Abstract
In this opinion, the antimicrobial resistant bacteria responsible for transmissible diseases that constitute a threat to the health of cattle have been assessed. The assessment has been performed following a methodology based on information collected by an extensive literature review and expert judgement. Details of the methodology used for this assessment are explained in a separate opinion. A global state of play on antimicrobial resistance in clinical isolates of Escherichia coli (non‐VTEC), Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Pasteurella multocida, Mannheimia haemolytica, Histophilus somni, Mycoplasma bovis, Moraxella bovis, Fusobacterium necrophorum and Trueperella pyogenes is provided. Among those bacteria, EFSA identified E. coli and S. aureus with ≥ 66% certainty as being the most relevant antimicrobial resistant bacteria in cattle in the EU based on the available evidence. The animal health impact of these most relevant bacteria, as well as their eligibility for being listed and categorised within the animal health law framework will be assessed in separate scientific opinions.
Keywords: antimicrobial resistance, animal health law, extensive literature review, cattle
1. Introduction
EFSA received a mandate from the European Commission to investigate the global state of play as regards resistant animal pathogens that cause transmissible animal diseases [Term of Reference (ToR) 1], to identify the most relevant bacteria in the EU (first part of ToR 2), to summarise the existing or potential animal health impact of those most relevant bacteria in the EU (second part of ToR 2), and to perform the assessment of those bacteria to be listed and categorised according to the criteria in Article 5, Appendix D according to Articles 8 and 9 within the Regulation (EU) 2016/429 on transmissible animal diseases (‘animal health law’)1 (ToR 3).
This scientific opinion presents the global state of play for resistant animal pathogens that cause transmissible animal diseases (ToR 1) and the results of the assessment of the most relevant bacteria in the EU (first part of ToR 2) for cattle following the methodology described in (EFSA AHAW Panel, 2021).
1.1. Background and Terms of Reference as provided by the requestor
The background and ToR as provided by the European Commission for the present document are reported in Sections 1.1 and 1.2 of the scientific opinion on the ad hoc method to be followed for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the animal health law (AHL) framework (EFSA AHAW Panel, 2021).
1.2. Interpretation of the Terms of Reference
The interpretation of the ToR is as in Sections 1.3.1 and 1.3.2 of the scientific opinion on the ad hoc method to be followed for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL framework (EFSA AHAW Panel, 2021).
The present document reports the results of the assessment of bacterial pathogens resistant to antimicrobials in cattle.
2. Data and methodologies
The methodology applied for this opinion is described in a dedicated document that details the ad hoc method for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL framework (EFSA AHAW Panel, 2021). Additional methods specific to this opinion (data collection by an extensive literature review) are detailed below.
2.1. Extensive literature review
The process to identify the bacterial species on which to focus in the extensive literature review (ELR) is described in Section 2.1.2 in the ad hoc method for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL (EFSA AHAW Panel, 2021). According to that methodology, the following target bacteria for cattle had been agreed upon by the EFSA working group: Escherichia coli (non‐VTEC), Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Pasteurella multocida, Mannheimia haemolytica, Histophilus somni, Mycoplasma bovis, Moraxella bovis, Fusobacterium necrophorum and Trueperella pyogenes. The ELR was carried out by the University of Copenhagen under the contract OC/EFSA/ALPHA/2020/02 – LOT 1.2 On 13 April 2021, two different search strings (Annex A) were applied in PubMed and Embase, respectively, resulting in a search result of 2,749 unique abstracts published since 2010. Upon importation into Rayyan software, these abstracts were screened by a senior scientist who followed the criteria described in the protocol for inclusion and exclusion of studies. When available, the full text of articles was downloaded into EndNote software. In addition, the national antimicrobial resistance (AMR) monitoring reports from Denmark, Finland, France, Ireland, Germany, Sweden, Switzerland and United Kingdom (written in English or German) were downloaded and used in the ELR.
Only the latest version of the AMR monitoring reports was included in the ELR as isolates included in these reports can be assumed to originate from the same sampled populations and most recent versions would therefore include the most up‐to‐date AMR data. The previous versions of the national AMR monitoring reports, i.e. up to the previous 5 years, were not included in the ELR but were downloaded and analysed separately to assess changes over time when possible. AMR data in the full texts of national reports were evaluated for eligibility applying the exclusion criteria as described in the ad hoc method followed for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL framework (EFSA AHAW Panel, 2021), with the following deviations from the standard methodology:
Exclusion criterion 8 (minimum number of isolates in a study to be considered acceptable): this number was set at 50 for E. coli and S. aureus and at the default of 10 for the other bacterial species (the minimum number is for the whole study, meaning that in one study there could be less than 50 E. coli from one country, but when isolates from different countries are added, the limit of 50 is applied; also, one study could have 25 E. coli isolates from one study period and 25 from another, and by merging those time periods, the limit of 50 isolates would be reached).
Exclusion criterion 6 (the same individual has been deliberately sampled more than once): This criterion was difficult to enforce in this opinion, as in many studies, it was reported that samples represented quarters of udders. Although these studies might have included more than one sample per animal, we decided to include them unless it was proven that more than one sample had been taken per animal (i.e. if the sample number was higher than the number of cattle sampled).
Exclusion criterion 16 (studies where AMR was only assessed genotypically): Studies in which mecA and/or mecC was used to infer the proportion of methicillin‐resistant S. aureus (MRSA) were considered eligible.
Year of bacterial isolation was neither extracted nor reported from the included studies, as in most studies, isolates had been collected over multiple years with no indication on the number of isolates per year. An exception to this rule was if only data from a certain time period within a study were extracted (in the case of national reports reporting multiple years, when only the last data points were considered).
Information extracted from the eligible assessed full‐text reports/publications is described in the scientific opinion on the ad hoc method applied in the assessment (EFSA AHAW Panel, 2021). Information on all the full‐text studies that were assessed, including the reason for exclusion for those that were excluded at the full‐text screening, is presented in Annex B. AMR was assessed for clinically relevant antibiotics according to the method detailed in Section 2.1.3 of the ad hoc method for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL (EFSA AHAW Panel, 2021). The list of clinically relevant antibiotics for each target bacterial species in cattle considered in this opinion are shown in Annex C. When more than one antimicrobial from a given class was considered eligible for inclusion in the report, the following order of preference for each antimicrobial class and bacterial pathogen was considered:
For methicillin in staphylococci, data for oxacillin, cefoxitin and presence of the mecA and mecC gene were accepted. If data for more than one of these antimicrobials were available in the same study, we included the one for which more isolates were tested. If the same number of isolates was tested for the different antimicrobials, the order of preference was mecA + mecC > cefoxitin > oxacillin.
For third‐generation cephalosporins (3GC) in Enterobacterales (as indicator of extended‐spectrum beta‐lactamase/AmpC), the order of preference was cefpodoxime > cefotaxime > ceftazidime > ceftriaxone > ceftiofur. If data for more than one of these antimicrobials were available in the same study, we included the one for which more isolates were tested. If resistance to at least one of these five 3GCs was not reported, we included instead – when available – other phenotypic data indicating the presence of ESBL/AmpC, typically data from a double disk synergy test (EUCAST, 2017).
The 3GC cefoperazone was reported separately for E. coli, Staphylococcus spp., S. dysgalactiae and S. uberis deriving from mastitis, as there is a mastitis‐specific clinical breakpoint for cefoperazone in these species.
For fluoroquinolones, the order of preference was enrofloxacin > ciprofloxacin, meaning that we always selected enrofloxacin if resistance data for both drugs were available.
For tetracyclines, the order of preference was tetracycline > oxytetracycline > doxycycline > chlortetracycline; hence, we always selected tetracycline if resistance data for all four drugs, or tetracycline + one of the other drugs, were present.
For each study, AMR data were extracted as percentages of resistant isolates (%R) and/or as percentages of non‐susceptible isolates by combining resistant and intermediate (I) isolates (%R + I). Moreover, the following decisions were made when evaluating data sets:
When no information on the I category was provided in a study, we considered that the reported %R only considered resistant isolates (i.e. I isolates had not been included in the R category).
When proportion of susceptibility (%S) was reported with no information on I, it was not possible to calculate %R. Instead, we calculated %R + I as 100% − %S.
When a study using ECOFFs reported %R, we considered this as %R + I, as the I category is always part of the non‐wild-type population.
When %I was reported separately, we extracted that along with %R and calculated %R + I.
For some drugs and presence of mecA/mecC, there is no I category for the bacterial species included, hence for those we could only report %R, irrespective of the assumptions mentioned above.
3. Assessment
3.1. ToR 1: global state of play for resistant bacterial animal pathogens that cause transmissible animal diseases
3.1.1. General overview of studies included and excluded
3.1.1.1. Data from the extensive literature review
After screening of the 2,750 abstracts, 491 publications were selected for evaluation according to the criteria under methods. Of these, 364 publications were excluded with the reasons for exclusion highlighted in columns D and E of Annex B. The reasons for exclusion of publications are listed in Table 1. The most common reason for exclusion (n = 108) was that an insufficient number of isolates had been investigated according to the inclusion criteria (≥ 50 for E. coli and S. aureus, ≥ 10 for the remaining species). The second most common reason for exclusion was that isolates were not clinical or that it was not possible to distinguish between clinical and non‐clinical isolates (n = 47); several of these publications had investigated milk samples but without specifying if they were from cows with mastitis or not.
Table 1.
Main reasons for exclusion of publications after full‐text evaluation affecting more than one publication (a publication could be excluded for more than one reason)(a)
| Reason | Code in Annex B | Number of publications |
|---|---|---|
| Fewer than the minimum number of isolates are included in the publication | 8 | 108 |
| Inclusion of non‐clinical isolates or isolates that cannot be distinguished from clinical isolates | 5 | 47 |
| Full text not available at server of the University of Copenhagen | 10 | 29 |
| Percentage of resistant isolates not reported | 7 | 27 |
| Criteria for selection of isolates unclear and/or high risk of data duplication | 14 | 26 |
| Same animals sampled repeatedly | 6 | 25 |
| Minimum inhibitory concentration data reported without interpretation | 12 | 22 |
| Publication does not follow a standard for antimicrobial susceptibility testing or a standard is not reported | 4 | 20 |
| AMR data included in another included publication | 9 | 15 |
| AMR assessed genotypically (except mecA used to infer methicillin resistance in staphylococci) | 16 | 11 |
| AMR data reported at bacterial genus level or above | 3 | 8 |
| AMR data from multiple host species (other than cattle) reported together | 2 | 7 |
| Biased data presented (only for drugs for which more resistance was found) | 17b | 7 |
| Antimicrobials tested are not among the ones of interest for this scientific opinion | 13 | 6 |
| All isolates in a publication originate from the same farm | 15 | 5 |
| Language (non‐English) | 11 | 2 |
| Publication investigating AMR in a subset of resistant clinical isolates | 17b | 2 |
| Data included in a more recent report published later | 17b | 2 |
After exclusion of these references, 127 eligible publications with information on AMR from clinical isolates were selected for data extraction. In addition, eight national reports representing Denmark, Finland, France, Germany, Ireland, Sweden, Switzerland and the UK were selected, as they contained eligible AMR data on clinical isolates from cattle according to the same set of eligibility criteria mentioned above (for a total of 135 references considered).
An overview of the number of eligible studies for each target bacterium is shown in Table 2.
Table 2.
Number of studies from which AMR data were extracted
| Bacterial species | Number of eligible studies for data extraction (n = 135)a |
|---|---|
| Staphylococcus aureus | 66 |
| Escherichia coli | 37 |
| Pasteurella multocida | 23 |
| Mannheimia haemolytica | 20 |
| Streptococcus uberis | 18 |
| Streptococcus dysgalactiae | 13 |
| Histophilus somni | 12 |
| Trueperella pyogenes | 8 |
| Mycoplasma bovis | 8 |
| Klebsiella pneumoniae | 5 |
| Moraxella bovis | 1 |
| Fusobacterium necrophorum | 0 |
A publication can provide information on more than one bacterial species.
Figure 1 below provides an overview of the 135 included studies (some with data on multiple bacterial species) sorted by year of publication.
Figure 1.
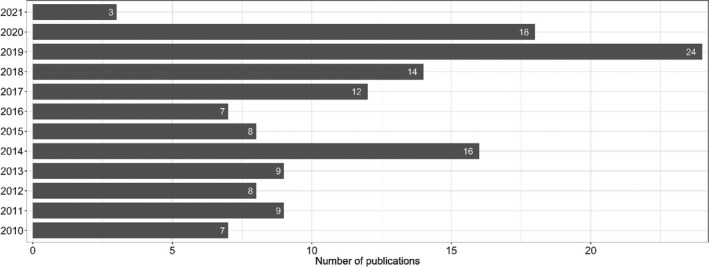
Date of publication of the 135 publications included in the extensive literature review
Considering geographical distribution, AMR data were reported in the following number of publications: Asia (53 publications), Europe (47), North America (13), Africa (11), South America (5) and Oceania (5) (Figure 2). One publication reported data from multiple continents. For publications including information from a single country, the country in which a higher number of publications were performed was China (26 publications) followed by Iran (8), Switzerland (7), USA (7), Canada (5), France (5), South Korea (5), South Africa (5) and Turkey (5). In addition, there were eight publications reporting data from multiple countries, of which six included a combination of European countries.
Figure 2.

Geographical distribution of the 135 included publications
Based on the type of isolates analysed in the publication, references included were divided into those based on the assessment of isolates from: (i) a clearly defined population of cattle in farms, hospitals or clinics; and (ii) those without – or with limited ‐ background information on sampled animals (comprising publications with isolates from a diagnostic laboratory or obtained in slaughterhouses). Ninety‐four publications had isolates obtained from samples actively collected in farms, whereas 29 publications had isolates from diagnostic laboratories and no publications were performed on samples collected exclusively at slaughterhouses. In four publications, isolates had a mixed origin (farm and diagnostic laboratory), and for the last eight publications, there was no information on sample and isolate origin, except they were clinical isolates from cattle.
3.1.1.2. Data from national AMR monitoring reports
Additional details/data on one or more of the pathogens of interest of this opinion that are provided in previous versions of eight national AMR monitoring reports retrieved (up to the previous 5 years), namely FINRES‐Vet – Finland, SWEDRES‐Svarm – Sweden, GERM‐VET – Germany, RESAPATH – France and UK‐VARSS – United Kingdom, DANMAP – Denmark, ANRESIS ARCH‐Vet – Switzerland and All‐Island Animal Disease Surveillance Report – Ireland, were also extracted and are presented in the following section (see Table 3). The same terminology used in the report (e.g. proportion of non‐susceptible or proportion of resistant isolates) based on the selected breakpoint for defining resistance/susceptibility in each report was used to describe the results provided.
Table 3.
AST methodology, bacterial species, host species, number of isolates and temporal coverage of the information on pathogens of interest from cattle provided in the eight national AMR monitoring reports (up to the last 5 years) reviewed in this opinion. When a monitoring programme does not include a pathogen of interest this is indicated in the table as ‘No’ marked in red
| Programme | UK‐VARSS | RESAPATH | DANMAP | All‐Islands | ANRESIS ARCH‐Vet | SWEDRES‐Svarm | FINRES‐Vet | GERM‐VET |
|---|---|---|---|---|---|---|---|---|
| Country | UK | France | Denmark | Ireland | Switzerland | Sweden | Finland | Germany |
| Laboratory method | Disk diffusion | Disk diffusion | Broth microdilution | Disk diffusion | Broth microdilution | Broth microdilution | Broth microdilution | Broth microdilution |
| AST interpretation | CBPsa | ECOFFsb | CBPs | CBPs | CBPs | ECOFFs | CBPs | CBPs |
| E. coli | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes |
| Origin (no. of isolates) | Mastitis 79–110/yearc | Mastitis/GI 504–4,222/year | Mastitis 17–23/year | Unknown 268 | Mastitis (54) | Mastitis/GI 29–117/year | GI/Mastitis (25–284/year) | |
| Years covered | 2015–2019 | 2014–2018 | 2018–2019 | 2018 | 2019 | 2012–2018 | 2014–1018 | |
| S. aureus | Yes | Nod | Yes | Yes | Yes | No | No | Yes |
| Origin (no. of isolates) | Mastitis (36–78/year) | Mastitis (12/year) | Mastitis (407) | Mastitis (56–60/year) | Mastitis (196–363/year) | |||
| Years covered | 2015–2019 | 2018–2019 | 2018 | 2016–2019 | 2015, 2017 | |||
| S. uberis | Yes | Yes | Yes | Yes | Yes | No | No | Yes |
| Origin (no. of isolates) | Mastitis 70–123/year | Mastitis (56–60/year) | Mastitis (16–17/year) | Mastitis (291) | Mastitis (56) | Mastitis (335–384/year) | ||
| Years covered | 2015–2019 | 2014–2018 | 2018–2019 | 2018 | 2019 | 2014, 2016 | ||
| S. dysgalactiae | Yes | Yes | Yes | No | No | No | No | Yes |
| Origin (no. of isolates) | Mastitis (18–41/year) | Mastitis (112–223/year) | Mastitis (19–20/year) | Mastitis (74–85/year) | ||||
| Years covered | 2015–2019 | 2014–2018 | 2018–2019 | 2014, 2016 | ||||
| K. pneumoniae | Yes | Yes | No | No | No | Yes | No | Yes |
| Origin (no. of isolates) | Mastitis (3–13/year) | Mastitis (44–90/year) | Mastitis (34–52/year) | Mastitis (58–97 per year) | ||||
| Years covered | 2016–2019 | 2014–2018 | 2014–2018 | 2014, 2015, 2016, 2018 | ||||
| P. multocida | Yes | Yes | No | Yes | No | Yes | Yes | Yes |
| Origin (no. of isolates) | Respiratory (42–76/year) | Respiratory (31–301/year) | Respiratory (181) | Respiratory (79–104/year) | Respiratory (135–267/year) | Respiratory (98–149/year) | ||
| Years covered | 2015–2019 | 2014–2018 | 2018 | 2016–2018 | 2015–2019 | 2014, 2016–2018 | ||
| M. haemolytica | Yes | Yes | No | Yes | No | No | Yes | Yes |
| Origin (no. of isolates) | Respiratory (28–70/year) | Respiratory (45–178/year) | Respiratory 150 | Respiratory (35–79/year) | Respiratory (65–81/year) | |||
| Years covered | 2015–2019 | 2014–2018 | 2018 | 2015–2019 | 2014, 2016–2018 | |||
| T. pyogenes | Yes | No | No | No | No | No | No | No |
| Origin (no. of isolates) | Mastitis 3–8/year | |||||||
| Years covered | 2015–2017 | |||||||
| H. somni | No | No | No | No | No | No | Yes | No |
| Origin (no. of isolates) | Respiratory (28–47) | |||||||
| Years covered | 2015–2019 |
Human breakpoints recommended by the British Society for Antimicrobial Chemotherapy when available and a uniform cut‐off point of 13 mm when not available.
Veterinary guidelines of the Antibiogram Committee of the French Society of Microbiology (CA‐SFM).
Data from 157 and 134 isolates from Scotland retrieved in 2018 and 2019 were also available.
Only data on ‘coagulase‐positive Staphylococcus’ are provided.
3.1.2. AMR frequency data
The figures and tables in the following pathogen‐specific sections summarise the AMR frequency data reported for cattle.
The AMR frequency data are extremely difficult to compare, as study design, selection criteria, study populations, sampling procedures, methods, interpretive criteria, etc., vary considerably between publications. The number of antimicrobial susceptibility testing (AST) results for any given antimicrobial extracted from the 135 selected references (total of 228,620, Annex B) was largely due to the number of results found for E. coli (95,407, 41.7% of the total number of AST), S. aureus (40,822, 17.9%), P. multocida (27,455, 12.0%), M. haemolytica (22,653, 9.91%) and S. uberis (18,693, 8.2%). Lower numbers of results were available for H. somni (9,217, 4.0%), S. dysgalactiae (5,510, 2.4%) and K. pneumoniae, Mycoplasma bovis, T. pyogenes and Moraxella bovis (< 4.000 and < 2% from each) and none for Fusobacterium necrophorum. The laboratory method most commonly used to determine the AST phenotype was disk diffusion (116,138 of all AST results obtained through this method, 50.8%) followed by broth microdilution (97,464, 42.6%), with the remaining being determined mostly through a combination of methods (Annex B).
Furthermore, the definition of AMR differed across publications, as the intermediate category defined by clinical breakpoints (CBPs) was included in the calculation of AMR frequencies in some publications, whereas it was omitted in others. Accordingly, in the figures with resistance data, we have illustrated for each study whether %R or %R + I was reported; hence, this should be taken into account when comparing publications. When presenting data obtained in the ELR in the text, the results are presented as proportion of resistant isolates irrespective of the cut‐off used except in specific cases. It is also important to mention that relatively few infection‐specific and host‐specific CBPs exist for bovine pathogens. This complicates interpretation of data, as for several publications, it was unclear if the CBPs used were adapted from other bacterial or animal species, from humans, or even ‘self‐invented’. In the present report, this issue is of particular relevance for mastitis, as this infection accounts for the vast majority of data and relatively few CBPs exist for this indication. Taken together, the outcomes of the present report should be interpreted and cited with caution, as not all specificities of individual publications can be taken into consideration. In order to support conclusions made from the figures or tables (e.g. a high proportion of resistance in a certain country/continent), it is strongly recommended that individual papers are consulted and checked in case results would be biased by previous antimicrobial treatment, sampling of animals in a certain environment, the use of certain diagnostic methods or breakpoints, or other factors.
For data included in the national AMR monitoring reports, details/data provided in previous versions of the reports from these monitoring programmes (up to the previous 5 years) were extracted and are presented at the end of each bacterium's specific section to assess the existence of changes over time in the proportion of non‐susceptible/resistant isolates when possible. The bacterial species most often included in the reports were E. coli (from mastitis, gastrointestinal samples or unknown origin), S. uberis (typically from mastitis cases) and P. multocida (from respiratory samples) (Table 3). Assessment of changes in AMR levels over time in the pathogens under evaluation based on the data in the reports is hampered in certain cases by the lack of consistent reporting over the years (i.e. only data from specific years were reported) and/or because data on isolates retrieved over several years were presented together. Between‐country comparisons must be performed carefully as different methodologies were applied to obtain the results presented in each report, number of isolates tested for certain species and countries was limited and results provided here are those presented in the reports (e.g. without accounting for the use of different breakpoints). A comparison of the methodology, bacterial pathogens, number of isolates and temporal coverage of the information provided in the last five reports of each monitoring programme is provided in Table 3.
3.1.3. Staphylococcus aureus
3.1.3.1. Results of the ELR by bacterium
Staphylococcus aureus is an opportunistic pathogen of the skin and mucosal membranes. As in other hosts, it may cause a variety of infections, but mastitis is by far the most important one in cattle. Although S. aureus survives well in the environment, transmission between cows mainly occurs during milking, via contaminated hands or equipment.
In total, 66 studies with ≥ 50 S. aureus isolates and results for one or more of the relevant antibiotics [cefoperazone, ceftiofur, enrofloxacin/ciprofloxacin, erythromycin, methicillin (cefoxitin, oxacillin or presence of mecA/mecC), neomycin, penicillin, penicillin–novobiocin, pirlimycin, sulfonamide–trimethoprim] were included. Those studies were distributed as follows: Africa (9), Asia (23), Europe (23), Oceania (3), North America (3) and South America (5).
The distribution of S. aureus isolates per site of infection is shown in Figure 3. For studies in which the origin was specified, the vast majority of isolates originated from milk/udder, meaning that isolate came from cases of either clinical or subclinical mastitis in dairy cattle. For non‐mastitis‐associated isolates, it was not possible to discriminate between other specific locations (e.g. wounds).
Figure 3.
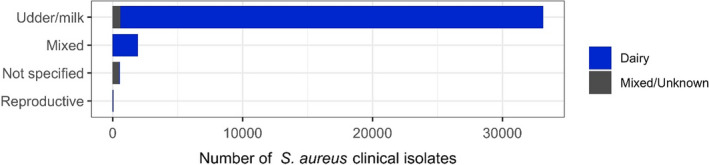
Distribution of Staphylococcus aureus isolates per site of infection
Figure 4 shows for each continent the proportion of resistance reported in individual studies with at least 50 S. aureus isolates. Information on proportion of resistance sorted by country is in Annex D.
Figure 4.

Staphylococcus aureus resistance data for each included study sorted by continent
Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (light blue circle), resistance merged with intermediate in isolates of dairy production origin (dark blue circle) or resistance in isolates of mixed or unknown origin (light grey circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of %R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
On average, the highest mean levels of resistance were observed for penicillin, but resistance proportions varied substantially between studies (Figure 4). In addition, there was a large difference between continents, e.g. the mean proportions of resistance in S. aureus from dairy cattle in Asia (64.2%) and Africa (57.7%) were substantially higher than in Europe (32.1%) (Table 4). In Europe, the lowest levels of penicillin resistance were generally observed in northern and central European countries, namely Sweden (4%), Denmark (17.5%), Austria (10%) and Switzerland (14%), whereas 63.1% of isolates were resistant in Italy even though the corresponding study reported that animals had not been subjected to antimicrobial treatment in the 3 weeks before sampling (Intorre et al., 2012).
Table 4.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Staphylococcus aureus for the target antimicrobials in each continent and sorted by production type. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC (Cefoperazone) | Europe | Dairy | 4 | 772 | 13.7 | 0 | 36.1 | 10.4 |
| 3GC (Cefoperazone) | South America | Dairy | 1 | 352 | 5 | 5 | 5 | NA |
| 3GC (Ceftiofur) | Africa | Dairy | 1 | 79 | 0 | 0 | 0 | NA |
| 3GC (Ceftiofur) | Asia | Dairy | 4 | 273 | 11.5 | 0 | 26.8 | 10.5 |
| 3GC (Ceftiofur) | Europe | Dairy | 4 | 317 | 6.9 | 0 | 41.5 | 15.5 |
| 3GC (Ceftiofur) | North America | Dairy | 2 | 1,630 | 0.1 | 0 | 0.1 | 0 |
| 3GC (Ceftiofur) | South America | Dairy | 3 | 539 | 0.2 | 0 | 0.3 | 0.1 |
| Erythromycin | Africa | Dairy | 6 | 483 | 22 | 0 | 62 | 25.4 |
| Erythromycin | Asia | Dairy | 9 | 1,309 | 30.9 | 1.2 | 79.9 | 26.3 |
| Erythromycin | Asia | Mixed/Unknown | 1 | 104 | 22.1 | 22.1 | 22.1 | NA |
| Erythromycin | Europe | Dairy | 10 | 1,066 | 5.5 | 0 | 41.7 | 13 |
| Erythromycin | North America | Mixed/Unknown | 1 | 123 | 0 | 0 | 0 | NA |
| Erythromycin | Oceania | Dairy | 1 | 782 | 28.8 | 28.8 | 28.8 | NA |
| Erythromycin | Oceania | Mixed/Unknown | 1 | 404 | 0.2 | 0.2 | 0.2 | NA |
| Erythromycin | South America | Dairy | 4 | 552 | 4.9 | 0 | 14.1 | 5.8 |
| Fluoroquinolones | Africa | Dairy | 4 | 303 | 6.1 | 0 | 14.3 | 6.3 |
| Fluoroquinolones | Asia | Dairy | 15 | 1,978 | 20.5 | 0 | 53.4 | 17.7 |
| Fluoroquinolones | Europe | Dairy | 6 | 582 | 7.9 | 0 | 36.9 | 14.9 |
| Fluoroquinolones | North America | Mixed/Unknown | 1 | 123 | 0 | 0 | 0 | NA |
| Fluoroquinolones | Oceania | Mixed/Unknown | 1 | 202 | 0 | 0 | 0 | NA |
| Fluoroquinolones | South America | Dairy | 4 | 824 | 0.8 | 0 | 2.5 | 0.8 |
| Methicillin | Africa | Dairy | 7 | 576 | 8.3 | 0 | 50 | 17.4 |
| Methicillin | Asia | Dairy | 21 | 2,944 | 19.1 | 0 | 60.7 | 16.6 |
| Methicillin | Asia | Mixed/Unknown | 1 | 96 | 13.7 | 13.7 | 13.7 | NA |
| Methicillin | Europe | Dairy | 13 | 1,984 | 9.9 | 0 | 27.1 | 10.8 |
| Methicillin | Oceania | Dairy | 1 | 733 | 2.3 | 2.3 | 2.3 | NA |
| Methicillin | Oceania | Mixed/Unknown | 1 | 202 | 0 | 0 | 0 | NA |
| Methicillin | South America | Dairy | 5 | 1,474 | 0.9 | 0 | 2.8 | 0.8 |
| Neomycin | Africa | Dairy | 3 | 233 | 3.9 | 0 | 6.3 | 2.8 |
| Neomycin | Europe | Dairy | 2 | 180 | 0.6 | 0 | 1.9 | 0.9 |
| Neomycin | North America | Dairy | 1 | 1,532 | 18.1 | 18.1 | 18.1 | NA |
| Neomycin | Oceania | Dairy | 1 | 103 | 8.9 | 8.9 | 8.9 | NA |
| Neomycin | South America | Dairy | 1 | 352 | 3.4 | 3.4 | 3.4 | NA |
| Penicillin | Africa | Dairy | 7 | 1,177 | 57.7 | 28.8 | 86 | 15.7 |
| Penicillin | Asia | Dairy | 15 | 1,837 | 64.2 | 11 | 97.1 | 28.9 |
| Penicillin | Europe | Dairy | 13 | 1,751 | 32.1 | 4 | 63.1 | 16 |
| Penicillin | North America | Mixed/Unknown | 1 | 123 | 26 | 26 | 26 | NA |
| Penicillin | Oceania | Dairy | 2 | 1,100 | 23.9 | 21.8 | 28 | 2.9 |
| Penicillin | Oceania | Mixed/Unknown | 1 | 202 | 12.4 | 12.4 | 12.4 | NA |
| Penicillin | South America | Dairy | 4 | 619 | 59.9 | 6.9 | 81.9 | 31.9 |
| Penicillin–novobiocin | Asia | Dairy | 1 | 52 | 0 | 0 | 0 | NA |
| Penicillin–novobiocin | Europe | Dairy | 1 | 78 | 0 | 0 | 0 | NA |
| Penicillin–novobiocin | North America | Dairy | 1 | 1,532 | 0.3 | 0.3 | 0.3 | NA |
| Penicillin–novobiocin | South America | Dairy | 1 | 115 | 1.7 | 1.7 | 1.7 | NA |
| Pirlimycin | Asia | Dairy | 1 | 52 | 0 | 0 | 0 | NA |
| Pirlimycin | Europe | Dairy | 2 | 160 | 25.6 | 0 | 41 | 19.9 |
| Pirlimycin | North America | Dairy | 1 | 1,532 | 1.9 | 1.9 | 1.9 | NA |
| Pirlimycin | South America | Dairy | 1 | 115 | 4.3 | 4.3 | 4.3 | NA |
| Sulfa/TMP | Africa | Dairy | 5 | 449 | 15.8 | 0.7 | 78.6 | 30.2 |
| Sulfa/TMP | Asia | Dairy | 7 | 1,041 | 37.9 | 0 | 91.8 | 34.8 |
| Sulfa/TMP | Europe | Dairy | 4 | 694 | 0.6 | 0 | 3.3 | 1.3 |
| Sulfa/TMP | North America | Dairy | 1 | 1,532 | 0.5 | 0.5 | 0.5 | NA |
| Sulfa/TMP | North America | Mixed/Unknown | 1 | 123 | 0 | 0 | 0 | NA |
| Sulfa/TMP | Oceania | Dairy | 1 | 364 | 0.5 | 0.5 | 0.5 | NA |
| Sulfa/TMP | Oceania | Mixed/Unknown | 1 | 202 | 0 | 0 | 0 | NA |
| Sulfa/TMP | South America | Dairy | 2 | 356 | 12.6 | 0.3 | 62 | 24.7 |
Resistance to other beta‐lactams was considerably less pronounced. For methicillin resistance (MR) in dairy cattle, this was uncommon in Oceania and South America (< 3%), whereas mean proportions were higher in Africa (8.8%), Europe (9.9%) and Asia (19.1%). Importantly, a study by Wu et al. (2019) illustrated that the MR indicator drugs we allowed in this report are not fully comparable, as 52.4% of isolates in that study were resistant to cefoxitin, whereas only ˜ 35% of the same isolates were resistant to oxacillin. It is also reasonable to argue that MR proportions based on the presence of mecA are not fully comparable with those based on both mecA and mecC. This was however not an issue, as only two studies screened for mecC, and both of them found none of the tested isolates to harbour this gene (Bonsaglia et al., 2018; Srednik et al., 2018). Resistance to the 3GCs cefoperazone and ceftiofur, for which mastitis‐specific CBPs exist, was even less pronounced in most continents (Table 4). Levels of resistance to these drugs were not always equal to MR despite being caused by the same resistance mechanism. For example, in two studies, proportions of resistance to ceftiofur were lower than MR (Costa et al., 2012; Dorneles et al., 2019). This means that using ceftiofur, clinical breakpoint for mastitis will sometimes result in treating MRSA infections with this drug, unless laboratories use an expert rule to classify MRSA isolates as resistant to all beta‐lactams. Penicillin–novobiocin appears to be effective for the treatment of mastitis caused by S. aureus with no or very little resistance observed in the four studies testing this combination (Figure 4).
Resistance to the lincosamide pirlimycin was generally low (< 5%), but a study from Austria stood out with 41% of 100 mastitis isolates being resistant (Wald et al., 2019). This contrasts with the 0% resistance (%R) observed 2 years later in 60 mastitis isolates from the neighbouring country Switzerland (ANRESIS ARCH‐Vet, 2020). Mean fluoroquinolone resistance levels were higher in Asia (20.5%) than in other continents (< 8%) (Table 2). Despite low mean levels in Europe, a study from Italy reported 36.9% of 122 isolates resistant to enrofloxacin (Intorre et al., 2012). This high proportion was observed in 2011 and reflected a significant increase over the years commencing with only 5.9% resistance in isolates from 2005 (Intorre et al., 2012). Resistance to neomycin was tested in relatively few studies and proportions were generally low. The highest proportion (18.3%) was observed in a study from Canada (Awosile et al., 2018), but this value is not fully comparable with most other studies, as the resistant and intermediate categories had been merged. The importance of the I category for this drug is evident in a South African study reporting 16.7% of S. aureus isolates as intermediate to neomycin (Schmidt, 2011). Most studies reported very low levels of resistance to sulfonamide–trimethoprim (Figure 4), but a few noteworthy exceptions were detected, and also for this drug, the highest mean resistance proportion (37.9%) was reported by studies from Asia (Table 4).
3.1.3.2. Results from the national AMR monitoring reports
Information on AMR in cattle clinical S. aureus isolates, typically originating from samples from cows with mastitis, was included in five national reports, although number of isolates and antimicrobials used for testing varied widely depending on the country. The base population represented in these data will also vary according to the source material for these tests.
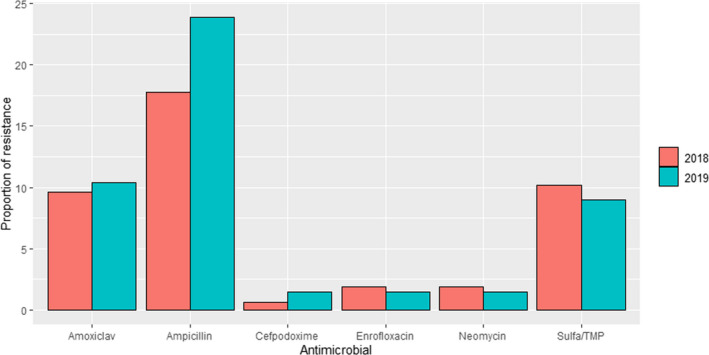
ANRESIS ARCH‐Vet (Switzerland): Data on AMR determined in 56 isolates in 2016–2017 (obtained through a pilot study) and 60 isolates in 2019 (coming from all the country) retrieved from mastitis cases, which can be detected in ˜ 57% of all dairy herds in Switzerland, were included in the last reports. Isolates were tested in both periods with five antimicrobials of interest for this opinion (ceftiofur, ciprofloxacin, erythromycin, penicillin and pirlimycin), and in addition, sulfonamide–trimethoprim was used in 2016–2017 and cefoperazone in 2019. The only antimicrobials for which non‐susceptible isolates were detected were ciprofloxacin and penicillin (Figure 5); although some changes are observed between the two periods for penicillin resistance, these should be interpreted with caution as they originated from different isolate populations.
Figure 5.
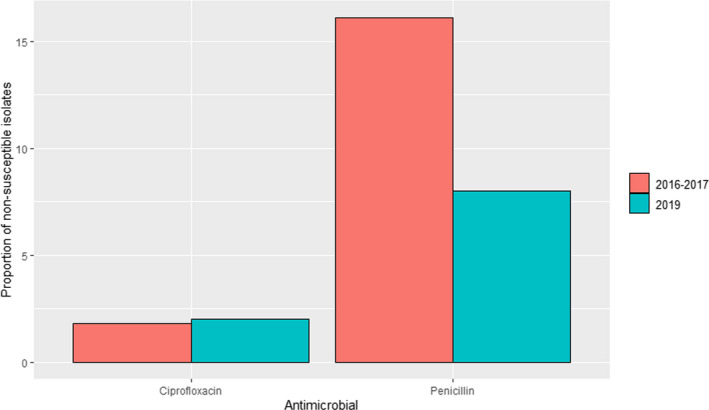
Proportion of clinical Staphylococcus aureus isolates non‐susceptible to ciprofloxacin and penicillin retrieved from mastitis cases reported by the ANRESIS ARCH‐Vet programme
All‐Islands Animal Disease Surveillance Report (Ireland): Detailed data on AMR obtained in clinical S. aureus are only provided for 407 isolates from mastitis cases in the 2018 report, providing results for sulfonamide‐trimethoprim with all isolates classified as susceptible (these data are already included in Figure 4 and Table 4).
DANMAP (Denmark): Resistance data from 12 clinical isolates submitted by veterinary clinics in 2018 and 2019 to the Technical University of Denmark (DTU) in relation to several research projects are included in the 2019 report. Isolates were tested for resistance to five antimicrobials of interest in this opinion (cefoxitin, ciprofloxacin, erythromycin, penicillin and sulfonamide–trimethoprim), and only one isolate resistant to penicillin and cefoxitin was found in 2018 and 2019, respectively.
UK‐VARSS (United Kingdom): Between 36 and 78 S. aureus isolates retrieved from mastitis cases in England and Wales were tested annually between 2015 and 2019 using two antimicrobials of interest for this opinion. Resistance levels were much higher for penicillin (12–35%), with values changing largely between years, than for neomycin (< 5%) (Figure 6).
Figure 6.
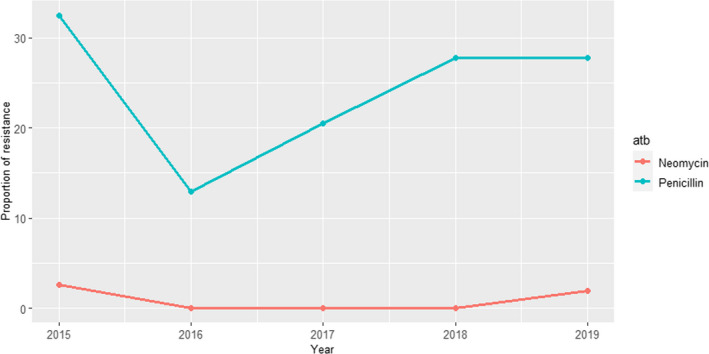
Proportion of clinical Staphylococcus aureus isolates retrieved from mastitis cases resistant to neomycin and penicillin reported by the UK‐VARSS programme
GERM‐VET (Germany): Resistance data from S. aureus isolates were reported in 2015 and 2017 with 363 and 196 isolates, respectively. All isolates were considered susceptible to trimethoprim/sulfamethoxazole and low levels of non‐susceptibility were detected for gentamicin (1–1.1%). Proportion of non‐susceptible isolates to ceftiofur (4.2% in 2015 and 14.3% in 2017), erythromycin (8.3% in 2015 and 4.1% in 2017), oxacillin (4.1% in 2015 and 13.8% in 2017) and pirlimycin (9.9% in 2015 and 5.1% in 2017) remained low, while for tetracycline non‐susceptibility levels between 14.6 and 17.3% were reported, and for penicillin between 24 and 25.9%.
3.1.4. Escherichia coli
3.1.4.1. Results of the ELR by bacterium
Escherichia coli is a commensal and an opportunistic pathogen residing in the intestinal microbiota of animals and humans. The environment can also constitute a reservoir for E. coli. A variety of infections can be caused by E. coli in cattle, but it is mostly known for causing intestinal or septicaemic infections in calves and mastitis in adult dairy cows. The former is a contagious disease, whereas the latter occurs through environmental contamination of the udder. Other less common presentations include peritonitis, cystitis/pyelonephritis, metritis, wound infections and meningitis derived from sepsis.
In total, 37 studies with ≥ 50 E. coli isolates and results for one or more of the relevant antibiotics (ampicillin/amoxicillin, amoxicillin‐clavulanic acid, apramycin, colistin, enrofloxacin/ciprofloxacin, gentamicin, neomycin, paromomycin, sulfonamide‐trimethoprim, tetracyclines, 3GC) were included. These were distributed as follows: Africa (2), Asia (12), Europe (19), Oceania (1), North America (3) and South America (0).
The distribution of E. coli isolates per site of infection is shown in Figure 7. Most isolates originated from mastitis in dairy cattle. Of note, clinical isolates included in this review from gastrointestinal tract/faeces were typically not subjected to typing to confirm their pathogenic nature, and therefore even though they were considered pathogenic in the references inclusion of a proportion of commensal strains cannot be ruled out.
Figure 7.

Distribution of Escherichia coli isolates per site of infection and type of production
Figure 8 shows for each continent the proportion of resistance reported in individual studies with at least 50 Escherichia coli isolates. Information on proportion of resistance sorted by country is in Annex D.
Figure 8.
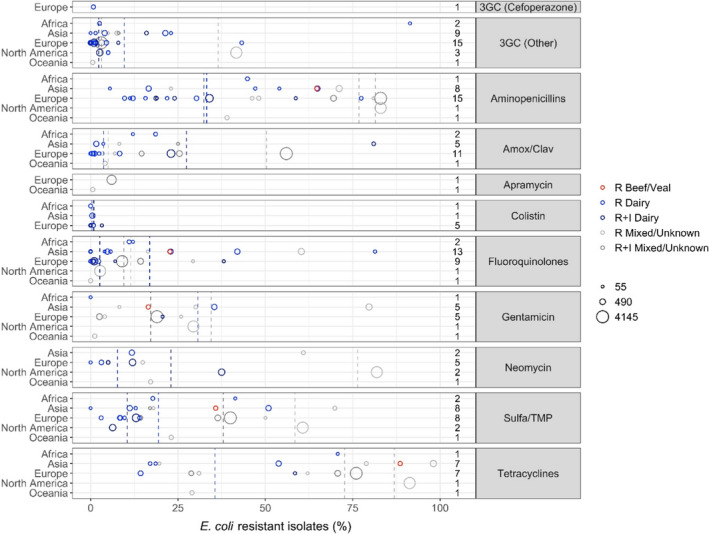
- Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (light blue circle), resistance merged with intermediate in isolates of dairy production origin (dark blue circle), resistance in isolates from beef/veal production (red circles), resistance in isolates of mixed or unknown origin (light grey circle) and resistance merged with intermediate in isolates of mixed or unknown origin (dark grey circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination
Before discussing results for E. coli, it should be noted that data for some of the antibiotics are reported selectively. This concerns gentamicin, apramycin and paromomycin, which are reported for all indications other than mastitis, according to clinical indications. Resistance data for tetracycline are also presented for non‐mastitis isolates. Conversely, cefoperazone is reported only for mastitis isolates, as a mastitis‐specific CBP exists for this drug. It must be highlighted that the route of administration may be different in cases of mastitis (intramammary or parenteral depending on the presentation) and gastrointestinal infections (oral or parenteral).
For 3GCs, there was a notable difference in resistance levels depending on the production type with a weighted mean proportion of 10.9% resistance in dairy isolates and 36.5% in isolates of mixed/unknown origin (Annex D). This is, however, strongly influenced by a large proportion of isolates (n = 3,360) in the latter category originating from calves in the USA where Cummings et al. (2014) found that 41.7% of these isolates were resistant to ceftiofur. One would expect an even higher proportion of resistance when merging the R and I categories, but this was not the case with only 3.1% of isolates with mixed/unknown origin being resistant to 3GCs. This low proportion is heavily influenced by the French monitoring system reporting only 3% of 4120 isolates resistant to ceftiofur (RESAPATH (ANSES), 2020) and could be due to the restriction in its use since 2016. Accordingly, weighted mean proportions sorted by production type should be interpreted critically taking into consideration other factors influencing the results. Specifically for Europe, 14 of 15 studies reported less than 8% of E. coli isolates resistant to 3GCs. The single exception was a study by Elias et al. (2020) who found 43.3% of 102 mastitis isolates in Ukraine to be resistant to ceftiofur. The authors stated that ‘this finding could potentially be explained by the unrestricted use of extended‐spectrum cephalosporins in rural farming of Ukraine, and more specifically by the preferred use of these antimicrobials for treatment of bovine mastitis’. The only included study testing cefoperazone susceptibility in mastitis E. coli isolates reported a resistance proportion of 0.8% among 135 isolates in France (Botrel et al., 2010).
For other beta‐lactams, resistance levels were generally high for aminopenicillins although with much variation between countries, irrespective of continent (Figure 6). Table 3 shows a large difference in susceptibility between isolates from dairy and other production types. This is even clearer when zooming in on the French and German monitoring reports; in France, 83% and 34% of E. coli from calf diarrhoea and mastitis, respectively, were resistant to amoxicillin (RESAPATH (ANSES), 2020). Corresponding figures in Germany (for ampicillin) were 81% and 12%, respectively. It therefore appears that E. coli causing gastrointestinal disorders are much more likely to be resistant to aminopenicillins than mastitis isolates. Although not described here in further detail, these two national reports showed the same trend for other antibiotics, namely amoxicillin–clavulanic acid, sulfonamide–trimethoprim and fluoroquinolones. As expected, mean resistance levels were somewhat lower for amoxicillin–clavulanic acid compared with ampicillin. The highest levels were detected in a Chinese study reporting resistance in 81 of 100 mastitis E. coli isolates (Cheng et al., 2019).
Mean proportions of fluoroquinolone resistance were low (Figure 6), although some rather large continent‐specific variations were observed. For example, the mean resistance proportions among isolates of dairy and unknown/mixed origin were 22% and 45%, respectively, in Asia, whereas corresponding values for Europe were 3% and 10%, respectively (Table 4). In Europe, two studies had a much higher proportion of fluoroquinolone resistance than others, namely Aasmäe et al. (2019) reporting 38.1% of Estonian dairy isolates of various origin non‐susceptible to ciprofloxacin, and GERM‐Vet (2020) reporting 29.3% of German isolates from calf diarrhoea resistant to ciprofloxacin based on (human) CBP.
Colistin‐resistant isolates were not found in four of the seven studies reporting data for this drug in E. coli. The remaining three studies showed resistance percentages between 0.5% and 3.2%, the highest in Estonia (Aasmäe et al., 2019).
For the aminoglycosides gentamicin and neomycin, higher mean resistance percentages were observed among isolates in Asia compared with Europe (Table 4). However, this is based on fewer studies compared to other drug classes. Even fewer studies reported data for apramycin; hence, geographical trends for this drug cannot be derived.
Similar to aminopenicillins, high average levels of resistance were observed for sulfonamide–trimethoprim and – especially – tetracyclines (Figure 6 and Table 3). As for most other drugs, the highest levels were observed in Asia compared with Europe. Specifically for Europe, the highest proportion of tetracycline resistance (79%) was reported by Cengiz and Adiguzel (2020) in calf diarrhoea isolates. A comparatively high proportion (76%, considering R + I) was observed in isolates of similar origin from France (RESAPATH (ANSES), 2020). Here, 40% and 69.9%, respectively, of the same isolates were resistant to sulfonamide–trimethoprim, therefore also among the highest proportions reported in Europe.
3.1.4.2. Results from the national AMR monitoring reports
Information on AMR in cattle clinical E. coli included in the National monitoring programmes originated from either samples from the gastrointestinal tract/faeces collected from young animals or from milk/mastitis samples. The cattle population from which isolates originated will also vary according to the source material for these tests.
ANRESIS ARCH‐Vet (Switzerland): Data on AMR in 54 E. coli isolates from mastitis cases tested with five antimicrobials of interest (ampicillin, cefotaxime, ceftiofur, ciprofloxacin and colistin) were reported in 2019 (these data are already included in Figure 8 and Table 5). Non‐susceptible isolates were only found for ampicillin (19%) and ciprofloxacin (7%).
Table 5.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Escherichia coli for the target antimicrobials in each continent, sorted by production type. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC (Cefoperazone) | Europe | Dairy | 1 | 135 | 0.8 | 0.8 | 0.8 | NA |
| 3GC (Other) | Africa | Dairy | 2 | 176 | 31.8 | 2.5 | 91.4 | 41.9 |
| 3GC (Other) | Asia | Dairy | 6 | 1,035 | 12.2 | 0 | 23 | 8.9 |
| 3GC (Other) | Asia | Mixed/Unknown | 3 | 250 | 7 | 4.9 | 8 | 1.2 |
| 3GC (Other) | Europe | Dairy | 14 | 2,767 | 4.3 | 0 | 43.3 | 10.6 |
| 3GC (Other) | Europe | Mixed/Unknown | 3 | 4,791 | 2.9 | 0.6 | 3.1 | 0.4 |
| 3GC (Other) | North America | Dairy | 2 | 814 | 2.9 | 2.6 | 5 | 0.8 |
| 3GC (Other) | North America | Mixed/Unknown | 1 | 3,360 | 41.7 | 41.7 | 41.7 | NA |
| 3GC (Other) | Oceania | Mixed/Unknown | 1 | 169 | 0.6 | 0.6 | 0.6 | NA |
| Aminopenicillins | Africa | Dairy | 1 | 118 | 44.9 | 44.9 | 44.9 | NA |
| Aminopenicillins | Asia | Beef/Veal | 1 | 176 | 64.8 | 64.8 | 64.8 | NA |
| Aminopenicillins | Asia | Dairy | 5 | 935 | 40.1 | 5.5 | 64.9 | 23.6 |
| Aminopenicillins | Asia | Mixed/Unknown | 2 | 691 | 66.9 | 23 | 71.1 | 13.7 |
| Aminopenicillins | Europe | Dairy | 13 | 2,575 | 31.1 | 9.7 | 77.4 | 15.7 |
| Aminopenicillins | Europe | Mixed/Unknown | 5 | 4,876 | 79.7 | 46.2 | 83 | 8.7 |
| Aminopenicillins | North America | Mixed/Unknown | 1 | 3,360 | 83 | 83 | 83 | NA |
| Aminopenicillins | Oceania | Mixed/Unknown | 1 | 169 | 39 | 39 | 39 | NA |
| Amox/Clav | Africa | Dairy | 2 | 176 | 16.5 | 12.1 | 18.6 | 3.1 |
| Amox/Clav | Asia | Dairy | 3 | 529 | 16.8 | 1.6 | 81 | 31 |
| Amox/Clav | Asia | Mixed/Unknown | 2 | 117 | 16.2 | 8.2 | 25 | 8.4 |
| Amox/Clav | Europe | Dairy | 9 | 2,418 | 13.3 | 0 | 23 | 10.3 |
| Amox/Clav | Europe | Mixed/Unknown | 5 | 5,078 | 49.1 | 3.4 | 56 | 14.8 |
| Amox/Clav | Oceania | Mixed/Unknown | 1 | 169 | 4.1 | 4.1 | 4.1 | NA |
| Apramycin | Europe | Mixed/Unknown | 1 | 2,057 | 6 | 6 | 6 | NA |
| Apramycin | Oceania | Mixed/Unknown | 1 | 169 | 0.6 | 0.6 | 0.6 | NA |
| Colistin | Africa | Dairy | 1 | 118 | 0 | 0 | 0 | NA |
| Colistin | Asia | Dairy | 1 | 374 | 0.5 | 0.5 | 0.5 | NA |
| Colistin | Europe | Dairy | 5 | 414 | 0.7 | 0 | 3.2 | 1.1 |
| Fluoroquinolones | Africa | Dairy | 2 | 176 | 11.4 | 11 | 12.1 | 0.5 |
| Fluoroquinolones | Asia | Beef/Veal | 1 | 176 | 22.7 | 22.7 | 22.7 | NA |
| Fluoroquinolones | Asia | Dairy | 8 | 1,433 | 22 | 0 | 81.4 | 20.8 |
| Fluoroquinolones | Asia | Mixed/Unknown | 4 | 880 | 45.2 | 0 | 60.3 | 24.3 |
| Fluoroquinolones | Europe | Dairy | 9 | 2,020 | 3 | 0 | 38.1 | 6.9 |
| Fluoroquinolones | Europe | Mixed/Unknown | 3 | 4,106 | 9.9 | 9 | 29.3 | 2.9 |
| Fluoroquinolones | North America | Mixed/Unknown | 1 | 3,360 | 2.7 | 2.7 | 2.7 | NA |
| Fluoroquinolones | Oceania | Mixed/Unknown | 1 | 169 | 0 | 0 | 0 | NA |
| Gentamicin | Africa | Dairy | 1 | 58 | 0 | 0 | 0 | NA |
| Gentamicin | Asia | Beef/Veal | 1 | 176 | 16.5 | 16.5 | 16.5 | NA |
| Gentamicin | Asia | Dairy | 1 | 379 | 35.4 | 35.4 | 35.4 | NA |
| Gentamicin | Asia | Mixed/Unknown | 3 | 824 | 66.4 | 8.2 | 79.7 | 24.5 |
| Gentamicin | Europe | Dairy | 1 | 63 | 20.6 | 20.6 | 20.6 | NA |
| Gentamicin | Europe | Mixed/Unknown | 4 | 4,785 | 17 | 2.5 | 25.9 | 5.5 |
| Gentamicin | North America | Mixed/Unknown | 1 | 3,354 | 29.4 | 29.4 | 29.4 | NA |
| Gentamicin | Oceania | Mixed/Unknown | 1 | 169 | 1.2 | 1.2 | 1.2 | NA |
| Neomycin | Asia | Dairy | 1 | 374 | 11.8 | 11.8 | 11.8 | NA |
| Neomycin | Asia | Mixed/Unknown | 1 | 133 | 60.9 | 60.9 | 60.9 | NA |
| Neomycin | Europe | Dairy | 4 | 1,168 | 9 | 0 | 12 | 4.3 |
| Neomycin | Europe | Mixed/Unknown | 1 | 99 | 14.9 | 14.9 | 14.9 | NA |
| Neomycin | North America | Dairy | 1 | 716 | 37.5 | 37.5 | 37.5 | NA |
| Neomycin | North America | Mixed/Unknown | 1 | 3,333 | 81.9 | 81.9 | 81.9 | NA |
| Neomycin | Oceania | Mixed/Unknown | 1 | 169 | 17.2 | 17.2 | 17.2 | NA |
| Sulfa/TMP | Africa | Dairy | 2 | 176 | 25.6 | 17.8 | 41.4 | 11.1 |
| Sulfa/TMP | Asia | Beef/Veal | 1 | 176 | 35.8 | 35.8 | 35.8 | NA |
| Sulfa/TMP | Asia | Dairy | 4 | 878 | 27.8 | 0 | 50.9 | 20.4 |
| Sulfa/TMP | Asia | Mixed/Unknown | 3 | 250 | 45.4 | 17 | 69.9 | 26.2 |
| Sulfa/TMP | Europe | Dairy | 7 | 2,050 | 12.6 | 3 | 40 | 7 |
| Sulfa/TMP | Europe | Mixed/Unknown | 4 | 4,983 | 38.4 | 14.2 | 50 | 6 |
| Sulfa/TMP | North America | Dairy | 1 | 716 | 6.3 | 6.3 | 6.3 | NA |
| Sulfa/TMP | North America | Mixed/Unknown | 1 | 3,343 | 60.7 | 60.7 | 60.7 | NA |
| Sulfa/TMP | Oceania | Mixed/Unknown | 1 | 169 | 23.1 | 23.1 | 23.1 | NA |
| Tetracyclines | Africa | Dairy | 1 | 58 | 70.7 | 70.7 | 70.7 | NA |
| Tetracyclines | Asia | Beef/Veal | 1 | 176 | 88.6 | 88.6 | 88.6 | NA |
| Tetracyclines | Asia | Dairy | 3 | 543 | 42.9 | 17 | 53.8 | 16.6 |
| Tetracyclines | Asia | Mixed/Unknown | 3 | 824 | 89.2 | 19.7 | 98.1 | 20.9 |
| Tetracyclines | Europe | Dairy | 2 | 343 | 22.4 | 14.3 | 58.5 | 17.1 |
| Tetracyclines | Europe | Mixed/Unknown | 5 | 4,867 | 71.8 | 28.8 | 76 | 12.3 |
| Tetracyclines | North America | Mixed/Unknown | 1 | 3,336 | 91.3 | 91.3 | 91.3 | NA |
| Tetracyclines | Oceania | Mixed/Unknown | 1 | 169 | 29 | 29 | 29 | NA |
All‐Islands Animal Disease Surveillance Report (Ireland): Detailed data on AMR obtained in clinical E. coli is only provided for 268 isolates of unknown origin in the 2018 report, providing results for sulfonamide–trimethoprim (14.2% non‐susceptible), amoxicillin–clavulanic acid (14.6% non‐susceptible) and tetracycline (28.8% non‐susceptible isolates) (these data are already included in Figure 8 and Table 5).
RESAPATH (France): AMR data from cattle clinical isolates are included in the annual reports from mastitis cases in adult cows and from digestive pathologies in young animals.
For cases from mastitis, AMR results from 504 to 1219 isolates tested with six antimicrobials annually are available for the period 2014–2018 (Figure 9); additionally, ceftazidime was also used on 39 isolates in 2014, yielding a 5% of non‐susceptible isolates. Proportions of non‐susceptible isolates were below 35% for all antimicrobials, with values above 8% recorded only for amoxicillin, amoxicillin + clavulanic acid and sulfonamides–trimethoprim, with higher values observed in the last 3 years, while resistance levels to enrofloxacin, ceftiofur and gentamicin were consistently below 4%. A decreasing trend can be seen for the resistance levels to critically important antimicrobials (CIA, i.e. enrofloxacin and ceftiofur) and an increasing trend for other molecules which could reflect a shift in antimicrobial use practices (EMA, 2020; RESAPATH (ANSES), 2020).
Figure 9.

Proportions of non‐susceptible clinical Escherichia coli isolates from cattle mastitis for six antimicrobials of interest from 2014 to 2018 reported by the RESAPATH monitoring programme
In the isolates from digestive cases in young animals (1,136–4,222 tested isolates each year during the 2014–2018 period), resistance levels were much higher, with values above 50% for amoxicillin, amoxicillin–clavulanic acid and tetracycline, and between 35 and 40% for sulfa/TMP (Figure 10). Resistance levels to enrofloxacin, apramycin and gentamicin ranged between 27% and 6%, with apparent decreasing trends for enrofloxacin and apramycin. Ceftiofur‐resistance decreased from 8% to 3% (Figure 10).
Figure 10.
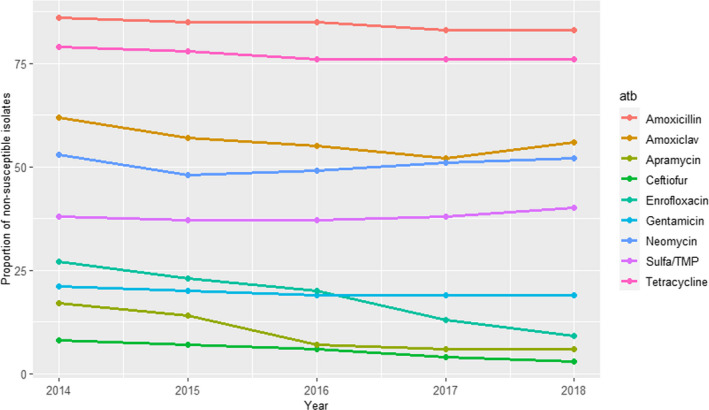
Proportion (%) of non‐susceptible clinical Escherichia coli isolates from cattle digestive cases for nine antimicrobials of interest reported by the RESAPATH monitoring programme
SWEDRES‐Svarm (Sweden): Data on AMR on isolates from two origins are included in the reports: isolates coming from faeces/gastrointestinal tract of young animals (a few weeks old) and those retrieved from clinical submissions of milk samples (i.e. probably coming from cows with clinical mastitis).
Between 74 and 113 isolates from mastitis were tested every year between 2014 and 2018 using four to five antimicrobials (colistin and cefotaxime were not used in isolates from 2014). Resistance levels were, in general, lower than those observed in isolates from faeces/gastrointestinal tract samples and the highest levels of resistance (9–27%) were observed for ampicillin and sulfonamide–trimethoprim while values ≤ 6% were recorded for all other antimicrobials and years (Figure 11).
Figure 11.
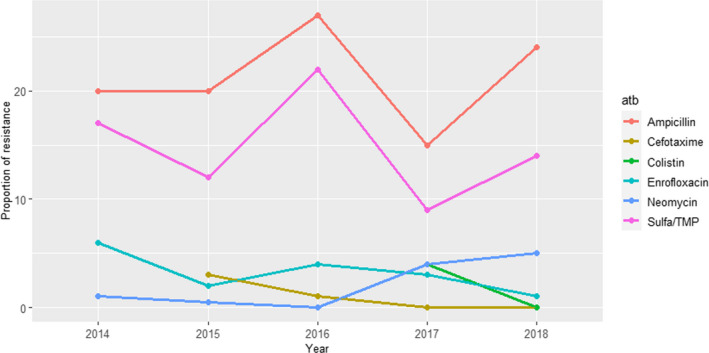
Proportion (%) of clinical Escherichia coli isolates retrieved from cattle mastitis cases resistant to six antimicrobials of interest reported by the SWEDRES‐Svarm monitoring programme
For the isolates from young animals, between 29 and 117 isolates from digestive samples were tested annually for resistance to seven or eight antimicrobials annually over the 2012–2018 period (ceftiofur was only used in isolates collected in 2012–2014 – 2% resistant isolates – and colistin and cefotaxime were not tested in isolates from those years). Over 30% of the isolates tested over the whole period were resistant to ampicillin and tetracyclines, while resistance to neomycin and sulfonamide–trimethoprim remained mostly between 10% and 30% and resistance levels < 10% were found for the remaining antimicrobials and periods (except enrofloxacin in 2012–2014) (Figure 12).
Figure 12.

Proportion (%) of clinical Escherichia coli isolates retrieved from cattle digestive samples resistant to eight antimicrobials of interest reported by the SWEDRES‐Svarm monitoring programme
DANMAP (Denmark): Resistance to 10 antimicrobials was determined in 23 and 17 isolates retrieved in 2018 and 2019, respectively, from mastitis cases. Between 4% and 6% of the isolates were resistant to amoxicillin–clavulanic acid, ampicillin, colistin or tetracycline in at least one of the sampling points (resistant isolates were only found in both years for ampicillin) (Figure 13), while all were susceptible to apramycin, cefotaxime, ceftiofur, ciprofloxacin, gentamicin and neomycin (data not shown).
Figure 13.
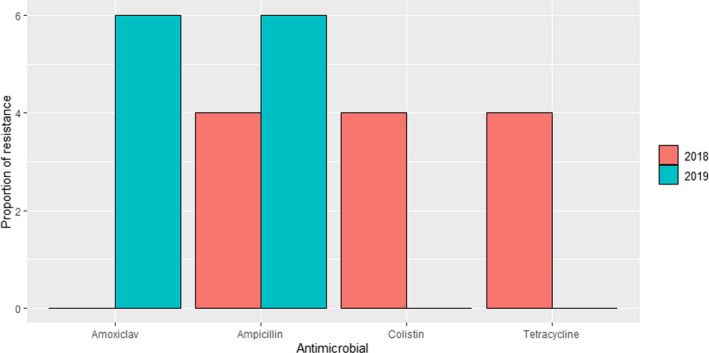
Proportion (%) of clinical Escherichia coli isolates retrieved from cattle mastitis samples resistant to four antimicrobials of interest reported by the DANMAP monitoring programme
UK‐VARSS (United Kingdom): Data on AMR from E. coli isolates retrieved from mastitis cases in England and Wales (between 79 and 110 cases annually during the 2015–2019 period) and Scotland (157 isolates in 2018 and 134 in 2019) are included in the last reports published. Isolates originating from England and Wales were more resistant to ampicillin (20–40% resistance) while resistance levels for the rest of antimicrobials tested remained below 10% after 2017 (Figure 14).
Figure 14.

Proportion (%) of clinical Escherichia coli isolates retrieved from cattle mastitis samples in England and Wales resistant to six antimicrobials of interest reported by the UK‐VARSS monitoring programme
For isolates from Scotland retrieved in 2018 and 2019, a similar pattern was observed (ampicillin > sulfonamide–trimethoprim = amoxicillin–clavulanic acid > remaining antimicrobials), although resistance to ampicillin remained at lower levels (18–24%) (Figure 15).
Figure 15.
Proportion (%) of clinical Escherichia coli isolates retrieved from cattle mastitis samples in Scotland resistant to six antimicrobials of interest reported by the UK‐VARSS monitoring programme
GERM‐VET (Germany): Sampling involved E. coli isolates from gastrointestinal disease in calves/young cattle for all years (2014–2018), for gastrointestinal disease in adult cattle (years 2015–2018) and for mastitis in adult cattle (2014, 2016, 2018). Antimicrobials tested and classified into susceptible and resistant (intermediate resistant and resistant) were ampicillin, amoxicillin/clavulanic acid, ciprofloxacin (2016–2018), doxycycline (only in 2018), gentamicin, tetracycline and sulfamethoxazole/trimethoprim. Isolates from the gastrointestinal tract (GIT) of calves/young cattle were 58–284/year, for GIT disease 34 isolates were analysed in 2015, 108 in 2016, 25 in 2017 and 39 in 2018 in adult cattle, and for the indication mastitis, 241 isolates were analysed in 2014, 275 in 2016 and 224 in 2018. Results are seen in Figure 16 for gastrointestinal disease in adults and calves and young cattle, and for adult cattle and mastitis in Figure 17.
Figure 16.

Proportion (%) of clinical Escherichia coli isolates from gastrointestinal disease in adult (top) and calves and young cattle (bottom) non‐susceptible to five antimicrobials of interest reported by the GERM‐Vet monitoring programme
Figure 17.

Proportion (%) of clinical Escherichia coli isolates from mastitis in cattle resistant to three antimicrobials of interest reported by the GERM‐Vet monitoring programme
3.1.5. Pasteurella multocida, Mannheimia haemolytica and Histophilus somni
3.1.5.1. Results of the ELR by bacterium
Pasteurella multocida, Mannheimia haemolytica and Histophilus somni are commensals of the bovine respiratory tract and among the several infectious agents involved in the bovine respiratory disease (BRD) complex. Calves and young bulls are particularly susceptible to BRD, and the disease is predisposed by factors affecting immunity like stable air pollutants, failure of passive transfer, nutritional deficiencies and several stressors related to management (e.g. transport, commingling, feed and water deprival, dehorning). Beside clinical outbreaks, a substantial number of calves also suffers from subclinical pneumonia (van Leenen et al., 2020, 2021). In addition, sporadic cases in adult animals have also been described (Dorso et al., 2021).
In total, 23, 20 and 12 studies with ≥ 10 P. multocida, M. haemolytica and H. somni isolates, respectively, were included. Each included study had results for one or more of the following relevant antibiotics: ampicillin/amoxicillin, enrofloxacin/ciprofloxacin/danofloxacin, erythromycin, florfenicol, gamithromycin, gentamicin, 3GC, penicillin, tetracyclines, tildipirosin, tilmicosin, tulathromycin and tylosin. Geographically, studies were distributed as follows: for P. multocida, Africa (0), Asia (4), Europe (11), Oceania (0), North America (8) and South America (0). For M. haemolytica, Africa (0), Asia (1), Europe (12), Oceania (0), North America (7) and South America (0). For H. somni, Africa (0), Asia (0), Europe (3), Oceania (1), North America (8) and South America (0).
The distribution of P. multocida, M. haemolytica, and H. somni isolates per site of infection is shown in Figure 18. Most isolates originated from respiratory infections. Of note, type of sampling was often defined very generally (e.g. ‘samples from respiratory cases’) and it was not always possible to differentiate isolates retrieved from lower or upper respiratory tract, or even from live vs. dead (i.e. necropsied) animals.
Figure 18.

Distribution of Pasteurella multocida, Mannheimia haemolytica and Histophilus somni isolates per site of infection and type of production
Figure 19 shows for each continent the proportion of resistance reported in individual studies with at least 10 P. multocida, M. haemolytica and H. somni isolates. Information on proportion of resistance sorted by country is in Annex D.
Figure 19.

- Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (blue circle), resistance merged with intermediate in isolates of dairy production origin (dark blue circle), resistance in isolates from beef/veal production (red circle), resistance merged with intermediate in isolates from beef/veal production (brown circle), resistance in isolates of mixed or unknown origin (light grey circle) and resistance merged with intermediate in isolates of mixed or unknown origin (dark grey circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % R or RI with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
For beta‐lactams, the vast majority of studies reported ≤ 2% ceftiofur resistance for the three species. One exception, an American study by Lamm et al. (2012), reported the highest levels of ceftiofur resistance for all three species (7.1–11.8%). However, this study on isolates obtained post‐mortem from bronchopneumonia in feedlot cattle, included only 11–17 isolates for each species, hence results must be interpreted with caution. Another exception was a Canadian study reporting 4.6% of H. somni isolates from respiratory infections in cattle resistant to ceftiofur. Average proportions of resistance to aminopenicillins were slightly higher than for ceftiofur (Tables 6, 77–8). The highest levels of resistance were observed in Germany with 39% and 63.5% of the M. haemolytica and P. multocida isolates, respectively, being resistant to ampicillin (GERM‐Vet, 2020). Interestingly, these proportions would have been 97.5% and 100% if resistance had been merged with the intermediate category. Such a high proportion of intermediate isolates suggests that data from other studies should be compared taking this into account, i.e. by not comparing %R from one study with %RI from another. Levels of penicillin resistance were generally in the range of that seen for aminopenicillins. Interestingly, the GERM‐Vet programme reported much less resistance to this drug compared to ampicillin, e.g. only 2% of P. multocida isolates were penicillin‐resistant (GERM‐Vet, 2020). By far, the highest proportion of penicillin resistance (65.2%) was reported among 46 H. somni respiratory isolates from heifers and beef steers in a Canadian study (Timsit et al., 2017).
Table 6.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Pasteurella multocida for the target antimicrobials in each continent, sorted by production type. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC | Asia | Mixed/Unknown | 2 | 379 | 0 | 0 | 0 | 0 |
| 3GC | Europe | Mixed/Unknown | 7 | 1,060 | 0.1 | 0 | 1.9 | 0.4 |
| 3GC | North America | Beef/Veal | 3 | 459 | 0.4 | 0 | 7.1 | 1.2 |
| 3GC | North America | Dairy | 1 | 1,146 | 0 | 0 | 0 | NA |
| 3GC | North America | Mixed/Unknown | 3 | 408 | 0.8 | 0 | 1.3 | 0.6 |
| Aminopenicillins | Asia | Mixed/Unknown | 2 | 379 | 7.1 | 5.9 | 9.2 | 1.6 |
| Aminopenicillins | Europe | Mixed/Unknown | 6 | 768 | 15.3 | 1 | 63.5 | 23.8 |
| Aminopenicillins | North America | Beef/Veal | 1 | 117 | 1.8 | 1.8 | 1.8 | NA |
| Aminopenicillins | North America | Dairy | 1 | 1,146 | 1 | 1 | 1 | NA |
| Erythromycin | North America | Mixed/Unknown | 1 | 238 | 58 | 58 | 58 | NA |
| Florfenicol | Asia | Mixed/Unknown | 2 | 379 | 0.5 | 0 | 0.8 | 0.4 |
| Florfenicol | Europe | Beef/Veal | 1 | 107 | 0 | 0 | 0 | NA |
| Florfenicol | Europe | Mixed/Unknown | 8 | 1,268 | 1.9 | 0 | 12.2 | 4.2 |
| Florfenicol | North America | Beef/Veal | 3 | 459 | 9.4 | 1.7 | 14.3 | 4.5 |
| Florfenicol | North America | Dairy | 1 | 1,145 | 4 | 4 | 4 | NA |
| Florfenicol | North America | Mixed/Unknown | 3 | 408 | 4.4 | 0 | 12.7 | 5.5 |
| Fluoroquinolones | Asia | Mixed/Unknown | 3 | 402 | 0 | 0 | 0 | 0 |
| Fluoroquinolones | Europe | Mixed/Unknown | 7 | 1,090 | 2.4 | 0 | 9 | 3.6 |
| Fluoroquinolones | North America | Beef/Veal | 3 | 459 | 6.3 | 0 | 8.8 | 4 |
| Fluoroquinolones | North America | Dairy | 1 | 1,145 | 1 | 1 | 1 | NA |
| Fluoroquinolones | North America | Mixed/Unknown | 2 | 170 | 2.4 | 0 | 6.7 | 3.2 |
| Gamithromycin | Europe | Mixed/Unknown | 1 | 134 | 1.5 | 1.5 | 1.5 | NA |
| Gamithromycin | North America | Dairy | 1 | 471 | 13 | 13 | 13 | NA |
| Gentamicin | Asia | Mixed/Unknown | 1 | 23 | 0 | 0 | 0 | NA |
| Gentamicin | Europe | Mixed/Unknown | 1 | 210 | 3 | 3 | 3 | NA |
| Gentamicin | North America | Beef/Veal | 1 | 117 | 8.5 | 8.5 | 8.5 | NA |
| Gentamicin | North America | Dairy | 1 | 1,145 | 3 | 3 | 3 | NA |
| Penicillin | Asia | Mixed/Unknown | 1 | 141 | 30.5 | 30.5 | 30.5 | NA |
| Penicillin | Europe | Mixed/Unknown | 3 | 414 | 1.7 | 0 | 5 | 1.8 |
| Penicillin | North America | Beef/Veal | 2 | 445 | 4.7 | 1.7 | 5.8 | 1.8 |
| Penicillin | North America | Dairy | 1 | 1,146 | 3 | 3 | 3 | NA |
| Penicillin | North America | Mixed/Unknown | 2 | 348 | 1.4 | 0 | 2.1 | 1 |
| Tetracyclines | Asia | Mixed/Unknown | 2 | 379 | 21.4 | 19.9 | 22.3 | 1.2 |
| Tetracyclines | Europe | Mixed/Unknown | 9 | 1,235 | 20.8 | 0 | 66.2 | 20.1 |
| Tetracyclines | North America | Beef/Veal | 3 | 459 | 46.4 | 42.7 | 57.1 | 5.9 |
| Tetracyclines | North America | Dairy | 1 | 1,145 | 36 | 36 | 36 | NA |
| Tetracyclines | North America | Mixed/Unknown | 4 | 582 | 30.3 | 5.5 | 80 | 28 |
| Tildipirosin | North America | Dairy | 1 | 516 | 19 | 19 | 19 | NA |
| Tilmicosin | North America | Beef/Veal | 2 | 131 | 42 | 41.9 | 42.9 | 0.3 |
| Tilmicosin | North America | Dairy | 1 | 1,144 | 23 | 23 | 23 | NA |
| Tilmicosin | North America | Mixed/Unknown | 3 | 473 | 17.4 | 12 | 43.3 | 10 |
| Tulathromycin | Europe | Mixed/Unknown | 3 | 469 | 5.3 | 0.5 | 14.1 | 6 |
| Tulathromycin | North America | Beef/Veal | 2 | 445 | 12.9 | 6.8 | 29.9 | 10.2 |
| Tulathromycin | North America | Dairy | 1 | 1,145 | 9 | 9 | 9 | NA |
| Tulathromycin | North America | Mixed/Unknown | 3 | 344 | 32.6 | 5.8 | 80.9 | 33.7 |
| Tylosin | North America | Beef/Veal | 1 | 117 | 99.1 | 99.1 | 99.1 | NA |
| Tylosin | North America | Dairy | 1 | 1,145 | 88 | 88 | 88 | NA |
Table 7.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Mannheimia haemolytica for the target antimicrobials in each continent, sorted by production type. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC | Asia | Mixed/Unknown | 1 | 310 | 0 | 0 | 0 | NA |
| 3GC | Europe | Mixed/Unknown | 8 | 763 | 0.2 | 0 | 1 | 0.4 |
| 3GC | North America | Beef/Veal | 3 | 554 | 0.7 | 0 | 11.8 | 2 |
| 3GC | North America | Dairy | 1 | 753 | 0.7 | 0.7 | 0.7 | NA |
| 3GC | North America | Mixed/Unknown | 3 | 352 | 0.6 | 0 | 1.1 | 0.5 |
| Aminopenicillins | Asia | Mixed/Unknown | 1 | 310 | 20.3 | 20.3 | 20.3 | NA |
| Aminopenicillins | Europe | Mixed/Unknown | 5 | 478 | 12.3 | 4.3 | 39 | 12.4 |
| Aminopenicillins | Mixed continents | Dairy | 1 | 54 | 22.2 | 22.2 | 22.2 | NA |
| Aminopenicillins | North America | Beef/Veal | 1 | 233 | 5.1 | 5.1 | 5.1 | NA |
| Aminopenicillins | North America | Dairy | 1 | 753 | 14 | 14 | 14 | NA |
| Erythromycin | North America | Mixed/Unknown | 1 | 187 | 52.9 | 52.9 | 52.9 | NA |
| Florfenicol | Asia | Mixed/Unknown | 1 | 310 | 0.3 | 0.3 | 0.3 | NA |
| Florfenicol | Europe | Beef/Veal | 1 | 44 | 0 | 0 | 0 | NA |
| Florfenicol | Europe | Mixed/Unknown | 8 | 888 | 0.8 | 0 | 3.7 | 1.2 |
| Florfenicol | North America | Beef/Veal | 3 | 554 | 8 | 4.3 | 47.1 | 7.3 |
| Florfenicol | North America | Dairy | 1 | 753 | 10 | 10 | 10 | NA |
| Florfenicol | North America | Mixed/Unknown | 3 | 352 | 10 | 0 | 34.7 | 15.7 |
| Fluoroquinolones | Asia | Mixed/Unknown | 1 | 310 | 18.7 | 18.7 | 18.7 | NA |
| Fluoroquinolones | Europe | Mixed/Unknown | 7 | 739 | 1.4 | 0 | 5 | 1.9 |
| Fluoroquinolones | Mixed continents | Dairy | 1 | 54 | 7.4 | 7.4 | 7.4 | NA |
| Fluoroquinolones | North America | Beef/Veal | 3 | 554 | 12.3 | 0 | 20.1 | 8.6 |
| Fluoroquinolones | North America | Dairy | 1 | 753 | 11 | 11 | 11 | NA |
| Fluoroquinolones | North America | Mixed/Unknown | 2 | 165 | 34.5 | 0 | 56.4 | 27.6 |
| Gamithromycin | Europe | Mixed/Unknown | 1 | 149 | 2.7 | 2.7 | 2.7 | NA |
| Gamithromycin | North America | Dairy | 1 | 291 | 13 | 13 | 13 | NA |
| Gentamicin | Europe | Mixed/Unknown | 1 | 117 | 14 | 14 | 14 | NA |
| Gentamicin | Mixed continents | Dairy | 1 | 54 | 16.7 | 16.7 | 16.7 | NA |
| Gentamicin | North America | Beef/Veal | 1 | 233 | 3.4 | 3.4 | 3.4 | NA |
| Gentamicin | North America | Dairy | 1 | 753 | 9 | 9 | 9 | NA |
| Penicillin | Europe | Mixed/Unknown | 3 | 229 | 21 | 12.5 | 24.4 | 4.5 |
| Penicillin | Mixed continents | Dairy | 1 | 54 | 33.3 | 33.3 | 33.3 | NA |
| Penicillin | North America | Beef/Veal | 2 | 537 | 25.3 | 7.2 | 39.1 | 15.8 |
| Penicillin | North America | Dairy | 1 | 753 | 19 | 19 | 19 | NA |
| Penicillin | North America | Mixed/Unknown | 2 | 251 | 4.8 | 1.6 | 5.9 | 1.9 |
| Tetracyclines | Asia | Mixed/Unknown | 1 | 310 | 24.8 | 24.8 | 24.8 | NA |
| Tetracyclines | Europe | Mixed/Unknown | 8 | 829 | 17.2 | 4.2 | 50 | 11.7 |
| Tetracyclines | Mixed continents | Dairy | 1 | 54 | 16.7 | 16.7 | 16.7 | NA |
| Tetracyclines | North America | Beef/Veal | 3 | 554 | 52.7 | 51.3 | 64.7 | 2.4 |
| Tetracyclines | North America | Dairy | 1 | 753 | 30 | 30 | 30 | NA |
| Tetracyclines | North America | Mixed/Unknown | 4 | 615 | 35.9 | 9.1 | 78.1 | 25.2 |
| Tildipirosin | North America | Dairy | 1 | 320 | 13 | 13 | 13 | NA |
| Tilmicosin | Europe | Mixed/Unknown | 4 | 467 | 5.3 | 1.2 | 16.3 | 5.8 |
| Tilmicosin | North America | Beef/Veal | 3 | 554 | 43.2 | 40.5 | 76.5 | 6.2 |
| Tilmicosin | North America | Dairy | 1 | 753 | 16 | 16 | 16 | NA |
| Tilmicosin | North America | Mixed/Unknown | 4 | 615 | 37.8 | 24.7 | 84.4 | 20.3 |
| Tulathromycin | Europe | Mixed/Unknown | 4 | 378 | 5.3 | 0 | 13.3 | 4.8 |
| Tulathromycin | Mixed continents | Dairy | 1 | 54 | 1.9 | 1.9 | 1.9 | NA |
| Tulathromycin | North America | Beef/Veal | 2 | 537 | 23.3 | 12.2 | 37.8 | 12.7 |
| Tulathromycin | North America | Dairy | 1 | 753 | 11 | 11 | 11 | NA |
| Tulathromycin | North America | Mixed/Unknown | 3 | 423 | 40.4 | 26.6 | 76.6 | 19 |
| Tylosin | Mixed continents | Dairy | 1 | 54 | 14.8 | 14.8 | 14.8 | NA |
| Tylosin | North America | Beef/Veal | 1 | 233 | 99.1 | 99.1 | 99.1 | NA |
| Tylosin | North America | Dairy | 1 | 753 | 99 | 99 | 99 | NA |
Table 8.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Histophilus somni for the target antimicrobials in each continent, sorted by production type. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC | Europe | Mixed/Unknown | 2 | 96 | 0 | 0 | 0 | 0 |
| 3GC | North America | Beef/Veal | 3 | 260 | 0.4 | 0 | 9.1 | 1.8 |
| 3GC | North America | Dairy | 1 | 458 | 2 | 2 | 2 | NA |
| 3GC | North America | Mixed/Unknown | 3 | 183 | 2.2 | 0 | 4.6 | 2.3 |
| 3GC | Oceania | Mixed/Unknown | 1 | 53 | 0 | 0 | 0 | NA |
| Aminopenicillins | North America | Beef/Veal | 1 | 75 | 11.9 | 11.9 | 11.9 | NA |
| Aminopenicillins | North America | Dairy | 1 | 459 | 2 | 2 | 2 | NA |
| Erythromycin | North America | Mixed/Unknown | 1 | 87 | 10.9 | 10.9 | 10.9 | NA |
| Florfenicol | Europe | Beef/Veal | 1 | 31 | 0 | 0 | 0 | NA |
| Florfenicol | Europe | Mixed/Unknown | 2 | 96 | 0 | 0 | 0 | 0 |
| Florfenicol | North America | Beef/Veal | 3 | 261 | 5.4 | 0 | 7.5 | 3 |
| Florfenicol | North America | Dairy | 1 | 459 | 3 | 3 | 3 | NA |
| Florfenicol | North America | Mixed/Unknown | 3 | 183 | 0 | 0 | 0 | 0 |
| Florfenicol | Oceania | Mixed/Unknown | 1 | 53 | 0 | 0 | 0 | NA |
| Fluoroquinolones | Europe | Mixed/Unknown | 2 | 96 | 0 | 0 | 0 | 0 |
| Fluoroquinolones | North America | Beef/Veal | 3 | 261 | 10.7 | 4 | 13.8 | 4.4 |
| Fluoroquinolones | North America | Dairy | 1 | 458 | 3 | 3 | 3 | NA |
| Fluoroquinolones | North America | Mixed/Unknown | 2 | 96 | 4.2 | 0 | 8 | 4 |
| Fluoroquinolones | Oceania | Mixed/Unknown | 1 | 53 | 0 | 0 | 0 | NA |
| Gamithromycin | Europe | Mixed/Unknown | 1 | 66 | 0 | 0 | 0 | NA |
| Gamithromycin | North America | Dairy | 1 | 187 | 6 | 6 | 6 | NA |
| Gentamicin | North America | Beef/Veal | 1 | 75 | 32 | 32 | 32 | NA |
| Gentamicin | North America | Dairy | 1 | 459 | 22 | 22 | 22 | NA |
| Penicillin | Europe | Mixed/Unknown | 1 | 30 | 0 | 0 | 0 | NA |
| Penicillin | North America | Beef/Veal | 2 | 249 | 7.6 | 5.2 | 13.3 | 3.7 |
| Penicillin | North America | Dairy | 1 | 464 | 13 | 13 | 13 | NA |
| Penicillin | North America | Mixed/Unknown | 2 | 133 | 24.8 | 3.4 | 65.2 | 29.5 |
| Tetracyclines | Europe | Mixed/Unknown | 2 | 96 | 3.1 | 3 | 3.3 | 0.1 |
| Tetracyclines | North America | Beef/Veal | 3 | 260 | 51.6 | 9.1 | 54.7 | 9 |
| Tetracyclines | North America | Dairy | 1 | 459 | 27 | 27 | 27 | NA |
| Tetracyclines | North America | Mixed/Unknown | 4 | 305 | 42.9 | 14.9 | 73.9 | 20.5 |
| Tetracyclines | Oceania | Mixed/Unknown | 1 | 53 | 1.9 | 1.9 | 1.9 | NA |
| Tildipirosin | North America | Dairy | 1 | 205 | 5 | 5 | 5 | NA |
| Tilmicosin | North America | Beef/Veal | 2 | 87 | 16.1 | 0 | 18.7 | 6.5 |
| Tilmicosin | North America | Dairy | 1 | 459 | 10 | 10 | 10 | NA |
| Tilmicosin | North America | Mixed/Unknown | 3 | 259 | 22.5 | 18 | 28 | 4.3 |
| Tilmicosin | Oceania | Mixed/Unknown | 1 | 53 | 0 | 0 | 0 | NA |
| Tulathromycin | Europe | Mixed/Unknown | 2 | 96 | 0 | 0 | 0 | 0 |
| Tulathromycin | North America | Beef/Veal | 2 | 249 | 19.7 | 19 | 21.3 | 1.1 |
| Tulathromycin | North America | Dairy | 1 | 458 | 10 | 10 | 10 | NA |
| Tulathromycin | North America | Mixed/Unknown | 3 | 215 | 18.2 | 2.2 | 27.1 | 8.7 |
| Tulathromycin | Oceania | Mixed/Unknown | 1 | 43 | 0 | 0 | 0 | NA |
| Tylosin | North America | Beef/Veal | 1 | 75 | 34.6 | 34.6 | 34.6 | NA |
| Tylosin | North America | Dairy | 1 | 464 | 10 | 10 | 10 | NA |
Low mean proportions of florfenicol resistance were reported for all three species. Two American studies stood out with fairly high levels of resistance, most notably Coetzee et al. (2019) with 34.7% of 101 M. haemolytica isolates being florfenicol‐resistant. Similarly, Lamm et al. (2012) found 43.3% resistance in the same species, although based on a collection of only 17 isolates.
Figure 19 and Tables 6, 77–8 show data for six different macrolides, namely erythromycin, gamithromycin, tilmicosin, tildipirosin, tulathromycin and tylosin. For H. somni, all three non‐American studies with data for macrolides showed 0% resistance to all agents within this drug class (Goldspink et al., 2015; El Garch et al., 2016; FINRES‐Vet, 2018). Somewhat higher levels of resistance (5–34.6%) were detected in the seven studies from North America. A similar clear tendency was observed for M. haemolytica and P. multocida with the highest macrolide resistance levels being reported by North American studies. For P. multocida, a (for Europe) relatively high proportion of tulathromycin resistance (14.1%) was observed among 149 isolates in Germany (GERM‐Vet, 2020).
For fluoroquinolones (enrofloxacin), resistance was close to the low levels observed for florfenicol in all three species (always ≤ 20%). It is worth noting that the few studies reporting %RI may not be comparable with studies reporting %R. This is illustrated by, e.g. Kong et al. (2014) who found none of Chinese P. multocida isolates resistant to enrofloxacin, but 60.9% of isolates were intermediate to this drug. Accordingly, the highest proportion of enrofloxacin resistance reported in Europe (9%, RESAPATH (ANSES) (2020)) may be `false high’ compared with other studies, as it represents merged R and I data.
The overall highest mean resistance levels in all three species were observed for tetracycline. As for most other drugs, the mean proportions of resistance were higher for isolates of North American origin when compared to European isolates (Figure 19 and Tables 6, 77–8). Despite this trend, two European studies reported a high proportion of tetracycline resistance in P. multocida: The British surveillance programme reported 66.2% tetracycline resistance among 74 P. multocida isolates from the UK (UK‐VARSS, 2019), whereas Van Driessche et al. (2018) found 46% of 100 P. multocida isolates in Belgium to be tetracycline resistant.
Very few studies tested susceptibility to gentamicin in these bacterial species (which could be due to the long withdrawal period of this drug for beef cattle, preventing their use in animals close to the slaughter age); therefore, continent‐associated trends are not easy to deduce for this agent. It should be noted that two studies found a high proportion of isolates (27.8% and 82.6%) in the intermediate category (Wang et al., 2017; Nefedchenko et al., 2019); hence, %R and %RI data should be compared with caution as for several other drugs.
3.1.5.2. Results from the national AMR monitoring reports
Information on AMR in clinical isolates belonging to one or more of the three species (P. multocida, M. haemolytica and/or H. somni) was included in five National monitoring programmes, typically with very little information on their origin (other than stating that they were recovered from respiratory samples).
All‐Islands Animal Disease Surveillance Report (Ireland): Detailed data on AMR obtained in clinical P. multocida and M. haemolytica are included in the 2018 report (181 and 150 isolates, respectively). Isolates were tested using ampicillin, florfenicol and tetracycline, and higher levels of non‐susceptible isolates in the first two antimicrobials were found in P. multocida (ampicillin: 6.6%; florfenicol: 12.2%) compared with M. haemolytica (4.7% and 0.7%), while the opposite was true for tetracycline (4.4% in P. multocida vs. 10% in M. haemolytica) (these data are already included in Figure 19 and Tables 6 and 7).
RESAPATH (France): Isolates from two species (P. multocida and M. haemolytica) are routinely monitored, although the number of isolates tested on average each year and the antimicrobials used slightly vary. For P. multocida, between 31 and 237 isolates were tested to eight antimicrobials of interest for this opinion (except in 2017 when seven were used). Proportions of non‐susceptibility were higher (mostly ≥ 25%) for doxycycline and tetracycline, while they remained ≤ 11% for the remaining antimicrobials (< 5% for amoxicillin, ceftiofur and florfenicol) (Figure 20).
Figure 20.
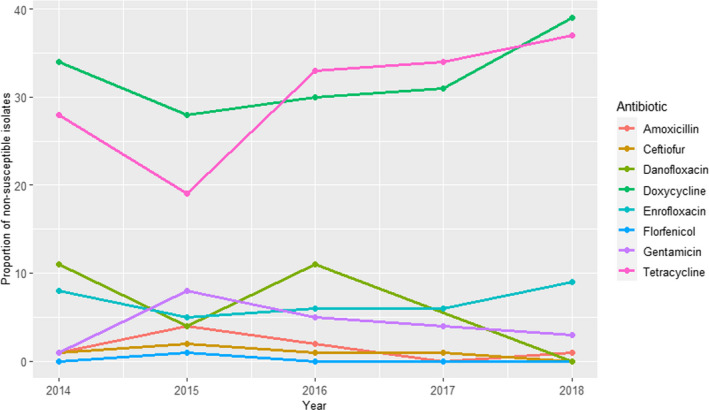
Proportion (%) of non‐susceptible clinical Pasteurella multocida isolates from cattle with respiratory pathology for eight antimicrobials of interest reported by the RESAPATH monitoring programme
For M. haemolytica, between 45 and 178 isolates from respiratory pathologies in young animals were tested for susceptibility to seven antimicrobials every year during the 2014–2018 period (additionally, susceptibility data for danofloxacin were available for 2014–2016, with 3–9% of non‐susceptible isolates). The proportion of non‐susceptible isolates was higher for tetracycline and doxycycline (from 15% to 50%, depending on the year), followed by gentamicin, amoxicillin and enrofloxacin (values between 4% and 18%), while the proportion of non‐susceptible isolates to florfenicol and ceftiofur were ≤ 2%) (Figure 21).
Figure 21.

Proportion (%) of non‐susceptible clinical Mannheimia haemolytica isolates from cattle with respiratory pathology for seven antimicrobials of interest reported by the RESAPATH monitoring programme
FINRES (Finland): AMR data from the three pathogens are routinely included in the reports published every year. For P. multocida, between 135 and 267 isolates were tested annually using six to eight antimicrobials of interest for this opinion in 2015–2019 (ampicillin was only used in 2015 and danofloxacin in 2015–2017). Resistance levels were < 2% for all antimicrobials except oxytetracycline (with values of 2–8%), and all isolates were susceptible to ceftiofur (Figure 22).
Figure 22.
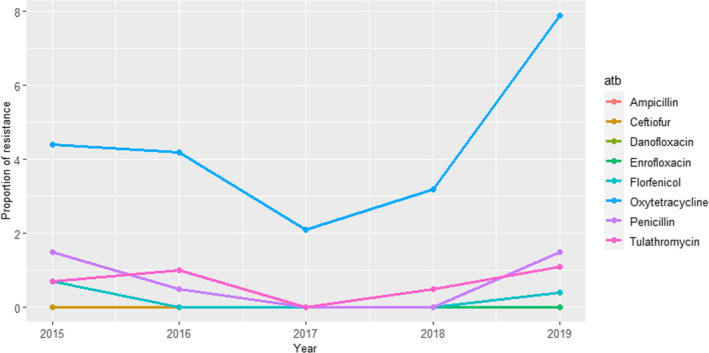
Proportion (%) of clinical Pasteurella multocida isolates retrieved from cattle respiratory samples resistant to eight antimicrobials of interest reported by the FINRES monitoring programme
For M. haemolytica, data on AMR were available for between 35 and 79 isolates tested annually using six to eight antimicrobials of interest for this opinion in 2015–2019 (ampicillin was only used in 2015 and danofloxacin in 2015–2017). All isolates were susceptible to ampicillin, ceftiofur, danofloxacin, enrofloxacin, florfenicol and tulathromycin (data not shown), while the proportion of resistant isolates to penicillin and oxytetracycline ranged between 1% and 17% (with higher values in most years for penicillin) (Figure 23).
Figure 23.
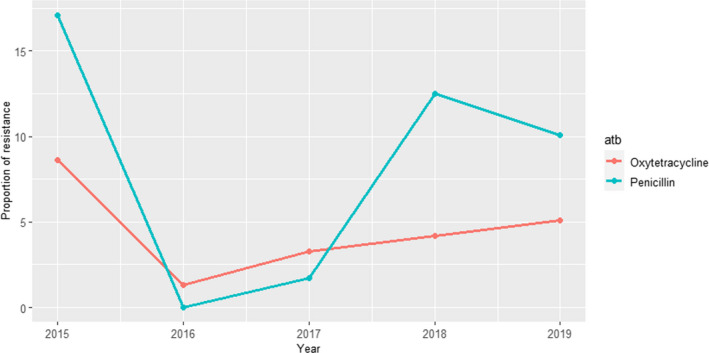
Proportion (%) of clinical Mannheimia haemolytica isolates retrieved from cattle respiratory samples resistant to oxytetracycline and penicillin reported by the FINRES monitoring programme
Finally, for H. somni between 28 and 47 isolates were tested annually using five (in 2015) or six (2016–2019) antimicrobials (oxytetracycline was missing in 2015). All isolates tested in 2015–2019 were susceptible to ceftiofur, enrofloxacin, florfenicol, penicillin and tulathromycin, while between 0 and 10.7% of the isolates in 2016–2019 were resistant to oxytetracycline (Figure 24).
Figure 24.

Proportion (%) of clinical Histophilus somni isolates retrieved from cattle respiratory samples resistant to oxytetracycline reported by the FINRES monitoring programme
SWEDRES‐Svarm (Sweden): Data on between 79 and 104 P. multocida isolates retrieved from respiratory samples (nasal swabs from calves with respiratory disease or lung samples collected during post‐mortem investigation) are provided in the annual reports for 2016–2018 (before, not all isolates were identified to the species level). Isolates were tested using five antimicrobials of interest, and resistance levels ranging between 2% and 13% were only found for penicillin and ampicillin (Figure 25), while all isolates were susceptible to enrofloxacin, florfenicol and tetracycline.
Figure 25.

Proportion (%) of clinical Pasteurella multocida isolates retrieved from cattle respiratory samples resistant to two antimicrobials of interest reported by the SWEDRES‐Svarm monitoring programme (the same values were reported for ampicillin and penicillin)
UK‐VARSS (United Kingdom): AMR data from P. multocida and M. haemolytica are included in the annual reports. For P. multocida, between 42 and 76 isolates were tested annually during the 2015–2019 period for resistance to five antimicrobials of interest for this opinion (a single isolate was also tested using tylosin in 2018). Resistance levels were much higher for tetracycline (˜40–68%) than for the remaining antimicrobials (these were below 3%, except in 2017 when 15% of all isolates tested were resistant) (Figure 26).
Figure 26.
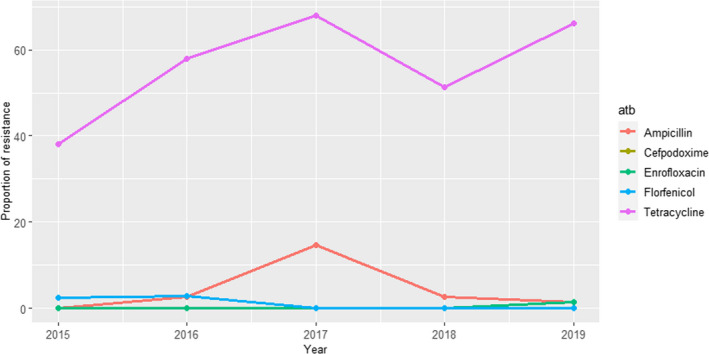
Proportion (%) of clinical Pasteurella multocida isolates retrieved from cattle respiratory samples resistant to five antimicrobials of interest reported by the UK‐VARSS monitoring programme
For M. haemolytica, between 28 and 70 isolates were tested each year during 2015–2019 using five antimicrobials of interest for this opinion. Again resistance levels were < 5% for all antimicrobials tested except for tetracycline, for which the proportion of resistant isolates increased from 0% to 50% over the 5‐year period (Figure 27).
Figure 27.

Proportion (%) of clinical Mannheimia haemolytica isolates retrieved from cattle respiratory samples resistant to five antimicrobials of interest reported by the UK‐VARSS monitoring programme
GERM‐VET (Germany): Sampling involved Mannheimia haemolytica and Pasteurella multocida in the years 2014, 2016, 2017 and 2018. Both bacterial species were isolated from respiratory disease. For M. haemolytica, isolates were divided into calves/young cattle and adult animals in 2014, and 106 and reported together in 2017 and 2018. Antimicrobials tested and classified into susceptible and resistant (intermediate resistant and resistant) were ampicillin (in the years 2017, 2018), ceftiofur (2016, 2017, 2018), enrofloxacin, florfenicol, penicillin, tetracycline, tilmicosin and tulathromycin. Results can be seen in Figure 28 for M. haemolytica and in Figure 29 for P. multocida.
Figure 28.
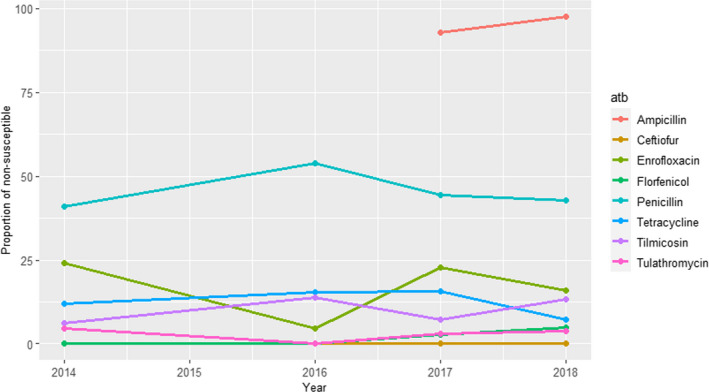
Proportion (%) of clinical Mannheimia haemolytica isolates from respiratory disease in cattle resistant to eight antimicrobials of interest reported by the GERM‐Vet monitoring programme
Figure 29.
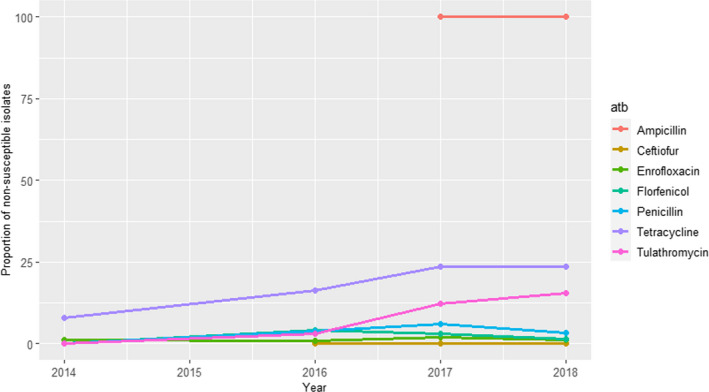
Proportion (%) of clinical Pasteurella multocida isolates from respiratory disease in cattle resistant to seven antimicrobials of interest reported by the GERM‐Vet monitoring programme
3.1.6. Streptococcus uberis and Streptococcus dysgalactiae
3.1.6.1. Results of the ELR by bacterium
Streptococcus uberis and S. dysgalactiae can be isolated from various sites in cattle, e.g. tonsils, mouth and the genital tract. From these sites, they may go to the environment, thereby facilitating transmission between cows. Streptococcus dysgalactiae may also persist in the mammary gland, thereby facilitating transmission during milking.
In total, 18 and 13 studies with ≥ 10 S. uberis and S. dysgalactiae isolates, respectively, were included. These studies had results for one or more of the following relevant antibiotics: cefoperazone, ceftiofur, enrofloxacin/ciprofloxacin, erythromycin, penicillin, penicillin–novobiocin, pirlimycin, spiramycin, sulfonamide–trimethoprim, tylosin. Geographically, these studies were distributed as follows: for S. uberis, Africa (0), Asia (1), Europe (13), Oceania (2), North America (2) and South America (0); for S. dysgalactiae, Africa (0), Asia (2), Europe (7), Oceania (2), North America (2) and South America (0).
All S. uberis and S. dysgalactiae isolates originated from mastitis (udder or milk samples) in dairy cattle.
Figure 30 shows for each continent the proportion of resistance reported in individual studies with at least 10 S. uberis and S. dysgalactiae isolates. Proportions of resistance sorted by country are in Annex D.
Figure 30.

- Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (light blue circle) and resistance merged with intermediate in isolates of dairy production origin (dark blue circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of %R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
Overall, resistance levels were fairly similar for the two streptococcal species. For the 3GCs cefoperazone and ceftiofur, < 7% resistance was observed in all studies, except in the Swiss national report which found 13% ceftiofur resistance among 56 S. uberis isolates (ANRESIS ARCH‐Vet, 2020). This value represents %RI and the fraction of intermediate isolates is unknown. Therefore, comparability to other studies reporting %R is unknown. For penicillin, the same picture was evident with overall low mean levels of resistance and the Swiss national report prominent with 44% in S. uberis. Again, this value includes the intermediate category and should be interpreted with caution. This is particularly evident from the VetPath study by Thomas et al. (2012) reporting no penicillin resistance among isolates from mixed European countries, but with 29.8% of isolates being intermediate. An earlier Swiss study found only 7.8% of 1,228 S. uberis isolates resistant to penicillin (Rüegsegger et al., 2014). As this value is also %RI, it appears as if there has been a national temporal increase in resistance over the 6–9 years between isolates were obtained in these two studies. It is, however, not clear if exactly the same breakpoints were used by the two studies, especially as there are no cattle‐specific penicillin breakpoints for streptococci and the breakpoints used must necessarily have been adapted from some other – unknown – animal species or humans. The very different number of isolates in the two studies and the different methods used (broth microdilution vs. agar dilution) are among other factors that may influence results and thereby comparability of these studies. Only one Canadian study had investigated susceptibility to penicillin–novobiocin, reporting 0.3% of 317 S. dysgalactiae and 0.3% of 1,171 S. uberis isolates resistant to that drug (Awosile et al., 2018).
For the macrolides erythromycin, spiramycin and tylosin, most studies reported less than 25% resistance. The most noteworthy exception was a Chinese study reporting erythromycin resistance in 47.7% of 88 S. dysgalactiae isolates. In Europe, the Swiss study by Rüegsegger et al. (2014) stood out with high proportions of S. uberis and S. dysgalactiae isolates (36.2% and 39.9%, respectively) being spiramycin resistant. As for other antimicrobials, this study reported %RI, meaning data may not be comparable with those of other studies, especially as a Portuguese study found a massive 30.8% of S. uberis isolates to be spiramycin intermediate (Simoes et al., 2020). However, it is more than twice the proportion (%RI) observed in for example France (RESAPATH (ANSES), 2020). The lincosamide pirlimycin was tested in very few studies. For this drug, the mean proportion of resistance observed in S. uberis from Europe was 17.6% (Table 9). Resistance to sulfonamide–trimethoprim was either absent or very uncommon (< 5%) in most studies, although studies from New Zealand (McDougall et al., 2014), France (RESAPATH (ANSES), 2020) and Thailand (Horpiencharoen et al., 2019) reported 12–22% resistance. Similar to other drugs, the French %RI data may have overestimated resistance levels compared with most other studies. Exactly the same issue exists for fluoroquinolones with RESAPATH (ANSES) (2020), reporting 37% and 47% of 1,068 S. uberis and 172 S. dysgalactiae isolates resistant to enrofloxacin. This is much higher than all other studies reporting < 10% of isolates resistant to this drug class, and the example illustrates again the problems of low data comparability. Apart from the difference between %R and %R + I, the French ECOFFs and methods differ in some aspects from the CLSI (and other) standards used by most studies.
Table 9.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Streptococcus uberis for the target antimicrobials in each continent. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | No. of papers | No. of isolatesa | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|
| 3GC (Cefoperazone) | Europe | 3 | 1,317 | 5.7 | 0 | 6 | 1 |
| 3GC (Ceftiofur) | Europe | 1 | 56 | 13 | 13 | 13 | NA |
| 3GC (Ceftiofur) | North America | 2 | 1,267 | 3.4 | 3.2 | 6.2 | 0.8 |
| Erythromycin | Asia | 1 | 12 | 0 | 0 | 0 | NA |
| Erythromycin | Europe | 7 | 1,974 | 15.6 | 5.7 | 25 | 3.3 |
| Erythromycin | Oceania | 1 | 703 | 8.9 | 8.9 | 8.9 | NA |
| Fluoroquinolones | Europe | 4 | 1,449 | 27.4 | 0 | 37 | 16.1 |
| Penicillin | Europe | 8 | 1,847 | 6.7 | 0 | 44 | 7.4 |
| Penicillin | Oceania | 2 | 817 | 8.2 | 1 | 9.2 | 2.7 |
| Penicillin‐novobiocin | North America | 1 | 1,171 | 0.3 | 0.3 | 0.3 | NA |
| Pirlimycin | Europe | 3 | 286 | 17.6 | 11.8 | 26 | 6.4 |
| Pirlimycin | North America | 1 | 1,171 | 17.9 | 17.9 | 17.9 | NA |
| Spiramycin | Europe | 4 | 2,716 | 26 | 7.7 | 36.2 | 9.4 |
| Sulfa/TMP | Europe | 5 | 1,803 | 15.2 | 0 | 22 | 9.4 |
| Sulfa/TMP | North America | 1 | 1,171 | 4.9 | 4.9 | 4.9 | NA |
| Sulfa/TMP | Oceania | 1 | 102 | 12.7 | 12.7 | 12.7 | NA |
| Tylosin | Europe | 2 | 831 | 19.5 | 2.9 | 21 | 5 |
All isolates were of dairy origin.
Table 10. Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Streptococcus dysgalactiae for the target antimicrobials in each continent. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Standard deviation |
|---|---|---|---|---|---|---|---|
| 3GC (Cefoperazone) | Europe | 1 | 213 | 4.2 | 4.2 | 4.2 | NA |
| 3GC (Ceftiofur) | North America | 2 | 414 | 0 | 0 | 0 | 0 |
| Erythromycin | Asia | 1 | 88 | 47.7 | 47.7 | 47.7 | NA |
| Erythromycin | Europe | 4 | 422 | 11.5 | 4.9 | 22 | 6.2 |
| Erythromycin | Oceania | 1 | 349 | 16.3 | 16.3 | 16.3 | NA |
| Fluoroquinolones | Asia | 1 | 14 | 7.1 | 7.1 | 7.1 | NA |
| Fluoroquinolones | Europe | 4 | 410 | 22.4 | 3.7 | 47 | 21 |
| Penicillin | Asia | 1 | 14 | 7.1 | 7.1 | 7.1 | NA |
| Penicillin | Europe | 5 | 321 | 4.6 | 0 | 7 | 3.3 |
| Penicillin | Oceania | 2 | 1,106 | 2.3 | 0 | 2.4 | 0.6 |
| Penicillin–novobiocin | North America | 1 | 317 | 0.3 | 0.3 | 0.3 | NA |
| Pirlimycin | North America | 1 | 317 | 7.9 | 7.9 | 7.9 | NA |
| Spiramycin | Europe | 3 | 579 | 19 | 4.3 | 39.9 | 16 |
| Sulfa/TMP | Asia | 1 | 14 | 14.3 | 14.3 | 14.3 | NA |
| Sulfa/TMP | Europe | 4 | 393 | 7.4 | 0 | 15 | 7 |
| Sulfa/TMP | North America | 1 | 317 | 0.3 | 0.3 | 0.3 | NA |
| Sulfa/TMP | Oceania | 1 | 64 | 17.2 | 17.2 | 17.2 | NA |
| Tylosin | Europe | 2 | 158 | 13.7 | 11.1 | 14 | 0.9 |
3.1.6.2. Results from the national AMR monitoring reports
Information on AMR in clinical isolates belonging to one or both streptococci and originating from mastitis cases/milk samples were included in five national monitoring programmes.
All‐Islands Animal Disease Surveillance Report (Ireland): Detailed data on AMR obtained in clinical S. uberis are included in the 2018 report, which provides the proportion of isolates non‐susceptible to sulfonamide–trimethoprim out of 291 isolates tested (6.2% resistant) (these data are already included in Figure 30 and Table 9).
ANRESIS ARCH‐Vet (Switzerland): Data on AMR to four antimicrobials of interest in this opinion determined in 56 mastitis S. uberis isolates were provided in 2019 (data already included in Table 9 and Figure 30), with values ranging between 13% and 44% (Figure 31), with levels of resistance to penicillin being higher than what was described in other studies, found in the ELR, what could be due at least in part by the joint reporting of resistant and intermediate categories in this report as mentioned before.
Figure 31.

Proportion (%) of clinical Streptococcus uberis isolates retrieved from cattle mastitis samples resistant to four antimicrobials of interest reported by the ANRESIS ARCH‐Vet monitoring programme
RESAPATH (France): Antimicrobial susceptibility results determined in clinical isolates from mastitis for both streptococci are included in the annual reports. For S. uberis, depending on the year (between 2014 and 2018) and the antimicrobial (considering those of interest in this opinion), from 707 to 1,523 AST results are provided. Proportions of non‐susceptibility were consistently higher for enrofloxacin (≥ 35%) compared with the rest of the antimicrobials, which remained below 35% (Figure 32). Furthermore, non‐susceptibility to oxacillin, used as a marker of non‐susceptibility to penicillin G, remained between 12 and 20% during the 2014–2018 period (data not shown).
Figure 32.
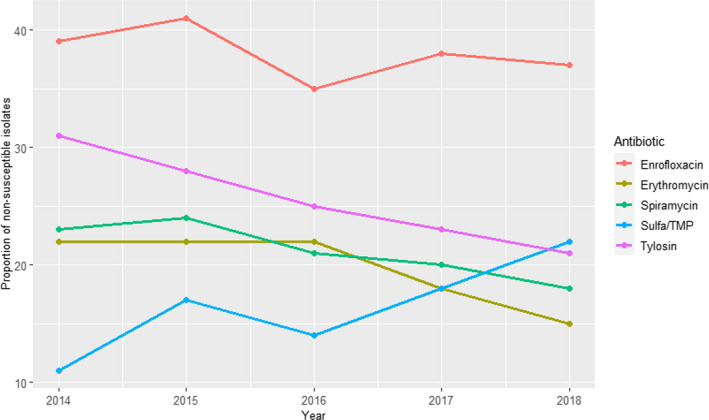
Proportion (%) of non‐susceptible clinical Streptococcus uberis isolates retrieved from cattle mastitis samples for five antimicrobials of interest reported by the RESAPATH monitoring programme
For S. dysgalactiae, depending on the year (between 2014 and 2018) and the antimicrobial (considering those of interest in this opinion), from 112 to 223 AST results are provided. Proportions of non‐susceptibility were high (≥ 44%) only for enrofloxacin, while values ranged between 4% and 25% for the remaining antimicrobials and years (Figure 33). Furthermore, non‐susceptibility to oxacillin, used as a marker of non‐susceptibility to penicillin G, remained ≤ 3% during the 2014–2018 period (data not shown).
Figure 33.

Proportion (%) of non‐susceptible clinical Streptococcus dysgalactiae isolates retrieved from mastitis samples for five antimicrobials of interest reported by the RESAPATH monitoring programme
DANMAP (Denmark): AMR data on mastitis isolates belonging to both streptococci are also available for the years 2018 and 2019. For S. uberis, 19 and 20 isolates were tested in 2018 and 2019, respectively, using four antimicrobials of interest in this opinion (ciprofloxacin, erythromycin, penicillin and sulfonamide–trimethoprim). One and three isolates found in 2018 and 2019, respectively, were resistant to erythromycin, while all isolates were susceptible to all the remaining antimicrobials. For S. dysgalactiae, 17 and 16 isolates were tested in 2018 and 2019, respectively, using four antimicrobials of interest in this opinion (ciprofloxacin, erythromycin, penicillin and sulfonamide–trimethoprim), and all isolates were susceptible except for one erythromycin‐resistant isolate retrieved in 2019.
UK‐VARSS (United Kingdom): Between 70 and 123 (S. uberis) and 18 and 41 (S. dysgalactiae) isolates from England and Wales were tested every year during the 2015–2019 period to determine the resistance to two antimicrobials of interest for this opinion (penicillin and tylosin). In both species, higher resistance levels were observed for tylosin than for penicillin, to which ≥ 99% of the isolates remained susceptible (Figures 34 and 35).
Figure 34.
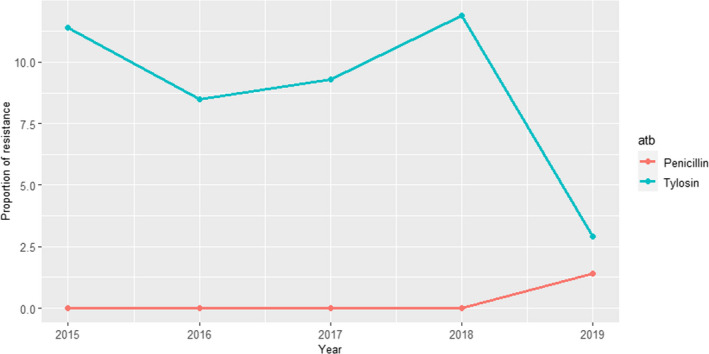
Proportion (%) of clinical Streptococcus uberis isolates retrieved from cattle mastitis samples resistant to two antimicrobials of interest reported by the UK‐VARSS monitoring programme
Figure 35.
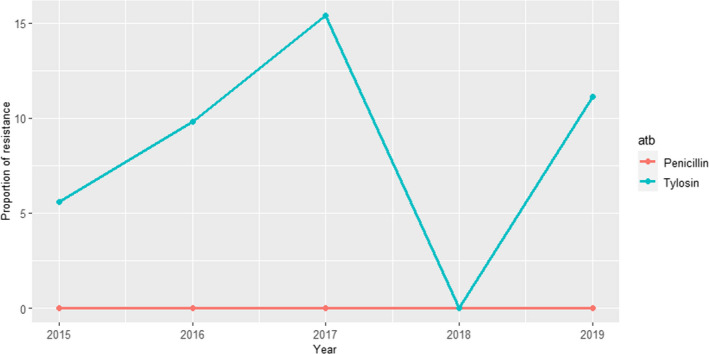
Proportion (%) of clinical Streptococcus dysgalactiae isolates retrieved from cattle mastitis samples resistant to two antimicrobials of interest reported by the UK‐VARSS monitoring programme
GERM‐VET (Germany): Sampling involved Streptococcus uberis and S. dysgalactiae from mastitis cases in 2014 and 2016. Antimicrobials tested and classified into susceptible and resistant (intermediate resistant and resistant) were ceftiofur (only in 2016 for S. uberis, with 4.2% of non‐susceptible isolates, and both years for S. dysgalactiae, with isolates being susceptible), erythromycin, penicillin and pirlimycin. Figure 36 shows the results for S. uberis and Figure 37 for S. dysgalactiae for the last three antimicrobials.
Figure 36.
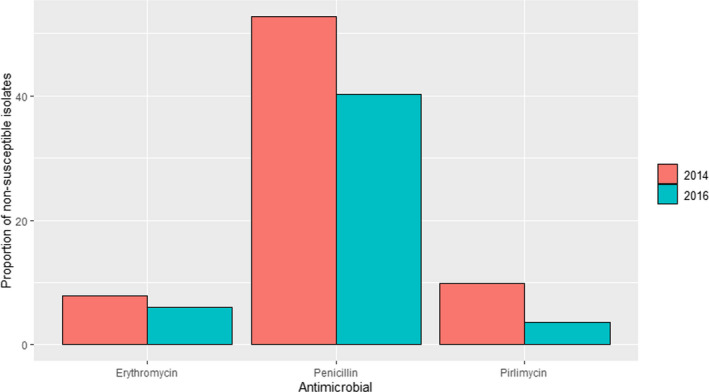
Proportion of clinical Streptococcus uberis isolates from cattle mastitis samples non‐susceptible to four antimicrobials of interest reported by the GERM‐Vet monitoring programme
Figure 37.

Proportion of clinical Streptococcus dysgalactiae isolates from cattle mastitis samples non‐susceptible to four antimicrobials of interest reported by the GERM‐Vet monitoring programme
3.1.7. Trueperella pyogenes
3.1.7.1. Results of the ELR by bacterium
Trueperella pyogenes (previously named Arcanobacterium pyogenes) resides in mucous membranes and is an opportunistic pathogen of many domestic animal species including cattle. It may cause a variety of purulent infections such as osteomyelitis, abscesses and lymphadenitis, and it is an aetiological agent of the summer mastitis complex involving also several other pathogens.
In total, eight studies with ≥ 10 T. pyogenes isolates and results for one or more of the relevant antibiotics (ampicillin/amoxicillin, 3GC, enrofloxacin/ciprofloxacin, erythromycin, penicillin, sulfonamide–trimethoprim, tetracyclines) were included. These were distributed as follows: Africa (0), Asia (5), Europe (2), Oceania (0), North America (1) and South America (0).
The distribution of T. pyogenes isolates per site of infection is shown in Figure 38. Most isolates originated from infections of the reproductive organs.
Figure 38.

Distribution of Trueperella pyogenes isolates per site of infection
Figure 39 shows for each continent the proportion of resistance reported in individual studies with at least 10 Trueperella pyogenes isolates. Information on proportion of resistance sorted by country is in Annex D.
Figure 39.
- Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (light blue circle), beef/veal production origin (red circle), mixed/unknown production origin (light grey circle), and resistance merged with intermediate in isolates of dairy production origin (dark blue circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
As there are no CBPs for T. pyogenes infections, interpretation of susceptibility data can be done in different ways. This was indeed the case for the studies included here, as they have used very different interpretive criteria, including S. aureus‐ and S. pneumoniae breakpoints adapted from human CLSI guidelines. The shortcoming of such diverse interpretation is of course that rational comparison of data between studies is nearly impossible.
Among the antibiotics tested, T. pyogenes was most frequently susceptible to beta‐lactams (Figure 39, Table 11). However, especially some of the Asian studies reported a high proportion of beta‐lactam resistance. For example, Zhang et al. (2014) found 71.9% and 53.1% of 32 Chinese isolates resistant to ceftiofur and ampicillin, respectively. Rezanejad et al. (2019) detected even higher proportions with more than 90% ampicillin resistance among a collection of 73 Iranian isolates. The limited data from Europe had been derived from Poland (Malinowski et al., 2011) and Polen/Belarus (Zastempowska and Lassa, 2012). Despite the obvious spatial relatedness of these studies, large differences in antimicrobial susceptibility were observed between them (Figure 39).
Table 11.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Trueperella pyogenes for the target antimicrobials in each continent. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | N of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Weighted standard deviation |
|---|---|---|---|---|---|---|---|---|
| 3GC | Asia | Mixed/Unknown | 2 | 97 | 23.7 | 0 | 71.9 | 34 |
| 3GC | Europe | Dairy | 1 | 55 | 0 | 0 | 0 | NA |
| 3GC | Europe | Mixed/Unknown | 1 | 161 | 4.2 | 4.2 | 4.2 | NA |
| 3GC | North America | Beef/Veal | 1 | 94 | 1.1 | 1.1 | 1.1 | NA |
| Aminopenicillins | Asia | Dairy | 2 | 123 | 59.3 | 6 | 95.9 | 44.3 |
| Aminopenicillins | Asia | Mixed/Unknown | 2 | 97 | 17.5 | 0 | 53.1 | 25.1 |
| Aminopenicillins | Europe | Dairy | 1 | 55 | 0 | 0 | 0 | NA |
| Aminopenicillins | Europe | Mixed/Unknown | 1 | 161 | 35.7 | 35.7 | 35.7 | NA |
| Aminopenicillins | North America | Beef/Veal | 1 | 94 | 1.1 | 1.1 | 1.1 | NA |
| Erythromycin | Asia | Dairy | 2 | 123 | 44.7 | 44 | 45.2 | 0.6 |
| Erythromycin | Asia | Mixed/Unknown | 1 | 65 | 36.9 | 36.9 | 36.9 | NA |
| Erythromycin | Europe | Dairy | 1 | 55 | 9.9 | 9.9 | 9.9 | NA |
| Fluoroquinolones | Asia | Dairy | 2 | 123 | 48 | 20 | 67.1 | 23.2 |
| Fluoroquinolones | Asia | Mixed/Unknown | 1 | 65 | 17 | 17 | 17 | NA |
| Fluoroquinolones | North America | Beef/Veal | 1 | 94 | 0 | 0 | 0 | NA |
| Penicillin | Asia | Dairy | 2 | 123 | 61.8 | 8 | 98.6 | 44.7 |
| Penicillin | Asia | Mixed/Unknown | 2 | 97 | 24.7 | 4.6 | 65.6 | 28.8 |
| Penicillin | Europe | Dairy | 1 | 55 | 0 | 0 | 0 | NA |
| Penicillin | Europe | Mixed/Unknown | 1 | 161 | 31.3 | 31.3 | 31.3 | NA |
| Penicillin | North America | Beef/Veal | 1 | 94 | 1.1 | 1.1 | 1.1 | NA |
| Sulfa/TMP | Asia | Dairy | 2 | 123 | 82.9 | 78.1 | 90 | 5.9 |
| Sulfa/TMP | Asia | Mixed/Unknown | 1 | 65 | 72.3 | 72.3 | 72.3 | NA |
| Tetracyclines | Asia | Dairy | 3 | 155 | 60.6 | 50.7 | 70 | 9.4 |
| Tetracyclines | Asia | Mixed/Unknown | 1 | 65 | 10.8 | 10.8 | 10.8 | NA |
| Tetracyclines | Europe | Dairy | 1 | 55 | 85.4 | 85.4 | 85.4 | NA |
| Tetracyclines | Europe | Mixed/Unknown | 1 | 161 | 63.7 | 63.7 | 63.7 | NA |
| Tetracyclines | North America | Beef/Veal | 1 | 94 | 94.7 | 94.7 | 94.7 | NA |
Resistance data for fluoroquinolones and erythromycin varied considerably between studies, whereas more consistently high resistance levels were observed for tetracyclines and – especially – sulfonamide–trimethoprim.
3.1.7.2. Results from the national AMR monitoring reports
Information on AMR in clinical T. pyogenes isolates was only included in the UK‐VARSS (United Kingdom) national monitoring programme, which provided resistance data to three antimicrobials of interest for this opinion determined on between three and eight isolates from England and Wales tested each year between 2015 and 2017. A higher proportion of resistant isolates were found for tetracyclines than for the other antimicrobials, although given the very small sample sizes data should be interpreted carefully (Figure 40).
Figure 40.

Proportion (%) of three to eight clinical Trueperella pyogenes isolates from cattle tested each year resistant to three antimicrobials of interest reported by the UK‐VARSS monitoring programme
3.1.8. Mycoplasma bovis
3.1.8.1. Results of the ELR by bacterium
Mycoplasma bovis is one of several infectious agents involved in the BRD complex. Calves are particularly susceptible, and the disease is predisposed by stress factors such as change of feed, transportation and changes of temperature and humidity in the near environment. Mycoplasma bovis is also able to cause other types of infections, such as mastitis, arthritis or otitis (Maunsell et al., 2011).
In total, eight studies with ≥ 10 M. bovis isolates and results for one or more of the relevant antibiotics (enrofloxacin/ciprofloxacin, erythromycin, florfenicol, tetracyclines, tilmicosin, tulathromycin and tylosin) were included. These were distributed as follows: Africa (0), Asia (1), Europe (5), Oceania (0), North America (2) and South America (0).
The distribution of M. bovis isolates per site of infection is shown in Figure 41. Most isolates originated from mixed infections.
Figure 41.

Distribution of Mycoplasma bovis isolates per site of infection
Figure 42 shows for each continent the proportion of resistance reported in individual studies with at least 10 M. bovis isolates. Information on the proportion of resistance sorted by country is in Annex D.
Figure 42.

Mycoplasma bovis resistance data for each included study sorted by continent
Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of beef/veal production origin (light red circle), mixed/unknown production origin (light grey circle), resistance merged with intermediate in isolates of beef/veal production origin (dark red circle) resistance merged with intermediate in isolates of beef/veal production origin (dark grey circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
As for T. pyogenes, there were no CBPs for M. bovis. Authors of the included studies have instead interpreted data in various ways, mostly using epidemiological cut‐off values derived from their data sets, or using CBPs of other cattle respiratory pathogens (typically Pasteurellaceae).
Figure 42 and Table 12 illustrate relatively low mean levels of resistance to florfenicol and fluoroquinolones across continents, whereas resistance to both macrolides and tetracyclines is much more pronounced. Two French studies are prominent, as they report resistance to tetracycline and macrolides in all tested isolates (Khalil et al. (2017), n = 43; Gautier‐Bouchardon et al. (2014), n = 26). Gautier‐Bouchardon et al. (2014) also found florfenicol resistance in 94% of isolates, whereas all isolates were intermediate to enrofloxacin. Based on a comparison with older M. bovis isolates, it was clear that resistance levels of M. bovis had markedly increased in the years before sampling (Khalil et al., 2017).
Table 12.
Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in M. bovis for the target antimicrobials in each continent. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | Production type | No. of papers | No. of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Weighted standard deviation |
|---|---|---|---|---|---|---|---|---|
| Erythromycin | Asia | Beef/Veal | 1 | 32 | 40.6 | 40.6 | 40.6 | NA |
| Florfenicol | Asia | Beef/Veal | 1 | 32 | 3.1 | 3.1 | 3.1 | NA |
| Florfenicol | Europe | Beef/Veal | 1 | 84 | 0 | 0 | 0 | NA |
| Florfenicol | Europe | Mixed/Unknown | 2 | 187 | 25.3 | 2.9 | 94 | 39.3 |
| Florfenicol | North America | Beef/Veal | 2 | 323 | 20.4 | 8.2 | 25.7 | 8 |
| Fluoroquinolones | Asia | Beef/Veal | 1 | 32 | 0 | 0 | 0 | NA |
| Fluoroquinolones | Europe | Mixed/Unknown | 3 | 379 | 3.5 | 0 | 6.6 | 2.9 |
| Fluoroquinolones | North America | Beef/Veal | 2 | 323 | 18 | 8 | 41.2 | 15.2 |
| Tetracyclines | Asia | Beef/Veal | 1 | 32 | 0 | 0 | 0 | NA |
| Tetracyclines | Europe | Mixed/Unknown | 4 | 331 | 43.1 | 0 | 100 | 41.7 |
| Tetracyclines | North America | Beef/Veal | 2 | 323 | 69.7 | 45.4 | 80.1 | 15.9 |
| Tilmicosin | Asia | Beef/Veal | 1 | 32 | 40.6 | 40.6 | 40.6 | NA |
| Tilmicosin | Europe | Mixed/Unknown | 3 | 190 | 97.5 | 95 | 100 | 2.5 |
| Tilmicosin | North America | Beef/Veal | 2 | 323 | 98.4 | 98.2 | 99 | 0.4 |
| Tulathromycin | Europe | Mixed/Unknown | 2 | 141 | 50.8 | 27 | 100 | 34.3 |
| Tulathromycin | North America | Beef/Veal | 2 | 323 | 89.4 | 83.5 | 92 | 3.9 |
| Tylosin | Asia | Beef/Veal | 1 | 64 | 40.6 | 40.6 | 40.6 | NA |
| Tylosin | Europe | Mixed/Unknown | 3 | 236 | 73.4 | 56.2 | 100 | 21 |
| Tylosin | North America | Beef/Veal | 2 | 323 | 96.2 | 92.8 | 97.7 | 2.2 |
3.1.9. Klebsiella pneumoniae
3.1.9.1. Results of the ELR by bacterium
As for E. coli, Klebsiella pneumoniae is a commensal and an opportunistic pathogen residing in the intestinal microbiota of animals and humans. In cattle, it is mostly known for causing environmental mastitis in dairy cows.
In total, five studies with ≥ 10 K. pneumoniae isolates and results for one or more of the relevant antibiotics (amoxicillin–clavulanic acid, 3GCs, colistin, enrofloxacin/ciprofloxacin, neomycin, sulfonamide–trimethoprim) were included. Among these, one and four included isolates from Africa and Europe, respectively.
All K. pneumoniae isolates originated from mastitis (udder/milk samples) in dairy cattle.
Figure 43 shows for each continent the proportion of resistance reported in individual studies with at least 10 K. pneumoniae isolates. Information on proportion of resistance sorted by country is in Annex D.
Figure 43.

- Each circle represents one study, and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates resistance in isolates of dairy production origin (light blue circle) and resistance merged with intermediate in isolates of dairy production origin (dark blue circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % R or %R + I with the same colour codes as used for the circles. The exact percentages these lines represent are listed in Annex E. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/continent combination.
Resistance levels were generally low (< 10%) for all tested antibiotics. The few exceptions were 22.1% and 16.1% resistance to amoxicillin–clavulanic acid and sulfonamide–trimethoprim among 77 isolates from Tunisia (Saidani et al., 2018), and 16% of non‐susceptibility for amoxicillin–clavulanic acid among 88 isolates from France (RESAPATH (ANSES), 2020). The latter proportion may even be an overestimation compared with other studies, as it represents also the intermediate category.
Table 13. Weighted arithmetic mean, minimum and maximum proportion of resistance (%R or %R + I) and weighted standard deviation (SD) in Klebsiella pneumoniae for the target antimicrobials in each continent. NA means that SD could not be calculated as only one study was included
| Antibiotic | Continent | No. of papers | N of isolates | Weighted arithmetic mean proportion of resistance (%) | Minimum resistance % observed | Maximum resistance % observed | Weighted standard deviation |
|---|---|---|---|---|---|---|---|
| 3GC | Africa | 1 | 77 | 7.8 | 7.8 | 7.8 | NA |
| 3GC | Europe | 3 | 141 | 0.5 | 0 | 1 | 0.5 |
| Amox/Clav | Africa | 1 | 77 | 22.1 | 22.1 | 22.1 | NA |
| Amox/Clav | Europe | 3 | 203 | 6.9 | 0 | 16 | 7.9 |
| Colistin | Africa | 1 | 77 | 1.3 | 1.3 | 1.3 | NA |
| Colistin | Europe | 2 | 70 | 0 | 0 | 0 | 0 |
| Fluoroquinolones | Africa | 1 | 77 | 6.5 | 6.5 | 6.5 | NA |
| Fluoroquinolones | Europe | 4 | 237 | 2.6 | 0 | 8 | 3 |
| Neomycin | Europe | 3 | 123 | 0 | 0 | 0 | 0 |
| Sulfa/TMP | Africa | 1 | 77 | 11.7 | 11.7 | 11.7 | NA |
| Sulfa/TMP | Europe | 3 | 234 | 3.8 | 0 | 8 | 3.3 |
3.1.9.2. Results from the national AMR monitoring reports
Resistance data on K. pneumoniae isolates originating from mastitis cases/clinical milk samples are included in three national monitoring programmes.
RESAPATH (France): depending on the year (between 2014 and 2018) and the antimicrobial (considering those of interest in this opinion), from 44 to 90 AST results are provided. Proportions of non‐susceptibility ranged between 11% and 17% for amoxicillin–clavulanic acid and staying ≤ 8% for the remaining antimicrobials (≤ 2% for enrofloxacin and ceftiofur) (Figure 44).
Figure 44.

Proportion (%) of non‐susceptible clinical Klebsiella pneumoniae isolates from cattle mastitis cases for five antimicrobials of interest reported by the RESAPATH monitoring programme
SWEDRES‐Svarm (Sweden): Data on AMR on isolates from clinical submissions of milk are provided for the period 2014–2019. Between 34 and 52 isolates were tested over that period with four or five antimicrobials (cefotaxime and colistin not included in the panel used in 2014). Resistance levels were below 10% for all antimicrobials and years except sulfonamide–trimethoprim in 2014 and enrofloxacin in 2016 (which still remained ≤ 17%) (Figure 45).
Figure 45.
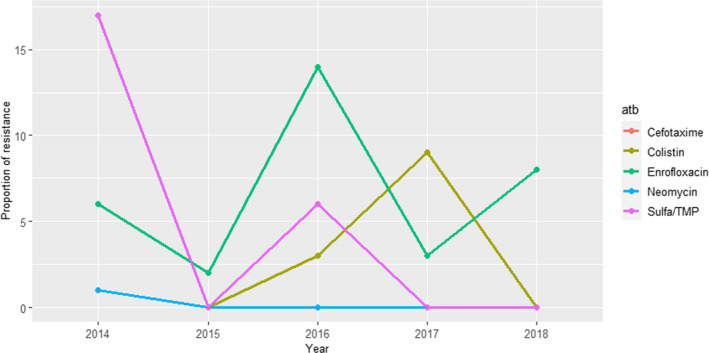
Proportion (%) of clinical Klebsiella pneumoniae isolates from cattle mastitis cases resistant to five antimicrobials of interest reported by the SWEDRES‐Svarm monitoring programme
UK‐VARSS (United Kingdom): AMR results determined for five antimicrobials of interest for this opinion in between 3 and 13 K. pneumoniae isolates from mastitis cases in England and Wales are provided in the reports. At least two‐thirds of all isolates tested remain susceptible every year to all antimicrobials, although values change, largely depending on the year and the antimicrobial (Figure 46). However, results must be interpreted carefully given the small sample size.
Figure 46.
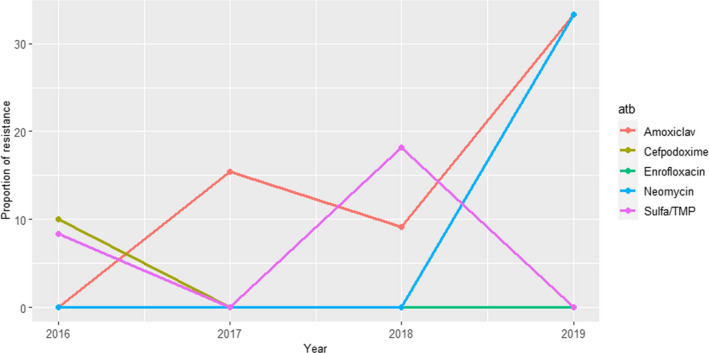
Proportion (%) of clinical cattle Klebsiella pneumoniae isolates from mastitis cases resistant to five antimicrobials of interest reported by the UK‐VARSS monitoring programme
GERM‐VET (Germany): In total, 58 (2014 and 2015), 90 (2016) and 97 (2018) Klebsiella isolates from mastitis cases identified only at the genus level (Klebsiella spp.) were tested using two antimicrobials of interest for this opinion (amoxicillin–clavulanic acid and sulfonamide–trimethoprim), with resistance levels ranging between 0% and 9% (Figure 47).
Figure 47.
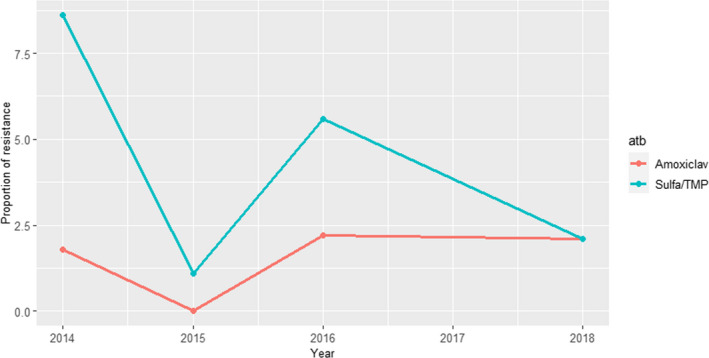
Proportion of clinical isolates of Klebsiella spp. for mastitis samples resistant to six antimicrobials of interest reported by the GERM‐Vet monitoring programme
3.1.10. Moraxella bovis
3.1.10.1. Results of the ELR by bacterium
Moraxella bovis is the cause of infectious keratoconjunctivitis in cattle, also known as ‘pink eye’. The bacterium can be transmitted via flies, aerosols and close contact, and clinical signs range from mild conjunctivitis to more serious disease including blindness.
Only one study from the USA was included (Loy and Brodersen, 2014). Due to the lack of M. bovis‐specific breakpoints, the authors claimed that interpretive criteria established for BRD or other Gram‐negative veterinary isolates as available were used’. Very low levels of resistance (≤ 6%) were detected for all tested antibiotics, namely florfenicol, oxytetracycline, penicillin, tilmicosin and tulathromycin.
3.2. ToR 2: identifying the most relevant bacteria in the EU
Following the methodology presented in the scientific opinion on the ad hoc method for the assessment of animal diseases caused by bacteria resistant to antimicrobials within the AHL framework (EFSA AHAW Panel, 2021), the evidence available was assessed individually by all working group members who provided individual judgements on the perceived relevance to cattle health of the antimicrobial‐resistant bacteria included in the list.
After discussion of the individual judgements for each bacterium, it was agreed with ≥ 66% certainty that the most relevant resistant bacteria in cattle for the EU were E. coli and S. aureus (Figure 48). The importance of antimicrobials to their treatment was highlighted by the very large number of references (Table 2) and AST results retrieved in the ELR, as well as by their frequent inclusion (especially for E. coli) in national AMR monitoring systems of European countries (Table 3). Escherichia coli causes serious health concerns both in young calves as a gastrointestinal pathogen and in dairy cows as a causative agent of mastitis. Antimicrobial therapy is often needed to treat gastrointestinal colibacillosis. For mastitis caused by E. coli, treatment of mild or moderate cases with antimicrobials is not recommended (NZVA, 2018), while treatment of acute cases with antimicrobials may be considered (NCAS, 2017; NZVA, 2018). Retrieved data suggest higher levels of resistance in isolates from gastrointestinal cases compared to mastitis cases for clinically important antimicrobials (Figures 9, 10, 1111–12 and 16–17). For gastrointestinal and other non‐mastitis infections, results of the ELR showed high levels of resistance to antimicrobial classes often used as first‐line options such as tetracyclines, aminopenicillins, potentiated sulfonamides, aminoglycosides, as well as resistance proportions that cannot be neglected for fluoroquinolones, although lower (Figure 8). This resulted in a high certainty on its inclusion among the most relevant cattle AMR pathogens, mainly due to its importance as a pathogen requiring antimicrobial treatment and often showing high resistance levels in non‐mastitis cases.
Figure 48.

Level of certainty for the inclusion of the selected antimicrobial resistant pathogens of cattle among the most relevant in the EU
For S. aureus, its importance as a very frequently isolated pathogen in clinical and subclinical mastitis, along with the high resistance levels to certain antimicrobial classes (e.g. beta‐lactams) and the results suggesting resistance to other antimicrobial classes (macrolides, fluoroquinolones) is common in clinical isolates from certain regions of the world (particularly Asia) (Figure 4) led to its inclusion among the most relevant antimicrobial resistant pathogens in cattle. Nevertheless, in this case, there was a larger uncertainty (reflected in a wider interval) derived from data suggesting strains are still typically susceptible to certain therapeutic options (e.g. penicillin–novobiocin).
Among the bacterial pathogens considered in this opinion associated with respiratory problems in young animals, Mycoplasma bovis was ranked the highest although it was not included among the most relevant AMR pathogens in cattle. Its importance is derived from its frequent occurrence in calves with respiratory diseases and its association with cases that are particularly challenging from a treatment standpoint. The relatively low number of studies retrieved through the ELR suggest in fact that M. bovis is often resistant to first‐line antimicrobials used for respiratory problems in young cattle, especially macrolides (e.g. tulathromycin or tilmicosin). Due to intrinsic β‐lactam resistance in Mycoplasma spp., effective alternatives to macrolides are limited to florfenicol, for which lower levels of AMR have been reported (Bokma et al., 2020), but concerns exist regarding side effects in young calves, and to fluoroquinolones which are CIAs and therefore not suitable for group medication, which is often used for control of M. bovis outbreaks. Even though there are vaccines available in the US and autovaccines are authorised in several EU Member States, there is no clear evidence about their efficacy. Thus, antimicrobial treatment is essential for the control of BRD outbreaks associated with M. bovis. However, due to the limited number of studies found (what could be due to the practical challenges for working with this bacterial species in a laboratory) and the lack of standardised methodologies and approved breakpoints to assess clinical resistance, there was a high uncertainty about the relevance of M. bovis, which resulted in its exclusion from the group of most relevant AMR pathogens in cattle.
Mannheimia haemolytica, Pasteurella multocida and Histophilus somni, the other pathogens most commonly associated with BRD, were also considered drivers of a very large proportion of the antimicrobial use in cattle, particularly in countries with a big feedlot production. While the ELR revealed medium to high levels of resistance to several antimicrobial classes commonly used to treat respiratory infections, particularly M. haemolytica, there were still several options (e.g. florfenicol) against which most isolates studied in the scientific literature were susceptible. Moreover, the review shows that resistance to macrolides such as tulathromycin and tilmicosin, which are often used for empiric treatment of bovine respiratory disease, is relatively low, especially in Europe. Similarly, data from the national monitoring reports also suggested that most clinical isolates were susceptible to several of the antimicrobials tested, although higher levels of AMR are usually reported in veal and feedlot production, where selective pressure due to antimicrobial use is higher. Treatment of BRD can be challenging and, in fact, treatment failure in a proportion of animals is not uncommon, but lack of response to the therapy may not always be due to AMR and, in fact, most animals typically respond within a few days when treatment is initiated early (Booker, 2020). Based on these data, none of these pathogens were included among the most relevant, although the uncertainty associated with the relevance of M. haemolytica was larger (and the judgement for H. somni indicated a lower relevance).
Among the remaining pathogens, S. uberis, a common cause of mastitis in dairy cattle, ranked highest although it was not included among the most relevant AMR pathogens in cattle due to the data found suggesting susceptibility to several first‐line antimicrobials (e.g. penicillin). However, reports suggesting the presence of intermediate levels of resistance to penicillins in this bacterial species (Haenni et al., 2010; Thomas et al., 2012) and results from certain monitoring programmes (ANRESIS ARCH‐Vet, 2020; GERM‐Vet, 2020) reporting higher resistance levels when the intermediate category is included compared with other studies, call for caution, and therefore, it could be particularly useful to monitor resistance trends to this antimicrobial class in the future. For S. dysgalactiae, another frequent cause of mastitis in cattle, resistance levels found were, in general, similar or lower, and therefore, its potential relevance was also judged as lower.
For the remaining pathogens considered (T. pyogenes, K. pneumoniae, Moraxella bovis and Fusobacterium necrophorum), the collective assessment concluded that, in spite of their potential importance as cattle pathogens, it was not likely (upper limit of the certainty ranges < 50%) that they were among the most relevant AMR pathogens in cattle. This was due to the limited evidence available (few or no studies were retrieved for all the bacteria in this group), suggesting that AMR was not a major concern for their treatment, and the results suggested that isolates were typically susceptible to the available therapeutic options.
4. Conclusions
In this opinion, EFSA presents the results of the assessment conducted to answer ToR 1 (global state of play of antimicrobial‐resistant animal bacteria) and the first part of ToR 2 (identifying the most relevant resistant bacteria in the EU) according to the ad hoc methodology (EFSA AHAW Panel, 2021). The second part of ToR 2 and ToR 3, namely the animal health impact of the selected species on cattle in the EU, and their eligibility for being listed and categorised as part of the AHL, will be assessed in the next step of this EFSA project.
The scientific assessment of the global state of play of the resistant bacterial pathogens of cattle included in this opinion and of their EU relevance was hampered by several important sources of uncertainty derived from the available data and the methodology followed in this assessment, as mentioned in Section 2.4 of EFSA AHAW Panel (2021) and in the preceding sections of this opinion:
Due to the scope of the ELR, only studies published in the last 10 years and in English were considered eligible (except for the GERM‐VET report, originally in German), therefore adding a possible selection bias.
Information on the rationale and study design for the references retrieved in the ELR was limited and very heterogeneous, making the detailed assessment of the representativeness of the isolates included in each study very difficult. For example, ~25% of the references (33/135) included isolates collected through the regular testing of veterinary diagnostic laboratories for which typically very limited information on representativeness is available. Moreover, they often originated from animals subjected to previous antimicrobial treatments, which may lead to higher levels of resistance in tested isolates, and several of the bacterial species included here can also be found in healthy animals (e.g. E. coli, P. multocida). Therefore, even if they originated from diseased animals, they may not be the causative agents in a proportion of cases that cannot be quantified. Finally, studies in which it was not clear if isolates were from diseased animals (i.e. if they were ‘clinical’ isolates) were excluded, but in some cases these could originate from subclinical (but not defined as such) infections (e.g. if isolates in milk were due to subclinical mastitis) and therefore could have been considered pathogenic, but due to the lack of precise information, it was not possible to make an informed decision. Similarly, sample type was often defined in a loose way (e.g. samples from respiratory disease) and thus typically it was not possible to differentiate isolates from different locations (e.g. lower vs. upper respiratory tract).
Even though only studies exceeding a minimum quality threshold were included (e.g. use of international or national standards), the methodology used was also diverse (e.g. use of disk diffusion or microdilution methods, CBP or ECOFFs, consideration or not of the intermediate category, etc.). Therefore, descriptive statistics provided here (average proportion of resistant isolates for bacterium, country and antimicrobial) should be considered carefully as they may not be representative of the true underlying situation, particularly in cases in which the sample size was small.
AMR data referring to one or more of the bacterial pathogens of interest were retrieved from six national AMR monitoring reports. However, comparison of data reported in the different countries is difficult due to differences in: (a) the bacterial species considered, (b) the geographical and temporal coverage of each report, (c) the choice of antimicrobials included in the panel for AST, (d) the methods for antimicrobial susceptibility determination (disk diffusion vs. broth microdilution, CBPs vs. ECOFFs) and (e) the limited sample sizes achieved and the potential biases associated with the process by which the panels of isolates were built.
EFSA has summarised the global state of play on AMR in cattle for the following bacteria: S. aureus, E. coli, P. multocida, M. haemolytica, S. uberis, S. dysgalactiae, H. somni, T. pyogenes, Mycoplasma bovis, K. pneumoniae, Moraxella bovis and F. necrophorum. Among those bacteria, based on the evidence available and expert opinion, EFSA identified E. coli and S. aureus as the most relevant antimicrobial‐resistant cattle pathogens in the EU with ≥ 66% certainty. Mycoplasma bovis was not selected in this group even though it is a frequent reason for group medication, and has limited therapeutic options due to its intrinsic resistance to β‐lactams and acquired resistance to alternative antimicrobials. Still, the assessment of AMR in this pathogen is hampered by the lack of approved interpretative criteria and standard procedures for susceptibility testing of Mycoplasma, leading to a large uncertainty in its assessment. Moderate resistance levels in M. haemolytica and to a lesser extent P. multocida to specific antimicrobials were found and these are frequent pathogens associated with BRD, driving a significant amount of antimicrobial use in cattle production. Still, results consistently suggested clinical strains are often susceptible to other therapeutic options leading to their exclusion from the most relevant antimicrobial resistant pathogens. Streptococcus uberis was typically described as susceptible to penicillins and thus was also excluded, although evidence suggesting the circulation of intermediate levels of resistance was also found.
Regarding the reports from national monitoring systems from European countries included in the assessment, small sample sizes make it difficult to draw clear conclusions in terms of AMR levels in cattle populations, although stable AMR trends were found for most pathogen–drug combinations and levels of resistance were in general low for most pathogen–antimicrobial combinations. Nevertheless, the significance of these observations should not be overinterpreted due to the above‐mentioned limitations.
As mentioned before, several major data gaps were identified, derived mainly from the lack of information from many countries in the world (and to a lesser extent from some regions in Europe), the insufficient information on the origins of the bacterial isolates tested (which could result in unknown selection biases) and the variety of antimicrobials, methodologies and breakpoints used to generate the data considered in this assessment.
The impact of the uncertainties deriving from these data gaps on the scientific assessment was incorporated into the results through expert opinion.
5. Recommendations
Data on AMR in bacterial pathogens are necessary to enhance animal health, promote the rational use of antimicrobials and identify specific therapeutic challenges attributable to AMR.
Therefore, there is a need for reliable data on pathogenic bacteria from cattle from different regions of the world obtained through the use of standardised methodologies that allow to make comparisons between locations and over time. This need is particularly critical for certain pathogens posing therapeutic challenges in which approved laboratory AST methods and/or interpretative criteria are missing such as Mycoplasma bovis. Furthermore, AST data should be accompanied by sufficient metadata to allow meaningful interpretations (such as previous antimicrobial treatments and details on clinical presentation).
National monitoring systems for AMR in diseased cattle are only available in certain countries and there are limitations that hamper the comparability of data reported by different countries (Mader et al., 2021). Assuming that sampling and methodological biases are relatively constant over time for a given monitoring programme, longitudinal data from national monitoring programmes can be helpful to detect the potential emergence of new antimicrobial resistant phenotypes of clinical importance or changes in resistance proportions in pathogens of cattle, and therefore help to guide antimicrobial stewardship. This may be particularly relevant for certain cases in which evidence and/or standardised methods are missing and that are associated with high antimicrobial usage on farm (e.g. M. bovis, M. haemolytica, P. multocida), and in cases in which decreased susceptibility has been reported (e.g. to ensure S. uberis isolates remain susceptible to penicillins).
In the future, standardisation and harmonisation of the methodology used by national monitoring programmes, including selection criteria for collecting bacterial isolates and performance of AST, or development of supra‐national monitoring systems, would allow more meaningful comparisons between countries (Mader et al., 2021). In addition, access to raw AST data generated by such programmes could enable analysis of data from different countries using the same laboratory methods and interpretive criteria (CBPs or ECOFFs), and facilitating identification of geographical differences in the distribution of specific antimicrobial resistance phenotypes of clinical relevance.
Abbreviations
- 3GC
third generation cephalosporin
- AHL
animal health law
- AST
antimicrobial susceptibility testing
- CLSI
Clinical and Laboratory Standards Institute
- ECOFF
epidemiological cut‐off
- ELR
extensive literature review
- ESBL
extended‐spectrum beta‐lactamase
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- I
intermediate
- MR
methicillin resistance
- MRSA
methicillin‐resistant Staphylococcus aureus
- R
resistant
- S
susceptible
Annex A – Search strings applied
A.1. Pubmed
Common search string “Antimicrobials”
((“antibiotic”[Title/Abstract] OR “antibiotics”[Title/Abstract] OR “antimicrobial”[Title/Abstract] OR “antimicrobials”[Title/Abstract] OR “Anti‐Bacterial Agents”[MeSH Terms:noexp]) AND (“resistan*”[Title/Abstract] OR “susceptib*”[Title/Abstract])) OR (“Microbial Sensitivity Tests”[MeSH Terms] OR “drug resistance, microbial”[MeSH Terms])
Host‐based strings:
“Cattle”[Title/Abstract] OR “cow”[Title/Abstract] OR “cows”[Title/Abstract] OR “bull”[Title/Abstract] OR “bulls”[Title/Abstract] OR “calf”[Title/Abstract] OR “calves”[Title/Abstract] OR “bovine”[Title/Abstract] OR “Cattle”[MeSH Terms]
“Bacterial species”
“Haemophilus somnus”[MeSH Terms] OR “Moraxella bovis”[MeSH Terms] OR “Mycoplasma bovis”[MeSH Terms] OR “Escherichia coli”[MeSH Terms] OR “Klebsiella pneumoniae”[MeSH Terms] OR “Mannheimia haemolytica”[MeSH Terms] OR “Staphylococcus aureus”[MeSH Terms] OR “Streptococcus uberis”[Supplementary Concept] OR “Corynebacterium pyogenes”[MeSH Terms] OR “Fusobacterium necrophorum”[MeSH Terms] OR “Pasteurella multocida”[MeSH Terms] OR “Streptococcus dysgalactiae”[Supplementary Concept] OR “Haemophilus somnus”[Title/Abstract] OR “Histophilus somni”[Title/Abstract] OR “Moraxella bovis”[Title/Abstract] OR “Mycoplasma bovis”[Title/Abstract] OR “Escherichia coli”[Title/Abstract] OR “Klebsiella pneumoniae”[Title/Abstract] OR “Mannheimia haemolytica”[Title/Abstract] OR “Staphylococcus aureus”[Title/Abstract] OR “Streptococcus uberis”[Title/Abstract] OR “Corynebacterium pyogenes”[Title/Abstract] OR “Fusobacterium necrophorum”[Title/Abstract] OR “Pasteurella multocida”[Title/Abstract] OR “Streptococcus dysgalactiae”[Title/Abstract]
A.2. Embase
Common search string “Antimicrobials”
antibiotic resistance/ or exp antibiotic sensitivity/ or exp drug resistance/
susceptib*.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
resistan*.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
2 or 3
antibiotic.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
antibiotics.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
antimicrobial.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
antimicrobials.mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
5 or 6 or 7 or 8
antibiotic agent/
10 or 9
11 and 4
12 or 1
Host‐based string:
bovine/
“calf (bovine)”/
(Cattle or cow or cows or bull or bulls or calf or calves or bovine).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
1 or 2 or 3
“Bacterial species”
Histophilus somni/
moraxella bovis/
mycoplasma bovis/
Escherichia coli/
Klebsiella pneumoniae/
Mannheimia haemolytica/
Staphylococcus aureus/
Streptococcus uberis/
Trueperella pyogenes/
Fusobacterium necrophorum/
Pasteurella multocida/
Streptococcus dysgalactiae/
(“Trueperella pyogenes” or “Haemophilus somnus” or “Histophilus somni” or “Moraxella bovis” or “Mycoplasma bovis” or “Escherichia coli” or “Klebsiella pneumoniae” or “Mannheimia haemolytica” or “Staphylococcus aureus” or “Streptococcus uberis” or “Corynebacterium pyogenes” or “Fusobacterium necrophorum” or “Pasteurella multocida” or “Streptococcus dysgalactiae”).mp. [mp=title, abstract, heading word, drug trade name, original title, device manufacturer, drug manufacturer, device trade name, keyword, floating subheading word, candidate term word]
1 or 2 or 3 or 4 or 5 or 6 or 7 or 8 or 9 or 10 or 11 or 12 or 13
Annex B – Excel file with information on all studies for full‐text screening
1.
Information on all the full‐text studies that were assessed, including the reason for exclusion for those that were excluded at the full‐text screening and the data extracted from the included studies, can be consulted at https://doi.org/10.5281/zenodo.5561163
Annex C – Clinically relevant antibiotics for which data were extracted
1.
| Bacterial species | Relevant resistance tested |
|---|---|
| Escherichia coli |
|
| Klebsiella pneumoniae |
|
| Staphylococcus aureus |
|
| Streptococcus uberis |
|
| Streptococcus dysgalactiae |
|
| Pasteurella multocida |
|
| Mannheimia haemolytica |
|
| Histophilus somni |
|
| Mycoplasma bovis |
|
| Moraxella bovis |
|
| Fusobacterium necrophorum |
|
| Trueperella pyogenes |
|
Annex D – Resistance proportion data sorted by country
1.
The figures below show for S. aureus, E. coli, P. multocida, M. haemolytica, S. uberis, S. dysgalactiae, H. somni, T. pyogenes, M. bovis and K. pneumoniae resistance proportion data sorted by country. Each circle represents one study and the size of each circle reflects how many isolates were included in the study. The colour of a circle illustrates whether the proportion represents resistance only (blue circle) or resistance merged with intermediate (red circle). The dashed lines indicate, for each antibiotic, the weighted arithmetic mean of % resistance, not taking into account the difference between %R and %R + I. Numbers written to the left of antibiotic names reflect the number of studies for a certain drug/country combination.
Annex E – Exact percentages of weighted arithmetic means of %R and %R + I, respectively, displayed as dashed lines in figures
1.
| Antibiotic | How resistance is reported (%R or %R + I) | Weighted arithmetic mean proportion of resistance (%) | Maximum resistance % observed | Minimum resistance % observed | Standard deviation (SD) | Bacterial species/genus |
|---|---|---|---|---|---|---|
| 3GC (Other) | R_Dairy | 10.9 | 91.4 | 0 | 17.4 | E. coli |
| 3GC (Other) | R_Mixed/Unknown | 36.5 | 41.7 | 0.6 | 13.3 | E. coli |
| 3GC (Other) | R + I_Dairy | 2.3 | 16 | 0 | 3.2 | E. coli |
| 3GC (Other) | R + I_Mixed/Unknown | 3.1 | 8 | 3 | 0.5 | E. coli |
| Aminopenicillins | R_Dairy | 34.9 | 77.4 | 5.5 | 23.4 | E. coli |
| Aminopenicillins | R_Mixed/Unknown | 76.8 | 83 | 23 | 13.2 | E. coli |
| Aminopenicillins | R + I_Dairy | 32.4 | 58.7 | 18.7 | 7.6 | E. coli |
| Aminopenicillins | R + I_Mixed/Unknown | 81.5 | 83 | 69.5 | 4.3 | E. coli |
| Amox/Clav | R_Dairy | 4.4 | 18.6 | 0 | 5.6 | E. coli |
| Amox/Clav | R_Mixed/Unknown | 5 | 8.2 | 3.4 | 1.8 | E. coli |
| Amox/Clav | R + I_Dairy | 27.4 | 81 | 23 | 15.4 | E. coli |
| Amox/Clav | R + I_Mixed/Unknown | 50.3 | 56 | 14.6 | 12.9 | E. coli |
| Colistin | R_Dairy | 0.4 | 0.8 | 0 | 0.3 | E. coli |
| Colistin | R + I_Dairy | 0.9 | 3.2 | 0 | 1.5 | E. coli |
| Fluoroquinolones | R_Dairy | 16.7 | 81.4 | 0 | 18.9 | E. coli |
| Fluoroquinolones | R_Mixed/Unknown | 11.5 | 60.3 | 0 | 20.2 | E. coli |
| Fluoroquinolones | R + I_Dairy | 2.6 | 38.1 | 0 | 7.5 | E. coli |
| Fluoroquinolones | R + I_Mixed/Unknown | 9.5 | 14.3 | 0 | 2.1 | E. coli |
| Gentamicin | R_Dairy | 30.7 | 35.4 | 0 | 12 | E. coli |
| Gentamicin | R_Mixed/Unknown | 34.5 | 79.7 | 1.2 | 19.4 | E. coli |
| Gentamicin | R + I_Mixed/Unknown | 17.2 | 19 | 2.5 | 5.2 | E. coli |
| Neomycin | R_Dairy | 7.7 | 11.8 | 0 | 4.7 | E. coli |
| Neomycin | R_Mixed/Unknown | 76.4 | 81.9 | 14.9 | 17.2 | E. coli |
| Neomycin | R + I_Dairy | 23 | 37.5 | 5 | 13.2 | E. coli |
| Sulfa/TMP | R_Dairy | 20.5 | 50.9 | 0 | 17.6 | E. coli |
| Sulfa/TMP | R_Mixed/Unknown | 58.5 | 69.9 | 18 | 9.6 | E. coli |
| Sulfa/TMP | R + I_Dairy | 10.5 | 13 | 6.3 | 3.3 | E. coli |
| Sulfa/TMP | R + I_Mixed/Unknown | 38 | 40 | 14.2 | 6.2 | E. coli |
| Tetracyclines | R_Dairy | 35.6 | 70.7 | 14.3 | 20.7 | E. coli |
| Tetracyclines | R_Mixed/Unknown | 86.9 | 98.1 | 19.7 | 17.3 | E. coli |
| Tetracyclines | R + I_Mixed/Unknown | 72.7 | 76 | 28.8 | 10.9 | E. coli |
| Erythromycin | R_Dairy | 18.8 | 47.7 | 4.9 | 18.4 | S. dysgalactiae |
| Erythromycin | R + I_Dairy | 16.2 | 16.3 | 16 | 0.1 | S. dysgalactiae |
| Fluoroquinolones | R_Dairy | 4.8 | 9.1 | 3.7 | 1.9 | S. dysgalactiae |
| Penicillin | R_Dairy | 0.5 | 7.1 | 0 | 1.9 | S. dysgalactiae |
| Penicillin | R + I_Dairy | 3.2 | 7 | 2.4 | 1.7 | S. dysgalactiae |
| Spiramycin | R + I_Dairy | 24.9 | 39.9 | 9 | 15.5 | S. dysgalactiae |
| Sulfa/TMP | R_Dairy | 5.2 | 17.2 | 0 | 7 | S. dysgalactiae |
| Sulfa/TMP | R + I_Dairy | 5.6 | 15 | 0.3 | 7.1 | S. dysgalactiae |
| 3GC | R_Beef/Veal | 1.2 | 9.1 | 0 | 3.1 | H. somni |
| 3GC | R_Mixed/Unknown | 0 | 0 | 0 | 0 | H. somni |
| Florfenicol | R_Beef/Veal | 1.1 | 1.3 | 0 | 0.5 | H. somni |
| Florfenicol | R_Mixed/Unknown | 0 | 0 | 0 | 0 | H. somni |
| Florfenicol | R + I_Beef/Veal | 6.4 | 7.5 | 0 | 2.7 | H. somni |
| Fluoroquinolones | R_Beef/Veal | 4.6 | 8.3 | 4 | 1.5 | H. somni |
| Fluoroquinolones | R_Mixed/Unknown | 1.6 | 8 | 0 | 3.2 | H. somni |
| Penicillin | R_Mixed/Unknown | 39.5 | 65.2 | 0 | 32.1 | H. somni |
| Tetracyclines | R_Beef/Veal | 48.9 | 54.7 | 9.1 | 15.3 | H. somni |
| Tetracyclines | R_Mixed/Unknown | 33.2 | 73.9 | 1.9 | 27 | H. somni |
| Tilmicosin | R_Beef/Veal | 16.1 | 18.7 | 0 | 6.5 | H. somni |
| Tilmicosin | R_Mixed/Unknown | 16 | 28 | 0 | 9.7 | H. somni |
| Tulathromycin | R_Mixed/Unknown | 11 | 27.1 | 0 | 11.2 | H. somni |
| 3GC | R_Dairy | 6.3 | 7.8 | 0 | 3.1 | K. pneumoniae |
| 3GC | R + I_Dairy | 0.6 | 1 | 0 | 0.5 | K. pneumoniae |
| Amox/Clav | R_Dairy | 8.9 | 22.1 | 0 | 10.9 | K. pneumoniae |
| Colistin | R_Dairy | 1.1 | 1.3 | 0 | 0.5 | K. pneumoniae |
| Fluoroquinolones | R_Dairy | 3.7 | 6.5 | 0 | 2.4 | K. pneumoniae |
| Fluoroquinolones | R + I_Dairy | 3.4 | 8 | 0 | 4 | K. pneumoniae |
| Neomycin | R + I_Dairy | 0 | 0 | 0 | 0 | K. pneumoniae |
| Sulfa/TMP | R_Dairy | 4.9 | 11.7 | 0 | 5 | K. pneumoniae |
| 3GC | R_Beef/Veal | 1.6 | 11.8 | 0.9 | 2.7 | M. haemolytica |
| 3GC | R_Mixed/Unknown | 0 | 0 | 0 | 0 | M. haemolytica |
| 3GC | R + I_Mixed/Unknown | 1.1 | 1.1 | 1 | 0 | M. haemolytica |
| Aminopenicillins | R_Dairy | 14.5 | 22.2 | 14 | 2.1 | M. haemolytica |
| Aminopenicillins | R_Mixed/Unknown | 20.5 | 39 | 4.3 | 10.2 | M. haemolytica |
| Aminopenicillins | R + I_Mixed/Unknown | 7.3 | 10 | 4.7 | 2.7 | M. haemolytica |
| Florfenicol | R_Beef/Veal | 7.2 | 47.1 | 4.3 | 10.8 | M. haemolytica |
| Florfenicol | R_Mixed/Unknown | 3.7 | 34.7 | 0 | 10.1 | M. haemolytica |
| Florfenicol | R + I_Beef/Veal | 7.5 | 8.6 | 0 | 2.9 | M. haemolytica |
| Florfenicol | R + I_Mixed/Unknown | 0.8 | 2 | 0 | 0.8 | M. haemolytica |
| Fluoroquinolones | R_Beef/Veal | 2.8 | 3 | 0 | 0.8 | M. haemolytica |
| Fluoroquinolones | R_Dairy | 10.8 | 11 | 7.4 | 0.9 | M. haemolytica |
| Fluoroquinolones | R_Mixed/Unknown | 11.1 | 56.4 | 0 | 16.8 | M. haemolytica |
| Gentamicin | R_Dairy | 9.5 | 16.7 | 9 | 1.9 | M. haemolytica |
| Penicillin | R_Dairy | 20 | 33.3 | 19 | 3.6 | M. haemolytica |
| Penicillin | R_Mixed/Unknown | 16.8 | 24.4 | 1.6 | 9 | M. haemolytica |
| Tetracyclines | R_Beef/Veal | 54.4 | 64.7 | 53.6 | 2.8 | M. haemolytica |
| Tetracyclines | R_Dairy | 29.1 | 30 | 16.7 | 3.3 | M. haemolytica |
| Tetracyclines | R_Mixed/Unknown | 30.7 | 78.1 | 4.2 | 23.2 | M. haemolytica |
| Tetracyclines | R + I_Mixed/Unknown | 18.4 | 25 | 9.1 | 7.6 | M. haemolytica |
| Tilmicosin | R_Beef/Veal | 46.4 | 76.5 | 44.2 | 8.1 | M. haemolytica |
| Tilmicosin | R_Mixed/Unknown | 23.6 | 84.4 | 1.2 | 24.8 | M. haemolytica |
| Tulathromycin | R_Dairy | 10.4 | 11 | 1.9 | 2.3 | M. haemolytica |
| Tulathromycin | R_Mixed/Unknown | 23.9 | 76.6 | 0 | 22.6 | M. haemolytica |
| Tylosin | R_Dairy | 93.4 | 99 | 14.8 | 21.1 | M. haemolytica |
| 3GC | R_Beef/Veal | 1.6 | 7.1 | 0.9 | 1.9 | P. multocida |
| 3GC | R_Mixed/Unknown | 0.1 | 1.9 | 0 | 0.4 | P. multocida |
| 3GC | R + I_Mixed/Unknown | 0.4 | 1.3 | 0 | 0.6 | P. multocida |
| Aminopenicillins | R_Mixed/Unknown | 27 | 63.5 | 1.4 | 27.4 | P. multocida |
| Aminopenicillins | R + I_Mixed/Unknown | 4.4 | 6.6 | 1 | 2.4 | P. multocida |
| Florfenicol | R_Beef/Veal | 3 | 14.3 | 1.7 | 3.9 | P. multocida |
| Florfenicol | R_Mixed/Unknown | 1.8 | 12.7 | 0 | 3.9 | P. multocida |
| Florfenicol | R + I_Beef/Veal | 9 | 11.9 | 0 | 5.1 | P. multocida |
| Florfenicol | R + I_Mixed/Unknown | 2.5 | 12.2 | 0 | 4.7 | P. multocida |
| Fluoroquinolones | R_Beef/Veal | 0 | 0 | 0 | 0 | P. multocida |
| Fluoroquinolones | R_Mixed/Unknown | 0.8 | 6.7 | 0 | 1.7 | P. multocida |
| Fluoroquinolones | R + I_Mixed/Unknown | 3.9 | 9 | 0 | 4.5 | P. multocida |
| Penicillin | R_Mixed/Unknown | 7.8 | 30.5 | 0 | 12.8 | P. multocida |
| Penicillin | R + I_Mixed/Unknown | 2.8 | 5 | 2.1 | 1.3 | P. multocida |
| Tetracyclines | R_Beef/Veal | 55.8 | 57.1 | 55.6 | 0.5 | P. multocida |
| Tetracyclines | R_Mixed/Unknown | 28.9 | 80 | 3.2 | 24.3 | P. multocida |
| Tetracyclines | R + I_Mixed/Unknown | 16.4 | 37 | 0 | 13.7 | P. multocida |
| Tilmicosin | R_Beef/Veal | 42 | 42.9 | 41.9 | 0.3 | P. multocida |
| Tilmicosin | R_Mixed/Unknown | 20 | 43.3 | 12 | 13.7 | P. multocida |
| Tulathromycin | R_Mixed/Unknown | 16.8 | 80.9 | 0.5 | 26.1 | P. multocida |
| Florfenicol | R_Beef/Veal | 18.9 | 25.7 | 3.1 | 9.1 | Mycoplasma bovis |
| Fluoroquinolones | R_Beef/Veal | 16.4 | 41.2 | 0 | 15.4 | Mycoplasma bovis |
| Fluoroquinolones | R_Mixed/Unknown | 1.6 | 4 | 0 | 2 | Mycoplasma bovis |
| Tetracyclines | R_Beef/Veal | 63.4 | 80.1 | 0 | 25.1 | Mycoplasma bovis |
| Tetracyclines | R_Mixed/Unknown | 75 | 100 | 50 | 25.1 | Mycoplasma bovis |
| Tilmicosin | R_Beef/Veal | 93.2 | 99 | 40.6 | 16.6 | Mycoplasma bovis |
| Tilmicosin | R_Mixed/Unknown | 97.5 | 100 | 95 | 2.5 | Mycoplasma bovis |
| Tulathromycin | R_Beef/Veal | 89.4 | 92 | 83.5 | 3.9 | Mycoplasma bovis |
| Tulathromycin | R_Mixed/Unknown | 50.8 | 100 | 27 | 34.3 | Mycoplasma bovis |
| Tylosin | R_Beef/Veal | 87 | 97.7 | 40.6 | 20.8 | Mycoplasma bovis |
| Tylosin | R + I_Mixed/Unknown | 67 | 98 | 56.2 | 18.3 | Mycoplasma bovis |
| 3GC (Cefoperazone) | R_Dairy | 4.8 | 5 | 4 | 0.4 | S. aureus |
| 3GC (Cefoperazone) | R + I_Dairy | 15.2 | 36.1 | 0 | 10.4 | S. aureus |
| 3GC (Ceftiofur) | R_Dairy | 3.3 | 41.5 | 0 | 10.2 | S. aureus |
| 3GC (Ceftiofur) | R + I_Dairy | 1.1 | 16 | 0 | 3.8 | S. aureus |
| Erythromycin | R_Dairy | 18 | 79.9 | 0 | 23.7 | S. aureus |
| Erythromycin | R_Mixed/Unknown | 3.8 | 22.1 | 0 | 8.1 | S. aureus |
| Erythromycin | R + I_Dairy | 23.1 | 62 | 0 | 16.1 | S. aureus |
| Fluoroquinolones | R_Dairy | 12.8 | 53.4 | 0 | 16.9 | S. aureus |
| Fluoroquinolones | R_Mixed/Unknown | 0 | 0 | 0 | 0 | S. aureus |
| Fluoroquinolones | R + I_Dairy | 14 | 36.9 | 0 | 14.7 | S. aureus |
| Methicillin | R_Dairy | 10.8 | 60.7 | 0 | 14.5 | S. aureus |
| Methicillin | R_Mixed/Unknown | 4.4 | 13.7 | 0 | 6.4 | S. aureus |
| Neomycin | R_Dairy | 2.9 | 6.3 | 0 | 2.1 | S. aureus |
| Neomycin | R + I_Dairy | 17.5 | 18.1 | 8.9 | 2.2 | S. aureus |
| Penicillin | R_Dairy | 52.5 | 97.1 | 6.4 | 29.4 | S. aureus |
| Penicillin | R_Mixed/Unknown | 17.5 | 26 | 12.4 | 6.6 | S. aureus |
| Penicillin | R + I_Dairy | 38.4 | 83.3 | 4 | 19.1 | S. aureus |
| Penicillin‐novobiocin | R_Dairy | 0.8 | 1.7 | 0 | 0.9 | S. aureus |
| Pirlimycin | R_Dairy | 17.2 | 41 | 0 | 18.5 | S. aureus |
| Pirlimycin | R + I_Dairy | 1.8 | 1.9 | 0 | 0.4 | S. aureus |
| Sulfa/TMP | R_Dairy | 23.4 | 91.8 | 0 | 32.9 | S. aureus |
| Sulfa/TMP | R_Mixed/Unknown | 0 | 0 | 0 | 0 | S. aureus |
| Sulfa/TMP | R + I_Dairy | 0.7 | 4 | 0 | 0.8 | S. aureus |
| 3GC | R_Mixed/Unknown | 11.5 | 71.9 | 0 | 22.8 | T. pyogenes |
| Aminopenicillins | R_Mixed/Unknown | 28.9 | 53.1 | 0 | 17.7 | T. pyogenes |
| Aminopenicillins | R + I_Dairy | 2.9 | 6 | 0 | 3 | T. pyogenes |
| Erythromycin | R + I_Dairy | 26.1 | 44 | 9.9 | 17.1 | T. pyogenes |
| Penicillin | R_Mixed/Unknown | 28.8 | 65.6 | 4.6 | 17.9 | T. pyogenes |
| Penicillin | R + I_Dairy | 3.8 | 8 | 0 | 4 | T. pyogenes |
| Sulfa/TMP | R_Dairy | 82.9 | 90 | 78.1 | 5.9 | T. pyogenes |
| Tetracyclines | R_Dairy | 56.2 | 68.7 | 50.7 | 8.3 | T. pyogenes |
| Tetracyclines | R_Mixed/Unknown | 48.5 | 63.7 | 10.8 | 24 | T. pyogenes |
| Tetracyclines | R + I_Dairy | 78.1 | 85.4 | 70 | 7.7 | T. pyogenes |
| 3GC (Cefoperazone) | R + I_Dairy | 5.8 | 6 | 2.6 | 0.8 | S. uberis |
| 3GC (Ceftiofur) | R + I_Dairy | 3.6 | 13 | 3.2 | 2 | S. uberis |
| Erythromycin | R_Dairy | 15.2 | 18.8 | 5.7 | 4.4 | S. uberis |
| Erythromycin | R + I_Dairy | 13.3 | 25 | 0 | 4 | S. uberis |
| Fluoroquinolones | R_Dairy | 0.5 | 0.7 | 0 | 0.3 | S. uberis |
| Penicillin | R_Dairy | 0.3 | 1.4 | 0 | 0.5 | S. uberis |
| Penicillin | R + I_Dairy | 9.1 | 44 | 3.9 | 5.9 | S. uberis |
| Pirlimycin | R + I_Dairy | 18.6 | 26 | 17.9 | 2 | S. uberis |
| Spiramycin | R_Dairy | 15.3 | 15.7 | 7.7 | 1.7 | S. uberis |
| Spiramycin | R + I_Dairy | 27.2 | 36.2 | 18 | 9.1 | S. uberis |
| Sulfa/TMP | R_Dairy | 2.9 | 12.7 | 0 | 5.3 | S. uberis |
| Sulfa/TMP | R + I_Dairy | 12.6 | 22 | 4.9 | 8.4 | S. uberis |
Suggested citation: EFSA AHAW Panel (EFSA Panel on Animal Health and Welfare) , Nielsen SS, Bicout DJ, Calistri P, Canali E, Drewe JA, Garin‐Bastuji B, Gonzales Rojas JL, Gortazar Schmidt C, Herskin M, Michel V, Miranda Chueca MA, Padalino B, Pasquali P, Roberts HC, Spoolder H, Stahl K, Velarde A, Viltrop A, Winckler C, Dewulf J, Guardabassi L, Hilbert F, Mader R, Baldinelli F and Alvarez J, 2021. Scientific Opinion on the assessment of animal diseases caused by bacteria resistant to antimicrobials: cattle. EFSA Journal 2021;19(12):6955, 89 pp. 10.2903/j.efsa.2021.6955
Requestor: European Commission
Question number: EFSA‐Q‐2021‐00576
Panel members: Søren Saxmose Nielsen, Julio Alvarez, Dominique Joseph Bicout, Paolo Calistri, Elisabetta Canali, Julian Ashley Drewe, Bruno Garin‐Bastuji, Jose Luis Gonzales Rojas, Christian Gortazar Schmidt, Mette Herskin, Virginie Michel, Miguel Angel Miranda Chueca, Barbara Padalino, Paolo Pasquali, Helen Clare Roberts, Hans Spoolder, Karl Stahl, Antonio Velarde, Arvo Viltrop and Christoph Winckler.
Declarations of interest: The declarations of interest of all scientific experts active in EFSA's work are available at https://ess.efsa.europa.eu/doi/doiweb/doisearch.
Acknowledgements: The AHAW Panel wishes to thank Peter Damborg, Carmen Espinosa‐Gongora, Steffen Lynge Jørgensen from the University of Copenhagen for conducting the extensive literature review under the contract OC/EFSA/ALPHA/2020/02 – LOT 1; Alberto Antoine Díez Guerrier from University Complutense Madrid, Raphael Guatteo from Oniris, Bart Pardon from Ghent University and Verena Oswaldi from EFSA for the support provided for this scientific output.
Adopted: 22 September 2021
This article was originally published on the EFSA website http://www.efsa.europa.eu on 3 November 2021 as part of EFSA’s publication procedures.
Notes
References
- Aasmäe B, Häkkinen L, Kaart T and Kalmus P, 2019. Antimicrobial resistance of Escherichia coli and Enterococcus spp. isolated from Estonian cattle and swine from 2010 to 2015. Acta Veterinaria Scandinavica, 61, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANRESIS ARCH‐Vet (Federal Office of Public Health and Federal Food Safety and Veterinary Office), 2020. Swiss Antibiotic Resistance Report 2020. Usage of Antibiotics and Occurrence of Antibiotic Resistance in Bacteria from Humans and Animals in Switzerland. Available online: https://www.anresis.ch/wp-content/uploads/2020/11/Swiss-Antibiotic-Resistance-Report-2020_def_WEB.pdf
- Awosile BB, Heider LC, Saab ME and McClure JT, 2018. Antimicrobial resistance in mastitis, respiratory and enteric bacteria isolated from ruminant animals from the Atlantic Provinces of Canada from 1994‐2013. Canadian Veterinary Journal, 59, 1099–1104. [PMC free article] [PubMed] [Google Scholar]
- Bokma J, Gille L, De Bleecker K, Callens J, Haesebrouck F, Pardon B and Boyen F, 2020. Antimicrobial susceptibility of mycoplasma bovis isolates from veal, dairy and beef herds. Antibiotics, 9, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsaglia ECR, Silva NCC, Rossi BF, Camargo CH, Dantas STA, Langoni H, Guimaraes FF, Lima FS, Fitzgerald JR, Fernandes AJ and Rall VLM, 2018. Molecular epidemiology of methicillin‐susceptible Staphylococcus aureus (MSSA) isolated from milk of cows with subclinical mastitis. Microbial Pathogenesis, 124, 130–135. 10.1016/j.micpath.2018.08.031 [DOI] [PubMed] [Google Scholar]
- Booker CW, 2020. Bovine respiratory disease treatment failure: definition and impact. Animal Health Research Reviews, 21, 172–174. 10.1017/S146625232000016X [DOI] [PubMed] [Google Scholar]
- Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY and Calavas D, 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône‐Alpes, France. Foodborne Pathog Dis, 7, 479–487. [DOI] [PubMed] [Google Scholar]
- Cengiz S and Adiguzel MC, 2020. Determination of virulence factors and antimicrobial resistance of E. coli isolated from calf diarrhea, part of eastern Turkey. Ankara Universitesi Veteriner Fakultesi Dergisi, 67, 365–371. [Google Scholar]
- Cheng J, Qu W, Barkema HW, Nobrega DB, Gao J, Liu G, De Buck J, Kastelic JP, Sun H and Han B, 2019. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. Journal of Dairy Science, 102, 2416–2426. [DOI] [PubMed] [Google Scholar]
- Coetzee JF, Magstadt DR, Sidhu PK, Follett L, Schuler AM, Krull AC, Cooper VL, Engelken TJ, Kleinhenz MD and O'Connor AM, 2019. Association between antimicrobial drug class for treatment and retreatment of bovine respiratory disease (BRD) and frequency of resistant BRD pathogen isolation from veterinary diagnostic laboratory samples. PLoS ONE, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa GM, Paiva LV, Figueiredo HC, Figueira AR, Pereira UP and Silva N, 2012. Population diversity of Staphylococcus aureus isolated from bovine mastitis in Brazilian dairy herds. Research in Veterinary Science, 93, 733–735. 10.1016/j.rvsc.2011.09.014 [DOI] [PubMed] [Google Scholar]
- Cummings KJ, Aprea VA and Altier C, 2014. Antimicrobial resistance trends among Escherichia coli isolates obtained from dairy cattle in the northeastern United States, 2004–2011. Foodborne Pathog Dis, 11, 61–67. [DOI] [PubMed] [Google Scholar]
- Dorneles EMS, Fonseca M, Abreu JAP, Lage AP, Brito M, Pereira CR, Brandão HM, Guimarães AS and Heinemann MB, 2019. Genetic diversity and antimicrobial resistance in Staphylococcus aureus and coagulase‐negative Staphylococcus isolates from bovine mastitis in Minas Gerais. Brazil. Microbiologyopen, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorso L, Rouault M, Barbotin C, Chartier C and Assié S, 2021. Infectious bovine respiratory diseases in adult cattle: an extensive necropsic and etiological study. Animals, 11, 2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA AHAW Panel (EFSA Panel on Animal Health and Welfare), Nielsen SS, Bicout DJ, Calistri P, Canali E, Drewe JA, Garin‐Bastuji B, Gonzales Rojas JL, Gortazar Schmidt C, Herskin M, Michel V, Miranda Chueca MA, Padalino B, Pasquali P, Roberts HC, Sihvonen LH, Spoolder H, Stahl K, Velarde A, Viltrop A, Winckler C, Dewulf J, Guardabassi L, Hilbert F, Mader R, Smith P, Aznar I, Baldinelli F and J A, 2021. Ad hoc method for the assessment of animal diseases caused by bacteria resistant to antimicrobials. EFSA Journal 2021;19(6):6645, 29 pp. 10.2903/j.efsa.2021.6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Garch F, de Jong A, Simjee S, Moyaert H, Klein U, Ludwig C, Marion H, Haag‐Diergarten S, Richard‐Mazet A, Thomas V and Siegwart E, 2016. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Veterinary Microbiology, 194, 11–22. 10.1016/j.vetmic.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Elias L, Balasubramanyam AS, Ayshpur OY, Mushtuk IU, Sheremet NO, Gumeniuk VV, Musser JMB and Rogovskyy AS, 2020. Antimicrobial susceptibility of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli isolated from mastitic dairy cattle in ukraine. Antibiotics (Basel), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA (European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption), 2020. Sales of veterinary antimicrobial agents in 31 European countries in 2018; Tenth ESVAC report. EMA/24309/2020. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2018-trends-2010-2018-tenth-esvac-report_en.pdf
- EUCAST , 2017. EUCAST guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance. Version 2.0 Edition. Place.
- FINRES‐Vet (Finnish Food Authority), 2018. Finnish Veterinary Antimicrobial Resistance Monitoring and Consumption of Antimicrobial Agents. 2669‐8307, Helsinki, Finland. Available online: https://www.ruokavirasto.fi/globalassets/viljelijat/elaintenpito/elainten-laakitseminen/antibioottiresistenssin_seuranta/finres-vet_2018_141119.pdf
- Gautier‐Bouchardon AV, Ferré S, Le Grand D, Paoli A, Gay E and Poumarat F, 2014. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE, 9, e87672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERM‐Vet , 2020. Bericht zur Resistenzmonitoringstudie 2018 Resistenzsituation bei klinisch wichtigen tierpathogenen Bakterien. Berlin, Germany, (BVL) BfVuL. Available online: https://www.bvl.bund.de/SharedDocs/Berichte/07_Resistenzmonitoringstudie/Bericht_Resistenzmonitoring_2018.pdf?__blob=publicationFile&v=4
- Goldspink LK, Mollinger JL, Barnes TS, Groves M, Mahony TJ and Gibson JS, 2015. Antimicrobial susceptibility of Histophilus somni isolated from clinically affected cattle in Australia. Vet J, 203, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni M, Saras E and Madec J‐Y, 2010. Demonstration of a shift towards penicillin resistance in the Streptococcus uberis population. Journal of Medical Microbiology, 59, 993–995. [DOI] [PubMed] [Google Scholar]
- Horpiencharoen W, Thongratsakul S and Poolkhet C, 2019. Risk factors of clinical mastitis and antimicrobial susceptibility test results of mastitis milk from dairy cattle in western Thailand: Bayesian network analysis. Prev Vet Med, 164, 49–55. 10.1016/j.prevetmed.2019.01.014 [DOI] [PubMed] [Google Scholar]
- Intorre L, Vanni M, Cerni D, Arrigoni N, Cammi G and Garbarino CA, 2012. Antimicrobial resistance of Staphylococcus aureus isolated from bovine milk in Italy. Journal of Veterinary Pharmacology and Therapeutics, 35, 108–109. [Google Scholar]
- Khalil D, Becker CAM and Tardy F, 2017. Monitoring the decrease in susceptibility to ribosomal RNAs targeting antimicrobials and its molecular basis in clinical mycoplasma Bovis isolates over time. Microbial Drug Resistance, 23, 799–811. [DOI] [PubMed] [Google Scholar]
- Kong LC, Gao D, Gao YH, Liu SM and Ma HX, 2014. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. Journal of Veterinary Medical Science, 76, 1655–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm CG, Love BC, Krehbiel CR, Johnson NJ and Step DL, 2012. Comparison of antemortem antimicrobial treatment regimens to antimicrobial susceptibility patterns of postmortem lung isolates from feedlot cattle with bronchopneumonia. Journal of Veterinary Diagnostic Investigation, 24, 277–282. [DOI] [PubMed] [Google Scholar]
- van Leenen K, Jouret J, Demeyer P, Van Driessche L, De Cremer L, Masmeijer C, Boyen F, Deprez P and Pardon B, 2020. Associations of barn air quality parameters with ultrasonographic lung lesions, airway inflammation and infection in group‐housed calves. Preventive Veterinary Medicine, 181. [DOI] [PubMed] [Google Scholar]
- van Leenen K, Jouret J, Demeyer P, Vermeir P, Leenknecht D, Van Driessche L, De Cremer L, Masmeijer C, Boyen F and Deprez P, 2021. Particulate matter and airborne endotoxin concentration in calf barns and their association with lung consolidation, inflammation, and infection. Journal of Dairy Science, 104, 5932–5947. [DOI] [PubMed] [Google Scholar]
- Loy JD and Brodersen BW, 2014. Moraxella spp. isolated from field outbreaks of infectious bovine keratoconjunctivitis: a retrospective study of case submissions from 2010 to 2013. Journal of Veterinary Diagnostic Investigation, 26, 761–768. [DOI] [PubMed] [Google Scholar]
- Mader R, Damborg P, Amat J‐P, Bengtsson B, Bourély C, Broens EM, Busani L, Crespo‐Robledo P, Filippitzi M‐E and Fitzgerald W, 2021. Building the European Antimicrobial Resistance Surveillance network in veterinary medicine (EARS‐Vet). Eurosurveillance, 26, 2001359. 10.2807/1560-7917.ES.2021.26.4.2001359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowski E, Lassa H, Markiewicz H, Kaptur M, Nadolny M, Niewitecki W and Zietara J, 2011. Sensitivity to antibiotics of Arcanobacterium pyogenes and Escherichia coli from the uteri of cows with metritis/endometritis. Vet J, 187, 234–238. 10.1016/j.tvjl.2009.12.010 [DOI] [PubMed] [Google Scholar]
- Maunsell F, Woolums A, Francoz D, Rosenbusch R, Step D, Wilson DJ and Janzen E, 2011. Mycoplasma bovis infections in cattle. Journal of Veterinary Internal Medicine, 25, 772–783. [DOI] [PubMed] [Google Scholar]
- McDougall S, Hussein H and Petrovski K, 2014. Antimicrobial resistance in Staphylococcus aureus, Streptococcus uberis and Streptococcus dysgalactiae from dairy cows with mastitis. New Zealand Veterinary Journal, 62, 68–76. [DOI] [PubMed] [Google Scholar]
- NCAS (National Centre for Antimicrobial Stewardship), 2017. Australian Veterinary prescribing Guidelines ‐ Cattle. Available online: https://cpb-ap-se2.wpmucdn.com/blogs.unimelb.edu.au/dist/d/257/files/2017/06/Bovine_flipbook-1ck2kcl.pdf
- Nefedchenko AV, Glotova TI and Glotov AG, 2019. Isolation and antimicrobial resistance of mannheimia haemolytica on dairy farms in Siberia. Bulgarian Journal of Veterinary Medicine, 22, 428–438. [Google Scholar]
- NZVA (New Zealand Veterinary Association), 2018. Antibiotic judicious use guidelines for the New Zealand veterinary profession | Dairy. Wellington, New Zealand. Available online: https://www.amrvetcollective.com/assets/guidelines/guide_dairy.pdf
- RESAPATH (ANSES) , 2020. French surveillance network for antimicrobial resistance in bacteria from diseased animals 2018 Annual report. Available online: https://www.anses.fr/fr/system/files/LABO-Ra-Resapath2018EN.pdf
- Rezanejad M, Karimi S and Momtaz H, 2019. Phenotypic and molecular characterization of antimicrobial resistance in Trueperella pyogenes strains isolated from bovine mastitis and metritis. BMC Microbiology, 19, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger F, Ruf J, Tschuor A, Sigrist Y, Rosskopf M and Hässig M, 2014. Antimicrobial susceptibility of mastitis pathogens of dairy cows in Switzerland. Schweizer Archiv fur Tierheilkunde, 156, 483–488. [DOI] [PubMed] [Google Scholar]
- Saidani M, Messadi L, Soudani A, Daaloul‐Jedidi M, Châtre P, Ben Chehida F, Mamlouk A, Mahjoub W, Madec JY and Haenni M, 2018. Epidemiology, antimicrobial resistance, and extended‐spectrum beta‐lactamase‐producing enterobacteriaceae in clinical Bovine Mastitis in Tunisia. Microb Drug Resist, 24, 1242–1248. [DOI] [PubMed] [Google Scholar]
- Schmidt T, 2011. In vitro antimicrobial susceptibility of Staphylococcus aureus strains from dairy herds in KwaZulu‐Natal. Journal of the South African Veterinary Association, 82, 76–79. [DOI] [PubMed] [Google Scholar]
- Simoes J, Branco M, Andrade J and Muller A, 2020. Phenotypic antimicrobial susceptibility of environmental bacteria from mastitic milk of pastured dairy cows of S. Miguel (Azores). Tropical Animal Health and Production, 52, 407–414. [DOI] [PubMed] [Google Scholar]
- Srednik ME, Usongo V, Lépine S, Janvier X, Archambault M and Gentilini ER, 2018. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. Journal of Dairy Research, 85, 57–63. [DOI] [PubMed] [Google Scholar]
- Thomas V, De Jong A, Simjee S, Maher K, Morrissey I, Butty P, Klein U, Marion H, Moyaert H, Rigaut D and Valle M, 2012. Antimicrobial susceptibility of mastitis pathogens isolated from diseased dairy cows across Europe: VetPath monitoring results. Clinical Microbiology and Infection, 18, 264–265. [Google Scholar]
- Timsit E, Hallewell J, Booker C, Tison N, Amat S and Alexander TW, 2017. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Veterinary Microbiology, 208, 118–125. 10.1016/j.vetmic.2017.07.013 [DOI] [PubMed] [Google Scholar]
- UK‐VARSS , 2019. Supplementary Material l (UK‐VARSS 2018). New Haw, Addlestone: Veterinary Medicines Directorate. Available online: www.gov.uk/government/collections/veterinary-antimicrobial-resistance-and-sales-surveillance
- Van Driessche L, Bokma J, Gille L, Ceyssens PJ, Sparbier K, Haesebrouck F, Deprez P, Boyen F and Pardon B, 2018. Rapid detection of tetracycline resistance in bovine Pasteurella multocida isolates by MALDI Biotyper antibiotic susceptibility test rapid assay (MBT‐ASTRA). Scientific Reports, 8, 13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wald R, Hess C, Urbantke V, Wittek T and Baumgartner M, 2019. Characterization of Staphylococcus species isolated from bovine quarter milk samples. Animals (Basel), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kong L‐C, Jia B‐Y, Liu S‐M and Jiang X‐Y, 2017. Aminoglycoside susceptibility of Pasteurella multocida isolates from bovine respiratory infections in China and mutations in ribosomal protein S5 associated with high‐level induced spectinomycin resistance. Journal of Veterinary Medical, Science, 17–0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li J, Qiao M, Meng D, Meng Q, Qiao J, Zhang X, Wang L, Cai K, Zhang J, Zhang Z, Yu W and Cai X, 2019. Characteristic profiles of biofilm, enterotoxins and virulence of Staphylococcus aureus isolates from dairy cows in Xinjiang Province. China. J Vet Sci, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastempowska E and Lassa H, 2012. Genotypic characterization and evaluation of an antibiotic resistance of Trueperella pyogenes (Arcanobacterium pyogenes) isolated from milk of dairy cows with clinical mastitis. Veterinary Microbiology, 161, 153–158. 10.1016/j.vetmic.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Zhang DX, Tian K, Han LM, Wang QX, Liu YC, Tian CL and Liu MC, 2014. Resistance to beta‐lactam antibiotic may influence nanH gene expression in Trueperella pyogenes isolated from bovine endometritis. Microbial Pathogenesis, 71–72, 20–24. 10.1016/j.micpath.2014.04.006 [DOI] [PubMed] [Google Scholar]


