Abstract
Thirty-eight bovine mammary Staphylococcus aureus isolates from diverse clinical, temporal, and geographical origins were genotyped by pulsed-field gel electrophoresis (PFGE) after SmaI digestion of prokaryotic DNA and by means of binary typing using 15 strain-specific DNA probes. Seven pulsed-field types and four subtypes were identified, as were 16 binary types. Concordant delineation of genetic relatedness was documented by both techniques, yet based on practical and epidemiological considerations, binary typing was the preferable method. Genotypes of bovine isolates were compared to 55 previously characterized human S. aureus isolates through cluster analysis of binary types. Genetic clusters containing strains of both human and bovine origin were found, but bacterial genotypes were predominantly associated with a single host species. Binary typing proved an excellent tool for comparison of S. aureus strains, including methicillin-resistant S. aureus, derived from different host species and from different databases. For 28 bovine S. aureus isolates, detailed clinical observations in vivo were compared to strain typing results in vitro. Associations were found between distinct genotypes and severity of disease, suggesting strain-specific bacterial virulence. Circumstantial evidence furthermore supports strain-specific routes of bacterial dissemination. We conclude that PFGE and binary typing can be successfully applied for genetic analysis of S. aureus isolates from bovine mammary secretions. Binary typing in particular is a robust and simple method and promises to become a powerful tool for strain characterization, for resolution of clonal relationships of bacteria within and between host species, and for identification of sources and transmission routes of bovine S. aureus.
Infections due to staphylococci are of major importance to veterinary and human medicine. Staphylococcus aureus is one of the most significant pathogens causing intramammary infections in dairy cattle worldwide (6, 15, 34). In humans, S. aureus is a major cause of community-acquired as well as nosocomial morbidity and mortality. In the most recent decades, the increasing prevalence of methicillin-resistant S. aureus (MRSA) strains has become an additional infection control problem in human medicine (4, 7, 25). Staphylococcal strains may vary considerably in virulence and epidemiological potential. To control the spread of staphylococcal infections, sources of contamination and mechanisms of transmission must be identified. Detailed pathogenetic and epidemiological studies depend on the availability of typing systems that differentiate between strains belonging to the same bacterial species.
In veterinary microbiology, many techniques have been applied for characterization of bovine S. aureus strains. Phenotypic methods include phage typing (3, 13, 37), biotyping (11, 23), and multilocus enzyme electrophoresis (MLEE) (12, 17). Genotypic methods include single-gene typing systems, such as detection of coagulase gene polymorphism (2, 36) and ribotyping (3, 12, 29), and whole-genome typing systems, such as arbitrarily primed PCR (12, 21, 24). Furthermore, plasmid pattern analysis has been used to differentiate among S. aureus isolates of bovine origin, based on the diversity of extrachromosomal DNA (3). In human microbiology, most of these procedures have been superseded by newer methods that have enhanced resolving powers, including pulsed-field gel electrophoresis (PFGE) of DNA macrorestriction fragments (5, 28, 33) and, more recently, binary typing (40). PFGE is a reliable and reproducible method with high discriminatory power. Drawbacks of this method are that it is laborious and expensive and that complex DNA patterns may be difficult to interpret, especially for large collections of isolates (38, 39). For clinical laboratories processing great numbers of samples, these limitations may be impediments to routine use. Binary typing is a highly reproducible and stable library typing method with excellent discriminatory abilities. It has the additional advantage of producing a simple binary output, facilitating interpretation and comparison of typing results, and it lacks experimentally unstable parameters, such as electrophoretic conditions (42). Recently, several authors have reported the use of PFGE for characterization of bovine isolates, but so far binary typing has not been applied to S. aureus isolates of bovine origin (3a, 21a).
The purpose of this study was to determine whether PFGE and binary typing are suitable techniques for the differentiation of isolates of S. aureus recovered from bovine mammary secretions. In addition, a collection of bovine isolates was compared to a collection of human isolates, including methicillin-resistant strains, to explore clonal relatedness of isolates from cattle and humans as determined by binary typing. Finally, associations of bacterial strains with clinical observations in cattle were examined to identify possible relations between genotypes and bacterial virulence or routes of spread.
MATERIALS AND METHODS
Bacterial isolates.
This study included 38 bovine S. aureus isolates collected from eight dairy herds in The Netherlands between May 1997 and February 1999. Three herds (I, II, and III) were involved in a longitudinal survey of population dynamics of intramammary infections. In those herds, milk samples were routinely collected from all four udder quarters of each cow at intervals of 3 weeks for 81 weeks. Samples from the other five herds were submitted to the diagnostic laboratory of the Animal Health Service, Deventer, The Netherlands, as part of a dairy health improvement scheme. Bacteria were cultured from milk samples according to National Mastitis Council standards (16) and identified at the species level as described previously (20). Isolates were stored frozen until further use. Isolates were selected to represent different geographical, temporal, and clinical origins (Table 1).
TABLE 1.
Summary of epidemiological data, PFGE typing data, and binary typing results for 38 bovine S. aureus isolates
| Herd no. | Isolate no. | Cow no.-quarter no.a | Collection period | Clinical characteristics | Clinical groupb | Pulsotype | Binary codec | Binary typed |
|---|---|---|---|---|---|---|---|---|
| I | 1 | 75-1 | August 1997 | Chronic subclinical, high SCC | 2 | A | 001111111111101 | 8189 |
| 2 | 75-2 | August 1997 | Chronic subclinical, high SCC | 2 | B | 001111111111101 | 8189 | |
| 3 | 75-4 | August 1997 | Chronic subclinical, high SCC | 2 | B | 001111111111101 | 8189 | |
| 5 | 78-3 | August 1997 | Chronic subclinical, high SCC | 2 | B.1 | 001111111111101 | 8189 | |
| 6 | 63-3 | June 1997 | Short duration, high SCC | 3 | B | 001111111111111 | 8191 | |
| 7 | 77-4 | October 1997 | Short duration, low SCC | 1 | C | 000010011000001 | 1217 | |
| 8 | 67-3 | January 1998 | Acute severe clinical disease | 4 | D | 101010011110011 | 21747 | |
| 9 | 74-4 | December 1997 | Acute severe clinical disease | 4 | D | 101010011110011 | 21747 | |
| 10 | 75-4e | December 1997 | Acute severe clinical disease | 4 | D | 101010011110001 | 21745 | |
| 12 | 25-4 | May 1997 | Short duration, high SCC | 3 | D | 101010011110011 | 21747 | |
| 13 | 25-4 | May 1998 | Short duration, high SCC | 3 | D | 101010011110001 | 21745 | |
| 14 | 18-4 | November 1998 | Chronic subclinical, high SCC | 2 | B.2 | 001111111110001 | 8177 | |
| 15 | 53-2 | November 1998 | Chronic (sub)clinical,f high SCC | 2 | B | 001111111110001 | 8177 | |
| 16 | 90-3 | November 1998 | Chronic (sub)clinical,f high SCC | 2 | B | 001111111110011 | 8179 | |
| II | 17 | 11-3 | July 1997 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 |
| 18 | 18-3 | July 1997 | Chronic subclinical, high SCC | 2 | E.1 | 010110011010011 | 11475 | |
| 19 | 99-2 | July 1997 | Chronic subclinical, high SCC | 2 | E | 000110011010001 | 3281 | |
| 20 | 108-3 | July 1997 | Chronic subclinical, high SCC | 2 | F | 001011011111111 | 5887 | |
| 21 | 47-3 | May 1998 | Mild clinical | 3 | D | 101010011010001 | 21713 | |
| 22 | 29-4 | August 1997 | Mild clinical | 3 | D | 101010011010011 | 21715 | |
| 23 | 70-3 | April 1998 | Mild clinical | 3 | D | 101010011010011 | 21715 | |
| 24 | 21-4 | May 1998 | Mild clinical | 3 | E | 000110011010011 | 3283 | |
| III | 25 | 25-3 | July 1997 | Chronic subclinical, low SCC | 1 | C | 000010011010011 | 1235 |
| 26 | 95-1 | July 1997 | Mild clinical | 3 | E | 000110011010011 | 3283 | |
| 27 | 46-4 | December 1997 | Chronic subclinical, low SCC | 1 | C | 000010011010011 | 1235 | |
| 28 | 29-1 | May 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 29 | 29-1 | July 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 30 | 31-3 | May 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 31 | 31-3 | July 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 32 | 86-2 | May 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 33 | 86-2 | July 1998 | Chronic subclinical, high SCC | 2 | E | 000110011010011 | 3283 | |
| 34 | NA | May 1998 | Bulk milk sample | E | 000110011010011 | 3283 | ||
| 35 | NA | July 1998 | Bulk milk sample | E | 000110011010011 | 3283 | ||
| IV | 36 | Ada126-4 | February 1999 | E | 000110011010011 | 3283 | ||
| V | 37 | 9363-3 | February 1999 | E.2 | 000111111110011 | 4083 | ||
| VI | 38 | 205-4 | February 1999 | E | 000110011010011 | 3283 | ||
| VII | 39 | 68-? | February 1999 | D | 101010011010011 | 21715 | ||
| VIII | 40 | Klara-4 | February 1999 | G | 001011111111101 | 6141 |
Udder quarter numbers: 1, right front; 2, left front; 3, right rear; 4, left rear.
1, subclinical infection with nonelevated SCC; 2, chronic subclinical infection with elevated SCC; 3, mild clinical disease or short-term subclinical disease with high SCC; 4, acute severe clinical disease.
Overall results after hybridization with 15 strain-specific DNA probes (AW-1 through AW-15) developed for typing of human S. aureus strains (42).
Binary type is the binary code transformed into a decimal number.
New animal assigned same cow number (75) as cow from which isolates 1, 2, and 3 were collected.
Subclinical disease with occasional mild clinical flare-ups (clots in milk).
Binary typing data of 55 human S. aureus isolates from diverse geographic and temporal origins in the United States and The Netherlands were used (Table 2). Human collections include MRSA strains (n = 37) and methicillin-susceptible S. aureus (MSSA) strains (n = 18) and have been described in detail before (38, 41, 42).
TABLE 2.
Characterization of human S. aureus collections from which binary types are used in this study
| Collection no. | Geographic origin | Isolate no. | Description of collection | Reference(s) |
|---|---|---|---|---|
| 1 | United States | 41-66 | Community-acquired MRSA strains from a New York City hospital (n = 26) | 41 |
| 2 | United States (CDCb) | 67-80 | Selection of geographically diverse strains from multicenter collection of MRSA strains (n = 5) and MSSA strains (n = 9) | 38, 42 |
| 3 | The Netherlands | 81-85 | MSSA strains isolated from healthy persistent nasal carriers (n = 5) | 42 |
| 4 | The Netherlands | 86-95 | MRSA strains (n = 6) and MSSA strains (n = 4) from outbreaks in Dutch hospitals and nursing homes | 42 |
Isolate numbers are as those used in Fig. 4.
CDC, Centers for Disease Control and Prevention.
Clinical and subclinical disease characteristics.
Detailed records of clinical observations were available for isolates 1 to 33 (Table 1). In addition, somatic cell counts (SCCs) of milk samples yielding isolates 1 to 33, with the exception of isolates 8 to 10, were determined by means of a Fossomatic cell counter (Foss Electric, Hillerød, Denmark). SCC is a measure of the leukocyte content of milk and is used as an indicator of infection. The threshold between noninfected and infected is commonly set at 200,000 cells/ml (10). Isolates 34 and 35 were cultured from bulk milk samples from farm III and disease classifications do not apply. For samples yielding isolates 36 to 40, SCC was determined, but detailed clinical data were not available.
Based on clinical symptoms and SCC, isolates 1 to 33 were classified as belonging to one of four clinical groups, in order of increasing severity of infection as follows: (i) subclinical infection with nonelevated SCC (median, 97 × 103 cells/ml; range, 11 to 152 × 103 cells/ml), (ii) chronic subclinical infection with elevated SCC (median, 1,278 × 103 cells/ml; range, 210 to 7,821 × 103 cells/ml), (iii) short duration mild clinical disease or short duration subclinical disease with high SCC (median, 4,560 × 103 cells/ml; range, 411 to 8,710 × 103 cells/ml), and (iv) acute severe clinical disease (Table 1). SCC was not determined for group 4 samples, because clot formation in mammary secretions interfered with SCC measurement.
PFGE.
PFGE was carried out as described by Struelens et al. (35). SmaI (Boehringer, Mannheim, Germany) was used for digestion of genomic DNA. PFGE of DNA digests was performed with a CHEF Mapper (Bio-Rad, Veenendaal, The Netherlands) through a 1% SeaKem agarose gel (FMC; SanverTECH, Heerhugowaard, The Netherlands) under the following conditions: initial switch time of 5 s to final switch time of 15 s, run time of 10 h, followed by initial switch time of 15 s to final switch time of 45 s for 14 h; linear ramping; 6 V cm−1; 120° angle (60° to −60°); 14°C; 0.5× Tris-borate-EDTA buffer. A lambda DNA polymer (Bio-Rad) was used as a molecular size marker. Gels were stained with ethidium bromide for 1 h, destained in water, and photographed under UV light with a charge-coupled device camera.
Macrorestriction patterns were analyzed both visually and by computer-aided methods. Visual interpretation of banding patterns was done following guidelines suggested by Bannerman et al. (5) and Tenover et al. (38, 39). Isolates with identical restriction profiles were assigned the same type and identified with a capital letter. Isolates that differed from main types by one to three band shifts consistent with a limited number of genetic events were assigned subtypes, indicated with a numeral suffix. Isolates with more than three such differences were considered to be different types. Banding patterns were digitized with a Hewlett-Packard Scanjet IIcx/T scanner and stored as TIFF files. Patterns were analyzed by using GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium) to calculate Dice coefficients of correlation and to generate a dendrogram by the unweighted pair group method using arithmetic averages (UPGMA) clustering.
Binary typing.
Macrorestriction fragments obtained through PFGE were Southern blotted onto Hybond N+ membranes (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). Cloned DNA fragments designed for binary typing of human S. aureus strains were used as probes (42). Labeling, hybridization, and detection of the probes were performed with ECL direct labeling and detection systems, according to the manufacturer's protocols (Amersham Life Science, Little Chalfont, Buckinghamshire, United Kingdom). Hybridization of 15 DNA probes was scored with a 1 or a 0 according to the presence or absence of a hybridization signal, resulting in a 15-digit binary code for each S. aureus isolate. Binary codes were transformed into decimal numbers to define binary types (BT), and a dendrogram was constructed by using hierarchical cluster analysis (SPSS 8.0 for Windows; SPSS Inc.).
Statistical analysis.
Log-normalized SCCs for clinical groups 1, 2, and 3 were compared by means of one-way ANOVA (SPSS 8.0 for Windows).
Fisher's exact test of the relationship between clinical groups of origin and strains was performed with analytical software (StatXact version 2.05; CYTEL Software Corporation, Cambridge, Mass.). Isolates 34 to 40 were excluded from this analysis because insufficient clinical data were available. Isolates 29, 31, and 33 were excluded because they represent the same infectious episodes as isolates 28, 30, and 32, respectively. For analysis of the association between clinical groups and pulsotypes, types that occurred only once (A and F) were excluded from analysis and subtypes (B.1, B.2, and E.1) were grouped together with their respective main types. For analysis of the association between clinical groups and binary types, BT clustering at 90% genetic similarity was used to define separate groups.
RESULTS
PFGE.
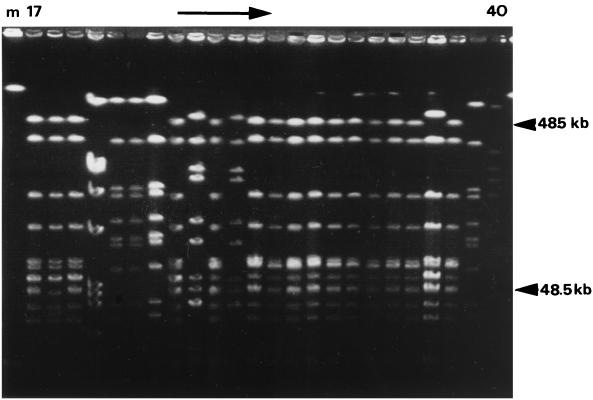
All bovine isolates were typeable by PFGE. Among 38 isolates, seven pulsed-field types and four subtypes were identified through visual interpretation of gels (Fig. 1; Table 1). Three pulsotypes (A, F, and G) and all subtypes (B.1, B.2, E.1, and E.2) were identified only once, while pulsotypes C, D, and E were found in two, three, and four herds, respectively.
FIG. 1.
Example of PFGE gel of SmaI macrorestriction fragments of bovine S. aureus isolates, showing isolates 17 to 40. Molecular sizes are indicated on the right.
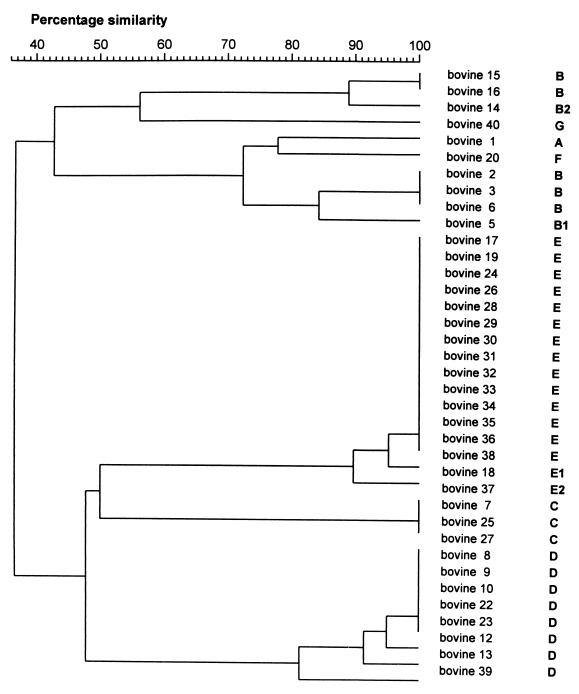
GelCompar analysis of PFGE results defined more clusters than visual interpretation. Depending on the level of genetic relatedness, 13, 11, and 8 clusters were identified for 95, 90, and 80% similarity, respectively (Fig. 2). The visually identified pulsotype B was divided into four (95%) or two (80%) separate clusters, while pulsotype D was divided into three (95%), two (90%), and one (80%) cluster(s). In the GelCompar analysis, visual pulsotypes E and E.1 were grouped together at 95% similarity and E, E.1, and E.2 were grouped together at 90% genetic similarity.
FIG. 2.
Dendrogram showing the level of similarity between SmaI macrorestriction patterns of 38 bovine S. aureus isolates as determined by PFGE and subsequent GelCompar analysis of digitized photographs. Scale indicates level of genetic relatedness within this set of strains. Capital letters indicate pulsotypes based on visual interpretation of PFGE results.
Binary typing.
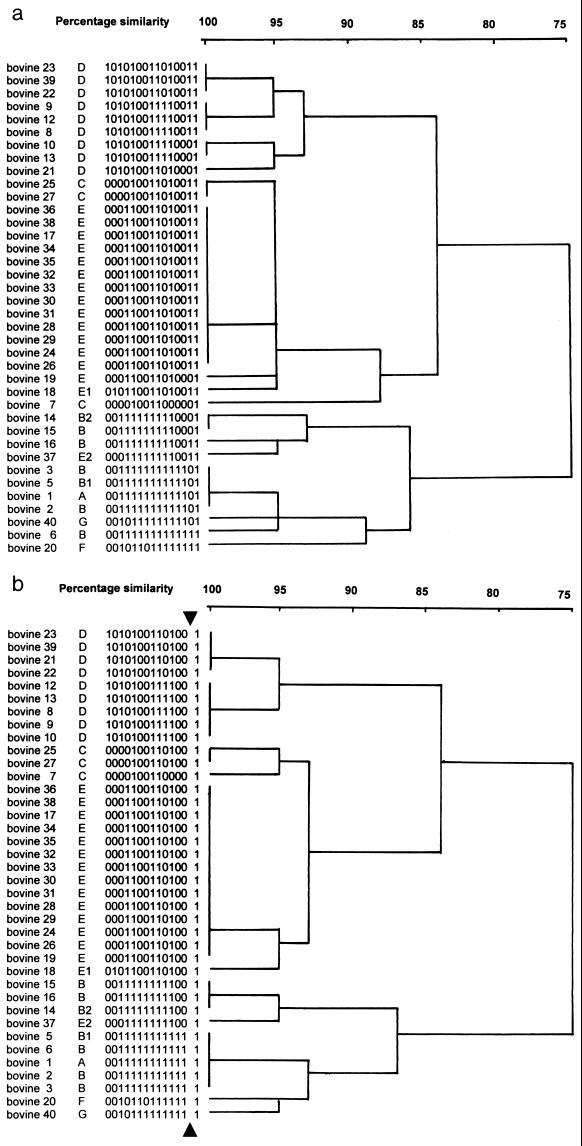
All bovine isolates were typeable by the binary method (Table 1). Out of 15 probes designed for typing of human S. aureus strains, four hybridized to all bovine isolates (AW-5, AW-8, AW-9, and AW-15), while all other probes hybridized to at least one bovine isolate. Genetic relatedness of isolates based on binary typing was depicted in a dendrogram (Fig. 3a). For 95, 90, and 85% genetic similarity, respectively, eight, six, and three clusters of strains were identified. Binding of probe AW-14 showed a low level of reproducibility among epidemiologically related isolates. Therefore, a separate dendrogram excluding AW-14 was constructed (Fig. 3b), reducing the number of clusters to six at 95% similarity.
FIG. 3.
Dendrogram showing the grouping of 38 bovine S. aureus strains on the basis of hybridization scores after binary typing with probes AW-1 to AW-15 (a) and after omission of probe AW-14, which is associated with hypervariable regions on the bovine staphylococcal genome (b). Isolate number, visual pulsotype, and binary code are given for all isolates. Scale indicates level of genetic relatedness within this set of strains.
Concordance between PFGE and binary typing.
Pulsotypes assigned to isolates were compared with binary types. General agreement was found between both techniques, but with some discrepancies. Several visually identified pulsotypes were grouped together by binary typing (e.g., A, three B isolates, B.1, and G at 95% binary similarity; E, E.1, and two out of three C isolates at 95% similarity; B, B.2, and E.2 at 90% similarity). Other pulsotypes were divided into multiple binary clusters that differed by two or three probes (e.g., B into two binary clusters at 90% similarity) (Fig. 3a). Concordance of delineation of genotypically related clusters as determined by PFGE and binary typing improved when probe AW-14 was excluded (Fig. 3b).
Within-herd and between-herd heterogeneity.
Genetic heterogeneity among S. aureus isolates recovered from bovine mammary secretions was observed within and between herds. Isolates from herd I (n = 16) were divided into four pulsotypes (A to D), and subclonal variation was observed in pulsotype B (subtypes B.1 and B.2). In herd II (n = 8), three pulsotypes (D to F) were identified, with subclonal variation in pulsotype E (subtype E.1). In herd III (n = 11), two pulsotypes (C and E) occurred. In isolates obtained from five herds that were not related to each other or herds I, II, and III, three pulsotypes and one subtype were identified (D, E, E.2, and G), demonstrating that both heterogeneity and homogeneity between herds exists.
Heterogeneity based on binary typing parallels heterogeneity of pulsotypes for all herds.
Comparison of bovine and human strains.
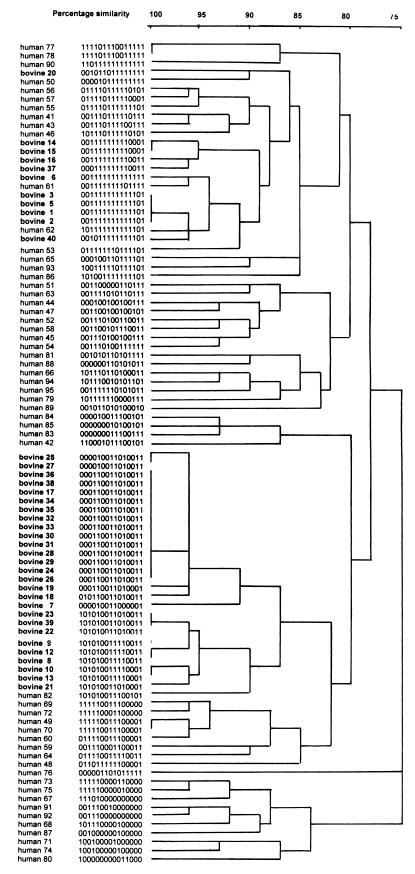
Binary types of bovine isolates from this study were compared to a well-defined collection of human S. aureus isolates that had been typed previously by the same method (41, 42). Most isolates clustered as host-specific clones, and full identity of the 15-digit binary codes of bovine and human isolates was never observed (Fig. 4). At 95% similarity, human isolate 61 clustered together with bovine isolate 6 (one-digit difference at probe AW-11), and human isolate 62 clustered with bovine isolates 1 to 5 (one-digit difference at probe AW-1) and bovine isolate 40 (two-digit difference). At 90% similarity, these bovine and human isolates formed one cluster that also included human isolate 53. Human isolates within this cluster differed from bovine isolates in the same cluster by three digits at most, with differences associated with 6 out of 15 DNA probes used. Human isolate 82 clustered together with all bovine type D isolates at 90% similarity, as did human isolate 50 with bovine isolate 20. Human isolates 50, 53, 61, and 62 were community-acquired MRSA strains from a New York City hospital (Table 2). Human isolate 82 was an MSSA strain isolated from a persistent nasal carrier in The Netherlands. Bovine isolates that clustered with human isolates originated from four Dutch dairy herds that were epidemiologically unrelated to each other or the human sources of S. aureus included in the comparison.
FIG. 4.
Dendrogram showing the grouping of 55 unrelated human S. aureus strains described previously (42) and 38 bovine S. aureus strains on the basis of hybridization scores after binary typing with 15 DNA probes. Isolate numbers and binary codes are shown for all isolates. Scale indicates level of genetic relatedness within this collection of strains.
Association with clinical characteristics.
Mean SCC of quarter milk samples differed with those of clinical groups for groups 1, 2, and 3 (F value = 45.63, P < 0.001, degrees of freedom [df] = 2). Subclinical infection with low SCC (clinical group 1) was associated with pulsotype C (Table 1; three C isolates in three group 1 samples). Binary typing discriminated between type C isolated from herd I (BT, 1217) and herd III (BT, 1235), in agreement with geographical clustering. Chronic subclinical infection with high SCC (clinical group 2) was associated with pulsotypes A and B in herd I. Pulsotypes A and B were not isolated from any samples belonging to clinical group 1 or 4 and only once from group 3. In herds II and III, clinical group 2 was associated with pulsotype E. Type E was also isolated from a group 3 sample, while one group 2 sample yielded pulsotype F. Clinical group 3 yielded several strains, categorized as B (herd I), D (herds I and II), or E (herds II and III). Acute severe clinical mastitis (clinical group 4) was associated with pulsotype D.
Associations between clinical groups and visually identified pulsotypes were statistically significant (Fisher statistic = 26.00, P = 0.002, df = 9). Associations between clinical groups and binary clusters were of borderline statistical significance when all probes were included in the analysis (Fisher statistic = 24.70, P = 0.05, df = 15). Associations were significant after exclusion of probe AW-14 (Fisher statistic = 19.10, P = 0.02, df = 15).
DISCUSSION
PFGE and binary typing.
Variation in gene content of staphylococcal chromosomes may be associated with the presence of nonessential but clinically or epidemiologically relevant genes (e.g., virulence genes, resistance genes) (23, 28). PFGE is a well-known and powerful method for detection of genetic variation in S. aureus populations (5, 35). Binary typing is a recently developed high-resolution molecular typing system that produces simple binary output and has the potential to become a technically simple and fast library typing system for S. aureus strains (42). In this study, PFGE profiles and binary codes for 38 isolates derived from bovine mammary secretions were determined and compared. After PFGE of SmaI macrorestriction fragments, seven main pulsotypes and four subtypes were identified visually. Computer-aided cluster analysis identified more distinct types, depending on the level of genetic similarity chosen as the cutoff value. What level of discrimination between clusters of strains is desired depends on the purpose of genotyping, and results of PFGE must be analyzed in light of the epidemiological background (5, 35). One visually identified pulsotype, type B, was subdivided over multiple clusters after UPGMA analysis, even at low genetic similarity levels (Fig. 2). Isolates were from similar clinical and geographical backgrounds, but subdivision may be related to different temporal origins of the samples (Table 1). Discrepancies between visual and computer-aided interpretation are a drawback of pulsed-field typing and limit its usefulness as a routine diagnostic technique for large numbers of samples.
In this study, binary typing was preceded by PFGE typing, but binary typing can be performed as a single typing technique (42). Binary typing yielded 16 binary codes clustered in three to eight clones, depending on levels of genomic similarity. The relevant level of discrimination and suitability of individual probes are subject of further study. However, interpretation of probe binding results is unequivocal. Probes AW-12 and AW-13 yielded identical results, while probes AW-5, -8, -9, and -15 hybridized to all bovine strains and did not contribute to the discriminatory power of the typing system. A larger collection of bovine isolates should be studied to determine the informative value of these probes for differentiation of bovine S. aureus strains.
Concordance between PFGE and binary typing.
Several pulsotypes were subdivided by binary typing. Binary codes within a pulsotype often differ by no more than one digit, and in many cases it was the digit associated with probe AW-14 (Table 1; Fig. 3a). The observed discrimination within pulsotypes may therefore be related to the detection of hypervariable domains on the genome of bovine S. aureus strains with probe AW-14. Similar hypervariability or inconsistent presence of probe-binding sequences has been described for epidemiologically and genetically related human S. aureus strains (40). The results could imply that probe AW-14, which is stable for typing of human S. aureus strains, is not stable for typing of S. aureus strains of bovine origin. On the other hand, probe AW-14 could be used to study short-term genome evolution in bacterial populations of bovine origin (41).
When binary code differences caused by probe AW-14 are ignored, closer agreement between binary typing and pulsed-field typing is obtained, but some one-digit differences within pulsotypes remain (Fig. 3b). Pulsotypes C isolated from herds I and III differ by one digit, which was associated with probe AW-11. This genotypic difference can be related to different geographical origins, but not to a difference in clinical course of infection. For pulsotype D, differences exist in geographical origin and in clinical course. Whether severity of disease is a herd effect (herd I versus herd II), a strain effect (BT 21745 and 21747 versus BT 21713 and 21715), a cow effect (mild cases in older animals, severe cases in heifers), or a chance effect is unknown.
Some binary clones are subdivided by PFGE (e.g., B and B.2 within BT 8177 and A, B, and B.1 within BT 8189). Since isolates within these binary types were of similar geographical, temporal, and clinical origin, binary typing seems the epidemiologically superior technique in these cases.
Within-herd and between-herd heterogeneity.
PFGE and binary typing differentiated strains within and between herds. Similar results were obtained by means of PCR-based DNA fingerprinting in the United States (24) and The Netherlands (21), through MLEE analysis of a worldwide collection of strains (17), by coagulase gene typing of European, American, and Asian isolates (2, 36), with a combination of techniques for bovine isolates from the United States and Ireland (12), and by PFGE of German isolates (3a). In all studies, including the present one, a limited number of predominant types was found both within herds, in agreement with the contagious nature of S. aureus mastitis (21), and between herds, suggesting that certain variants present in the environment may have a predilection for causing intramammary infections (2, 12, 36).
Subclonal heterogeneity within herds may be due to temporal evolution. Herds were selected for inclusion in the longitudinal survey based on a history of the presence of the pathogen in the herd for more than 1 year. The study period covered an additional 18 months, allowing for further genetic diversification (41). Similar subclonal genetic variation over time has been described for DNA macrorestriction patterns from human S. aureus isolates (27).
Comparison of bovine and human strains.
Out of 55 human isolates and 38 bovine isolates, five human and 16 bovine isolates belonged to clusters sharing 90 to 95% similarity, as determined by binary typing. At higher similarity levels, all clones were host species specific. Similar results were obtained by Kapur et al. (17) and by Lopes et al. (22). The results are consistent with the concept of host specificity among S. aureus clones and imply that successful transfer of bacteria between humans and cattle is not a frequent event (17). However, several studies are available that suggest that transfer of bacteria between humans and cows is possible (13, 30, 37). Those studies mostly focus on the role of humans as a source of infection for dairy cattle. Another reason to be concerned about interspecies transfer of S. aureus is the routine use of antibiotics in dairy herd management (15, 32, 34). In farms with S. aureus mastitis problems, oxacillin is used as a dry cow treatment for all animals (8). Resistance to the closely related antibiotic methicillin is rare in bovine S. aureus (22) but has been reported in New York State (29), Europe (9), and Japan (cited in reference 18). Widespread use of oxacillin could promote the selection of resistant clones (7). If interspecies transfer occurs, methicillin resistance in bovine strains may contribute to increasing prevalence of MRSA strains in humans. Since binary typing is a library system that can be applied to S. aureus isolates originating from humans and cattle, it is a useful tool in monitoring origins of MRSA strains and interspecies transfer of S. aureus. Addition of probes to test for the presence of the mecA gene in the bovine typing system would furthermore allow monitoring of MRSA prevalence in veterinary diagnostic laboratories.
Association with clinical characteristics.
A limited number of isolates were included in statistical analyses, and the interdependence of within-herd observations was not taken into account. Thus, results of the analyses must be interpreted with care. However, in this study there was a significant correlation between S. aureus strains and disease characteristics observed in vivo. Such information is rarely available because most studies focus on clinical or subclinical mastitis only (21, 21a, 37) or don't contain information on the clinical background of samples (2, 12, 36). Matthews et al. (24) observed heterogeneity between subclinical and clinical isolates based on arbitrarily primed PCR, but heterogeneity within the group of subclinical isolates and overlap between genotypes isolated from both groups precluded firm associations. Kenny et al. (19) reported enterotoxin production by bovine mammary isolates of S. aureus and suggested that enterotoxin production may be associated with the clinical course of disease. Matsunaga et al. (23) attempted to relate toxin production and other virulence factors to the severity of clinical disease. They concluded that the properties of S. aureus strains isolated from peracute cases were different from those of acute and chronic isolates. No obvious differences between acute and chronic isolates were observed. The first conclusion is in agreement with our finding that all group 4 cases (peracute cases) are attributable to a specific pulsotype and binary type. In addition, our results suggest a difference between acute (clinical) and chronic (subclinical) cases, as shown by the associations between pulsotype C and clinical group 1, pulsotypes B and E and group 2, and pulsotype D and group 3, respectively.
Pulsotypes C and E differed in binding of probe AW-4 only but were associated with a clearly distinguishable leukocyte response in vivo (low versus high SCC). Differences in leukocyte response in vitro have been described by Aarestrup et al. (1) for different coagulase types isolated from cases of subclinical mastitis. Probe AW-4 has been shown to be homologous to a mobile genetic element, IS257 (42). IS257, also known as IS431, is a common insertion sequence in the staphylococcal chromosome and plasmids and can be associated with several resistance determinants, including methicillin resistance (7).
It must be emphasized that associations between clinical outcome of disease, pulsotypes, binary types, and specific probes in the binary typing system are as yet speculative, and more typing needs to be done. If associations are confirmed, binary typing can be used for the identification of unusual and more virulent strains, allowing for further pathogenetic studies and for tailored advice to farmers on the management of specific cases.
An aspect of the association between genotype and epidemiological background that merits attention is the relation between pulsotype D and its origin. Pulsotype D was isolated from all group 4 samples, all of which were obtained from heifers before first calving. The occurrence of S. aureus mastitis in preparturient heifers is a widely reported phenomenon (14, 26). Based on biotyping, antibiograms, and phage typing, Roberson et al. (31) concluded that milk from the dairy herd and heifer body sites are the most likely sources of infections. In their study, the environment was a possible source of infection in 17 out of 61 cases but never the sole possible source. In contrast, our results show that the predominant S. aureus genotype in the milking herd (pulsotype B, BT 8177 and 8189) is different from the genotype found in heifer mastitis isolates (pulsotype D, BT 21745 and 21747). This implies that the dairy herd is not the most likely source of heifer infections. In herd II, all type D cases occurred at a time when no other infected animals were present in the milking herd, as determined by routine samplings taken every 3 weeks (data not shown). Though not conclusive, this observation also suggests that the environment is a more likely source of infection than the dairy herd. Determination of reservoirs, including environmental sources, is considered an important step when attempting to control S. aureus in a dairy herd (30, 32). The genotyping techniques presented in this paper can be helpful in elucidating the relative importance of environmental sources in the farm level ecology of S. aureus.
Conclusion and future developments.
This study shows that both PFGE and binary typing can be successfully applied to characterize S. aureus isolates of bovine mammary origin. Binary output was easier to interpret than banding patterns generated by PFGE, and binary typing seemed superior to PFGE in clustering isolates from similar epidemiological backgrounds. As a library typing system, binary typing facilitates the comparison of S. aureus isolates of bovine and human origins from worldwide collections, analysis of clonal relatedness and host specificity, and monitoring of interspecies transfer. In this study, genetically related clusters of strains of human, bovine, and mixed origins occurred. For isolates obtained from bovine mammary secretions, associations between bacterial strains and clinical characteristics of infection in vivo were observed, as was a tentative association between strains and sources of infection. Those observations need further validation through the study of larger strain collections or infection experiments. We conclude that binary typing in particular is a technique that is suitable for use in veterinary clinical microbiology and may contribute to the development of case-specific and farm-specific recommendations for the management of S. aureus problems in bovine medicine.
REFERENCES
- 1.Aarestrup F M, Scott N L, Sordillo L M. Ability of Staphylococcus aureus to resist neutrophil bactericidal activity and phagocytosis. Infect Immun. 1994;62:5679–5682. doi: 10.1128/iai.62.12.5679-5682.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aarestrup F M, Dangler C A, Sordillo L M. Prevalence of coagulase gene polymorphism in Staphylococcus aureus isolates causing bovine mastitis. Can J Vet Res. 1995;59:124–128. [PMC free article] [PubMed] [Google Scholar]
- 3.Aarestrup F M, Wegener H C, Rosdahl V T. Evaluation of phenotypic and genotypic methods for epidemiological typing of Staphylococcus aureus isolates from bovine mastitis in Denmark. Vet Microbiol. 1995;45:139–150. doi: 10.1016/0378-1135(95)00043-a. [DOI] [PubMed] [Google Scholar]
- 3a.Annemüller C, Lämmler C, Zschock M. Genotyping of Staphylococcus aureus isolated from bovine mastitis. Vet Microbiol. 1999;69:217–224. doi: 10.1016/s0378-1135(99)00117-0. [DOI] [PubMed] [Google Scholar]
- 4.Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 5.Bannerman T L, Hancock G A, Tenover F C, Miller J M. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J Clin Microbiol. 1995;33:551–555. doi: 10.1128/jcm.33.3.551-555.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkema H W, Schukken Y H, Lam T J G M, Beiboer M L, Wilmink H, Benedictus G, Brand A. Incidence of clinical mastitis in dairy herds grouped in three categories by bulk milk somatic cell count. J Dairy Sci. 1998;81:411–419. doi: 10.3168/jds.S0022-0302(98)75591-2. [DOI] [PubMed] [Google Scholar]
- 7.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummins K A, McCaskey T A. Multiple infusions of cloxacillin for treatment of mastitis during the dry period. J Dairy Sci. 1987;70:2658–2665. doi: 10.3168/jds.S0022-0302(87)80336-3. [DOI] [PubMed] [Google Scholar]
- 9.Devriese L A, Hommez J. Epidemiology of methicillin-resistant Staphylococcus aureus in dairy herds. Res Vet Sci. 1975;19:23–27. [PubMed] [Google Scholar]
- 10.Dohoo I R, Leslie K E. Evaluation of changes in somatic cell counts as indicators of new intramammary infections. Prev Vet Med. 1993;10:225–237. [Google Scholar]
- 11.Farah I O, Pedersen E, Halgaard C, Bruhn K. Comparative characterization and biotyping of Staphylococcus aureus isolates from human and bovine sources. Acta Vet Scand. 1988;29:303–310. doi: 10.1186/BF03548622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald J R, Meaney W J, Hartigan P J, Smyth C J, Kapur V. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol Infect. 1997;119:261–269. doi: 10.1017/s0950268897007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox L K, Gershman M, Hancock D D, Hutton C T. Fomites and reservoirs of Staphylococcus aureus causing intramammary infections as determined by phage typing: the effect of milking time hygiene practices. Cornell Vet. 1991;81:183–193. [PubMed] [Google Scholar]
- 14.Fox L K, Chester S T, Hallberg J W, Nickerson S C, Pankey J W, Weaver L D. Survey of intramammary infections in dairy heifers at breeding age and first parturition. J Dairy Sci. 1995;78:1619–1628. doi: 10.3168/jds.S0022-0302(95)76786-8. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez R N, Jasper D E, Farver T B, Bushnell R B, Franti C E. Prevalence of udder infections and mastitis in 50 California dairy herds. J Am Vet Med Assoc. 1988;193:323–328. [PubMed] [Google Scholar]
- 16.Harmon R J, Eberhart R J, Jasper D E, Langlois B E, Wilson R A. Microbiological procedures for the diagnosis of bovine udder infection. Arlington, Va: National Mastitis Council; 1990. [Google Scholar]
- 17.Kapur V, Sischo W M, Greer R S, Whittam T S, Musser J M. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J Clin Microbiol. 1995;33:376–380. doi: 10.1128/jcm.33.2.376-380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawano J, Shimizu A, Saitoh Y, Yagi M, Saito T, Okamoto R. Isolation of methicillin-resistant coagulase-negative staphylococci from chickens. J Clin Microbiol. 1996;34:2072–2077. doi: 10.1128/jcm.34.9.2072-2077.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny K, Reiser R F, Bastida-Corcuera F D, Norcross N L. Production of enterotoxins and toxic shock syndrome toxin by bovine mammary isolates of Staphylococcus aureus. J Clin Microbiol. 1993;31:706–707. doi: 10.1128/jcm.31.3.706-707.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam T J G M, Pengov A, Schukken Y H, Smit J A H, Brand A. The differentiation of Staphylococcus aureus from other Micrococcaceae isolated from bovine mammary glands. J Appl Bacteriol. 1995;79:69–72. doi: 10.1111/j.1365-2672.1995.tb03125.x. [DOI] [PubMed] [Google Scholar]
- 21.Lam T J G M, Lipman L J A, Schukken Y H, Gaastra W, Brand A. Epidemiological characteristics of bovine clinical mastitis caused by Staphylococcus aureus and Escherichia coli studied by DNA fingerprinting. Am J Vet Res. 1996;57:39–42. [PubMed] [Google Scholar]
- 21a.Lange C, Cardoso M, Senczek D, Schwarz S. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet Microbiol. 1999;67:127–141. doi: 10.1016/s0378-1135(99)00031-0. [DOI] [PubMed] [Google Scholar]
- 22.Lopes C A D M, Moreno G, Curi P R, Gottschalk A F, Modolo G R, Horacio A, Corrêa A, Pavan C. Characteristics of Staphylococcus aureus from subclinical bovine mastitis in Brazil. Br Vet J. 1990;146:443–448. doi: 10.1016/0007-1935(90)90033-y. [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga T, Kamata S, Kakiichi N, Uchida K. Characteristics of Staphylococcus aureus isolated from peracute, acute and chronic bovine mastitis. J Vet Med Sci. 1993;55:297–300. doi: 10.1292/jvms.55.297. [DOI] [PubMed] [Google Scholar]
- 24.Matthews K R, Kumar S J, O'Conner S A, Harmon R J, Pankey J W, Fox L K, Oliver S P. Genomic fingerprints of Staphylococcus aureus of bovine origin by polymerase chain reaction-based DNA fingerprinting. Epidemiol Infect. 1994;112:177–186. doi: 10.1017/s095026880005754x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musser J M, Kapur V. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J Clin Microbiol. 1992;30:2058–2063. doi: 10.1128/jcm.30.8.2058-2063.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myllys V. Staphylococci in heifer mastitis before and after parturition. J Dairy Res. 1995;62:51–60. doi: 10.1017/s0022029900033665. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien F G, Pearman J W, Gracey M, Riley T V, Grubb W B. Community strain of methicillin-resistant Staphylococcus aureus involved in a hospital outbreak. J Clin Microbiol. 1999;37:2858–2863. doi: 10.1128/jcm.37.9.2858-2862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prevost G, Jaulhac B, Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivas A L, Gonzalez R N, Wiedmann M, Bruce J L, Cole E M, Bennett G J, Schulte H F, Wilson D J, Mohammed H O, Batt C A. Diversity of Streptococcus agalactiae and Staphylococcus aureus ribotypes recovered from New York dairy herds. Am J Vet Res. 1997;58:482–487. [PubMed] [Google Scholar]
- 30.Roberson J R, Fox L K, Hancock D D, Gay J M, Besser T E. Ecology of Staphylococcus aureus isolated from various sites on dairy farms. J Dairy Sci. 1994;77:3354–3364. doi: 10.3168/jds.S0022-0302(94)77277-5. [DOI] [PubMed] [Google Scholar]
- 31.Roberson J R, Fox L K, Hancock D D, Gay J M, Besser T E. Sources of intramammary infections from Staphylococcus aureus in dairy heifers at first parturition. J Dairy Sci. 1998;81:687–693. doi: 10.3168/jds.S0022-0302(98)75624-3. [DOI] [PubMed] [Google Scholar]
- 32.Saperstein G, Hinckley L S, Post J E. Taking the team approach to solving staphylococcal mastitis infection. Vet Med. 1988;83:940–947. [Google Scholar]
- 33.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sischo W M, Heider L E, Miller G Y, Moore D A. Prevalence of contagious pathogens of bovine mastitis and use of mastitis control practices. J Am Vet Med Assoc. 1993;202:595–600. [PubMed] [Google Scholar]
- 35.Struelens M J, Deplano A, Godard C, Maes N, Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992;30:2599–2605. doi: 10.1128/jcm.30.10.2599-2605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su C, Herbelin C, Frieze N, Skardova O, Sordillo L M. Coagulase gene polymorphism of Staphylococcus aureus isolates from dairy cattle in different geographical areas. Epidemiol Infect. 1999;122:329–336. doi: 10.1017/s0950268899002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swartz R, Jooste P J, Novello J C. Bacteriophage typing of Staphylococcus aureus strains isolated from Bloemfontein dairy herds. J S Afr Vet Assoc. 1985;56:69–73. [PubMed] [Google Scholar]
- 38.Tenover F C, Arbeit R, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hébert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Leeuwen W, Sijmons M, Sluijs J, Verbrugh H, van Belkum A. On the nature and use of randomly amplified DNA from Staphylococcus aureus. J Clin Microbiol. 1996;34:2770–2777. doi: 10.1128/jcm.34.11.2770-2777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Leeuwen W, van Belkum A, Kreiswirth B, Verbrugh H. Genetic diversification of methicillin-resistant Staphylococcus aureus as a function of prolonged geographic dissemination and as measured by binary typing and other genotyping methods. Res Microbiol. 1998;149:497–507. doi: 10.1016/s0923-2508(98)80004-1. [DOI] [PubMed] [Google Scholar]
- 42.van Leeuwen W, Verbrugh H, van der Velden J, van Leeuwen N, Heck M, van Belkum A. Validation of binary typing for Staphylococcus aureus strains. J Clin Microbiol. 1999;37:664–674. doi: 10.1128/jcm.37.3.664-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]