Immune activation is an important feature in the development and progression of NAFLD. Macrophages are the most abundant immune cell type in the liver and are implicated in this process. However, intrahepatic macrophages are a heterogeneous and dynamic population. They can be classified based on ontogeny into: yolk sac-derived tissue-resident macrophages, commonly referred to as Kupffer cells (KCs), and bone marrow (BM) monocyte-derived macrophages. In health, embryonic KCs (referred to as ResKCs) are the predominant population, capable of self-renewal, and located in the sinusoids, where they can interact with hepatocytes as well as nonparenchymal cells, including LSECs and HSCs. During liver injury, for example in NASH, macrophages are recruited from circulating monocytes and can have both a detrimental and supportive role.(1)
Remmerie et al.(2) characterized the dynamics and Remmerie et al. characterized the dynamics and subsets in a mouse model of NAFLD induced by feeding them a Western diet (WD) for up to 36 weeks. First, using cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) and an unbiased approach, discrete immune cell types were identified in livers from mice on WD compared to a standard diet and across time. These clusters were based on differentially expressed genes along with protein expression profiles and standard cell identity genes. For instance, macrophage populations were classified based on the expression of C-type lectin domain family 4 member F (CLEC4F; a C-type lectin) to identify KCs, which were further categorized based on expression of T-cell immunoglobulin and mucin domain-containing 4 (TIM4; a receptor for eat-me signals of apoptotic cells) to identify resident KCs or lymphocyte antigen 6 complex (Ly6C) to identify monocytes. After 24 weeks on the WD, the numbers and proportions of CLEC4F+TIM4− KCs (terme–d monocyte-derived KCs [MoKCs]) and CLEC4F monocyte-derived recruited macrophages increased, with a concomitant reduction in the proportion of CLEC4F+TIM4+ KCs (ResKCs), thus confirming that intrahepatic macrophages are repopulated by BM-derived monocytes in NAFLD. Annotation into three subsets, ResKC, MoKC, and CLEC4F− macrophages, was confirmed by mapping signatures from bulk RNA-sequencing of these three subsets onto a Signature Finder algorithm (Fig. 1).
FIG. 1.
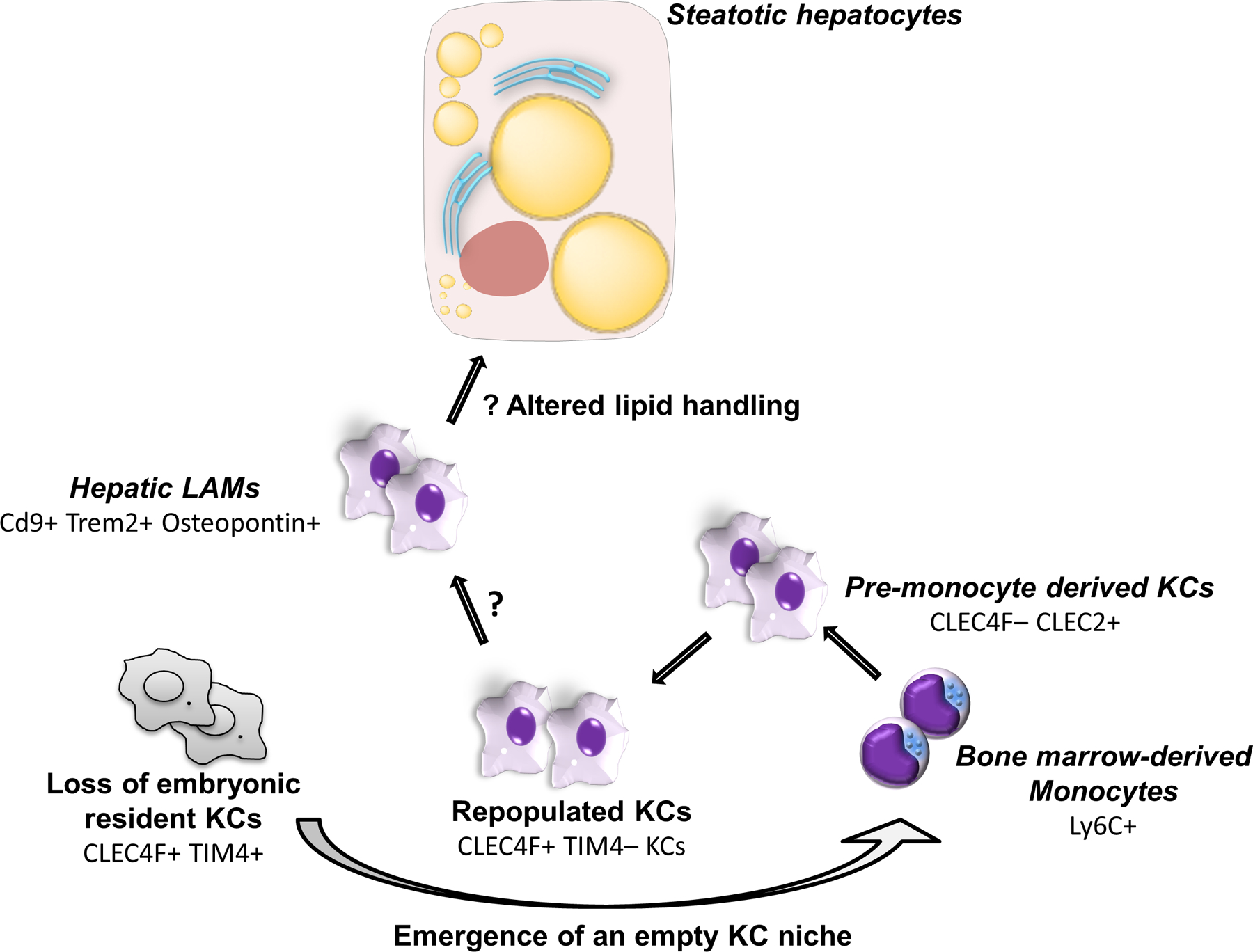
Heterogeneity of monocyte-derived macrophages in NASH. In a mouse model of NAFLD, resident yolk sac-embryonic KCs are gradually lost, and this niche is subsequently repopulated by BM-derived monocytes. The resulting intrahepatic macrophage population is heterogeneous, consisting of many subsets including pre-monocyte-derived KCs and hepatic LAMs. Hepatic LAMs express osteopontin, are located in fibrotic zones, and possess a unique transcriptomic profile. What signals drive the differentiation of monocyte-derived KCs into hepatic LAMs is unclear. Whereas hepatic LAMs had a distinct ability to metabolize lipid compared to KCs, whether this has significance in their crosstalk with steatotic hepatocytes is also undefined.
Intriguingly, stimulated proliferation of KCs was preserved, albeit attenuated in WD-fed mice, which may contribute to ResKC loss. This process remains incompletely understood given that no deficiency of signals from LSECs or HSCs was observed. These signals from LSECs, HSCs, and also hepatocytes together imprint the liver-specific macrophage identity within the KC niche. To further investigate the response of these niche-defining cells, single-cell (sc) RNA-sequencing analysis of live cells of these populations was performed. In NAFLD, proliferation of LSECs, HSCs and cholangiocytes, based on expression of marker of proliferation Ki-67, was increased. Hepatocytes increased in size and had greater gene expression of Cd36 and fatty-acid–binding protein 4, reflecting an increased lipid load. Confocal microscopy identified that recruited macrophages in NAFLD occupied similar locations as KCs in health—between the portal and central veins and atleast partly, in contact with LSECs and HSCs.(3) However, recruited CLEC4F− macrophages were the main subset in fibrotic zones, which were identified on the basis of increased desmin expression.
Secondary clustering of macrophage and monocyte cells in the CITE-seq data identified several clusters that varied between diet and time points, but two subsets were specific to WD-fed mice and were termed Mac1 and Mac2. The gene expression profile of Mac1 was similar to monocytes en route to becoming MoKCs, demonstrating the expression of C-type lectin domain family 2 (CLEC2), considered an early marker of MoKCs.(4) On the other hand, Kyoto Encyclopedia of Genes and Genomes path way analysis of Mac2 suggested similarities with CD9 and triggering receptor expressed on myeloid cells 2 (TREM2)-positive lipid-associated macrophages (LAMs) in adipose tissue,(5) as well as scar-associated macrophages (SAMs) in fibrotic human livers.(6) Therefore, Mac1 was termed “pre-moKCs” and Mac2 “hepatic LAMs.” Reanalysis of the CITE-seq data and a flow cytometric mRNA assay identified secreted phosphoprotein 1, encoding the chemokine, osteopontin, as a specific marker enriched in hepatic LAMs. In separate studies, osteopontin has been implicated in NASH and fibrosis.
Functionally, LAMs, monocyte-derived KCs, and resident KCs differed in their expression of lipid metabolism genes, and pre-moKCs were intermediate between KCs and the LAMs. Lipid species composition differed between these subsets, and both LAMs and MoKCs contained less lipid than ResKCs. Whether these differences have functional consequences on liver injury or are simply a reflection of lesser time spent in their respective niches are unclear. In contrast, there were no differences in the metabolome, neutral lipid volume, or composition of resident KCs between diets and time points. Furthermore, immune activation of resident KCs was not noted at any stage of NAFLD in the scRNA-sequencing or bulk RNA-sequencing analysis. Put together, Remmerie et al. provide a careful description of macrophage subsets in a mouse model of NAFLD—beginning with loss of resident embryonic KCs, which are subsequently repopulated by monocytes, which are themselves also a heterogenous population. One subset of these recruited macrophages, termed hepatic LAMs, express osteopontin, are located in fibrotic zones, and possess a unique transcriptomic profile.
These findings can be reconciled with two other recent studies. In mice fed a methionine- and choline-deficient diet to induce NASH, Tran et al. similarly demonstrated that loss of ResKCs led to repopulation by monocyte-derived KCs, which was associated with decreased hepatic triglyceride storage and increased inflammation.(4) Consistently, Seidman et al. identified distinct transcriptional and phenotypic states of recruited macrophages, driven by a strong niche-specific effect.(7) Notably, they also identified a subset of monocyte-derived KCs that share phenotypic similarities to SAMs. Putting these studies together, the relationship between SAMs and LAMs, and their mechanistic role in NASH progression and fibrosis, is incompletely defined. Several intriguing questions arise: What triggers ResKC death, and how are recruited macrophages protected from the same? What precise downstream effects do different subsets of recruited KCs have on parenchymal and nonparenchymal cells in the liver? Macrophage heterogeneity likely reflects a dynamic state, with subsets capable of transitioning phenotypes and functions rather than destined to remain in defined subsets. To speculate, sterile inflammation in NASH may be related to imbalances between different subsets with discrete and tonic functions—perhaps a weakened adaptive subset versus a dominant maladaptive subset. More fundamentally, whether this heterogeneity in macrophages in NASH is a teleological phenomenon or a consequence of spatial gradients of niche cells is also unclear. Last, differences between murine and human macrophage phenotypes and functions are likely, and will need to be defined in future studies. Nevertheless, the work of Remmerie et al. is invaluable in expanding our knowledge of the complex fate of macrophages in NASH.
It is intuitive to target macrophages as mediators of inflammation in NASH. Indeed, cenicriviroc (CVC), a dual C-C chemokine receptors type 2 and 5 dual antagonist that inhibits macrophage infiltration and with convincing antifibrotic effects in numerous animal models of NASH, was evaluated in the CENTAUR phase 2b trial. Treatment for 52 weeks was associated with histological fibrosis improvement—20% versus 10% compared to placebo.(8) It is therefore being further evaluated in a phase 3 study (AURORA, NCT02217475) and a phase 2 combination trial with the farnesoid X receptor ligand, tropifexor (TANDEM, NCT03517540). Findings from the recent studies in Immunity highlight the fact that macrophages have diverse roles in NASH. Characterizing their different subsets and functions is the first step toward understanding how and when different mechanisms are important in NASH pathobiology. Indeed, in the CVC trial, select targeting of SAMs might explain the findings of fibrosis improvement. Thus, greater understanding of macrophage heterogeneity will only further aid in our ability to specifically target pathogenic and perhaps even beneficial subsets of these critical immune cells.
Acknowledgments
Supported by NIH grant DK111378 (to H.M.) and the Mayo Foundation.
Footnotes
Potential conflict of interest: Nothing to report.
REFERENCES
- 1).Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 2014;60:1090–1096. [DOI] [PubMed] [Google Scholar]
- 2).Remmerie A, Martens L, Thoné T, Castoldi A, Seurinck R, Pavie B, et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 2020;53:641–657.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 2019;51:638–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Tran S, Baba I, Poupel L, Dussaud S, Moreau M, Gélineau A, et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity 2020;53:627–640. [DOI] [PubMed] [Google Scholar]
- 5).Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 2019;178:686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019;575: 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 2020;52:1057–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Friedman SL, Ratziu V, Harrison SA, Abdelmalek MF, Aithal GP, Caballeria J, et al. A randomized, placebo-controlled trial of cenicriviroc for treatment of nonalcoholic steatohepatitis with fibrosis. Hepatology 2018;67:1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]


