Abstract
Supplementary information: Supplementary data are available at Bioinformatics online.
With the publication of high-quality genome-scale metabolic models (GSMs) for several organisms, the Systems Biology community has developed a plethora of algorithms for their analysis making use of ever-growing omics data (Heirendt et al., 2019). In particular, in human metabolism, the reconstruction of the first genome-scale model RECON1 (Duarte et al., 2007) promoted the development of Context-Specific Model (CS-Model) reconstruction methods (Opdam et al., 2017). This family of algorithms aims to identify the catalogue of metabolic reactions involved in a cell in a given condition using omics data, commonly gene expression levels. CS-Models are particularly suitable for studying metabolic differences between human tissues and are widely used in the area of human health (Uhlen et al., 2016), with applications including the prediction of metabolic vulnerabilities in cancer (Agren et al., 2014) and the inference of biomarkers in Alzheimer and diabetes (Geng and Nielsen, 2017), among others. Currently, we have dozens of these CS-Models available in different public repositories, usually stored under the Systems Biology Markup Language standard (Hucka et al., 2003).
An essential component of CS-Model reconstruction algorithms are gene-protein-reaction (GPR) rules, which include the information about how genes relate to protein complexes and isozymes as well as the reactions they catalyze. GPR rules are expressed in the form of logical equations and allow us to define active/inactive reactions by mapping expression data onto them. This set of reaction states is used by reconstruction algorithms to extract the final CS-Model, which involves the reactions mapped as active plus a small set of inactive reactions automatically added to fill gaps in the network (gap-filling reactions). The relevance of GPR rules for CS-Model reconstruction algorithms can be more clearly observed in the Supplementary Information, where a general workflow is depicted.
Different CS-Model reconstruction algorithms have their own strengths and weaknesses depending on the problem under study and omics data available. In this direction, Opdam and collaborators performed an extensive benchmark of CS-Model algorithms and found that no particular method outperforms the others (Opdam et al., 2017). However, after careful inspection, we found that all of these algorithms share a common ‘bug’ in the way GPR rules and gene expression data are treated when reconstructing CS-Models.
The first issue we encountered is related to how gap-filling reactions are managed in the reconstruction process. Model extraction algorithms may add reactions classified as inactive to fill gaps in the CS-Model. Figure 1A represents a toy model with four reactions: two categorized as active, {1, 4}, and two as inactive, {2, 3}. In this example, the CS-model includes the active reactions and the reaction 2 for gap filling. Importantly, the decision to include the gap-filling reaction 2 implies to update our assumption about the state of genes involved in such reaction, which in this case means to update the state of gene B to active. In addition, if the state of gene B is updated, this change must be propagated through the CS-model. Following the example in Figure 1A, propagating the change on the state of gene B implies that reaction 3 becomes active and thus it should be included in the consolidated CS-Model. We found that this consolidation step is not performed by the published CS-Model reconstruction algorithms, and that neglecting the GPR consolidation step will worsen the model predictions by overestimating the effect of gene knockouts.
Fig. 1.
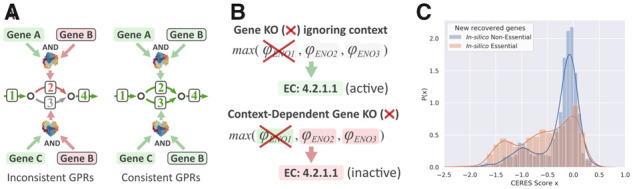
Identified errors in context-specific metabolic model reconstruction. (A) Illustration of the consequences of not managing the gap-filling reactions properly. (B) Illustration of the consequences of not taking the molecular context into account. (C) The probability distribution of CERES essentiality scores (Meyers et al., 2017) for recovered essential (orange) and non-essential (blue) genes. The recovered essential genes are those genes which are predicted non-essential by standard GIMME (Becker and Palsson, 2008) but essential when the errors in (A) and (B) are amended. The recovered non-essential genes are those genes which are predicted essential by standard GIMME (Becker and Palsson, 2008) but non-essential when the errors in (A) and (B) are amended. Red and green coloring refers to inactive and active genes, respectively. The green arrows correspond to active reactions, the red arrow refers to an inactive reaction which has been selected as gap-filling and the gray arrow corresponds to an inactive reaction which has not been selected as gap-filling
The second and more striking issue is that the molecular context used to reconstruct the CS-model is usually not included as part of the CS-model. If the expression confidences used to infer the reaction states (active/inactive) are left aside after the reaction mapping, part of the information used to reconstruct the CS-Model is lost and, therefore, the model formulation will be incomplete. In the example of Figure 1B, the enolase reaction was set as active based on its expression scores and GPR rule. However, if the GPR of enolase is evaluated without the expression scores, it is not possible to guess which genes are supporting the enolase activity (in Fig. 1B ENO1). Moreover, without the information about gene expression, the default hypothesis will be that all the genes included in the CS-Model are active. Thus, analyzing a CS-Model without considering the context can affect the predictions. Specifically, we found that this inconsistent treatment of the CS-Model leads to under-estimating the effect of gene knockouts. This can be observed in Figure 1B, where the deletion of ENO1 should lead to disrupt the enolase reaction and this is not the case if the context is not taken into account.
In order to evaluate the effect of this flaw, we reconstructed ∼400 CS-Models for cell lines from the Cancer Cell Line Encyclopedia (Barretina et al., 2012) and conducted a gene essentiality analysis. The CS-Models were reconstructed using Gene Expression Barcode to classify genes as active/inactive (McCall et al., 2014), Recon3D as the reference model (Brunk et al., 2018) and GIMME as the extraction algorithm (Becker and Palsson, 2008). We amended the output of GIMME to account for the issues discussed above, using a simple and effective approach, which is fully detailed in the Supplementary Information. Summarizing the results over all the cell lines, our approach found 3160 essential genes not predicted by standard GIMME (∼8 per model) and discarded 1061 essential genes predicted by standard GIMME (∼2.5 per model) (Supplementary Fig. S1). These results clearly indicate that ignoring the molecular context has drastic effects on the in-silico predictions.
To validate these results, for each cell line we gathered CRISPR–Cas9 essentiality data from DepMap (Tsherniak et al., 2017) corrected using CERES essentiality score (Meyers et al., 2017). Figure 1C shows the CERES scores’ probability distributions for the aforementioned 3160 non-essential genes becoming essential (orange curve) and 1061 essential genes becoming non-essential when GIMME considered context (blue curve). This same result is shown in Supplementary Figure S2 using absolute frequencies instead of probability density. As expected, the first group is significantly enriched in DepMap essential genes (one tailed Mann-Whitney test p-value = 9.96·10-41), demonstrating the practical importance of the correct treatment of GPR rules and molecular context. The same analysis was performed with other model extraction methods, i.e. FastGapFill (Thiele et al., 2014) and FASTCORMICS (Pacheco et al., 2015) finding similar results (Supplementary Fig. S3).
Altogether, our results illustrate the importance of the errors introduced during the GPR translation in many of the published metabolic reconstructions. It is worth to note that, despite the main results were obtained using a particular gene expression thresholding, the problem of the inconsistent treatment of GPR rules is independent of the gene expression thresholding approach. To overcome this issue, we advocate for a strict control of the specific molecular context during the translation of the GPR rules to CS-Models. To that end, the existing CS-Model reconstruction algorithms and storage standards should be modified to be GPR consistent and provide the molecular context. Here, we showed the positive results in the performance of GIMME when this limitation was corrected and similar results are expected with other algorithms.
Funding
I.A. was supported by a Basque Government predoctoral grant [PRE_2018_2_0065]. This work was supported by the Minister of Economy and Competitiveness of Spain [BIO2013-48933, BIO2016-77998-R] and by the European Commission under the INFORE project [H2020-ICT- 825070].
Conflict of Interest: none declared.
Supplementary Material
References
- Agren R. et al. (2014) Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol. Syst. Biol., 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J. et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S.A., Palsson B.O. (2008) Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk E. et al. (2018) Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat. Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte N.C. et al. (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA, 104, 1777–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nielsen J. (2017) In silico analysis of human metabolism: reconstruction, contextualization and application of genome-scale models. Curr. Opin. Syst. Biol., 2, 29–38. [Google Scholar]
- Heirendt L. et al. (2019) Creation and analysis of biochemical constraint-based models: the COBRA Toolbox v3.0. Nat. Protoc., 8, 321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M. et al. (2003) The systems biology markup language (SBML): A medium for representation and exchange of biochemical network models. Bioinformatics, 19, 524–531. [DOI] [PubMed] [Google Scholar]
- McCall M.N. et al. (2014) The Gene Expression Barcode 3.0: improved data processing and mining tools. Nucleic Acids Res., 42, 938–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers R.M. et al. (2017) Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet., 49, 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdam S. et al. (2017) A systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst., 4, 318–329.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco M.P. et al. (2015) Integrated metabolic modelling reveals cell-type specific epigenetic control points of the macrophage metabolic network. BMC Genomics, 16, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I. et al. (2014) FASTGAPFILL: efficient gap filling in metabolic networks. Bioinformatics, 30, 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsherniak A. et al. (2017) Defining a cancer dependency map. Cell, 170, 564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M. et al. (2016) Transcriptomics resources of human tissues and organs. Mol. Syst. Biol., 12, 862–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


