Abstract
For epidemiological tracing of the thermotolerant Campylobacter species C. jejuni and C. coli, reliable and highly discriminatory typing techniques are necessary. In this study the genotyping techniques of flagellin typing (flaA typing), pulsed-field gel electrophoresis (PFGE), automated ribotyping, and amplified fragment length polymorphism (AFLP) fingerprinting were compared. The following aspects were compared: computer-assisted analysis, discriminatory power, and use for epidemiological typing of campylobacters. A set of 50 campylobacter poultry isolates from The Netherlands and neighboring countries was analyzed. Computer-assisted analysis made cluster analysis possible and eased the designation of different genotypes. AFLP fingerprinting was the most discriminatory technique, identifying 41 distinct genotypes, while PFGE identified 38 different types, flaA typing discriminated 31 different types, and ribotyping discriminated 26 different types. Furthermore, AFLP analysis was the most suitable method for computer-assisted data analysis. In some cases combining the results of AFLP fingerprinting, PFGE, and flaA typing increased our ability to differentiate strains that appeared genetically related. We conclude that AFLP is a highly discriminatory typing method and well suited for computer-assisted data analysis; however, for optimal typing of campylobacters, a combination of multiple typing methods is needed.
The thermotolerant Campylobacter species Campylobacter jejuni and Campylobacter coli are a major cause of human acute enteritis all over the world (18). A main source of human infection is thought to be the consumption of contaminated poultry meat (20, 21). To date, however, infection routes of broiler flocks are still unknown. Reliable and powerful typing methods for Campylobacter are necessary in order to gain more insight into these infection routes.
Traditionally, phenotyping methods such as serotyping, phage typing, and biotyping have been used. The drawbacks of these methods are their restricted resolutions, the lack of specific reagents for serotyping, and a large portion of untypeable strains.
To resolve these problems, attention has turned to genotyping methods that are more generally available and applicable. Several techniques have been developed and are in general use already, such as flagellin typing (flaA typing) (1, 12), pulsed-field gel electrophoresis (PFGE) (2, 6, 27), and ribotyping (5, 15). These methods were an improvement in comparison to the older phenotyping techniques; however, none of these combines high resolution, high throughput, and simple, reliable data analysis. In order to fulfill these needs, the amplified fragment length polymorphism (AFLP) technique has been adjusted for use with Campylobacter (4, 10).
Interpretation of data is an extremely important aspect of genotyping techniques. Results of phenotyping techniques are often “black or white” or “present or absent,” whereas results of genotyping techniques are often complicated banding patterns. Band presence or absence, position, and intensity are relevant input data in comparison analyses. Analysis of genotyping data in a numeric manner requires computer assistance. Furthermore, computer-assisted analysis allows data sharing and can ease the processing of large numbers of samples.
The purpose of this study was to establish an optimal typing system for Campylobacter with regard for the above-mentioned considerations. We performed a comparative analysis of flaA typing, PFGE, ribotyping, and AFLP fingerprinting using a set of 50 poultry isolates. Advantages and disadvantages of computer-assisted data analysis and the discriminatory powers of the four different techniques were compared. With these results, the potential for automated analysis in epidemiological typing of Campylobacter was examined.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Fifty C. jejuni and C. coli strains isolated from poultry from dispersed places in The Netherlands (46 strains) and neighboring countries (4 strains) over a period of time (1990, 1992, 1993, and 1997) were used in this study (Table 1). Strains were grown on heart infusion plates containing 5% sheep blood for 48 h (24 h for preparations of PFGE plugs) at 42°C under microaerobic conditions. Alternatively, for storage and sample preparation for ribotyping, strains were grown overnight in heart infusion broth at 37°C under microaerobic conditions with gentle shaking (100 rpm). Strains were stored at −80°C in heart infusion broth containing 15% glycerol.
TABLE 1.
Campylobacter strains used in this study
| Straina | Speciesb | Yr of isolation | Farmc | Straina | Speciesb | Yr of isolation | Farmc | |
|---|---|---|---|---|---|---|---|---|
| C144 | C. jejuni | 1990 | B | C2446 | C. jejuni | 1992 | ||
| C350 | C. jejuni | 1990 | A | C2450 | C. jejuni | 1992 | ||
| C356 | C. jejuni | 1990 | A | C2461 | C. jejuni | 1992 | ||
| C591 | C. jejuni | 1990 | A | C2466 | C. coli | 1992 | ||
| C626 | C. coli | 1990 | B | C2470 | C. coli | 1992 | ||
| C690 | C. jejuni | 1990 | B | C2476 | C. jejuni | 1992 | ||
| C2143 | C. jejuni | 1992 | C2481 | C. jejuni | 1992 | |||
| C2146 | C. jejuni | 1992 | C2485 | C. coli | 1992 | B | ||
| C2150 | C. jejuni | 1992 | C2498 | C. coli | 1992 | |||
| C2152 | C. coli | 1992 | C2505 | C. coli | 1992 | |||
| C2155 | C. coli | 1992 | C2515 | C. jejuni | 1992 | |||
| C2246 | C. jejuni | 1992 | C2520 | C. coli | 1992 | |||
| C2264 | C. jejuni | 1992 | C2535 | C. jejuni | 1992 | |||
| C2345 | C. jejuni | 1992 | C2540 (G) | C. coli | 1992 | |||
| C2355 | C. coli | 1992 | C2545 | C. coli | 1992 | |||
| C2360 | C. jejuni | 1992 | C | C2551 (Dk) | C. coli | 1992 | ||
| C2362 | C. jejuni | 1992 | C | C2555 | C. jejuni | 1992 | ||
| C2375 | C. coli | 1992 | C2605 | C. coli | 1992 | |||
| C2380 | C. coli | 1992 | C2609 (B) | C. jejuni | 1992 | |||
| C2385 | C. coli | 1992 | C2641 | C. jejuni | 1992 | |||
| C2390 | C. coli | 1992 | C2651 | C. jejuni | 1993 | |||
| C2400 (B) | C. coli | 1992 | C4596 | C. coli | 1997 | D | ||
| C2412 | C. jejuni | 1992 | C4601 | C. coli | 1997 | D | ||
| C2436 | C. coli | 1992 | C4602 | C. coli | 1997 | D | ||
| C2441 | C. jejuni | 1992 | C4611 | C. coli | 1997 | D |
All strains were isolated in The Netherlands (9), except those indicated with (B), (G), or (Dk), which were isolated in Belgium, Germany, or Denmark, respectively (9).
Strains were tested according to the multiplex PCR described by van de Giessen et al. (23).
Strains with the same letter are from the same farm.
Genomic DNA isolation.
Genomic DNAs were extracted from 48-h-old cultures using a Wizard genomic DNA purification kit (Promega, Madison, Wis.).
flaA typing.
PCR mixtures for flagellin A (flaA) typing contained 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 0.01% (wt/vol) gelatin, 2 mM MgCl2, 0.2 μM each deoxynucleoside triphosphate, 50 pmol of the flaA primer (5′-CGTATTAACACAAATGTTGCAGC-3′, adapted from reference 1), 50 pmol of the flaR primer (5′-GATTTGTTATAGCAGTTTCTGCTATATCC-3′, adapted from reference 1), 50 pmol of the template (genomic DNA), and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), with a total reaction volume of 50 μl. Reaction conditions were 94°C for 60 s, followed by 45 cycles of 45 s of 94°C, 45 s of 55°C, and 2 min of 72°C, and ended with 5 min of 72°C. After verification of the PCR product, 12.5 μl of the amplicon was digested for 2 h at 37°C using 10 U of DdeI (Boehringer Mannheim, GmbH, Mannheim, Germany) in a total volume of 15 μl. After digestion, restriction fragments were separated on an agarose gel containing 2.0% (wt/vol) NuSieve (FMC, Rockland, Maine) agarose, by using 0.5% (wt/vol) multipurpose agarose (Boehringer Mannheim) in 1× TAE (17) for 4 h at 80 V.
PFGE.
Preparation of DNA-containing agarose blocks for PFGE was adapted from the work of On et al. (14). Cells grown for 24 hours were resuspended in Pett IV buffer (1 M NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA) at an optical density at 420 nm of 1.5 and heated to 50°C. Three hundred microliters was mixed with 700 μl of warm (50°C) 1% Resove Low (Biozym, Landgraaf, The Netherlands) agarose. The mixture was cast into molds (Bio-Rad, Richmond, Calif.) and solidified for 10 min at 4°C. Plugs were incubated in 3 ml of ESP lysis solution (0.5 M EDTA, 1.0% N-lauroyl sarcosine, 1 mg of proteinase K per ml) at 50°C for 48 h. The plugs were washed three times for 20 min each time in 2 ml of TE buffer (Tris-HCl 10 mM [pH 8.0], 1 mM EDTA) with 1.5 mM phenylmethylsulfonyl fluoride, followed by three washings of 20 min each with TE buffer. Subsequently, the plugs were equilibrated with 1× restriction buffer for 48 h at room temperature and DNA was finally cut for 4 h at 25°C in 250 μl of restriction buffer containing 20 U of SmaI (Boehringer Mannheim).
Digested DNA plugs were loaded on a 1% SeaKem genetic technology grade agarose (FMC) gel and separated on a contour-clamped homogeneous electric field DR-III apparatus (Bio-Rad) in 0.5× TBE buffer (17) for 22 h at 14°C. Electrophoresis conditions were 6 V/cm, the included angle was 120 degrees, and ramp times were 5 to 10 s over 4 h, 10 to 40 s over 14 h, and 50 to 60 s over 4 h. After electrophoresis, gels were stained in a 1-mg/ml ethidium bromide solution and destained in electrophoresis buffer and bands were visualized under UV light.
Automated ribotyping.
Automated ribotyping was performed on a RiboPrinter (Qualicon, Wilmington, Del.) with the restriction enzyme PstI, according to the instructions of the manufacturer. Shortly, the cell suspension was lysed and chromosomal DNA was isolated, digested with PstI, electrophoresed, and simultaneously blotted in an automated manner. Subsequently, the Southern blot was hybridized with a chemiluminescently labeled 16 to 23S rRNA primer. Bands were detected and analyzed with RiboPrinter software.
AFLP fingerprinting.
AFLP analysis was performed as previously described (4). Shortly afterward, genomic DNAs were digested with HindIII and HhaI. Simultaneously, site-specific adapters were ligated to the restriction fragments. A preselective PCR amplification was followed by a selective PCR using a labeled HindIII primer containing a selective nucleotide (A) and an HhaI primer containing a selective A nucleotide. Final products were analyzed on a 7.3% denaturing sequence gel on an ABI 373 automated DNA sequencer.
Data analysis.
Patterns obtained by flaA typing and PFGE were photographed using a digital camera (Minolta RD-175) and saved as TIFF files for use with GelCompar version 4.1 software (Applied Maths, Kortrijk, Belgium). Normalization was done according to molecular weight standards on each gel, with one molecular weight standard being used for every four samples (flaA typing) or for every six samples (PFGE). AFLP patterns were collected with Genescan software (PE Applied Biosystems), and densitometric curves were transferred to GelCompar version 4.1. AFLP gels were normalized according to internal size standards added to each lane. Ribotyping patterns as obtained from Qualicon software were exported as txt files, converted using Gelconvert 1.01 (Qualicon), and imported into GelCompar version 4.1 as int files. Normalization was done by the Qualicon software according to molecular weight standards on each gel (one molecular weight standard for every two samples).
Construction of similarity matrices was carried out with GelCompar version 4.1. For AFLP analysis the Pearson product-moment correlation coefficient was used, whereas for flaA typing, PFGE, and ribotyping data the band-based Dice coefficient was used. In all cases the unweighted-pair group method using average linkages (UPGMA) was used to cluster the patterns. Bands for analysis with the Dice coefficient were assigned manually, according to densitometric curves and the accompanying hard-copy photograph.
Species discrimination between C. jejuni and C. coli.
For discrimination between C. jejuni and C. coli, a Campylobacter species-discriminating multiplex PCR (23) was performed. Primers were based on the nucleotide sequences of species-specific probes selected from C. jejuni and C. coli DNA fragment libraries (24).
RESULTS
Cutoff value.
The experimental variation between duplicate experiments was determined for six replicate experiments using six Campylobacter strains. For flaA typing these data were used to establish a cutoff value of 90% for typing identical strains with identical outputs. In a similar way, the cutoff values for PFGE and AFLP analysis were determined to be 90%. The reproducibility of ribotyping had been determined with RiboPrinter software, and only one pattern from each strain was imported into GelCompar version 4.1. The cutoff value could therefore not be determined and was arbitrarily chosen to be 90%.
flaA typing.
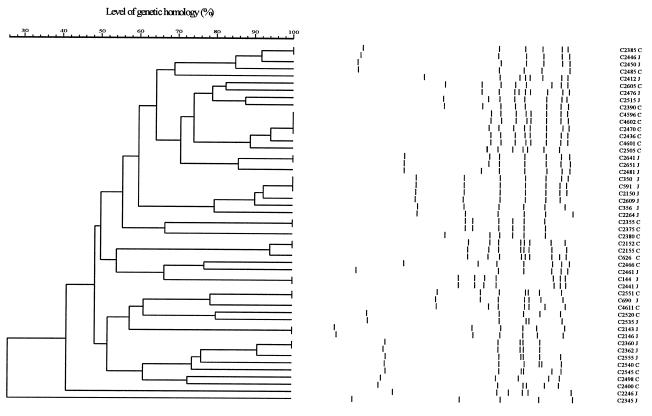
In this study flaA typing, using a cutoff of 90%, discriminated 31 different patterns out of 50 strains (Fig. 1). Bands could be reliably assigned down to 60 bp. The total number of bands ranged from 5 to 9 (Fig. 1). C. jejuni and C. coli isolates are randomly distributed within the dendrogram, indicating that flaA typing does not discriminate these species. Moreover, some C. jejuni and C. coli isolates, namely, C690 and C2551 and C2385, C2446, and C2450, share the same flagellin type (Fig. 1 and Table 2).
FIG. 1.
Dendrogram showing the assigned bands of the flagellin patterns. Levels of similarity were calculated with the Dice coefficient, and for cluster analysis the UPGMA was used. In GelCompar version 4.1 a position tolerance of 1.00% and an optimization of 0.50% were used. The species of the strains are indicated behind the strain number, with J indicating C. jejuni and C indicating C. coli.
TABLE 2.
Schematic representation of the strains with more than 90% genetic homology as determined by analysis with GelCompar version 4.1
| Method and straina |
|---|
| AFLP analysis |
| C4596, C4601, C4602 |
| C2390, C2400, C2436 |
| C2355, C2375 |
| C2264, C2360, C2362 |
| C350, C356 |
| C2143, C2146 |
| flaA typing |
| C4596, C4601, C4602, C2470, C2436 |
| C2360, C2362, C2555 |
| C2385, C2446, C2450 |
| C350, C356, C2150, C591, C2609 |
| C2152,b C2155b |
| C2641, C2651 |
| C144, C2441 |
| C2355, C2375 |
| C2143, C2146 |
| C2551, C690 |
| PFGE |
| C4596, C4601, C4602 |
| C350, C356 |
| C2360, C2362 |
| C2143, C2146 |
| C2152,b C2155b |
| C2380, C2390, C2400, C2436, C2470 |
| C2355, C2375 |
| Ribotyping |
| C626, C2152,b C2155,b C2385, C2466 |
| C2380, C2390, C2400, C2470, C2498, C2505 |
| C2355, C2375, C2545 |
| C4596, C4601, 4602, C2605 |
| C4611, C2540, C2551 |
| C690, C2143, C2146 |
| C144, C2441 |
| C2345, C2446 |
| C2264, C2450 |
| C350, C356 |
| C2412, C2515 |
| C2360, C2362 |
Underlined strains are >90% similar by all methods, and italicized and underlined strains are >90% similar by all techniques except flaA typing.
Strains showing 87% homology by AFLP analysis due to background in the banding pattern of C2155 but showing more than 90% homology by all other methods.
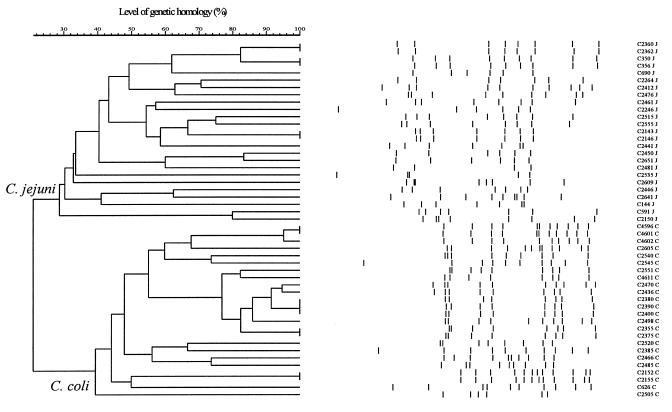
PFGE analysis.
Using a cutoff of 90%, PFGE analysis discriminated 38 different patterns (Fig. 2) and one isolate proved untypeable under the conditions used. The number of bands in PFGE patterns ranged from 4 to 12 (Fig. 2). As previously described (27), PFGE analysis allowed discrimination of C. jejuni and C. coli (Fig. 2).
FIG. 2.
Dendrogram of the PFGE patterns with designated bands. Cluster analysis was performed as described for Fig. 1. The clusters representing C. jejuni and C. coli are indicated, and the species are indicated behind the strain number, with J indicating C. jejuni and C indicating C. coli. Isolate C2345, which was untypeable, is not shown.
PFGE data are usually analyzed according to the guidelines of Tenover et al. (22). However, these criteria could not be used for our strains since they cannot be applied to populations of strains collected over periods of more than 1 year or to patterns consisting of less than 10 distinct fragments.
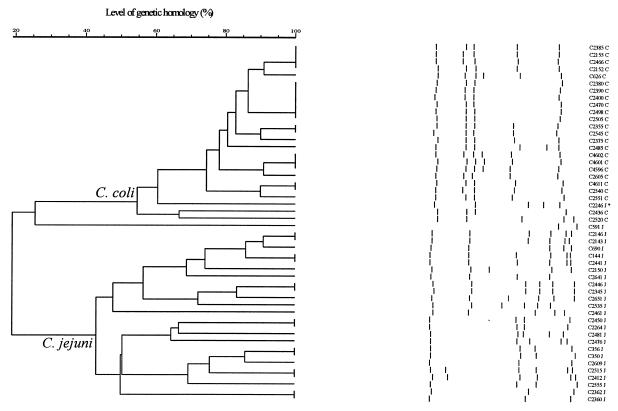
Automated ribotyping.
With the use of PstI, an enzyme that does not cut within the 16S rRNA gene of Campylobacter, ribotyping produced three to six bands and discriminated 26 different types (Fig. 3) when a cutoff of 90% was used. As previously described (5, 19), ribotyping discriminated C. jejuni from C. coli (Fig. 3). C. jejuni isolate C2246, however, clustered within a number of C. coli isolates near the border between the C. jejuni and C. coli isolates (Fig. 3), which indicates that species discrimination according to automated ribotyping is not completely reliable.
FIG. 3.
Dendrogram of ribotyping data with designated bands. Cluster analysis was performed as described for Fig. 1. The clusters representing C. jejuni and C. coli are indicated, and the species are indicated behind the strain number, with J indicating C. jejuni and C indicating C. coli. ∗ indicates a C. jejuni strain that is clustered among the C. coli strains.
The number of types identified by analysis with GelCompar version 4.1 is somewhat lower than the number of ribotypes determined with the RiboPrinter software (31 ribotypes, data not shown), indicating that automated ribotyping can be best analyzed with a RiboPrinter. However, cluster analysis is not possible with the Riboprinter software.
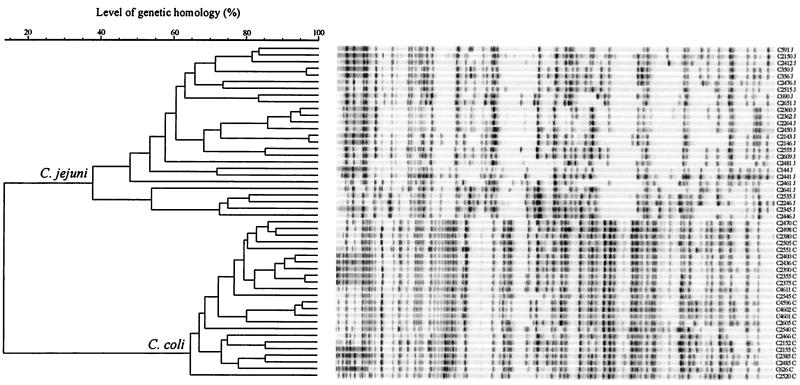
AFLP typing.
AFLP fingerprints consisted of 40 to 70 bands in the range of 50 to 500 bp (Fig. 4). Using a cutoff of 90% (4), we identified 41 distinct patterns. There was a clear distinction between AFLP fingerprints from C. jejuni and C. coli strains, indicating that AFLP analysis is capable of discriminating between these species.
FIG. 4.
Dendrogram of AFLP patterns. Cluster analysis was performed with GelCompar version 4.1 by using the UPGMA and the Pearson product-moment correlation coefficient. The clusters representing C. jejuni and C. coli are indicated, and the species are indicated behind the strain number, with J indicating C. jejuni and C indicating C. coli.
Comparison of the levels of discrimination obtained by the used methods.
Comparison of the results of the four techniques identified several strains that are genetically related by all methods. These results are depicted in Table 2. Several strains with related genotypes were isolated from the same farm; examples are C350 and C356, C2360 and C2362, and C4596, C4601, and C4602 (Tables 1 and 2). However, strains isolated from different places, like C2143 and C2146 and C2355 and C2375 (Table 2) were also related. Different flaA patterns but genetically related AFLP, PFGE, and ribotyping patterns were found for C2390 and C2400 (Table 2). In contrast, identical flaA patterns but different AFLP, PFGE, and ribotyping patterns were found for isolates C591 versus C350, C356, C2150, and C2609 (Table 2).
DISCUSSION
In this study a set of 50 Campylobacter isolates from poultry was typed by four genotyping techniques. The values of the routinely used genotyping techniques flaA typing, PFGE, and ribotyping and of the newly introduced AFLP analysis were compared for differentiation of strains and for computer-assisted analysis. All four genotyping techniques produce genetic fingerprints, although they differ in experimental approach and in levels of genetic discrimination. flaA typing is based on only one locus, the flaA gene. Ribotyping is based on three rRNA gene clusters and their flanking regions as opposed to the one locus of flaA typing. AFLP, only recently adjusted for typing Campylobacter species (4, 10), is based on a subset of small fragments (50 to 500 bp) from the whole genome. In PFGE the complete genome is cut into a small number of large fragments. The obtained numbers of bands differed substantially between the different techniques. The calculation of levels of similarity between patterns was highly influenced by the number of bands; the smaller the number of bands in a pattern, the larger the effect of one distinct band. AFLP analysis therefore appeared less subject to influences of individual band differences than ribotyping.
A distinction can be made in the processing of genetic fingerprints. Band assignment is necessary for methods that are analyzed by a band-based analysis such as that of the Dice coefficient but not for methods that are analyzed by a correlation-based analysis such as that of the Pearson correlation coefficient. The band-based Dice coefficient method is based on the comparison of designated band positions and divides the number of matching bands between patterns by the total number of bands, thereby emphasizing the matching bands (3). The Pearson correlation coefficient method compares the whole densitometric curves of patterns and is independent of band definition (16). It is largely independent of relative pattern intensities but is sensitive to differences in background. This makes the Pearson correlation coefficient method insensitive to peak-shoulder mismatches often found with band-matching coefficients. Differences in background intensities were observed with flaA typing, PFGE, and ribotyping. These differences influenced the Pearson coefficient analysis, which could therefore not be used. Instead, the band-based Dice coefficient was used for the analysis of flaA typing, PFGE, and ribotyping data. AFLP data were complex, band assignment was very laborious, and relatively minor differences in background levels occurred. Therefore, the correlation-based Pearson coefficient method was preferred.
Both analyses, band-based and correlation-based analyses, are largely influenced by the settings at which they are performed; different settings lead to different clusterings. The settings should therefore be carefully selected and should be kept constant within a comparison study.
Computer-assisted analysis, in theory, enables data transfer between different labs. Thus far the RiboPrinter method is the only method adapted for data exchange since it uses a standardized method and standardized materials. With GelCompar version 4.1, data exchange of the other three techniques between labs is possible with the data-sharing module. However, the methods will have to be standardized to enable valid comparisons between the exchanged data sets.
The discriminatory powers of the four techniques were examined according to calculated similarities. AFLP fingerprinting was the most discriminatory technique, followed by PFGE, flaA typing, and ribotyping. Discriminatory power, however, is not the only criterion on which a technique should be judged for usefulness in epidemiological typing. Ease of use, availability and price of materials and consumables, and the amount of throughput are also important factors. flaA typing is inexpensive, fairly quick, and the easiest method to perform in a laboratory. Drawbacks of this method are the risk of possible recombination events of the flagellin gene (8, 11, 25) and lack of species discrimination. Ribotyping has the advantage of being an automated, high-throughput process. However, the apparatus and consumables are expensive, it has only limited resolution, and due to the highly automated process, it is difficult to interfere with the identification or settings. The RiboPrinter is not capable of cluster analysis. Automated ribotyping can therefore be used only in situations in which a low resolution is satisfactory and cluster analysis is not necessary. PFGE is currently the most accepted method for typing campylobacters due to its high resolution (15, 19). However, it demands a specialized PFGE apparatus, is time-consuming and laborious, and is therefore unsuitable for typing large numbers of samples. In our study AFLP analysis was the most discriminatory technique; it was also capable of typing large numbers of samples, and it was best suited for computer-assisted analysis due to the easy transfer of data from the automatic sequencer to GelCompar version 4.1. Automated sequence equipment is desirable but not essential. Manual sequence equipment in conjunction with labeled isotopes is possible, but the easy transfer of data between the automated sequencer and GelCompar version 4.1 is lost. Furthermore, internal markers in every lane cannot be used and it is difficult to standardize the background intensities.
Combining the results of all four methods provided additional information about the studied strains. For example, clones consisting of isolates sharing identical patterns by all four methods and of common geographical origins could be discriminated (Tables 1 and 2). Other strains that were genetically similar by all methods did not share geographical relationships (Tables 1 and 2), indicating possible dispersion of clones. Indications for flagellin-specific recombination (8, 11, 25) were also found in this set of 50 strains, e.g., C2390 and C2400 possess the same AFLP, PFGE, and ribotyping patterns but have different flaA patterns (Fig. 1 and Table 2). In contrast, C591 versus C2150, C350, and C356 show the same flaA pattern but have different AFLP, PFGE, and ribotyping patterns (Table 2).
As observed previously (8, 11), flagellin patterns can be shared between C. jejuni and C. coli strains, e.g., between C. coli isolate C2385 and two C. jejuni strains (C2446 and C2450) and between C. coli strain C2551 and C. jejuni strain C690 (Fig. 1). The most likely explanation is the lack of discriminatory power of flaA typing because of the use of a single restriction enzyme. Genomic recombinations need to be considered when genotyping techniques are applied to Campylobacter. Recently there have been reports indicating genomic recombination (7, 13, 26), but the effect of recombination on genotyping methods is not yet known. The possible influence of recombination, combined with the finding that multiple techniques result in better discrimination and identification of strains, supports the use of multiple genotyping techniques, including AFLP fingerprinting, for optimal epidemiological typing of Campylobacter.
ACKNOWLEDGMENTS
This work was partly funded by the Product Boards for livestock, meat, and eggs, Rijswijk, The Netherlands.
We thank Trudy Wassenaar for critically reading the manuscript.
REFERENCES
- 1.Ayling R D, Woodward M J, Evans S, Newell D G. Restriction fragment length polymorphism of polymerase chain reaction products applied to the differentiation of poultry campylobacters for epidemiological investigations. Res Vet Sci. 1996;60:168–172. doi: 10.1016/s0034-5288(96)90013-2. [DOI] [PubMed] [Google Scholar]
- 2.Chang N, Taylor D E. Use of pulsed-field agarose gel electrophoresis to size genomes of Campylobacter species and to construct a SalI map of Campylobacter jejuni UA580. J Bacteriol. 1990;172:5211–5217. doi: 10.1128/jb.172.9.5211-5217.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dice L R. Measures of the amount of ecological association between species. J Ecol. 1945;26:297–302. [Google Scholar]
- 4.Duim B, Wassenaar T M, Rigter A, Wagenaar J. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol. 1999;65:2369–2375. doi: 10.1128/aem.65.6.2369-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald C, Owen R J, Stanley J. Comprehensive ribotyping scheme for heat-stable serotypes of Campylobacter jejuni. J Clin Microbiol. 1996;34:265–269. doi: 10.1128/jcm.34.2.265-269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson J, Lorenz E, Owen R J. Lineages within Campylobacter jejuni defined by numerical analysis of pulsed-field gel electrophoretic DNA profiles. J Med Microbiol. 1997;46:157–163. doi: 10.1099/00222615-46-2-157. [DOI] [PubMed] [Google Scholar]
- 7.Hänninen M L, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs-Reitsma W F, Bolder N M, Mulder R W. Caecal carriage of Campylobacter and Salmonella in Dutch broiler flocks at slaughter: a one-year study. Poult Sci. 1994;73:1260–1266. doi: 10.3382/ps.0731260. [DOI] [PubMed] [Google Scholar]
- 10.Kokotovic B, On S L. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol Lett. 1999;173:77–84. doi: 10.1111/j.1574-6968.1999.tb13487.x. [DOI] [PubMed] [Google Scholar]
- 11.Meinersmann R J, Helsel L O, Fields P I, Hiett K L. Discrimination of Campylobacter jejuni isolates by fla gene sequencing. J Clin Microbiol. 1997;35:2810–2814. doi: 10.1128/jcm.35.11.2810-2814.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nachamkin I, Bohachick K, Patton C M. Flagellin gene typing of Campylobacter jejuni by restriction fragment length polymorphism analysis. J Clin Microbiol. 1993;31:1531–1536. doi: 10.1128/jcm.31.6.1531-1536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.On S L. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol Lett. 1998;165:341–346. doi: 10.1111/j.1574-6968.1998.tb13167.x. [DOI] [PubMed] [Google Scholar]
- 14.On S L, Nielsen E M, Engberg J, Madsen M. Validity of SmaI-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol Infect. 1998;120:231–237. doi: 10.1017/s0950268898008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen R J, Sutherland K, Fitzgerald C, Gibson J, Borman P, Stanley J. Molecular subtyping scheme for serotypes HS1 and HS4 of Campylobacter jejuni. J Clin Microbiol. 1995;33:872–877. doi: 10.1128/jcm.33.4.872-877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson K. On the coefficient of racial likeliness. Biometrika. 1926;18:105–117. [Google Scholar]
- 17.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 18.Skirrow M B, Blaser M J. Clinical and epidemiologic considerations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 3–9. [Google Scholar]
- 19.Stanley J, Linton D, Sutherland K, Jones C, Owen R J. High-resolution genotyping of Campylobacter coli identifies clones of epidemiologic and evolutionary significance. J Infect Dis. 1995;172:1130–1134. doi: 10.1093/infdis/172.4.1130. [DOI] [PubMed] [Google Scholar]
- 20.Stern N J. Reservoirs for Campylobacter jejuni and approaches for intervention in poultry. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 49–60. [Google Scholar]
- 21.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 22.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Giessen A W, Tilburg J J, Ritmeester W S, van der Plas J. Reduction of Campylobacter infections in broiler flocks by application of hygiene measures. Epidemiol Infect. 1998;121:57–66. doi: 10.1017/s0950268898008899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Plas J, Hofstra H, Huis in't Veld J H J, et al. DNA probe assays for detection, identification and typing of Campylobacter species and Helicobacter pylori. Microb Ecol Health Dis. 1993;4:S60. [Google Scholar]
- 25.Wassenaar T M, Fry B N, van der Zeijst B A M. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology. 1995;141:95–101. doi: 10.1099/00221287-141-1-95. [DOI] [PubMed] [Google Scholar]
- 26.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan W, Chang N, Taylor D E. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J Infect Dis. 1991;163:1068–1072. doi: 10.1093/infdis/163.5.1068. [DOI] [PubMed] [Google Scholar]