials; Y.X., Y.C., F.L., and X.C. did the experiments; Y.X., H.C., J.H.T.P., J.W., S.J.R., and Y.L. analyzed the data; Y.X., S.J.R., and Y.L. wrote the article.
Data Availability
A new RH1 sequence has been deposited to GenBank, with the accession number MW288702. The accession numbers for all gene sequences studied are listed in supplementary table S1, Supplementary Material online. All phenotypic data of rhodopsin discussed are available in Supplementary Material online.
References
- Ala-Laurila P, Kolesnikov AV, Crouch RK, Tsina E, Shukolyukov SA, Govardovskii VI, Koutalos Y, Wiggert B, Estevez ME, Cornwall MC.. 2006. Visual cycle: dependence of retinol production and removal on photoproduct decay and cell morphology. J Gen Physiol. 128(2):153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrow S, Meade R, Marrinan M, McKeogh E, O’Brien J.. 2018. First confirmed sighting of Sowerby’s beaked whale (Mesoplodon bidens (Sowerby, 1804)) with calves in the Northeast Atlantic. Mar Biodivers Rec. 11(1):20. [Google Scholar]
- Borges R, Khan I, Johnson WE, Gilbert MT, Zhang G, Jarvis ED, O’Brien SJ, Antunes A.. 2015. Gene loss, adaptive evolution and the co-evolution of plumage coloration genes with opsins in birds. BMC Genomics. 16:751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Kuemmel C, Birge RR, Knox BE.. 2012. Rapid release of retinal from a cone visual pigment following photoactivation. Biochemistry 51(20):4117–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H, Cui Y, Rossiter SJ, Liu Y.. 2020. Convergent spectral shifts to blue-green vision in mammals extends the known sensitivity of vertebrate M/LWS pigments. Proc Natl Acad Sci U S A. 117(15):8303–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crescitelli F. 1990. Adaptations of visual pigments to the photic environment of the deep sea. J Exp Zool Suppl. 5:66–75. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WI, Collin SP, Hunt DM.. 2012. Molecular ecology and adaptation of visual photopigments in craniates. Mol Ecol. 21(13):3121–3158. [DOI] [PubMed] [Google Scholar]
- Doyle TK, Houghton JD, O’Súilleabháin PF, Hobson VJ, Marnell F, Davenport J, Hays GC.. 2008. Leatherback turtles satellite-tagged in European waters. Endang Species Res. 4:23–31. [Google Scholar]
- Dungan S. 2018. The molecular evolution of cetacean dim-light vision: in vitro studies of rhodopsin over a macroevolutionary transition [doctoral thesis]. Toronto: University of Toronto.
- Dungan SZ, Chang BS.. 2017. Epistatic interactions influence terrestrial-marine functional shifts in cetacean rhodopsin. Proc R Soc B. 284:20162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerling CA, Huynh HT, Nguyen MA, Meredith RW, Springer MS.. 2015. Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc R Soc B. 282:20151817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrens DL, Khorana HG.. 1995. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J Biol Chem. 270(10):5073–5076. [DOI] [PubMed] [Google Scholar]
- Fasick JI, Robinson PR.. 2000. Spectral-tuning mechanisms of marine mammal rhodopsins and correlations with foraging depth. Vis Neurosci. 17(5):781–788. [DOI] [PubMed] [Google Scholar]
- Gavryushkina A, Heath TA, Ksepka DT, Stadler T, Welch D, Drummond AJ.. 2017. Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst Biol. 66(1):57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez EA, Castiglione GM, Morrow JM, Schott RK, Loureiro LO, Lim BK, Chang BSW.. 2018. Functional shifts in bat dim-light visual pigment are associated with differing echolocation abilities and reveal molecular adaptation to photic-limited environments. Mol Biol Evol. 35(10):2422–2434. [DOI] [PubMed] [Google Scholar]
- Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw. 33:1–22.20808728 [Google Scholar]
- Hadfield JD, Heap EA, Bayer F, Mittell EA, Crouch NMA.. 2013. Disentangling genetic and prenatal sources of familial resemblance across ontogeny in a wild passerine. Evolution 67(9):2701–2713. [DOI] [PubMed] [Google Scholar]
- Hauser FE, Ilves KL, Schott RK, Castiglione GM, Lopez-Fernandez H, Chang BSW.. 2017. Accelerated evolution and functional divergence of the dim light visual pigment accompanies cichlid colonization of Central America. Mol Biol Evol. 34(10):2650–2664. [DOI] [PubMed] [Google Scholar]
- Heerah K, Andrews-Goff V, Williams G, Sultan E, Hindell M, Patterson T, Charrassin JB.. 2013. Ecology of Weddell seals during winter: influence of environmental parameters on their foraging behaviour. Deep Sea Res 2 Top Stud Oceanogr. 88 − 89:23–33. [Google Scholar]
- Kirk JTO. 1977. Attenuation of light in natural waters. Mar Freshwater Res. 28(4):497–508. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson DH. 2004. The visual pigments of diving tetrapods: genetic and electroretinographic investigations of pinnipeds, cetaceans, sea turtles, and penguins [doctoral thesis]. [San Diego (CA)]: University of California.
- Levenson DH, Ponganis PJ, Crognale MA, Deegan JF II, Dizon A, Jacobs GH.. 2006. Visual pigments of marine carnivores: pinnipeds, polar bear, and sea otter. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 192(8):833–843. [DOI] [PubMed] [Google Scholar]
- Levenson DH, Schusterman RJ.. 1997. Pupillometry in seals and sea lions: ecological implications. Can J Zool. 75(12):2050–2057. [Google Scholar]
- Levenson DH, Schusterman RJ.. 1999. Dark adaptation and visual sensitivity in shallow and deep-diving pinnipeds. Mar Mammal Sci. 15(4):1303–1313. [Google Scholar]
- Liu Y, Chi H, Li L, Rossiter SJ, Zhang S.. 2018. Molecular data support an early shift to an intermediate-light niche in the evolution of mammals. Mol Biol Evol. 35(5):1130–1134. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cui Y, Chi H, Xia Y, Liu H, Rossiter SJ, Zhang S.. 2019. Scotopic rod vision in tetrapods arose from multiple early adaptive shifts in the rate of retinal release. Proc Natl Acad Sci U S A. 116(26):12627–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe JN, Dartnall HJA.. 1970. A “deep sea rhodopsin” in a mammal. Nature 227(5261):955–956. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR.. 2019. Mesquite: a modular system for evolutionary analysis. Version 3.61. Available from: http://www.mesquiteproject.org.
- McFarland WN. 1971. Cetacean visual pigments. Vision Res. 11(10):1065–1076. [DOI] [PubMed] [Google Scholar]
- McGowen MR, Tsagkogeorga G, Williamson J, Morin PA, Rossiter SJ.. 2020. Positive selection and inactivation in the vision and hearing genes of cetaceans. Mol Biol Evol. 37(7):2069–2083. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS.. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9(4):e1003432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamikawa S, Iwasaki T, Kishiro T.. 2007. Diving behaviour of a Baird’s beaked whale, Berardius bairdii, in the slope water region of the western North Pacific: first dive records using a data logger. Fisheries Oceanogr. 16(6):573–577. [Google Scholar]
- Niemiller ML, Fitzpatrick BM, Shah P, Schmitz L, Near TJ.. 2013. Evidence for repeated loss of selective constraint in rhodopsin of amblyopsid cavefishes (Teleostei: Amblyopsidae). Evolution 67(3):732–748. [DOI] [PubMed] [Google Scholar]
- Pan H, Cole TL, Bi X, Fang M, Zhou C, Yang Z, Ksepka DT, Hart T, Bouzat JL, Argilla LS, et al. 2019. High-coverage genomes to elucidate the evolution of penguins. GigaScience 8(9):giz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter T, Peichl L.. 2008. Structure and function of the retina in aquatic tetrapods. In: Thewissen JGM, Nummela S, editors. Sensory evolution on the threshold: adaptations in secondarily aquatic vertebrates. Berkeley (CA: ): University of California Press. p. 149−172. [Google Scholar]
- Rey C, Gueguen L, Semon M, Boussau B.. 2018. Accurate detection of convergent amino-acid evolution with PCOC. Mol Biol Evol. 35(9):2296–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice MR, Balazs GH.. 2008. Diving behavior of the Hawaiian green turtle (Chelonia mydas) during oceanic migrations. J Exp Mar Biol Ecol. 356(1 − 2):121–127. [Google Scholar]
- Robinson PW, Costa DP, Crocker DE, Gallo-Reynoso JP, Champagne CD, Fowler MA, Goetsch C, Goetz KT, Hassrick JL, Huckstadt LA, et al. 2012. Foraging behavior and success of a mesopelagic predator in the northeast Pacific Ocean: insights from a data-rich species, the northern elephant seal. PLoS One 7(5):e36728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Imamoto Y, Yamashita T, Shichida Y.. 2010. Functional analysis of the second extracellular loop of rhodopsin by characterizing split variants. Photochem Photobiol Sci. 9(11):1490–1497. [DOI] [PubMed] [Google Scholar]
- Schorr GS, Falcone EA, Moretti DJ, Andrews RD.. 2014. First long-term behavioral records from Cuvier’s beaked whales (Ziphius cavirostris) reveal record-breaking dives. PLoS One 9(3):e92633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall KD, Oliver GW, Lewis JW, Le Boeuf BJ, Levenson DH.. 2002. Visual pigment sensitivity in three deep diving marine mammals. Mar Mammal Sci. 18(1):275–281. [Google Scholar]
- Vacquie-Garcia J, Royer F, Dragon AC, Viviant M, Bailleul F, Guinet C.. 2012. Foraging in the darkness of the Southern Ocean: influence of bioluminescence on a deep diving predator. PLoS One 7(8):e43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hazel I, Dungan SZ, Hauser FE, Morrow JM, Endler JA, Chang BS.. 2016. A comparative study of rhodopsin function in the great bowerbird (Ptilonorhynchus nuchalis): spectral tuning and light-activated kinetics. Protein Sci. 25(7):1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nynatten A, Castiglione GM, Gutierrez EA, Lovejoy NR, Chang BSW.. 2021. Recreated ancestral opsin associated with marine to freshwater croaker invasion reveals kinetic and spectral adaptation. Mol Biol Evol. 38(5):2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrant EJ, Locket NA.. 2004. Vision in the deep sea. Biol Rev Camb Philos Soc. 79(3):671–712. [DOI] [PubMed] [Google Scholar]
- Watanuki Y, Kato A, Naito Y, Robertson G, Robinson S.. 1997. Diving and foraging behaviour of Adelie penguins in areas with and without fast sea-ice. Polar Biol. 17:296–304. [Google Scholar]
- Wienecke B, Robertson G, Kirkwood R, Lawton K.. 2007. Extreme dives by free-ranging emperor penguins. Polar Biol. 30(2):133–142. [Google Scholar]
- Wilson RP. 1985. The Jackass Penguin (Spheniscus demersus) as a pelagic predator. Mar Ecol Prog Ser. 25:219–227. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L.. 2008. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci U S A. 105(36):13480–13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A new RH1 sequence has been deposited to GenBank, with the accession number MW288702. The accession numbers for all gene sequences studied are listed in supplementary table S1, Supplementary Material online. All phenotypic data of rhodopsin discussed are available in Supplementary Material online.