Abstract
Vertebrate genomes contain endogenous retroviruses (ERVs) that represent remnants of past germline infections by ancient retroviruses. Despite comprising 8% of the human genome, the human ERVs (HERVs) do not encode a replication competent retrovirus. However, some HERV genes have been co-opted to serve host functions, most notably the viral envelope-derived syncytins involved in placentation. Here, we identify the oldest HERV intact gag gene with an open reading frame, gagV1. Its provirus contains an intact env, envV1, and the first open reading frame found in an HERV gag leader, pre-gagV1, which encodes a novel protein. This HERV is linked to a related gag gene, gagV3, and these three genes all show patterns of evolutionary conservation in primates. gagV1 and pre-gagV1 orthologs are present in all simian primate lineages indicating that this HERV entered the germline of the common simian primate ancestor at least 43 Ma, whereas gagV3 is found in Old and New World monkeys. gagV1 and gagV3 have undergone recurrent gene conversion events and positive selection. Expression of gagV1, gagV3, and pre-gagV1 is restricted to the placenta in humans and macaques suggesting co-option for placenta-specific host functions. Transcriptomic analysis of human tumors also found upregulated levels of gagV1 transcripts in diffuse large B-cell lymphomas. These findings suggest that these HERV-V genes may be useful markers for the most common type of non-Hodgkin’s lymphoma and that they may have contributed to the successive domestications of env and gag genes in eutherians involved in the ongoing ERV-driven evolution of the placenta.
Keywords: endogenous retroviruses, HERV-V group of human retroviruses, endogenous retroviruses with placenta-specific expression, co-opted human/primate retroviral gag gene, B‐cell lymphoma marke
Introduction
As part of their replication cycle, retroviruses use a virally encoded reverse transcriptase to convert their RNA genome into double-stranded DNA which is subsequently integrated into and becomes a permanent part of the host cell genome (Coffin et al. 1997). On rare occasions, when retroviruses infect germline cells or their precursors, an integrated copy of the retroviral genome can be passed on to the next generation in a Mendelian manner (Dewannieux and Heidmann 2013; Johnson 2019). These integrated viral copies, termed endogenous retroviruses (ERVs), make up approximately 8% of the human genome (Lander et al. 2001). Throughout millions of years of primate evolution, in the absence of selection pressures to keep them intact or the presence of selective forces to neutralize them, the vast majority of the human endogenous retroviruses (HERVs) have been inactivated by mutations, insertions, and deletions, leaving only remnants of past infections behind.
Although no HERV is known to encode a fully functional retrovirus, HERVs with individually intact viral genes, gag, pol, and env, have been identified (Johnson 2019). Most of the HERV genes with an intact open reading frame (ORF) represent recent integrations and have no known function attributed to them (Villesen et al. 2004; Vargiu et al. 2016). However, studies in the past two decades identified some HERV genes that have been co-opted by their human hosts for physiological functions (Mi et al. 2000; Blaise et al. 2003; Heidmann et al. 2017). The most prominent examples of these are two different HERV envelope (env) genes (syncytin-1 and syncytin-2) that have retained an intact ORF in hominoids and simian primates, respectively (Mi et al. 2000; Blaise et al. 2003; Cheynet et al. 2005). These Env proteins are highly expressed in placental syncytiotrophoblasts, are fusogenic, and serve an important role in placenta formation (Mi et al. 2000; Blaise et al. 2003; Cheynet et al. 2005). Notably, syncytin-like genes derived from other, often unrelated ERVs have also been co-opted for placentation in other mammalian lineages in a remarkable example of convergent evolution (Dupressoir et al. 2005; Heidmann et al. 2009; Dupressoir et al. 2011; Cornelis et al. 2012, 2013, 2014, 2015, 2017; Redelsperger et al. 2014). Since the discovery of syncytins, additional intact, expressed HERV env genes have been identified, but much less is known about the function of these Env proteins (Herve et al. 2004; Villesen et al. 2004; Aagaard et al. 2005; Blaise et al. 2005; Kjeldbjerg et al. 2008; Esnault et al. 2013; Blanco-Melo et al. 2017; Heidmann et al. 2017).
The retroviral gag gene encodes a polyprotein which is cleaved during maturation by the viral protease to release the major viral structural proteins: matrix (MA), capsid (CA), and nucleocapsid (NC). Gag proteins function in viral assembly, RNA packaging, and particle formation (Coffin et al. 1997; Bell and Lever 2013). Despite its multiple functional motifs required for viral replication and its multidomain organization, there have only been a few reported examples of the co-option of ERV gag genes in mammals. The best-known example of a co-opted gag gene from an ERV is the rodent restriction factor Fv1 (Best et al. 1996). Originally discovered in laboratory mice as a postentry inhibitor of murine leukemia virus (MLV) infection (Lilly 1967), Fv1 is a remnant of an ancient retroviral insertion related to the ERV-L group that likely entered the rodent genome 45–50 Ma (Best et al. 1996; Benit et al. 1997; Boso et al. 2018). A much more recently endogenized ERV with an intact gag ORF acts as a transdominant inhibitor to block the release of the betaretrovirus JSRV (Jaagsiekte sheep retrovirus) and is found in the genus Ovis, which includes domestic sheep (Mura et al. 2004; Arnaud et al. 2007; Sistiaga-Poveda and Jugo 2014; Cumer et al. 2019).
Previous systematic searches for HERV genes with intact ORFs were either aimed at generating a large-scale database of HERVs or concentrated on the identification of env ORFs (Villesen et al. 2004; Nakagawa and Takahashi 2016; Ueda et al. 2020). In this study, we combined computational similarity searches with phylogenomic analyses to probe the human genome for the presence of intact HERV gag ORFs. We extracted 41 HERV gag genes of which the oldest with an intact, full-length gag ORF entered the genome of simian primates at least 43 Ma (Steiper and Young 2006; Perelman et al. 2011). This ancient gag gene is conserved in all simian lineages, and remarkably, the large leader sequence of this ancient HERV contains another ORF, here termed pre-gagV1, that encodes a novel protein that is also highly conserved in all simian primate lineages. A second provirus with a related intact gag gene is found downstream of this ancient ERV in Old World and New World monkeys. Phylogenetic analysis of these gag genes revealed evidence of recurrent gene conversion and positive selection. Expression of both pre-gag and gag transcripts is highly restricted to the placenta in both humans and rhesus macaques as is also the case for their associated env genes (Blaise et al. 2005; Esnault et al. 2013). This suggests that all of these genes may have been co-opted for placenta-specific functions. Moreover, transcriptomic analyses of human tumors revealed that this co-opted gag gene is also significantly upregulated in diffuse large B-cell lymphomas (DLBCLs), providing a possible marker for this type of cancer.
Results
Screening for gag ORFs in the Human Genome
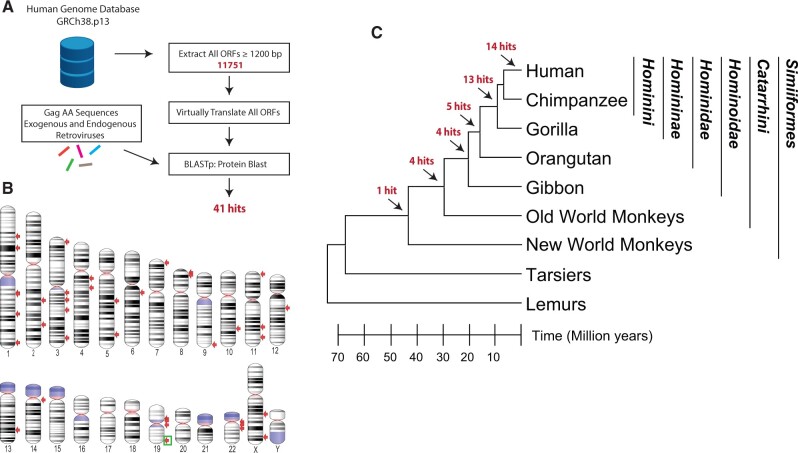
To identify the intact gag genes of HERVs, we screened the latest human genome assembly (GRCh38.p13) for ORF sequences longer than 1,200 bp flanked by start and stop codons (fig. 1A). We virtually translated these ORFs and queried the resulting amino acid sequences using BlastP to identify matches to Gag sequences from various exogenous and endogenous retroviruses (supplementary table S1, Supplementary Material online). We identified 41 hits on 17 chromosomes, named with the chromosome and sequential numbers (fig. 1B and table 1). Similarity searches of protein domain databases via InterProScan (Jones et al. 2014) for these hits showed that 32 of them had at least two of the three Gag domains: matrix (MA), capsid (CA), and nucleocapsid (NC), whereas nine hits represent partial Gags with only one of these domains. Six hits also included a protease and/or a reverse transcriptase domain (table 1). Two hits with partial Gag sequences (HSA14_gag1 and HSA22_gag1) were almost identical and represent the fusion of a partial retroviral Gag to the ORF2 of a LINE1 element (table 1). These results indicate that our screening methodology was sensitive enough to capture HERV-derived ORFs with only short gag-like sequences.
Fig. 1.
Computational similarity screen for HERV Gag ORFs. (A) Workflow of the in silico screen for identification of HERV Gag ORFs. (B) Human chromosome ideogram is shown with the location of the screen hits pointed out with red arrows. Light blue indicates variable regions and pink indicates centromeres. Location of the oldest identified Gag ORF is indicated with a green box. (C) A cladogram indicating the evolutionary relationship between the major primate lineages is shown. Branch lengths are proportional to time as indicated at the bottom. The predicted position of the insertions of the ERVs that represent the origin of each Gag ORF hit is shown with arrows. Family, subfamily, and tribe classifications under apes are shown on the right. The species tree was generated with TimeTree (Hedges et al. 2015).
Table 1.
Properties of Gag ORFs Identified in a Screen of Human Genome Assembly.
| Hit ID | Dfam Designation of Gag ORF HERV Groupa | Provirusb | Location of Gag ORFc | Strand | Orthology | ORF Conservation |
|---|---|---|---|---|---|---|
| HSA1_gag1 | HERV-H (partial gag) | 55023242–55024501 | + | Homininae | Human Specific | |
| HSA1_gag2 | HERV-K (HML-2) | 1p31.1 | 75378614–75380197 | + | Human Specific | Human Specific |
| HSA1_gag3 | HERV-9NC | 154673229–154674548 | + | Human Specific | Human Specific | |
| HSA1_gag4 | HERV-K (HML-2) | 1q22 | 155632734–155634038 | − | Human Specific | Human Specific |
| HSA1_gag5 | HERV-H (partial gag) | 183613715–183614944 | + | Homininae | Human Specific | |
| HSA1_gag6 | HERV-K13 | 237917591–237919054 | + | Homininae | Hominini | |
| HSA2_gag1 | HERV-E (pol, | 158882965–158884236 | + | Hominidae | Homininae | |
| partial gag) | ||||||
| HSA2_gag2 | HERV-9 | 202498491–202499780 | + | Hominidae | Human Specific | |
| HSA3_gag1 | HERV-K (HML-2) (gag, dUTPase) | 3p25.3 | 9851020–9853113 | − | Hominidae | Human Specific |
| HSA3_gag2 | HERV-K (HML-2) | 3q12.3 | 101693005–101695008 | + | Homininae | Homininae |
| HSA3_gag3 | HERV-K (HML-2) | 3q13.2 | 113030248–113032332 | − | Human Specific | Human Specific |
| HSA3_gag4 | HERV-K11 (gag, dUTPase, pro) | 130449221–130451068 | − | Hominidae | Human Specific | |
| HSA3_gag5 | HERV-K (HML-2) (gag, dUTPase, pro) | 3q27.2 | 185567810–185570893 | − | Human Specific | Human Specific |
| HSA4_gag1 | HERV-K11 (pol, partial gag) | 64145040–64146809 | − | Hominoidae | Human Specific | |
| HSA4_gag2 | HERV-17 (pol, partial gag) | 85353157–85354584 | − | Catarrhini | Human Specific | |
| HSA5_gag1 | HERV-S71 | 95710726–95712081 | − | Hominoidea | Hominini | |
| HSA5_gag2 | HERV-K (HML-2) | 5q33.3 | 156663774–156665774 | − | Human Specific | Human Specific |
| HSA6_gag1 | HERV-K | 77723263–77725263 | − | Human Specific | Human Specific | |
| HSA7_gag1 | HERV-K(HML-2) | 7p22.1a | 4588786–4590090 | − | Human Specific | Human Specific |
| HSA8_gag1 | HERV-K(HML-2) | 8p23.1a | 7504291–7506234 | − | Human Specific | Human Specific |
| HSA8_gag2 | HERV-K(HML-2) | 8p23.1b | 8198347–8199615 | + | Homininae | Human Specific |
| HSA8_gag3 | HERV-K(HML-2) | 8p23.1c | 12223547–12224818 | − | Homininae | Human Specific |
| HSA8_gag4 | HERV-K(HML-2) | 8p23.1d | 12466057–12467328 | − | Homininae | Human Specific |
| HSA9_gag1 | HERV-E | 135442339–135443550 | − | Hominoidae | Human Specific | |
| HSA10_gag1 | HERV-K(HML-2) | 10q24.2 | 99825872–99827437 | − | Human Specific | Human Specific |
| HSA11_gag1 | HERV-Fc2 | 5929216–5930448 | + | Homininae | Human Specific | |
| HSA11_gag2 | HERV-K(HML-2) | 11q22.1 | 101696870–101698174 | + | Human Specific | Human Specific |
| HSA11_gag3 | HERV-K(HML-2) | 11q23.3 | 118727068–118728381 | − | Homininae | Homininae |
| HSA12_gag1 | HERV-K(HML-2) | 12q14.1 | 58333804–58336072 | − | Human Specific | Human Specific |
| HSA13_gag1 | HERV-H (partial gag) | 86358566–86359789 | + | Homininae | Human Specific | |
| HSA14_gag1 | HERV-I (partial gag) | 18349411–18350967 | + | Gag: Catarrhini | Human Specific | |
| LINE1 ORF2 | LINE1: human | |||||
| HSA19_gag1 | HERV-9 (pol, partial gag) | 18580605–18581828 | + | Hominoidea | Homininae | |
| HSA19_gag2 | HERV-FH21 | 20094153–20095379 | − | Homininae | Human Specific | |
| HSA19_gag3 | HERV-K13(HML-2) | 19p12.c | 21886667–21889372 | − | Hominidae | Human Specific |
| HSA19_gag4 | HERV-K(HML-2) | 19q11 | 27644310–27646310 | − | Human Specific | Human Specific |
| HSA19_gag5 | MER50 | 53009305–53010798 | + | Simiiformes | Simiiformes | |
| HSA22_gag1 | HERV-I (partial gag) | 15280221–15281648 | + | Gag: Catarrhini | Human Specific | |
| LINE1 ORF2 | LINE1: human | |||||
| HSA22_gag2 | HERV-K(HML-2) | 22q11.21 | 18939785–18941785 | + | Human Specific | Human Specific |
| HSA22_gag3 | HERV-K(HML-2) | 22q11.23 | 23538874–23540745 | + | Homininae | Homininae |
| HSAX_gag1 | HERV-Fc2 | 97842167–97843579 | + | Homininae | Hominini | |
| HSAX_gag2 | HERV-K3 | 141197579–141198778 | − | Catarrhini | Hominini |
Hits that contain only partial gag sequences and those with more than just gag sequences are indicated.
Nineteen gag genes were identifiable by map location and sequence as specific HML-2 proviruses (Subramanian et al. 2011).
Genomic location coordinates are for GRCh38.p13.
To characterize these hits, we searched the Dfam repeat database (Storer et al. 2021) for each ORF, which placed more than half of them (25/41) in the HML (human MMTV-like) superfamily of class II (betaretrovirus) HERVs (HERV-K) (table 1). Moreover, at least three of these HML ORFs (HSA3_gag1, HSA3_gag4, and HSA3_gag5) included a dUTPase-like domain, a common feature of betaretroviruses with a similar genomic position in the 3′-end of gag (table 1) (Hizi and Herzig 2015). Nineteen of the 25 HERV-K gag genes represent segments of previously identified HML-2 proviruses that have full-length gag genes (table 1) (Subramanian et al. 2011). The rest of the hits are members of the various families of the class I (gammaretrovirus) HERVs, including three hits with a partial gag sequence that belong to HERV-H (HSA1_gag1, HSA1_gag5 and HSA13_gag1), two hits that belong to HERV-Fc2 (HSA11_gag1 and HSAX_gag1), and two hits that belong to HERV-E (HSA2_gag1 and HSA9_gag1) (table 1). Next, we determined the phylogenetic relationships of these HERV gag ORFs and known HERV families. A phylogenetic tree including only the hits with full-length Gag along with known exogenous and endogenous retroviruses (supplementary data set 1 and supplementary fig. S1, Supplementary Material online) is consistent with the Dfam search results (table 1) (Storer et al. 2021). Hits identified as HML clustered with betaretroviruses and class II HERVs, whereas other hits clustered with gammaretroviruses and class I HERVs (supplementary fig. S1, Supplementary Material online).
To determine the evolutionary history of these gag containing HERVs, we searched for orthologs in nonhuman primates. The majority (27/41) of these HERVs entered the primate lineage after the branch leading to modern orangutans split from the family Hominidae, approximately 15 Ma (table 1, fig. 1C and supplementary methods, Supplementary Material online) (Steiper and Young 2006; Perelman et al. 2011). Among these, 14 HERVs are human specific, whereas 13 are found in orthologous locations in the chimpanzee and gorilla genomes (fig. 1C). Notably 13 of 14 human-specific HERVs as well as 7 of 13 HERVs found in the subfamily Homininae belong to the HERV-K group (table 1). Among the rest of the HERVs (27 of 41), five are found in the orangutan genome, including three that belong to the HERV-K group, one member of the HERV-E group, and another belonging to HERV-9. Another four HERVs have gibbon orthologs including two with partial gag sequences that belong to HERV-K11 and HERV-9, suggesting that they were endogenized at least 15 and 20 Ma, respectively (Steiper and Young 2006; Perelman et al. 2011). Only five HERVs had orthologs in Old World monkeys of which four were likely acquired by the common ancestor of Old World monkeys and apes at least 30 Ma, and a single HERV (HSA19_gag5) has a New World monkey ortholog (fig. 1C). Notably, 10/41 HERVs contained a full-length gag ORF in nonhuman primates, but only a single gag ortholog with an ORF (HSA19_gag5) was found outside the subfamily Homininae (table 1, fig. 1C). Thus, the oldest HERV with an intact gag ORF in the human genome (HSA19_gag5) is also found in the genomes of apes and Old and New World monkeys, suggesting that it was endogenized at least 43 Ma (table 1) (Steiper and Young 2006; Perelman et al. 2011). Hence, we decided to further investigate this remarkably conserved gag gene (HSA19_gag5).
An Additional ORF in the Unusually Long Leader of the Oldest HERV gag ORF
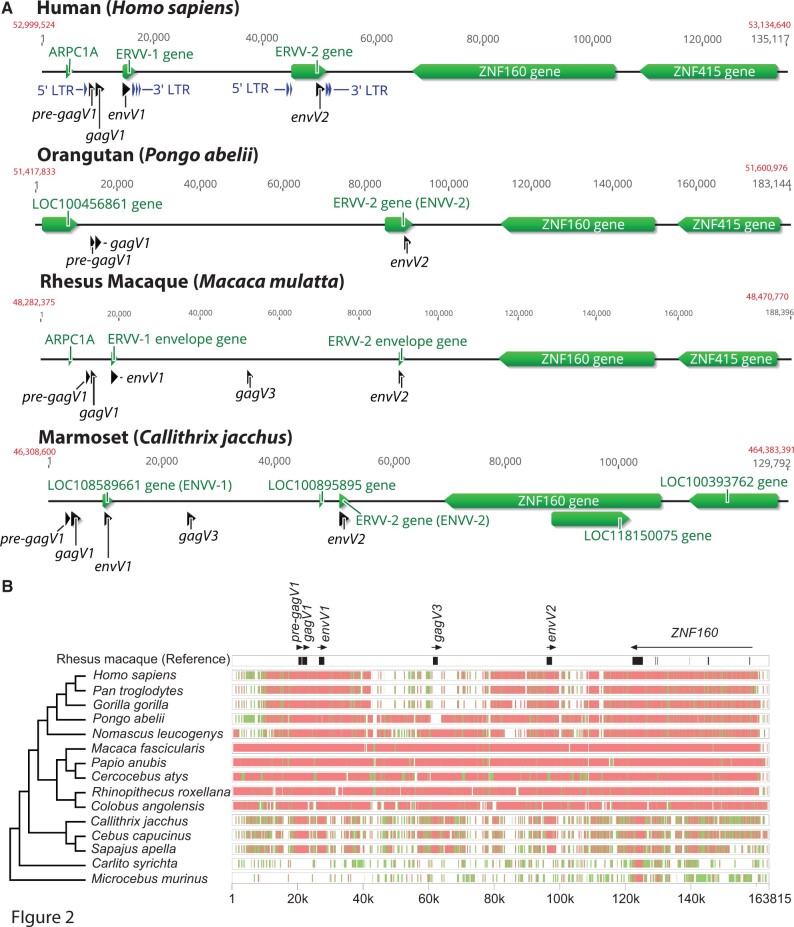
The chromosomal region around the HSA19_gag5 gag ORF contains two previously identified HERV proviruses separated by approximately 40 kb termed HERV-V1 and HERV-V2 with associated env genes envV1 and envV2 (fig. 2A) (Blaise et al. 2005; Kjeldbjerg et al. 2008). Accordingly, the HSA19_gag5 gag ORF just upstream of the envV1 gene was named gagV1. We did not find an intact gag ORF as a part of HERV-V2, and only the remnants of pol and 5′ and 3′ long terminal repeats (LTRs) can be detected in both HERV-V1 and HERV-V2 (supplementary fig. S2, Supplementary Material online). Although LTR sequence divergence can be used to estimate the age of an ERV, a comparison of 5′- and 3′-LTRs of human and macaque ERV-V1 revealed ancient insertion dates (88–71 Ma for human, 95–76 Ma for macaque), which would, inaccurately, put endogenization of this ERV before the split from the common ancestor of primates (Steiper and Young 2006; Perelman et al. 2011). It is possible that the conserved nature of this ERV ORF disconnects it from the evolution of the two LTRs or that the LTRs themselves may have undergone recombination with similar elements leading to an increase in divergence between them. Moreover, the 3′-LTR of HERV-V1 and both HERV-V2 LTRs are interrupted by several Alu elements which may prevent the accurate determination of the age of this HERV by LTR divergence (supplementary fig. S2, Supplementary Material online).
Fig. 2.
Conserved synteny of the HERV-V locus in primates. (A) Chromosomal context of the HERV-V locus is shown for human (GRCh38.p13, chromosome 19), orangutan (Susie_PABv2, chromosome 19), rhesus macaque (Mmul_10, chromosome 19), and marmoset (Callithrix_jacchus_cj1700_1.1, chromosome 22) genomes. Refseq annotated genes are shown in green. Location of the pre-gagV1, gagV1, gagV3, envV1, and envV2 ORFs are indicated in black. Starting and ending coordinates of the depicted region for each species are indicated in red. The figure was created using Geneious (Kearse et al. 2012). (B) Genomic segments containing the ERV-V locus and ZNF160 starting with 20k bases upstream of the pre-gagV1 sequence were extracted from the NCBI genome database for each of the indicated species. Cross-species homologies were analyzed using the MultiPipMaker alignment tool (Elnitski et al. 2010). ORFs of pre-gagV1, gagV1, gagV3, envV1, and envV2 as well as exons of ZNF160 are shown in black boxes in the rhesus macaque reference assembly. Regions with less than 75% and more than 50% identity are shown as green boxes. Regions with more than 75% identity are shown as red boxes. A cladogram representing the phylogenetic relationships between the primate species is shown on the left. The species tree was generated with TimeTree (Hedges et al. 2015).
A search of the Pfam (El-Gebali et al. 2019) and Supfam (Pandurangan et al. 2019) protein domain databases using the predicted amino acid sequence of gagV1 revealed an MA-like domain, a CA-like domain containing a major homology region, and a C-terminal zinc finger-like domain common to NC proteins (supplementary fig. S3A, Supplementary Material online) (Coffin et al. 1997). GagV1 also contains a consensus myristoylation signal (MGxxxS) at the second methionine codon (supplementary fig. S3A, Supplementary Material online), a common feature of retroviral Gag proteins that is important for Gag–membrane interactions (Resh 2013; Dick and Vogt 2014) suggesting that this methionine is likely the translation initiation site for GagV1. This HERV has a long leader sequence upstream of gag (∼1,600 bp) that contains an additional ORF of 981 bp, here termed pre-gagV1, that is not in frame with the gag ORF (fig. 2A). Analysis of the putative amino acid sequence of this ORF using Phobius and TMHMM membrane topology prediction software suggests the presence of a transmembrane domain close to the N-terminal end of the protein (supplementary fig. S3B, Supplementary Material online), but pre-gagV1 shows no sequence homology to any known protein. Comparably long leader sequences are not found in exogenous viruses, and although a few other HERVs have long leaders, only a subset of class I HERVs, including HERV-V, contain very long leaders (>1,500 bp) (Jern et al. 2005; Blanco-Melo et al. 2017; Grandi et al. 2020) (supplementary fig. S4, Supplementary Material online). There are a few examples of exogenous and endogenous retroviruses with ORFs in their leader sequences, but none is related to pre-gagV1 (Prats et al. 1989; Holzschu et al. 1995; Kambol et al. 2003; Jern et al. 2005; Blanco-Melo et al. 2017; Grandi et al. 2020).
Conservation of pre-gagV1 and gagV1 ORFs in Simiiformes
The region of human chromosome 19 that contains the pre-gagV1, gagV1, and envV1 ORFs is located downstream of the ZNF160 and ZNF415 genes (fig. 2A). Three divergent primate species belonging to hominoids (orangutan), Old World monkeys (rhesus macaque), and New World monkeys (marmoset) were selected for genomic comparisons. The orthologous regions in the orangutan, rhesus macaque, and marmoset genomes contain the gagV1 and pre-gagV1 ORFs (fig. 2A). Notably, both rhesus macaque and marmoset, but not orangutan, have an additional ORF approximately halfway between the envV1 and envV2 genes. A BLAST search of this ORF revealed more than 95% identity to the gagV1 ORFs in the respective species, and therefore, we labeled this ORF gagV3 (fig. 2A). The presence of a third HERV-V element in this region was previously reported without identification of an intact gag gene as part of this HERV (Kjeldbjerg et al. 2008).
To examine the sequence conservation of this region in primates, we extracted and aligned the orthologous genomic segments from apes, Old World and New World monkeys, and prosimians (fig. 2B). This genomic region is strongly conserved among Old World monkeys and apes although the high similarity between the members of the parvorders Catarrhini (Old World monkeys and apes) and Platyrrhini (New World monkeys) is clustered around the ORFs of gagV1, pre-gagV1, envV1, and envV2. The gagV3 sequence is missing in great apes (family Hominidae); however, the sequence immediately surrounding gagV3 shows relatively high similarity in Pongo abelii (Sumatran orangutan) and Old World monkeys (fig. 2B). This suggests that gagV3 was likely lost after the Hominidae and Hylobatidae families diverged, and the rest of the provirus was deleted in the subfamily Homininae. However, the gagV3 in Nomascus leucogenys (northern white-cheeked gibbon) lacks an intact ORF suggesting that more recent selection pressures may have eliminated this ERV in some lineages.
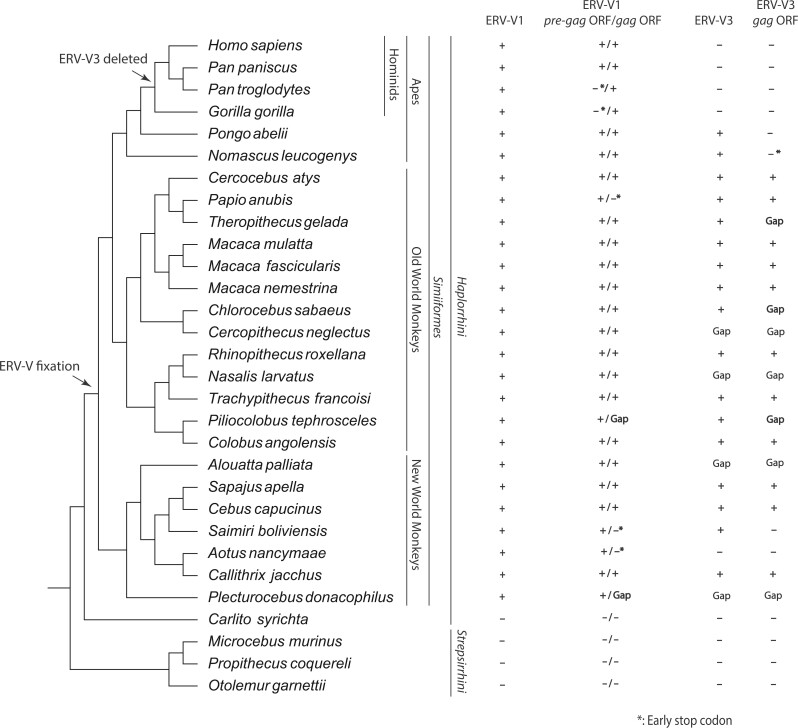
To describe the evolutionary history of the ERV-V gag and pre-gag ORFs, we searched the primate genomes for human gagV1 and pre-gagV1 sequences. These ORFs were found in all lineages basal to the infraorder Simiiformes (fig. 3). Although some species contain HERV-V1 and HERV-V2 orthologs in a single scaffold/chromosome, other species have a gap in the predicted location of gagV1 and gagV3 (fig. 3). Although some species have early stop codons in gagV1 or pre-gagV1, there is remarkable conservation of the gag and pre-gag ORFs within the remnants of an ancient retrovirus (fig. 3). BLAST searches failed to find gagV1 and pre-gagV1 in Tarsiiformes (tarsiers) or Strepsirrhini (prosimians) (fig. 3), and we did not find ERV-V orthologs in Carlito syrichta (Philippine tarsier) or Microcebus murinus (grey mouse lemur) (fig. 2B). Thus, the ancient retroviral progenitor of ERV-V1, ERV-V2, and ERV-V3 invaded the genome of a common ancestor of Simiiformes at least 43 Ma (Steiper and Young 2006; Perelman et al. 2011).
Fig. 3.
pre-gagV1 and gagV1 orthologs with an intact ORF are present in all simian primate lineages. A cladogram illustrating the evolutionary relationship between the primate species with a genome assembly in the NCBI database is shown. The species tree was generated with TimeTree (Hedges et al. 2015). Phylogenetic classifications are shown on the right. + indicates the presence of ERV-V1, ERV-V3 orthologs or gagV1, gagV3, or pre-gagV1 orthologs with ORFs as identified via BLAST searches of each genome assembly. * indicates the presence of an early stop codon. “Gap” indicates the presence of a sequencing gap in the relevant region of the indicated genome. Arrows suggest the location of the ERV-V insertion and the ERV-V3 deletion according to the orthology analysis.
These findings prompted us to search for additional HERV-V gag and pre-gag sequences in the human genome. BLAST searches identified five other gag-like HERV sequences that had the same gagV1 Dfam repeat designation (table 2). We also found remnants of a pre-gag sequence upstream of the gag-like sequence in at least two of these HERVs (table 2). Four of these HERVs have orthologs in both New and Old World monkeys, and one has orthologs in Old World monkeys suggesting that ERV-V-like viruses invaded the genome of a common ancestor of simian primates. These findings also indicate that the long leader sequence and the pre-gag ORF were likely present in the ancient virus that generated the pre-gagV1 provirus.
Table 2.
Locations of Other HERV-V Elements Found in the Human Genome.
| Gene | Locationa | Orientation | Orthology | Dfam designation |
|---|---|---|---|---|
| gag | ChrX: 83161089–83161913 | + | Simiiformes | MER50 |
| pre-gag | ChrX: 84708729–84709383 | − | Catarrhini | MER50 |
| gag | ChrX: 84707334–84708555 | − | Catarrhini | MER50 |
| pre-gag | ChrX: 98578387–98578970 | − | Simiiformes | MER50 |
| gag | ChrX: 98577295–98577791 | − | Simiiformes | MER50 |
| gag | Chr5: 59311710–59313003 | + | Simiiformes | MER50 |
| gag | Chr20: 36984411–36985174 | + | Simiiformes | MER50 |
Genomic location coordinates are for GRCh38.p13 (hg38).
pre-gagV1 and gagV1 Show Placenta Specific Expression
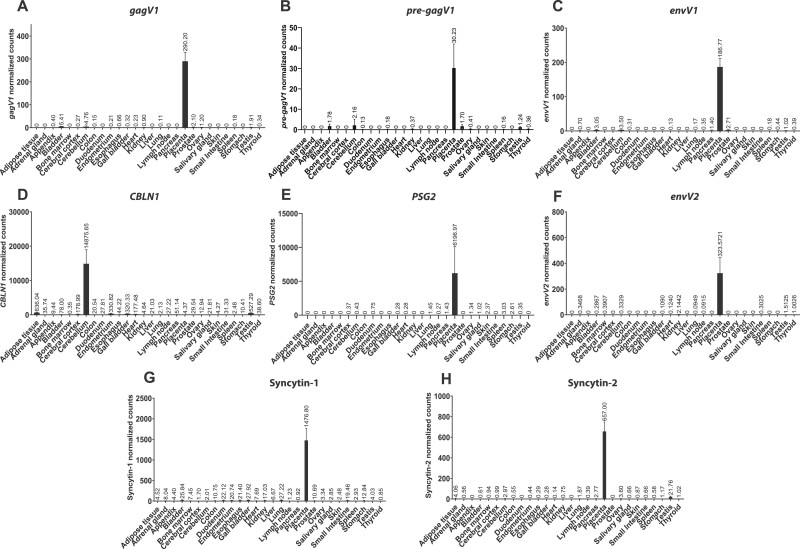
A BLAST search of the NCBI nucleotide database using the gagV1 ORF as a probe revealed the presence of a previously reported RNA (GenBank accession number: AK127846.1) that encompasses this region of the human genome. Data for this RNA from the Expression Atlas show that it is expressed highly in the placenta with very low levels of expression in other tissues such as cerebellum and prostate (Papatheodorou et al. 2020). To provide a more complete description of the tissue-specific expression of gagV1 and pre-gagV1, we performed an in silico analysis of RNA sequencing data from a large-scale study that includes placenta and 26 other tissues from 95 individuals (Fagerberg et al. 2014). Since this study does not include cerebellum, we also examined RNAseq data from two other studies that include cerebellum (Prudencio et al. 2017; Tan et al. 2017) (fig. 4 and supplementary methods, Supplementary Material online). Because neither of these genes is annotated in the human genome, we remapped the extracted raw RNA sequencing reads to the latest human genome assembly (GRCh38.p13) and used a custom annotation that included gagV1 and pre-gagV1 ORFs. This analysis showed that the expression of both gagV1 and pre-gagV1 is highly restricted to the placenta with very low-level expression of both transcripts in kidney, bladder, prostate, and cerebellum (fig. 4A and B). We also confirmed the previously observed placenta-specific expression of the envV1 and envV2 transcripts (Blaise et al. 2005; Esnault et al. 2013) (fig. 4C and F). Notably, we also found low levels of expression of the co-opted placenta-specific HERV envelope genes, syncytin-1 and syncytin-2, in various other tissues (fig. 4G and H) which match the expression profiles of these genes in the Expression Atlas (Papatheodorou et al. 2020).
Fig. 4.
Human pre-gagV1 and gagV1 expression is restricted to the placenta. In silico analysis of RNA sequencing data from 27 human tissues is shown for (A) gagV1, (B) pre-gagV1, (C) envV1, (D) CBLN1, (E) PSG2, (F) envV2, (G) Syncytin-1, and (H) Syncytin-2. The mapping accuracy of the RNA sequencing reads, and the normalization of the read counts was confirmed using PSG2 (placenta specific) and CBLN1 (cerebellum specific) as tissue-specific controls. Raw RNA sequencing data extracted from SRA projects PRJEB4337, PRJNA279249, and PRJNA237340 were mapped to the human genome assembly and the counts that map to the coding region of each gene were extracted and normalized using DEseq2.
Next, to describe the expression profile of gagV1 and pre-gagV1 in another primate, we analyzed RNA sequencing data from 13 macaque tissues. Like their human orthologs, macaque gagV1, pre-gagV1, and envV1 show a placenta-specific expression profile (supplementary fig. S5, Supplementary Material online). Also, the expression of gagV3, which is not found in the human genome, is highly restricted to the macaque placenta (supplementary fig. S5, Supplementary Material online). Collectively, these findings indicate that in both humans and macaques, gagV1 and pre-gagV1 transcripts are placenta specific.
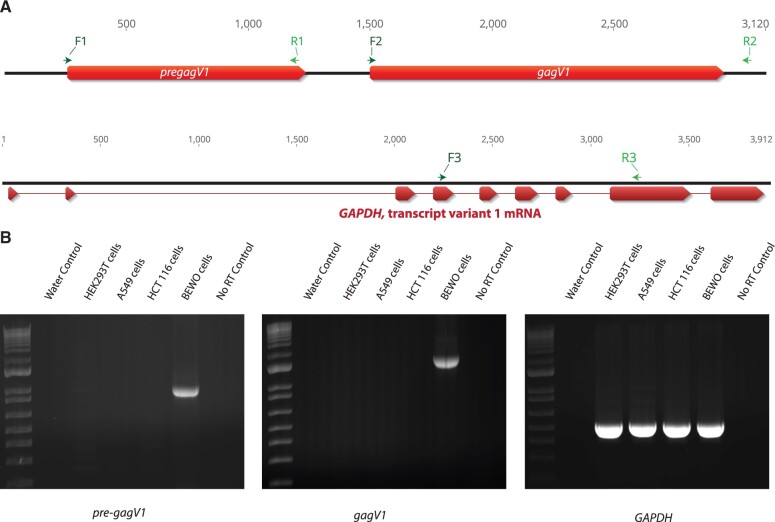
We used 5′ rapid amplification of cDNA ends (RACE)-PCR and reverse transcription (RT)-PCR to determine the structure of the transcripts that encode GagV1 and Pre-GagV1. First, we probed for pre-gagV1 and gagV1 ORFs in total RNA extracted from cell lines developed from different human tissues via RT-PCR using the primers shown in figure 5A. Consistent with our RNA sequencing data, transcripts were only detected in BEWO cells, derived from a placenta-derived choriocarcinoma (fig. 5B) (Pattillo et al. 1968). Next, RT-PCR identified multiply spliced products in BEWO cell RNA for gagV1 as well as an unspliced product that contains both pre-gagV1 and gagV1 ORFs (supplementary fig. S6, Supplementary Material online). Sequencing of these products showed them to be identical to the corresponding sequence at the human reference genome assembly (GRCh38.p13) (supplementary data set 4, Supplementary Material online). Notably, alignment of the orthologous region showed that the splice acceptor and donor sites as well as the surrounding sequence is highly conserved in various simian primates apart from a splice donor site in the alternative splice product in the New World monkeys (supplementary fig. S7, Supplementary Material online), indicating that this RNA splicing scheme for gagV1 may be conserved throughout simian primate evolution. Sequencing of the splice products also revealed that the splice acceptor sequence for gagV1 is immediately upstream of the start codon with the myristoylation signal (supplementary fig. S3A, Supplementary Material online) establishing that translation initiates at this codon and not at the ATG that is 42 bases upstream. In addition, 5′ RACE-PCR analysis showed that the spliced gagV1 and envV1 transcripts and the unspliced transcript for pre-gagV1 all share a start site that is 180 bases upstream of the predicted primer binding site (PBS) and is thus within the 5′ LTR of HERV-V1 (supplementary fig. S6, Supplementary Material online). The 3′ end of the transcripts containing pre-gagV1 and/or gagV1 was not immediately detectable via 3′ RACE-PCR due to the presence of several A-rich regions in the multiple Alu elements inserted 1,500–2,500 bp downstream of the gagV1 stop codon (supplementary figs. S2 and S6, Supplementary Material online). However, RT-PCR and sequencing indicate that the 3′-end extends at least 1,400 bases downstream of the gagV1 stop codon suggesting there is a very long 3′ untranslated region for transcripts that contain gagV1 (supplementary fig. S6, Supplementary Material online).
Fig. 5.
Human pre-gagV1 and gagV1 expression in human cell lines. (A) At the top is a schematic of the genomic region that contains pre-gagV1 and gagV1 ORFs. Locations of the primers used in RT-PCR are indicated above the diagram. At the bottom of the panel is a schematic of the genomic region that contains human GAPDH. Exons of GAPDH are shown in red. Locations of the primers used in RT-PCR are indicated above each diagram. Primers that were designed for different exons of GAPDH were used to confirm the absence of DNA contamination as well as uniform RNA loading. The figure was created using Geneious (Kearse et al. 2012). (B) Agarose gels showing the RT-PCR products of pre-gagV1, gagV1, and GAPDH that were amplified from the indicated human cell lines with the primers shown in the upper panel. A control done without an RT step (only Taq polymerase) was included to confirm the absence of HERV DNA contamination. Gels are representative of at least two independent experiments.
Evidence of Purifying Selection in pre-gagV1
Once fixed in the genome of the host, ERV-derived genes that serve a host function are subject to the same evolutionary pressures as any other host gene. Such genes either evolve under diversifying/positive selection which leads to adaptive changes in the amino acid sequence, or purifying/negative selection to retain their physiological function (Meyerson and Sawyer 2011; Johnson 2013). Such selection pressures are detected by the ratio of the rate of nonsynonymous (dN) and synonymous (dS) changes in related species (Yang and Bielawski 2000). Among the known co-opted ERVs, env genes with a syncytin-like fusion function, including envV2, are evolving under purifying selection (dN/dS < 1) (Kjeldbjerg et al. 2008; Esnault et al. 2013; Lavialle et al. 2013; Heidmann et al. 2017).
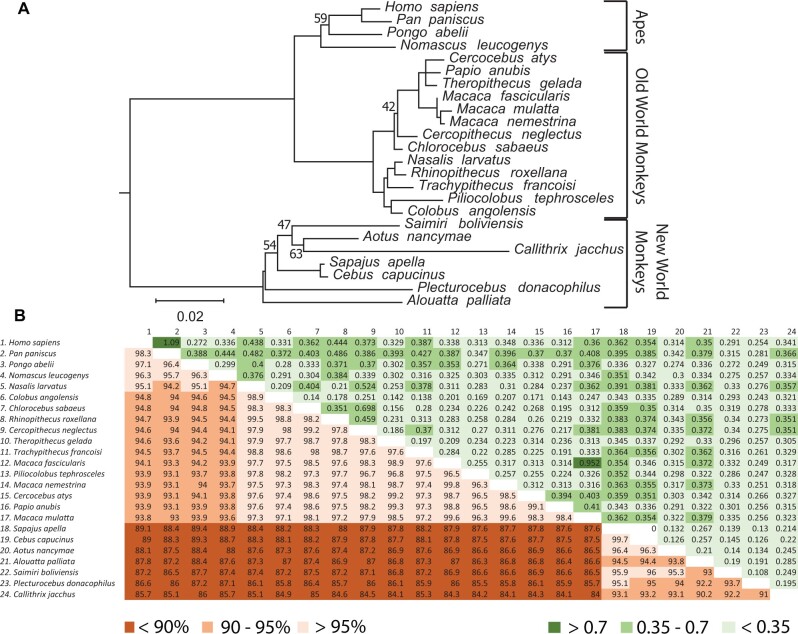
To identify evolutionary pressures that have shaped primate pre-gagV1, we extracted and aligned pre-gagV1 ORFs from simian primates and generated a maximum likelihood tree. This tree shows high bootstrap support and clear separation of lineages of Catarrhini (apes, Old World and New World monkeys) (fig. 6A). Moreover, the pre-gagV1 nucleotide sequence shows high similarity (84–99%) among simian primates (fig. 6B) despite being derived from an ancient ERV. Comparisons of all pairs of species revealed dN/dS values much lower than unity for most (<0.35) except for some very closely related species (e.g., human/bonobo and pig-tailed macaque/rhesus macaque) (fig. 6B). To determine whether any specific residues of pre-gagV1 are subject to recurrent positive selection, we used the maximum likelihood models in the codeml program of PAML4 (Yang 2007). No evidence of positive selection was found at any sites (table 3), which was confirmed using the SLAC, FEL, and REL programs from the datamonkey webserver (Weaver et al. 2018). These findings indicate that pre-gagV1 has been under strong purifying selection during simian primate evolution.
Fig. 6.
Evolution of pre-gagV1 in Simiiformes. (A) pre-gagV1 ORFs from the indicated primate species were aligned (supplementary data set 2, Supplementary Material online) and a maximum likelihood tree was generated using RaxML with 500 replicates. Bootstrap values that are below 70 are shown at the relevant nodes. The tree was midrooted. Phylogenetic classifications are shown on the right. Scale bar represents 0.02 nucleotide substitutions per site. (B) Pairwise percentage nucleotide identity (lower triangle) and the pairwise dN/dS values (upper triangle) of pre-gagV1 of the indicated simian primate species are shown. Color coding for each series is shown below the figure.
Table 3.
PAML Analysis of pre-gagV1 and gagV1 from Simian Primates.
| Gene | Codon Frequency | ω0 | M1–M2 |
M7–M8 |
Tree Lengtha | dN/dS (%) | Residuesb with dN/dS of >1 and pr of >0.95 | ||
|---|---|---|---|---|---|---|---|---|---|
| 2δ | P Value | 2δ | P Value | ||||||
| pre-gagV1 | f3 × 4 | 0.3 | 1.01 | 0.604 | 2.91 | 0.233 | 1.4524 | N.S. | N.S. |
| f3 × 4 | 1.7 | 1.01 | 0.604 | 2.91 | 0.233 | 1.4524 | N.S. | N.S. | |
|
gagV1
(Segment A) |
f3 × 4 | 0.3 | 13.1 | 0.0014 | 16.25 | 0.0003 | 1.6499 | 2.78 (11.5) | 9Q, 45K |
| f3 × 4 | 1.7 | 13.1 | 0.0014 | 16.25 | 0.0003 | 1.6499 | 2.78 (11.5) | 9Q, 45K | |
|
gagV1
(Segment B) |
f3 × 4 | 0.3 | 5.33 | 0.0694 | 8.412 | 0.0149 | 1.2764 | N.S. | N.S. |
| f3 × 4 | 1.7 | 5.25 | 0.0724 | 8.486 | 0.0143 | 1.2764 | N.S. | N.S. | |
Note.—ωo, the initial seed value of ω used; 2δ, two times the difference of the natural log values of the maximum likelihood from pairwise comparisons of the different models; pr, posterior probability.
Tree length is defined as the sum of the nucleotide substitutions per codon at each branch.
Residue numbers are based on the human pre-gagV1 and gagV1 sequences.
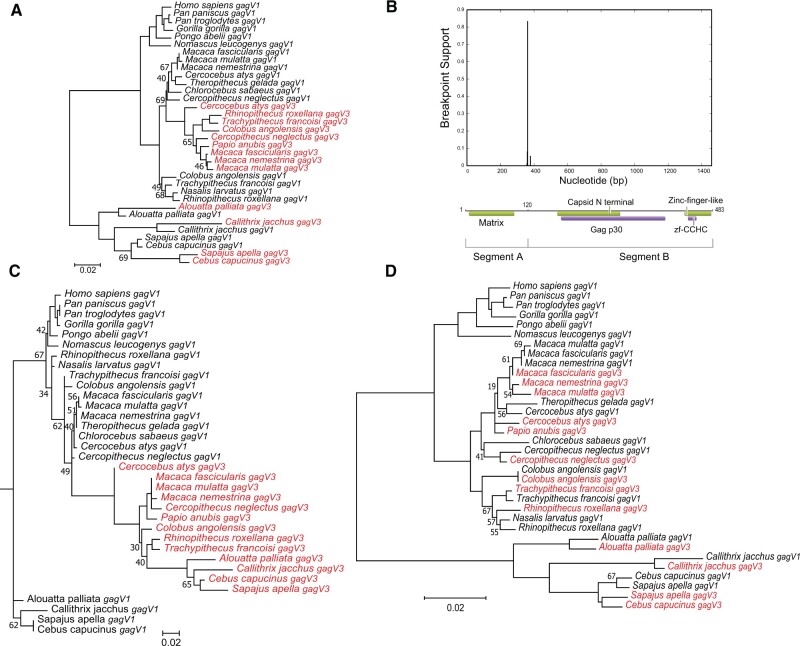
Our phylogenomic analysis showed that, unlike pre-gagV1 which is present in a single copy, some species carry another co-opted gag gene with an intact ORF and high similarity to gagV1, gagV3. We therefore generated a phylogenetic tree using the ORFs from both gagV1 and gagV3 (fig. 7A). These genes show high sequence similarity (86–99%) in simian primates (supplementary fig. S8A, Supplementary Material online). Since the orthology analysis indicates that these genes were fixed before the divergence of simians and prosimians (43 Ma) (Steiper and Young 2006; Perelman et al. 2011), we expected the gagV1 and gagV3 genes to evolve as monophyletic groups that cluster separately in a maximum likelihood tree. However, the gagV1 and gagV3 genes of different lineages did not fall into this expected grouping but instead were intermingled (fig. 7A). In Old World monkeys, the gagV3 genes clustered together but are more closely related to the gagV1 genes from the same species than to their gagV3 orthologs in New World monkeys (fig. 7A). Similarly, the gagV1 and gagV3 genes of the New World monkeys did not cluster into separate groups (fig. 7A). These results indicate that, unlike pre-gagV1, gagV1 and gagV3 have more complex evolutionary histories.
Fig. 7.
Evolution of gagV1 and gagV3 in Simiiformes. (A) gagV1 and gagV3 ORFs from the indicated primate species were aligned (supplementary data set 3, Supplementary Material online) and a maximum likelihood tree was generated using RaxML with 500 replicates. Bootstrap values that are below 70 are shown at the relevant nodes. The tree was midrooted. gagV3 genes are shown in red. (B) Plot with the model-averaged support (y-axis) for recombination breakpoints as calculated by GARD is shown above the human GagV1 protein. Nucleotide position of human gagV1 is indicated on the x-axis. Domains predicted via SupFam (green) and Pfam (purple) are indicated on GagV1. Maximum likelihood trees are generated using RaxML with aligned gagV1 and gagV3 nucleotide sequences from (C) GARD segment A (first 360 nucleotides) or (D) GARD segment B. Bootstrap values that are below 70 are shown at the relevant nodes. The tree was midrooted. gagV3 genes are shown in red.
Recurrent Gene Conversion between gagV1 and gagV3
These findings prompted us to investigate whether gene conversion events impacted the evolution of the gagV1 and gagV3 genes. Use of the genetic algorithm for recombination detection (GARD) program (Kosakovsky Pond et al. 2006; Weaver et al. 2018) revealed strong evidence of recombination between these genes with significant support (P = 0.0002) for a recombination breakpoint at the N-terminal end between the predicted MA and the CA domains creating two segments (A and B) with distinct evolutionary histories (fig. 7B). In a phylogenetic tree of segment A, the gagV3 genes from Old and New World monkeys cluster together (fig. 7C), although the gagV1 and gagV3 genes still did not separate into monophyletic clades. Although most nodes in this phylogenetic tree showed strong bootstrap support, we observed low support values in a few nodes which may be caused by high sequence similarity between gagV1 and gagV3 orthologs in segment A as well as the short length of this sequence. Because of the loss of gagV3 in the ape lineage, we produced a segment A tree from only Old and New World monkeys which showed complete segregation of gagV1 and gagV3 genes into two separate clades (supplementary fig. S8B, Supplementary Material online). This discrepancy may be explained by the similarity of gagV1 and gagV3, short length of segment A, and the small number of species outside of Old World monkeys used for this analysis. In contrast to these results, a phylogenetic tree of segment B revealed several instances of gene conversion events between gagV1 and gagV3 (fig. 7D), most notably in C. angolensis, in which segment B of gagV1 and gagV3 is identical.
Recombination events can lead to the detection of false selection signatures (Anisimova et al. 2003). Because we found significant evidence of recombination between gagV1 and gagV3, we carried out our selection analysis individually on each GARD segment. Segment A shows significant recurrent positive selection (P < 0.001) with two residues under positive selection with a posterior probability of >0.95 (table 3). One of these residues (9Q) was also identified as evolving under positive selection using FEL and REL (Weaver et al. 2018). In contrast, segment B shows no evidence of positive selection (P > 0.01) (table 3).
GagV1 or Pre-GagV1 Does Not Impact Postentry Stages of HIV-1 or MoMLV Infection
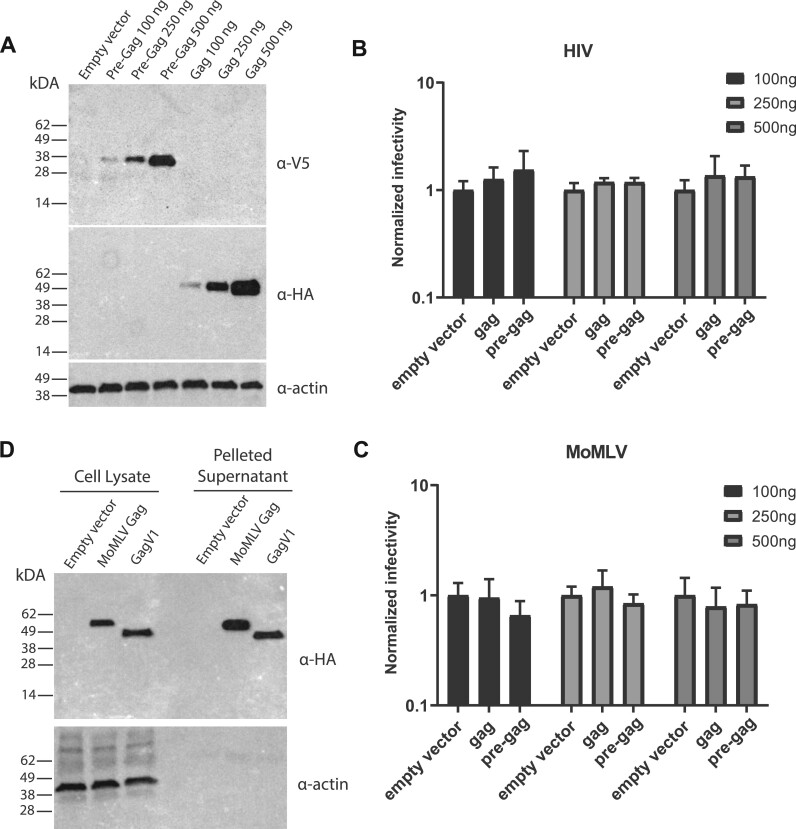
Previous studies showed that coexpression of HIV-1 with a reconstituted HERV-K gag gene in the same cells leads to coassembly of viral particles and decreased HIV-1 infectivity (Monde et al. 2012). To determine whether gagV1 can similarly restrict assembly and release of exogenous retroviruses, we expressed human gagV1 and pre-gagV1 ORFs with C-terminal HA and V5 tags, respectively, in 293T cells (fig. 8A). Co-transfections with constructs that produce Moloney MLV (MoMLV) or VSV-G-pseudotyped HIV-1 with luciferase reporters show that neither gagV1 nor pre-gagV1 alters the infectivity of either the MLV or HIV-1 particles (fig. 8B). Thus, although the HERV-derived genes gagV1 and pre-gagV1 can express stable proteins under a constitutive promoter, neither restricts HIV-1 or MLV assembly or release.
Fig. 8.
Expression and evaluation of the antiretroviral activity of GagV1 and Pre-GagV1. (A) Immunoblotting analysis of HEK293T cells transfected with increasing amounts of plasmids expressing empty vector, GagV1-HA and Pre-GagV1-V5 was done using the indicated antibodies. (B, C) HIV-RenLuc or MLV-Luc produced in HEK293T cells following transfection with the indicated amounts of plasmids expressing GagV1-HA or Pre-GagV1-V5 was used to infect HELA (HIV-RenLuc) or NIH3T3 (MLV-Luc) cells. Normalized (B) Renilla or (C) firefly luciferase reporter values are shown relative to empty vector control. Error bars indicate standard deviation. Values represent the average of three independent experiments. (D) Immunoblotting analysis of whole cell lysate and pelleted supernatant from HEK293T cells transfected with GagV1-HA, MLV Gag-HA, or empty vector. Immunoblots are representative of at least two independent experiments.
The most well-known co-opted gag gene of an ERV is the rodent restriction factor Fv1 which blocks replication of certain retroviruses postentry but before integration (Yang et al. 1980; Best et al. 1996). To test whether GagV1 can restrict the postentry stage of exogenous retroviruses in a single-cycle infection assay, we ectopically expressed different dosages of C-terminally HA-tagged human gagV1 or a C-terminally V5-tagged postentry restriction factor, the owl monkey TrimCyp (omTRIMCyp) as a positive control (supplementary fig. S9A, Supplementary Material online). As expected, omTRIMCyp expression led to a strong inhibition of VSV-G pseudotyped HIV-1 but had no impact on VSV-G pseudotyped MoMLV (supplementary fig. S9B, Supplementary Material online). In contrast, ectopic expression of GagV1 had no impact on infection by VSV-G pseudotyped HIV-1 or MoMLV. These findings duplicate the previously reported restriction profile of omTRIMCyp (Sayah et al. 2004; Boso et al. 2019) and indicate that GagV1 does not restrict the postentry stages of HIV-1 or MoMLV.
GagV1 Is Released from Human Cells
A common feature of retroviral Gag proteins is that even in the absence of a functional protease, Gag expression can lead to virion assembly at the cell membrane and the release of virus-like particles (Coffin et al. 1997; Bell and Lever 2013). To determine whether GagV1 has retained this function, we expressed human GagV1 and MLV Gag with a C-terminal HA tag in 293T cells and probed for the presence of Gag proteins in the pelleted culture supernatant. As shown in figure 8D, both MLV Gag and GagV1 were detected in the cell lysate as well as in the pelleted supernatant. These findings indicate that despite being endogenized more than 40 Ma, the oldest co-opted HERV gag gene, gagV1, can be released into the cell supernatant when ectopically expressed.
gagV1 and envV1 Expression in Human Tumors
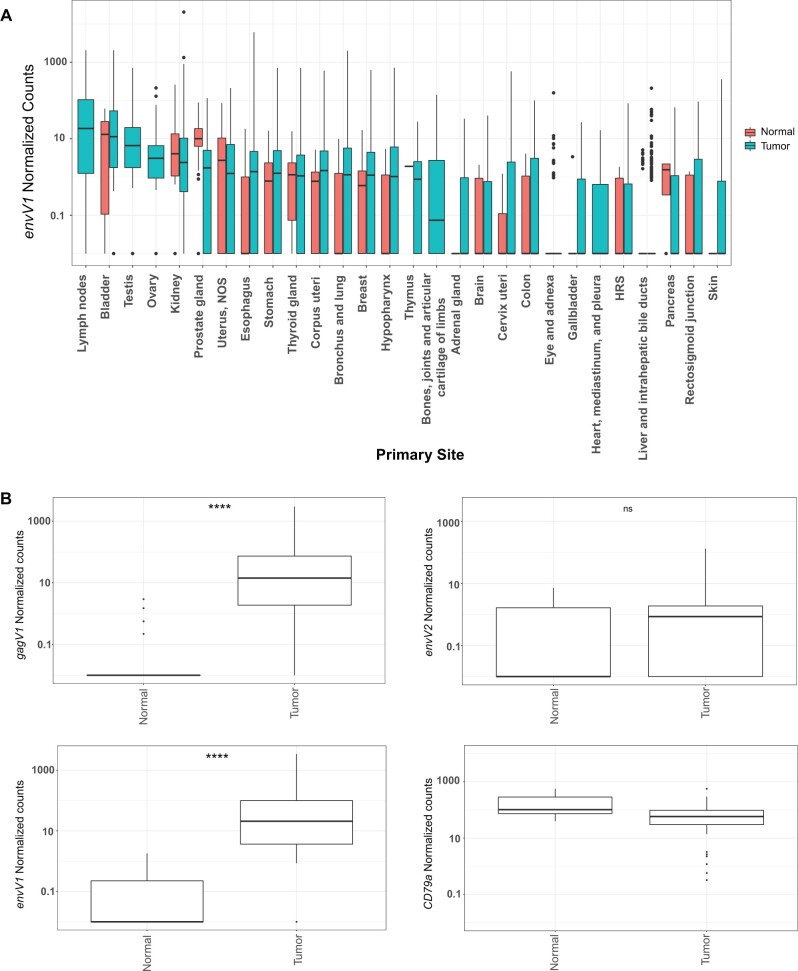
Numerous reports have described increased expression of various HERVs in human tumors and suggested that this expression may serve as diagnostic markers for specific cancers (Wang-Johanning et al. 2007, 2014; Maliniemi et al. 2013; Wallace et al. 2014; Perot et al. 2015; Ma et al. 2016; Zhou et al. 2016; Heidmann et al. 2017). To determine the expression levels of gagV1 and envV1 genes in human tumors, we extracted RNA sequencing data from 16,495 patient samples that span 56 different cancer projects from the National Cancer Institute Genomic Data Commons (NCI-GDC) data portal (Grossman et al. 2016). Since gagV1 is not annotated in the human genome database, expression data for this gene are not included in the raw count data from NCI-GDC, so instead, we analyzed envV1 expression data across various tumors, as these genes share a transcription start site (supplementary fig. S6, Supplementary Material online). As shown in figure 9A, the highest level of envV1 expression is found in lymph node tumors. High levels are also observed in bladder tumors but correspond to levels in normal bladder (fig. 9A), whereas there is no detectable expression of envV1 in normal lymph nodes (fig. 5). Moreover, transcriptomic analysis of envV2, which has an expression profile similar to envV1 (fig. 5), did not show upregulated expression levels in lymph-node-derived tumors (supplementary fig. S10A, Supplementary Material online). Almost all tumor samples (517/519) from patient lymph nodes in the NCI-GDC data set are identified as DLBCLs, a type of non-Hodgkin’s lymphoma. To further identify the specific type of cancer associated with increased expression of envV1, we analyzed gene expression data from the individual projects in this data set (supplementary fig. S10B, Supplementary Material online); this revealed that samples in all three DLBCL projects had higher envV1 transcript levels than the projects that analyzed other tumor types.
Fig. 9.
Expression of gagV1 and envV1 is upregulated in DLBCL samples. (A) Htseq count data from RNA sequencing samples were extracted from the NCI-GDC data portal and normalized using DEseq2. Box plot with the normalized count values for envV1 (log scale) in the y-axis and the primary isolation site of the normal or tumor samples in the x-axis is shown. (B) Raw RNA sequencing data extracted from six different SRA projects (see supplementary methods, Supplementary Material online, for project numbers) that represent 74 primary DLBCL tumor samples and 33 naïve and germinal B-cell samples were mapped to the human genome assembly and the counts that map to the coding region of each indicated gene were extracted via featurecounts and normalized using DEseq2. CD79a is a B-cell-specific marker and was used to confirm the accuracy of mapping and normalization (Mason et al. 1995). Box plots of normal and tumor samples (x-axis) with the normalized count values for the indicated genes (log scale) in the y-axis are shown. Samples with 0 count values were changed to 0.01 for visualization in the log scale. Statistical significance comparing tumor and normal counts for each indicated gene was estimated with the Welch two-sample t-test. ****P-value < 0.0001. ns, not significant. Box plots were generated using the ggplot2 R package.
Since this data set does not contain matched normal controls for DLBCL samples, we expanded our analysis to include RNA sequencing samples from additional studies. We extracted and analyzed RNA sequencing data from 74 primary DLBCL tumor samples and 33 naïve and germinal B cells as normal controls that span six different projects from the NCBI SRA database. As shown in figure 9B, these DLBCL samples showed significantly higher expression of gagV1 and envV1 than normal B cells. Notably, 29/33 naïve and germinal B-cell samples had no detectable gagV1 expression, and the low levels of envV2 transcript in both DLBCL and normal B-cell samples showed no significant difference between tumor and normal samples (fig. 9B). Taken together, these results indicate that gagV1 and envV1 transcripts are significantly upregulated in DLBCL.
Discussion
Although ERVs make up about 8% of the human genome, the discovery that these ERVs can be co-opted by their hosts to serve important functions has shifted our understanding of these elements from “junk DNA” to important players that serve as sources of genetic diversity. In this study, we identified the oldest HERV that expresses an intact gag gene with a full-length protein-coding sequence: HERV-V1. The full length ORF of this gag gene, gagV1, is conserved in simian primates and is part of a provirus that includes another evolutionarily conserved ORF in the leader sequence of HERV-V1, pre-gagV1; this provirus also contains a previously reported env gene, envV1, which is also conserved in simian primates (Blaise et al. 2005; Kjeldbjerg et al. 2008). This ERV is closely linked to the previously described related ERV (Blaise et al. 2005; Kjeldbjerg et al. 2008; Esnault et al. 2013), HERV-V2, that also contains an intact env gene, envV2, that is highly similar to envV1. A third related provirus, ERV-V3, approximately halfway between ERV-V1 and ERV-V2 contains the second gag ORF in this region, gagV3, which is conserved in Old and New World monkeys. Although ERV-V3 is lost in the genomes of hominids, its remnants can be found in orangutans and gibbons. Thus, although the human genome contains a small number of HERV-V copies, two of these have not only become fixed within a span of 100 kb but have conserved ORFs.
The remarkable conservation of three large ORFs in the single HERV-V1 provirus in all simian primate lineages for more than 40 My (Steiper and Young 2006; Perelman et al. 2011) suggests exaptation of each of these genes for a host function, and this is coupled with the surprising observation that ORFs of env or gag are found in all three of the linked ERV-V copies in primate genomes. Even more surprising is the fact that all four of the ORF-containing genes examined here, envV1, envV2, gagV1, and pre-gagV1, show placenta-specific expression as shown previously for the two env genes (Blaise et al. 2005; Esnault et al. 2013). Independent domestication of ERV envelope genes for a placental function in various lineages throughout vertebrate evolution is a remarkable example of convergent evolution and indicates that ERVs have played an important role in the evolution of this transient organ at the materno-fetal interface. In humans, trophoblast fusion in the placenta is affected by the HERV Env proteins, syncytin-1 and syncytin-2, which were derived from HERV-W and HERV-FRD ERVs, respectively (Mi et al. 2000; Blaise et al. 2003). In other mammalian lineages, different ERV env genes have been co-opted to serve this same function (Blaise et al. 2005; Kudaka et al. 2008; Kammerer et al. 2011; Esnault et al. 2013; Heidmann et al. 2017; Bergallo et al. 2020). Although the human EnvV1 and EnvV2 proteins do not have fusogenic properties, the EnvV2 proteins of some Old and New World monkeys can fuse human and feline cell lines in vitro (Esnault et al. 2013) suggesting that this may have been a primate syncytin that was deactivated in humans. This also suggests that captured syncytins can be replaced in diverging lineages by env genes from newly acquired ERVs that have a selective advantage in placentation, perhaps accounting for syncytin diversity in mammals and loss of this particular syncytin in humans.
Our study suggests that the progressive evolution of the placenta has been mediated by successive ERV exaptations. We identified the first placenta-specific expression of a co-opted HERV gag gene in humans and for gagV1 and gagV3 in rhesus macaques. Although this expression pattern suggests a physiological role for pre-gagV1, gagV1, and gagV3 in placental formation or function, it remains to be determined what function these retroviral gag-like genes might perform in the placenta. Previous studies demonstrated that mammalian genomes carry several expressed genes that resemble retrotransposon gag genes including ARC which encodes a neuronal RNA transport protein (Ashley et al. 2018; Pastuzyn et al. 2018) as well as a set of 12 genes derived from the gag-pol regions of the suchi-ichi LTR retrotransposon family (SIRH), three of which are involved in placentation and/or embryogenesis (Kaneko-Ishino and Ishino 2015). Peg10/Sirh1, Peg11/Sirh2/Rtl1r, and Sirh7/Ldoc1 are expressed in placenta and some embryonic tissues, and knockouts of each of these genes result in fetal death or abnormal development (Ono et al. 2001; Naruse et al. 2014; Kitazawa et al. 2017). These three genes have been assigned roles in placenta formation (Peg10), development of fetal capillaries at the feto-maternal interface (Peg11), and regulation of placental hormones (Sirh7/Ldoc1) (Ono et al. 2001, 2006; Sekita et al. 2008; Naruse et al. 2014; Kitazawa et al. 2017). All are highly conserved, with Peg10 found in mammals and marsupials whereas the other two were fixed later, in eutherian mammals (Kaneko-Ishino and Ishino 2015). Their conservation throughout mammalian evolution and their placental roles suggest they might have been involved in the initial development of the placenta, and their successive acquisition may have contributed to the diversification of placental structures in mammals. The co-option of the known syncytins, HERV-V env and gag genes, which are restricted to primates, and the subsequent decommissioning of envV1 and envV2 in humans is further suggestive evidence of an ongoing evolutionary refinement of placental morphology and physiology, with co-opted ERV genes, like HERV-V, playing leading roles in this process.
It is also possible that the HERV-V genes described here serve functions unrelated to placentation. During the retroviral life cycle, Gag proteins are targeted to the plasma membrane where they assemble into virions and package viral RNA (Coffin et al. 1997). Some of the features involved in these functions are still intact in gagV1 including the N-terminal myristoylation site, involved in membrane targeting (Spearman et al. 1994), the C terminal zinc finger-like CCHC motif, required for RNA binding (Coffin et al. 1997), and the major homology region which functions in capsid formation (Strambio-de-Castillia and Hunter 1992; Alin and Goff 1996a, 1996b; Orlinsky et al. 1996). Moreover, we found that GagV1 is released from human cells when ectopically expressed (fig. 8D), suggesting that the ability of this ancient gag gene to form virus like particles may be preserved despite the original virus being endogenized more than 40 Ma. The preservation of Gag motifs important for viral replication is also suggestive of a possible protective role in reproduction based on several examples of endogenous gag genes interfering with infectious virus replication. A reconstituted HERV-K gag gene was shown to interfere with the assembly and release of HIV-1 particles (Monde et al. 2012), although no HERV-K gag genes in the human genome have yet been found to be restrictive. In other species, the recently acquired gag gene of a JSRV ERV in sheep (Mura et al. 2004; Arnaud et al. 2007; Monde et al. 2012; Sistiaga-Poveda and Jugo 2014; Cumer et al. 2019) blocks replication of the JSRV betaretrovirus, whereas in muroid rodents, the retroviral restriction factor, Fv1, a remnant of the gag gene of an ERV-L element, inhibits the replication of some MLVs and other retroviruses (Best et al. 1996; Benit et al. 1997; Boso et al. 2018). It is possible that gagV1 and pre-gagV1 might function to limit retroviral infection and/or amplification in developing embryos, but although we found no effect of gagV1 expression on HIV-1 and MLV infectivity (fig. 8B, and supplementary fig. S9, Supplementary Material online), such restrictive activity might be limited to specific retroviral families.
The conservation of gagV1 and gagV3 genes among different primates suggests that evolutionary pressures acted to preserve these genes. Although there is evidence of positive selection acting on the N-terminal segment of ERV-V gag genes, we found only two residues evolving under adaptive evolution, and most of the gene is under purifying selection suggesting it serves an important function that does not tolerate polymorphisms (table 3). In contrast, the co-opted gag gene Fv1 evolved under strong positive selection (Yan et al. 2009; Boso et al. 2018) consistent with its function as a host restriction factor that is in evolutionary conflict with retroviruses. Also, although Fv1 has been lost or pseudogenized in several rodent lineages possibly due to nonuniform selective pressures from retroviral challenge, the gagV1 ORF is as old as Fv1 but is more stable in simian primates with only three species containing an early stop codon that interrupts the ORF (fig. 3). Notably, the NCBI dbSNP database (https://www.ncbi.nlm.nih.gov/snp/, last accessed August 23, 2021) shows that the region of the human genome that encompasses HERV-V1 (supplementary fig. S2, Supplementary Material online) contains large numbers of single-nucleotide polymorphisms (SNPs), but few are in the gagV1 and pre-gagV1 ORF segments and none of these introduces stop codons or has clinical associations. Further study of the allele frequencies of these SNPs as well as their possible impact on gagV1 and/or pre-gagV1 function may shed light on the ongoing evolution of these ancient retroviral remnants in human populations. It is also important to note that the sequence surrounding the predicted start codons of pre-gagV1 and gagV1 does not match the consensus Kozak sequence identified for human genes (GCCGCCACCatgGCG) (supplementary fig. S6 and supplementary data set 4, Supplementary Material online) (Grzegorski et al. 2014). Hence, analysis of the translation efficiency of these genes in physiological conditions in both humans and other primates may be useful in further understanding their evolution and co-option.
HERV-V1 is unusual among retroviruses in encoding a large ORF in the leader sequence between the PBS and the gagV1 start codon. Preservation of this pre-gagV1 ORF in all simian primates with strong purifying selection for more than 40 My (Steiper and Young 2006; Perelman et al. 2011) suggests that the putative protein that is expressed by this ORF has a physiological function that is beneficial to the host. Unusually long leader sequences with ORFs are found in some exogenous and endogenous retroviruses (Prats et al. 1989; Holzschu et al. 1995; Jern et al. 2005; Blanco-Melo et al. 2017; Grandi et al. 2020). The most prominent example of this is glyco-gag which is found in a subset of exogenous gammaretroviruses, including MLVs (Prats et al. 1989). Glyco-gag is not a separate ORF but is an N-terminally extended form (∼88 aa) of the viral Gag protein that is expressed from an alternative start codon (CUG) located in the leader sequence (Prats et al. 1989). In contrast, pre-gagV1 is fully contained in the leader sequence and not in frame with the gagV1 ORF. In addition to the gammaretroviruses, the presence of a long leader sequence with a putative ORF was also reported in a few groups of class I gammaERVs including ERV-W, HERV-H, and HERV-T (Jern et al. 2005; Blanco-Melo et al. 2017; Grandi et al. 2020) and epsilonERVs in frogs (Kambol et al. 2003). The presence of this ORF in the gag leader region among some other class I ERVs implies a possible functional role in the replication of the original ancient viruses, but the variable size of these genes, sequence and structural variations and linkage to gag are not suggestive of any shared function.
Closely linked paralogous genes have an increased chance of gene conversion events (Ezawa et al. 2006; Tareen et al. 2009; Mitchell et al. 2015; Molaro et al. 2020), and we found strong evidence for recurrent gene conversion between gagV1 and gagV3, two co-opted gag genes from the same ERV group in close genomic proximity for millions of years. Interestingly, the first reported gene conversion between two large HERV-derived genes also involves this same set of ERVs, specifically envV1 and envV2 (Kjeldbjerg et al. 2008). For gagV1 and gagV3, the N-terminal portion of the encoded proteins containing the putative matrix domain is more divergent than the rest of the protein. Since these genes are otherwise highly similar in sequence, they may have redundant functions. Hence, gene conversion may have led to the homogenization of the GARD segment B (fig. 7B) by limiting the divergence and increasing gene dosage in some lineages. This may also explain the loss of gagV3 in hominoids, because, in the absence of gene conversion, further divergence from gagV1 may have led to a decrease in evolutionary pressures to maintain this gene. Alternatively, further divergence from gagV1 may have interfered with gene convergence eliminating any evolutionary pressure to maintain the gene.
Our transcriptomic analyses revealed an increased expression of gagV1 and envV1 transcripts in DLBCL which is the most common type of non-Hodgkin’s lymphoma (Armitage et al. 2017). As they make up about 8% of the human genome (Lander et al. 2001), transcription from LTRs of HERVs can be highly regulated (Hurst and Magiorkinis 2017). The highly restricted expression of gagV1 and envV1 in the placenta driven from the same LTR represents a good example of this type of regulation. On the other hand, a common feature found in cancer cells is transcriptional deregulation via global hypomethylation (Ehrlich 2009). Thus, over the years, several studies have shown activation of different HERVs in tumor samples including lymphomas, melanomas, as well as ovarian and breast cancers (Wang-Johanning et al. 2007, 2014; Maliniemi et al. 2013; Wallace et al. 2014; Perot et al. 2015; Ma et al. 2016; Zhou et al. 2016; Heidmann et al. 2017). For example, anti-HERV-K antibodies and gag mRNA can be detected in early-stage breast cancer indicating a potential diagnostic value (Wang-Johanning et al. 2014). Considering the complete lack of expression of gagV1 and envV1 in most tissues (fig. 4), these genes represent a strong potential diagnostic marker for DLBCL. Additional transcriptomic as well as immunological analysis of large numbers of DLBCL and matched control samples may determine whether gagV1 and/or envV1 expression can be used as a reliable marker for a specific disease state in DLBCL.
This study describes the discovery of the oldest HERV with an intact gag gene and the presence of a novel protein expressed from an independent ORF located in the leader sequence of the same HERV, all potentially involved in placentation. The discovery of the oldest co-opted gag gene, as well as the first description of a co-opted pre-gag gene in the human genome, will likely spur new avenues of research to further uncover the complex roles HERVs played during primate evolution.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Statistical Analysis
The Welch two-sample t-test was utilized to calculate statistical significance as implemented in the ggpubr R package.
Supplementary Material
Acknowledgments
This work was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (Grant No. AI000300-38) to C.A.K.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files, Supplementary Material online.
References
- Aagaard L, Villesen P, Kjeldbjerg AL, Pedersen FS.. 2005. The approximately 30-million-year-old ERVPb1 envelope gene is evolutionarily conserved among hominoids and Old World monkeys. Genomics 86(6):685–691. [DOI] [PubMed] [Google Scholar]
- Alin K, Goff SP.. 1996a. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222(2):339–351. [DOI] [PubMed] [Google Scholar]
- Alin K, Goff SP.. 1996b. Mutational analysis of interactions between the Gag precursor proteins of murine leukemia viruses. Virology 216(2):418–424. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Nielsen R, Yang Z.. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164(3):1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JO, Gascoyne RD, Lunning MA, Cavalli F.. 2017. Non-Hodgkin lymphoma. Lancet 390(10091):298–310. [DOI] [PubMed] [Google Scholar]
- Arnaud F, Murcia PR, Palmarini M.. 2007. Mechanisms of late restriction induced by an endogenous retrovirus. J Virol. 81(20):11441–11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T.. 2018. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 172(1–2):262–274.e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell NM, Lever AM.. 2013. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol. 21(3):136–144. [DOI] [PubMed] [Google Scholar]
- Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T.. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 71(7):5652–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergallo M, Marozio L, Botta G, Tancredi A, Dapra V, Galliano I, Montanari P, Coscia A, Benedetto C, Tovo PA.. 2020. Human endogenous retroviruses are preferentially expressed in mononuclear cells from cord blood than from maternal blood and in the fetal part of placenta. Front Pediatr. 8:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP.. 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382(6594):826–829. [DOI] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Benit L, Heidmann T.. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A. 100(22):13013–13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise S, de Parseval N, Heidmann T.. 2005. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology 2(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Gifford RJ, Bieniasz PD.. 2017. Co-option of an endogenous retrovirus envelope for host defense in hominid ancestors. Elife 6:e22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boso G, Buckler-White A, Kozak CA.. 2018. Ancient evolutionary origin and positive selection of the retroviral restriction factor Fv1 in muroid rodents. J Virol. 92(18):e00850–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boso G, Shaffer E, Liu Q, Cavanna K, Buckler-White A, Kozak CA.. 2019. Evolution of the rodent Trim5 cluster is marked by divergent paralogous expansions and independent acquisitions of TrimCyp fusions. Sci Rep. 9(1):11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheynet V, Ruggieri A, Oriol G, Blond JL, Boson B, Vachot L, Verrier B, Cosset FL, Mallet F.. 2005. Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. J Virol. 79(9):5585–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM, Hughes SH, Varmus HE.. 1997. Retroviruses. Cold Spring Harbor (NY: ): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Cornelis G, Funk M, Vernochet C, Leal F, Tarazona OA, Meurice G, Heidmann O, Dupressoir A, Miralles A, Ramirez-Pinilla MP, et al. 2017. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc Natl Acad Sci U S A. 114(51):E10991–E11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Veron G, Mulot B, Dupressoir A, Heidmann T.. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci U S A. 109(7):E432–E441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Heidmann O, Degrelle SA, Vernochet C, Lavialle C, Letzelter C, Bernard-Stoecklin S, Hassanin A, Mulot B, Guillomot M, et al. 2013. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc Natl Acad Sci U S A. 110(9):E828–E837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, et al. 2015. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A. 112(5):E487–E496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Vernochet C, Malicorne S, Souquere S, Tzika AC, Goodman SM, Catzeflis F, Robinson TJ, Milinkovitch MC, Pierron G, et al. 2014. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci U S A. 111(41):E4332–E4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumer T, Pompanon F, Boyer F.. 2019. Old origin of a protective endogenous retrovirus (enJSRV) in the Ovis genus. Heredity (Edinb). 122(2):187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T.. 2013. Endogenous retroviruses: acquisition, amplification and taming of genome invaders. Curr Opin Virol. 3(6):646–656. [DOI] [PubMed] [Google Scholar]
- Dick RA, Vogt VM.. 2014. Membrane interaction of retroviral Gag proteins. Front Microbiol. 5:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T.. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A. 102(3):725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T.. 2011. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A. 108(46):E1164–E1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics 1(2):239–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Burhans R, Riemer C, Hardison R, Miller W.. 2010. MultiPipMaker: a comparative alignment server for multiple DNA sequences. Curr Protoc Bioinformatics. Chapter 10:Unit10 14. [DOI] [PubMed] [Google Scholar]
- Esnault C, Cornelis G, Heidmann O, Heidmann T.. 2013. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 9(3):e1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezawa K, OOta S, Saitou N, SMBE Tri-National Young Investigators. 2006. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol Biol Evol. 23(5):927–940. [DOI] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. 2014. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 13(2):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi N, Pisano MP, Demurtas M, Blomberg J, Magiorkinis G, Mayer J, Tramontano E.. 2020. Identification and characterization of ERV-W-like sequences in Platyrrhini species provides new insights into the evolutionary history of ERV-W in primates. Mob DNA. 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, Staudt LM.. 2016. Toward a shared vision for cancer genomic data. N Engl J Med. 375(12):1109–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorski SJ, Chiari EF, Robbins A, Kish PE, Kahana A.. 2014. Natural variability of Kozak sequences correlates with function in a zebrafish model. PLoS One 9(9):e108475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S.. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 32(4):835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O, Beguin A, Paternina J, Berthier R, Deloger M, Bawa O, Heidmann T.. 2017. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc Natl Acad Sci U S A. 114(32):E6642–E6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann O, Vernochet C, Dupressoir A, Heidmann T.. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herve CA, Forrest G, Lower R, Griffiths DJ, Venables PJ.. 2004. Conservation and loss of the ERV3 open reading frame in primates. Genomics 83(5):940–943. [DOI] [PubMed] [Google Scholar]
- Hizi A, Herzig E.. 2015. dUTPase: the frequently overlooked enzyme encoded by many retroviruses. Retrovirology 12:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschu DL, Martineau D, Fodor SK, Vogt VM, Bowser PR, Casey JW.. 1995. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 69(9):5320–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst TP, Magiorkinis G.. 2017. Epigenetic control of human endogenous retrovirus expression: focus on regulation of long-terminal repeats (LTRs). Viruses 9(6):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Sperber GO, Ahlsen G, Blomberg J.. 2005. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J Virol. 79(10):6325–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE. 2013. Rapid adversarial co-evolution of viruses and cellular restriction factors. Curr Top Microbiol Immunol. 371:123–151. [DOI] [PubMed] [Google Scholar]
- Johnson WE. 2019. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat Rev Microbiol. 17(6):355–370. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambol R, Kabat P, Tristem M.. 2003. Complete nucleotide sequence of an endogenous retrovirus from the amphibian, Xenopus laevis. Virology 311(1):1–6. [DOI] [PubMed] [Google Scholar]
- Kammerer U, Germeyer A, Stengel S, Kapp M, Denner J.. 2011. Human endogenous retrovirus K (HERV-K) is expressed in villous and extravillous cytotrophoblast cells of the human placenta. J Reprod Immunol. 91(1–2):1–8. [DOI] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Ishino F.. 2015. Mammalian-specific genomic functions: newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals. Proc Jpn Acad Ser B Phys Biol Sci. 91(10):511–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Tamura M, Kaneko-Ishino T, Ishino F.. 2017. Severe damage to the placental fetal capillary network causes mid- to late fetal lethality and reduction in placental size in Peg11/Rtl1 KO mice. Genes Cells. 22(2):174–188. [DOI] [PubMed] [Google Scholar]
- Kjeldbjerg AL, Villesen P, Aagaard L, Pedersen FS.. 2008. Gene conversion and purifying selection of a placenta-specific ERV-V envelope gene during simian evolution. BMC Evol Biol. 8:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD.. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22(24):3096–3098. [DOI] [PubMed] [Google Scholar]
- Kudaka W, Oda T, Jinno Y, Yoshimi N, Aoki Y.. 2008. Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 29(3):282–289. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. ; International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T.. 2013. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 368(1626):20120507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly F. 1967. Susceptibility to two strains of Friend leukemia virus in mice. Science 155(3761):461–462. [DOI] [PubMed] [Google Scholar]
- Ma W, Hong Z, Liu H, Chen X, Ding L, Liu Z, Zhou F, Yuan Y.. 2016. Human endogenous retroviruses-K (HML-2) expression is correlated with prognosis and progress of hepatocellular carcinoma. Biomed Res Int. 2016:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliniemi P, Vincendeau M, Mayer J, Frank O, Hahtola S, Karenko L, Carlsson E, Mallet F, Seifarth W, Leib-Mosch C, et al. 2013. Expression of human endogenous retrovirus-w including syncytin-1 in cutaneous T-cell lymphoma. PLoS One 8(10):e76281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DY, Cordell JL, Brown MH, Borst J, Jones M, Pulford K, Jaffe E, Ralfkiaer E, Dallenbach F, Stein H.. 1995. CD79a: a novel marker for B-cell neoplasms in routinely processed tissue samples. Blood 86(4):1453–1459. [PubMed] [Google Scholar]
- Meyerson NR, Sawyer SL.. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol. 19(6):286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403(6771):785–789. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Young JM, Emerman M, Malik HS.. 2015. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog. 11(12):e1005304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Malik HS, Bourc'his D.. 2020. Dynamic evolution of de novo DNA methyltransferases in rodent and primate genomes. Mol Biol Evol. 37(7):1882–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde K, Contreras-Galindo R, Kaplan MH, Markovitz DM, Ono A.. 2012. Human endogenous retrovirus K Gag coassembles with HIV-1 Gag and reduces the release efficiency and infectivity of HIV-1. J Virol. 86(20):11194–11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura M, Murcia P, Caporale M, Spencer TE, Nagashima K, Rein A, Palmarini M.. 2004. Late viral interference induced by transdominant Gag of an endogenous retrovirus. Proc Natl Acad Sci U S A. 101(30):11117–11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takahashi MU.. 2016. gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database (Oxford). 2016:baw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse M, Ono R, Irie M, Nakamura K, Furuse T, Hino T, Oda K, Kashimura M, Yamada I, Wakana S, et al. 2014. Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 141(24):4763–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F.. 2001. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73(2):232–237. [DOI] [PubMed] [Google Scholar]
- Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, et al. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 38(1):101–106. [DOI] [PubMed] [Google Scholar]
- Orlinsky KJ, Gu J, Hoyt M, Sandmeyer S, Menees TM.. 1996. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J Virol. 70(6):3440–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan AP, Stahlhacke J, Oates ME, Smithers B, Gough J.. 2019. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 47(D1):D490–D494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou I, Moreno P, Manning J, Fuentes AM, George N, Fexova S, Fonseca NA, Fullgrabe A, Green M, Huang N, et al. 2020. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 48(D1):D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV, McCormick J, Yoder N, Belnap DM, Erlendsson S, Morado DR, et al. 2018. The neuronal gene Arc encodes a repurposed retrotransposon Gag protein that mediates intercellular RNA transfer. Cell 172(1–2):275–288.e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattillo RA, Gey GO, Delfs E, Mattingly RF.. 1968. Human hormone production in vitro. Science 159(3822):1467–1469. [DOI] [PubMed] [Google Scholar]
- Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, Kessing B, Pontius J, Roelke M, Rumpler Y, et al. 2011. A molecular phylogeny of living primates. PLoS Genet. 7(3):e1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perot P, Mullins CS, Naville M, Bressan C, Huhns M, Gock M, Kuhn F, Volff JN, Trillet-Lenoir V, Linnebacher M, et al. 2015. Expression of young HERV-H loci in the course of colorectal carcinoma and correlation with molecular subtypes. Oncotarget 6(37):40095–40111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats AC, De Billy G, Wang P, Darlix JL.. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 205(2):363–372. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Gonzales PK, Cook CN, Gendron TF, Daughrity LM, Song Y, Ebbert MTW, van Blitterswijk M, Zhang YJ, Jansen-West K, et al. 2017. Repetitive element transcripts are elevated in the brain of C9orf72 ALS/FTLD patients. Hum Mol Genet. 26(17):3421–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redelsperger F, Cornelis G, Vernochet C, Tennant BC, Catzeflis F, Mulot B, Heidmann O, Heidmann T, Dupressoir A.. 2014. Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J Virol. 88(14):7915–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh MD. 2013. Covalent lipid modifications of proteins. Curr Biol. 23(10):R431–R435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayah DM, Sokolskaja E, Berthoux L, Luban J.. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430(6999):569–573. [DOI] [PubMed] [Google Scholar]
- Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, Hino T, Suzuki-Migishima R, Kohda T, Ogura A, et al. 2008. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat Genet. 40(2):243–248. [DOI] [PubMed] [Google Scholar]
- Sistiaga-Poveda M, Jugo BM.. 2014. Evolutionary dynamics of endogenous Jaagsiekte sheep retroviruses proliferation in the domestic sheep, mouflon and Pyrenean chamois. Heredity (Edinb). 112(6):571–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman P, Wang JJ, Vander Heyden N, Ratner L.. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol. 68(5):3232–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiper ME, Young NM.. 2006. Primate molecular divergence dates. Mol Phylogenet Evol. 41(2):384–394. [DOI] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF.. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA. 12(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Hunter E.. 1992. Mutational analysis of the major homology region of Mason-Pfizer monkey virus by use of saturation mutagenesis. J Virol. 66(12):7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM.. 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, et al. ; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. 2017. Dynamic landscape and regulation of RNA editing in mammals. Nature 550(7675):249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M.. 2009. An expanded clade of rodent Trim5 genes. Virology 385(2):473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda MT, Kryukov K, Mitsuhashi S, Mitsuhashi H, Imanishi T, Nakagawa S.. 2020. Comprehensive genomic analysis reveals dynamic evolution of endogenous retroviruses that code for retroviral-like protein domains. Mob DNA. 11:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargiu L, Rodriguez-Tome P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J.. 2016. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P, Aagaard L, Wiuf C, Pedersen FS.. 2004. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace TA, Downey RF, Seufert CJ, Schetter A, Dorsey TH, Johnson CA, Goldman R, Loffredo CA, Yan P, Sullivan FJ, et al. 2014. Elevated HERV-K mRNA expression in PBMC is associated with a prostate cancer diagnosis particularly in older men and smokers. Carcinogenesis 35(9):2074–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Johanning F, Li M, Esteva FJ, Hess KR, Yin B, Rycaj K, Plummer JB, Garza JG, Ambs S, Johanning GL.. 2014. Human endogenous retrovirus type K antibodies and mRNA as serum biomarkers of early-stage breast cancer. Int J Cancer. 134(3):587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Johanning F, Liu J, Rycaj K, Huang M, Tsai K, Rosen DG, Chen DT, Lu DW, Barnhart KF, Johanning GL.. 2007. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int J Cancer. 120(1):81–90. [DOI] [PubMed] [Google Scholar]
- Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL.. 2018. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Mol Biol Evol. 35(3):773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Buckler-White A, Wollenberg K, Kozak CA.. 2009. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci U S A. 106(9):3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WK, Kiggans JO, Yang DM, Ou CY, Tennant RW, Brown A, Bassin RH.. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc Natl Acad Sci U S A. 77(5):2994–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski JP.. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol. 15(12):496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Li M, Wei Y, Lin K, Lu Y, Shen J, Johanning GL, Wang-Johanning F.. 2016. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 7(51):84093–84117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files, Supplementary Material online.