Abstract
In angiosperms, the α/β hydrolase DWARF14 (D14), along with the F-box protein MORE AXILLARY GROWTH2 (MAX2), perceives strigolactones (SL) to regulate developmental processes. The key SL biosynthetic enzyme CAROTENOID CLEAVAGE DIOXYGENASE8 (CCD8) is present in the moss Physcomitrium patens, and PpCCD8-derived compounds regulate moss extension. The PpMAX2 homolog is not involved in the SL response, but 13 PpKAI2LIKE (PpKAI2L) genes homologous to the D14 ancestral paralog KARRIKIN INSENSITIVE2 (KAI2) encode candidate SL receptors. In Arabidopsis thaliana, AtKAI2 perceives karrikins and the elusive endogenous KAI2-Ligand (KL). Here, germination assays of the parasitic plant Phelipanche ramosa suggested that PpCCD8-derived compounds are likely noncanonical SLs. (+)-GR24 SL analog is a good mimic for PpCCD8-derived compounds in P. patens, while the effects of its enantiomer (−)-GR24, a KL mimic in angiosperms, are minimal. Interaction and binding assays of seven PpKAI2L proteins pointed to the stereoselectivity toward (−)-GR24 for a single clade of PpKAI2L (eu-KAI2). Enzyme assays highlighted the peculiar behavior of PpKAI2L-H. Phenotypic characterization of Ppkai2l mutants showed that eu-KAI2 genes are not involved in the perception of PpCCD8-derived compounds but act in a PpMAX2-dependent pathway. In contrast, mutations in PpKAI2L-G, and -J genes abolished the response to the (+)-GR24 enantiomer, suggesting that PpKAI2L-G, and -J proteins are receptors for moss SLs.
The study of moss PpKAI2L receptors for strigolactones and related compounds highlights MORE AXILLARY GROWTH2-dependent and -independent pathways for the perception of these compounds.
Introduction
Strigolactones (SLs) are butenolide compounds with dual roles in plants: exuded in soil, SLs signal the presence of a host to arbuscular mycorrhizal (AM) fungi (Akiyama et al., 2005; Besserer et al., 2006) and thus favor the establishment of symbiosis; as endogenous compounds, they (or derived compounds) play a hormonal role in developmental programs (Gomez-Roldan et al., 2008; Umehara et al., 2008; for reviews see Lopez-Obando et al., 2015; Waters et al., 2017). SLs exuded from plant roots also act as signaling molecules in the rhizosphere, inducing parasitic plant seed germination (Cook et al., 1966; for review see Delavault et al., 2017). SLs have been found in most land plants, including bryophytes, lycophytes, gymnosperms, and angiosperms (Yoneyama et al., 2018b). However, their synthesis and signaling pathways have mainly been described in angiosperms where core enzyme pathways involving DWARF27 (D27) isomerase and two CAROTENOID CLEAVAGE DIOXYGENASEs (CCD7 and CCD8) convert carotenoids into carlactone (CL). To date, CL is the reported precursor of all known SLs (Alder et al., 2012) and the substrate for further enzymes such as the cytochrome-P450 MORE AXILLARY GROWTH1 (MAX1; for review see Al-Babili and Bouwmeester, 2015). Depending on the plant species, CL is converted into canonical or noncanonical SLs. These differ in the structure attached to the conserved enol ether-D ring moiety, which is shared by all SLs and essential for biological activity (Yoneyama et al., 2018a; Yoneyama, 2020). In angiosperms, SLs are perceived by the α/β hydrolase DWARF14 (D14)/DECREASED APICAL DOMINANCE2 (DAD2)/RAMOSUS 3 (RMS3; Arite et al., 2009; Hamiaux et al., 2012; de Saint Germain et al., 2016), which interacts with the F-box protein MORE AXILLARY GROWTH2 (MAX2) to target SUPPRESSOR OF MAX2-LIKE (SMXL) repressor proteins for proteasome degradation (Soundappan et al., 2015; Waters et al., 2015a). An unusual aspect of SL perception is that the D14 protein is both a receptor and an enzyme that cleaves its substrate (and covalently binds part of the SL) in a signaling mechanism that is still under debate (Yao et al., 2016; de Saint Germain et al., 2016; Shabek et al., 2018; Seto et al., 2019). In all cases, the pocket of the α/β hydrolase appears to be essential for substrate/ligand (SL) interactions (for review see Bürger and Chory, 2020).
The evolutionary origins of SLs, and in particular whether their primary role is that of a hormone or rhizospheric signal, are still unclear. The identification and quantification of SLs are a challenge in many species due to the very low amounts of the molecules present in plant tissues or exudates and their high structural diversification (Xie, 2016; Yoneyama et al., 2018b). Therefore, the occurrence of SLs in a species was often inferred from the presence of the core biosynthesis enzymes encoded in its genome (Delaux et al., 2012; Walker et al., 2019) or from germination assays using plant exudates on parasitic plant seeds (Yoneyama et al., 2018b). Recently, SLs were proposed to only be produced in land plants (Walker et al., 2019). Evidence of a signaling pathway ancestral to the SL pathway came from the identification of an ancient D14 paralog named KARRIKIN INSENSITIVE2/HYPOSENSITIVE TO LIGHT (KAI2/HTL) during a screening of Arabidopsis thaliana mutants (Waters et al., 2012). Like D14, KAI2 is also an α/β hydrolase that interacts with the MAX2 F-box protein in a pathway regulating Arabidopsis seed germination and seedling development (Nelson et al., 2011; Waters et al., 2012). KAI2 is also involved in stress tolerance, drought tolerance, and AM symbiosis (Gutjahr et al., 2015; Wang et al., 2018; Villaecija-Aguilar et al., 2019; Li et al., 2020). However, the endogenous signal perceived by KAI2 remains unknown and is reported thus far as the KAI2-Ligand (KL; Conn and Nelson, 2015).
To gain insight into SL signaling evolution, we focused our studies on a model for nonvascular plants, Physcomitrium (Physcomitrella) patens. Along with hornworts and liverworts, mosses such as P. patens belong to the bryophytes (Bowman et al., 2019). Bryophytes are currently described as a monophyletic group of embryophytes sharing an ancestor with vascular plants (Puttick et al., 2018; Harris et al., 2020). Therefore, comparing signaling pathways between extant vascular plants and extant bryophytes can provide insights into the evolutionary origin of these pathways (Guillory and Bonhomme, 2021b). Furthermore, studying extant bryophytes may provide clues for understanding how the first plants have been able to survive out of water, and conquer land, 450 million years ago (Bowman et al., 2019; Blázquez et al., 2020; Harris et al., 2020).
In P. patens, several D27 homologs, as well as both CCD enzymes (PpCCD7 and PpCCD8) required for SL synthesis are found (Proust et al., 2011), and CL has been detected as the product of PpCCD8 (Decker et al., 2017). The extended phenotype of Ppccd8 mutant plants indicates that PpCCD8-derived molecules are required for regulating moss filament growth. These molecules also act as a growth-limiting signal between neighboring moss plants, as they are exuded into the medium (Proust et al., 2011). PpCCD8-derived molecules also appear to play a role in rhizoid elongation and gametophore shoot branching (Delaux et al., 2012; Coudert et al., 2015). The application of the artificial SL (±)-GR24 complemented the Ppccd8 mutant phenotype, suggesting that PpCCD8-derived molecules are indeed SL-like compounds (Proust et al., 2011). However, the exact nature of PpCCD8-derived molecules is still elusive (Yoneyama et al., 2018b), and the absence of MAX1 homologs in P. patens suggests that the biosynthesis pathway in this moss may differ from that of vascular plants. Nevertheless, phylogenetic analysis of MAX1 homologs highlights the presence of this gene in other mosses and suggests that the biosynthesis pathway is otherwise conserved in land plants (Walker et al., 2019).
SL signaling also seems to differ between angiosperms and P. patens. Indeed, contrary to its angiosperm homolog, PpMAX2 is likely not involved in the response to PpCCD8-derived molecules, as the corresponding mutant does respond to (±)-GR24 (Lopez-Obando et al., 2018). The PpMAX2 F-box protein appears to be involved in a light-dependent pathway required for early moss development and the regulation of gametophore number and size. No true homolog for the D14 SL receptor is found in the P. patens genome, whereas between 11 and 13 PpKAI2-LIKE (PpKAI2L) candidate genes were described, depending on which version of the P. patens genome was searched. These genes, first called PpD14La to PpD14Lk (Delaux et al., 2012), were renamed PpKAI2L-A to PpKAI2L-M (Lopez-Obando et al., 2016a) and grouped into four subclades. Hereafter, for simplicity, we renamed the subclades as eu-KAI2 (previous clade i, including PpKAI2L-A to -E); (FK), including PpKAI2L-F and -K (previous clade ii); (HIL), including PpKAI2L-H, -I, - (previous clade i.i–i.ii); (GJM), including PpKAI2L-G, -J, and -M (previous clade iii). A comprehensive phylogenetic assessment placed the P. patens clades (FK), (HIL), and (GJM) into a super clade called DDK (D14/DLK2/KAI2) containing spermatophyte (angiosperm and gymnosperm) D14 clades, while the eu-KAI2 clade is highly conserved and common to all land plants (Bythell-Douglas et al., 2017). Nevertheless, moss proteins from the DDK clade were found to be as different from D14 as from KAI2 (Bythell-Douglas et al., 2017).
Prediction of PpKAI2L protein structures found various pocket sizes, as observed for D14 and KAI2 from vascular plants (Lopez-Obando et al., 2016a). Larger pocket sizes were predicted for PpKAI2L-F and -K, while smaller pockets were predicted for eu-KAI2 clade proteins. Consequently, these proteins could be receptors with diverse substrate preferences and might bind to either PpCCD8-derived compounds or the elusive KL. Accordingly, in our previous study of PpMAX2, we hypothesized that this F-box protein might be involved in a putative P. patens KL signaling pathway (Lopez-Obando et al., 2018). However, the involvement of PpKAI2L proteins in the PpMAX2 pathway remains an open question. The crystal structures of PpKAI2L-C, -E, and -H were recently published (Bürger et al., 2019). In vitro purified PpKAI2L proteins -C, -D, and -E (eu-KAI2 clade) were destabilized by (−)-5-deoxystrigol, a canonical SL with nonnatural stereochemistry, but the binding affinity for the pure enantiomer was not determined. In contrast, PpKAI2L proteins -H, -K, and -L could bind to the karrikin KAR1 (Bürger et al., 2019). Proteins from the (GJM) clade were not studied, and no evidence for a role of one (or several) PpKAI2L as receptors for PpCCD8-derived molecules was provided, nor were experiments carried out in P. patens to validate the results. Moreover, the involvement of PpKAI2L proteins in the putative PpMAX2-dependent KL signaling pathway remains to be explored.
The aim of the present study was to investigate the nature of the PpCCD8-derived molecules in moss and to identify the moss receptors for these compounds. In an attempt to shed light on the type of SLs derived from PpCCD8, we first tested the activity of P. patens as a stimulant for seed germination of the root parasitic plant Phelipanche ramosa, for which one SL receptor, PrKAI2d3, has been recently characterized (de Saint Germain et al., 2021b). We then looked for mimics of SL and KL that we could use in assays on P. patens. So far, the racemic (±)-GR24 has been used as a SL analog, but recent reports in angiosperms found that the different enantiomers present in this synthetic mixture do not have the same effect (Scaffidi et al., 2014). Indeed, (+)-GR24, (also called GR245DS) with a configuration close to the natural strigol is mostly perceived by D14 and mimics CCD8-derived SLs (e.g. CL). On the other hand, as for (−)-5-deoxystrigol, the configuration of (−)-GR24 (also called GR24ent-5DS) has so far not been found in natural SLs. However, KAI2 perceives the (−)-GR24 analog better than D14 proteins, and (−)-GR24 has therefore been described as a KL mimic (Scaffidi et al., 2014; Zheng et al., 2020). We tested both (+)-GR24 and (−)-GR24 isomers in phenotypic assays. Then, refining and supplementing the work of Bürger et al. (2019), we fully characterized seven PpKAI2L proteins in vitro by testing their cleavage activity and binding to pure GR24 enantiomers. We showed that stereoselectivity of most of the PpKAI2L proteins for GR24 enantiomers is weak, except for the eu-KAI2 clade, which shows preferential affinity for (−)-GR24. We highlighted the stronger (compared to the other PpKAI2L) and nonselective enzyme activity of PpKAI2L-H. We expressed these proteins in the Arabidopsis d14-1 kai2-2 double mutant to examine conservation of the SL and/or KL perception function. Finally, we used CRISPR-Cas9 technology to generate several P. patens multiple mutants affected in all four PpKAI2L clades. By coupling analysis of these mutants’ phenotypes and responses to pure GR24 enantiomers with our biochemistry results, we provide strong evidence that eu-KAI2 clade PpKAI2L proteins could be moss KL receptors that function in a PpMAX2-dependent pathway, while (GJM) clade PpKAI2L proteins would function as moss SL receptors in a PpMAX2-independent pathway.
Results
PpCCD8-derived compounds induce the germination of a hemp-specific population of P. ramosa
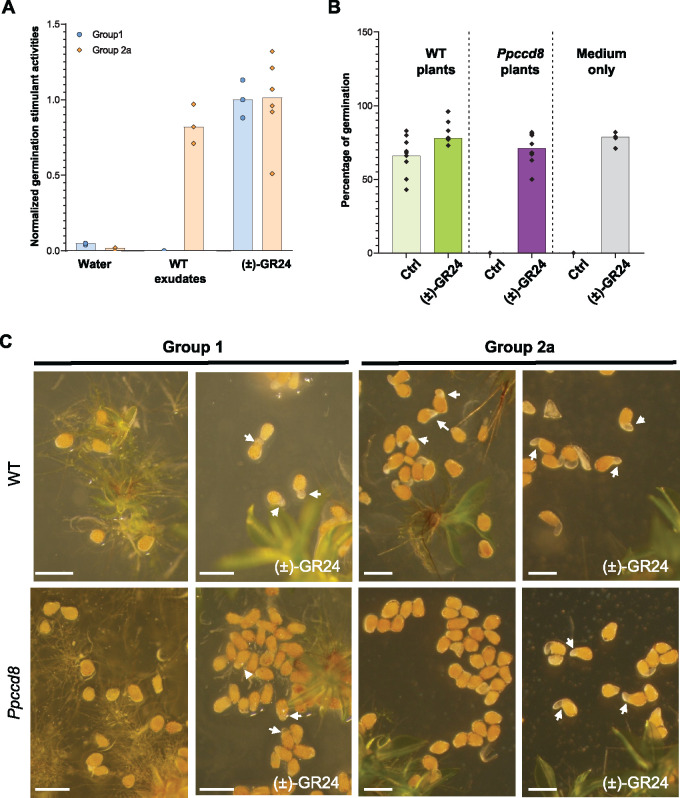
A recent report (Yoneyama et al., 2018b) indicated that canonical SLs previously identified in P. patens tissues (Proust et al., 2011) could be contaminants. However other evidence suggests that P. patens does synthesize SL-like products derived from CL. Indeed, PpCCD8 shows carlactone synthase activity (Decker et al., 2017), and both the synthetic SL analog (±)-GR24 and CL do complement the Ppccd8 phenotype (Proust et al., 2011; Decker et al., 2017). Still, quantification of SL and related compounds is a challenge in many species (Boutet-Mercey et al., 2018; Yoneyama et al., 2018b; Rial et al., 2019; Floková et al., 2020), and so far no known SL has been identified from P. patens (Yoneyama et al., 2018b). Here we tested the ability of P. patens exudates to induce the germination of parasitic seeds. Parasitic plants such as P. ramosa can parasitize various host plants in response to specific exuded germination stimulants (GSs). Different genetic groups of P. ramosa seeds have been identified, depending on the crop grown in the field where the seeds were collected (Huet et al. 2020). Seeds from two populations of P. ramosa harvested in hemp (Cannabis sativa; P. ramosa group 2a) and oilseed rape (Brassica napus; P. ramosa group 1) fields (Stojanova et al., 2019; Huet et al., 2020) were assayed with wild-type (WT) moss exudates (Figure 1). As a control, both groups of seeds were germinated in the presence of (±)-GR24 (Figure 1A). WT moss exudates induced the germination of P. ramosa group 2a seeds but not P. ramosa group 1 seeds (Figure 1A). In another assay, P. ramosa seeds were added to culture plates close to WT or Ppccd8 plants, with and without (±)-GR24 (Figure 1, B and C). Phelipanche ramosa group 2a but not group1 seeds germinated on WT moss plates, while no germination was observed in the vicinity of Ppccd8 plants. In all cases (WT and Ppccd8), the addition of (±)-GR24 to the medium restored seed germination. Thus, PpCCD8-derived compounds induce the germination of a specific population of P. ramosa seeds, responding to not yet identified GS exuded by hemp.
Figure 1.
PpCCD8-derived compounds are GSs of a specific group of P. ramosa. A, Germination stimulant activities of P. patens exudates on P. ramosa group 1 and 2a seeds relative to 0.1-μM (±)-GR24 (n = 6). B, Percentage of germinated seeds of P. ramosa group 2a on plates with P. patens WT plants, Ppccd8 plants, or culture medium only, with or without 0.1-μM (±)-GR24 (n = 6). C, Seeds from P. ramosa group 1 (left) and group 2a (right) on plates with WT (top) or Ppccd8 plants (bottom), with or without 0.1-μM (±)-GR24. Arrows indicate germinating seeds. Scale bar = 0.5 mm.
Physcomitrium patens responds strongly to (+)-GR24 and carlactone application but poorly to (−)-GR24 and KAR2 in the dark
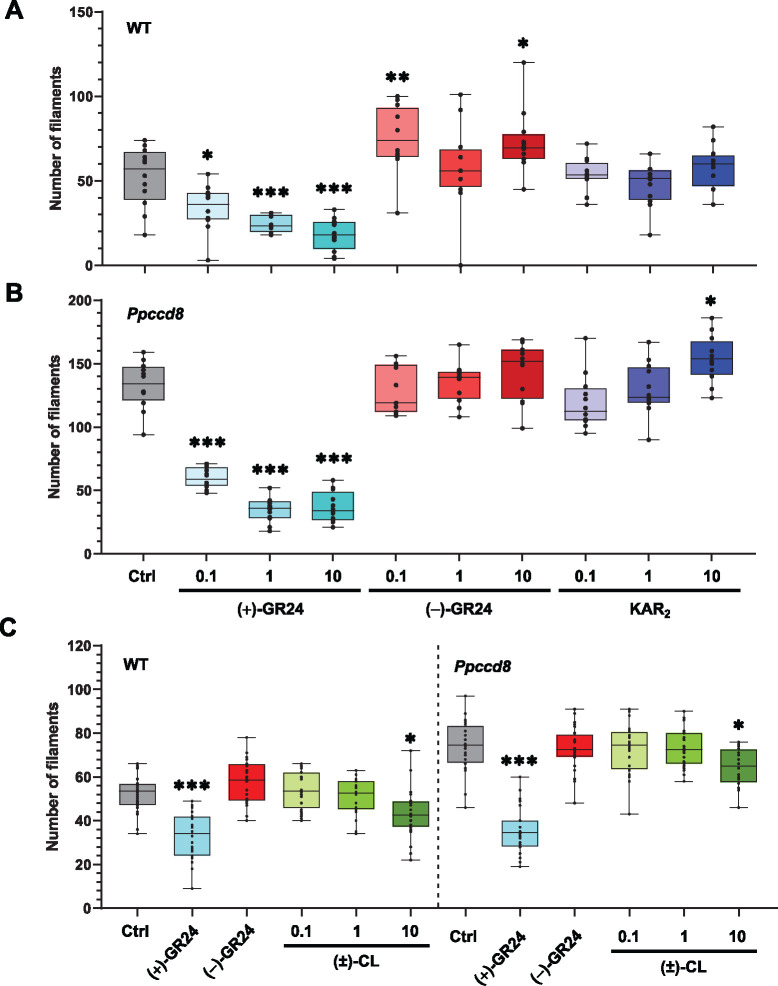
With the aim of determining which molecules could be used to mimic the yet unknown PpCCD8-derived compounds and moss KL, we tested both enantiomers of GR24 ((+)-GR24 and (−)-GR24), the SL precursor CL as racemic form, and the karrikin KAR2. To quantify the phenotypic response of P. patens to SL, we counted the number of caulonemal filaments per plant grown in the dark (Guillory and Bonhomme, 2021a; Figure 2). The number of caulonema filaments decreased following the application of (+)-GR24 in the WT and Ppccd8 in a dose-dependent manner (Figure 2, A and B). A dose of 0.1 µM was sufficient to observe a clear and significant response in both genotypes (Figure 2). No significant changes in caulonema filament number were observed with (−)-GR24, except in the WT, for which the 0.1- and 10-µM doses led to a significant increase (Figure 2A). However, in further assays (see below, Figure 9), this effect of (−)-GR24 was not repeatable.
Figure 2.
Phenotypic response to (+)- and (−)-GR24 enantiomers and natural compounds: number of caulonema filaments. Caulonema filaments were counted for the WT (A) and the SL synthesis mutant Ppccd8 (B) grown for 10 days vertically in the dark with increasing concentrations (0.1, 1, and 10 μM) of (+)-GR24 (blue boxes), (−)-GR24 (red boxes), and KAR2 (dark blue boxes). Control: 0.01% DMSO. C, Caulonema filament numbers in the WT and Ppccd8 grown for 10 days vertically in the dark with increasing concentrations (0.1, 1, and 10 μM) of (±)-CL (green boxes). Control (Ctrl): 0.01% DMSO. (+)-GR24 (blue boxes) and (−)-GR24 (red boxes) were applied at 1 μM. Significant differences were detected between control and treated plants within a genotype based on an ANOVA, followed by a Dunnett post hoc test for multiple comparisons: ***P < 0.001; **P < 0.01; *P < 0.05; For each genotype/treatment combination, n = 24 plants were grown in three different well-plates. Whiskers refer to minimum and maximum values, bars inside the boxplot to the median.
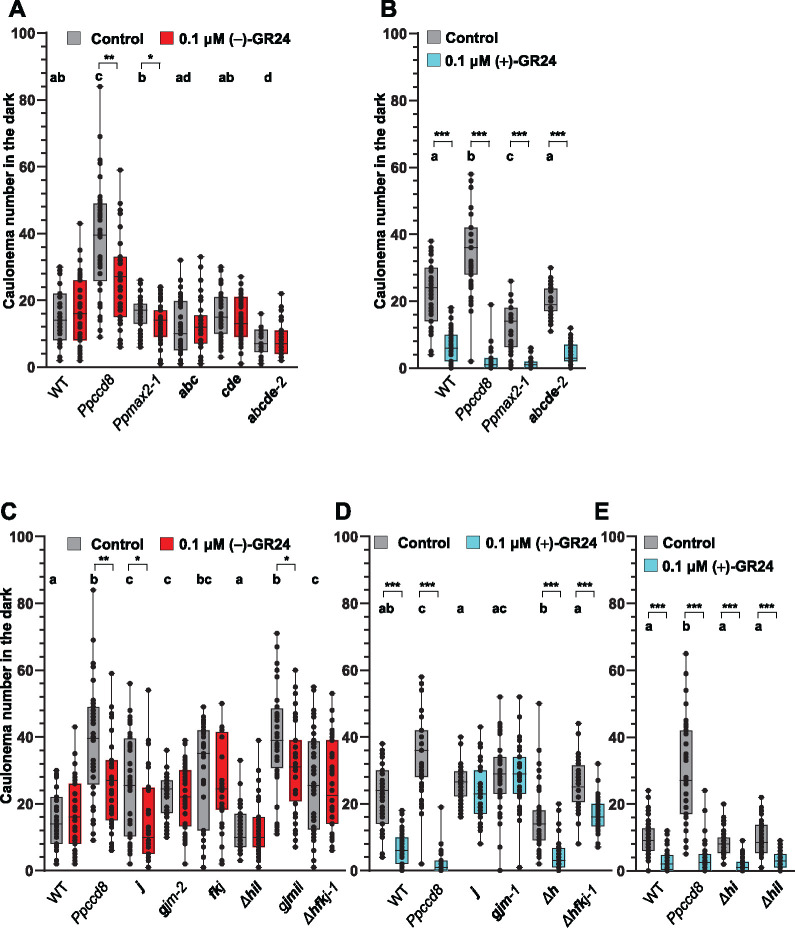
Figure 9.
Phenotypic responses of Ppkai2L mutants to (−)-GR24 and (+)-GR24 application: caulonema number in the dark. A, B, Caulonema numbers from mutants affecting eu-KAI2 clade genes following the application of 0.1-μM (−)-GR24 (in red, A) or 0.1-μM (+)-GR24 (in turquoise, B). DMSO was applied as a control treatment (dark grey). WT and both the Ppccd8 and Ppmax2-1 mutants were used as control genotypes. C–E, Caulonema numbers from mutants affecting (GJM) clade genes: j; gjm-2; gjm-1; (FK) and (GJM) clade genes: fkj, (HIL) clade genes: Δh, Δhi, Δhil, (HIL) and (GJM) clade genes: gjmil, or all three clades genes: Δhfkj-1, following the application of 0.1-μM (−)-GR24 (in red, C) or 0.1-μM (+)-GR24 (in turquoise, D, E). 0.01% DMSO was applied as a control treatment (dark grey). WT and Ppccd8 were used as control genotypes. Mutant genotypes carry mutations described in Supplemental Figure S10 and Supplemental Table S1. Bold letters indicate null mutations. For each genotype/treatment combination, caulonema were counted after 2 weeks in the dark, from 24 individuals (n = 24). Whiskers refer to minimum and maximum values, bars inside the boxplot to the median. Letters indicate statistical significance of comparisons between all genotypes under control conditions, based on a Kruskal–Wallis test followed by a Dunn post hoc test (P < 0.05). Significant differences were detected between control and treated plants within a genotype based on a Kruskal–Wallis test, followed by a Dunn post hoc test for multiple comparisons: ***P < 0.001; **P < 0.01; *P < 0.5.
Recent biochemistry experiments (Bürger et al., 2019) showed that some PpKAI2L proteins could bind to KAR1 (PpKAI2L-H, K and L), while a previous study concluded that P. patens was insensitive to KAR1 (Hoffmann et al., 2014). In the present work, we tested the KAR2 molecule, described as being more active than KAR1 in Arabidopsis (Waters et al., 2015a; Yao et al., 2021; Figure 2, A and B). KAR2 has an unmethylated butenolide group, unlike KAR1. In the WT, no significant effect on caulonema number was observed following the application of increasing doses of KAR2 (Figure 2A). In Ppccd8, we observed an increase in filament number at 10 µM (Figure 2B). To conclude, the phenotypic effects of KAR2 on P. patens were slight and not clearly dose responsive.
We also tested CL, described as the natural product of PpCCD8 in P. patens (Decker et al., 2017). As previously reported (Decker et al., 2017), racemic CL application had a negative effect on caulonema filament number for both WT and Ppccd8 plants; however, in our assays, the effect was only significant at 10 µM for both genotypes (Figure 2C).
Overall, findings from these phenotypic assays suggest that GR24 enantiomers have distinct effects in P. patens, as observed in Arabidopsis (Scaffidi et al., 2014). Indeed, the (+)-GR24 analog mimics the effects of CL, although it is far more potent, and can thus be used to mimic the effects of PpCCD8-derived compounds. On the other hand, the (−)-GR24 analog and KAR2 have slight phenotypic effects that are not always consistent or even tend to be opposite those of PpCCD8-derived compounds.
All PpKAI2L genes are expressed at relatively low levels and putatively encode proteins with a conserved catalytic triad
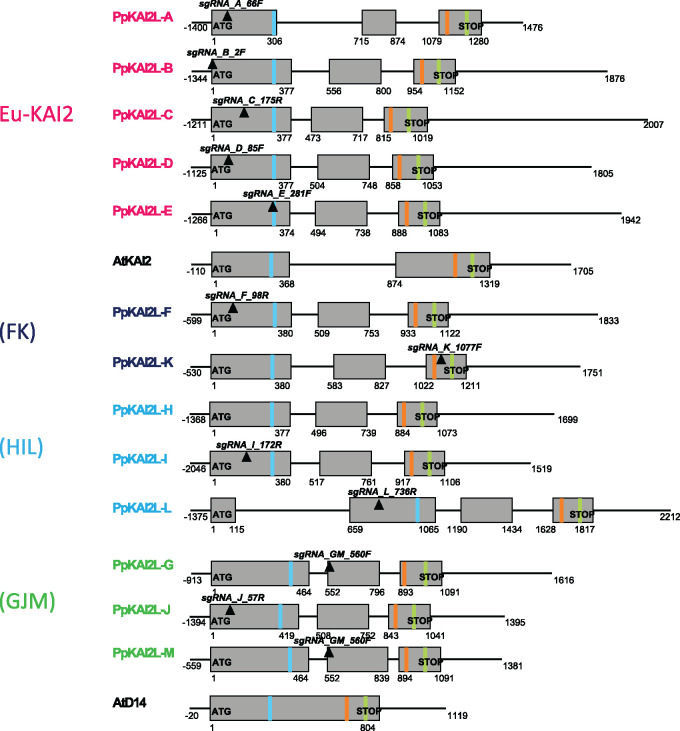
Like D14 and KAI2 genes, all 13 PpKAI2L genes encode a catalytic triad (Ser, His, Asp; Figure 3; Supplemental Figure S1). To test if the high number of PpKAI2L genes hints at different spatial and temporal expression profiles, we obtained the expression patterns of all PpKAI2L genes in P. patens, using a cDNA library from various organs/tissues (Supplemental Figure S2). PpKAI2L genes transcripts were detected in all tested tissues, at relatively low levels compared to the control genes. Quantitative reverse transcription polymerase chain reaction (RT-qPCR) could not be used to assess the expression of PpKAI2L-M, as its predicted transcript is almost identical to that of PpKAI2L-G, and the observed transcript levels are attributable to both PpKAI2L-G and -M. In spores, PpKAI2L-F and -J had higher transcript levels than other PpKAI2L genes. In protonema and gametophores, however, PpKAI2L-D from eu-KAI2 clade showed the highest transcript levels among PpKAI2L genes (Supplemental Figure S2A). The data are consistent with those previously reported (Ortiz-Ramirez et al., 2016; Perroud et al., 2018).
Figure 3.
Gene models of the PpKAI2L gene family showing the catalytic triad and position of crRNAs. Genes are presented as organized in subclades. Exons are displayed as grey boxes, introns and UTRs are depicted as thin black lines. Start and stop codons are written in bold, while plain text indicates the start/end position for each feature relative to the start codon. Only 5′-UTRs are not represented true to scale. Transcript versions that were used are V3.1 (downloaded from the Phytozome website in September 2019) for all PpKAI2L genes except for PpKAI2L-B, PpKAI2L-H, and PpKAI2L-M (V3.2). Regions targeted by crRNAs are indicated as black inverted triangles, with their names written in bold italics. Light blue, orange, and light green bands represent the codons for the S, D, and H residues of the catalytic triad, respectively (see Supplemental Table S3 for reference sequences).
The PpKAI2L-C, -D, and -E proteins are destabilized by (−)-GR24, as observed for AtKAI2; PpKAI2L-F, -K, -L, and -H interact weakly with GR24 enantiomers
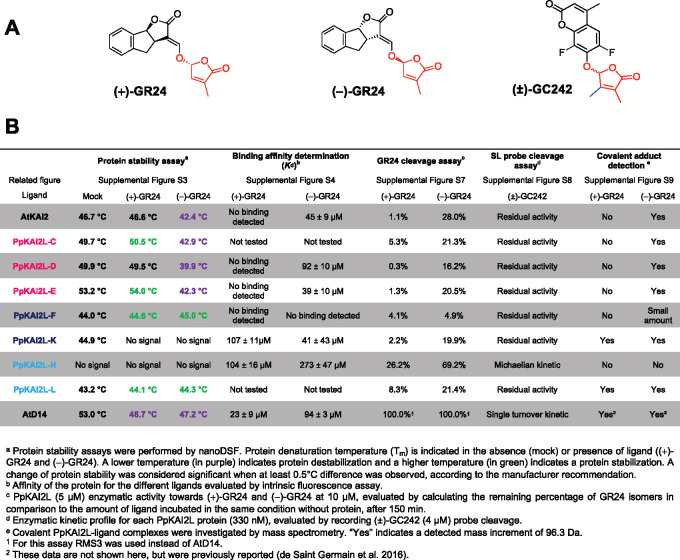
To investigate whether the PpKAI2L proteins behave similarly to AtD14 or AtKAI2 in vitro, we cloned the coding sequences (CDS) of all PpKAI2L genes and overexpressed them in Escherichia coli. After the successful purification and solubilization of seven PpKAI2L proteins (-C, -D, -E, -F, -H, -K, and -L), we investigated their interactions with SL analogs and their potential enzymatic activities. Unfortunately, due to low solubility, the six other PpKAI2L proteins could not be purified in sufficient amounts to ensure their quality. We tested the interactions of the purified PpKAI2L proteins with SL analogs via nano differential scanning fluorimetry (nanoDSF; Figure 4; Supplemental Figure S3). (+)-GR24 and (−)-GR24 enantiomers destabilized AtD14 (Supplemental Figure S3A), while AtKAI2 was destabilized by (−)-GR24 addition only (Supplemental Figure S3B), as previously reported (Waters et al., 2015a). All tested eu-KAI2 clade proteins (PpKAI2L-C -D and -E) were destabilized when (−)-GR24 was added, as was AtKAI2 (Supplemental Figure S3, C–E), in accordance with the reported stereoselectivity for unnatural (−)-5DS (Bürger et al., 2019). Puzzlingly, the PpKAI2L-C, and -E proteins showed a tendency to be stabilized by (+)-GR24 at high concentrations (Supplemental Figure S3, C–E), which was not reported when using (+)-5DS. PpKAI2L-F and PpKAI2L-L also showed a slight increase in Tm following the addition of both (−)-GR24 and (+)-GR24 (≤ 1°C), suggesting a slight stabilization (Figure 4; Supplemental Figure S3, F and I). None of the GR24 isomers affected the stability of the PpKAI2L-K or PpKAI2L-H proteins (Supplemental Figure S3, G and H).
Figure 4.
Biochemical characterization of the PpKAI2L proteins. A, Chemical structures of the (+)-GR24 and (−)-GR24 enantiomers, and the (±)-GC242 profluorescent probe. B, Summary of biochemical assays for testing interactions between PpKAI2Ls and SL analogs and probe.
Since we sometimes observed opposite effects of different isomers on the stability of the PpKAI2L proteins, we reasoned that their binding affinity had to be assessed for the GR24 isomers, rather than for (±)-GR24, as previously reported (Bürger et al., 2019). Binding affinities were quantified with Kd affinity calculations following intrinsic tryptophan fluorescence measurements (Figure 4; Supplemental Figure S4). The affinity for (+)-GR24 could be evaluated for PpKAI2L-K and PpKAI2L-H only (Kd superior to 100 µM), and was weaker than that of AtD14 (23 µM). No Kd value for (+)-GR24 could be determined for AtKAI2, eu-KAI2, or PpKAI2L-F, suggesting a very weak affinity for (+)-GR24 or a lack of binding. With (−)-GR24, comparable affinities were found for AtKAI2 (45 µM), AtD14 (94 µM), PpKAI2L-D (92 µM), PpKAI2L-E (39 µM), and PpKAI2L-K (41 µM). A much weaker affinity for (−)-GR24 was found for PpKAI2L-H (273 µM).
PpKAI2L-C, -D, -E, -K, and -L preferentially cleave (−)-GR24, while PpKAI2L-H cleaves both (−)-GR24 and (+)-GR24
As all PpKAI2L proteins contain the conserved catalytic triad, and since we found that most of them were able to bind to at least one of the GR24 isomers, we tested their enzyme activity against SL analogs. First, when incubated with the generic substrate for esterases, 4-nitrophenyl acetate (p-NPA), AtKAI2 and all tested PpKAI2L proteins showed enzyme activity (Supplemental Figure S5A-B), consistent with a previous report (Bürger et al., 2019). Kinetic constants were in the same range for all proteins but one, and similar to that of AtKAI2. The exception was PpKAI2L-H, which had a higher Vmax and KM, highlighting faster catalysis and a better affinity for p-NPA than all the others (Supplemental Figure S5C). We further characterized the enzyme activity of PpKAI2L proteins on the GR24 isomers. We compared their substrate bias to that of the pea (Pisum sativum) SL receptor RMS3/PsD14 and to that of AtKAI2 (Figure 4; Supplemental Figure S6). None of the PpKAI2L proteins showed as high an enzyme activity as RMS3/PsD14 (100% cleavage of (+)-GR24 and (−)-GR24 (de Saint Germain et al., 2016)). Only PpKAI2L-H showed a relatively high catalytic activity toward the GR24 isomers, especially toward (−)-GR24 (almost 70%, Supplemental Figure S6). AtKAI2 and all other tested PpKAI2L proteins except one selectively cleaved the (−)-GR24 enantiomer, but with low activity (less than 28%, observed for AtKAI2). Finally, PpKAI2L-F showed very low enzyme activity toward both isomers (less than 5%).
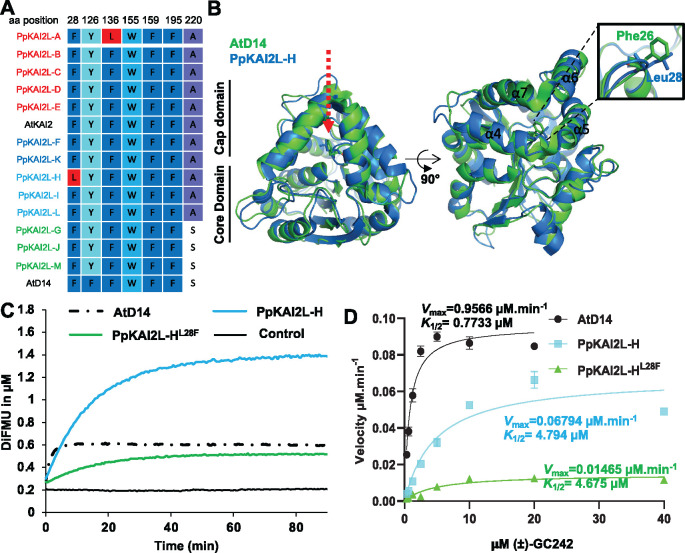
The higher cleavage activity of PpKAI2L-H on synthetic SL analogs is explained by the presence of a specific leucine residue
The higher enzyme activity of PpKAI2L-H (Supplemental Figure S6) and its lack of thermal shift when incubated with GR24 isomers (Supplemental Figure S3H) set this protein apart from other PpKAI2L proteins. To better characterize PpKAI2L-H enzyme activity, we used a pro-fluorescent probe as substrate ((±)-GC242), in which the ABC rings of GR24 are replaced by a coumarine-derived moiety (DiFMU; de Saint Germain et al. 2016). (±)-GC242 is bioactive on moss, as it reduced the number of caulonema filaments in the dark in a dose-responsive manner (evaluated on the Ppccd8 mutant, Supplemental Figure S7A). The use of (±)-GC242 as a substrate confirmed the relatively high enzyme activity of PpKAI2L-H compared to all the other PpKAI2L proteins (Supplemental Figure S7B). Indeed, after 2 h, PpKAI2L-H catalyzed the formation of 1-µM DiFMU, while other PpKAI2L activities were indistinguishable from background noise. However, the PpKAI2L-H enzymatic profile did not show the biphasic curve (a short burst phase, quickly followed by a plateau phase), which characterizes the single turnover activity of AtD14 (Supplemental Figure S7B; de Saint Germain et al., 2016). The lack of a plateau for PpKAI2L-H suggested that this protein acted as a Michaelian enzyme on the SL analog.
To try to understand this singularity, we compared the solvent-exposed residues in the binding pocket of the PpKAI2L proteins and noticed that PpKAI2L-H harbors a leucine28 (Leu28) residue instead of the phenylalanine found in AtD14 (Phe26), AtKAI2, and all other PpKAI2L proteins (Figure 5A). The Phe residue is located at the junction between helix α4 and α5, near the catalytic site (Figure 5B), and the crystal structure of PpKAI2L-H (Bürger et al., 2019) indicates that this residue interacts with the D-ring of the SL. Furthermore, a mutant PpKAI2L-H protein where Leu28 is changed to Phe showed a single turnover profile similar to AtD14, both reaching a plateau at 0.4-µM DiFMU, correlating with the protein concentration (Figure 5, C and D). PpKAI2L-H and PpKAI2L-HLeu28Phe proteins had comparable affinity toward (±)-GC242 (K1/2= 4.794 µM versus 4.675 µM) but showed different Vmax values (Vmax=0.06794 µM·min−1 vs. 0.01465 µM·min−1), suggesting that the Leu28 residue affects the velocity of catalytic activity (Figure 5, C and D).
Figure 5.
PpKAI2L-H enzymatic activity exhibits a special feature. A, Sequence alignment of active site amino acid residues in PpKAI2L proteins. Amino acids that differ from AtKAI2 are colored in red. A fully expanded alignment can be found in Supplemental Figure S1. B, Superimposition of the AtD14 and PpKAI2L-H structures showing the positions of Phe28 and Leu28 residues. A close-up view of helices α4 and α5 is shown. C, Enzyme kinetics for PpKAI2L-H, PpKAI2L- HLeu28Phe, and AtD14 incubated with (±)-GC242. Progress curves during probe hydrolysis, monitored at 25°C (λem 460 nm). The protein catalyzed hydrolysis when 330 nM of protein and 20 μM of probe were used. These traces represent one of the three replicates, and the experiments were repeated twice. D, Hyperbolic plot of pre-steady-state kinetics reaction velocity with (±)-GC242. Initial velocity was determined with pro-fluorescent probe concentrations from 0.310 to 40 μM and protein at 400 nM. Points are the mean of three replicates, and error bars represent se. Experiments were repeated at least three times.
Moss PpKAI2L proteins, like vascular plant receptors, covalently link GR24 enantiomers
To further investigate whether PpKAI2L proteins play roles as receptors of SLs and related compounds, we examined the covalent attachment of the GR24 isomers to the PpKAI2L proteins (Figure 4; Supplemental Figure S8). Mass spectrometry (MS) analyses revealed 96 Da increments (corresponding to the D ring mass) when AtKAI2 and PpKAI2L-C, -D, -E, -F, or -L were incubated with (−)-GR24. Strikingly, 96 Da increments were also observed when PpKAI2L-E, -F, -L, and -K were incubated with the other isomer (+)-GR24, in contrast to other reports (Bürger et al., 2019). However, for PpKAI2L-E, the intensity peak was much lower with (+)-GR24 than with (−)-GR24, confirming the better affinity for the latter (Figure 4; Supplemental Figure S8). PpKAI2L-H did not covalently bind the D ring following incubation with either enantiomer, further suggesting that it displays Michaelian enzyme activity.
Thus, poor interactions were observed with (+)-GR24, which was reported to mimick SLs and had the most potent effect on P. patens in our phenotypic assays (Figure 2). Strikingly, all eu-KAI2 clade proteins tested showed the strongest affinity for the (−)-GR24 enantiomer (Figure 4), which is reported to be a good mimic for the putative KL in vascular plants (Scaffidi et al., 2014; Zheng et al., 2020). We then carried out in planta studies to investigate if these PpKAI2L homologs are necessary for SL or KL perception.
None of the PpKAI2L genes complement the Arabidopsis d14-1 kai2-2 double mutant
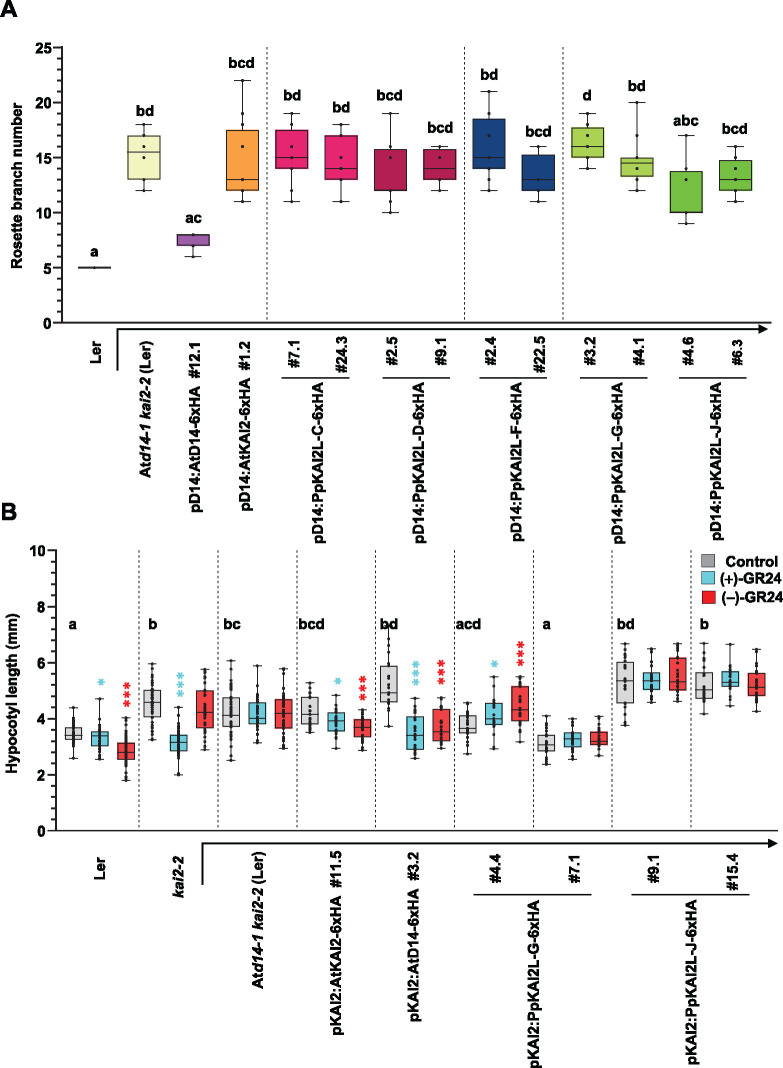
We used cross species complementation assays to test whether any of the PpKAI2L proteins could carry out similar functions in SL and/or KL signaling to that of AtD14 and/or AtKAI2 in Arabidopsis (Figure 6). CDS of the PpKAI2L-C, -D, -F, -G, and -J genes were cloned downstream of the AtD14 or AtKAI2 promoter, and the resulting constructs were expressed in the Arabidopsis double mutant Atd14-1 kai2-2, which shows both a hyperbranched phenotype and elongated hypocotyls (Supplemental Figure S9). As controls, the double mutant was transformed with the AtKAI2 or AtD14 CDS under the control of endogenous promoters. Only lines expressing AtD14 under the control of the AtD14 promoter fully restored rosette branching to WT (Ler) values (Figure 6A). Under the control of the AtD14 promoter, neither AtKAI2 nor any of the PpKAI2L genes significantly restored the branching phenotype of Atd14-1 kai2-2. We conclude that none of the tested PpKAI2L genes can fully complement the function of AtD14 in shoot branching.
Figure 6.
Complementation assays of the Arabidopsis Atd14-1 kai2-2 double mutant with PpKAI2L genes. Complementation assays of the Atd14-1 kai2-2 mutant (in the Ler background), transformed with the AtD14 promoter (A) or AtKAI2 promoter (B) controlling the expression of AtD14, AtKAI2 (controls), or PpKAI2L CDS (as noted below the graph). Ler (WT), kai2-2, and Atd14-1 kai2-2 are shown as controls. A, Number of rosette axillary branches per plant. Results are mean of n = 12 plants per genotype, except for Ler and lines pD14:AtD14 #12.1 and pD14:PpKAI2L-C#24.3 (n = 11). Different letters indicate significantly different results between genotypes based on a Kruskal–Wallis test (P < 0.05, Dunn post hoc test). B, Hypocotyl length under low light on 1/2 MS medium with DMSO (control, grey bars) 1-μM (+)-GR24 (turquoise bars) or 1-μM (−)-GR24 (red bars). Results of n = 20–24 seedlings. Whiskers refer to minimum and maximum values, bars inside the boxplot to the median. Different letters indicate significantly different results between genotypes under control conditions based on a Kruskal–Wallis test (P < 0.05, Tukey post hoc test). Asterisks in turquoise and red give the statistical significance of the response to (+)-GR24 and (−)-GR24, respectively (Mann–Whitney tests, *0.01 ≤ P < 0.05; ***P ≤ 0.001).
We also examined possible complementation of the AtKAI2 function in the Atd14-1 kai2-2 mutant by monitoring hypocotyl length under low light conditions, with or without 1 µM (+)-GR24 or (−)-GR24 in the culture medium (Figure 6B; Supplemental Figure S9). Compared to the WT, the double mutant showed longer hypocotyls under control conditions, as did the single kai2-2 mutant. The addition of (+)-GR24 or (−)-GR24 had no effect on the phenotype of the double mutant (Figure 6B). In contrast, (+)-GR24 treatment led to shorter kai2-2 hypocotyls, likely due to perception and transduction by the AtD14 protein, which is still active in the single mutant. As expected from the loss of AtKAI2 function, (−)-GR24 addition had no effect on kai2-2 hypocotyls. Surprisingly, expressing AtKAI2 under the control of the AtKAI2 promoter did not fully restore the hypocotyl length of the double mutant under our control conditions, but it did restore the response to (−)-GR24, as anticipated (Figure 6B).
Under control conditions, almost all double mutant lines expressing AtD14 or one PpKAI2L protein showed long hypocotyls (Figure 6B; Supplemental Figure S9B). Even longer hypocotyl phenotypes (compared to the double mutant) were found under control conditions for one line expressing PpKAI2L-C (#16.2) and one line expressing PpKAI2L-D (#1.4; Supplemental Figure S9B). This unexpected effect of the introduced α/β hydrolases will be discussed below. In contrast, a line expressing PpKAI2L-G (#7.1) had short hypocotyls under control conditions, suggesting that the expressed protein had indeed complemented the AtKAI2 function (Figure 6B). In a separate assay, in addition to shorter hypocotyls, all three lines expressing PpKAI2L-G in the double mutant background showed significantly larger cotyledons, further hinting at the restoration of AtKAI2 function (Supplemental Figure S9C).
When either (+)-GR24 or (−)-GR24 was added (Figure 6B), short hypocotyls (similar to the WT) were observed in the pKAI2:AtD14 expressing line, indicating that AtD14-mediated signal transduction of both enantiomers occurs when AtD14 is present in tissues where AtKAI2 is normally active. However, adding GR24 enantiomers to the medium had no such effect on most lines expressing PpKAI2L proteins (Figure 6B; Supplemental Figure S9B). Overall, these assays showed that PpKAI2L-G may mediate KL signaling in Arabidopsis hypocotyls, even though it could not fully ensure the function of AtKAI2 in this process.
Multiplex editing of PpKAI2L genes
Multiplex gene editing using CRISPR-Cas9 allowed us to isolate P. patens mutants affected in one or several PpKAI2L genes (Lopez-Obando et al. 2016b; Figure 3; Supplemental Figure S10). For the eu-KAI2 clade, two triple (abc, cde) and two quintuple mutants (abcde) were chosen for further analysis (Supplemental Table S1). The remaining (HIL), (FK), and (GJM) clades were targeted in separate experiments using combinations of specific CRISPR RNAs (crRNAs) for each gene. As biochemistry experiments suggested a purely enzymatic role for PpKAI2L-H, a single deletion mutant was obtained for the PpKAI2L-H gene through homologous recombination, where the full CDS was removed from the moss genome (Δh mutant; Supplemental Figures S10 and S11). This Δh mutant was employed in further transformation experiments with crRNAs from the same (HIL) clade and/or from the (FK) and (GJM) clades, generating the Δhil and Δhfkj mutants (Supplemental Table S1). Eventually, a 7X mutant was obtained (Δhifkgjm) where all the mutations except those in PpKAI2L-J and -M were null (Supplemental Table S1 and see below). For convenience, null mutations are hereafter indicated in bold letters.
The eu-KAI2 clade quintuple mutants phenocopy Ppmax2-1 in white light, while mutants in other clades are more similar to the WT or Ppccd8
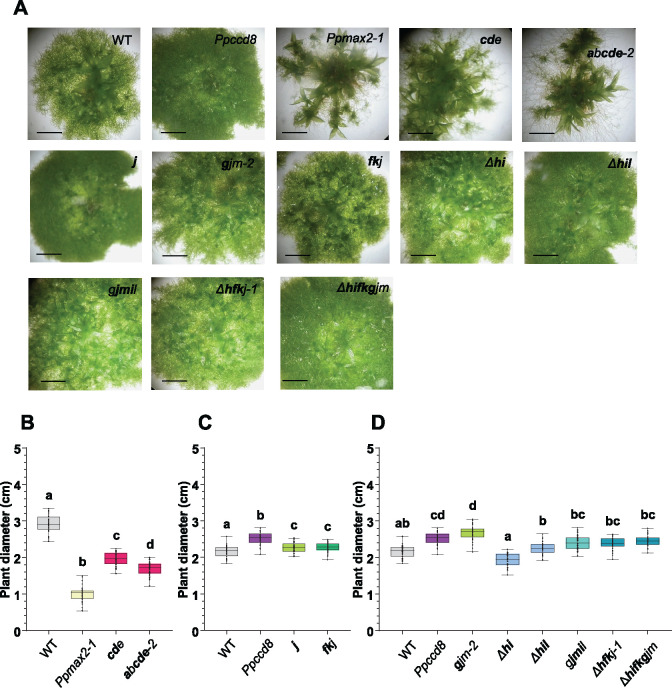
Our rationale was that a mutant affected in the response to PpCCD8-derived compounds should show a phenotype similar to that of the SL biosynthesis mutant Ppccd8. We first performed a phenotypic analysis of the mutants in the light. After 4 weeks of culture, Ppccd8 plants were slightly bigger than the WT (Proust et al., 2011; Figure 7), whereas Ppmax2-1 plants were smaller, with fewer but bigger gametophores (Lopez-Obando et al., 2018; Figure 7). The diameter of mutants in eu-KAI2 clade genes was significantly smaller than that of Ppccd8 and the WT and slightly larger than that of Ppmax2-1 (Figure 7, A and B). The phenotype of the eu-KAI2 clade mutants, with early and large gametophores, resembled that of Ppmax2-1, although not as pronounced (Figure 7A). To the naked eye, 3-week-old plants from mutants in genes from the (FK), (GJM), and (HIL) clades were indistinguishable from WT (Figure 7A). After a month of growth, however, all mutants affecting genes from the (FK) and (GJM) clades showed a slightly larger diameter, intermediate between that of the WT and Ppccd8 (Figure 7D). Mutants in the (HIL) clade such as Δh, Δhi, and Δhil were comparable to the WT (Figure 7D). Thus, based on mutant phenotypes in the light, eu-KAI2 clade genes would be involved in a PpMAX2-dependent pathway, while (FK) and (GJM) clade genes would instead be involved in the PpCCD8-derived SLs pathway. Genes from the (HIL) clade would not be involved in either pathway.
Figure 7.
Phenotypes of the Ppkai2L mutants in the light. A, Three-week-old plants. Scale bar = 2 mm. (B) and (C) Diameters of 30-day-old plants (n > 40) and (D) 28-day-old plants (n = 30). B–D, All plants were grown on cellophane disks. Whiskers refer to minimum and maximum values, bars inside the boxplot to the median. Letters indicate statistical significance of comparisons between all genotypes based on a Kruskal–Wallis test followed by a Dunn post hoc test (P < 0.05). Mutant genotypes carry mutations as described in Supplemental Figure S10 and Supplemental Table S1. Bold letters indicate null mutations.
Like Ppmax2, eu-KAI2 clade quintuple mutants are affected in photomorphogenesis
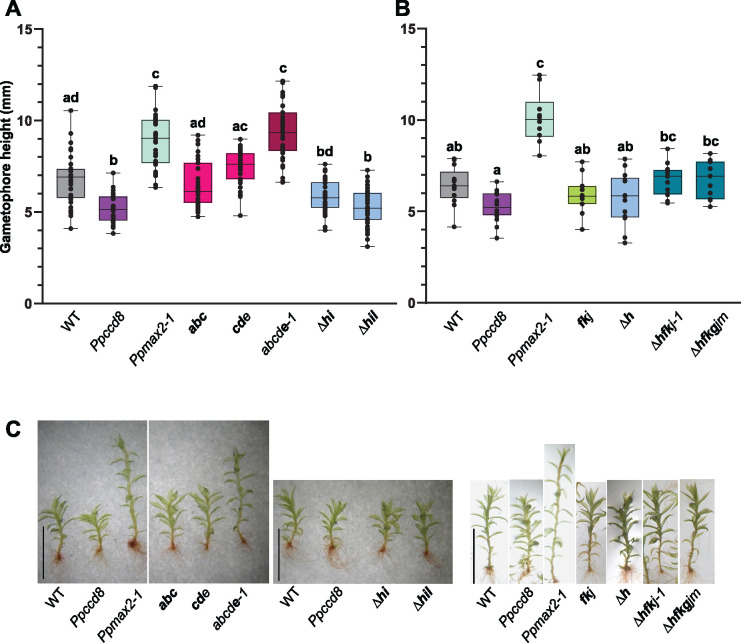
Mutants in eu-KAI2 clade genes showed the typical phenotype of the Ppmax2-1 mutant in white light. We previously showed that the Ppmax2-1 mutant is affected in photomorphogenesis under red light (Lopez-Obando et al., 2018). After 2 months of growth under red light, Ppmax2-1 gametophores were much more elongated than WT gametophores, whereas Ppccd8 gametophores were shorter (Figure 8). Among eu-KAI2 mutants, gametophores of both triple mutants abc and cde were a similar height to WT. Interestingly, the quintuple mutant (abcde-1) showed significantly elongated gametophores, similar to Ppmax2-1 (Figure 8, A and C). The other tested quintuple mutant (abcde-2) also had elongated gametophores under red light, intermediate between WT and Ppmax2-1 (Supplemental Figure S12). The weak phenotypes of both triple mutants (Figure 8A) suggest a functional redundancy among genes of the eu-KAI2 clade, as knockout (KO) mutations for PpKAI2L-A, B, C, and/or D did not result in plants with gametophores as elongated as those of the Ppmax2-1 mutant.
Figure 8.
Ppkai2L gametophores in red light. A, Gametophore height of Ppkai2L affecting eu-KAI2 clade genes (abc; cde; abcde-1) and clade (HIL) genes (Δhiand Δhil) compared to that of the WT, Ppccd8, and Ppmax2-1 following 2 months of growth under red light. Box plots of n = 32–36 gametophores, grown in three magenta pots, harboring between 15 and 25 leaves. Whiskers refer to minimum and maximum values, bars inside the boxplot to the median. Letters indicate statistical significance of comparisons between all genotypes based on a Kruskal–Wallis test followed by a Dunn post hoc test (P < 0.05). B, Gametophore heights of Ppkai2L mutants affecting clade (HIL) gene (Δh), both clades (FK) and (JGM) (fkj), and all three clades (HIL), (FK), and (JGM) (Δhfkj-1 and Δhfkgjm). Box plots of n = 11–15 gametophores, grown in three Magenta pots, harboring between 15 and 25 leaves. Whiskers refer to minimum and maximum values, bars inside the boxplot to the median. Letters indicate statistical significance of comparisons between all genotypes based on a Kruskal–Wallis test followed by a Dunn post hoc test (P < 0.05). C, Examples of gametophores following 2 months of growth under red light, from the WT, Ppccd8, Ppmax2-1, and Ppkai2L as shown in (A) and (B). Scale bar = 5 mm. Mutant genotypes carry mutations as described in Supplemental Figure S10 and Supplemental Table S1. Bold letters indicate null mutations.
Gametophores from mutants where genes from the (FK) and/or (GJM) clades were mutated (fkj, Δhfkj-1, and Δhfkgjm) were similar in height to WT, suggesting that genes from clade (FK) and clade (GJM) have no role in photomorphogenesis in red light (Figure 8, B and C; Supplemental Figure S12). (HIL) clade triple mutant Δhil showed shorter gametophores under red light, similar to Ppccd8 (Figure 8A). In contrast, the gametophores of both the single Δh and the double Δhi mutants were intermediate in height between the WT and Ppccd8 (Figure 8, A and B). This suggests a specific role for the (HIL) clade genes, with an opposite impact on gametophore development compared to that of the PpMAX2 pathway.
In conclusion, the phenotypes of the Ppkai2L mutants in red light allowed us to differentiate the functions of the eu-KAI2 clade genes. These genes are likely to be involved in a PpMAX2-dependent pathway related to photomorphogenesis, whereas genes from the three other clades (DDK superclade) are more likely to play roles independent of PpMAX2.
Mutants in the (GJM) clade no longer respond to (+)-GR24 application
To determine which of the Ppkai2L mutants carry mutations in potential receptors for PpCCD8-compounds (SL-related) or other (KL-related) compounds, we tested their phenotypic responses to GR24 enantiomers at 0.1 µM (Figure 9). In the assays reported above on the WT and Ppccd8 in the dark (Figure 2), (−)-GR24 appeared to be a poor KL mimic in moss. However, the stereospecificity of the PpKAI2L proteins for (−)-GR24 led us to pursue our assays with this enantiomer.
For the eu-KAI2 clade, under control conditions, all mutants showed an equivalent number of filaments to WT, except for the quintuple mutant. The latter tended to have fewer filaments under control conditions, another similar phenotype to Ppmax2-1 (Figure 9A; Supplemental Figure S13A). The addition of 0.1-µM (−)-GR24 had no significant effect on eu-KAI2 clade mutants or the WT (Figure 9A). However, in this assay, both Ppccd8 and Ppmax2-1 showed a significant decrease in caulonema filament number in response to 0.1-µM (−)-GR24. In a separate experiment, a dose of 1 µM of (−)-GR24 had opposite effects on the WT and Ppccd8 filament number, with an increase and decrease, respectively (Supplemental Figure S13A; Figure 2 above), but had no significant effect on the Ppmax2-1 mutant, although a tendency toward a decrease was observed. At this higher dose, the quintuple eu-KAI2 clade mutant showed a significant decrease in caulonema filament number, like Ppccd8, but in contrast to the WT. Thus, similar to PpMAX2 loss of function, mutating eu-KAI2 genes did not abolish a response to the (−)-GR24 enantiomer. The application of (+)-GR24 had a significant negative effect on the number of filaments for the quintuple abcde mutant, like for WT and the Ppccd8 and Ppmax2-1 mutants (Figure 9B). Thus, mutating any of the eu-KAI2 clade genes did not hamper the response to (+)-GR24, and therefore it likely does not hamper the responses to PpCCD8-derived compounds.
We then tested the effects of GR24 enantiomers on Ppkai2L mutants from the three other clades: (FK), (GJM), and (HIL). Strikingly, under control conditions, like Ppccd8, all the mutants had more filaments than WT, except for mutants in the (HIL) clade, which were similar to the WT (Figure 9, C–E) or tended to have fewer filaments (Supplemental Figure S13). Both the single j mutant and the quintuple mutant gjmil showed a significant response to (−)-GR24 (fewer caulonema), as did Ppccd8 (Figure 9C; Supplemental Figure S13, A and B). No clear response to (−)-GR24 was seen in mutants with KO mutations in PpKAI2L-F, -K, -H, -G, -M, -I, or -L, like for the WT (Figure 9C; Supplemental Figure S13, A and B). Finally, we examined the response to (+)-GR24 for mutants of the (FK), (GJM), and (HIL) clades (Figure 9, D and E; Supplemental Figure S13, C and D). The number of caulonema was clearly reduced in the WT and Ppccd8 (as shown above, Figure 2), and in mutants carrying the Δh mutation, alone or in combination with f, k, i, or l null mutations. Thus, genes from the (FK) and (HIL) clades do not play a role in the response to (+)-GR24. However, the response to the (+)-GR24 enantiomer was abolished in all mutants where the PpKAI2L-J gene was knocked out (Figure 9D:j and gjm-1; Supplemental Figure S13C: gjmil and Δhfkj-3 and Supplemental Figure S13D: gjm-3 and gjm-5). Interestingly, in the two lines where the j mutation was not null but the PpKAI2L-G gene was knocked out (7x Δhifkgjm and gjm-4 mutants), the response to (+)-GR24 was also abolished (Supplemental Figure S13, C and D). Thus, based on phenotypic assays of mutant caulonema, both the PpKAI2L-J and -G genes appear to be necessary for the response to (+)-GR24 and are therefore the best candidates for receptors to PpCCD8-derived SLs.
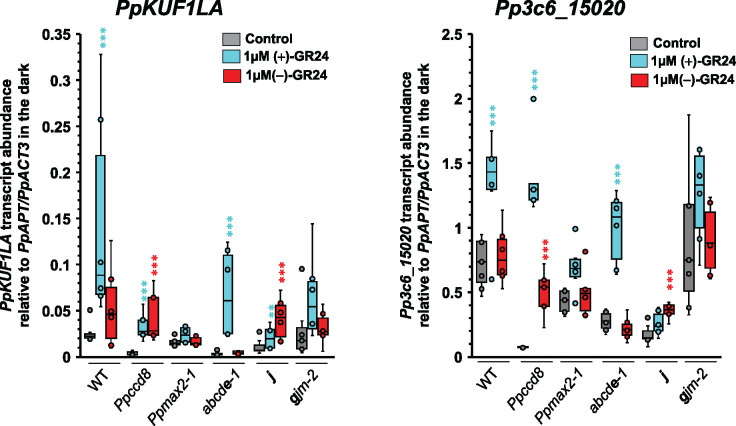
To further test if PpKAI2L-J and -G could be receptors for PpCCD8-derived SLs, we measured the transcript levels of SL-responsive genes in the corresponding mutants (Figure 10). We previously reported that in the WT and Ppccd8 plants, PpKUF1LA gene transcript abundance increases 6 h after plant transfer onto medium containing 3-µM (±)-GR24 and that this response is enhanced in the dark (Lopez-Obando et al., 2018). We used this marker along with the Pp3c6_15020 gene (encoding a putative histidine kinase) that was previously found to be upregulated by (±)-GR24 (Supplemental Figure S14). Using GR24 enantiomers, we confirmed that the transcript levels of both genes increased following 1-µM (+)-GR24 addition in the WT and Ppccd8, but not in Ppmax2-1. Strikingly, an increase in transcript levels following (−)-GR24 application was observed for both markers in Ppccd8, and for PpKUF1LA only in the WT (Figure 10). In the quintuple eu-KAI2 mutant, the addition of (+)-GR24 but not (−)-GR24 increased PpKUF1LA and Pp3c6_15020 transcript levels. In contrast, in the j mutant, the transcript levels of both genes increased following (−)-GR24 addition and were slightly increased (PpKUF1LA) or unchanged (Pp3c6_15020) in response to (+)-GR24 application. In the gjm-2 mutant, the response marker transcript levels were unchanged following either (+)-GR24 or (−)-GR24 addition. Thus, the transcriptional responses of the tested mutants confirm the notion that eu-KAI2 genes are not involved in the response to (+)-GR24, while this response is impaired in (GJM) clade mutants. Only the Ppccd8 and j mutants showed a clear and significant transcriptional response (for both markers) to (−)-GR24 addition.
Figure 10.
Transcriptional responses of the Ppkai2L mutant to (+)-and (−)-GR24. Analysis of the transcript abundance of the SL-responsive genes PpKUF1LA and Pp3c6_15020, in the WT, Ppccd8, Ppmax2-1, and Ppkai2L mutants abcde-1 (eu-KAI2 clade), j, and gjm-2 ([GJM] clade), after a 6-h treatment in the dark with DMSO (control, grey), 1-μM (+)-GR24 (turquoise), or 1-μM (−)-GR24 (red). Box plots of at least four biological repeats are shown (n ≥ 4), relative to mean (PpAPT-PpACT3) transcript abundance. Whiskers refer to lower and upper quartiles, bars inside the box to the median. Kruskal–Wallis test followed by a Dunnett post hoc test (asterisks indicate significant differences between the treatment and the corresponding DMSO control; **P < 0.01 and ***P < 0.001).
Discussion
Are PpCCD8-derived products noncanonical SLs?
P. patens exudates were previously reported to induce the germination of Orobanche ramosa (old name for Phelipanche ramosa) seeds (Decker et al., 2017), but the origin of the tested population was not specified. Here, we further demonstrated that PpCCD8-derived products are GSs of P. ramosa group 2a seeds that were harvested in a hemp field but do not induce the germination of P. ramosa group 1 seeds collected from an oilseed rape field. Differences in the susceptibility of root parasitic weeds can be attributed to the chemical nature of host plant exudates (Yoneyama et al., 2018b). Our results suggest that the PpCCD8-derived products share similarities with hemp secondary metabolites. So far, no known canonical SLs have been isolated from hemp (Huet et al., 2020). Since P. patens likely produces CL (Decker et al., 2017) but lacks a true MAX1 homolog (Proust et al., 2011), we hypothesize that PpCCD8-derived compounds correspond to noncanonical SLs derived from CL or hydroxyl CLs (Yoneyama, 2020). Further supporting this hypothesis, we previously showed that GR5, a noncanonical SL analog, was as bioactive as (±)-GR24 on P. patens caulonema length (Hoffmann et al., 2014). As mimics of SLs in the present study, we used the (+)- and (−)-GR24 artificial analogs that were available at the time of our study (see below). Of note, both isomers are active on P. ramosa group 1 and group 2a seeds. However, the (+)-GR24 isomer, which has a canonical SL structure, has similar germination stimulating activity to (±)-GR24, while the (−)-GR24 isomer is far less active (de Saint Germain et al., 2019). For further identification of PpCCD8-derived SLs in the future, noncanonical SL analogs such as the recently described methyl phenlactonoates (Jamil et al., 2020) may be more appropriate (see also below).
Looking for the best mimic of SLs or KL
The (−)-GR24 analog has a non-natural configuration that has never been found in plant exudates, contrary to the (+)-GR24 enantiomer, which bears similarity to 5-deoxystrigol ((+)-5DS) and strigol-type canonical SLs (Scaffidi et al., 2014). In our bioassays of moss phenotypes, CL application decreased the number of caulonema of both WT and Ppccd8 at 10 µM. A similar (but much stronger) effect was observed with (+)-GR24, which we thus consider to be the best mimic of PpCCD8-derived compounds available so far. It is not surprising that (+)-GR24 is more potent than CL, as the assays were carried out in aqueous medium, and natural SLs were described as being far less stable than synthetic analogs in aqueous medium (Akiyama et al., 2010; Boyer et al., 2012). Moreover, we cannot exclude the possibility that CL needs to be transformed into a more bioactive noncanonical SL in planta to trigger the effects observed here. In contrast, the effects of (−)-GR24 are weak, not dose responsive, and sometimes contradictory in the WT versus Ppccd8. Indeed, in several assays, we observed a significant increase in caulonema number in the WT (Figure 2A; Supplemental Figure S13A) while this number consistently decreased in Ppccd8, mimicking the result of SL application (Figure 9; Supplemental Figure S13, A and B).
Interestingly, we also observed an increase in caulonema number when testing KAR2, although this increase was only significant at 10 µM for Ppccd8 (Figure 2B). So far we had not observed any effect of karrikins (KAR1) on the phenotypes of P. patens (Hoffmann et al., 2014), and this is thus the first hint of a possible effect of some karrikins on moss, which needs to be confirmed. We hypothesize that the increase in caulonema filament number is the effect triggered by the as yet unidentified moss KL (see also below and Figure 11). It is puzzling, however, that the effect of KAR2 is more easily observed in Ppccd8 (thus in the absence of SLs) than in the WT, while the same effect of (−)-GR24 (increasing of filament number) is only seen in the WT. We propose (Figure 11 and see below) that (GJM) clade PpKAI2L proteins perceive (−)-GR24, which would explain the apparent contrary effects of this enantiomer. In the absence of endogenous SL (in the Ppccd8 mutant), (−)-GR24 would thus trigger the SL pathway. This could also explain the transcriptional response to (−)-GR24 observed in Ppccd8 (Figure 10). Thus, (−)-GR24 is not a very robust mimic of the as yet unidentified moss KL, and KAR2 is likely not a potent KL mimic either. This conclusion also suggests that P. patens KL may be quite different from that of angiosperms. Recently, desmethyl butenolides were reported to be more potent and to function as specific ligands for KAI2, both in vitro and in plants, including the lycophyte Selaginella moellendorffii and the bryophyte Marchantia polymorpha (Yao et al., 2021). Thus, testing desmethyl butenolides, and in particular the (−)-desmethyl GR24 enantiomer (dGR24ent5DS) on P. patens plants and proteins is a priority in the search for a KL mimic.
Figure 11.
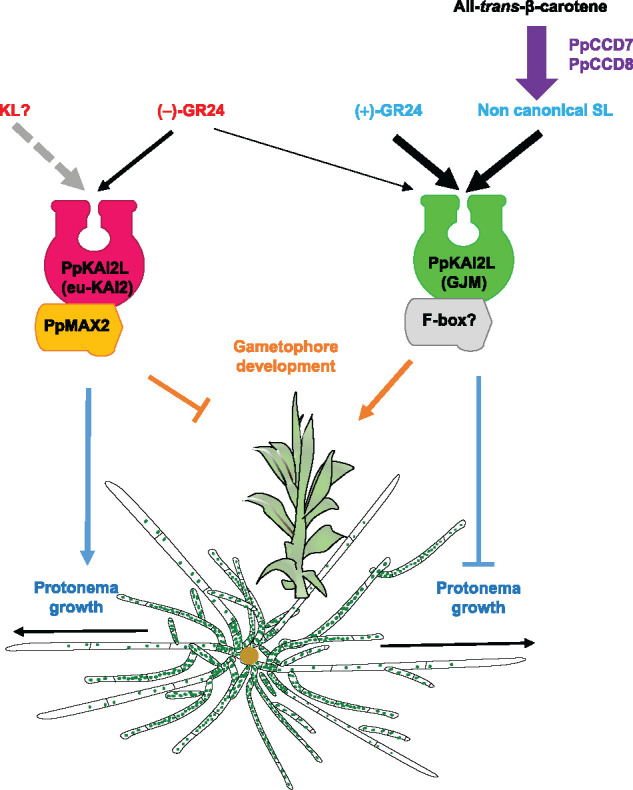
Current model for SL (PpMAX2-independent) and KL (PpMAX2-dependent) perception in P. patens. Eu-KAI2 clade PpKAI2L proteins perceive KL compounds, which promote protonema growth and inhibit gametophore development through a PpMAX2-dependent pathway. (GJM) clade PpKAI2L proteins perceive PpCCD8-derived compounds (noted noncanonical SL), which inhibit protonema growth while promoting gametophore development, in a PpMAX2-independent manner. (+)-GR24 mimics the effects of PpCCD8-derived compounds and is perceived by (GJM) clade PpKAI2L proteins. (−)-GR24 is perceived by both eu-KAI2 clade and (GJM) clade PpKAI2L proteins, making it a poor mimic of moss KL. Grey dotted arrow indicates nondemonstrated effect. Thickness of black arrows indicates the strength of the compound’s effect.
Biochemical characterization suggests that PpKAI2L proteins from the eu-KAI2 clade act as KL receptors
Among the analogs examined, we observed the best binding of the (−)-GR24 enantiomer to AtKAI2 and all tested PpKAI2L proteins from the eu-KAI2 clade. This confirms a recent report (Bürger et al., 2019) of the preferential binding of (−)-5DS by PpKAI2L-C, -D, and -E. In addition, we found that the (+)-GR24 enantiomer interacts poorly with these PpKAI2L proteins, but not with AtKAI2, indicating less stringency for moss eu-KAI2 clade proteins. Still, this result suggests that eu-KAI2 proteins share a perception mechanism with AtKAI2, and furthermore, they may recognize KL-like compounds. The moss KL compound(s) may, however, differ from the angiosperm KL, since the expression of PpKAI2L-C or -D did not complement the kai2-2 hypocotyl phenotype in Arabidopsis, or more likely, the protein partners in Arabidopsis are different (see also below).
It should be noted that AtKAI2 is degraded following KAR perception, in a MAX2-independent manner (Waters et al., 2015a), and this could be tested on eu-KAI2 clade proteins. As with the two other tested clades, (FK) and (HIL), none of the interaction assays revealed any preferential binding of GR24 isomers, despite the use of pure enantiomers. Indeed, only PpKAI2L-K showed similar binding affinity with both (−)-GR24 and (+)-GR24, but no stereoselectivity. Unfortunately, none of the PpKAI2L proteins from the (GJM) clade could be purified for interaction assays, which was also reported by Bürger et al. (2019). In the future, overexpression in P. patens or in other heterologous systems (yeast, insect cells) may be a solution for producing these proteins and carrying out biochemistry studies.
PpKAI2L-H is the most efficient hydrolase among PpKAI2L proteins
The PpKAI2L-H protein showed stronger cleavage activity toward both GR24 stereoisomers as well as toward the synthetic probe (±)-GC242 compared to any other PpKAI2L protein, but also compared to the Arabidopsis AtKAI2 and AtD14 proteins. Mutating the Leu28 residue into a Phe was sufficient to reduce the efficiency of this activity (strong reduction of the kcat and of the Vmax), but had no effect on the K1/2 toward (±)-GC242. The efficient cleavage activity of PpKAI2L-H is therefore not likely due to a stronger affinity of this protein for the substrate. It has been hypothesized that the Leu28 residue (like the Phe181 residue), which is unique to PpKAI2L-H, does not particularly enlarge the pocket size of PpKAI2L-H (Bürger et al., 2019). Therefore, our results may highlight the ability of a residue to control the exit of the cleavage reaction product. Indeed, the conserved Phe28 residue in D14/KAI2 proteins may act as a gate keeper, and this could explain the single turnover kinetics observed with some SL analogs (de Saint Germain et al., 2016). We hypothesize that the high velocity of PpKAI2L-H may be linked to the lack of this gate keeper residue, allowing a high substrate turnover. The stronger enzyme activity of PpKAI2L-H could have a specific role in P. patens, perhaps as a catabolic enzyme, to regulate the levels of bioactive signaling molecules (Seto et al., 2019).
When only the PpKAI2L-H gene was mutated (Δh mutant), no striking phenotype was observed (Figure 7), and in particular, the phenotypic response to (+)-GR24, which mimics PpCCD8-derived SLs, was similar to that of WT plants (Figure 9D). In red light, however, the gametophores of Δhi and Δhil mutants were less elongated than WT gametophores, which was also observed in Ppccd8 (Figure 8). However, in Ppccd8, this could be related to the higher number of filaments, leading to the initiation of more (but smaller) gametophores, whereas the number of filaments in the dark was not higher in mutants from the (HIL) clade (Figure 9 and Table 1). In addition, these mutants even tended to have fewer filaments than the WT (Figure 9, D and E; Supplemental Figure S13A). Thus, mutants from the (HIL) clade are quite different from Ppccd8, and the corresponding genes PpKAI2L-H-I and -L likely do not encode receptors for PpCCD8-derived compounds. Notably, our results do not support a likely role for PpKAI2L-H in the KL pathway, even though the binding of KAR1 by this protein was reported (Bürger et al. 2019). Indeed, the Δh and higher order mutants (Δhi and Δhil) tended to be smaller than WT in white light (Figure 7D and Table 1), but also showed a slight reduction in gametophore height in red light (Figure 8A), in contrast to Ppmax2-1 and eu-KAI2 clade mutants (Table 1). These findings suggest that (HIL) clade genes play a specific role in P. patens development. The association of this role with PpKAI2L-H enzyme activity remains to be discovered.
Table 1.
Overview of Ppkai2L mutant phenotypes
| Genotypes | Diameter | Gametophore Size | Caulonema Number | Response to (+)-GR24 |
Response to (−)-GR24 |
||
|---|---|---|---|---|---|---|---|
| Caulonema number | Transcriptional markers | Caulonema number | Transcriptional markers | ||||
| (Figure 7) | (Figure 8, Supplemental Figure S12) | (Figures 2 and 9; Supplemental Figure S13) | (Figures 2 and 9; Supplemental Figure S13) | (Figure 10) | (Figures 2 and 9; Supplemental Figure S13) | (Figure 10) | |
| WT | WT | WT | WT | ↘ | ↗ | ↗ or = | = |
| Ppccd8 | >WT | <WT | >WT | ↘ | ↗ | = or ↘ | ↗ |
| Ppmax2-1 | <WT | >WT | <WT | ↘ | = | = or ↘ | = |
| eu-kai2 | <WT | >WT | ≤WT | ↘ | ↗ | = or ↘ | = |
| (fk) a | ≥WT | ≈WT | ≥WT | ↘ | Not tested | = | Not tested |
| (hil) | ≤WT | ≤WT | ≤WT | ↘ | Not tested | = | Not tested |
| (gjm) | ≥WT | ≤WT | >WT | = | ↗ or = | = or ↘ | ↗ or = |
In combination with j and/or h mutations.
The Arabidopsis Atd14-1 kai2-2 mutant complementation assay provides important clues about PpKAI2L activity
Using the endogenous AtD14 promoter, we confirmed that expression of the PpKAI2L-C protein does not complement the Arabidopsis D14 function in rosette branching, as previously reported using the 35S promoter (Bürger et al., 2019). We can now extend this observation to PpKAI2L-D, PpKAI2L-F, PpKAI2L-G, and PpKAI2L-J. Using the endogenous AtKAI2 promoter, we also confirmed that PpKAI2L-C cannot complement the kai2-2 mutation, as observed by (Bürger et al., 2019). Furthermore, we extended this observation to PpKAI2L-D and PpKAI2L-J. However, expressing moss PpKAI2L-G reduced the hypocotyl size and led to larger cotyledons than in the double mutant Atd14-1 kai2-2 (Supplemental Figure S9C), suggesting that PpKAI2L-G may be able to perceive and transduce the endogenous KL signal, even though it does not respond to (−)-GR24. Similarly, when expressed in Arabidopsis, one of the two KAI2 homologs from S. moellendorffi (SmKAI2b) partially complemented the kai2 mutant, whereas the other homolog and both the KAI2 homologs from M. polymorpha did not restore the kai2 mutant phenotypes (Waters et al., 2015b). Strikingly, when expressed under the control of the AtKAI2 promoter in the Arabidopsis Atd14-1 kai2-2 mutant, the moss PpKAI2L-C or -D proteins exacerbated the defect induced by the kai2-2 mutation by leading to even more elongated hypocotyls (for each construct, in one line out of two, Supplemental Figure S9B). This suggests a putative interaction of these proteins with the Arabidopsis KAI2/KL pathway, which should be further investigated. Still, it is clear that none of the PpKAI2L proteins fully complements the AtD14 or KAI2 function, likely due to defective interactions with AtMAX2 and/or other components of SL/KL pathways.
Genetic analysis suggests that genes from the eu-KAI2 clade are involved in the PpMAX2-dependent pathway
Mutant phenotypes clearly distinguish the eu-KAI2 clade from the three other clades (Table 1). Indeed, the quintuple eu-KAI2 mutant shows a phenotype in white light quite similar to that of Ppmax2-1. It also has elongated gametophores under red light, and few caulonema filaments in the dark, suggesting that eu-KAI2 and PpMAX2 proteins could be members of the same pathway (Figure 11). As eu-KAI2 proteins preferentially bind to the (−)-GR24 enantiomer, we expected the mutants in this clade to be blind to (−)-GR24 application. This is what we observed when transcriptional response markers were examined, with transcript levels remaining unchanged following (−)-GR24 application in both the Ppmax2-1 and the quintuple eu-KAI2 clade mutant, but increasing in Ppccd8 (but not in the WT; Figure 10 and Table 1). However, as mentioned above, (−)-GR24 does not appear to be a perfect mimic of the unknown moss KL, and other transcriptional response markers need to be found that would better reflect the moss KL response. As for the phenotypic response, the application of 0.1-µM (−)-GR24 had no effect on caulonema number in the dark in the eu-KAI2 clade mutants or in the WT, but it significantly decreased caulonema number in both Ppccd8 and Ppmax2-1 (Figure 9A). Strikingly, a 1 µM concentration of (−)-GR24, which had the opposite effect on the WT (increased caulonema number) and Ppccd8 (decreased caulonema number), produced no response in the Ppmax2-1 mutant, while the quintuple eu-KAI2 mutant showed a significant decrease in caulonema number under this treatment (Supplemental Figure S13). Thus, the quintuple eu-KAI2 mutant is still able to perceive (−)-GR24, as is the Ppmax2-1 mutant. As mentioned above, we hypothesize that other PpKAI2L proteins (presumably members of the (GJM) clade, see below) may perceive (−)-GR24, triggering the PpCCD8-derived compounds pathway (Figure 11). This does not rule out the possibility that eu-KAI2 clade and PpMAX2 proteins function in the same pathway. In addition, (−)-GR24 was reported to have a dual effect, promoting both the KAI2 and the D14 pathways in Arabidopsis roots (Villaecija-Aguilar et al., 2019).
Finally, both phenotypic responses in the dark and transcriptional responses to the (+)-GR24 enantiomer were unaffected in the eu-KAI2 clade mutants (Table 1), indicating that eu-KAI2 proteins are not receptors for PpCCD8-derived compounds.
PpKAI2L- G, -J, and -M mediate PpCCD8-derived (SL-related) responses
In white light (Figure 7), mutants affecting (FK) and (GJM) clades showed phenotypes intermediate between those of WT and Ppccd8. In the dark under control conditions (Figure 9), the caulonema numbers of mutants in the (FK) and (GJM) clades were also intermediate between those of the WT and Ppccd8 (see also Table 1). Based on the hypothesis that synthesis and response mutants show similar phenotypes, genes from the (FK) and (GJM) clades are thus the best candidates for PpCCD8-derived compound receptors. When the phenotypic responses of these mutants to (+)-GR24 application were examined, plants with KO mutations in PpKAI2L-J or PpKAI2L-G/M no longer responded to this compound (Figure 9, C and D, j and gjm-1; Supplemental Figure S13D, gjm-3, gjm-5, and gjm-4). In contrast, both Δhfkj-1 and Δhfkj-2 mutants showed a significant response to (+)-GR24 application (Figure 9D; Supplemental Figure S13C), indicating that KO mutations in both PpKAI2L-F and PpKAI2L-K, or the deletion of PpKAI2L-H, do not abolish the response to the PpCCD8-derived compound mimic, not even additively. The absence of a response in higher-order mutants where either PpKAI2L-J or PpKAI2L-G were knocked out confirms the prominent roles of both genes in the response to (+)-GR24. However, while as expected, the transcript levels of the Pp3c6_15020 response marker gene did not change in either j and gjm-2 mutants following (+)-GR24 application (Figure 10), the transcript levels of the PpKUF1LA gene increased in the j mutant, suggesting a response to the SL analog. Thus, while the KO mutation of either PpKAI2L-J or PpKAI2L-G was sufficient to abolish the phenotypic response in the dark, it did not completely abolish the transcriptional response to (+)-GR24. Presumably, the mutation of both PpKAI2L-J and -G or even all three genes (-G-J-M) is necessary to fully impair this response. Another possibility is that the transcriptional markers, first identified using (±)-GR24 (Lopez-Obando et al., 2016a; Lopez-Obando et al., 2018; Supplemental Figure S14), are not fully specific for assaying the responses to enantiomers. This result could also suggest that the transcriptional response, which was assessed far earlier than the phenotypic response (6 h versus 15 days), is perhaps more sensitive to a very slight activation of the PpCCD8-derived SL pathway by PpKAI2L proteins.
PpKAI2L proteins are likely receptors in two separate pathways, one dependent and the other independent of the F-box protein PpMAX2
Our previous study of the F-box protein PpMAX2 indicated that, in contrast to its homolog in angiosperms, it is not involved in the response to PpCCD8-derived compounds (Lopez-Obando et al., 2018). Like MAX2 in angiosperms, however, PpMAX2 plays a role in early gametophore development and photomorphogenesis. We suggested that PpMAX2 could play a role in the moss KL signaling pathway, but we lacked evidence for other actors in this pathway in P. patens. The present study suggests that eu-KAI2 clade proteins are α/β hydrolases involved in the same pathway as PpMAX2, since mutating these genes resulted in similar light-related phenotypes to those of Ppmax2. Specific mimics for the moss KL are, however, still missing, preventing us from obtaining further evidence that eu-KAI2 clade proteins are receptors of the moss KL (Figure 11). Still, these results are consistent with the view that the KL pathway is ancestral relative to the SL pathway, and that the ancestral role of MAX2 in the land plants lineage is the transduction of the KL signal (Bythell-Douglas et al., 2017; Walker et al., 2019). Such a KL pathway has been recently reported in M. polymorpha, further supporting this view (Mizuno et al., 2021).
PpKAI2L-J and PpKAI2L-G proteins are likely receptors of PpCCD8-derived compounds, which we suspect to be noncanonical SLs. Strikingly, these receptors are not particularly more similar to D14 than other PpKAI2L proteins. As hypothesized earlier (Lopez-Obando et al., 2016a; Bythell-Douglas et al., 2017), the expansion of the PpKAI2L family in P. patens (and not in other bryophytes such as M. polymorpha, which contains two MpKAI2 genes), as in parasitic angiosperms (Conn et al., 2015; Toh et al., 2015), may have allowed the emergence of SL sensitivity (de Saint Germain et al., 2021b). Neofunctionalization of additional KAI2 copies in P. patens ancestry toward SL perception is therefore a possible explanation for our findings in this moss and would reveal a parallel evolutionary process, relative to the emergence of D14 in seed plants. We can also imagine that these neofunctionalized PpKAI2L proteins lost the ability to interact with MAX2 in P. patens, and established a different protein network that potentially integrates new factors such as an alternative F-box protein (Figure 11). The remaining question is therefore to determine how SL signal transduction is achieved downstream of perception by (GJM) clade PpKAI2L proteins.
Consequently, the search for proteins that interact with P. patens KL and PpCCD8-derived compound receptors should be a priority in the near future. In particular, since SMXL proteins are key members of both the SL and KL pathways in angiosperms and of the KL pathway in M. polymorpha (Soundappan et al., 2015; Wang et al., 2015; Khosla et al., 2020; Mizuno et al., 2021), the specific involvement of PpSMXL homologs (four genes) is currently under investigation by our group.
Materials and methods
Plant materials and growth conditions
The P. patens Gransden WT strain was grown as previously described (Hoffmann et al., 2014; Lopez-Obando et al., 2018) under long day (16 h) conditions. Unless otherwise stated in the legends, the experiments were always carried out on PpNO3 medium (minimal medium described by Ashton et al., 1979) under the following control conditions: 25°C during the day and 23°C at night, 50% humidity, quantum irradiance of ∼80 µmol·m−2·s−1 using OSRAM L 36W/865 LUMILUX Cool daylight fluorescent tubes. Tissues from young protonema fragments were multiplied prior to every experiment under the same conditions but using medium with higher nitrogen content (PpNH4 medium, PpNO3 medium supplemented with 2.7-mM NH4 tartrate). For red light experiments, plants were grown on PpNO3 medium in Magenta pots at 25°C, in continuous red light (∼45 µmol·m−2·s−1).
Germination assays of P. ramosa seeds
Physcomitrium patens WT plants were grown on PpNH4 plates with cellophane disks for 2 weeks. The plants (and cellophane) were transferred onto low-phosphate PpNO3 medium (phosphate buffer was replaced with 1 g·L−1 of MES buffer and the pH adjusted to 5.8) for another 2 weeks. Physcomitrium patens exudates were collected by transferring the plants (still on cellophane disks) onto plates containing 10-mL distilled water and incubating them in the growth chamber with gentle agitation. After 48 h, the exudates were pipetted and filtered (0.2 µm). The exudates (Figure 1A) were diluted twice prior to testing their germination-stimulating activity on preconditioned seeds of parasitic P. ramosa plants, as described previously (Pouvreau et al., 2013, 2021). Distilled water was used as a negative control and 0.1-µM (±)-GR24 as a positive control. For each P. ramosa group, GS activities were normalized with the respective negative and positive controls ([GS−GS−]/[GS+−GS−]; with GSs: Germination Stimulant activity of a bioassay, GS−: mean of GS activities of negative controls and GS+: mean of GS activities of positive controls). For germination assays on plates (Figure 1, B and C), WT and Ppccd8 were cultivated as above, and preconditioned P. ramosa seeds were placed onto the plates after 10 days of phosphate starvation. Germinated and nongerminated seeds were counted on three plates, with 7–10 microscope fields per plate, 5 days after adding P. ramosa seeds. Results are expressed as percentage of germination.
CRISPR-Cas9 mediated mutagenesis and homologous recombination in P. patens
Physcomitrium patens mutants were obtained as described in (Lopez-Obando et al., 2016b), using CRISPR-Cas9 technology. PpKAI2L coding sequences were used to search for CRISPR RNA (crRNA) contiguous to a PAM motif recognized by Streptococcus pyogenes Cas9 (NGG), using the webtool CRISPOR V4 against the P. patens genome Phytozome V9 (http://crispor.tefor.net/). Guide crRNAs were chosen in each PpKAI2L gene, preferably in the first exon, to ideally obtain the earliest nonsense mutation possible. When no guide could be designed in the first exon, it was alternatively chosen to recognize a region in close proximity to the codon for one of the last two residues of the catalytic triad (Figure 3). The same crRNA was used to target PpKAI2L-G and -M. Small constructs containing each crRNA fused to either the proU6 or the proU3 snRNA promoters (Collonnier et al., 2017) between the attB1/attB2 Gateway recombination sequences were synthesized by Twist Biosciences. These inserts were then cloned into pDONR207 vectors (Invitrogen). Polyethylene glycol-mediated protoplast transformation was performed with multiple pDONR207-sgRNA as described previously (Lopez-Obando et al., 2016b). Mutations in the PpKAI2L genes were confirmed by PCR amplification of PpKAI2L loci around the recognition sequence of each guide RNA and by sequencing the PCR products.
The PpKAI2L-H (Δh) deletion mutant was obtained through homologous recombination. The full coding sequence of PpKAI2L-H from the ATG to stop was replaced with a resistance cassette. A 550 bp PpKAI2L-H 5′ CDS flanking sequence was cloned into the pBNRF vector (Thelander et al., 2007) cut with BstBI/XhoI. Then a 500-bp PpKAI2L-H 3′ CDS flanking sequence was cloned into the BNRF-PpKAI2L-H 5′ construct digested with BcuI, so that the kanamycin resistance cassette of the vector was flanked by the PpKAI2L-H 5′ and 3′ sequences. Physcomitrium patens WT protoplasts were transformed with the resulting construct as described previously (Lopez-Obando et al., 2016b) and transformants selected on 50 mg·L−1 Geneticin/G418. Transient expression of the CRE recombinase (Trouiller et al., 2006) in a confirmed transformant removed the resistance cassette to obtain the Ppkai2L-Δh mutant, as described in Supplemental Figures S10 and S11.
Phenotypic assays of P. patens
To measure diameters in the light, plants were grown in Petri dishes (six to eight plates per genotype), starting from very small pieces of protonema, with five plants per plate, on PpNO3 medium (Ashton et al., 1979) covered with cellophane disks as previously described (Hoffmann et al., 2014). Thus, between 30 and 40 plants were grown per genotype. After 5 weeks of growth, pictures of the plates were taken, and the ImageJ (http://imagej.nih.gov/ij/) manual selection tool was used for each plant, allowing the area and Feret diameter to be measured. Analysis of caulonema growth in the dark was performed in 24-well plates, starting from very small pieces of protonema. For each genotype/treatment combination, 24 plants were grown, dispatched in three different plates. These plants were grown for ∼2 weeks under control conditions before being placed vertically (± treatment) in the dark for ∼10 days. A single picture of each well was taken under Axio Zoom microscope (Zeiss) with a dedicated program. Caulonema were counted and measured using ImageJ (http://imagej.nih.gov/ij/; see also Guillory and Bonhomme, 2021a).
Chemicals
Racemic and pure enantiomers of GR24 and the (±)-GC242 probe were produced by F-D Boyer (ICSN, France; de Saint Germain et al., 2021a). Racemic CL was kindly provided by A. Scaffidi (University of Western Australia, Perth, Australia). Chemicals were diluted in DMSO or acetone as indicated in the figure legends. KAR2 was purchased from Chiralix.
RT-qPCR analysis
Freshly ground WT (Gransden) tissues were inoculated in Petri dishes containing PpNO3 medium, overlaid with a cellophane sheet. Protonema tissues were harvested after 6 days, 10 days, or 15 days of growth under long-day conditions (see above). To obtain gametophores and spores, plants regenerated from spores were cultivated for 2 weeks, transferred to Magenta pots containing PpNO3 medium (nine plants per pot), and cultivated under the same conditions as above. Gametophores were harvested after 35 days. Both protonema and gametophore samples were flash frozen in liquid nitrogen and stored at −80°C until RNA extraction. At 35 days, the remaining pots were transferred to short-day conditions (15°C, 100% hygrometry, quantum irradiance of 15 µmol m−2 s−1) for approximately 2 months until capsule maturity. Capsules were surface sterilized (5 min in a mixture of 90% pure ethanol and 10% sodium hypochlorite solution) and rinsed with sterile water. Each of the four biological replicates consisted of 10–20 capsules from which spores were freed by mechanical disruption and separated from capsule debris by filtering through a 25-µm nylon mesh. Spores were kept in sterile water, flash frozen in liquid nitrogen, and stored at −80°C until RNA extraction. For all samples except spores, tissues were ground in liquid nitrogen using a mortar and pestle and RNA was extracted from the samples and treated with DNAse I using a Plant RNeasy Mini extraction kit (Qiagen) following the manufacturer’s instructions. Spores were recovered in 1-mL TRIzol reagent (Invitrogen) and crushed manually using fine pestles. RNA was separated from cell debris and protein using chloroform, precipitated with isopropanol, and washed with 70% ethanol. The spore RNA pellets were dissolved in RLT buffer from a Qiagen Plant RNeasy Mini kit and treated with DNAse I on columns following the manufacturer’s instructions. About 500 ng of each RNA sample was used for reverse-transcription using RevertAid H Minus Reverse Transcriptase from ThermoFisher. The quality of the obtained cDNA extracts was verified by RT-PCR using the reference gene PpAPT. RT-qPCR was carried out in a total volume of 5 µL using SsoAdvanced Universal SYBR Green Supermix from BioRad and the following program on QuantStudio 5 (ThermoFisher Scientific): initial denaturation at 95°C for 3 min, then 45 cycles of 95°C for 10 s, and 60°C for 30 s. Using the CTi (for the genes of interest) and CTref (mean for the two reference genes) values obtained, relative expression (RE) was calculated as .
Constructs and generation of transgenic Arabidopsis lines
The expression vectors for transgenic Arabidopsis were constructed using a MultiSite Gateway Three-Fragment Vector Construction kit (Invitrogen). All of the PpKAI2L constructs were tagged with a 6xHA epitope tag at their C-terminus. The transgenic lines were resistant to hygromycin. The AtD14 native promoter (0.8 kb) and AtKAI2 native promoter (0.7 kb) were amplified by PCR from Col-0 genomic DNA and cloned into pDONR-P4P1R, using Gateway recombination (Invitrogen) (see Supplemental Table S2 for primers). The AtD14 CDS and AtKAI2 CDS were PCR amplified from Col-0 cDNA, and the PpKAI2L CDS were PCR amplified from P. patens cDNA and recombined into pDONR221 (Invitrogen). 6xHA with a linker (gift from U. Pedmale, Cold Spring Harbor Laboratory) was cloned into pDONR-P2RP3 (Invitrogen). The suitable combination of promoters, CDS, and 6xHA were cloned into the pH7m34GW final destination vectors using the three fragment recombination system (Karimi et al., 2007) and named pD14:CDS-6xHA or pKAI2:CDS-6xHA. The Arabidopsis d14-1 kai2-2 double mutant in the Landsberg background (gift from M. Waters, University of Western Australia Perth) was transformed following the conventional floral dip method (Clough and Bent, 1998), with Agrobacterium strain GV3101. For all constructs, more than 12 independent T1 lines were isolated, and between two and four representative single-insertion lines were selected in the T2 generation. Two lines per construct were shown in these analyses. The phenotypic analysis shown in Figure 6 and Supplemental Figure S9 was performed on segregating T3 homozygous lines.
Protein extraction and immunoblotting
Total protein extract was prepared from eight to ten 10-day-old Arabidopsis seedlings in Laemmli buffer and boiled for 5 min. Total protein were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane (Bio-Rad) probed with anti-HA primary antibody (1:10,000; SIGMA H9658-100UL Lot#128M4789V) and then anti-mouse-IgG-HRP secondary antibody (1:10,000; SIGMA A9044-2ML-100UL Lot#029M4799V). As a loading control, a Ponceau staining or a Tubulin detection with anti-Tubulin primary antibody (1:10,000; SIGMA T5168-2ML Lot#04M4760V) and then anti-mouse-IgG-HRP secondary antibody (1:10,000; SIGMA A9044-2ML-100UL Lot#029M4799V) was used.
Arabidopsis hypocotyl elongation and cotyledon expansion assays
Arabidopsis seeds were surface sterilized by consecutive treatments of 5 min in 70% (v/v) ethanol and 0.05% (w/v) sodium dodecyl sulfate, and 5 min in 95% (v/v) ethanol. Then seeds were sown on 0.25× Murashige and Skoog medium (Duchefa Biochemie) containing 1% agar, supplemented with 1-μM (+)-GR24, (−)-GR24 or with 0.01% DMSO (control). For the hypocotyl elongation assay, seeds were stratified at 4°C (2 days in the dark), exposed to white light (125–180 µmol·m-2·s−1) for 3 h, transferred to the dark for 21 h, and exposed to low light (25–30 µmol·m-2·s−1) for 10 days at 21°C. For the cotyledon expansion assay, seedlings were grown in white light (125–180 µmol·m-2·s−1) for 4 days. Cotyledon area and hypocotyl lengths were quantified using ImageJ (http://imagej.nih.gov/ij/)
Quantification of Arabidopsis branching
Experiments were carried out in the summer in a greenhouse under long photoperiods (15–16 h per day); daily temperatures fluctuated between 18°C and 25°C. Peak PAR levels were between 700 and 1,000 μmol m−2 s−1. Plants were watered twice a week with tap water. The number of rosette branches longer than 5 mm was counted when the plants were 40 days old.
Protein expression and purification
AtD14, RMS3, and AtKAI2 with cleavable GST tags were expressed and purified as described in de Saint Germain et al. (2016). For PpKAI2L protein expression, the full-length CDS from P. patens were amplified by PCR using cDNA template and specific primers (see Supplemental Table S2) containing a protease cleavage site for tag removal, and subsequently cloned into the pGEXT-4T-3 expression vector. For PpKAI2L-L, the N-terminal 47 amino acids were removed. The expression and purification of PpKAI2L proteins followed the same method as for AtD14 and AtKAI2.
Site-directed mutagenesis
Site-directed mutagenesis of PpKAI2L-H was performed using a QuickChange II XL Site Directed Mutagenesis kit (Stratagene), performed on pGEX-4T-3-PpKAI2L-H (see Supplemental Table S2 for primers). Mutagenesis was verified by DNA sequencing.
Enzymatic degradation of GR24 isomers by purified proteins
The ligand (10 µM) was incubated with and without purified RMS3/AtKAI2/PpKAI2L proteins (5 µM) for 150 min at 25°C in phosphate buffer saline (PBS; 0.1 mL, pH = 6.8) in the presence of (±)-1-indanol (100 µM) as an internal standard. The solutions were acidified to pH = 1 by adding trifluoroacetic acid (2 µL) to quench the reaction and centrifuged (12 min, 12,000 rpm). The samples were then subjected to RP-UPLC-MS analyses. The instrument used for all the analyses was an Ultra Performance Liquid Chromatography system equipped with a PDA and Triple Quadrupole mass spectrometer Detector (Acquity UPLC-TQD, Waters, USA). RP-UPLC (HSS C18 column, 1.8 μm, 2.1 mm × 50 mm) was carried out with 0.1% formic acid in CH3CN and 0.1% formic acid in water (aq. FA, 0.1%, v/v, pH 2.8) as eluents (10% CH3CN, followed by a linear gradient from 10% to 100% of CH3CN [4 min]) at a flow rate of 0.6 mL·min-1. The detection was performed by PDA and using the TQD mass spectrometer operated in electrospray ionization (ESI) positive mode at 3.2-kV capillary voltage. The cone voltage and collision energy were optimized to maximize the signal, with a cone voltage of 20 V and collision energy of 12 eV. The collision gas was argon at a pressure maintained near 4.5 × 10−3 mBar.
Enzymatic kinetic assays
Enzyme assays with pro-fluorescent probes and p-nitrophenyl acetate were performed as described in de Saint Germain et al. (2016), using a TriStar LB 941 Multimode Microplate Reader from Berthold Technologies.
nanoDifferential scanning fluorimetry
Proteins were diluted in PBS (100-mM phosphate, pH 6.8, 150-mM NaCl) to ∼10 μM concentration. Ligand was tested at the concentration of 200 µM. The intrinsic fluorescence signal was measured as a function of increasing temperature in a Prometheus NT.48 fluorimeter (Nanotemper), with 55% excitation light intensity and 1°C·min-1 temperature ramp. Analyses were performed on capillaries filled with 10 µL of the respective samples. The intrinsic fluorescence signal was expressed as the 350 nm/330 nm emission ratio, which increases as the proteins unfold, and was plotted as a function of temperature. The plots show one of the three independent data collections that were performed for each protein.
Intrinsic tryptophan fluorescence assays
Intrinsic tryptophan fluorescence assays and determination of the dissociation constant Kd were performed as described in de Saint Germain et al. (2016), using a Spark Multimode Microplate Reader from Tecan.
Direct ESI-MS of PpKAI2L proteins under denaturing conditions
Mass spectrometry measurements were performed with an electrospray Q-TOF mass spectrometer (Waters) equipped with a Nanomate device (Advion, Inc.). The HD_A_384 chip (5 μm I.D. nozzle chip, flow rate range 100−500 nL·min−1) was calibrated before use. For ESI-MS measurements, the Q-TOF instrument was operated in RF quadrupole mode with the TOF data being collected between m/z 400 and 2,990. Collision energy was set to 10 eV, and argon was used as the collision gas. PpKAI2L proteins (50 µM) in 50-mM ammonium acetate (pH 6.8) in the presence or absence of GR24 enantiomers (500 µM) were incubated for 10 min at room temperature before denaturation in 50% acetonitrile and 1% formic acid. The solutions were directly injected for Mass spectra acquisition or digested before LC−MS/MS analyses. Mass Lynx version 4.1 (Waters) and Peakview version 2.2 (Sciex) software were used for data acquisition and processing, respectively. Deconvolution of multiply charged ions was performed by applying the MaxEnt algorithm (Sciex). The average protein masses were annotated in the spectra, and the estimated mass accuracy was ± 2 Da. External calibration was performed with NaI clusters (2 μg·μL−1, isopropanol/H2O 50/50, Waters) in the acquisition m/z mass range.
Homology modeling
Figure 5B showing a superimposition model was prepared using PyMOL (DeLano Scientific) with the crystal structures of AtD14 (PDB ID 4IH4) and PpKAI2L-H (PDB ID 6AZD).
Statistical analyses
ANOVA, Kruskal–Wallis, Mann–Whitney, post hoc Dunnett, post hoc Dunn, and Tukey multiple comparisons tests (details in figure legends) were carried out either in R version 3.6.3 or in GraphPad Prism version 8.4.2. Unless otherwise defined, the statistical significance scores used were as follows: *0.01≤P < 0.05, **0.001≤P < 0.01, ***P < 0.001. Same letters scores indicate that P ≥ 0.05 (nonsignificant differences). All statistical analyses are described in Supplemental File S1.
Accession numbers
Sequence data from this article can be found in the GenBank/Phytozome libraries under the accession numbers described in Supplemental Table S3.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Sequence alignment of P. patens (Pp) PpKAI2L proteins with AtD14 and AtKAI2 proteins from A. thaliana (At).
Supplemental Figure S2 . Expression of PpKAI2L genes throughout P. patens vegetative development.
Supplemental Figure S3 . nanoDSF analysis of the PpKAI2L protein response to GR24 isomers.
Supplemental Figure S4 . Intrinsic tryptophan fluorescence shows that GR24 isomers bind to PpKAI2L proteins with different affinities.
Supplemental Figure S5 . PpKAI2L hydrolysis activity toward p-NPA.
Supplemental Figure S6 . PpKAI2L enzymatic activities confirm stereoselectivity.
Supplemental Figure S7 . Characterization of (±)-GC242 profluorescent probe activity on moss and PpKAI2L enzymatic activities toward (±)-GC242.
Supplemental Figure S8 . Mass spectrometry characterization of covalent PpKAI2L-ligand complexes.
Supplemental Figure S9 . Complementation assays of the Arabidopsis Atd14-1 kai2-2 double mutant.
Supplemental Figure S10 . Mutations obtained in all 13 PpKAI2L genes.
Supplemental Figure S11 . The Ppkai2L-Δh mutant.
Supplemental Figure S12 . Gametophore heights of Ppkai2L mutants in red light.
Supplemental Figure S13 . Phenotypic response of Ppkai2L mutants to (-)-GR24 and (+)-GR24 application: caulonema number in the dark.
Supplemental Figure S14 . Transcriptional response of WT and Ppccd8 to (±)-GR24 application: use of the Pp3c6_15020 marker gene.
Supplemental Table S1 . Mutants used in this study.
Supplemental Table S2 . Oligonucleotides used in this study.
Supplemental Table S3 . List of gene sequences used in this study.
Supplemental File S1 . Results of statistical tests.
Supplementary Material
Acknowledgments
The authors thank Adrian Scaffidi (University of Western Australia, Perth, Australia) for the gift of carlactone, and Mark Waters (University of Western Australia, Perth, Australia) for Arabidopsis kai2-2 and d14-1 kai2-2 mutants. We are grateful to Jean-Paul Pillot (IJPB) for precious help with Arabidopsis branching assays, to Pauline Rode for help with genotyping, and to Fabien Nogué (IJPB) for stimulating discussions. MLO thanks Eva Sundberg for her support on his SL/KL’s activities at SLU.
Funding
This research was supported by the Agence Nationale de la Recherche (contract ANR-12-BSV6-004-01). The IJPB benefits from the support of the Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). This work was supported by a “Infrastructures en Biologie Santé et Agronomie” grant to SICAPS platform of the Institute for Integrative Biology of the Cell, and CHARM3AT Labex program (ANR-11-LABX-39) for ICSN. A.d.S.G. is the recipient of an AgreenSkills award from the European Union in the framework of the Marie-Curie FP7 COFUND People Programme and fellowship from Saclay Plant Sciences (ANR-17-EUR-0007). This work has benefited from the facilities and expertise of the I2BC proteomic platform (Proteomic-Gif, SICaPS) supported by IBiSA, Ile de France Region, Plan Cancer, CNRS and Paris-Sud University.
Conflict of interest statement. None declared.
S.B., A.d.S.G., M.L.-O., and C.R. designed the project. M.L.-O., A.G., F.-D.B., D.C., B.H., P.L.B., J.-B.P., A.d.S.G., and S.B. conducted the experiments. A.G., M.L.-O., F.-D.B., D.C., P.L.B., J.-B.P., P.D., C.R., A.d.S.G., and S.B. analyzed the data. A.G. and S.B. wrote the manuscript, with essential contributions from M.L.-O., F.-D.B., P.D., C.R., and A.d.S.G.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors ((https://academic.oup.com/plcell) are: Sandrine Bonhomme (Sandrine.Bonhomme@inrae.fr) and Alexandre de Saint Germain (Alexandre.De-Saint-Germain@inrae.fr).
References
- Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827 [DOI] [PubMed] [Google Scholar]
- Akiyama K, Ogasawara S, Ito S, Hayashi H (2010) Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol 51:1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186 [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al-Babili S (2012) The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 335:1348–1351 [DOI] [PubMed] [Google Scholar]
- Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S, Kyozuka J (2009) d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50:1416–1424 [DOI] [PubMed] [Google Scholar]
- Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144:427–435 doi:10.1007/bf00380118 [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pages V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Becard G, Sejalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Nelson DC, Weijers D (2020) Evolution of plant hormone response pathways. Annu Rev Plant Biol 71:327–353 [DOI] [PubMed] [Google Scholar]
- Boutet-Mercey S, Perreau F, Roux A, Clave G, Pillot JP, Schmitz-Afonso I, Touboul D, Mouille G, Rameau C, Boyer FD (2018) Validated method for strigolactone quantification by ultra high-performance liquid chromatography - electrospray ionisation tandem mass spectrometry using novel deuterium labelled standards. Phytochem Anal 29:59–68 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Briginshaw LN, Florent SN (2019) Evolution and co-option of developmental regulatory networks in early land plants. Curr Top Dev Biol 131:35–53 [DOI] [PubMed] [Google Scholar]
- Boyer FD, de Saint Germain A, Pillot JP, Pouvreau JB, Chen VX, Ramos S, Stevenin A, Simier P, Delavault P, Beau JM, et al. (2012) Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol 159:1524–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Chory J (2020) The many models of strigolactone signaling. Trends Plant Sci 25:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürger M, Mashiguchi K, Lee HJ, Nakano M, Takemoto K, Seto Y, Yamaguchi S, Chory J (2019) Structural basis of karrikin and non-natural strigolactone perception in Physcomitrella patens. Cell Rep 26:855–865.e855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R, Rothfels CJ, Stevenson DWD, Graham SW, Wong GK, Nelson DC, Bennett T (2017) Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol 15:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743 [DOI] [PubMed] [Google Scholar]
- Collonnier C, Guyon-Debast A, Maclot F, Mara K, Charlot F, Nogué F (2017) Towards mastering CRISPR-induced gene knock-in in plants: survey of key features and focus on the model Physcomitrella patens. Methods 121–122:103–117 [DOI] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, Yoshida S, Whittington B, Westwood JH, Shirasu K, Bond CS, Dyer KA, Nelson DC (2015) PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349:540–543 [DOI] [PubMed] [Google Scholar]
- Conn CE, Nelson DC (2015) Evidence that KARRIKIN-INSENSITIVE2 (KAI2) Receptors may perceive an unknown signal that is not karrikin or strigolactone. Front Plant Sci 6:1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH (1966) Germination of Witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154:1189–1190 [DOI] [PubMed] [Google Scholar]
- Coudert Y, Palubicki W, Ljung K, Novak O, Leyser O, Harrison CJ (2015) Three ancient hormonal cues co-ordinate shoot branching in a moss. eLife 4:e06808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clave G, Badet-Denisot MA, Pillot JP, Cornu D, Le Caer JP, Burger M, Pelissier F, Retailleau P, Turnbull C, et al. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat Chem Biol 12:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Clavé G, Boyer FD (2021a) Synthesis of profluorescent strigolactone probes for biochemical studies. Methods Mol Biol 2309: 219–231 [DOI] [PubMed] [Google Scholar]
- de Saint Germain A, Jacobs A, Brun G, Pouvreau J-B, Braem L, Cornu D, Clavé G, Baudu E, Steinmetz V, Servajean V, et al. (2021b) A Phelipanche ramosa KAI2 protein perceives enzymatically strigolactones and isothiocyanates. Plant Commun 2: 100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Retailleau P, Norsikian S, Servajean V, Pelissier F, Steinmetz V, Pillot JP, Rochange S, Pouvreau JB, Boyer FD (2019) Contalactone, a contaminant formed during chemical synthesis of the strigolactone reference GR24 is also a strigolactone mimic. Phytochemistry 168:112112. [DOI] [PubMed] [Google Scholar]
- Decker EL, Alder A, Hunn S, Ferguson J, Lehtonen MT, Scheler B, Kerres KL, Wiedemann G, Safavi-Rizi V, Nordzieke S, et al. (2017) Strigolactone biosynthesis is evolutionarily conserved, regulated by phosphate starvation and contributes to resistance against phytopathogenic fungi in a moss, Physcomitrella patens. New Phytol 216:455–468 [DOI] [PubMed] [Google Scholar]
- Delaux PM, Xie X, Timme RE, Puech-Pages V, Dunand C, Lecompte E, Delwiche CF, Yoneyama K, Becard G, Sejalon-Delmas N (2012) Origin of strigolactones in the green lineage. New Phytol 195:857–871 [DOI] [PubMed] [Google Scholar]
- Delavault P, Montiel G, Brun G, Pouvreau JB, Thoiron S, Simier P (2017) Communication between host plants and parasitic plants. Adv Bot Res 82:55–82 [Google Scholar]
- Floková K, Shimels M, Andreo Jimenez B, Bardaro N, Strnad M, Novák O, Bouwmeester HJ (2020) An improved strategy to analyse strigolactones in complex sample matrices using UHPLC-MS/MS. Plant Methods 16:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pages V, Dun EA, Pillot JP, Letisse F, Matusova R, Danoun S, Portais JC, et al. (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194 [DOI] [PubMed] [Google Scholar]
- Guillory A, Bonhomme S (2021a) Methods for medium-scale study of biological effects of strigolactone-like molecules on the moss Physcomitrium (Physcomitrella) patens. Methods Mol Biol 2309: 143–155 doi:10.1007/978-1-0716-1429-7_12 [DOI] [PubMed] [Google Scholar]
- Guillory A, Bonhomme S (2021b) Phytohormone biosynthesis and signaling pathways of mosses. Plant Mol Biol. doi:10.1007/s11103-021-01172-6 [DOI] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, Riemann M, Johnston MG, Summers W, Carbonnel S, Mansfield C, Yang SY, Nadal M, et al. (2015) Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350: 1521–1524 [DOI] [PubMed] [Google Scholar]
- Hamiaux C, Drummond RS, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC (2012) DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol 22: 2032–2036 [DOI] [PubMed] [Google Scholar]
- Harris BJ, Harrison CJ, Hetherington AM, Williams TA (2020) Phylogenomic evidence for the monophyly of bryophytes and the reductive evolution of stomata. Curr Biol 30:2001–2012. e2002 [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Proust H, Belcram K, Labrune C, Boyer FD, Rameau C, Bonhomme S (2014) Strigolactones inhibit caulonema elongation and cell division in the moss Physcomitrella patens. PLoS One 9:e99206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet S, Pouvreau J-B, Delage E, Delgrange S, Marais C, Bahut M, Delavault P, Simier P, Poulin L (2020) Populations of the parasitic plant Phelipanche ramosa influence their seed microbiota. Front Plant Sci 11: 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil M, Kountche BA, Wang JY, Haider I, Jia KP, Takahashi I, Ota T, Asami T, Al-Babili S (2020) A new series of carlactonoic acid based strigolactone analogs for fundamental and applied research. Front Plant Sci 11:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Bleys A, Vanderhaeghen R, Hilson P (2007) Building blocks for plant gene assembly. Plant Physiol 145:1183–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla A, Morffy N, Li Q, Faure L, Chang SH, Yao J, Zheng J, Cai ML, Stanga JP, Flematti GR, et al. (2020) Structure-function analysis of SMAX1 reveals domains that mediate its karrikin-induced proteolysis and interaction with the receptor KAI2. Plant Cell. doi:10.1105/tpc.19.00752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, Watanabe Y, Osakabe Y, Sato M, Toyooka K, Seo M, Tian L, Tian C, et al. (2020) Comparative functional analyses of DWARF14 and KARRIKIN INSENSITIVE 2 in drought adaptation of Arabidopsis thaliana. Plant J. doi:10.1111/tpj.14712 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S (2016a) Structural modelling and transcriptional responses highlight a clade of PpKAI2-LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta 243:1441–1453 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, de Villiers R, Hoffmann B, Ma L, de Saint Germain A, Kossmann J, Coudert Y, Harrison CJ, Rameau C, Hills P, et al. (2018) Physcomitrella patens MAX2 characterization suggests an ancient role for this F-box protein in photomorphogenesis rather than strigolactone signalling. New Phytol 219:743–756 [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M, Hoffmann B, Géry C, Guyon-Debast A, Téoulé E, Rameau C, Bonhomme S, Nogué F (2016b) Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 (Bethesda) 6:3647–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Obando M, Ligerot Y, Bonhomme S, Boyer F-D, Rameau C (2015) Strigolactone biosynthesis and signaling in plant development. Development 142:3615–3619 doi:10.1242/dev.120006 [DOI] [PubMed] [Google Scholar]
- Mizuno Y, Komatsu A, Shimazaki S, Naramoto S, Inoue K, Xie X, Ishizaki K, Kohchi T, Kyozuka J (2021) Major components of the KARRIKIN INSENSITIVE2-dependent signaling pathway are conserved in the liverwort Marchantia polymorpha. Plant Cell 33: 2395–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A 108: 8897–8902 doi:10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Hernandez-Coronado M, Thamm A, Catarino B, Wang M, Dolan L, Feijo JA, Becker JD (2016) A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol Plant 9:205–220 [DOI] [PubMed] [Google Scholar]
- Perroud PF, Haas FB, Hiss M, Ullrich KK, Alboresi A, Amirebrahimi M, Barry K, Bassi R, Bonhomme S, Chen H, et al. (2018) The Physcomitrella patens gene atlas project: large-scale RNA-seq based expression data. Plant J 95:168–182 [DOI] [PubMed] [Google Scholar]
- Pouvreau JB, Gaudin Z, Auger B, Lechat MM, Gauthier M, Delavault P, Simier P (2013) A high-throughput seed germination assay for root parasitic plants. Plant Methods 9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau JB, Poulin L, Huet S, Delavault P (2021) Strigolactone-like bioactivity via parasitic plant germination bioassay. Methods Mol Biol 2309: 59–73 [DOI] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Nogue F, Rameau C (2011) Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138:1531–1539 [DOI] [PubMed] [Google Scholar]
- Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, et al. (2018) The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol 28:733–745.e732 [DOI] [PubMed] [Google Scholar]
- Rial C, Varela RM, Molinillo JMG, López-Ráez JA, Macías FA (2019) A new UHPLC-MS/MS method for the direct determination of strigolactones in root exudates and extracts. Phytochem Anal 30:110–116 [DOI] [PubMed] [Google Scholar]
- Scaffidi A, Waters MT, Sun YK, Skelton BW, Dixon KW, Ghisalberti EL, Flematti GR, Smith SM (2014) Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol 165:1221–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Yasui R, Kameoka H, Tamiru M, Cao M, Terauchi R, Sakurada A, Hirano R, Kisugi T, Hanada A, et al. (2019) Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat Commun 10:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N, Ticchiarelli F, Mao H, Hinds TR, Leyser O, Zheng N (2018) Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 563:652–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson D (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27:3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova B, Delourme R, Duffé P, Delavault P, Simier P (2019) Genetic differentiation and host preference reveal non-exclusive host races in the generalist parasitic weed Phelipanche ramosa. Weed Res 59:107–118 [Google Scholar]
- Thelander M, Nilsson A, Olsson T, Johansson M, Girod PA, Schaefer DG, Zryd JP, Ronne H (2007) The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol Biol 64:559–573 [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350:203–207 [DOI] [PubMed] [Google Scholar]
- Trouiller B, Schaefer DG, Charlot F, Nogue F (2006) MSH2 is essential for the preservation of genome integrity and prevents homeologous recombination in the moss Physcomitrella patens. Nucleic Acids Res 34:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455:195–200 [DOI] [PubMed] [Google Scholar]
- Villaecija-Aguilar JA, Hamon-Josse M, Carbonnel S, Kretschmar A, Schmidt C, Dawid C, Bennett T, Gutjahr C (2019) SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet 15:e1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CH, Siu-Ting K, Taylor A, O'Connell MJ, Bennett T (2019) Strigolactone synthesis is ancestral in land plants, but canonical strigolactone signalling is a flowering plant innovation. BMC Biol 17:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27:3128–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Waters MT, Smith SM (2018) Karrikin-KAI2 signalling provides Arabidopsis seeds with tolerance to abiotic stress and inhibits germination under conditions unfavourable to seedling establishment. New Phytol 219:605–618 [DOI] [PubMed] [Google Scholar]
- Waters MT, Gutjahr C, Bennett T, Nelson DC (2017) Strigolactone signaling and evolution. Annu Rev Plant Biol 68:291–322 [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139:1285–1295 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Flematti G, Smith SM (2015a) Substrate-induced degradation of the alpha/beta-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol Plant 8:814–817 [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SL, Sun YK, Flematti GR, Smith SM (2015b) A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27:1925–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X (2016) Structural diversity of strigolactones and their distribution in the plant kingdom. J Pestic Sci 41:175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Scaffidi A, Meng Y, Melville KT, Komatsu A, Khosla A, Nelson DC, Kyozuka J, Flematti GR, Waters MT (2021) Desmethyl butenolides are optimal ligands for karrikin receptor proteins. New Phytol 230:1003–1016 [DOI] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, Li S, Wang F, Ma S, Yu C, Yang M, Chen L, Chen L, et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536:469–473 [DOI] [PubMed] [Google Scholar]
- Yoneyama K (2020) Recent progress in the chemistry and biochemistry of strigolactones. J Pestic Sci 45: 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K, Mori N, Sato T, Yoda A, Xie X, Okamoto M, Iwanaga M, Ohnishi T, Nishiwaki H, Asami T, et al. (2018a) Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol 218:1522–1533 [DOI] [PubMed] [Google Scholar]
- Yoneyama K, Xie X, Yoneyama K, Kisugi T, Nomura T, Nakatani Y, Akiyama K, McErlean CSP (2018b) Which are the major players, canonical or non-canonical strigolactones? J Exp Bot 69:2231–2239 [DOI] [PubMed] [Google Scholar]
- Zheng J, Hong K, Zeng L, Wang L, Kang S, Qu M, Dai J, Zou L, Zhu L, Tang Z, et al. (2020) Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell 32:2780–2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.