Abstract
Human centromeres are mainly composed of alpha satellite DNA hierarchically organized as higher-order repeats (HORs). Alpha satellite dynamics is shown by sequence homogenization in centromeric arrays and by its transfer to other centromeric locations, for example, during the maturation of new centromeres. We identified during prenatal aneuploidy diagnosis by fluorescent in situ hybridization a de novo insertion of alpha satellite DNA from the centromere of chromosome 18 (D18Z1) into cytoband 15q26. Although bound by CENP-B, this locus did not acquire centromeric functionality as demonstrated by the lack of constriction and the absence of CENP-A binding. The insertion was associated with a 2.8-kbp deletion and likely occurred in the paternal germline. The site was enriched in long terminal repeats and located ∼10 Mbp from the location where a centromere was ancestrally seeded and became inactive in the common ancestor of humans and apes 20–25 million years ago. Long-read mapping to the T2T-CHM13 human genome assembly revealed that the insertion derives from a specific region of chromosome 18 centromeric 12-mer HOR array in which the monomer size follows a regular pattern. The rearrangement did not directly disrupt any gene or predicted regulatory element and did not alter the methylation status of the surrounding region, consistent with the absence of phenotypic consequences in the carrier. This case demonstrates a likely rare but new class of structural variation that we name “alpha satellite insertion.” It also expands our knowledge on alphoid DNA dynamics and conveys the possibility that alphoid arrays can relocate near vestigial centromeric sites.
Keywords: alpha satellite, structural variation, ancestral centromere
Introduction
Alpha satellite is a class of highly repetitive DNA defined by a group of related, highly divergent AT-rich repeats or “monomers,” each approximately 171 bp in length. Alpha satellite, also named alphoid DNA, comprises up to 10% of the human genome and is mostly found tandemly repeated within constitutive heterochromatin at centromeres and pericentromeric regions. At centromeric regions, satellite monomers are hierarchically organized into larger repeating units, in which a defined number of monomers have been homogenized. These units, which are named “higher-order repeats” (HORs), are tandemly arranged into chromosome-specific, megabase-sized satellite arrays with limited nucleotide differences between repeat copies (Willard and Waye 1987; Durfy and Willard 1989; Schueler et al. 2001; McNulty and Sullivan 2018; Miga et al. 2020; Sullivan and Sullivan 2020).
The centromere is the chromosomal locus where sister chromatids attach and the kinetochore is assembled, which is essential for proper chromosome segregation during cell division. While alpha satellite DNA constitutes the sequence of all mature centromeres, it is not sufficient nor necessary for centromere identity. This is demonstrated by dicentric chromosomes that assemble the kinetochore at only one of two alpha-satellite regions (Earnshaw and Migeon 1985) and analphoid chromosomes that possess fully functional centromeres (Voullaire et al. 1993). Centromere function appears to be epigenetically established and maintained by local enrichment of the CENP-A histone H3 variant within nucleosomes rather than the presence of alphoid DNA (Palmer et al. 1991; Karpen and Allshire 1997; Panchenko and Black 2009; McKinley and Cheeseman 2016). This function can be inactivated at an original site and moved to a new position along the chromosome (Montefalcone et al. 1999). It is similarly turned off after a chromosomal fusion to ensure the stability of the derived dicentric chromosome. These events determine the emergence of evolutionary new centromeres and the appearance of recognizable genomic regions where the centromere used to be positioned in the past (Amor and Choo 2002; Rocchi et al. 2009). Insights into the molecular steps of centromere repositioning from the birth of a new centromere to its maturity were uncovered by studying fly, primate, and equid chromosomes (Marshall et al. 2008; Piras et al. 2010). These analyses showed that new centromeres are first epigenetically specified and then mature by acquiring the satellite DNA array, in some cases going through intermediate configurations bearing DNA amplification (Kalitsis and Choo 2012; Nergadze et al. 2018).
Besides the main pericentromeric and centromeric locations, smaller regions of alpha satellite DNA are located in the human genome >5 Mbp from the centromeres, with around 100 blocks annotated in the reference by the RepeatMasker program (Rudd and Willard 2004; Feliciello et al. 2020). For example, three large blocks, 11-, 8-, and 13-kbp long, are located within cytoband 2q21 with SVA (SINE/VNTR/Alu) and LINE elements intervening between them. These alphoid sequences are the relics of an ancestral centromere that became inactive ∼5 Mya (million years ago) after the fusion of two ancestral chromosomes in the human lineage compared to big apes (IJdo et al. 1991; Avarello et al. 1992; Baldini et al. 1993; Chiatante et al. 2017).
Here, we report an individual with a de novo insertion of an alpha satellite DNA array from the centromere of chromosome 18 into chromosome 15q26, the first observation of insertion of satellite DNA array outside centromeric and pericentromeric regions of the human genome that we are aware of. This case brings to light a probably rare and new class of structural variation and expands our knowledge on the spread and dynamics of alpha satellite.
Results
Prenatal, Postnatal, and Family Investigations
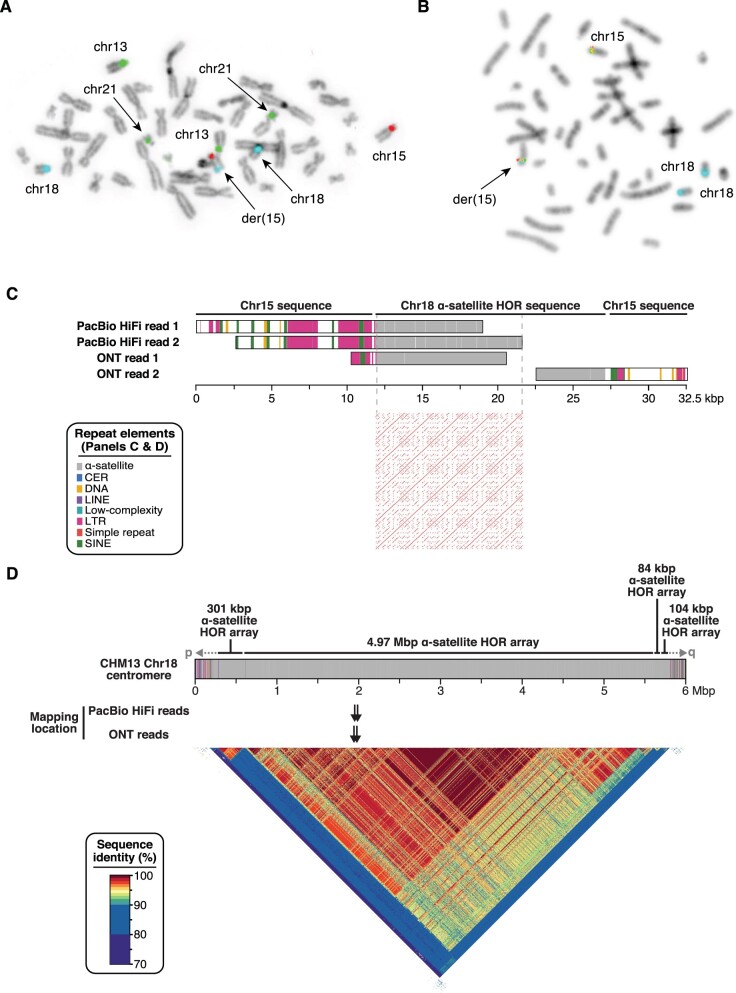
Amniocentesis was performed at 15 weeks’ gestation in a 35-year-old gravida woman for her sixth pregnancy. She already had two healthy children, one miscarriage, and two pregnancies terminated due to fetal trisomy 21. Interphase fluorescent in situ hybridization (FISH) on uncultured amniocytes with probes for main aneuploidies showed the presence of three signals for the alpha satellite DNA probe of chromosome 18 (D18Z1) in all cells (150/150) and three signals for chromosome 21-specific probe in 29 out of 121 cells (24%), suggesting a trisomy 18 and a mosaic trisomy 21. Karyotyping of cultured cells confirmed the presence of the mosaic trisomy 21 at 19% (12/62 cells) but showed the presence of two normal chromosomes 18. Metaphase FISH on cultured cells revealed the aberrant hybridization of the D18Z1 probe at chromosome 15q26 (fig. 1A andsupplementary fig. S1, Supplementary Material online). Chromosomal microarray did not show any imbalances, except the mosaic trisomy 21 (13%). FISH analysis of both parents showed that the alphoid DNA insertion was de novo. Pregnancy sonographic follow-up was normal. The proband, a healthy male baby, was born at term with normal birth parameters. Postnatal karyotype and FISH confirmed the mosaic trisomy 21 (6/33 cells; 18%) and the presence of the insertion of chromosome 18 alpha satellite on the long arm of a chromosome 15. At 1-year old, growth clinical examination (weight 10.6 kg, +1 standard deviation [SD]; height 75 cm, +1 SD; occipito-frontal circumference 46.5 cm, +1 SD) and psychomotor development were normal, consistent with low level mosaic trisomy 21, suggesting that the alpha satellite insertion had no phenotypic consequences.
Fig. 1.
D18Z1 alpha satellite de novo insertion. (A) FISH results of cultured amniocytes using alpha satellite DNA probes of chromosomes 15 (D15Z1, Texas-Red), 13/21 (D13/21Z1, green), and 18 (D18Z1, aqua), showing the hybridization of D18Z1 at 15q26. (B) FISH results of cultured amniocytes using the 15q25 BAC probes RP11-635O8 (red) and RP11-752G15 (green) flanking the ancestral centromere and the D18Z1 (aqua) probe. (C) Read length, repeat composition (color code in inset), and mapping location of the four selected HiFi and ONT reads (top). Dot plot (window size 20) of the 10-kb alpha satellite sequence from the centromere of chromosome 18 showing its tandem repetitive structure (bottom). (D) Schematic representation of the CHM13-T2T chromosome 18 centromere with its repeat composition (top). A heatmap representation of sequence identity over the region is presented below. The mapping location of the PacBio HiFi and ONT reads is pinpointed by black arrows.
Structural Characterization of the Rearrangement
To characterize the alphoid insertion at the sequence level, we performed whole genome sequencing (WGS) of the proband using the short-read Illumina platform. We first analyzed these data using a routine clinical analysis pipeline that did not identify any structural variant at chromosome 15q26. We then followed a customized approach, mapping reads to a library made up of the entire chromosome 15 and chromosome 18 centromeric alpha satellite DNA sequences. We isolated high-quality discordant paired reads mapped to both sequences, as well as chimeric reads anchored to chromosome 15 and containing alpha satellite DNA. These reads allowed us to define the positions of the proximal and distal breakpoints of the insertion at chr15:92359068 and chr15:92361920 (GRCh38), respectively. These coordinates, both subsequently validated by PCR, revealed the deletion of a 2,851-bp segment that was replaced by the insertion. We noted that the target site was ∼10-Mbp distal from the position where an ancestral centromere was seeded and was shown to be active ∼25 Mya in the common ancestor of Old World monkeys and apes and was then inactivated sometime between 20 and 25 Mya in the common ancestor of the Hominoids (lesser apes, great apes, and humans) (Ventura et al. 2003). This was further confirmed by the cohybridization of the D18Z1 probe with two BAC probes flanking the ancestral centromere locus (RP11-752G15 and RP11-635O8) (Giannuzzi et al. 2013). This experiment showed, at metaphase resolution, that the satellite probe signal colocalized with both BAC probes on the derivative chromosome 15 (fig. 1B). The intensity and size of the FISH signal was similar to the ones of the BAC probes, suggesting that the inserted satellite DNA was ∼50–300-kbp long.
We then sought to better characterize the rearrangement by generating long-read sequence information. We employed two technologies, ONT (Oxford Nanopore Technologies) with selective sampling via Read Until (Loose et al. 2016), targeting 50 kb of sequence on either side of the insertion, and PacBio HiFi sequencing. We sequenced the proband (∼11.5× coverage at the targeted region), father (∼20.1×), and mother (∼19.8×) using readfish (Payne et al. 2021; Miller et al. 2021) on an ONT GridION, and the proband’s genome on one PacBio SMRT cell (∼6.5× coverage). We confirmed the insertion breakpoints and the 2.8-kbp deletion but were unable to assemble a contiguous sequence spanning the entire insertion. To determine which parental chromosome the event occurred on, we phased the proband, father, and mother’s ONT reads and searched for diagnostic single-nucleotide variants that differed between the maternal and paternal haplotypes. The proband is hemizygous for two maternal variants mapping within the deleted region while the father is homozygous for the alternative allele. Conversely, the proband harbored one paternal variant on the haplotype with the insertion that is absent in his mother. This demonstrated that the rearrangement occurred on the paternal chromosome. Analysis of the junctions showed that, besides the aforementioned deletion, no further rearrangements, such as a target site duplication, occurred at the boundaries. At the proximal junction, a short sequence stretch of four nucleotides (CAAA) was identified that could not uniquely be assigned to the chromosome 15 or the satellite DNA. However, due to its small size, it is unlikely that this stretch of homologous sequence had a role in the rearrangement mechanism, particularly in the determination of the target site.
We analyzed the content of interspersed repeats in 5-kb segments upstream and downstream of the rearrangement breakpoints as well as in the deleted segment on chromosome 15 sequence. These segments were enriched for content in endogenous retrovirus long terminal repeats (LTRs) when compared to the human genome average, as assessed by simulation for the entire 13-kb segment (4.34-fold, P = 0.035, table 1).
Table 1.
Content in interspersed repeat elements of the rearranged target site on chromosome 15.
| Repeat elements | Sequence upstream of the insertion (5 kb) (%) | Deletion (2,851 bp) (%) | Sequence downstream of the insertion (5 kb) (%) | Entire region (%) | Human genome average (%) | E, P ± SE |
|---|---|---|---|---|---|---|
| SINEs | 9 | 0 | 12 | 8 | 12 | 0.65, 0.57 ± 0.005 |
| LINEs | 0 | 0 | 0 | 0 | 19 | 0, 1 |
| LTR elements | 62 | 32 | 13 | 36 | 8 | 4.34, 0.035 ± 0.002 |
| DNA elements | 0 | 0 | 9 | 4 | 3 | 1.21, 0.3 ± 0.005 |
The “E” value is the enrichment coefficient that was calculated by dividing the observed value by the mean of 10,000 genome-wide permutations (human genome average).
Structural Characterization of the Alpha Satellite DNA Insertion
While we were unable to assemble the full sequence of the insertion, we investigated its structural properties by identifying reads with the longest content in alpha satellite DNA and unequivocally derived from this site, that is, chimeric reads anchored to chromosome 15 sequence on either side of the insertion, spanning one breakpoint, and containing chromosome 18 centromeric alpha satellite sequences.
We selected two PacBio HiFi reads (PacBio HiFi read 1 and PacBio HiFi read 2) containing 7,199 and 9,821 bp of satellite DNA and having 99.95% and 99.48% estimated accuracy, respectively, both transitioning over the proximal junction. We also selected an ONT read with 8,618 bp of satellite DNA at the proximal junction (ONT read 1) and an ONT read with 4,583 bp of satellite DNA at the distal junction (ONT read 2) (fig. 1C). Best alignments to the human genome reference (GRCh38) of alpha satellite segments from these four sequences showed identity with centromere reference models of chromosome 18 (Miga et al. 2014; Rosenbloom et al. 2015). Alignments to the CHM13-T2T (telomere-to-telomere) genome (Miga et al. 2020; Logsdon et al. 2021; Nurk et al. 2021) resulted in unique locations for each read and pointed the origin of the insertion to a precise 10-kb region in the centromere of chromosome 18 (chr18:17500488–17510699) (fig. 1D). HiFi reads showed 99.6% and 99.1% identity with this region, with the remaining divergence not explained by sequencing errors (∼0.4%) probably reflecting interindividual differences in centromeric sequences. ONT reads showed lower identity values with the same region (94.5% and 94.4%), mainly due to errors in their sequence. As the estimated size of the inserted segment (order of hundreds kbp) is bigger than the size of the corresponding interval within chromosome 18 centromeric sequence, we hypothesize that this region is variable among humans and likely expanded in the proband or alternatively in his paternal lineage. Overall, these results confirmed that the insertion originated from chromosome 18 centromeric DNA and suggest that the CHM13 and our proband’s centromeres are structurally different in their sequence and size.
We next analyzed the repetitive structure of the satellite insertion using the corresponding T2T chromosome 18 centromeric sequence with ∼99.99% accuracy. As chromosome 18 centromere is composed of two alpha satellite families, family I (D18Z1) and family II (D18Z2), both belonging to the suprachromosomal family 2 (SF2), whose arrays have a dimeric structure based on D1 and D2 monomers (Alexandrov et al. 1991), we assessed the similarity with deposited sequences representing both families. Local pairwise alignments showed 98.8% and 81.7% identity, respectively, with D18Z1 (M65181.1) and D18Z2 (M38466.1) sequences. Similar results, that is, higher similarity with family I sequence, were obtained for both PacBio HiFi reads and the ONT read 2 transitioning over the distal breakpoint. These results indicate a closer relationship of the inserted satellite DNA to the D18Z1 family.
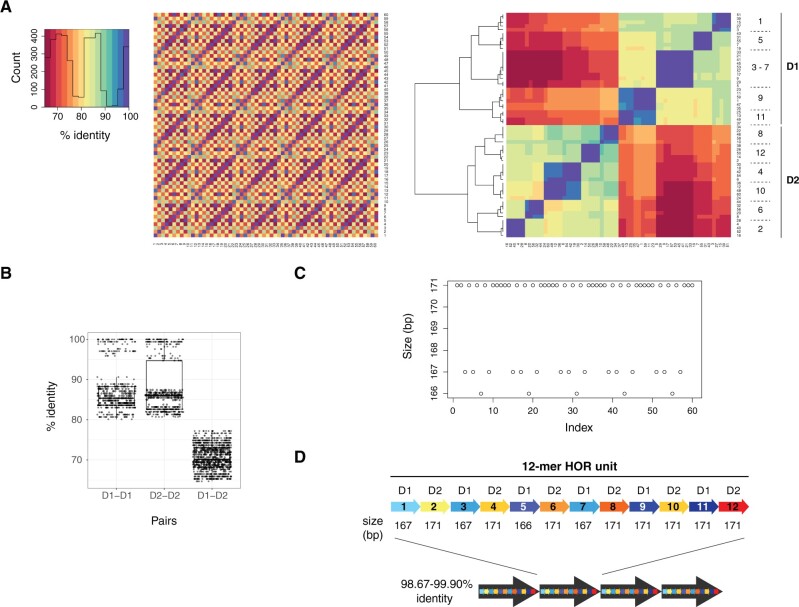
The T2T centromeric sequence showed a higher density of matches every ∼2,000 bp when assessed using the re-DOT-able tool (https://www.bioinformatics.babraham.ac.uk/projects/redotable/) (fig. 1C, bottom panel). To further assess this periodicity, we extracted 60 monomers, built a multiple sequence alignment, and visualized all pairwise identity percentages by creating two heatmaps. The first one shows monomers ordered according to their position in the array, while the second heatmap depicts monomers ordered according to the dendrogram determined by the hierarchical clustering of identity percentages (fig. 2A). In the dendrogram-based heatmap, the monomers cluster into two main clades formed by D1 and D2 monomers, as expected from the dimeric structure of the D18Z1 array. D1 and D2 monomers further group into 11 clades in agreement with their organization in a HOR unit of 12 monomers, with D1 monomers at positions 3 and 7 that are homogenized and form a single clade. D1 monomers at positions 9 and 11 and D2 monomers at positions 4 and 10 are also partly homogenized (fig. 2A, right panel). These results are consistent with a 12-mer HOR structure, matching the known organization of the D18Z1 satellite array (McNulty and Sullivan 2018). D1–D1/D2–D2 sequence identity ranges from 80.12% to 100% (median 85.96%); D1–D2 identity ranges from 64.67% to 77.19% (median 70.18%) (fig. 2B). While most monomers have a size of 171 bp, some D1 monomers are 166- or 167-bp long. The monomer size distribution is not random but follows a precise pattern in the HOR unit, showing another feature of the HOR hierarchical organization (fig. 2C) (Wu and Manuelidis 1980). Lastly, we grouped every 12 monomers into ∼2-kb units and obtained four HOR repeats with 98.67–99.90% pairwise sequence identity (fig. 2D).
Fig. 2.
Organization of the alpha satellite array. (A) Heatmaps of identity percentages between the 60 alpha satellite monomers derived from the T2T chr18:17500488–17510699 sequence, with monomers ordered either according to their position in the sequence (left) or as determined by clustering (right). In the latter, the position (1–12) in the 12-mer unit and type (D1 or D2) of the monomers are shown. (B) Boxplots of identity percentages between D1–D1, D2–D2, and D1–D2 monomer pairs. (C) Plot of monomer size with monomers ordered according to their position in the original sequence. (D) Schematic representation of the 12-mer HOR units.
Functional Profiling of the Rearranged Site
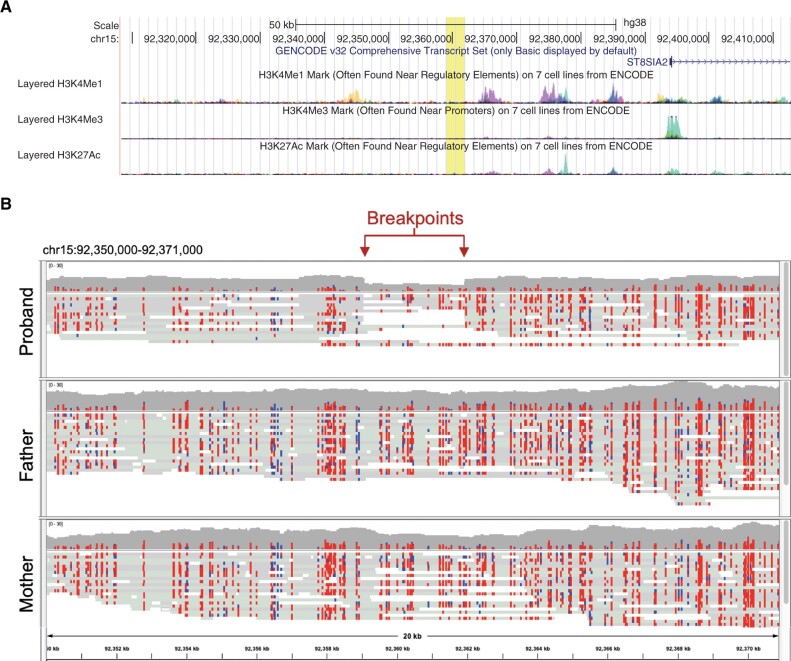
To assess whether this structural change is likely to have functional impact, we examined gene annotation (GENCODE v32) at the insertion breakpoints as well as in the deleted region. We find that the rearrangement did not directly disrupt any gene, with the closest one (ST8SIA2) annotated 32 kb distally (fig. 3A). We then evaluated whether the rearrangement affected other functional elements, such as regulatory DNA. To this end, we leveraged publicly available data from the ENCODE consortium of chromatin activity measured by chromatin immunoprecipitation sequencing (ChIP-seq) for three histone modifications, that is, methylated histone 3 at lysine 4 (H3K4me1), tri-methylated histone 3 at lysine 4 (H3K4me3), and acetylated histone 3 at lysine 27 (H3K27ac), on seven cell lines. These epigenetic marks are associated with poised enhancers (H3K4me1), promoters (H3K4me3), and active enhancers (H3K27ac). Neither the deleted segment nor the breakpoints overlapped any of these chromatin features, suggesting that the rearrangement did not disrupt a regulatory element (fig. 3A).
Fig. 3.
Functional profiling of the rearrangement site. (A) UCSC view of the 100-kbp region surrounding the rearrangement at 15q26.1. The deleted region is highlighted in yellow, with deletion extremes corresponding to the satellite insertion positions. The GENCODE v32 and ENCODE regulation (H3K4me1, H3K4me3, and H3K27ac) tracks are shown (hg38). No gene and no enrichment of epigenetic marks found near regulatory elements are annotated in the deleted region. The closest gene, ST8SIA2, is mapped 32 kb distally. (B) Methylation pattern of the insertion site in the family trio obtained from the ONT selective sequencing. Methylated (red) and unmethylated (blue) CpGs are shown. The methylation profiles are similar among the family trio.
Next, we assessed whether the insertion of centromeric satellite DNA, which comes from a heterochromatic locus, modifies the epigenetic status of the 15q26 target region. We leveraged CpG methylation data of the 20-kb genomic segment surrounding the insertion site using the ONT data of the proband and his parents. Cytosine methylation is an epigenetic modification often found in CpG dinucleotides that contributes to the formation of heterochromatic regions and leads to transcriptional modulation, in particular silencing. Comparison of the proband mutated allele with unrearranged ones (i.e., his maternal allele and the four alleles of his parents) revealed no major difference in the methylation patterns, indicating that the satellite insertion did not alter the methylation status of the surrounding region (fig. 3B). The absence of functional elements (gene or likely regulatory element) at the site and the maintenance of the methylation profile of the broader region suggest that the rearrangement itself has had no functional consequences. This is in line with the absence of clinical features in the proband that could not be explained by his trisomy 21 mosaicism.
Immuno-FISH with Anti-CENP-A and CENP-B Antibodies
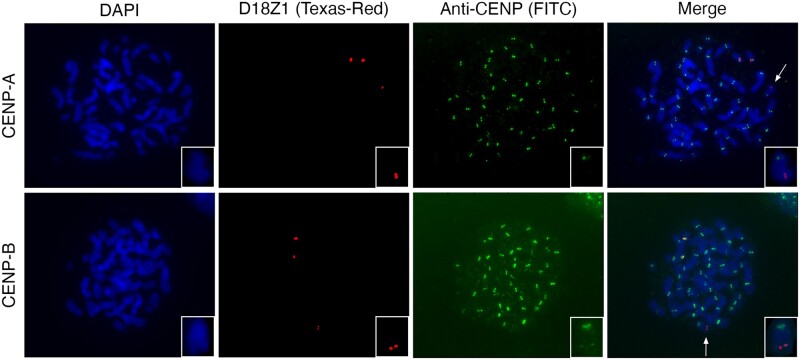
Cytogenetic evaluation of the derived chromosome 15 revealed no chromosomal constriction at the position where the satellite DNA sequence was inserted, suggesting that this site did not acquire properties of a functional centromere. To further demonstrate this lack of epigenetically defined centromeric function, we performed an immuno-FISH experiment with an antibody against the CENP-A protein. We observe no colocalization of the D18Z1 probe and CENP-A staining at the satellite insertion locus on the derivative chromosome 15 (fig. 4). We also assessed by immuno-FISH the binding of the CENP-B box by the CENP-B protein. In 20 out of 25 mitoses, we observe a faint pattern of staining of the CENP-B antibody corresponding to the satellite insertion, whereas in the remaining five we observed no signal (fig. 4). Such faint signals may derive either from the smaller size of the satellite insertion compared to a centromeric satellite array or to a weaker binding of the CENP-B protein. Nevertheless, these results suggest that CENP-B proteins recognize and bind the CENP-B box on the satellite monomers of the inserted sequence. Although CENP-B is not necessary and sufficient to confer centromeric function, it was shown that it creates epigenetic chromatin states permissive for CENP-A or heterochromatin assembly (Otake et al. 2020).
Fig. 4.
CENP-A and CENP-B immuno-FISH. Cohybridization of the D18Z1 probe (red) with antibodies against CENP-A (top) and CENP-B (bottom) proteins (green) on chromosome metaphases from the proband. The arrows point at the derivative chromosome 15 that is also shown in larger magnification in the insets.
Discussion
During routine prenatal testing for aneuploidy by FISH, we serendipitously identified an individual carrying a de novo insertion of alpha satellite DNA from the centromere of chromosome 18 into cytoband 15q26 (fig. 5A). Long-read sequencing and alignment to the CHM13-T2T genome showed that this segment originates from a precise location in the main chromosome 18 centromeric HOR array. Analysis of the repetitive structure showed novel features of chromosome 18 12-mer HOR, such as the presence of a regular pattern in monomer size, homogenization of D1 monomers at positions 3 and 7, partial homogenization of D1 monomers at positions 9 and 11, and partial homogenization of D2 monomers at positions 4 and 10.
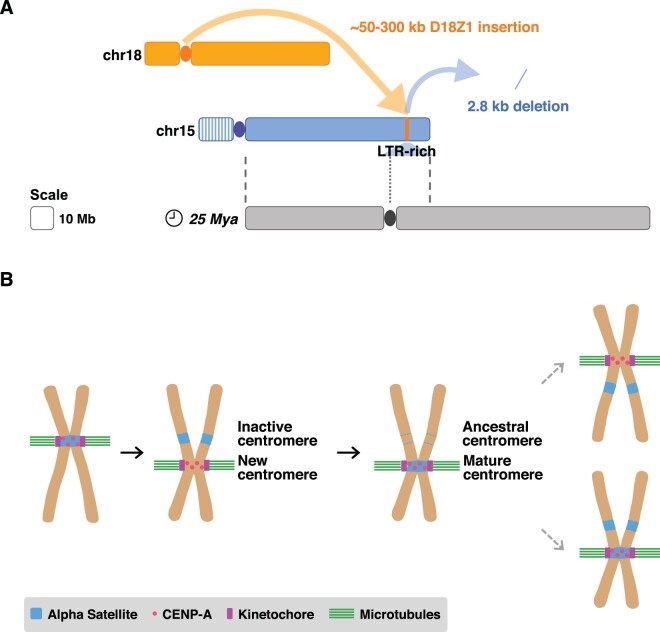
Fig. 5.
(A) Schematic overview of the rearrangement. An alphoid array ∼50–300 kbp in size from the centromere of chromosome 18 inserted into an LTR-rich region of chromosome 15q26, ∼10 Mbp distally from the site where an ancestral centromere was seeded ∼25 Mya. This insertion was coupled with a 2.8-kbp deletion. Dashed lines pinpoint the boundaries of the synteny between chromosome 15 and the ancestral submetacentric chromosome; the dotted line indicates the position of the ancestral centromere. (B) Possible inferred model of alphoid DNA dynamics relative to centromere repositioning. Following a centromere repositioning event, the new centromere is epigenetically specified by CENP-A binding and subsequently acquires alphoid DNA. It is possible that not only centromeric function but also the presence of alphoid DNA can be resurrected at ancestral centromeric sites.
Our report expands our knowledge on alpha satellite dynamics and proposes a new class of structural variation that we call “alpha satellite insertion” (ASI). While our study describes an alphoid insertion into a noncentromeric/pericentromeric region, several prenatal FISH diagnostic reports describe the crosshybridization of chromosome-specific centromeric alpha satellite probes to centromeric or pericentromeric regions of nontargeted chromosomes, that is, the centromeres of chromosomes 19 and 22, the heterochromatin of chromosomes 1 and 9, and the pericentromeric region of chromosome 2 (Thangavelu et al. 1998; Winsor et al. 1999; Wei et al. 2007; Musilova et al. 2008; Collin et al. 2009).
While the presence of some alphoid blocks outside centromeric and pericentromeric regions in the human genome reference (Rudd and Willard 2004) can be explained by the evolutionary history of the locus and past presence of a centromere, the existence of the others could result from fixed satellite insertion events. The maturation process of new centromeres that switch from satellite-free to satellite-rich regions is telltale of the dynamism of alphoid DNA (Kalitsis and Choo 2012; Nergadze et al. 2018). Novel localizations of alphoid DNA were reported in the white-cheeked gibbon, a lesser ape with an extensively rearranged karyotype when compared to the ancestral primate karyotype. In this species, alpha satellite DNA is found not only at centromeres but also at telomeres and interstitial positions corresponding to some evolutionary breakpoints (Cellamare et al. 2009).
Although the mechanisms governing alphoid insertion are not well understood, they are likely to involve a nonreciprocal transfer of DNA from a mature centromere via recombination, transposable elements, and/or rolling circle replication and reinsertion (Plohl et al. 2008; Kalitsis and Choo 2012). Our structural characterization of the rearrangement provides some insights on its mechanism of origin. The coordinated deletion suggests the involvement of double-strand breakage of DNA, as inferred for duplication events (Cantsilieris et al. 2020). It may be noteworthy that we also identified an enrichment of LTR elements in the long-range acceptor site. LTR retrotransposon activity is currently very limited or fully absent in humans (IHGSC 2001) and therefore is unlikely to have directly driven the rearrangement. Such repeat-rich regions have been noted to be deleted as part of the duplication events associated with the new insertion of large (>100 kbp) blocks of segmental duplication (Johnson et al. 2006; Cantsilieris et al. 2020). Similarly, such coordinated deletions often (but not always) occur in gene-poor regions of the genome minimizing functional impacts of such massive new insertions on the fitness of the zygote/fetus.
The poor identification of alphoid DNA insertions outside centromeric regions until now could be linked either to the fact that they are extremely rare and/or because current sequencing-based methodologies and analytical approaches aimed at genotyping structural variants are opaque to these events due to their size and highly repetitive nature. Likewise, as only centromeric probes of chromosomes 18, X, and Y are routinely used to screen for aneuploidies prenatally, the ASI of other centromeric satellites and ASI smaller than the standard FISH resolution (∼10 kb) could not be serendipitously found. While a designated analysis was performed to detect mobile element insertions (MEIs, including insertions of Alu, L1, and SVA) in 2,504 human genomes (Sudmant et al. 2015), the insertion of satellite DNA has not been specifically assessed in diverse human genomes. Similarly, our standard WGS diagnostic pipeline failed to identify the variant we describe.
Finally, the ASI location at 15q26 is noteworthy as a centromere resided at chromosome 15q25 in our past, ∼10 Mbp away from the insertion site, and became inactive sometime between 20 and 25 Mya in the common ancestor of the ape lineage (Ventura et al. 2003; Giannuzzi et al. 2013). This feature raises the intriguing possibility that the alphoid array did not move to a random repeat-rich location in the genome, but instead revisited an evolutionary favored site mapping close to an ancestral centromere. Such a scenario together with the aforementioned prenatal reports proposes that alphoid DNA might preferentially move to other extant or past centromeric locations (fig. 5B). It also recalls previous studies suggesting that certain regions of the genome may have a potential to host centromeric function possibly because they are gene poor and/or enriched in high-copy or low-copy repeats. As observed for the satellite insertion reported here, several analphoid clinical new centromeres seeded ∼1–14 Mb from an ancestral centromeric site or a region that is orthologous to evolutionary new centromeres in other primate lineages (Ventura et al. 2003; Ventura et al. 2004; Cardone et al. 2006; Capozzi et al. 2008, 2009). This suggests that centromeric function and satellite array evolution may be restricted to region rather than precise chromosomal location.
The variant we identified hints that an alternative route to centromere formation might exist, where the region first acquires the satellite array and then the epigenetically defined centromeric function emerges. Support for the latter comes from the observation that introduction of alpha satellite arrays in human cells can result in the formation of functional new centromeres (Harrington et al. 1997; Ebersole et al. 2000).
Lastly, this case further highlights the risk of identifying false-positive aneuploidies of chromosomes 18, X, and Y when depending solely on centromeric satellite probes in rapid interphase FISH. Thus, it is critically important to follow up and confirm them by karyotyping.
Materials and Methods
Short-Read Sequencing and Data Analysis
We extracted genomic DNA from cultured amniocytes of the proband using QIAamp DNA mini kit (Qiagen, Hilden, Germany). We performed 150-bp paired-end WGS using the short-read Illumina platform. We aligned reads to the hg38 version of the human genome using BWA-MEM version 0.7.10 (Li and Durbin 2009), run the BreakDancer version 1.4.5 (Chen et al. 2009) and ERDS version 1.1 (Zhu et al. 2012), and visually inspected the 15q24-26 region using the Integrative Genomics Viewer (IGV) tool. As we identified no structural variant, we realigned reads to a custom library made of chromosome 15 sequence (hg38) and a deposited sequence of alpha satellite family 1 of chromosome 18 (M65181.1) (Alexandrov et al. 1991) using BWA version 0.7.17. To identify read pairs mapping at the insertion breakpoints, we selected discordant pairs with one end mapping on chromosome 15 and the other one on the satellite sequence and mapping quality (MAPQ) > 0. We removed soft and hard clipped reads and those mapping at the pericentromeric region of chromosome 15. We next identified chimeric reads spanning the breakpoints among the soft clipped reads using the IGV tool (Robinson et al. 2011).
Long-Read Sequencing and Data Analysis
We isolated peripheral blood mononuclear cells from the blood of the proband and both parents. We extracted DNA from approximately 1–2 million cells of actively growing culture by first pelleting the cells and resuspending them in 1.0 ml Cell Lysis Solution (Qiagen). The samples were incubated with RNase A solution at 37 °C for 40 min. Protein Precipitation Solution (Qiagen) was added at 0.33× and mixed. After a 10-min incubation on ice, the precipitate was pelleted (3 min, 15,000 rpm, 4 °C). The supernatant was transferred to new tubes, and DNA was precipitated with an equal volume of isopropanol. The DNA was pelleted (2 min, 15,000 rpm, 4 °C) and the pellet was washed three times with 70% EtOH. The clean DNA was rehydrated with DNA Hydration Solution (Qiagen) and left for 2 days to resuspend.
We generated a PacBio HiFi library from the proband’s genomic DNA using g-TUBE shearing (Covaris) and the Express Library Prep Kit v2 (PacBio), size selecting on the SageELF platform (Sage Science) to give a tight fraction of around 23 kbp by FEMTO Pulse analysis (Agilent). The library was sequenced on one SMRT Cell 8M using v2 chemistry, and we obtained 20.5 Gbp of HiFi reads with the mean length of 20.9 kbp and the median quality of Q27. We assembled the data with HiCanu (Nurk et al. 2020) and Hifiasm (Cheng et al. 2021) and aligned reads to the GRCh38 (hg38) reference genome using pbmm2 (https://github.com/PacificBiosciences/pbmm2, last accessed March 4, 2021).
Adaptive sampling was performed on an ONT GridION (one flow cell per sample) using readfish (Payne et al. 2021). For each sample, 1.5 μg of DNA was used to prepare an LSK-109 library according to the manufactures protocol. DNA was sheared in a Covaris g-TUBE at 6k rpm for 2 min. The region targeted was chr15:92309068–92411920 (hg38 coordinates). ONT FAST5 files were base-called using guppy 4.0.11 using the high-accuracy model. FASTQ files were pooled and aligned to hg38.no_alt.fa using both minimap2 (Li 2018) and ngmlr (Sedlazeck et al. 2018). We identified reads spanning the breakpoints (located at chr15:92359068 and chr15:92361920) by manual inspection of the 15q26 read alignments in IGV v2.4.16 (Robinson et al. 2011). We called and phased variants using Longshot (Edge and Bansal 2019) and called CpG methylation using Nanopolish (Simpson et al. 2017). Selected PacBio and ONT reads were aligned to the CHM13-T2T genome using pbmm2 and minimap2, respectively.
Analysis of Repeat Element Content
We assessed the content in repeat elements in the deleted segment and in the 5-kb segments upstream and downstream the insertion breakpoints by using the annotation of the GRCh38 RepeatMasker track (Smit et al. 2013–2015). The null distributions were generated by performing 10,000 permutations of the entire 12,851-bp segment, excluding gaps and centromeres, by using BEDTools version v2.30.0 (Quinlan and Hall 2010). R v4.0.3 (R Core Team 2020) was used to compute empirical P values. Standard error (SE) was estimated using the formula SE = sqrt(P × (1 − P)/10,000).
Satellite Monomer and HOR Analysis
Dot plots were created using the re-DOT-able tool (https://www.bioinformatics.babraham.ac.uk/projects/redotable/, last accessed February 2, 2021). We extracted satellite monomers by blast alignment (Altschul et al. 1990) with D1 monomer sequence (AJ130751.1). We performed multiple sequence alignments of monomers using Muscle (Edgar 2004) with default options. We created heatmaps and plots using the gplots v3.1.0 (https://CRAN.R-project.org/package=gplots) and ggplot2 v.2.2.1 (Wickham 2009) packages in the R software environment (R Core Team 2020).
FISH and Immuno-FISH
FISH on uncultured amniocytes was performed with the Aquarius FAST FISH Prenatal kit (Cytocell, Cambridge, UK) (DXZ1, DYZ3, D18Z1, RB1, DYRK1A probes) according to manufacturer’s instructions. Metaphase spreads were prepared from amniotic fluid cells and lymphocytes according to standard procedures. FISH was further performed using BAC probes localized in 15q25.2, RP11-752G15 (FITC) (chr15:82627211–82802988, hg38) and RP11-635O8 (chr15:82023617–82178139) (TRITC) (RainbowFish, Empire Genomics, Buffalo, NY, USA) and alpha-satellites probes for chromosomes 15 (D15Z1, Texas-Red), 18 (D18Z1, Aqua), and 13/21 (D13/21Z1, Green) (Cytocell).
Immuno-FISH was performed on lymphoblastoid cells from the patient. Metaphase cell spreads were prepared according to a protocol adapted from Jeppesen (Jeppesen 2000). Briefly, lymphoblastoid cells were harvested after 44-h culture and incubated at 37 °C with colchicine (0.2 µg/ml final concentration) during 2 h and then in a 75 mM KCl hypotonic solution during 25 min. After centrifugation, cell pellet was resuspended in 75 mM KCl/0.1% Tween 20 and then cytocentrifuged 5 min at 1,000 rpm. The slides were transferred to a Coplin jar containing KCMc solution (120 mM KCl, 20 mM NaCl, 10 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 0.1% [v/v] Triton X-100) and incubated 15 min at room temperature. Then, immuno-FISH was performed with a protocol derived from Solovei et al. (2002) using as primary antibodies mouse anti-CENP-A (Abcam, Ab13939) (1/200) and mouse anti-CENP-B (5E6C1 clone, generous gift from Hiroshi Masumoto, Japan) (1/200); AlexaFluor conjugated goat antimouse as secondary antibody (1/1,000) and D18Z1 probe (Texas-Red) (Cytocell). Images were captured under a Zeiss AxioImager Z2 fluorescence microscope equipped with a CoolCube Camera.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
G.G. is the recipient of a Pro-Women Scholarship from the Faculty of Biology and Medicine, University of Lausanne. This work was supported by the Swiss National Science Foundation (grant number 31003A_182632) and the Jérôme Lejeune Foundation to A.R.; the National Institutes of Health (grant number HG010169 to E.E.E.); and a grant from the Brotman Baty Institute for Precision Medicine to D.E.M. and E.E.E. We wish to thank Emilie Chopin (Cell Biotechnology Center, Hospices Civils de Lyon) for providing lymphoblastoid cell lines, as well as Patrick Lomonte for his kind gift of antibodies and his advice.
Data Availability
The data underlying this article are available in the SRA database (https://www.ncbi.nlm.nih.gov/sra) and can be accessed with the BioProject accession number PRJNA714231.
References
- Alexandrov IA, Mashkova TD, Akopian TA, Medvedev LI, Kisselev LL, Mitkevich SP, Yurov YB.. 1991. Chromosome-specific alpha satellites: two distinct families on human chromosome 18. Genomics. 11(1):15–23. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amor DJ, Choo KH.. 2002. Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet. 71(4):695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M.. 1992. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 89(2):247–249. [DOI] [PubMed] [Google Scholar]
- Baldini A, Ried T, Shridhar V, Ogura K, D'Aiuto L, Rocchi M, Ward DC.. 1993. An alphoid DNA sequence conserved in all human and great ape chromosomes: evidence for ancient centromeric sequences at human chromosomal regions 2q21 and 9q13. Hum Genet. 90(6):577–583. [DOI] [PubMed] [Google Scholar]
- Cantsilieris S, Sunkin SM, Johnson ME, Anaclerio F, Huddleston J, Baker C, Dougherty ML, Underwood JG, Sulovari A, Hsieh P, et al. 2020. An evolutionary driver of interspersed segmental duplications in primates. Genome Biol. 21(1):202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Purgato S, D'Addabbo P, Archidiacono N, Battaglia P, Baroncini A, Capucci A, Stanyon R, Della Valle G, Rocchi M.. 2009. Evolutionary descent of a human chromosome 6 neocentromere: a jump back to 17 million years ago. Genome Res. 19(5):778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi O, Purgato S, Verdun di Cantogno L, Grosso E, Ciccone R, Zuffardi O, Della Valle G, Rocchi M.. 2008. Evolutionary and clinical neocentromeres: two faces of the same coin? Chromosoma. 117(4):339–344. [DOI] [PubMed] [Google Scholar]
- Cardone MF, Alonso A, Pazienza M, Ventura M, Montemurro G, Carbone L, de Jong PJ, Stanyon R, D'Addabbo P, Archidiacono N, et al. 2006. Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 7(10):R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellamare A, Catacchio CR, Alkan C, Giannuzzi G, Antonacci F, Cardone MF, Della Valle G, Malig M, Rocchi M, Eichler EE, et al. 2009. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol Biol Evol. 26(8):1889–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. 2009. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 6(9):677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Concepcion GT, Feng X, Zhang H, Li H.. 2021. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 18(2):170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiatante G, Giannuzzi G, Calabrese FM, Eichler EE, Ventura M.. 2017. Centromere destiny in dicentric chromosomes: new insights from the evolution of human chromosome 2 ancestral centromeric region. Mol Biol Evol. 34(7):1669–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin A, Sladkevicius P, Soller M.. 2009. False-positive prenatal diagnosis of trisomy 18 by interphase FISH: hybridization of chromosome 18 alpha-satellite probe (D18Z1) to chromosome 2. Prenat Diagn. 29(13):1279–1281. [DOI] [PubMed] [Google Scholar]
- Durfy SJ, Willard HF.. 1989. Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics. 5(4):810–821. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR.. 1985. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 92(4):290–296. [DOI] [PubMed] [Google Scholar]
- Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B.. 2000. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet. 9(11):1623–1631. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge P, Bansal V.. 2019. Longshot enables accurate variant calling in diploid genomes from single-molecule long read sequencing. Nat Commun. 10(1):4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciello I, Pezer Ž, Kordiš D, Bruvo Mađarić B, Ugarković Đ.. 2020. Evolutionary history of alpha satellite DNA repeats dispersed within human genome euchromatin. Genome Biol Evol. 12(11):2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannuzzi G, Pazienza M, Huddleston J, Antonacci F, Malig M, Vives L, Eichler EE, Ventura M.. 2013. Hominoid fission of chromosome 14/15 and the role of segmental duplications. Genome Res. 23(11):1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF.. 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet. 15(4):345–355. [DOI] [PubMed] [Google Scholar]
- IHGSC. 2001. Initial sequencing and analysis of the human genome. Nature. 409:860–921. [DOI] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA.. 1991. Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A. 88(20):9051–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. 2000. Immunofluorescence in cytogenetic analysis: method and applications. Genet Mol Biol. 23(4):1107–1014. [Google Scholar]
- Johnson ME, Cheng Z, Morrison VA, Scherer S, Ventura M, Gibbs RA, Green ED, Eichler EE; National Institute of Health Intramural Sequencing Center Comparative Sequencing Project. 2006. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc Natl Acad Sci U S A. 103(47):17626–17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Choo KH.. 2012. The evolutionary life cycle of the resilient centromere. Chromosoma. 121(4):327–340. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Allshire RC.. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13(12):489–496. [DOI] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, Hsieh P, Mao Y, Liskovykh MA, Koren S, Nurk S, Mercuri L, Dishuck PC, Rhie A, et al. 2021. The structure, function and evolution of a complete human chromosome 8. Nature. 593(7857):101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose M, Malla S, Stout M.. 2016. Real-time selective sequencing using nanopore technology. Nat Methods. 13(9):751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH.. 2008. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 82(2):261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM.. 2016. The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol. 17(1):16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty SM, Sullivan BA.. 2018. Alpha satellite DNA biology: finding function in the recesses of the genome. Chromosome Res. 26(3):115–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Koren S, Rhie A, Vollger MR, Gershman A, Bzikadze A, Brooks S, Howe E, Porubsky D, Logsdon GA, et al. 2020. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 585(7823):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ.. 2014. Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24(4):697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DE, Sulovari A, Wang T, Loucks H, Hoekzema K, Munson KM, Lewis AP, Fuerte EPA, Paschal CR, Thies J, University of Washington Center for Mendelian Genomics, et al. 2021. Targeted long-read sequencing identifies missing disease-causing variation. Am J Hum Genet. 108(8):1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefalcone G, Tempesta S, Rocchi M, Archidiacono N.. 1999. Centromere repositioning. Genome Res. 9(12):1184–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musilova P, Rybar R, Oracova E, Vesela K, Rubes J.. 2008. Hybridization of the 18 alpha-satellite probe to chromosome 1 revealed in PGD. Reprod Biomed Online. 17(5):695–698. [DOI] [PubMed] [Google Scholar]
- Nergadze SG, Piras FM, Gamba R, Corbo M, Cerutti F, McCarter JGW, Cappelletti E, Gozzo F, Harman RM, Antczak DF, et al. 2018. Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 28(6):789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, et al. 2021. The complete sequence of a human genome. bioRxiv. 2021.2005.2026.445798.
- Nurk S, Walenz BP, Rhie A, Vollger MR, Logsdon GA, Grothe R, Miga KH, Eichler EE, Phillippy AM, Koren S.. 2020. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 30(9):1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake K, Ohzeki JI, Shono N, Kugou K, Okazaki K, Nagase T, Yamakawa H, Kouprina N, Larionov V, Kimura H, et al. 2020. CENP-B creates alternative epigenetic chromatin states permissive for CENP-A or heterochromatin assembly. J Cell Sci. 133(15):. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DK, O'Day K, Trong HL, Charbonneau H, Margolis RL.. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 88(9):3734–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchenko T, Black BE.. 2009. The epigenetic basis for centromere identity. Prog Mol Subcell Biol. 48:1–32. [DOI] [PubMed] [Google Scholar]
- Payne A, Holmes N, Clarke T, Munro R, Debebe BJ, Loose M.. 2021. Readfish enables targeted nanopore sequencing of gigabase-sized genomes. Nat Biotechnol. 39(4):442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras FM, Nergadze SG, Magnani E, Bertoni L, Attolini C, Khoriauli L, Raimondi E, Giulotto E.. 2010. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 6(2):e1000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrovic N, Mantovani B.. 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 409(1–2):72–82. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. Available from: https://www.R-project.org/.
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP.. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi M, Stanyon R, Archidiacono N.. 2009. Evolutionary new centromeres in primates. Prog Mol Subcell Biol. 48:103–152. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43(Database issue):D670–681.25428374 [Google Scholar]
- Rudd MK, Willard HF.. 2004. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 20(11):529–533. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF.. 2001. Genomic and genetic definition of a functional human centromere. Science. 294(5540):109–115. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz MC.. 2018. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 15(6):461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W.. 2017. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 14(4):407–410. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P.. 2013. –2015. RepeatMasker Open-4.0.
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T.. 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp Cell Res. 276(1):10–23. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, et al. ; 1000 Genomes Project Consortium. 2015. An integrated map of structural variation in 2,504 human genomes. Nature. 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Sullivan BA.. 2020. Genomic and functional variation of human centromeres. Exp Cell Res. 389(2):111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavelu M, Chen PX, Pergament E.. 1998. Hybridization of chromosome 18 alpha-satellite DNA probe to chromosome 22. Prenat Diagn. 18(9):922–925. [DOI] [PubMed] [Google Scholar]
- Ventura M, Mudge JM, Palumbo V, Burn S, Blennow E, Pierluigi M, Giorda R, Zuffardi O, Archidiacono N, Jackson MS, et al. 2003. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 13(9):2059–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Weigl S, Carbone L, Cardone MF, Misceo D, Teti M, D'Addabbo P, Wandall A, Bjorck E, de Jong PJ, et al. 2004. Recurrent sites for new centromere seeding. Genome Res. 14(9):1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voullaire LE, Slater HR, Petrovic V, Choo KH.. 1993. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 52(6):1153–1163. [PMC free article] [PubMed] [Google Scholar]
- Wei S, Siu VM, Decker A, Quigg MH, Roberson J, Xu J, Adeyinka A.. 2007. False-positive prenatal diagnosis of trisomy 18 by interphase FISH: hybridization of chromosome 18 alpha-satellite DNA probe (D18Z1) to the heterochromatic region of chromosome 9. Prenat Diagn. 27(11):1064–1066. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Willard HF, Waye JS.. 1987. Hierarchical order in chromosome-specific human alpha satellite DNA. Trends Genet. 3:192–198. [Google Scholar]
- Winsor EJ, Dyack S, Wood-Burgess EM, Ryan G.. 1999. Risk of false-positive prenatal diagnosis using interphase FISH testing: hybridization of alpha-satellite X probe to chromosome 19. Prenat Diagn. 19(9):832–836. [DOI] [PubMed] [Google Scholar]
- Wu JC, Manuelidis L.. 1980. Sequence definition and organization of a human repeated DNA. J Mol Biol. 142(3):363–386. [DOI] [PubMed] [Google Scholar]
- Zhu M, Need AC, Han Y, Ge D, Maia JM, Zhu Q, Heinzen EL, Cirulli ET, Pelak K, He M, et al. 2012. Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet. 91(3):408–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the SRA database (https://www.ncbi.nlm.nih.gov/sra) and can be accessed with the BioProject accession number PRJNA714231.