Abstract
Reconstruction of the Tree of Life is a central goal in biology. Although numerous novel phyla of bacteria and archaea have recently been discovered, inconsistent phylogenetic relationships are routinely reported, and many inter-phylum and inter-domain evolutionary relationships remain unclear. Here, we benchmark different marker genes often used in constructing multidomain phylogenetic trees of bacteria and archaea and present a set of marker genes that perform best for multidomain trees constructed from concatenated alignments. We use recently-developed Tree Certainty metrics to assess the confidence of our results and to obviate the complications of traditional bootstrap-based metrics. Given the vastly disparate number of genomes available for different phyla of bacteria and archaea, we also assessed the impact of taxon sampling on multidomain tree construction. Our results demonstrate that biases between the representation of different taxonomic groups can dramatically impact the topology of resulting trees. Inspection of our highest-quality tree supports the division of most bacteria into Terrabacteria and Gracilicutes, with Thermatogota and Synergistota branching earlier from these superphyla. This tree also supports the inclusion of the Patescibacteria within the Terrabacteria as a sister group to the Chloroflexota instead of as a basal-branching lineage. For the Archaea, our tree supports three monophyletic lineages (DPANN, Euryarchaeota, and TACK/Asgard), although we note the basal placement of the DPANN may still represent an artifact caused by biased sequence composition. Our findings provide a robust and standardized framework for multidomain phylogenetic reconstruction that can be used to evaluate inter-phylum relationships and assess uncertainty in conflicting topologies of the Tree of Life.
Keywords: Tree of Life, phylogenetic uncertainty, Gracilicutes, Terrabacteria, taxon sampling, concatenated phylogenetic trees, Patescibacteria
Introduction
Due to the lack of informative morphological characters and a limited fossil record, phylogenies of bacteria and archaea have historically relied on molecular sequences (Altermann and Kazmierczak 2003; Battistuzzi et al. 2004). Woese and collaborators proposed the use of the small subunit ribosomal RNA genes (SSU) due to their “molecular chronometer” nature and fast- and slow-evolving positions (Woese and Fox 1977; Doolittle 1999). This allowed the reconstruction of a universal Tree of Life (TOL) that included bacteria, archaea, and eukaryotes (Woese 1987). Although single genes like 16S rRNA have had a tremendous value for the study of prokaryotes phylogeny over the last decades, their use is often problematic owing to PCR-amplification bias, saturation derived from the use of nucleotides sequences, and a limited number of alignment positions that may be insufficient for resolving evolutionary relationships among divergent lineages (Lerat et al. 2003; Konstantinidis and Tiedje 2007; Rajendhran and Gunasekaran 2011). Recently, the application of high-throughput sequencing methodologies has allowed for the recovery of a vast amount of genomic data that have improved taxonomic sampling across bacteria and archaea and enabled for “whole-genome phylogenies” that is, trees inferred from the concatenation of numerous marker genes. These advances, together with improvements in computational power now permit analyses of concatenated alignments that include thousands of characters belonging to a broad diversity of taxa (Ciccarelli et al. 2006; Klenk and Göker 2010; Segata et al. 2013; Hug et al. 2016; Parks et al. 2017; Coleman et al. 2021).
Despite these advances, it remains unclear if the inclusion of more genes and genomes necessarily improves the quality of resulting trees, and, if not, which marker gene sets and taxon sampling strategies produce the most robust phylogenies. The concatenation of multiple genes may improve accuracy due to an increase in the number of phylogenetically informative characters in relation to noise sites (Gadagkar et al. 2005; de Queiroz and Gatesy 2007). Still, some genes may have undergone horizontal gene transfer (HGT) and will therefore have an evolutionary history distinct from other genes in the alignment, introducing phylogenetic noise and complicating the interpretation of results. In addition, different genes evolve at different rates, and the inclusion of many disparate protein families can introduce a heterotacheous signal that may lead to long-branch attraction and other phylogenetic artifacts (Philippe and Laurent 1998; Gribaldo and Philippe 2002; Bleidorn 2017). Moreover, traditional approaches used to assess phylogenetic confidence, such as the bootstrap, were developed for the analysis of single-gene trees and often provide misleadingly high support when applied to trees constructed from multigene concatenations because support increases artificially with alignment length (Delsuc et al. 2005; Jeffroy et al. 2006; Salichos et al. 2014; Simmons and Gatesy 2016; Simon 2020; Stott and Bobay 2020). Lastly, given the highly biased taxonomic composition of the sequenced genome collection, different taxonomic groups can be sampled to dramatically different depths. Several studies have noted that taxon sampling can impact phylogenetic results (Rokas and Carroll 2005; Nasir et al. 2016; Cunha et al. 2017), but the overall impact of markedly different taxon sampling between phylogenetic groups remains unclear.
Given the complications associated with the construction of phylogenetic trees from concatenated alignments, it is not surprising that many recent studies have reported conflicting results concerning the placement of deep-branching groups of bacteria and archaea. For example, several studies have reported the Patescibacteria (also known as the Candidate Phyla Radiation, or CPR) as basal-branching in bacteria, but recent studies have suggested that this group is a sister phylum to the Chloroflexota (Coleman et al. 2021; Taib et al. 2020). Controversy has also surrounded the placement of the Asgard archaea, with some studies showing they are placed near the TACK superphylum (comprised of the Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota) and are closely related to eukaryotes, and other studies reporting placement within the Euryarchaeota (Cunha et al. 2017; Zaremba-Niedzwiedzka et al. 2017; Da Cunha et al. 2018; Williams et al. 2020). Further, some studies have even suggested that the long branch between bacteria and archaea precludes any robust generation of a multidomain TOL, further complicating the identification of basal-branching groups from any domain (Gaucher et al. 2010; Coleman et al. 2021).
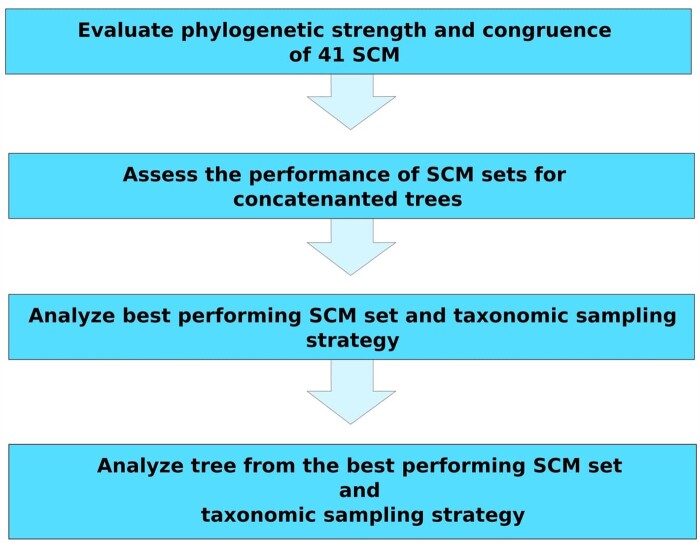
In this study, we benchmarked different single-copy marker genes (SCMs) commonly used in multidomain bacterial and archaeal phylogenetics and using recently-developed tree certainty (TC) metrics we identify a set of SCMs that performs best for phylogenetic trees derived from concatenated alignments (workflow in fig. 1). Moreover, we benchmark different taxon sampling strategies and demonstrate that uneven representation of phyla can dramatically impact the resulting trees and lower their overall TC. Using the best-performing marker gene set and balanced taxon sampling across bacteria and archaea, we then reconstructed a high-resolution tree that clarifies the phylogenetic relationships between several phyla and identifies several deep-branching nodes where the true topology remains unclear. Our results provide a robust and standardized framework for phylogenetic reconstruction of bacteria and archaea that quantifies the certainty and limitations of concatenated gene trees for resolving deep branching nodes.
Fig. 1.
Schematic summary of the methodological workflow used in this study. SCM, single-copy marker.
Results and Discussion
Evaluating the Phylogenetic Congruence of Individual Marker Genes Used for TOL Reconstruction
Given the large phylogenetic distance encompassed by bacteria and archaea, there are few SCMs that are suitable for inter-domain phylogenetic reconstruction (Berkemer and McGlynn 2020). Nevertheless, several independent studies have found 30–40 orthologous protein families that can be used for this purpose (Ciccarelli et al. 2006; Wu and Eisen 2008; Williams et al. 2012); these include RNA polymerase subunits, ribosomal proteins, tRNA synthetases, and proteins annotated as involved in intracellular trafficking. Using a set of 41 SCMs that encompasses this set and has been previously used for this purpose (Sunagawa et al. 2013), we first evaluated the occurrence of these SCMs in a curated set of 1,650 bacterial and archaeal genomes derived from the Genome Taxonomy Database (GTDB, see Materials and Methods) (Chaumeil et al. 2019). Our results confirmed that these SCMs are broadly found in diverse bacterial and archaeal lineages as single-copy genes: RNA polymerase subunits, ribosomal proteins, tRNA synthetases, and intracellular trafficking proteins showed a high occurrence (83–97%) and low presence of multiple copies (0.1–0.8%) (supplementary table S15, Supplementary Material online). In contrast, other genes that have been used in the past as SCMs were found in either a lower fraction of the genomes (e.g., recA, found in only 71% of the genomes surveyed), or were often not found as single-copy (e.g., EF-Tu, found as multicopy in 26% of the genome surveyed) (supplementary table S15, Supplementary Material online). The β and β’ subunits of RNA Polymerase (RNAP, COG0085 and COG0086, respectively) are known to be fragmented into multiple individual genes (6.42% and 3.52% for COG0085 and COG0086, respectively) (Werner and Grohmann 2011), which can lead to the erroneous conclusion that paralogs of these genes are present, but a concatenation of the gene fragments ameliorates this issue (supplementary table S16, Supplementary Material online, see Materials and Methods).
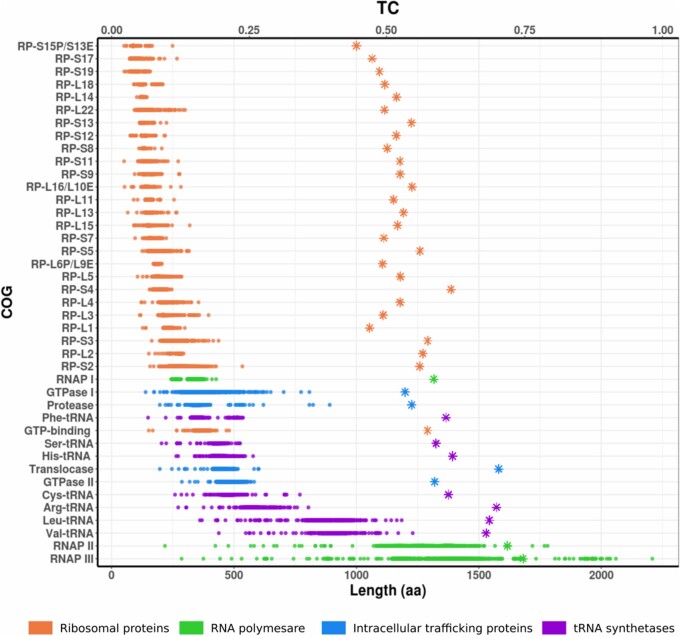
We developed a bioinformatic tool called MarkerFinder to easily identify different marker gene sets from bacterial and archaeal genomes and produce a concatenated alignments that can be used for phylogenetic reconstruction (https://github.com/faylward/markerfinder). MarkerFinder also identifies fragmented RNAP subunits and concatenates them together, thereby obviating the difficulty in including genomes with fragmented RNAP subunits (see Materials and Methods). Using this bioinformatic framework we first benchmarked the phylogenetic signal and congruence of each SCM individually using the TC metric. The TC represents the mean of all the “Internode Certainty” values (IC), an estimate that assesses the degree of conflict of each internal node in a given tree (Salichos and Rokas 2013; Kobert et al. 2016). In contrast to other support estimates like bootstrap or posterior probabilities, the IC index reflects the conflict of a given bipartition by comparing its frequency with a set of conflicting bipartitions in a collection of replicate trees (Kobert et al. 2016). Our results show a clear relationship between SCM length and TC estimates (fig. 2), consistent with the view that longer SCMs tend to have higher phylogenetic signal. The β and β’ subunits of RNAP have the highest phylogenetic signal and represent the longest genes, followed by several tRNA-synthetases (fig. 2, supplementary table S2, Supplementary Material online). Ribosomal proteins (RPs) were among the shortest SCMs in our analysis and tended to have low phylogenetic signal, indicating that these genes, when used individually, generally perform poorly as phylogenetic markers.
Fig. 2.
Tree Certainty (TC) and length of marker genes used for the reconstruction of prokaryotic phylogenies. Circles represent the length of each sequence used to reconstruct each COG/protein tree and asterisks denote TC estimates.
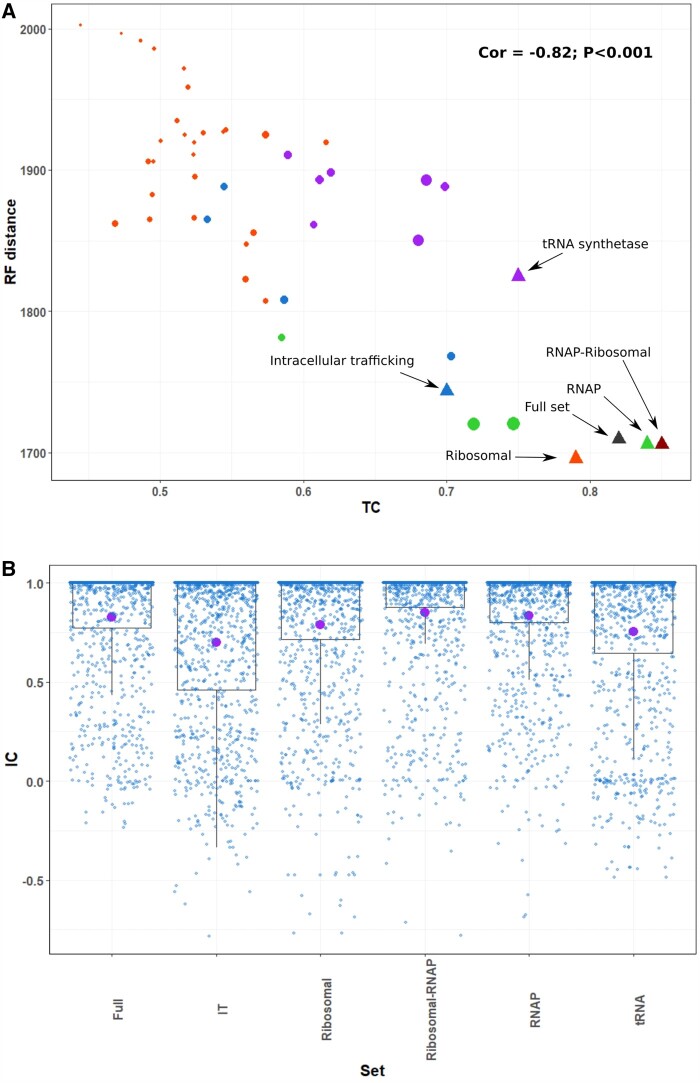
Because the concatenation of multiple SCMs with disparate evolutionary histories will lead to ambiguous results in the resulting tree, it is also critical to assess the level of phylogenetic congruence between different SCMs before concatenation. To do this we compared the TC values of each SCM against the mean Robinson-Foulds distance (RF) of each SCM’s tree against those of all other SCMs. The mean RF distances can be taken as a measure of how consistent the phylogenetic signal of each SCM is compared to all others (Robinson and Foulds 1981). The resulting plot showed a negative correlation between mean RF distance and TC (fig. 3A, Pearson’s Rho −0.82, P < 0.001), consistent with the view that SCMs with high phylogenetic signal tend to provide more consistent topologies because they provide more robust phylogenetic reconstruction. This was most clearly evidenced for the RNAP β and β’ subunits, which had the lowest RF distances and highest TC, consistent with their length.
Fig. 3.
Relationship between tree certainty (TC) and Robinson–Foulds distance for individual markers and markers sets and internode certainty (IC) estimated for marker sets. (A) Pearson correlation of mean RF distance vs. TC for Individual markers trees (circles) and marker sets trees (triangles). Colors: orange, ribosomal proteins; green, RNAP proteins; blue, intracellular trafficking; purple, tRNA proteins; gray, full set; red, RNAP-ribosomal set. Size of circles is equivalent to the median length of each COG or length of the final alignment (shortest = 98 aa; longest = 1392 aa). RF distance values represent the mean pairwise distance (B) IC (blue) and TC (purple) estimates for the maximum likelihood built from the concatenation of single-copy markers.
High mean RF distances are most likely the result of either orthologous gene displacement (OGD), which will lead to contrasting evolutionary histories in SCMs, or low phylogenetic signal, which will lead to topological differences in SCM trees that are merely the result of inadequate information for tree construction. Distinguishing between these two scenarios is critical because SCMs with low phylogenetic signal can still be used in concatenated alignments, where their phylogenetic signal can be considered additive rather than conflicting. Comparison of mean RF distances and TC values offers a possible way of distinguishing between OGD and low phylogenetic signal. For example, the tRNA-synthetase SCMs exhibited RF distances that are higher than expected given their relatively high TC values (fig. 3A, supplementary table S2, Supplementary Material online). This pattern is consistent with the higher incidence of both ancient and recent OGD events that have previously been noted in tRNA-synthetases (Wolf et al. 1999; Creevey et al. 2011; Fournier et al. 2015), which would result in a decoupling of the TC and RF values because they would have an evolutionary history distinct from the other SCMs (Wolf et al. 1999; Creevey et al. 2011). Indeed, an inspection of individual SCM phylogenies revealed that tRNA-synthetases have experienced several inter-domain and inter-phylum OGD events (supplementary fig. S3, Supplementary Material online). In contrast to tRNA-synthetase genes, the relatively high RF values recovered for ribosomal proteins are likely due to their short length and low individual phylogenetic signal for these SCMs, rather than high incidence of OGD. The shortest ribosomal proteins had the lowest TC and the highest RF distances of all SCMs. In general, our TC and RF results are in agreement with the “Complexity Hypothesis” (Jain et al. 1999), which proposes that genes of the same structural system are involved in informational processes (i.e., RNAP and ribosomal proteins) tend to undergo fewer OGD events.
Identifying the Best-Performing SCM Set for Interdomain Phylogenetic Reconstruction
We next evaluated trees constructed using concatenated alignments made from different SCM sets (table 1). We evaluated alignments of all 41 SCMs (Full set) and SCM sets divided according to functional categories: ribosomal proteins (RP), RNAP subunits (RNAP), intracellular trafficking (IT), and tRNA synthetases (tRNA). Moreover, we also evaluated a concatenated set of both, ribosomal proteins and RNAP subunits (RNAP-RP set) because these SCMs had high phylogenetic congruence according to our previous analysis, and both belong to large multimeric complexes where OGD is less likely. All SCMs sets had high median bootstrap support (99–100%, table 1), demonstrating the insufficiency of this metric for assessing differences between concatenated alignments. This finding is consistent with a previous report that bootstrap support provides misleadingly high confidence values for trees based on concatenated alignments (Salichos and Rokas 2013). TC values provide a more robust metric for evaluating the tree quality (table 1; fig. 3A and B): the tree built using the Full set and the RNAP-RP set had the highest TC values, whereas the most uncertain trees were obtained for the IT and tRNA genes sets. As expected, although individual ribosomal trees showed low certainty values, their concatenation in the RP set showed higher congruence (table 1, fig. 2A and B), consistent with the view that these SCMs have low phylogenetic signal independently but can be effectively concatenated due to their consistent evolutionary histories. Importantly, the RNAP-RP set outperformed the full set of 41 markers despite having a shorter overall alignment length (table 1, figs. 2A and B), likely because the IT and tRNA sets incorporate phylogenetic signals incongruent with the other SCMs. This is consistent with studies that have noted that these genes have higher rates of HGT than the other SCMs (Ciccarelli et al. 2006; Creevey et al. 2011). Overall, these results identify RNAP-RP as the best-performing SCM set for multidomain phylogenetic reconstruction, underscore the importance of evaluating phylogenetic congruence when choosing SCMs for a concatenated alignment, and demonstrate that the inclusion of additional SCMs does not necessarily improve phylogenetic accuracy.
Table 1.
Statistics of Phylogenetic Trees Built Using the Concatenation of SCMs.
| Marker Set | Alignment Length (aa) | Number of Proteins |
Modela |
TCb |
Median Bootstrap |
|---|---|---|---|---|---|
| Full set | 16,141 | 41 | LG+R10 | 0.82 | 100 |
| Intracellular trafficking | 1,964 | 4 | LG+F+R10 | 0.70 | 99 |
| Ribosomal | 5,197 | 27 | LG+R10 | 0.79 | 100 |
| tRNA | 4,993 | 7 | LG+R10 | 0.75 | 100 |
| RNAP | 3,987 | 3 | LG+F+R10 | 0.84 | 100 |
| Ribosomal-RNAP | 9,184 | 30 | LG+R10 | 0.85 | 99 |
Best-performing substitution model according to the BIC criterion.
TC = Tree certainty based on best ML tree vs. bootstrap replicates.
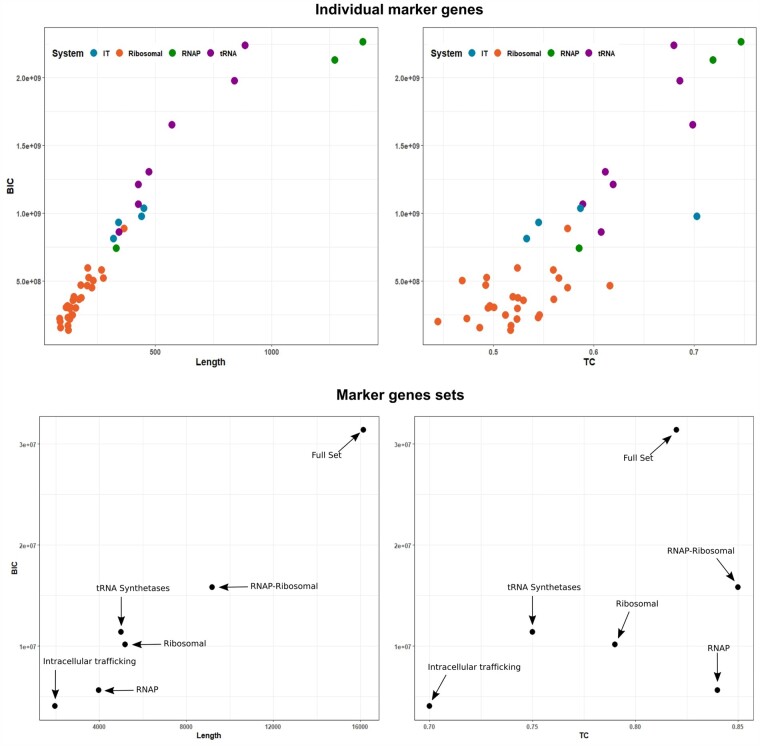
We also evaluated the fit of different substitution models to see if this could explain our results. For individual SCMs the Bayesian Information Criterion (BIC) of the best-fit substitution model increased linearly with protein length, as expected given that alignment length is used directly in the calculation of BIC. Similarly, BIC and TC were correlated, indicating once again that longer SCMs tend to have a higher phylogenetic signal. Interestingly, for concatenated alignments, the correlation between alignment length and BIC was upheld, but the relationship between BIC and TC was not evident (fig. 4). The RNAP-Ribosomal, Ribosomal, and RNAP alignments all had higher TC than would be expected given their alignment length, indicating that the concatenation of SCMs with congruent phylogenetic signal effectively boosts the accuracy of phylogenetic reconstruction (fig. 4). Analysis of the relationship between BIC, alignment length, and TC may therefore represent a complementary method for identifying sets of marker genes with congruent phylogenetic signals.
Fig. 4.
Relationship between substitution model fit based on the Bayesian Information Criterion (BIC) and alignment length, and BIC and tree certainty for individual marker genes and marker genes sets.
Evaluating the Effect of Taxon Sampling in Tree Topology
Several studies have shown that an increase in taxon sampling can improve phylogenetic accuracy (Pollock et al. 2002; Zwickl and Hillis 2002; Jeffroy et al. 2006), and this strategy is commonly used as a solution to resolve unstable nodes in the TOL (Young and Gillung 2020). In some cases it has been suggested that conflict among reported trees may result from differences in taxonomic representation, however (Nasir et al. 2016; Cunha et al. 2017), and it remains unclear to what extent the oversampling of some taxa relative to others can deleteriously affect tree reconstruction. To test the effect of relative taxon sampling and taxonomic level selection on the certainty and topology of multidomain phylogenies, we compared multidomain trees constructed using different taxon evenness across phyla (supplementary table S1, Supplementary Material online), with the Gini index used to assess evenness at the phylum level (supplementary fig. S6, Supplementary Material online, see Materials and Methods). We performed this analysis on three genome sets in which representative genomes were selected at the Order, Family, and Genus level, according to the classifications of the GTDB. For these three genome sets (unbalanced data sets) we employed two strategies to increase taxonomic evenness: 1) we removed poorly represented phyla only (fewer than five genomes present for a given phylum) (partially unbalanced data sets), and 2) we removed both poorly represented phyla and also down-sampled over-represented phyla (balanced data sets) (supplementary fig. S6, Supplementary Material online; see Materials and Methods for details).
Our results demonstrate that taxon sampling markedly affects both TC values (table 2; supplementary table S16, Supplementary Material online) and overall tree topology (fig. 6, supplementary figs. S7–S14, Supplementary Material online). Similar to our benchmarking of different SCM sets, the trees could not be distinguished based on bootstrap support (100% median support for all trees). The TC values of the trees at all the taxonomic levels improved when both poorly-represented taxa are removed and overrepresented groups are downsampled (table 2; supplementary table S16, Supplementary Material online). Importantly, in all cases, we found an increase in TC values when both poorly-represented groups were removed and over-represented phyla were down-sampled compared to if only poorly-sampled groups were removed (table 2; supplementary table S16, Supplementary Material online). This demonstrates that perhaps counterintuitively, removal of some genomes can actually improve tree quality, and that larger genome sets do not necessarily improve phylogenetic inference. We surmise that taxon oversampling lowers TC values in part because of complications that arise at the alignment stage; an alignment that is highly over-represented in certain groups would not necessarily be expected to align homologous regions in all taxa equally well, especially if some were highly divergent compared to the over-sampled groups. The incorporation of poorly sampled groups, or the oversampling of some groups relative to others, may therefore lead to long-branch artifacts that can influence the placement of other groups in the tree (Felsenstein 1978; Bergsten 2005). Balanced sampling improved TC values at all levels (Order, Family, and Genus), underscoring the important effect that balanced taxon sampling has when studying deeply divergent nodes. For studies specifically focusing on lineages for which only few genomes are available, we recommend including these genomes in an otherwise balanced tree. This approach would represent a compromise that would both mitigate the deleterious effects of unbalanced taxon sampling while still allowing for phylogenetic placement of the lineage under examination.
Table 2.
Statistics of Phylogenetic Trees Built Using Balanced, Partially Unbalanced, and Unbalanced Genomes Data Sets.
| Sampling Strategy | Genomes Included | Alignment Length (aa) | TCa | TC10b | Gini Index | Median Bootstrap |
|---|---|---|---|---|---|---|
| Order balanced | 620 | 9,203 | 0.83 | 0.83 | 0.44 | 100 |
| Partially unbalanced orderc | 722 | 9,146 | 0.75 | 0.71 | 0.5 | 100 |
| Order unbalanced | 834 | 9,298 | 0.77 | 0.71 | 0.70 | 100 |
| Family balanced | 1,650 | 9,625 | 0.86 | 0.83 | 0.59 | 100 |
| Partially unbalanced familyc | 1,925 | 9,407 | 0.81 | 0.77 | 0.64 | 100 |
| Family unbalanced | 2,023 | 9,407 | 0.78 | 0.72 | 0.75 | 100 |
| Genus balanced | 4,340 | 9,845 | 0.88 | 0.79 | 0.67 | 100 |
| Partially unbalanced genusc | 7,260 | 9,731 | 0.8 | 0.77 | 0.78 | 100 |
| Genus unbalanced | 7,325 | 9,730 | 0.84 | 0.8 | 0.84 | 100 |
TC = Tree certainty based on best ML tree vs. bootstrap replicates.
TC calculated based on the 10% closest nodes to the root.
Partially unbalanced trees have had low-abundance phyla removed, but over-represented phyla have not been down-sampled.
An RNAP-ribosomal concatenated alignment was used to reconstruct all trees. All trees were built using the LG+R10 substitution model after selection according to the BIC criterion.
Fig. 6.
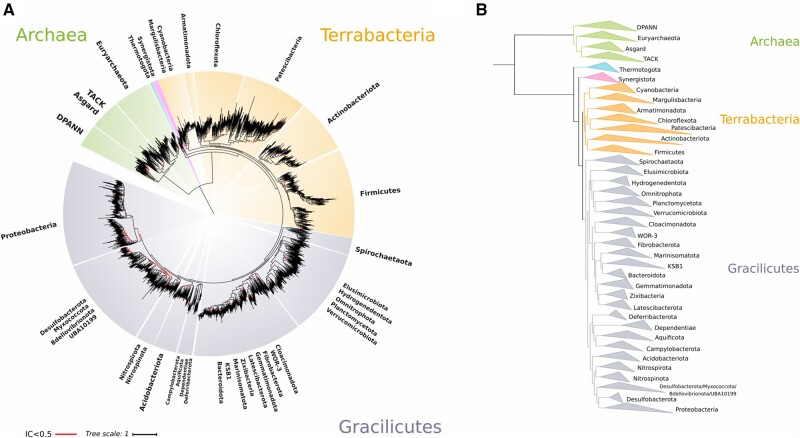
Rooted interdomain tree built using a balanced taxonomic representation. (A) Maximum likelihood tree built using the concatenation of 30 RNAP subunits and ribosomal protein sequences and the substitution model LG+R10. A balanced sampling strategy was used to select genomes at the family level according to the GTDB (see Materials and Methods for details). An IC > 0.5 indicates that more than 80% of the bootstrap replicate trees support the node shown (Salichos et al. 2014). Euryarchaeota is represented by three different phyla on the GTDB; Methanobacteriota, Thermoplasmatota, and Halobacteriota, TACK Archaea by one phylum; Thermoproteota, and the DPANN superphylum by the phyla Iainarchaeota and Nanoarchaeota. (B) Detailed phylogenetic relationships among the phyla studied.
In addition to evaluating overall TC values for our order-, family-, and genus-level trees, we also sought to examine which contained the highest IC values specifically for deep-branching nodes, and would therefore be most appropriate for examining inter-phyla evolutionary relationships. For this, we estimated the TC values of our trees based on only 10% of the nodes closest to the root (TC10 metric, table 2; supplementary table S16, Supplementary Material online). Our estimates show that although the balanced Genus tree had the highest TC when considering all nodes (TC of 0.88), the balanced Order and Family trees showed the highest certainty in deep nodes (TC10 of 0.83, table 2; supplementary table S16, Supplementary Material online), indicating the latter two trees are more appropriate for the analysis of inter-phylum relationships.
Analysis of the Highest Quality Multidomain Tree
After benchmarking the best-performing SCM set and the appropriate level of taxon sampling for accurate phylogenetic reconstruction, we sought to examine the evolutionary relationships revealed by our best-performing trees. The balanced Family and Order trees both had the highest TC10 values, but we focus our analysis primarily on the former because this phylogeny contains the largest representation of bacterial and archaeal lineages (fig. 6A and B). The salient features we discuss below were also shared with the balanced Order-level tree, however (supplementary fig. S9, Supplementary Material online).
Our phylogeny indicates that the root in Bacteria lies between Thermotogota and the rest of the bacterial phyla (fig. 6A and B; supplementary table S16, Supplementary Material online). Although some analyses state that the basal-branching placement of Thermotogota may be an artifact derived from the transfer of archaeal and Actinobacterial genes related with thermophily (Nesbo et al. 2001; Zhaxybayeva et al. 2009; Cavalier-Smith 2010), the topology we report is in agreement with previous studies of the early-branching position of the group (Woese 1987; Bachleitner et al. 1989; Woese et al. 1990), and the potential hyperthermophile of early life (Gaucher et al. 2010). Although early phylogenetic studies showed that Aquificota is a deep-branching group (Burggraf et al. 1992; Boussau et al. 2008), analyses based on the presence and absence of conserved signature indels in highly conserved genes have suggested that Aquificota is a late-branching group within bacteria (Griffiths and Gupta 2004; Rosenberg et al. 2014), which agrees with their placement in our tree (fig. 6A and B). In addition to the Thermotogota, the Synergistota were also basal-branching in our tree, and all other bacteria could be divided into two groups corresponding to the superphyla Terrabacteria and Gracilicutes (Battistuzzi et al. 2004; Cavalier-Smith 2006). Terrabacteria include the Cyanobacteria, Firmicutes, Actinobacteriota, Patescibacteria, and Chloroflexota, among other phyla, while the Gracilicutes include a wide variety of lineages including the Proteobacteria, Acidobacteria, Bacteriodota, Spirochaetes, Planctomycetota, and Verrucomicrobiota (fig. 6A and B).
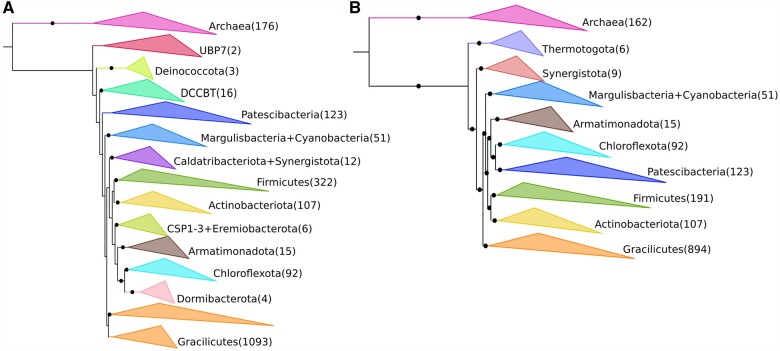
Our results indicate that the Patescibacteria (also called the CPR) are a derived group that is sister to the Chloroflexota, consistent with two recent studies (Coleman et al. 2021). This is in contrast to other studies that placed this group as either basal-branching or falling outside of the Terrabacteria (Hug et al. 2016; Parks et al. 2017; Castelle et al. 2018; Méheust et al. 2019). Previous studies have suggested that the inclusion of a distant outgroup like archaea may explain the artifactual basal branching of the Patescibacteria (Taib et al. 2020; Coleman et al. 2021), but our result indicates that it is possible to obtain a derived placement of this group even with the inclusion of archaea. Early findings of a basal branching placement of the Patescibacteria may have been influenced by unbalanced taxon sampling, indeed, our unbalanced family tree showed that Patescibacteria was placed near to the root (fig. 5A; supplementary table S16, Supplementary Material online) in contrast to our balanced tree which had the same number of Patescibacteria genomes (fig. 5B), and our partially unbalanced order tree showed the Patescibacteria as basal branching but with a low certainty (supplementary fig. S10 and table S16, Supplementary Material online). In support of our result, a recent study found similar genomic signatures of monoderm envelope structure in the Patescibacteria and Chloroflexota and attributed this to the possible transition from diderm to monoderm envelope structures in their common ancestor (Taib et al. 2020).
Fig. 5.
Comparison of the phylogenetic placement of Terrabacteria phyla using different sampling strategies. (A) Maximum likelihood phylogenetic tree built using an unbalanced taxonomic representation at the family level. (B) Maximum likelihood phylogenetic tree built using a balanced taxonomic representation at the family level. Abbreviations: DCCBT; Dictyoglomota, Coprothermobacterota, Caldisericota, Bipolaricaulota, Thermotogota. Number of genomes used for each group is indicated in parentheses. Black circles over branches indicate IC > 0.5.
Our balanced Family-level tree places the root of Archaea between the DPANN and the rest of Archaea (fig. 6A and B), and this placement has been reported previously in a study based on gene tree-species tree reconciliation (Williams et al. 2017). Since the discovery of the first DPANN representative (Rinke et al. 2013), the placement of this group in the archaeal TOL has been uncertain because of their extreme genome reduction and long branch lengths (Dombrowski et al. 2019). The Patescibacteria, another lineage with small genomes and long branches, was also reported to have basal placement in the TOL (Hug et al. 2016), but subsequent work and our findings here suggest that they are a sister lineage to the Chloroflexota (Taib et al. 2020; Coleman et al. 2021). The basal placement of DPANN must therefore be treated with some caution. Although the basal branching position and monophyly of the DPANN shows high certainty, the overall low taxon sampling available for archaea relative to bacteria gives us cause for doubt of this result, and it is possible that this is an artifact caused by similar substitution bias and homoplasies that may cause the grouping of unrelated lineages and Long Branch Attraction (LBA) (Brochier et al. 2005; Philippe and Roure 2011; Petitjean et al. 2014; Gouy et al. 2015; Aouad et al. 2018). This remains a distinct possibility because most of the DPANN described so far share similarities in their host-dependent ectosymbiotic lifestyle and residence in deep subsurface environments (He et al. 2021). Previous studies have shown that the phylogenetic resolution of DPANN is sensitive to the taxa included (Williams et al. 2017; Dombrowski et al. 2019), and we therefore speculate that additional sequencing of archaeal diversity will be necessary to increase the genomic representation of this domain and clarify the placement of the DPANN.
For the rest of the Archaea, our tree recovered the monophyly of Euryarchaeota obtained previously (Petitjean et al. 2014; Williams et al. 2017), which contrasts with a study that suggests a paraphyletic origin of the group (Raymann et al. 2015). Interestingly, all our unbalanced trees showed paraphyly in Euryarchaeota (supplementary figs. S8, S11, and S13 and table S16, Supplementary Material online), raising the possibility that the topology of this group is sensitive to taxon sampling. The placement of Asgard archaea at the base of TACK showed the maximal certainty (fig. 6A; IC = 1) and was found in all our trees independently of the sampling strategy (fig. 6A and B, supplementary figs. S7–S14, Supplementary Material online). This finding is supported by recent studies (Spang et al. 2015; Adam et al. 2017; Williams et al. 2017; Zaremba-Niedzwiedzka et al. 2017). Some studies have suggested that the placement of Asgard archaea near TACK may be due in part to unbalanced taxon sampling (Nasir et al. 2016; Cunha et al. 2017), but our results are inconsistent with this view.
By using TC metrics we are also able to identify deep-branching nodes for which the topology remains uncertain, and for which additional analyses will be necessary to resolve evolutionary relationships. In particular we observed high uncertainty in the cluster formed by Desulfobacterota (paraphyletic), Myxococcota, Bdellovibrionota (which was paraphyletic in our tree), and UBA10199 (fig. 6A and B). These newly established phyla previously belonged to the Proteobacteria, therefore it is possible that these groups need revision or reclassification.
Conclusions
Whole-genome phylogenies have become increasingly common in recent years owing to the large volume of genomic data that is now available for diverse bacteria and archaea, and it is now common to use hundreds of concatenated protein sequences to infer the evolutionary relationships of microbial taxa. There are several reasons to doubt that the use of more genes and genomes necessarily improves the quality of the resulting trees, however. For example, some genes may have undergone OGD, and their evolutionary history will therefore conflict with the other genes in the concatenated alignment, in effect creating phylogenetic noise. Moreover, the common phylogenetic confidence metrics, such as bootstrap support, provide misleadingly high values when applied to long concatenated alignments, and it is therefore unclear whether adding more genes truly enhances phylogenetic accuracy or merely artifactually increases support values. Lastly, given the different number of genomes available for different phyla of bacteria and archaea, it is unclear if the relative oversampling of some lineages over others can negatively affect the quality of phylogenetic inference.
Our findings show that both SCM selection and taxon sampling strategies are critical considerations that impact the quality of multidomain phylogenetic trees constructed from concatenated alignments. We find that selecting SCMs with congruent phylogenetic signals improves the performance of resulting trees generated from concatenated alignments and that more SCMs do not necessarily improve tree quality. Moreover, we found that taxon sampling can dramatically impact the topology of resulting trees and that over-sampling of some lineages relative to others can introduce topological inconsistencies and yield nodes with low certainty. Taken together, these results show that more genes and genomes do not necessarily improve phylogenetic inference, and that the use of phylogenetically congruent SCMs on a balanced taxon set is likely to yield the best results. Many of these issues have been previously recognized in phylogenomic analyses of eukaryotes, in particular animals and yeast, suggesting these are common issues that arise in evolutionary analyses of different groups at disparate phylogenetic scales (Rokas and Carroll 2005; Nishihara et al. 2007; Philippe et al. 2011; Salichos and Rokas 2013).
Our results have several implications for investigations of the TOL. Firstly, our finding that more genes and genomes do not necessarily improve phylogenetic accuracy is important considering that phylogenies constructed from large taxon and SCM sets can hinder the use of complex models and effectively restrict researchers to the use of a small set of tools that are optimized for speed (Price et al. 2010). Although this issue will only become more pronounced in the future as more genomes continue to be sequenced, our results indicate that down-sampling over-represented groups will both alleviate computational burdens, allow for more complex phylogenetic models to be employed, and ultimately improve tree quality, in particular at deep-branching nodes. Secondly, our results show that phyla for which only few genomes are available will likely have uncertain phylogenetic placement given the inability to include them in balanced trees. This is unavoidable to a large extent, and underscores the importance of diversity-based sequencing efforts that expand the genomic representation of poorly-characterized phyla. Thirdly, although it has been proposed that the inclusion of both domains may lead to artifacts due to the evolutionary distance between bacteria and archaea (Coleman et al. 2021), our analysis shows that high fidelity multidomain trees can be constructed using certain SCM sets and taxon sampling strategies. Lastly, it has recently been suggested that small SCM sets that include many ribosomal proteins are undesirable in multidomain phylogenetic analyses due to their large inter-domain divergence (Zhu et al. 2019), but our results conflict with this view and suggest that the addition of more genes with potentially discordant evolutionary histories will often increase noise and reduce tree quality. Indeed, the long interdomain distance between some SCMs has long been considered to be a signature of their presence in the LUCA (Woese 1998; Forterre 2006), which would make them particularly useful markers for analysis of ancient diversification events.
Although our analyses addressed several difficulties that arise in the generation of phylogenetic trees containing both bacteria and archaea (i.e., SCM selection and taxa sampling), other biological factors may still limit the accuracy of phylogenetic inference. For example, substitution models may have difficulty dealing with high evolutionary rates or biased amino acid composition of SCMs, which may in turn lead to long-branch artifacts. We suspect these issues are at play in the DPANN group, which may lead to their artifactual placement at the base of the archaea in both our trees and those of other studies (Rinke et al. 2013; Hug et al. 2016; Parks et al. 2017; Williams et al. 2017; Dombrowski et al. 2019). Further work will therefore be needed to address these complications, potentially through the developments of additional statistical models that account for these possible biases or through detailed analyses of indels or other phylogenetic markers that are useful for the placement of specific lineages.
Materials and Methods
Assessing the Congruence of Individual Marker Genes
In order to evaluate the phylogenetic certainty of 41 marker genes commonly used to build prokaryotic phylogenies (supplementary table S2, Supplementary Material online), we compiled a genomic data set encompassing a broad diversity of bacteria and archaea. We obtained one representative genome for each family available on the Genome Taxonomy Database (GTDB) (Release 05-RS95; 17th July 2020) (Chaumeil et al. 2019). The selection criteria included genome completeness, contamination, N50 contig size, and the presence of all the marker genes tested, totaling 1119 families (supplementary table S1, Supplementary Material online). The open reading frames (ORF) obtained from the GTDB were compared to the HMMs of the 41 marker genes using the hmmsearch tool available in HMMER v. 3.2.1 (Eddy 2011) with a specific score cutoff for each marker gene (supplementary table S2, Supplementary Material online). To generate a reproducible workflow and address the fragmentation of COG0085 and COG0086 orthologs into multiple genes, we developed a custom python program (MarkerFinder; https://github.com/faylward/markerfinder) (supplementary table S16, Supplementary Material online). We previously used an earlier version of this tool to resolve evolutionary relationships in the Thaumarchaeota (Aylward and Santoro 2020). Once annotated, marker genes were aligned using Clustal Omega v. 1.2.3 with the default parameters (Sievers and Higgins 2018) and trimmed with trimAl v1.4.rev15 (-gt 0.1) (Capella-Gutiérrez et al. 2009). Maximum likelihood phylogenetic trees were estimated using IQ-TREE v1.6.12 (Nguyen et al. 2015) with the options -MFP to find the best-fitting substitution model available under the BIC criterion (Kalyaanamoorthy et al. 2017) (the best model for each marker gene is reported in supplementary table S2, Supplementary Material online), and -bb 1,000 to obtain 1,000 ultrafast bootstraps (Minh et al. 2013). The resulting trees were manually inspected on interactive Tree of Life (iTOL) (Letunic and Bork 2019) to identify topologies suggestive of LGT (supplementary fig. S3, Supplementary Material online). We additionally analyzed the prevalence of these marker genes in 1,650 genomes (balanced family data set, supplementary table S1, Supplementary Material online) as well as recA (COG0468), elongation factor G (COG0480), and elongation factor TU (COG0050) (supplementary table S15, Supplementary Material online).
The congruence of each marker gene tree was assessed by calculating the “TC” metric. The TC represents the mean of all the “IC” values, an estimate that assesses the degree of conflict of each internal node in a given tree by calculating Shannon's Measure of Entropy (Shannon 1948; Salichos and Rokas 2013; Kobert et al. 2016). In contrast to other congruence and support estimates alternative to the bootstrap, the IC index reflects the degree to which the most favored bipartition is contested (Kobert et al. 2016). Estimates of IC and TC indices were achieved with raxmlHPC implemented on RAxML v8.2.X (Stamatakis 2014) with the parameters -f i and -m GTRCAT and the files .treefile (-t) and .ufboot (-z) obtained from IQ-TREE as input files. Additionally, we estimated the Robinson and Foulds distance (RF) for each pair of trees (Robinson and Foulds 1981) using IQ-TREE (Nguyen et al. 2015). The RF metric calculates the distance between phylogenetic trees by counting the number of topological changes needed to convert one tree into the other (Robinson and Foulds 1981).
Phylogenetic Congruence of Concatenated Marker Genes Sets
In addition to the assessment of each SCM' congruence, we analyzed the phylogenetic congruence of the maximum likelihood phylogenetic tree built based on the concatenation of all the SCMs, as well as phylogenetic trees built from the alignment of subsets of SCMs with related functions. Individual trimmed sequences resulting from the previous step were concatenated and maximum likelihood trees and certainty values were estimated as described above. The best model for each phylogenetic is reported in table 2.
Balanced Sampling across Prokaryotes Diversity
To evaluate the effect of taxon sampling on the certainty and topology of the prokaryotic TOL, we constructed three genomic data sets by selecting representative genomes from the GTDB at the Order, Family, and Genus level. Representative genomes for each taxonomic level were chosen from the GTDB based on its estimated completeness, contamination, and N50 contig length. Genome representatives were filtered based on a completeness cutoff of 70% and the presence of at least 25 out of the 30 marker genes belonging to the RNAP + Ribosomal marker set. In addition, we only used genomes where both COG0086 and COG0085 could be found, because these RNAP subunits are particularly long and have a strong phylogenetic signal (fig. 2), and their absence would therefore have a pronounced impact on alignment quality. To assess the evenness of the these sets, genomes were grouped at the phylum level, and phylum-level distributions were evaluated using the Gini Index (GI). The GI is a widely used metric for equality that varies from 0 (full equality) to 1 (fully unequal) (Gini 1912). Thus, if applied to genome sets, the GI describes the level of taxonomic evenness. Once we calculated the GI on the initial Order, Family, and Genus-level genome sets (referred here as the unbalanced data sets), we increased taxonomic evenness by using two methods: 1) we removed phyla that contained <5 representatives (the partially unbalanced data sets), and 2) we both removed phyla that contained <5 representatives and down-sampled the most over-represented phyla (referred to as the balanced data sets). In the second case, we performed phylum-level downsampling using the following equation:
where N represents the number of genomes for each phylum and Quant represents the 0.9 quantile of the taxonomic genome counts. In this case Quant was derived by using the quantile function in R, with a list of the phylum-level genome counts used as input. We refer to these data sets as “balanced datasets” (supplementary fig. S6, Supplementary Material online).
In our analysis, S represents the final number of genomes for each downsampled phyla. Genomes for downsampled phyla were selected randomly. Our final data consisted of three unbalanced, three balanced, and three partially unbalanced datasets (two for each taxonomic level evaluated) (supplementary table S1, Supplementary Material online). Marker genes belonging to the RNAP-RP SCM set were identified and concatenated as described previously, and maximum likelihood trees were built using the parameters -m MFP and -bb 1,000. Additionally, TC values were estimated and each tree topology was explored manually using iTOL.
Assessment of Model Fit on TC
In order to assess if TC values could be inflated due to substitution model misspecifications, we explored the relationship between TC, alignment length, and substitution model fit (BIC) for both individual trees and marker gene sets. We plotted TC vs. BIC and BIC vs. median length of individual marker genes and alignment length for marker sets (fig. 4), which suggest that TC is not directly affected by substitution model fit. In addition, we assessed the impact of model complexity on the TC of our SCM sets by repeating the model selection analysis but including the C10–C60 mixture models (Le et al. 2008). The BIC indicated that the C60 mixture model was the best fitting model for all the SCM sets except the RNAP set, for which the LG+R10 model was the best fit. We obtained a site-frequency matrix according to the PMSF method (Wang et al. 2018) using the C60 mixture model and then ran five independent maximum likelihood trees on each SCM alignment. Results of this analysis are presented in supplementary table S4, Supplementary Material online. Our results showed that the RNAP-RP set once again had the highest TC, supporting our previous findings that the RNAP-RP is the best SCM set tested in our study (supplementary table S4, Supplementary Material online). Similarly, we tested whether the results observed in our balanced sampling analysis were caused by model misspecifications by using the C60 mixture model through the PMSF approach (Wang et al. 2018) on our balanced, partially unbalanced, and unbalanced sets at the order level (best-fitting model according to the BIC criterion). Although the TC of unbalanced trees improved when using a more complex model, the balanced tree still showed the highest TC, suggesting that our results are consistent and independent of the substitution model used (supplementary table S5, Supplementary Material online). Lastly, we reran our balanced family tree using the C60 model without the PMSF approach to confirm that substitution model complexity did not adversely affect tree topology, and we did not observe topological changes or improvements in the TC value (TC identical to our LG+R10 model tree) that modify our results and conclusions (supplementary fig. S17, Supplementary Material online).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge the use of the Virginia Tech Advanced Research Computing Center for bioinformatic analyses performed in this study. This work was supported by grants from the Institute for Critical Technology and Applied Science and the NSF (IIBR-1918271) and a Simons Early Career Award in Marine Microbial Ecology and Evolution to F.O.A. We thank members of the Aylward lab for helpful comments on an earlier version of this manuscript.
Data Availability
Raw alignments, iTOL datasets, and treefile files can be accessed through Figshare (doi.org/10.7294/16543164.v1).
References
- Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S.. 2017. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11(11):2407–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann W, Kazmierczak J.. 2003. Archean microfossils: a reappraisal of early life on earth. Res Microbiol. 154(9):611–617. [DOI] [PubMed] [Google Scholar]
- Aouad M, Taib N, Oudart A, Lecocq M, Gouy M, Brochier-Armanet C.. 2018. Extreme halophilic archaea derive from two distinct methanogen Class II lineages. Mol Phylogenet Evol. 127:46–54. [DOI] [PubMed] [Google Scholar]
- Aylward FO, Santoro AE.. 2020. Heterotrophic Thaumarchaea with small genomes are widespread in the dark ocean. Ecol Evol Sci. 5:e00415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachleitner M, Ludwig W, Stetter KO, Schleifer KH.. 1989. Nucleotide sequence of the gene coding for the elongation factor Tu from the extremely thermophilic eubacterium Thermotoga maritima. FEMS Microbiol Lett. 48(1):115–120. [DOI] [PubMed] [Google Scholar]
- Battistuzzi FU, Feijao A, Hedges SB.. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol Biol. 4(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsten J. 2005. A review of long-branch attraction. Cladistics. 21(2):163–193. [DOI] [PubMed] [Google Scholar]
- Berkemer SJ, McGlynn SE.. 2020. A new analysis of Archaea–bacteria domain separation: variable phylogenetic distance and the tempo of early evolution. Mol Biol Evol. 37(8):2332–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleidorn C. 2017. Sources of error and incongruence in phylogenomic analyses. In: Bleidorn C, editor. Phylogenomics: an introduction. Cham: Springer International Publishing. p. 173–193. [Google Scholar]
- Boussau B, Guéguen L, Gouy M.. 2008. Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria. BMC Evol Biol. 8:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Gribaldo S, Zivanovic Y, Confalonieri F, Forterre P.. 2005. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 6(5):R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S, Olsen GJ, Stetter KO, Woese CR.. 1992. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 15(3):352–356. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF.. 2018. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol. 16(10):629–645. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2006. Rooting the tree of life by transition analyses. Biol Direct. 1:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 2010. Deep phylogeny, ancestral groups and the four ages of life. Philos Trans R Soc Lond B Biol Sci. 365(1537):111–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH.. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36(6):1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P.. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311(5765):1283–1287. [DOI] [PubMed] [Google Scholar]
- Coleman GA, Davín AA, Mahendrarajah TA, Szánthó LL, Spang A, Hugenholtz P, Szöllősi GJ, Williams TA.. 2021. A rooted phylogeny resolves early bacterial evolution. Science 372(6542):eabe0511. [DOI] [PubMed] [Google Scholar]
- Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P.. 2011. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One. 6(8):e22099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha VD, Da Cunha V, Gaia M, Gadelle D, Nasir A, Forterre P.. 2017. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 13(6):e1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Cunha V, Gaia M, Nasir A, Forterre P.. 2018. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 14(3):e1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H.. 2005. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 6(5):361–375. [DOI] [PubMed] [Google Scholar]
- Dombrowski N, Lee J-H, Williams TA, Offre P, Spang A.. 2019. Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. FEMS Microbiol Lett. 366(2):fnz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284(5423):2124–2128. [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7(10):e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst Biol. 27(4):401–410. [Google Scholar]
- Forterre P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117(1):5–16. [DOI] [PubMed] [Google Scholar]
- Fournier GP, , AndamCP, , Gogarten JP.. 2015. Ancient horizontal gene transfer and the last common ancestors. BMC Evol Biol. 15(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Rosenberg MS, Kumar S.. 2005. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zool B Mol Dev Evol. 304(1):64–74. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Kratzer JT, Randall RN.. 2010. Deep phylogeny–how a tree can help characterize early life on Earth. Cold Spring Harb Perspect Biol. 2(1):a002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gini C. 1912. Variabilita e mutabilita. Studi economicoaguridici delle facoltta di giurizprudenza dell. Universite di Cagliari III Parte II.
- Gouy R, Baurain D, Philippe H.. 2015. Rooting the tree of life: the phylogenetic jury is still out. Philos Trans R Soc Lond B Biol Sci. 370(1678):20140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Philippe H.. 2002. Ancient phylogenetic relationships. Theor Popul Biol. 61(4):391–408. [DOI] [PubMed] [Google Scholar]
- Griffiths E, Gupta RS.. 2004. Signature sequences in diverse proteins provide evidence for the late divergence of the Order Aquificales. Int Microbiol. 7(1):41–52. [PubMed] [Google Scholar]
- He C, Keren R, Whittaker ML, Farag IF, Doudna JA, Cate JHD, Banfield JF.. 2021. Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat Microbiol. 6(3):354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. 2016. A new view of the tree of life. Nat Microbiol. 1:16048. [DOI] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA.. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA. 96(7):3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H.. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22(4):225–231. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk H-P, Göker M.. 2010. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol. 33(4):175–182. [DOI] [PubMed] [Google Scholar]
- Kobert K, Salichos L, Rokas A, Stamatakis A.. 2016. Computing the internode certainty and related measures from partial gene trees. Mol Biol Evol. 33(6):1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM.. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 10(5):504–509. [DOI] [PubMed] [Google Scholar]
- Le SQ, , GascuelO, , Lartillot N.. 2008. Empirical profile mixture models for phylogenetic reconstruction. Bioinformatics. 24(20):2317–2323. [DOI] [PubMed] [Google Scholar]
- Lerat E, Daubin V, Moran NA.. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-proteobacteria. PLoS Biol. 1(1):e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méheust R, Burstein D, Castelle CJ, Banfield JF.. 2019. The distinction of CPR bacteria from other bacteria based on protein family content. Nat Commun. 10(1):4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, , NguyenMAT, , von Haeseler A.. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir A, Kim KM, Da Cunha V, Caetano-Anollés G.. 2016. Arguments reinforcing the three-domain view of diversified cellular life. Archaea 2016:1851865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbo CL, L'Haridon S, Stetter KO, Doolittle WF.. 2001. Phylogenetic analyses of two “Archaeal” genes in Thermotoga maritima reveal multiple transfers between Archaea and bacteria. Mol Biol Evol. 18(3):362–375. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Okada N, Hasegawa M.. 2007. Rooting the eutherian tree: the power and pitfalls of phylogenomics. Genome Biol. 8(9):R199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW.. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2(11):1533–1542. [DOI] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D.. 2014. Rooting the domain Archaea by phylogenomic analysis supports the foundation of the new kingdom Proteoarchaeota. Genome Biol Evol. 7(1):191–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D.. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9(3):e1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Laurent J.. 1998. How good are deep phylogenetic trees? Current Opinion in Genetics & Development. 8(6):616–623. [DOI] [PubMed] [Google Scholar]
- Philippe H, Roure B.. 2011. Difficult phylogenetic questions: more data, maybe; better methods, certainly. BMC Biol. 9:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock DD, Zwickl DJ, McGuire JA, Hillis DM.. 2002. Increased taxon sampling is advantageous for phylogenetic inference. Syst Biol. 51(4):664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, , DehalPS, , Arkin AP.. 2010. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz A, Gatesy J.. 2007. The supermatrix approach to systematics. Trends Ecol Evol. 22(1):34–41. [DOI] [PubMed] [Google Scholar]
- Rajendhran J, Gunasekaran P.. 2011. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res. 166(2):99–110. [DOI] [PubMed] [Google Scholar]
- Raymann K, Brochier-Armanet C, Gribaldo S.. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci USA. 112(21):6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499(7459):431–437. [DOI] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR.. 1981. Comparison of phylogenetic trees. Mathemat Biosci. 53(1–2):131–147. [Google Scholar]
- Rokas A, Carroll SB.. 2005. More genes or more taxa? The relative contribution of gene number and taxon number to phylogenetic accuracy. Mol Biol Evol. 22(5):1337–1344. [DOI] [PubMed] [Google Scholar]
- Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. 2014. The prokaryotes: other major lineages of bacteria and the Archaea. Berlin, Heidelberg: Springer. [Google Scholar]
- Salichos L, Rokas A.. 2013. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497(7449):327–331. [DOI] [PubMed] [Google Scholar]
- Salichos L, Stamatakis A, Rokas A.. 2014. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol Biol Evol. 31(5):1261–1271. [DOI] [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC, Huttenhower C.. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 4:2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J. 27(3):379–423. [Google Scholar]
- Sievers F, Higgins DG.. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27(1):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Gatesy J.. 2016. Biases of tree-independent-character-subsampling methods. Mol Phylogenet Evol. 100:424–443. [DOI] [PubMed] [Google Scholar]
- Simon C. 2020. An evolving view of phylogenetic support. Syst Biol. 0(0):1–8. [DOI] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG.. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521(7551):173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott CM, Bobay L-M.. 2020. Impact of homologous recombination on core genome phylogenies. BMC Genomics. 21(1):829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, et al. 2013. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods. 10(12):1196–1199. [DOI] [PubMed] [Google Scholar]
- Taib N, Megrian D, Witwinowski J, Adam P, Poppleton D, Borrel G, Beloin C, Gribaldo S.. 2020. Genome-wide analysis of the Firmicutes illuminates the diderm/monoderm transition. Nat Ecol Evol. 4(12):1661–1672. [DOI] [PubMed] [Google Scholar]
- Wang H-C, Minh BQ, Susko E, Roger AJ.. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst Biol. 67(2):216–235. [DOI] [PubMed] [Google Scholar]
- Werner F, Grohmann D.. 2011. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 9(2):85–98. [DOI] [PubMed] [Google Scholar]
- Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM.. 2020. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 4(1):138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM.. 2012. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 279(1749):4870–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Szöllősi GJ, Spang A, Foster PG, Heaps SE, Boussau B, Ettema TJG, Embley TM.. 2017. Integrative modeling of gene and genome evolution roots the archaeal tree of life. Proc Natl Acad Sci USA. 114(23):E4602–E4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. 1998. The universal ancestor. Proc Natl Acad Sci U S A. 95(12):6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiol Rev. 51(2):221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE.. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 74(11):5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML.. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 87(12):4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Aravind L, Grishin NV, Koonin EV.. 1999. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 9(8):689–710. [PubMed] [Google Scholar]
- Wu M, Eisen JA.. 2008. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9(10):R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AD, Gillung JP.. 2020. Phylogenomics — principles, opportunities and pitfalls of big‐data phylogenetics. Syst Entomol. 45(2):225–247. [Google Scholar]
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541(7637):353–358. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT, Nelson KE, Nesbø CL, Doolittle WF, Gogarten JP, et al. 2009. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci USA. 106(14):5865–5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, , MaiU, , PfeifferW, , JanssenS, , AsnicarF, , SandersJG, , Belda-FerreP, , Al-GhalithGA, , KopylovaE, , McDonald D, . et al. 2019. Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea. Nat Commun. 10(1):5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM.. 2002. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 51(4):588–598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw alignments, iTOL datasets, and treefile files can be accessed through Figshare (doi.org/10.7294/16543164.v1).