Abstract
Plasmids are extrachromosomal genetic elements in prokaryotes that have been recognized as important drivers of microbial ecology and evolution. Plasmids are found in multiple copies inside their host cell where independent emergence of mutations may lead to intracellular genetic heterogeneity. The intracellular plasmid diversity is thus subject to changes upon cell division. However, the effect of plasmid segregation on plasmid evolution remains understudied. Here, we show that genetic drift during cell division—segregational drift—leads to the rapid extinction of novel plasmid alleles. We established a novel experimental approach to control plasmid allele frequency at the levels of a single cell and the whole population. Following the dynamics of plasmid alleles in an evolution experiment, we find that the mode of plasmid inheritance—random or clustered—is an important determinant of plasmid allele dynamics. Phylogenetic reconstruction of our model plasmid in clinical isolates furthermore reveals a slow evolutionary rate of plasmid-encoded genes in comparison to chromosomal genes. Our study provides empirical evidence that genetic drift in plasmid evolution occurs at multiple levels: the host cell and the population of hosts. Segregational drift has implications for the evolutionary rate heterogeneity of extrachromosomal genetic elements.
Keywords: bacterial population genetics, extrachromosomal genetic elements, experimental evolution, multilevel genetic drift, heteroplasmy
Introduction
Extrachromosomal genetic elements are ubiquitous in all domains of life. Examples are prokaryotic plasmids and the eukaryotic organelles—mitochondria and plastids—that replicate autonomously within the host cell; their copy number typically outnumbers that of the chromosomes and depends on copy number control mechanisms (Novick 1969; Robin and Wong 1988; Satoh and Kuroiwa 1991; Nordström 2006). Extrachromosomal genetic elements residing within a host cell may be genetically heterogeneous, a phenomenon which has been termed heteroplasmy (Novick 1987; Birky 2001). The segregation of extrachromosomal element alleles during cell division is considered to occur in the absence of selection, hence the evolution of their genetic diversity at the level of a single host cell is likely governed by drift. Previous studies hypothesized that genetic drift during cell division has an effect on the evolutionary rate of extrachromosomal elements (Birky 2001). However, empirical evidence for multiple levels of genetic drift in the evolution of extrachromosomal elements is lacking.
Here, we study the consequences of genetic drift for the allele dynamics of prokaryotic plasmids. Plasmid acquisition may lead to the rapid evolution of the organism phenotype and niche occupation (e.g., Martínez-Martínez et al. 1998; Heuer et al. 2011; Gullberg et al. 2014; Stoesser et al. 2016); hence they are considered major drivers of lateral gene transfer in prokaryote evolution (Smalla et al. 2015; Hall et al. 2017). The copy number of natural plasmids ranges between 1 and 15, yet high-copy plasmids may reach up to 200 copies in the cell (Bazaral and Helinski 1968; Projan et al. 1987). Plasmid copy number is an important determinant of plasmid mutational supply and hence of plasmid heteroplasmy because the independent emergence of novel plasmid alleles contributes to the intracellular plasmid genetic heterogeneity (San Millan et al. 2016). The replication of plasmid copies in a host cell occurs in an unbiased manner (i.e., at random) (Rownd 1969; Cullum and Broda 1979; Nordström and Dasgupta 2006) and is governed by plasmid copy number control mechanisms (Solar et al. 1998). The segregation of plasmid alleles depends on plasmid partitioning mechanisms that can be either active, as typically observed in low-copy plasmids, or passive, where segregation depends on the plasmid physical distribution during cell division, which is common for high-copy plasmids (Reyes-Lamothe et al. 2012). The distribution of plasmid alleles into the daughter cells may be biased by plasmid localization at the cell poles or by temporal plasmid clustering (Pogliano et al. 2001; Münch et al. 2015; Wang et al. 2016; Hsu and Chang 2019). Plasmid interaction with DNA-binding proteins can lead to clustered plasmid allele segregation as well; examples are active partition mechanisms (Hyland et al. 2014) and transcriptional regulatory elements (Wu et al. 1992). Notwithstanding, plasmid segregation into the daughter cells is random (Reyes-Lamothe et al. 2014), hence it occurs in the absence of selection on the plasmid alleles. Consequently, plasmid allele dynamics comprise a component of random genetic drift—termed segregational drift (Ilhan et al. 2019)—which has been suggested to decrease the fixation probability of plasmid alleles (Ilhan et al. 2019; Santer and Uecker 2020).
Previous computer simulations predicted that the evolution of plasmid alleles is characterized by higher loss frequency and extended fixation time in comparison to chromosomal alleles (Ilhan et al. 2019). Nonetheless, empirical evidence for the workings of segregational drift in plasmid evolution is lacking. To study the effect of segregational drift, we compared the dynamics of plasmid and chromosomal alleles under nonselective conditions using an experimental evolution approach. For that purpose, we established a system that enables the introduction of a novel plasmid allele into a bacterial population and the documentation of the allele segregation over time. In our system, we used the naturally competent model organism Acinetobacter baylyi (Young et al. 2005) where a novel plasmid allele can be introduced into the population with known frequencies at the intracellular and population level. The dynamics of allele segregation were compared between two plasmid assortment modes: random or clustered assortment, where daughter plasmid replicons are inherited together. Our results reveal that plasmid alleles are rapidly lost in the population in comparison to chromosomal alleles, regardless of the plasmid assortment mode, thus supplying experimental evidence for the workings of segregational drift.
Results
An Experimental System for Tracing Plasmid Allele Dynamics in the Population
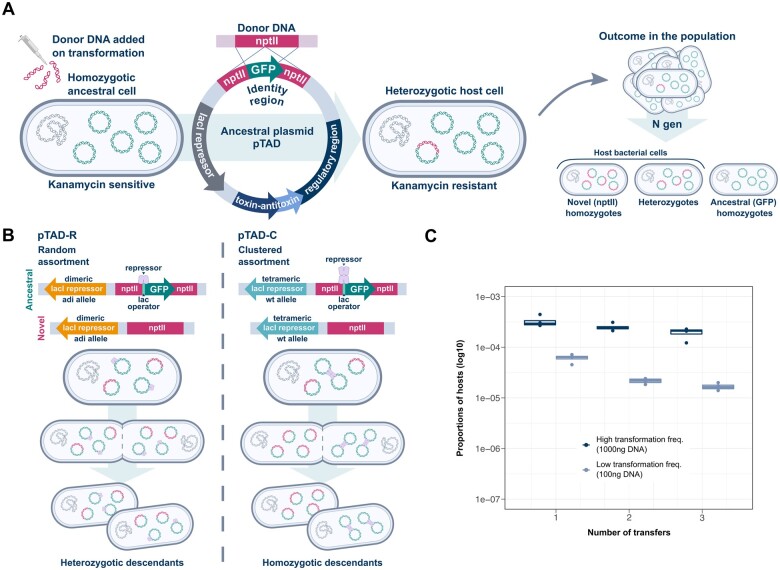
To study plasmid allele dynamics, we established an experimental system that allows us to introduce a predefined frequency of a novel plasmid allele into a host population and follow it over time. Our model plasmid pTAD (8.6 kb) is a derivative of pVRL1 (Lucidi et al. 2018), which was constructed from the native cryptic plasmid pWH1277 from A. calcoaceticus, for which the mode of synchronization of replication and segregation is unknown. The plasmid pWH1277 has no active partition mechanism and is putatively mobilizable; it is stably maintained due to a toxin–antitoxin system and has a narrow host range within Acinetobacter (Hunger et al. 1990). The model plasmid pTAD encodes a green fluorescence protein (gfp), whose transcription is under the control of a lacI repressor. The gfp gene constitutes the ancestral allele in our system; hence, the ancestral population comprises homozygotes of the gfp plasmid alleles (fig. 1A).
Fig. 1.
An experimental system for tracing plasmid allele dynamics. (A) The model plasmid pTAD and the genetic system. The novel plasmid allele (nptII gene) is introduced into the population by natural transformation via homologous recombination with the model plasmid. The initial population is constituted by hosts of the novel plasmid allele and nonhosts. Hosts are followed for several generations by tracing the presence of the novel allele. (B) An illustration of the two model plasmids; pTAD-R and pTAD-C that are characterized by random and clustered assortment modes, respectively. The model plasmids differ in the lacI repressor allele. The lacI repressor allele in pTAD-C encodes for a LacI repressor that binds two plasmids and thus reduces the unit of segregation of the ancestral plasmid allele. (C) A demonstration of the ability to control the host frequency in the initial population. The frequency of hosts is proportional to the donor DNA concentration. Using 1,000 ng donor DNA for the initial transformation generates a host frequency of 1–5 × 10−4 (∼0.01%), whereas using 100 ng DNA generates a host frequency of 1–5 × 10−5 (∼0.001%).
The introduction of a novel plasmid allele into the ancestral population was achieved by natural transformation that is followed by homologous recombination of the novel allele and the model plasmid. As the novel plasmid allele, we chose nptII, which encodes for kanamycin resistance. The gfp locus in pTAD is flanked by two nonfunctional fragments of the nptII gene that are the target site for the recombination of the introduced nptII allele. A successful recombination of the novel allele leads to the emergence of heterozygotes carrying both ancestral and novel alleles; these cells are fluorescent and kanamycin-resistant. Plasmid segregation in our experiment is thus expected to give rise to three types of cells: hosts of the novel allele, including heterozygotes and homozygotes to the nptII allele, as well as ancestral homozygotes for the gfp allele (fig. 1A).
To explore the effect of different plasmid assortment modes, we constructed two variants of our model plasmid. The mode of plasmid assortment—random or clustered—is determined by two different LacI repressor alleles. The wild-type LacI repressor is a tetrameric protein complex that binds to lac operator sequences present in the DNA. Note that the natural form of regulation in Escherichia coli presents two lac operator sequences. Each dimer binds to one operator and in some cases the tetrameric repressor can bind two distinct DNA sequences (Krämer et al. 1987; Oehler et al. 1990). When only one operator is present in a plasmid, the tetrameric LacI repressor can bind two distinct plasmid molecules prior to plasmid assortment (Wu et al. 1992). In contrast to the wild-type LacI, the LacIadi mutant of the repressor generates a dimeric protein that can bind to only a single lac operator (Brenowitz et al. 1991; Hsieh and Brenowitz 1997). The two model plasmid variants in our system contain one lac operator sequence and differ in the LacI allele. In pTAD-C, the wild-type LacI repressor can bind two plasmid molecules at the same time, thus decreasing the segregational units of the ancestral plasmid type and leading to clustered plasmid allele segregation during cell division. Whereas in pTAD-R, the LacIadi repressor can only bind a single plasmid, hence molecules of that model plasmid will segregate independently leading to random allele segregation during cell division (fig. 1B).
For the establishment of the ancestral homozygotic population, we introduced the model plasmids pTAD-R and pTAD-C into a naïve A. baylyi strain BD413 (ADP1). Quantifying the plasmid stability in the population revealed no measurable loss (0 ± 0%; n = 4, for both plasmids), hence both pTAD variants are stably maintained in the population. The average plasmid copy number of both pTAD variants is 15 (15.7 ± 3.44, n = 10 Ancestral allele carrying pTAD; 13.7 ± 3.75, n = 8 Novel allele carrying pTAD). The novel allele was introduced into the ancestral population by natural transformation that creates heterozygotic host cells for the novel nptII plasmid allele (fig. 1A). For simplicity, here, we use the term “host” for cells harboring the novel nptII plasmid allele (i.e., hosts are resistant to kanamycin). Natural transformation in A. baylyi follows one-hit kinetics (Overballe-Petersen et al. 2013). Hence, the transformation frequency, that is, the number of created hosts of the novel allele, is directly proportional to the concentration of donor DNA (fig. 1C and supplementary fig. 1, Supplementary Material online). Increasing DNA concentration has no effect on the probability of uptake of DNA molecules per host cell (supplementary fig. 1, Supplementary Material online), with only a single donor DNA molecule successfully recombining with only one of the copies of the resident ancestral plasmid in the vast majority of cells (97%) (see Materials and Methods and the Supplementary material online). This ensures an intracellular frequency of a single plasmid carrying the novel allele per host. The frequency of hosts in our system is proportional to the donor DNA concentration and is stable over short time scales (fig. 1C), thus providing high reproducibility and control over the frequency of hosts in the ancestral population.
The Dynamics of Plasmid Allele Segregation under Nonselective Conditions
To study the effect of segregational drift on plasmid allele dynamics, we performed an evolution experiment of the two model plasmids using serial transfer applying a constant daily dilution, that is, a bottleneck size of 1:100 (∼107 cells). The experiment was conducted with eight replicates for each model plasmid under conditions that are nonselective for the plasmid maintenance. The ancestral populations were established by introducing the novel plasmid nptII allele, which generates a subpopulation of hosts with a proportion of approximately 5 × 10−5 within the total population (i.e., ∼5 × 104 cells). At that stage, all hosts are heterozygotes that contain the novel allele in one copy out of the 15 resident plasmids (PCN) (as in fig. 1C). We evolved the populations over 23 transfers, which correspond to approximately 160 generations. The frequency of hosts containing the novel nptII allele was estimated according to their phenotype via selective plating on kanamycin and observing their fluorescence. Plasmid allele segregation creates three distinct cell types; heterozygote hosts, containing both ancestral and novel plasmid alleles, homozygote hosts in which the novel plasmid allele nptII has been fixed, and ancestral homozygotes harboring only the ancestral plasmid allele gfp. Since different plasmid alleles may have different effect on the host fitness, we tested for a fitness difference between homozygotes to the ancestral and novel plasmid alleles. Performing pairwise competition experiments revealed no measurable fitness differences between the ancestral and novel homozygotes (H0: w = 1, P = 0.0781 [pTAD-R] and 0.2744 [pTAD-C] using Wilcoxon test, n = 16; supplementary fig. 2, Supplementary Material online). Hence, changes in the novel plasmid allele frequency over time in our experiment are due to neutral processes rather than fitness differences between the ancestral and novel homozygotes.
The results of the evolution experiments of both model plasmids show a stable persistence of the novel plasmid hosts within the population during the first approximately 15 transfers. At the same time, we observed evidence for plasmid allele segregation: the frequency of heterozygote hosts constantly decreased and the frequency of homozygote hosts increased (fig. 2A). After approximately 15 transfers, we observed a decrease in the frequency of hosts of the novel allele, with the exception of replicate R3. At the end of the experiment the novel plasmid allele went extinct in the majority of populations. To test if the dynamics of the novel plasmid allele are the result of competition between hosts and nonhosts, we compared the growth dynamics of the host population and the total population. Our results show that prior to the decrease in the proportion of hosts, the proportion of hosts in the total population was constant for both model plasmids, hence fitness differences between hosts and nonhosts in our experiment were negligible (wpTAD-R = 0.973; wpTAD-C = 1; supplementary fig. 4, Supplementary Material online). Consequently, we conclude that the novel plasmid allele dynamics observed during the evolution experiment are the result of plasmid allele segregation rather than solely processes at the population level.
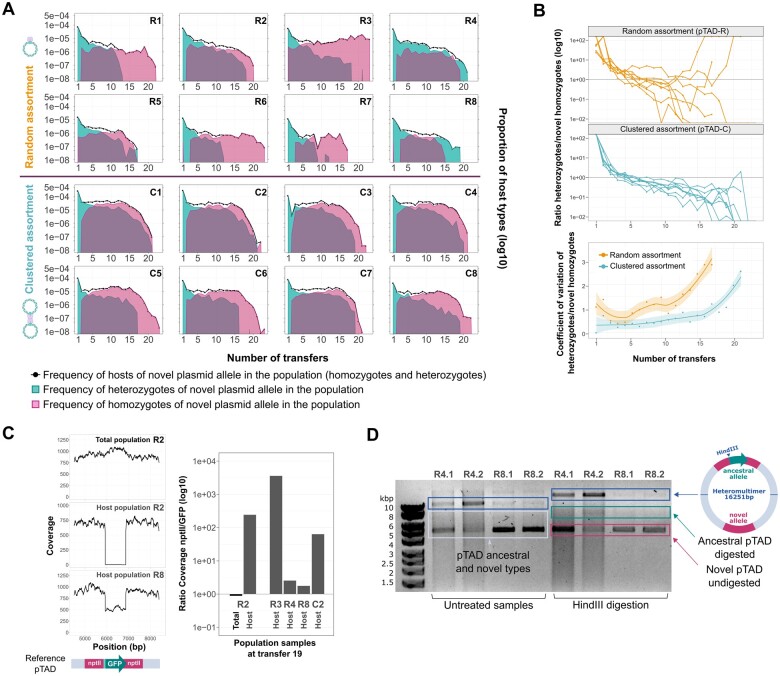
Fig. 2.
Plasmid allele dynamics in nonselective conditions. (A) The dynamics of pTAD-R (top) and pTAC-C (bottom) over approximately 160 generations. The novel allele was introduced at the beginning of the experiment employing a concentration of 100 ng/ml donor DNA. Host frequencies in the total population are shown in black, heterozygote frequencies are shown in green and novel homozygotes in pink. Performing the evolution experiment with higher donor DNA concentrations and subsequently higher host proportions revealed similar dynamics (supplementary fig. 3, Supplementary Material online). (B) A comparison of plasmid allele dynamic variability between the two model plasmids (i.e., assortment modes). Top: host type ratios for each model plasmid are plotted, where a ratio of 1 indicates equal frequencies of the two host types. Bottom: the coefficient of variation per day is shown. Lines present the trendline and the shaded area corresponds to the confidence interval. (C) Sequencing coverage data of evolved selected populations. The sequencing coverage of the plasmid identity region is shown on the left. The ratios of the novel and ancestral alleles (nptII and GFP) coverage are shown on the right. (D) Analysis of plasmid content in heterozygotic populations (R4 and R8) by 0.7% agarose gel electrophoresis gel. Untreated samples (i.e., circular plasmid) are shown on the left; plasmids treated with HindIII (i.e., linear plasmids) are shown on the right (note the HindIII site in the gfp gene). Both analyses show the presence of ancestral and novel plasmid types, accompanied by a heteromultimer of both plasmid types (see illustration).
To describe the novel plasmid allele segregation in detail, we focus on the dynamics of the randomly segregating model plasmid pTAD-R (fig. 2A). At the beginning of the evolution experiment, the hosts were predominantly heterozygotes due to the introduction of the novel plasmid allele via natural transformation. The emergence of homozygotes for the novel plasmid allele is evidence for plasmid allele segregation within the host population. Notably, the frequency of homozygote and heterozygote hosts during the first approximately five transfers had a similar pattern in most replicates. As the experiment progresses, the frequency of novel homozygotes increased until the novel plasmid allele reached fixation within the subpopulation of hosts in most replicates (i.e., heterozygote hosts were no longer observed). Replicates revealing exceptional plasmid allele dynamics within the host population comprised two replicates where the heterozygotes, rather than the homozygotes, comprise the majority of the host subpopulation prior to the host extinction (replicates R4, R8 in fig. 2A). The observed variability in the ratio of homozygote and heterozygote hosts across replicates is as expected under segregational drift of the novel plasmid allele.
Plasmid Assortment Mode Is a Determinant of Plasmid Allele Dynamics
Comparing the dynamics of host genetic diversity in the evolution of our model plasmids pTAD-R and pTAD-C demonstrates the effect of plasmid assortment mode on the evolution of plasmid allele dynamics. The clustered segregation of pTAD-C alleles is expected to decrease the segregational units of the ancestral plasmid allele upon division into the daughter cells. In other words, the chances of the ancestral and novel alleles to be inherited together are decreased, thus the clustered mode of inheritance is expected to accelerate the segregation of homozygotes (fig. 1B). Although the plasmid allele dynamics of the randomly segregating pTAD-R were characterized by stochastic behavior, the pTAD-C evolution revealed more deterministic plasmid allele dynamics (i.e., the outcome in all replicates is nearly identical). The emergence of equal homozygote and heterozygote frequencies after the fifth transfer stands out as a highly stable consequence of the clustered plasmid assortment mode. A comparison of the relative frequency of heterozygotes and novel homozygotes during the evolution experiment further reveals a high variability in the evolution of pTAD-R and homogeneity in the evolution of pTAD-C (fig. 2B). The stochastic pTAD-R allele dynamics are further characterized by increasingly high variability in the ratio of heterozygotes and novel homozygotes among replicate populations during the evolution experiment. In comparison, the variability among pTAD-C replicate populations is moderate throughout the experiment (fig. 2B). Consequently, we conclude that plasmid assortment mode has an effect on plasmid evolution via its effect on the dynamics of plasmid alleles.
To further test if the plasmid allele dynamics in our experiment reflect vertical inheritance of the alleles only, we quantified the extent of lateral transfer of the novel plasmid allele during the evolution experiment. Testing for the extent of transformation with the novel plasmid allele (nptII) showed that DNA is readily present in the spent media (supplementary table 2, Supplementary Material online). Nonetheless, the low transformation frequencies observed indicate that the extent of transformation by released DNA of previously transformed cells is neglectable. To examine the effect of transformation on the long-term plasmid allele dynamics, we conducted an evolution experiment of pTAD-C, with or without repeated treatment with DNase, that digests free DNA in the media. Our results show that the dynamics of the novel plasmid allele over the experiment duration were not markedly different between the treatments (supplementary fig. 5, Supplementary Material online). Consequently, we conclude that the segregation of plasmid alleles in our experiment is only due to plasmid assortment during cell division (i.e., vertical inheritance).
Plasmid Fusion Maintains Plasmid Heterogeneity in the Host Population
The stochastic dynamics of pTAD-R segregation reveal several replicate populations that stand out as exceptions to the overall similar allele dynamics. Considering dynamics at the level of the whole population, this includes one replicate where the hosts persist instead of going extinct (replicate R3 in fig. 2A). Another two replicates show exceptional plasmid allele dynamics within the host population, where the heterozygotes reach fixation within the host subpopulation prior to their extinction (replicates R4, R8 in fig. 2A). To gain further insight into the genetic basis of the exceptional replicates, selected evolved host populations were chosen for genome sequencing. The analysis of the evolved host genomes revealed several single nucleotide variants, however none of those reached fixation in the populations analyzed. Hence, no evidence for genetic variants or genome rearrangements that may play a role in the observed allele dynamics could be found in either the chromosome or the plasmid (supplementary table 3, Supplementary Material online). Using the sequencing results, we furthermore examined the sequencing coverage of the plasmid allele identity regions. To estimate the relative frequency of the novel and ancestral alleles, we calculated the ratio of the mean nptII (novel allele) to gfp (ancestral allele) coverage (fig. 2C and supplementary fig. 6, Supplementary Material online). In host populations where the novel homozygotes are predominant (higher nptII coverage), we would expect high ratio values. However, host populations that are predominantly heterozygotic would have an expected ratio of approximately 2 (since nptII is found in both ancestral and novel types; see fig. 1A). The calculated ratios are in agreement with our observations based on colony counts (fig. 2B); hence the sequencing coverage serves as an independent estimation for the relative frequency of homozygote and heterozygote hosts in the population (fig. 2C). The coverage of the allele identity region shows a decreased coverage at the GFP locus (ancestral allele) in those evolved populations where the novel homozygotes dominate the host population (see replicate R2 in fig. 2C). Notably, the ratio of novel to ancestral plasmid alleles in replicates where the host population is dominated by heterozygotes (R4 and R8) reveals nearly equal frequencies of both alleles (fig. 2C), hence, we conclude that both ancestral and novel alleles are indeed maintained in those populations.
To elucidate how the plasmid heterozygotic state is maintained, we examined the plasmid conformation in the heterozygotic populations (n = 4; sampling two colonies from each population). Agarose gel analysis of the undigested sample showed the expected two plasmid types, ancestral and novel plasmids in different proportions. Notably, the presence of a high molecular weight band that is usually associated with plasmid multimers was observed (fig. 2D). The formation of plasmid multimers is typically the result of homologous recombination between sister plasmids (i.e., recombination between either the ancestral or novel plasmids) or nonsister plasmids (i.e., recombination of ancestral with the novel plasmid). Although recombination between sister plasmids can give rise to dimers and higher multimers, recombination between nonsister plasmids creates heteromultimeric plasmids. We treated the plasmid isolates with nicking and restriction enzymes to observe relaxed open circular (oc) plasmid molecules (supplementary fig. 7, Supplementary Material online) and the specific digestion products of multimers (fig. 2D). Enzymatic digestion showed that each heterozygotic colony we analyzed contained a mixture of ancestral, novel, and heteromultimeric plasmids. The presence of the heteromultimer formed by the two fused plasmid backbones was further verified by PCR reaction (supplementary table 1, Supplementary Material online). Thus, we conclude that the observed heterozygotes in our experiment are the result of recombination and fusion between the two plasmid types, yielding a single plasmid replicon in which both ancestral and novel alleles are encoded. To test the heritability of the observed heteromultimeric plasmid, we isolated the entire plasmid content from replicates R4 and R8, and introduced the plasmids into a plasmid-free population by electroporation. An examination of the resulting populations revealed the presence of heterozygotic cells (50% of the R4 descendant population and 25% of the R8 descendant population). We further validated the heteromultimeric plasmid heritability by performing single colony streaks of heterozygote colonies on solid media, which revealed the emergence of heterozygotic and homozygotic colonies. Our results thus show that the heterozygotic state is stably maintained in the population while, at the same time, plasmid alleles segregate such that homozygotes are generated. Further examination of the plasmid copy number using quantitative polymerase chain reaction (qPCR) showed no changes in the PCN and further validated the presence of both plasmid alleles in the population. In conjunction with the sequencing results that showed no relevant mutations or rearrangements in the plasmid genome, we conclude that the heteromultimers are composed of the intact two plasmid types: ancestral and novel. Borrowing the nomenclature from population genetics of diploid organisms (Graur 2016), the evolution of heterozygotes in our experiment is akin to permanent heterozygosity, where the heterozygotic state is maintained by the presence of the two alleles on the same replicon.
Novel Plasmid Alleles Are Rapidly Lost Due to Genetic Drift at Multiple Levels
To further examine the contribution of drift at the population level to rapid extinction of the novel plasmid allele, we compared the observed plasmid allele dynamics to the dynamics of a novel neutral allele present in the bacterial chromosome. For that purpose, we performed an evolution experiment of a chromosomal allele under the same serial transfer conditions as our plasmid evolution experiment (23 transfers with a 1:100 dilution, i.e., bottleneck size 107 cells). In the chromosomal allele dynamics experiment, the novel allele comprised the trpE gene present in A. baylyi BD4 (tryptophan prototroph wild-type), whereas the ancestral allele was the trpE27 allele present in A. baylyi BD413 (auxotrophic for tryptophan). The trpE27 allele is neutral for A. baylyi in rich media (as demonstrated via pairwise competition experiments; H0: w = 1, P = 0.9799 using Wilcoxon test, n = 16; supplementary fig. 8, Supplementary Material online). The chromosomal allele dynamics experiment was conducted with two initial frequencies of the novel chromosomal allele. These were determined as high and low bounds of the initial plasmid allele relative frequency (∼5 × 10−5) to be either 10−5 (low-bound) or 10−4 (high-bound). The experiment was conducted with six replicates for each of the initial frequencies. The frequency of hosts of the novel chromosomal allele was followed over the experiment duration by selective plating on M9 minimal media.
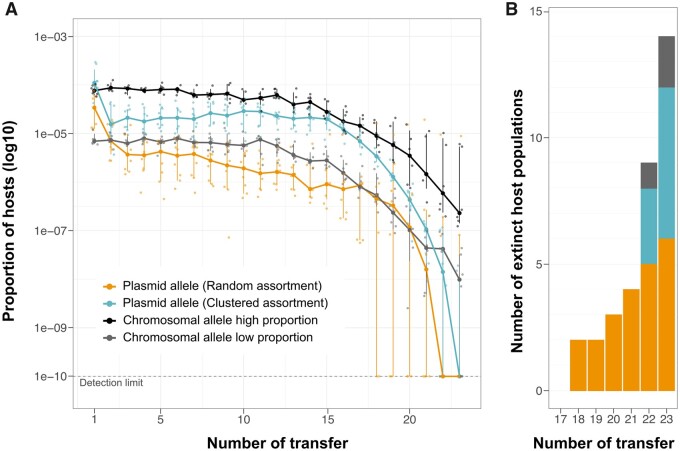
The results of the chromosomal allele dynamics experiment show a stable maintenance of the novel chromosomal allele (i.e., host cells) during the first approximately 15 transfers (fig. 3A). Subsequently, a decrease in the proportion of hosts of the novel allele was observed for both initial novel allele frequencies (High and Low), as expected under genetic drift (Bodmer and Cavalli-Sforza 1976). The differences between the high- and low-bound populations reflect the different initial allele frequencies. A comparison of the novel chromosomal allele dynamics to the pTAD-R and pTAD-C allele dynamics reveals a similar level of persistence during the first approximately 15 transfers. The comparison further shows that the rate of novel plasmid allele loss is higher in comparison to the chromosomal allele and leads to a rapid extinction of the novel allele with both model plasmids (fig. 3B). We note that the loss of the novel plasmid allele for pTAD-R starts earlier in comparison to the late but fast extinction for pTAD-C. As both chromosomal and plasmids novel alleles possess no fitness advantage or disadvantage in comparison to their ancestral version, the differences between the loss dynamics of chromosomal and plasmid alleles supply experimental evidence for the importance of segregational drift in plasmid evolution. Our results thus demonstrate that under nonselective conditions, plasmid allele segregation during cell division accelerates the extinction of novel plasmid alleles.
Fig. 3.
A comparison between chromosomal and plasmid allele dynamics. (A) The dynamics of a neutral chromosomal allele over approximately 160 generations. The two chromosomal allele host proportions (high and low) are depicted together with the dynamics of model plasmid pTAD-R and pTAC-C. Points correspond to the proportion of hosts in each population and sampling time. Lines show the median proportion of hosts per sampling time with error bars that correspond to the median confidence interval (bootstrap percentile method; Carpenter and Bithell 2000). A dotted line indicating the detection limit in our study; host proportions below the detection limit document allele extinction. (B) The number of replicate populations per transfer where the allele went extinct. The bar colors correspond to the legend in (A).
The Model Plasmid pWH1277 Is of Ancient Origin and Is Highly Conserved
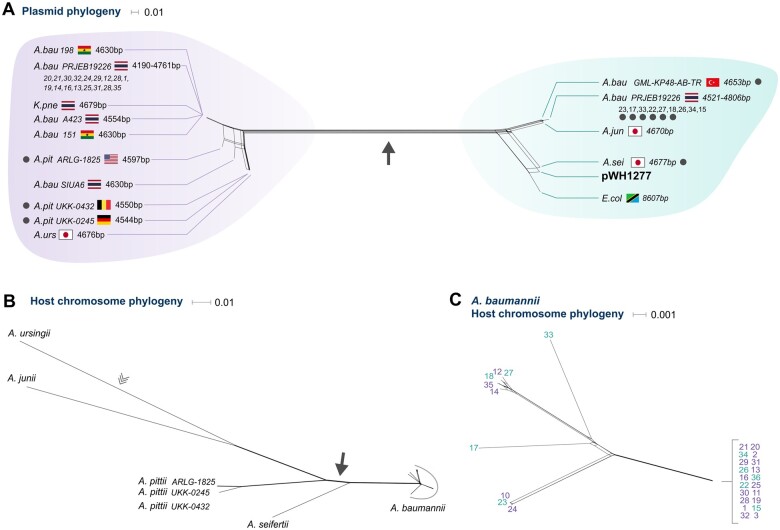
The results from the evolution experiment support the role of segregational drift in the elimination of novel alleles and thus raise the hypothesis that the rate of plasmid evolution is lower than the expected from their mutational supply. To gain further insights into the evolution of our model plasmid in natural environments, we surveyed genomic databases for homologous plasmid backbones of the cryptic plasmid pWH1277 (Hunger et al. 1990; Lucidi et al. 2018). Searching for pWH1277 derivatives using sequence similarity to our model plasmid yielded several related plasmid sequences found in isolates from diverse geographical locations. Most of the plasmid hosts are clinical isolates reported as multi drug resistant bacteria with a majority of Acinetobacter species (fig. 4A and supplementary table 4, Supplementary Material online). Additionally, we found evidence for pWH1277-homologs in one E. coli and one Klebsiella pneumoniae isolates. The plasmid phylogeny indicates A. baumannii as the donor of the K. pneumoniae host and an A. calcoaceticus-like donor of the E. coli host (fig. 4A).
Fig. 4.
Evolutionary reconstruction of plasmid pWH1277. Phylogenetic network of pWH1277 homologous plasmids (A) and chromosome (B, C) of the hosting isolates. (A) The plasmid phylogenetic network was inferred from a multiple alignment of the full-length plasmid genome sequence. For simplicity, closely related Acinetobacter baumannii isolates within the same bioproject are abbreviated with a single label. Strains marked with gray dots were included in the evolutionary rate comparison. Corresponding splits in the plasmid and chromosome phylogenies are marked by a filled arrow. Isolate label includes country of origin and plasmid contig length. We note that the plasmid is not expected to replicate in Escherichia and Klebsiella. Further validation of the host taxonomic classification according to the genomic data (see Materials and Methods) confirmed their reported taxonomy; we consider the observation of pWH1277 in those hosts a rare event. (B) The chromosome phylogenetic network was inferred from a concatenated alignment of 1,617 universal single-copy gene families shared among all Acinetobacter isolates identified as pWH1277-derivative hosts. The root position is marked by an open arrowhead as previously inferred (Mateo-Estrada et al. 2019). (C) Phylogenetic network of A. baumannii isolates was inferred from 2,645 chromosomal universal genes. The color of the labels corresponds to the plasmid groups observed in (A). Plasmids from the purple and blue groups are intermixed in the chromosomal phylogeny, indicating mixed plasmid origins in within A. baumannii. Isolate serial number in (A) and (C) identifies the isolates as in supplementary table 4, Supplementary Material online.
Quantifying the evolutionary rate of pWH1277 genome requires to disentangle the contribution of vertical inheritance and lateral transfer to pWH1277 evolution. For that purpose, we compared the phylogenies of the plasmid and host chromosomes. The plasmid phylogeny reveals two groups of diverged plasmid sequences that are separated by a deep split within the Acinetobacter isolates (fig. 4A). The Acinetobacter host phylogeny reconstructed from chromosomal gene families conforms the recognized phylogenetic relation within the genus (Mateo-Estrada et al. 2019), where A. ursingii has a basal position (fig. 4B). A comparison between the plasmid and chromosome phylogenies reveals multiple discordant splits. The presence of A. baumannii on both sides of the deep split in the plasmid phylogeny may correspond either to deep divergence within A. baumannii or a lateral transfer event in the plasmid evolutionary history. To distinguish between these two possibilities, we extended the phylogenetic reconstruction of A. baumannii host chromosomes. The resulting A. baumannii chromosome phylogeny (fig. 4C) reveals no evidence for the deep split observed in the plasmid phylogeny; thus, the A. baumannii plasmids branching with A. pittii (see purple group in fig. 4A) is likely evidence for the plasmid transfer from an A. pittii donor to A. baumannii. Furthermore, the topology of A. ursingii and A. junii in the plasmid phylogeny does not correspond to their basal position in the Acinetobacter phylogeny (Mateo-Estrada et al. 2019) (fig. 4B). Consequently, we conclude that plasmid phylogeny shows evidence for two additional lateral transfer events: from A. pittii to A. ursingii and from A. baumannii to A. junii. Taken together, the deep split in plasmid phylogeny indicates that the pWH1277 derivatives share a common ancestor that dates back to the split between A. seifertii and A. pittii (fig. 4B). The plasmid likely evolved by vertical inheritance following that divergence, which was accompanied by several transfer events.
To further examine the evolutionary rate of plasmid-encoded genes, we compared the nucleotide substitution rate between the plasmid and chromosome universal gene families while excluding the isolates where plasmid acquisition by lateral transfer was identified. Our results reveal that the number of substitutions per site is significantly different between the plasmid and chromosome-encoded genes, with the plasmid substitution rates smaller than that of the chromosome (Median chromosome total branch length = 0.332; Median plasmid total branch length = 0.196; P = 0.035, using bootstrapped median comparisons; supplementary fig. S9, Supplementary Material online). To examine the effect of purifying selection on the plasmid and chromosome evolutionary rates, we compared the ratio of nonsynonymous to synonymous substitution rate (dN/dS) between genes encoded in both replicon types. Performing the comparison of dN/dS among isolates in a pairwise manner, we observed a majority of comparisons (53 out of 55) where the dN/dS ratio was either not significantly different between the plasmid and chromosome or higher for plasmid genes (supplementary fig. S10, Supplementary Material online). These results indicate that the difference in evolutionary rate between the plasmid and chromosome cannot be explained by higher purifying selection acting on the plasmid-encoded genes. Consequently, we conclude that the evolutionary rate of the pWH1277-encoded genes is lower than that of the host chromosome-encoded genes, despite the plasmid higher mutational supply.
Discussion
Extrachromosomal genetic elements are typically found in multiple copies in their host cells, which may contribute to the emergence of intracellular genetic diversity. The evolution of extrachromosomal elements is consequently influenced by processes at two levels: the host cell and the population of hosts. Indeed, previous studies show evidence for multilevel selection in the evolution of viruses (e.g., Buskirk et al. 2020; Meir et al. 2020) and the eukaryotic mitochondria (e.g., Taylor et al. 2002; Gitschlag et al. 2016, 2020). Evidence for multilevel genetic drift has been so far reported in clonal multicellular organisms (e.g., Kuntz et al. 2020; Yu et al. 2020). Focusing on bacterial plasmids, we supply here empirical data on the effect of genetic drift on plasmid allele dynamics at multiple levels: segregational drift within the host and genetic drift of hosts within the bacterial population.
The effect of segregational drift on plasmid alleles may vary according to various factors that govern the plasmid replication and inheritance. This includes the coordination of replication time, spatial plasmid distribution within the cell, and the presence of active partition systems (Pinto et al. 2012). Here, we tested for the effect of plasmid assortment mode by altering the plasmid unit of inheritance (illustrated in fig. 1B). Our results reveal that the random plasmid assortment leads to stochastic plasmid allele segregation dynamics, in comparison to the clustered assortment, which leads to deterministic allele segregation dynamics (fig. 2B). The clustered plasmid assortment accelerates the segregation of plasmid alleles in heterozygote hosts and leads to rapid emergence of homozygotes (fig. 2). The difference between the two assortment modes is further apparent in the dynamics of plasmid allele extinction, which is gradual for the random plasmid assortment and rapid for the clustered plasmid assortment (fig. 3B). Consequently, we conclude that plasmid assortment mode has far reaching implications for the dynamics of plasmid alleles; the presence of genetic elements that alter the plasmid unit of inheritance, for example, regulatory elements or partition systems, may therefore have an effect on plasmid evolution.
Previous studies show that heterozygote hosts for two alternative plasmid alleles can be maintained in the population depending on selective conditions for the plasmid alleles (Rodriguez-Beltran et al. 2018). Our results reveal that the emergence of plasmid fusions (plasmid heteromultimers) can maintain plasmid heterogeneity in the host, and within the population under nonselective conditions. Permanent heterozygosity is comparable to gene duplication and functional divergence in chromosomes (Graur 2016). Plasmid recombination is not an infrequent phenomenon, as plasmid oligomers are formed as part of the plasmid lifecycle (James et al. 1982; Chédin et al. 1997; Dionisio et al. 2019). Mosaic plasmids composed of genetic elements from distinct sources, are frequently observed in nature (Chaconas and Norris 2013; Pesesky et al. 2019). Whereas the formation of homomultimers occurs due to homologous recombination between sister plasmids molecules during or after replication, heteromultimer formation is due to recombination of nonsister plasmid molecules. This process is similar to events of gene duplication due to unequal crossing over of sister chromatids (e.g., Ohta 1984). We note that the emergence of permanent heterozygosity was only observed in the evolution experiment of plasmid pTAD-R that has a random segregation mode. The formation of heteromultimers is less likely to occur in the clustered segregation mode since the nonsister plasmid molecules are less likely to be spatially linked in order to recombine. Current literature often links the emergence of permanent heterozygosity with selection regimes entailing heterozygote advantage (e.g., segregation avoidance; Spofford 1969; Hahn 2009). Indeed, the fusion of multiple beneficial plasmid alleles, for example, genes encoding for various antibiotics resistance, may be advantageous for the plasmid host (depending on the environmental conditions; Hülter et al. 2020). Here, we show that plasmid fusions can be formed and stably inherited also under nonselective conditions. Plasmid fusion thus has the capacity to maintain plasmid heterogeneity in the population.
According to the neutral theory, most emerging mutations are expected to be neutral or slightly deleterious (Ota and Kimura 1971). In our study, we compared the dynamics of plasmid and chromosomal alleles under non selective conditions. Notwithstanding, mutations in the plasmid genome may have a fitness effect on the plasmid only, for example, via modification of plasmid stability (Wein et al. 2020), or have a fitness effect on the host, for example, due a modification of beneficial plasmid functions (Rodríguez-Beltrán et al. 2020). The novel allele we traced in our experiment (nptII) has no effect on the plasmid stability, and furthermore, the evolution experiment was performed under nonselective conditions for kanamycin resistance. The comparison between the dynamics of the novel plasmid and chromosomal alleles reveals marked differences between the allele loss rate depending on the replicon type. We note that both plasmid and chromosomal novel alleles experience population bottleneck events that are inherent to experimental evolution studies (Barrick and Lenski 2013). Thus, we conclude that dynamics of the novel plasmid allele in our experiment are subject to genetic drift at two levels: within the host and within the population. Our experimental results are in agreement with the theoretical predictions for the effect of segregational drift (Ilhan et al. 2019; Santer and Uecker 2020). The rapid extinction of the plasmid allele is best understood by the combined effect of multilevel genetic drift. At the same time, one exceptional replicate in our experiment (R3; fig. 2A) reveals the possibility of rare events where plasmid alleles that are maintained at the host level may further persist in the population. Taken together, our study shows that segregational drift of plasmid alleles interferes with the fixation of novel plasmid alleles that are neutral to the host fitness.
Plasmids are expected to evolve under strong selection pressure on genomic loci related to plasmid fitness—that is—functions that are related to the plasmid life cycle, including replication and segregation (Hülter et al. 2020; Garoña and Dagan 2021). In small plasmids such functional loci may account for a high proportion of the plasmid genome. The comparative genomics of pWH1277 backbone as observed in natural isolates reveals a high sequence conservation of the plasmid genome. Plasmid genome sequences may remain conserved over millions of years across isolates and habitats (Mindlin et al. 2020), yet plasmid sequence conservation is often interpreted as evidence for long-range lateral plasmid transfer (e.g., Petersen et al. 2019). Indeed, lateral transfer events have the potential to scramble the genomic record for evolutionary reconstructions (Dickerson 1980). Nonetheless, the presence of a deep split in pWH1277 phylogeny indicates an ancient origin of the plasmid, hence the high sequence conservation cannot be explained solely by lateral transfer events. A more parsimonious inference is thus slow evolutionary rate of pWH1277 derivatives. Our results show that segregational drift counteracts the elevated mutational supply of multicopy plasmids via its effect on the intracellular plasmid genetic heterogeneity. The presence of extrachromosomal elements within a host cell generates a partitioned population structure where their genome is shaped by the interplay of evolutionary forces at multiple levels.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
The A. baylyi strain BD413 (DSM No. 588, German Collection of Microorganisms and Cell Cultures, DSMZ) also known as strain ADP1 (GenBank accession number NC_005966.1) was used as the model organism in all experiments. Either A. baylyi BD413 or E. coli DH5α were used during plasmid constructions. Plasmids and primers used in this study are listed in supplementary tables (supplementary table 1, Supplementary Material online). Escherichia coli DH5α was routinely grown at 37 °C in lysogeny broth (LB) medium at 250 rpm shaking or on LB-agar plates. Acinetobacter baylyi was propagated at 30 °C in LB medium in liquid shaking cultures or deep-well plates. For molecular cloning and plating, antibiotics for the selection of plasmid carrying cells were used at the following concentrations: kanamycin 10 μg/ml, chloramphenicol 10 μg/ml, trimethoprim 125 µg/ml, and gentamicin 5 μg/ml. IPTG (Isopropyl β-d-1-thiogalactopyranoside) was added to the media to a final concentration of 1 mM when derepression of the LacI-repressed pTrc promoter was desired. Plasmids were extracted using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific). DNA quantification was done using the Multiskan GO spectrophotometer instrument (Thermo Fisher Scientific).
The model plasmid pTAD was derived from plasmid pVRL1 (Lucidi et al. 2018; GenBank accession number MG462882), a cloning vector derived from the cryptic Acinetobacter plasmid pWH1277 (Hunger et al. 1990), by PCR-amplifying its backbone (5,400 bp) containing the origin of replication, an addiction system (toxin–antitoxin) and a gentamicin resistance gene (aacC1) for selection using primer pair NH140/NH137. For the construction of pTAD-C, the PCR-fragment of pVRL1 was assembled using primer pair NH141_GA.FOR/NH142_GA.REV by the Gibson assembly technique (NEBuilder protocol; New England Biolabs) with a cassette comprising the lacIq gene and a nptII gene into which a gfp gene (gfpmut3.1 fused to the LacI-repressible Ptrc promoter) was inserted into middle of the open reading frame of nptII (fig. 1; see Supplementary Material online for the construction of the cassette). For the construction of pTAD-R, pTAD-C was subjected to PCR-mediated site-directed mutagenesis using the primer pair (AG58/AG60) that introduced a one nucleotide deletion in the lacI gene. This deletion causes a frameshift mutation that changes amino acid residue 330 in the LacI protein from Leu to Trp, creating a premature stop codon that results in a functional dimeric LacI repressor instead of a tetrameric one (Brenowitz et al. 1991; Hsieh and Brenowitz 1997).
Natural Transformation of Acinetobacter baylyi
The preparation of competent cells of A. baylyi carrying pTAD-C or -R was performed as described previously (Kickstein et al. 2007). Briefly, the cells were grown at 30 °C overnight with shaking in 2 ml of LB medium and then used for the inoculation (1:100) of fresh cultures. The cultures were grown until early stationary phase (∼1 × 109 cells/ml), cooled down, and then stored as concentrated stocks (1 × 1010 cell/ml) at −80 °C.
For each transformation performed in the study, the frozen competent cells were thawed on ice and diluted to a cell density of 2.5 × 108 cells/ml. The donor DNA was a PCR fragment (see Supplementary Material online) containing the nondisrupted nptII as the single segment of sequence identity to the recipient plasmid. After 90 min of incubation at 30 °C with shaking, diluted aliquots of the culture were plated on LB medium to estimate the titer of the population and transformants were scored on selective media. Transformation frequencies were calculated as transformants per recipient. Natural transformation kinetics were verified experimentally alongside with the specificity of the homologous recombination event (see Supplementary Material online).
Vertical Inheritance of the Introduced Allele
To determine whether the observed plasmid allele frequencies are influenced by ongoing transformation during the experiment, we studied the extent of transformation, as during normal growth cells secrete plasmid and chromosomal DNA that could potentially transform neighboring cells causing subsequent recombination events. The employed resident plasmid is also nonconjugative so that it cannot be transferred between cells. We first evaluated whether the secreted DNA from the cells during growth was able to transform naturally competent cultures, performing a series of transformation tests employing spent media (supplementary table 2, Supplementary Material online). Next, we performed a repetition of the experimental evolution experiment with pTAD-C employing the same conditions to be used in our experimental set up, serial transfer with 1:100 bottleneck size (supplementary fig. 5, Supplementary Material online). The experiment was performed with two treatments, with and without DNase 1 (0.1 mg/ml) (n = 8 per treatment).
Evolution Experiments
The plasmid allele dynamics evolution experiment was conducted under nonselective conditions with ancestral plasmid carrying A. baylyi populations. The ancestral plasmid present varied, with pTAD-R for random segregation (n = 8) and pTAD-C (n = 8) for clustered segregation. The experiments started with the natural transformation of A. baylyi strain BD413, containing the ancestral plasmid. Competent cells were prepared as previously described (see Natural transformation of A. baylyi), and the donor DNA was added to a final concentration of 100 ng/ml.
After 90 min of incubation at 30 °C with shaking, a fraction of the cells was plated to determine the total cell population and the transformation frequency. The viable titer of the cultures was obtained from the cells plated on LB media (50 µl of 10−4, 10−5, and 10−6 dilutions), whereas the transformation efficiency was calculated by plating on LB + kanamycin (5 µg/ml) (10°, 10−1 and 10−2 dilutions). Cells that had restored the nptII gene on one of copies of the recipient plasmid survived the kanamycin treatment. The cultures were then serially passaged daily into fresh medium using a 1:100 dilution factor, corresponding to a bottleneck size of 107 cells. The transfer regime was then maintained for 23 transfers, which corresponds to approximately 160 generations (∼6–7 generations per transfer in liquid medium). Every day the population sizes were estimated, the total population size was estimated from the plating on LB media and the proportion of hosts in the population was determined by plating on selective media, LB supplemented with kanamycin. The differentiation between heterozygotes and homozygotes of the novel allele (nptII) is possible by the reporter construct. Heterozygotes for the novel allele formed green colonies, whereas homozygotes of the novel allele formed white colonies on selective plates.
The chromosomal allele experiment was performed by employing two isogenic A. baylyi populations of two strains that differed only in carrying either the wild-type trpE gene (strain BD4) or the trpE27 mutation (strain BD413). The experiment was initiated with six replicate colonies of each strain. The replicate colonies were grown overnight in LB medium at 30 °C with shaking. At the onset of the experiment, the replicate cultures of the two strains were diluted and mixed at a ratio of 1:100,000 (i.e., 10−5; low-bound) and 1:10,000 (i.e., 10−4; high-bound), respectively, with strain BD4 being the proportionally less abundant genotype in the mixture. The mixtures were then used to inoculate 1 ml cultures with a starting density of in total 107 cells per ml. The experimental set up was identical as the one employed for the plasmid allele dynamics experiment and serially passaged for 23 transfers with a 1:100 dilution factor. At t = 0 (after inoculation) and thereafter every day, the proportions of the two genotypes (BD4/BD413), were determined as previously described using M9 minimal media supplemented with glucose (2 mg/ml) as selective media.
Plasmid Copy Number Determination
The PCN of the plasmid types was determined using qPCR as described in (Škulj et al. 2008). Samples of overnight cultures were lysed by 10 min incubation at 98 °C followed by 10 min at −20 °C. The primers employed targeted both the chromosome and the plasmid. The chromosomal primers were complementary to the alkB gene (Alkane 1-monooxygenase) of A. baylyi BD413 (NH72/NH73). The plasmid primers targeted the regulatory region of pVRL (AG21/AG22), making them a universal primer pair to study all the plasmids involved in the experiments, regardless of their variations in the reporter construct.
The qPCR reactions were performed in a total volume of 10 μl containing 1× iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories), 100 nM of each primer (final concentration), and 1 μl sample. All qPCR reactions including positive and nontemplate controls were performed in technical replicates on a CFX Connect Real-Time PCR Detection System (Bio-Rad Laboratories) using the following cycling conditions: 95 °C for 3 min, 40 cycles of 10 s at 95 °C and 1 min at 59 °C. Primer specificity and efficiency were determined using standard-curve and melt-curve analyses. The ratio between the number of plasmid amplicons and chromosome amplicons is defined as the PCN (comparative CT method) and was calculated considering the amplification efficiencies of both primer pairs (Škulj et al. 2008).
Plasmid Stability Experiments
To determine the stability or segregational loss of the model plasmid variants pTAD-R and pTAD-C, a serial transfer experiment without selection was performed during 6 days (∼42 generations), showing a 100% stability for the plasmid (no measurable loss 0 ± 0%; n = 4).
Fitness Experiments
The relative fitness (w) of the ancestral allele carrying strain versus the novel allele carrying strain was estimated by direct pairwise competition experiments, with 16 replicate populations per replicon type. The competition experiments were initiated with a 1:1 ratio of the competing strains, diluted 1:100 (∼107 cells transferred) from overnight cultures, preconditioned to growth in liquid media for two days. Then, cultures were serially transferred in a 1:100 bottleneck for five transfers and the competitors’ population sizes measured every transfer (24 h). Relative fitness of the competing genotypes was determined by calculating population sizes of both competitors via selective plating using the formulation as described in Lenski et al. (1991) and Starikova et al. (2013). The two competitors are distinguished by plating on nonselective (LB supplemented with IPTG 1 mM for allele carrying plasmids or LB for chromosomal allele strains) and selective media (LB supplemented with kanamycin 10 μg/ml for allele carrying plasmids or M9 minimal medium supplemented with glucose 2 mg/ml for chromosomal allele containing strains).
Genome Sequencing
Population sequencing was used to study plasmid host genotype frequencies and to detect genetic variants present either on the plasmid or the chromosome. Total DNA, containing both plasmid and chromosomal DNA, was isolated from 1 ml culture using the Wizard Genomic DNA Purification Kit (Promega). The concentration and quality of the extracted DNA were determined using the NanoDrop (Thermo Fisher Scientific) and Qubit (Invitrogen by Life Technologies). Illumina sequencing was performed to sequence the samples, the sample libraries were prepared with TruSeq Nano DNA Low Throughput Library Prep Kit (Illumina, Inc.), and the sequencing was performed with paired-end reads on the Miseq system (Illumina, Inc.).
Sequencing reads were trimmed to remove low-quality bases using the program Trimmomatic v.0.38 (Bolger et al. 2014) (parameters: CROP:250 HEADCROP:5 LEADING:20 TRAILI NG:20 SLIDINGWINDOW:4:20 MINLEN:36). For the reference sequence, we joined the genome of A. baylyi ADP1 (GenBank accession number NC_005966.1) with the pTAD plasmid sequence (GenBank accession number: MW757345). The sequencing reads were mapped to the reference genomes using BWA-MEM v.0.7.17 (Li 2013). Subsequent indexing and local realignment of sequencing reads were performed using SAMtools v1.11 (Li et al. 2009). Next, variants were called using LoFreq v2.1.5 (Wilm et al. 2012). All variants with an allele frequency higher than 0.05 were considered for the analysis. The coverage distribution was used to visualize plasmid allele variants.
Comparative Genomics and Phylogeny of Plasmid pWH1277 Backbone
The search for similar plasmid sequences was performed using the interactive NCBI BLAST tool (Boratyn et al. 2013) against the RefSeq database (Pruitt et al. 2005). The hallmark of the plasmid pWH1277 backbone is the regulatory region that contains the putative origin of replication (fig. 1). Searching for similar plasmids using BlastN with the regulatory region as the query sequence returned only cloning vectors (e.g., pVRL derivatives). Consequently, we extended our search to sequences flanking the regulatory region, which include the toxin–antitoxin module and the putative repB protein. Similar protein-coding genes were searched using BlastP with the amino acids sequence as a query. Only hits of >90% identical amino acids were retained. A total of 38 contigs where all three pTAD components were found were further aligned with the pWH1277 backbone sequence and the global sequence similar to the plasmid genome was manually inspected (supplementary table S4, Supplementary Material online). The full genomes of these 38 isolates were downloaded from NCBI (ver. 08/12/2020). To validate the taxonomical classification of the E. coli and K. pneumoniae data sets, we observed the 16S rRNA sequences and found that they corresponded to the designated species. Clusters of homologous proteins were reconstructed based on amino acids sequence similarity. First, we performed a pairwise BLAST (Camacho et al. 2009) search among all isolate pairs (including all contigs) using a threshold of E-value ≤1e-10. At the next step, reciprocal best BLAST hits were identified and a global pairwise alignment of those pairs of protein sequences was performed using the Needleman–Wunsch algorithm with parasail (Daily 2016). Pairs having ≥30% amino acids sequence identity were retained and used to reconstruct clusters of homologous protein sequences using MCL (Enright et al. 2002) with an inflation factor = 2.
Phylogenetic networks and trees were inferred for the plasmid and chromosome independently. For the chromosome analysis, we identified 1,617 universal single-copy protein families that were used for the phylogenetic reconstruction. The plasmid phylogenetic reconstruction comprised five universal single-copy protein families. All protein families were aligned with MAFFT using the automatic option (Katoh and Standley 2013) and were converted into codon alignments using Pal2Nal (Suyama et al. 2006). Phylogenetic networks were inferred with SplitsTree ver. 4.14.6 (Huson and Bryant 2006) using Hamming distances. The chromosome phylogenetic network was reconstructed from a concatenated alignment including the codon alignments of all 1,617 protein families. The plasmid network was reconstructed from an alignment of the whole plasmid genome (i.e., nucleotide sequence). Additionally, we reconstructed a phylogenetic network for only A. baumannii based on a concatenated alignment of 2,645 universal genes.
For the comparison of evolutionary rates, multiple alignments of the protein families including a selection of 11 strains (fig. 4A and supplementary table S4, Supplementary Material online) were reconstructed using MAFFT (automatic option) and phylogenies were inferred for each protein family using IQTREE (Minh et al. 2020) using a general model for codon evolution (Goldman and Yang 1994) and specifying the bacterial genetic code. Additionally, to infer genome pairwise dN/dS for each gene phylogeny, we used the program codeml, from the PAML package (Yang 2007). To calculate the statistical significance of differences in the median tree size and pairwise dN/dS between the plasmid and chromosome, we used a bootstrap analysis (Whitlock and Schluter 2009). This methodology was chosen due to the large difference in sample size between the two data sets which comprised five genes for the plasmid group and 2,315 genes for the chromosome group. The empirical distribution of median values was established from the median tree size in 1,000 bootstrap replicates of five chromosomal protein families each. The P value for differences in tree size was calculated as the frequency of bootstrap replicates that resulted in a median tree size equal or lower than the plasmid median tree size. Similarly, P value for differences in genome pairwise dN/dS was calculated as the frequency of bootstrap replicates that resulted in a median dN/dS pair genome pair equal or larger (or smaller) than the plasmid median dN/dS (using an α of 0.05).
Statistical Analysis
All statistical tests and data analysis were performed in RStudio version 1.2.1327. Confidence intervals for medians were calculated using the package “rcompanion” and the “groupwiseMedian” function for grouped data with n = 1,000 replicates for the bootstrap percentile method (Carpenter and Bithell 2000). Comparisons between groups (e.g., fitness calculations) were performed with two-sided Wilcoxon signed-rank test (Whitlock and Schluter 2009) by the “wilcox.test” function.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Lea Grahl and Maya Schmidt for their assistance in the experimental work. We thank Marina Khachaturyan and Maxime Godfroid for their advice in the computational analysis. We thank Erik Brinks and Adrian Prager for their assistance with the genome sequencing. We also thank Thorsten Reusch, Hildegard Uecker, Tanita Wein, Giddy Landan, and Fabian Nies for critical comments on the manuscript and the entire genomic microbiology group for their help and discussions. A.G. was supported by the International Max Planck Research School (IMPRS) for Evolutionary Biology and D.R.P. was supported by the DFG Collaborative Research Center (CRC) 1182 “Origin and Function of Metaorganisms.”
Author Contributions
A.G., N.F.H., and T.D. conceived the study and designed the experiments. N.F.H. designed the experimental system. A.G. and N.F.H. established and performed the experimental work and analyzed the data. D.R.P. performed the comparative genomics and phylogenetic analysis. T.D. supported the data analysis. All authors interpreted the results and wrote the manuscript.
Data Availability
The source data underlying figures 1–3 and supplementary figures 1–5 and 8, Supplementary Material online, are provided as a Source Data file. Data of the comparative genomics (alignments, trees, and networks) are available at www.uni-kiel.de/genomik/ressourcen (last accessed September 27, 2021). Model plasmids are available under GenBank accession number MW757345. All sequencing reads are available at the NCBI SRA database under the accession number PRJNA714632.
References
- Barrick JE, Lenski RE.. 2013. Genome dynamics during experimental evolution. Nat Rev Genet. 14(12):827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaral M, Helinski DR.. 1968. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 36(2):185–194. [DOI] [PubMed] [Google Scholar]
- Birky CW. 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu Rev Genet. 35:125–148. [DOI] [PubMed] [Google Scholar]
- Bodmer WF, Cavalli-Sforza LL.. 1976. Genetics, evolution, and man. San Francisco: W.H. Freeman. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41(Web Server issue):W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz M, Mandal N, Pickar A, Jamison E, Adhya S.. 1991. DNA-binding properties of a lac repressor mutant incapable of forming tetramers. J Biol Chem. 266(2):1281–1288. [PubMed] [Google Scholar]
- Buskirk SW, Rokes AB, Lang GI.. 2020. Adaptive evolution of nontransitive fitness in yeast. Elife 9:e62238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J, Bithell J.. 2000. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 19(9):1141–1164. [DOI] [PubMed] [Google Scholar]
- Chaconas G, Norris SJ.. 2013. Peaceful coexistence amongst Borrelia plasmids: getting by with a little help from their friends? Plasmid 70(2):161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédin F, Dervyn R, Ehrlich SD, Noirot P.. 1997. Apparent and real recombination frequencies in multicopy plasmids: the need for a novel approach in frequency determination. J Bacteriol. 179(3):754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullum J, Broda P.. 1979. Rate of segregation due to plasmid incompatibility. Genet Res. 33(1):61–79. [Google Scholar]
- Daily J. 2016. Parasail: SIMD C library for global, semi-global, and local pairwise sequence alignments. BMC Bioinformatics 17(1):81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson RE. 1980. Evolution and gene transfer in purple photosynthetic bacteria. Nature 283(5743):210–212. [DOI] [PubMed] [Google Scholar]
- Dionisio F, Zilhão R, Gama JA.. 2019. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid 102:29–36. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoña A, Dagan T.. 2021. Darwinian individuality of extrachromosomal genetic elements calls for population genetics tinkering. Environ Microbiol Rep. 13(1):22–26. [DOI] [PubMed] [Google Scholar]
- Gitschlag BL, Kirby CS, Samuels DC, Gangula RD, Mallal SA, Patel MR.. 2016. Homeostatic responses regulate selfish mitochondrial genome dynamics in C. elegans. Cell Metab. 24(1):91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitschlag BL, Tate AT, Patel MR.. 2020. Nutrient status shapes selfish mitochondrial genome dynamics across different levels of selection. eLife 9:e56686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z.. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11(5):725–736. [DOI] [PubMed] [Google Scholar]
- Graur D. 2016. Molecular and genome evolution. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI.. 2014. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio 5(5):e01918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 100(5):605–617. [DOI] [PubMed] [Google Scholar]
- Hall JPJ, Brockhurst MA, Harrison E.. 2017. Sampling the mobile gene pool: innovation via horizontal gene transfer in bacteria. Philos Trans R Soc B. 372:20160424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H, Schmitt H, Smalla K.. 2011. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 14(3):236–243. [DOI] [PubMed] [Google Scholar]
- Hsieh M, Brenowitz M.. 1997. Comparison of the DNA association kinetics of the lac repressor tetramer, its dimeric mutant LacI adi, and the native dimeric Gal repressor. J Biol Chem. 272(35):22092–22096. [DOI] [PubMed] [Google Scholar]
- Hsu T-M, Chang Y-R.. 2019. High-copy-number plasmid segregation—single-molecule dynamics in single cells. Biophys J. 116(5):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülter NF, Wein T, Effe J, Garoña A, Dagan T.. 2020. Intracellular competitions reveal determinants of plasmid evolutionary success. Front Microbiol. 11:2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger M, Schmucker R, Kishan V, Hillen W.. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87(1):45–51. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D.. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 23(2):254–267. [DOI] [PubMed] [Google Scholar]
- Hyland EM, Wallace EWJ, Murray AW.. 2014. A model for the evolution of biological specificity: a cross-reacting DNA-binding protein causes plasmid incompatibility. J Bacteriol. 196(16):3002–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhan J, Kupczok A, Woehle C, Wein T, Hülter NF, Rosenstiel P, Landan G, Mizrahi I, Dagan T.. 2019. Segregational drift and the interplay between plasmid copy number and evolvability. Mol Biol Evol. 36(3):472–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AA, Morrison PT, Kolodner R.. 1982. Genetic recombination of bacterial plasmid DNA: analysis of the effect of recombination-deficient mutations on plasmid recombination. J Mol Biol. 160(3):411–430. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickstein E, Harms K, Wackernagel W.. 2007. Deletions of recBCD or recD influence genetic transformation differently and are lethal together with a recJ deletion in Acinetobacter baylyi. Microbiology 153(Pt 7):2259–2270. [DOI] [PubMed] [Google Scholar]
- Krämer H, Niemöller M, Amouyal M, Revet B, von Wilcken-Bergmann B, Müller-Hill B.. 1987. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 6(5):1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz KLV, Kitchen SA, Conn TL, Vohsen SA, Chan AN, Vermeij MJA, Page C, Marhaver KL, Baums IB.. 2020. Juvenile corals inherit mutations acquired during the parent’s lifespan. bioRxiv:2020.10.19.345538.
- Lenski RE, , Rose MR, , Simpson SC, , Tadler SC.. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 138(6):1315–1341. [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio].
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucidi M, Runci F, Rampioni G, Frangipani E, Leoni L, Visca P.. 2018. New shuttle vectors for gene cloning and expression in multidrug-resistant Acinetobacter species. Antimicrob Agents Chemother. 62(4):e02480–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Martínez L, Pascual A, Jacoby GA.. 1998. Quinolone resistance from a transferable plasmid. Lancet 351(9105):797–799. [DOI] [PubMed] [Google Scholar]
- Mateo-Estrada V, Graña-Miraglia L, López-Leal G, Castillo-Ramírez S.. 2019. Phylogenomics reveals clear cases of misclassification and genus-wide phylogenetic markers for Acinetobacter. Genome Biol Evol. 11(9):2531–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir M, Harel N, Miller D, Gelbart M, Eldar A, Gophna U, Stern A.. 2020. Competition between social cheater viruses is driven by mechanistically different cheating strategies. Sci Adv. 6(34):eabb7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindlin S, Beletsky A, Rakitin A, Mardanov A, Petrova M.. 2020. Acinetobacter plasmids: diversity and development of classification strategies. Front Microbiol. 11:588410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch KM, Müller J, Wienecke S, Bergmann S, Heyber S, Biedendieck R, Münch R, Jahn D.. 2015. Polar fixation of plasmids during recombinant protein production in Bacillus megaterium results in population heterogeneity. Appl Environ Microbiol. 81(17):5976–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K. 2006. Plasmid R1—replication and its control. Plasmid 55(1):1–26. [DOI] [PubMed] [Google Scholar]
- Nordström K, Dasgupta S.. 2006. Copy-number control of the Escherichia coli chromosome: a plasmidologist’s view. EMBO Rep. 7(5):484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. 1969. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 33(2):210–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. 1987. Plasmid incompatibility. Microbiol Mol Biol Rev. 51(4):381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehler S, Eismann ER, Krämer H, Müller-Hill B.. 1990. The three operators of the lac operon cooperate in repression. EMBO J. 9(4):973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1984. Some models of gene conversion for treating the evolution of multigene families. Genetics 106(3):517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Kimura M.. 1971. On the constancy of the evolutionary rate of cistrons. J Mol Evol. 1(1):18–25. [DOI] [PubMed] [Google Scholar]
- Overballe-Petersen S, Harms K, Orlando LAA, Mayar JVM, Rasmussen S, Dahl TW, Rosing MT, Poole AM, Sicheritz-Ponten T, Brunak S, et al. 2013. Bacterial natural transformation by highly fragmented and damaged DNA. Proc Natl Acad Sci U S A. 110(49):19860–19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesky MW, Tilley R, Beck DAC.. 2019. Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid 102:10–18. [DOI] [PubMed] [Google Scholar]
- Petersen J, Vollmers J, Ringel V, Brinkmann H, Ellebrandt-Sperling C, Spröer C, Howat AM, Murrell JC, Kaster A-K.. 2019. A marine plasmid hitchhiking vast phylogenetic and geographic distances. Proc Natl Acad Sci U S A. 116(41):20568–20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto UM, Pappas KM, Winans SC.. 2012. The ABCs of plasmid replication and segregation. Nat Rev Microbiol. 10(11):755–765. [DOI] [PubMed] [Google Scholar]
- Pogliano J, Ho TQ, Zhong Z, Helinski DR.. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc Natl Acad Sci U S A. 98(8):4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Projan SJ, Monod M, Narayanan CS, Dubnau D.. 1987. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J Bacteriol. 169(11):5131–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR.. 2005. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33(Database Issue):D501–D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Nicolas E, Sherratt DJ.. 2012. Chromosome replication and segregation in bacteria. Annu Rev Genet. 46:121–143. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Tran T, Meas D, Lee L, Li AM, Sherratt DJ, Tolmasky ME.. 2014. High-copy bacterial plasmids diffuse in the nucleoid-free space, replicate stochastically and are randomly partitioned at cell division. Nucleic Acids Res. 42(2):1042–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin ED, Wong R.. 1988. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 136(3):507–513. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Beltran J, Hernandez-Beltran JCR, DelaFuente J, Escudero JA, Fuentes-Hernandez A, MacLean RC, Peña-Miller R, San Millan A.. 2018. Multicopy plasmids allow bacteria to escape from fitness trade-offs during evolutionary innovation. Nat Ecol Evol. 2(5):873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Beltrán J, Sørum V, Toll-Riera M, Vega C, de la Peña-Miller R, Millán ÁS.. 2020. Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria. Proc Natl Acad Sci U S A. 117(27):15755–15762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R. 1969. Replication of a bacterial episome under relaxed control. J Mol Biol. 44(3):387–402. [DOI] [PubMed] [Google Scholar]
- San Millan A, Escudero JA, Gifford DR, Mazel D, MacLean RC.. 2016. Multicopy plasmids potentiate the evolution of antibiotic resistance in bacteria. Nat Ecol Evol. 1(1):10. [DOI] [PubMed] [Google Scholar]
- Santer M, Uecker H.. 2020. Evolutionary rescue and drug resistance on multicopy plasmids. Genetics 215(3):847–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Kuroiwa T.. 1991. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res. 196(1):137–140. [DOI] [PubMed] [Google Scholar]
- Škulj M, Okršlar V, Jalen Š, Jevševar S, Slanc P, Štrukelj B, Menart V.. 2008. Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microb Cell Fact. 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalla K, Jechalke S, Top EM.. 2015. Plasmid detection, characterization, and ecology. Microbiol Spectr. 3(1):PLAS-0038-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solar G, del Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R.. 1998. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 62(2):434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford JB. 1969. Single-locus modification of position-effect variegation in Drosophila melanogaster. II. Region 3c loci. Genetics 62(3):555–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starikova I, , Al-HaroniM, , WernerG, , RobertsAP, , SørumV, , NielsenKM, , Johnsen PJ.. 2013. Fitness costs of various mobile genetic elements in Enterococcus faecium and Enterococcus faecalis. J Antimicrob Chemother. 68(12):2755–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesser N, Sheppard AE, Pankhurst L, Maio ND, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, et al. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7(2):e02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P.. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34(Web Server issue):W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DR, Zeyl C, Cooke E.. 2002. Conflicting levels of selection in the accumulation of mitochondrial defects in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 99(6):3690–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Penkul P, Milstein JN.. 2016. Quantitative localization microscopy reveals a novel organization of a high-copy number plasmid. Biophys J. 111(3):467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein T, Wang Y, Hülter NF, Hammerschmidt K, Dagan T.. 2020. Antibiotics interfere with the evolution of plasmid stability. Curr Biol. 30(19):3841–3847.e4. [DOI] [PubMed] [Google Scholar]
- Whitlock M, Schluter D.. 2009. The analysis of biological data. Greenwood Village (CO: ): Roberts and Co. Publishers. [Google Scholar]
- Wilm A, Aw PPK, Bertrand D, Yeo GHT, Ong SH, Wong CH, Khor CC, Petric R, Hibberd ML, Nagarajan N.. 2012. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 40(22):11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-Y, Lau K, Liu LF.. 1992. Interlocking of plasmid DNAs due to lac repressor-operator interaction. J Mol Biol. 228(4):1104–1114. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Young DM, Parke D, Ornston LN.. 2005. Opportunities for genetic investigation afforded by Acinetobacter baylyi, a nutritionally versatile bacterial species that is highly competent for natural transformation. Annu Rev Microbiol. 59:519–551. [DOI] [PubMed] [Google Scholar]
- Yu L, Boström C, Franzenburg S, Bayer T, Dagan T, Reusch TBH.. 2020. Somatic genetic drift and multilevel selection in a clonal seagrass. Nat Ecol Evol. 4(7):952–962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data underlying figures 1–3 and supplementary figures 1–5 and 8, Supplementary Material online, are provided as a Source Data file. Data of the comparative genomics (alignments, trees, and networks) are available at www.uni-kiel.de/genomik/ressourcen (last accessed September 27, 2021). Model plasmids are available under GenBank accession number MW757345. All sequencing reads are available at the NCBI SRA database under the accession number PRJNA714632.