Abstract
Neurodegenerative diseases are becoming major socio-economic burdens. However, most of them still have no effective treatment. Growing evidence indicates excess exposure to pesticides are involved in the development of various forms of neurodegenerative and neurological diseases through trigger epigenetic changes and inducing disruption of the epigenome. This review summaries studies on epigenetics alterations in nervous systems in relation to different kinds of pesticides, highlighting potential mechanism in the etiology, precision prevention and target therapy of various neurodegenerative diseases. In addition, the current gaps in research and future areas for study were also discussed.
Keywords: Neurodegenerative disease, Epigenetic, Nervous system, Environmental pollution, Pesticide, DNA methylation, Histone modifications, Non-coding RNA, RNA methylation, N6-methyladenosine
Introduction
With aging of the population in worldwide, neurodegenerative diseases, especially Alzheimer’s disease (AD) and Parkinson’s disease (PD) are becoming major socio-economic burdens. Their increasing prevalence and without effective treatment mean these diseases will be a challenge for future generations [1]. Although several cellular mechanisms and genes have been proved implicated in the onset and progression of the disease, the precise molecular underpinnings of these diseases remain unclear [2].
Epigenetics is generally defined as a heritable change in gene function, which can influence gene expression and subsequent protein expression levels without altering DNA sequence [3]. The epigenetic molecular factors include DNA methylation [4], histone modifications [5], non-coding RNA [6], chromatin structure [7], and RNA methylation [8]. Epigenetic dysregulation may induce the development of neurological disorders like Parkinson’s disease, Huntington’s disease, and mood disorders (including depression and anxiety) [9]. Growing evidence indicates that environmental neurotoxicants are involved in the development of various forms of neurodegenerative and neurological diseases through trigger epigenetic changes and inducing disruption of the epigenome [10–13]. Some sources of environmental pollutants were related to neurotoxic manifestations, such as metals, pesticides, solvent and some other environmental pollutions [14–16]. Chemicals could regulate gene expression by influencing gene transcription, mRNA degradation and translation, etc. Abnormal changes in DNA or RNA methylation, non-coding RNA, histone modification can serve as biomarkers for environmental pollutant-induced neurotoxicity [17–21]. Consequently, insight into the epigenetic mechanisms by which environmental contaminants causes neurotoxicity is key to modelling targeted preventions and treatments [22].
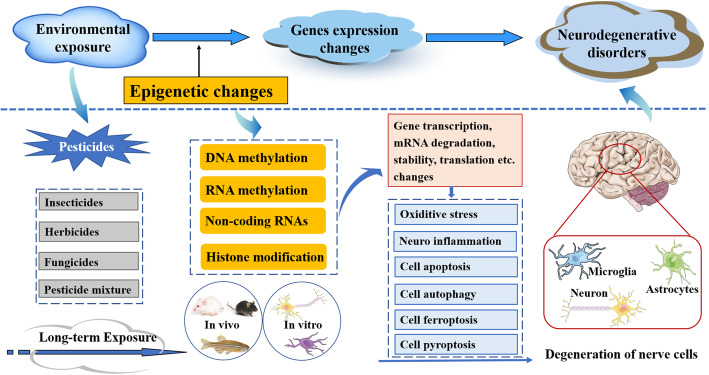
Pesticides, including insecticides, herbicides and fungicides, are widely used in agriculture for preventing, destroying, repelling or mitigating harmful or unwanted insects, weeds and fungi. However, most pesticides are not highly selective and generally toxic to many nontarget species, including humans [23]. Particularly, insecticides, which kill insects by disrupting their nervous system, exert neurotoxic effects in humans as well. Neurotoxicity can be induced upon high acute exposure, or by chronic exposure at low doses. Multiple studies have proved chronic exposure to pesticides at a low dose is a risk factor for the development of neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (ALS), and attention deficit hyperactivity disorder (ADHD) etc. [24–30]. However, nowadays there are still about 25 million workers experience unintentional pesticide poisoning each year, due to inhalation or skin absorption [31]. Therefore, it is very important to study the pathogenic mechanism and preventive measures of neurodegenerative and neurological diseases induced by pesticides. For the past few years, many researchers have concerned the potential role of epigenetic mechanism in pesticide induced neurotoxicity (as shown in Fig. 1).
Fig. 1.
Schematic representation of the mechanism of epigenetic alterations and neurodegenerative disorders due to exposure to different kind of pesticides
Here we review details studies on epigenetics alterations in nervous systems in relation to pesticides, highlighting potential mechanism in the etiology of various neurodegenerative diseases (summarized Table 1). We also highlight current gaps and future areas for the studies upon epigenetic neurotoxicity induced by pesticides.
Table 1.
Summary of epigenetic changes induced by pesticisdes
| Pesticides | Model | Exposure route | Exposure dose | Exposure length | Neurological damage | Epigenetic change | Ref. |
|---|---|---|---|---|---|---|---|
| Insecticides | |||||||
| Chlorpyrifos | Long Evans rats | Subcutaneous injection | 3 or 10 mg/kg | 21 days | Associated with problems in cognitive function. | MiR-132 and miR-212 in the CPF-exposed rat hippocampus were up-regulated. | [32] |
| SH-SY5Y cells | Cell culture | 0, 25, 50, 100, and 200 μM | 24 h | Cell viability was decreased. Pyroptosis related proteins, ROS levels, as well as level of caspase-1 and the TUNEL positive cells were all significantly up-regulated. | Expression of miR-181 was enhanced. | [33] | |
| Chlorpyrifos-oxon | Zebrafish | Embryonic exposure | 0.01 and 50 μg/L | 4–120 h post fertilization | Even low dose exposures can have transgenerational effects in neurological activity. | Induce alterations in global DNA methylation. | [34] |
| Dieldrin | C57BL/6 mice | Oral intake | 0.3 mg/kg/3 days | 30 days | Induce impairment of dopaminergic neuron development and maintenance. | Dieldrin-induced differential methylation was sex-specific. | [35] |
| C57BL/6 J mice | Intraperitoneal injection | 5.0 mg/kg every other day | 30 days | Induce apoptotic cell death in dopaminergic neuronal Cells. | Induce a time-dependent increase in the acetylation of core histones H3 and H4. | [36] | |
| Permethrin | Wistar rats | Oral intake | 34.05 mg/kg | From PND6 to PND21 | Early life permethrin exposure in rats, at a dose close to No Observed Adverse Effect Level (NOAEL) during neonatal brain development leads to its accumulation long after exposure. | DNMT1, DNMT3a were increased. The aggregation of DNMT3b and α-synuclein was enhanced. | [37]. |
| Wistar rats | Oral intake | 34.05 mg/kg | From PND6 to PND21 | Low dosage exposure to permethrin during neonatal brain development leads to dopamine decrease in rat striatum nucleus, oxidative stress and behavioural changes linked to the development of Parkinson’s like neurodegeneration later in life. | Global 5mC and 5hmC levels were increased. Methylation levels at H3K9me3 position at both Th and Nurr1 promoter regions were reduced. | [38] | |
| Wistar rats | Oral intake | 34.05 mg/kg | From PND6 to PND21 | An intergenerational permethrin-induced damage on progenies has been identified. | Global genome-wide DNA methylation was decreased in mothers exposed in early life to permethrin as well as in their offspring. | [39] | |
| Wistar rats | Oral intake | 34.05 mg/kg | From PND6 to PND21 | Parental exposure leads to a significant reduction in dopamine level in the offspring (F1) born from parents or just mothers early-life treated. | Early-life exposure to the stressor is associated with changes in global DNA methylation and hydroxymethylation in adult age. | [40] | |
| Paraquat | N27 dopaminergic cells | Cell culture | 400 mM | 12, 24 or 36 h. | Cell culture models of Parkinson’s disease. | Histone H3 acetylation was induced in a time dependent manner. | [41] |
| hNPCs | Cell culture | 0, 5, 10, 20, 40, or 80 μM | 24 h | Induce developmental neurotoxicity. | Induced differentially miRNAs expression. | [42] | |
| hNPCs | Cell culture | 10 and 100 μM | 24 h | Lead to the alteration of several neurodevelopment related key biological processes and crucial pathways. | Alter mRNAs and miRNAs expression. | [43] | |
| hNPCs | Cell culture | 0, 25, 50 and 100 μM | 24 h | PQ dramatically suppressed neural cell differentiation ability. | Direct binding effect between CTNNB1 and miR-200a existed following PQ exposure. | [44] | |
| Neuro-2a cells | Cell culture | 100 μM | 48 h | CaM and p21 were involved in PQ-induced toxicity. | Nrf2-regulated miR-380-3p inhibited cell proliferation and enhanced the PQ-induced toxicity in N2a cells potentially by blocking the translation Sp3 mRNA. | [45] | |
| SH-SY5Y cells | Cell culture | 50 μM | 24 h | Impairs Nrf2/ARE defense network. | Cause miR153 to bind to and target Nrf2 3′ UTR thereby weakening the cellular antioxidant Defense. | [46] | |
| Neuro-2a cells | Cell culture | 300 | 48 h | Cell culture models of Parkinson’s disease. | The expression of miR-17-5p was downregulated, DNA methylation level was upregulated after PQ exposure. | [17] | |
| Neuro-2a cells | Cell culture | 300 | 48 h | Induce Parkinson’s disease pathology. | The miR-17-5p was expressed at lower levels in PQ-treated Neuron-2a cells, overexpression of miR17-5p in Neuro-2a cells could enhance cell proliferation, suppresse apoptosis and promote S phase transition of the cell cycle after PQ treatment. | [47] | |
| ICR mice | Intraperitoneal injection | 5, 10 mg/kg/2 days | 10 times | Animal models of Parkinson’s disease. | PQ caused lncRNA expression profiling alteration in the substantia nigra (SN) through an interaction with Nrf2, thus changing the NR_027648/Zc3h14/Cybb and NR_030777/Zfp326/Cpne5 mRNA pathways. | [48] | |
| ICR mice | Intraperitoneal injection | 5 mg/kg/3 days, 10 mg/kg/2 days, | 10 times | Inhibited microglia and dopaminergic cells proliferation and microglia migration. | PQ-induced low expression of AK039862 rescued microglia proliferation and migration inhibition via the AK039862/Pafah1b1/Foxa1 pathway, AK039862 also participated in the interaction between microglia and dopaminergic cells with PQ treatment. | [49] | |
| BV2 cells and MN9d cells | Co-culture | 0, 50, 100, 150 μM | 36 h | ||||
| Neuro-2a cells | Cell culture | 100, 300 μM | 24, 36 h | Induce neurotoxicity | LncRNA NR_030777 has a vital protective role by regulating the expression of Zfp326 and Cpne 5 in neurotoxicity induced by PQ. | [50] | |
| MN9D cells | |||||||
| Primary cortical neuron | |||||||
| Neuro-2a cells | Cell culture | 200 μM | 3 h | Induce oxidative stress | m6A participated in a specific regulatory network of circRNAs to modulate the expression of downstream genes in response to PQ-induced oxidative stress | [51] | |
| Avermectin | Pigeons | Oral intake | 20, 40, 60 mg/kg | 30, 60, 90 days | AVM exhibits significant cytotoxicity in pigeon brain nerve cells. | Global DNA hypomethylation and down-regulation of DNMT mRNA expression occurred in a dose-time-dependent manner in pigeon brains. | [52] |
| Fipronil | Zebrafish | Embryonic exposure | 40 μg/L | From 6 to 96 h post fertilization | R-fipronil exhibited more intense neurotoxicity compared with S-fipronil. | The fipronil-conducted enantioselective neurotoxicity likely applied its enantioselectivity by the dysregulation of DNA methylation. | [53] |
| Atrazine (ATR) | Carp | Embryonic exposure | 4.28, 42.8 and 428 μg/L | 40 days | Not mentioned. | The MBD2 mRNA expression was up-regulated in the brain, the DNMTs mRNA expression was down-regulated | [54] |
| Chlorpyrifos (CPF) | 1.16, 11.6 and 116 μg/L | ||||||
| Combined ATR/CPF | 1.13, 11.3 and 113 μg/L | ||||||
| Deltamethrin | C57BL/6 N mice | Oral intake | 3 mg/kg | During gestation, lactation, and weaning at postnatal day (PND) 21 | Deltamethrin insecticide and stress exposure during neurodevelopment leads to alterations in dopamine function (PND21–60). | Hypermethylation of Nr3c1 is in response to dual deltamethrin and corticosterone exposures during development. | [55] |
| Corticosterone | Drinking water | 25 μg/mL | From adolescence through adulthood | ||||
Mechanism of epigenetics
DNA methylation
DNA methylation is an epigenetic mechanism involving the transfer of a methyl group onto the C5 position of the cytosine to form 5-methylcytosine [56]. Methyl groups to cytosine are added by the DNA methyltransferase (DNMT) enzyme family, namely DNMT3A, DNMT3B, and DNMT1 [57–59]. DNA methylation is the most well studied epigenetic regulators in relation to environmental exposures, which can result in altered global and gene-specific DNA methylation [60]. In the nervous system, neuronal activity can also modulate DNA methylation in response to physiological and environmental stimuli [61, 62].
RNA methylation
In addition to genomic DNA modifications, various modifications of nucleosides which form the basis for RNA (including tRNA, rRNA, mRNA etc.) were also appreciated [63–65]. N6-methyladenosine (m6A) in mRNA has been the best-characterized mRNA modification so far, with roles in modulating mRNA transcript processing and regulation [66, 67]. The m6A effectors include “writers”, “erasers” and “readers” that respectively install, remove and interpret the methylation [68]. The ‘writer’ methyltransferase enzymes (including METTL3, METTL14, WTAP, and KIAA1429) add a methyl group to the N6 position of adenosine in RNA, and removed by the “erasers” demethylases (including FTO and ALKBH5). The methyl group can be recognized by “reader” proteins (HNRNPC, HNRNPA2B1, YTHDF2, YTHDF1, and eIF3), and influence almost all steps of RNA metabolism, including mRNA translation, degradation, splicing, export and folding, consequently altering target gene expression, influencing the corresponding cell processes and physiological function [69, 70].
Histone modification
Histones (H3, H4, H2A, H2B and H1) are the most abundant proteins in the eukaryotic nuclear DNA structure [71]. Most of the amino acids reside on the N-terminal tails of the histone proteins are subjected to modifications, including acetylation, methylation and ubiquitination on lysine, methylation and citrullination on arginine, etc. [72]. Histone modification can influence all DNA-based processes, including chromatin compaction, nucleosome dynamics, and transcription [73]. Moreover, histone core modifications can also directly regulate transcription, and influence processes of DNA repair, replication, stemness, and changes in cell state [74]. In general, active enhancers are related to the enrichments of both monomethylated H3K4 (H3K4me1) and H3K27ac. Gene bodies of actively transcribed genes are associated with trimethylated H3K36 (H3K36me3), and transcription start sites of actively transcribed genes can be identified by trimethylated H3K4 (H3K4me3) and acetylated H3K27 (H3K27ac) [72].
Non-coding RNAs
Non-coding RNAs (ncRNA) are a relatively recently described but significant subpopulation of the transcriptome [75]. According to their size, non-coding RNAs can be classified to short RNAs which are < 200 nucleotides (nts) in length and long non-coding RNAs (lncRNAs) are longer than 200 nts. Short non-coding RNAs include PIWI-interacting RNAs (piRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), circular RNA (circRNA) and small nuclear RNAs (snRNAs). In the last decade, gene expression has been proved to be largely regulated by ncRNAs. A growing number of studies have investigated the involvement of ncRNAs in various physiological processes [75]. Moreover, miRNAs, lncRNAs and circRNAs have been proved to be involved in transcriptional regulation at different level. Therefore, ncRNA is a burgeoning area for all the biology fields, including neurotoxicology [76].
Pesticide induced epigenetic neurotoxic effect
Insecticides
Organophosphate pesticide
Organophosphate pesticide has been known as the most widely used pesticides during the past half century. Number of literatures regard their association with neurodegenerative and neurodevelopmental disorders with respect to epigenetic mechanisms [77, 78].
Chlorpyrifos (CPF) is one of the most widely used organophosphorus pesticide (OP) in the world. However, CPE and its active metabolite chlorpyrifos-oxon (CPO) have been proved involving in several neurodevelopmental disorders. Lee’s work identifies the potential epigenetic mechanism of hlorpyrifos neurotoxicity, founding that miR132/212 was elevated in the CA1 hippocampal region, disrupting the neurotrophin mediated cognitive processes after CPF administration [32]. Zhao’s team observed chlorpyrifos could activate cell pyroptosis and increases susceptibility on oxidative stress-induced toxicity by miR-181/SIRT1/PGC-1α/Nrf2 signaling pathway [33]. In addition, the adverse effects of developmental exposure to the active CPO have been proved to persist into adulthood even future generations [79]. Schmitt et al. demonstrated that early life stage exposures to CPO can lead to epigenetic changes in neurological activity, which may lead to alterations in response to CPO in future generations [34]. Liu’ team also investigated H3K4me3 and DNA methylation levels of the PPARγ gene in the placenta was associated with prenatal chlorpyrifos exposure, and could effect on birth outcomes and neurodevelopment [80].
Organochlorine pesticide
Organochlorine pesticide is another kind of insecticides. Nowadayes, some OCPs are banned in most industrialized countries. However, due to the very long half-life in humans, the circulating levels of the breakdown product of dchlorodiphenyltrichloroethane (DDT), p,p′-DDE (1, 1-dichloro2, 2-bis (p-chlorophenyl) ethylene) are still found in almost all humans in the industrialized world. Moreover, some OCPs, such as DDT is still used to combat malaria in Asia and Africa [81]. Researches in epidemiology showed workers with Parkinson’s disease were related with exposure to organochlorines [77].
DDT is a common environmental organochlorine pesticide with a long half-life. Because it can passes through the placental barrier, DDT exposure during pregnancy may heavily influence lifetime health of the offspring.. An epidemiological study showed that prenatal exposure to DDT is associated with fetal genome-wide DNA methylation [82]. Another cohort study also investigated prenatal exposure to persistent organic pollutants can induce DNA methylation of LINE-1 and imprint genes in placenta [83].
Dieldrin is also a highly toxic organochlorine pesticide. Although it was phased out of commercial use in the 1970s, dieldrin was still persisted in the environment and easily accumulated in lipid-rich tissues like the brain, due to its high stability and lipophilicity [84]. Previous epidemiology studies have shown a positive association between dieldrin exposure and PD [85, 86]. Experimental researches also investigated that developmental dieldrin exposure could alter DNA methylation at genes related to dopaminergic neuron development and Parkinson’s disease [35]. Song’ research revealed that dieldrin induced a time-dependent increase in the acetylation of core histones H3 and H4 in neuronal cells. Moreover, the histone acetylation appeared within 10 min after exposure to dieldrin, suggesting that acetylation is an early event in dieldrin neurotoxicity [36].
Pyrethroid
Pyrethroid insecticides contain natural pyrethrins which are extracted from pyrethrum flowers, and their synthetic derivatives, pyrethroids [87]. Pyrethroids with low mammalian toxicity are now commonly used for household and post-harvest insect control [88]. However, pyrethroid pesticide exposures may be also associated with disruption of neurological functioning [89–92]. Neonatal exposure to permethrin can induce a Parkinson-like disease.
Permethrin is a synthetic pyrethroid widely used as anti-woodworm agent and for indoor and outdoor pest control. However, early life permethrin exposure induces long-term brain changes [93]. These long-term changes were regulated by early impairment of epigenetic pattern in neurodegeneration, such as DNA methylation or histone alterations [37, 38]. In addition, on the prospective intergenerational effect of this pesticide, parental exposure also leads to global DNA methylation changes and hydroxymethylation impairment in their offspring, providing pivotal evidences on intergenerational effects of postnatal exposure to permethrin [39, 40]. Tevoltage-gated sodium channel (VGSC), a motoneuronal transport protein, is the target of all pyrethroids [94]. In addition, Kubik et al. found miRNA-33 modulates permethrin induced toxicity by regulating VGSC transcripts [95].
Herbicide
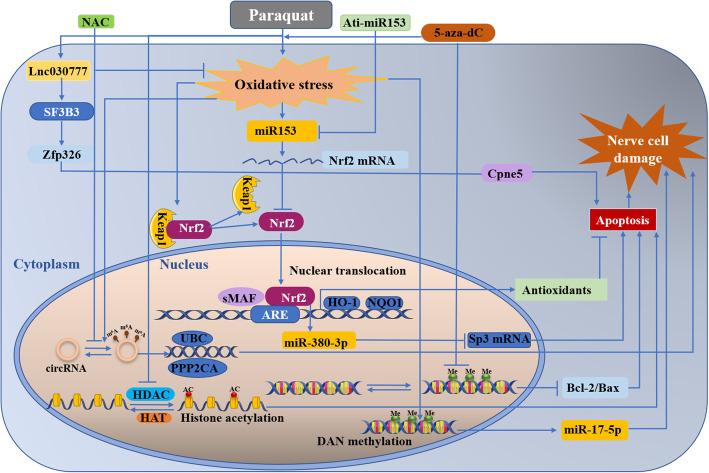
Herbicides are essential tool in weed management. The first commercial herbicides were released in the 1940s and hundreds more since then [96]. Among all the herbicides, the neurotoxicity paraquat (PQ) are probably the most conclusive [97]. Paraquat exposure has been linked to an increased risk for Parkinson’s disease [98], and has been used for modeling sporadic Parkinson’s Disease [99]. However, the impacts of PQ exposure on the central nervous system remain unclear. In recent years, epigenetic mechanisms involved paraquat induced neurons damage have been extensively investigated in order to explore new preventive and therapeutic targets of Parkinson’s disease. Our team also do a lot of exploration there [17, 30, 45, 47–51, 100]. The epigenetic molecular mechanisms were summarized in Fig. 2.
Fig. 2.
Epigenetic regulation in the context ofparaquat induced nerve cell damage. DAN methylation, m6A modification of circRNA,histone acetylation, miRNA, and lncRNAs were involved in the epigenetic regulation process. The details of the molecular mechanisms were descripted in the following text
As shown in Fig. 2, DNA methylations were found to be causative factors in paraquat induced neurons damage. When pretreated with methyltransferase inhibitor 5′-aza-dC, the level of reactive oxygen species (ROS) increased more significantly. The rate of bcl-2/bax decreased, consequently enhancing the cell apoptosis induced by PQ. This work demonstrated the interaction of DNA methylation and paraquat, providing additional new insights into the pathogenic mechanisms of PD [101].
A major epigenetic change in chromatin can regulate gene expression. Song et al. found that exposure to paraquat could induced histone H3 acetylation in N27 dopaminergic cells in a time dependent manner. These changes were associated with decreased total histone deacetylase (HDAC) activity and HDAC4 and 7 protein expression levels. Additionally, the inhibition of histone acetyltransferase (HAT) activity by anacardic acid can protect against apoptotic cell death induced by PQ, indicating that histone acetylation may represent key epigenetic changes during neurotoxic insults [41].
Evidence indicates that miRNA also play a key role in PQ involved neurodegenerative diseases. Zhou’s team analyzed the impacts of PQ on the miRNome of human neural progenitor cells (hNPCs) during proliferation. Upon PQ treatment, 66 miRNAs were identified as differentially expressed in proliferating hNPCs. With further analysis, the target genes were found enriched in regulation of cell proliferation and differentiation, cell cycle and apoptosis as well as tumor protein 53 (p53), Wnt, Notch and mitogen-activated protein kinases (MAPK) signaling pathways [42]. Furtherly, they found mRNAs and miRNAs expression changes induced by PQ in hNPCs could lead to the alteration of several neurodevelopment related key biological processes and crucial pathways, especially Wnt signaling pathway. The results suggested that PQ could downregulate Wnt signaling pathway via miRNA to induce developmental neurotoxicity [43]. In addition, they also found miR-200a was down regulated in the PQ treated neural stem cell. This process subsequently decreased cell viability, increased epithelial-mesenchymal transition process and the inhibited differential through CTNNB1 pathway [44]. The transcription factor Nrf2 has been proved to regulate the expression of many miRNAs, our previous work also found that Nrf2 can promote the expression of miR-380-3p, which blocks the translation of Sp3 protein and enhances paraquat-induced toxicity in mouse neuroblastoma N2a cells [45]. Interestingly, Narasimhan et al. explained the relationship between Nrf2 and miRNA from a different perspective. Their study showed that PQ induced miR153 dependent hydrogen peroxide (H2O2), and then caused miR153 to bind to and target Nrf2 3′ UTR thereby weakening the cellular antioxidant defense [46].
Emerging evidence showing that oxidative stress and DNA methylation can alter miRNA expression. Our previous work also investigated that PQ can induce DNA methylation variations through ROS production, leading to the downregulation of miR-17-5p expression [17]. The downregulation of miR-17-5p expression contributes to PQ-induced dopaminergic neurodegeneration through influencing the cell proliferation, cell apoptosis and S phase transition of the cell cycle [47]. Subsequently, we found long noncoding RNAs also play a crucial part in PQ induced neurotoxicity. The alteration of the lncRNA expression profile were found in PQ treated mouse, suggesting lncRNAs were involved in PQ- induced neurotoxicity. Further studies displayed PQ caused lncRNA expression profiling alteration in the substantia nigra (SN) through an interaction with Nrf2, thus changing the NR_027648/Zc3h14/ Cybb and NR_030777/Zfp326/Cpne5 mRNA pathways [48]. Latterly, we proved PQ-induced low expression of AK039862 rescued microglia proliferation and migration inhibition via the AK039862/Pafah1b1/Foxa1 pathway [49]. LncRNA NR_030777 is another lncRNA involved in PQ induced neurotoxicity. It is highly conserved among species and was firstly confirmed by our team. Evidence illustrated NR_030777 7 has a vital protective role in neurotoxicity induced by PQ through regulating the expression of Zfp326 and Copine 5 [50]. These novel discoveries suggested lncRNAs could be a potential target for the prevention and treatment of PQ-induced neurodegenerative disorders such as PD. Recently, our team successfully established a positive link between the alteration of circRNAs driven by m6A modification and PQ-induced oxidative stress. Moreover, the alteration of m6A methylated circRNAs upon PQ exposure could be partially reversed by N-acetylcysteine (NAC) pretreatment, providing a new preventive or therapeutic tools for PQ-induced neurodegenerative disorders [51].
Other pesticides
In addition to the pesticides described above, there are still some other pesticides, such as biological insecticides, combination of various pesticides and some other new low toxicity chemical pesticides. The epigenetic mechanisms of their neurotoxicity were also investigated.
Avermectin
Avermectin (AVM) is an effective insecticidal and nematicidal agent, it has been extensively used in agriculture and stock farming areas. Subsequently, the residues of AVM or its active metabolites present a toxic threat through epigenetic mechanisms. Cao et al. reported subchronic exposure to AVM could decrease the mRNA expression levels of DNA methyltransferases (DNMT1 and DNMT3a/3b), and increase the mRNA levels of demethylase methyl-CpG-binding domain protein 2 (MBD2), consequently down regulated the global DNA methylation level decreased in pigeon brain tissues [52].
Fipronil
Fipronil is a broad-spectrum chiral insecticide. It has been documented to induce significant neurotoxicity to nontarget aquatic species. Qian et al. firstly discussed the enantioselective toxicity of chiral pesticide fipronil in central nervous system, and find R-fipronil exhibited more intense neurotoxicity compared with S-fipronil. Further research revealed that R-fipronil disrupted five signaling pathways including MAPK, Calcium signaling, Neuroactive ligand-receptor interaction, Purine metabolism, and Endocytosis. These pathways revealed greater extent than S-fipronil through the hypermethylation of several important neuro-related genes. This study indicated that the fipronil-conducted enantioselective neurotoxicity by the dysregulation of DNA methylation, providing a novel epigenetic insight into the study of enantioselective biological effects [74].
Pesticide mixture
Nowadays, mixed formulation of pesticide is commonly used to reduce insect resistance, achieve-control efficiency and/or economic concers [102]. Xing et al. examined the effect of atrazine (ATR), chlorpyrifos (CPF) and combined ATR/CPF exposure on DNA methylation in the brain of zebra fish. Compared to the control fish, a significant global DNA hypomethylation was observed in the common carp exposed to ATR, CPF and their mixture. Moreover, the MBD2 mRNA expression was up-regulated and the DNMTs mRNA expression was down-regulated in the brain and gonad of the common carp exposed to ATR, CPF and their mixture [75]. Nuclear receptor subfamily 3 group C member 1 (Nr3c1) is a transcription factors necessary for proper dopamine neuron development. Vester et al. also reported combined neurodevelopmental exposure to deltamethrin and corticosterone could induce Nr3c1 hypermethylation in the midbrain of adult male mice [76].
Concluding remarks
The underlying cause of many neurological and neurodegenerative diseases are still unclear. However, it is now widely accepted that the environmental toxicants contribute to many of these disorders. Increasing evidence indicates that the epigenome may be targeted on people at risk for neurodegenerative disorders. Multiple studies have indicated chronic exposure to pesticides at a low dose could increase the risk of neurodegenerative diseases. Epigenetic disruptions induced by pesticides, which can induce neurotoxicity were summarized in this work. Epidemiologic studies and experimental researches performed both in vivo and in vitro provide evidence that exposure to pesticides even at a low dose have long-term effects on central nervous systems, possibly via epigenetic modifications. Individuals, exposure to neurotoxic in the early life and even in the womb could induce epigenetic alterations in adulthood, the aged or even next generations, moreover some of the alterations are sex-depended. Meanwhile, compared with genetic changes, epigenetic aberrations could be more easily reversible. Drugs, even some nutrients, which target the specific epigenetic mechanisms involved in the regulation of gene expression could be emerging preventive or therapeutic tools for disease. In this review, there are some evidences illustrated inhibits the epigenetic modification changes by compounds or regulates the expression of non-coding RNA by genetic tools might alleviate pesticide induced neurological and neurodegenerative impairments. These will shed new light on the precise prevention and targeted treatment for the neurodegenerative diseases. However, a great deal of further investigations is still needed. For instance (1) explore how pesticides exposure leads to epigenetic alteration in specific genes; (2) identify the susceptible genes of human in the epigenetic alterations induced by exposure to pesticides; (3) prove whether epigenetic changes can be used as biomarkers for the early detection and treatment target for neurodegenerative disorders; (4) develop novel epigenetic modification inhibitors; (5) explore the epigenetic mechanisms of some novel and mixtures of pesticides.
Acknowledgements
Not applicable.
Abbreviations
- AD
Alzheimer’s disease
- ADHD
Attention deficit/hyperactivity disorder
- ALS
Amyotrophic lateral sclerosis
- AVM
Avermectin
- circRNA
Circular RNA
- CNS
Central nervous system
- CPF
Chlorpyrifos
- CPO
Chlorpyrifos-oxon
- DDT
Dchlorodiphenyltrichloroethane
- DNMT
DNA methyltransferase
- HAT
Histone acetyltransferases
- HDACs
Histone deacetylases
- hNPCs
Human neural progenitor cells
- lncRNAs
Long non-coding RNAs
- m6A
N6-methyladenosine
- MAPK
Mitogen-activated protein kinases
- MBD2
Methyl-CpG-binding domain protein 2
- miRNA
Micrornas
- ncRNA
Non-coding RNAs
- NAC
N-acetylcysteine
- OP
Organophosphorus pesticide
- PD
Parkinson’s disease,
- piRNAs
PIWI-interacting RNAs
- PQ
Paraquat
- ROS
Reactive oxygen species
- siRNAs
Small interfering RNAs
- SN
Substantia nigra
- snRNAs
Small nuclear RNAs
Authors’ contributions
G. Yu wrote the manuscript. Q. Su, Y. Chen and L. Wu collected information. Prof. S. Wu and H. Li designed and guided the work. The author(s) read and approved the final manuscript.
Funding
The financial support from the National Natural Science Foundation of China (grant number: 81973083, 81903352, 22006019, 82173553), the Joint Funds for the innovation of science and Technology, Fujian province (grant number: 2017Y9105, 2018Y9098, 2019Y9020), the Provincial Natural Science Foundation of Fujian Province (key project) (grant number: 2020 J02021),the Provincial Natural Science Foundation of Fujian Province (grant number: 2019 J05081), Natural Science Foundation of Zhejiang Province (grant number: LQ19H260002), Ningbo Natural Science Foundation (grant number: 2018A610317).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have approved the publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Siying Wu, Email: fmuwsy@163.com.
Huangyuan Li, Email: lhy@fjmu.edu.cn.
References
- 1.Dugger BN, Dickson DW. Pathology of Neurodegenerative Diseases. Cold Spring Harb Perspect Biol. 2017;9(7). 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed]
- 2.Pavlou MAS, Outeiro TF. Epigenetics in Parkinson's disease. Adv Exp Med Biol. 2017;978:363–390. doi: 10.1007/978-3-319-53889-1_19. [DOI] [PubMed] [Google Scholar]
- 3.Inbar-Feigenberg M, Choufani S, Butcher DT, Roifman M, Weksberg R. Basic concepts of epigenetics. Fertil Steril. 2013;99(3):607–615. doi: 10.1016/j.fertnstert.2013.01.117. [DOI] [PubMed] [Google Scholar]
- 4.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187(4173):226–232. doi: 10.1126/science.187.4173.226. [DOI] [PubMed] [Google Scholar]
- 5.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54(1):21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaniv M. Chromatin remodeling: from transcription to cancer. Cancer Genet. 2014;207(9):352–357. doi: 10.1016/j.cancergen.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24(15):1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8(1):57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babenko O, Kovalchuk I, Metz GA. Epigenetic programming of neurodegenerative diseases by an adverse environment. Brain Res. 2012;1444:96–111. doi: 10.1016/j.brainres.2012.01.038. [DOI] [PubMed] [Google Scholar]
- 11.Berson A, Nativio R, Berger SL, Bonini NM. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41(9):587–598. doi: 10.1016/j.tins.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coppedè F, Mancuso M, Siciliano G, Migliore L, Murri L. Genes and the environment in neurodegeneration. Biosci Rep. 2006;26(5):341–367. doi: 10.1007/s10540-006-9028-6. [DOI] [PubMed] [Google Scholar]
- 13.Brown RC, Lockwood AH, Sonawane BR. Neurodegenerative diseases: an overview of environmental risk factors. Environ Health Perspect. 2005;113(9):1250–1256. doi: 10.1289/ehp.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin-Chan M, Navarro-Yepes J, Quintanilla-Vega B. Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front Cell Neurosci. 2015;9:124. doi: 10.3389/fncel.2015.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqubal A, Ahmed M, Ahmad S, Sahoo CR, Iqubal MK, Haque SE. Environmental neurotoxic pollutants: review. Environ Sci Pollut Res Int. 2020;27(33):41175–41198. doi: 10.1007/s11356-020-10539-z. [DOI] [PubMed] [Google Scholar]
- 16.Yu G, Guo Z, Li H. Analyses of epigenetic modification in environmental pollutants-induced neurotoxicity. Methods Mol Biol. 2021;2326:123–141. doi: 10.1007/978-1-0716-1514-0_9. [DOI] [PubMed] [Google Scholar]
- 17.Zhan Y, Guo Z, Zheng F, Zhang Z, Li K, Wang Q, Wang L, Cai Z, Chen N, Wu S, Li H. Reactive oxygen species regulate miR-17-5p expression via DNA methylation in paraquat-induced nerve cell damage. Environ Toxicol. 2020;35(12):1364–1373. doi: 10.1002/tox.23001. [DOI] [PubMed] [Google Scholar]
- 18.Forster VJ, McDonnell A, Theobald R, McKay JA. Effect of methotrexate/vitamin B (12) on DNA methylation as a potential factor in leukemia treatment-related neurotoxicity. Epigenomics. 2017;9(9):1205–1218. doi: 10.2217/epi-2016-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neal M, Richardson JR. Epigenetic regulation of astrocyte function in neuroinflammation and neurodegeneration. Biochim Biophys Acta Mol basis Dis. 2018;1864(2):432–443. doi: 10.1016/j.bbadis.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logan S, Jiang C, Yan Y, Inagaki Y, Arzua T, Bai X. Propofol alters long non-coding RNA profiles in the neonatal mouse Hippocampus: implication of novel mechanisms in anesthetic-induced developmental neurotoxicity. Cell Physiol Biochem. 2018;49(6):2496–2510. doi: 10.1159/000493875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu C, Wen Y, Wu L, Wang Y, Wu Q, Wang D, Wang Y, Liu Q, Zhang J. Arsenite-induced transgenerational glycometabolism is associated with up-regulation of H3K4me2 via inhibiting spr-5 in caenorhabditis elegans. Toxicol Lett. 2020;326:11–17. doi: 10.1016/j.toxlet.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Jose CC, Wang Z, Tanwar VS, Zhang X, Zang C, Cuddapah S. Nickel-induced transcriptional changes persist post exposure through epigenetic reprogramming. Epigenetics Chromatin. 2019;12(1):75. doi: 10.1186/s13072-019-0324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa LG, Giordano G, Guizzetti M, Vitalone A. Neurotoxicity of pesticides: a brief review. Front Biosci. 2008;13(13):1240–1249. doi: 10.2741/2758. [DOI] [PubMed] [Google Scholar]
- 24.Paul KC, Chuang YH, Cockburn M, Bronstein JM, Horvath S, Ritz B. Organophosphate pesticide exposure and differential genome-wide DNA methylation. Sci Total Environ. 2018;645:1135–1143. doi: 10.1016/j.scitotenv.2018.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan D, Zhang Y, Liu L, Shi N, Yan H. Pesticide exposure and risk of Parkinson's disease: dose-response meta-analysis of observational studies. Regul Toxicol Pharmacol. 2018;96:57–63. doi: 10.1016/j.yrtph.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Martins R, Carruthers M. Testosterone as the missing link between pesticides, Alzheimer disease, and Parkinson disease. JAMA Neurol. 2014;71(9):1189–1190. doi: 10.1001/jamaneurol.2014.795. [DOI] [PubMed] [Google Scholar]
- 27.Richardson JR, Roy A, Shalat SL, von Stein RT, Hossain MM, Buckley B, Gearing M, Levey AI, German DC. Elevated serum pesticide levels and risk for Alzheimer disease. JAMA Neurol. 2014;71(3):284–290. doi: 10.1001/jamaneurol.2013.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernard A. Elevated serum DDE and risk for Alzheimer disease. JAMA Neurol. 2014;71(8):1055–1056. doi: 10.1001/jamaneurol.2014.432. [DOI] [PubMed] [Google Scholar]
- 29.Mostafalou S, Abdollahi M. The link of organophosphorus pesticides with neurodegenerative and neurodevelopmental diseases based on evidence and mechanisms. Toxicology. 2018;409:44–52. doi: 10.1016/j.tox.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Chen N, Guo Z, Luo Z, Zheng F, Shao W, Yu G, Cai P, Wu S, Li H. Drp1-mediated mitochondrial fission contributes to mitophagy in paraquat-induced neuronal cell damage. Environ Pollut. 2021;272:116413. doi: 10.1016/j.envpol.2020.116413. [DOI] [PubMed] [Google Scholar]
- 31.van der Plaat DA, de Jong K, de Vries M, van Diemen CC, Nedeljković I, Amin N, Kromhout H, Vermeulen R, Postma DS, van Duijn CM, Boezen HM, Vonk JM. Occupational exposure to pesticides is associated with differential DNA methylation. Occup Environ Med. 2018;75(6):427–435. doi: 10.1136/oemed-2017-104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YS, Lewis JA, Ippolito DL, Hussainzada N, Lein PJ, Jackson DA, Stallings JD. Repeated exposure to neurotoxic levels of chlorpyrifos alters hippocampal expression of neurotrophins and neuropeptides. Toxicology. 2016;340:53–62. doi: 10.1016/j.tox.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao MW, Yang P, Zhao LL. Chlorpyrifos activates cell pyroptosis and increases susceptibility on oxidative stress-induced toxicity by miR-181/SIRT1/PGC-1α/Nrf2 signaling pathway in human neuroblastoma SH-SY5Y cells: implication for association between chlorpyrifos and Parkinson's disease. Environ Toxicol. 2019;34(6):699–707. doi: 10.1002/tox.22736. [DOI] [PubMed] [Google Scholar]
- 34.Schmitt C, Peterson E, Willis A, Kumar N, McManus M, Subbiah S, Crago J. Transgenerational effects of developmental exposure to chlorpyrifos-oxon in zebrafish (DANIO RERIO) Toxicol Appl Pharmacol. 2020;408:115275. doi: 10.1016/j.taap.2020.115275. [DOI] [PubMed] [Google Scholar]
- 35.Kochmanski J, VanOeveren SE, Patterson JR, Bernstein AI. Developmental dieldrin exposure alters DNA methylation at genes related to dopaminergic neuron development and parkinson's disease in mouse midbrain. Toxicol Sci. 2019;169(2):593–607. doi: 10.1093/toxsci/kfz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77(4):621–632. doi: 10.1124/mol.109.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fedeli D, Montani M, Bordoni L, Galeazzi R, Nasuti C, Correia-Sá L, Domingues VF, Jayant M, Brahmachari V, Massaccesi L, Laudadio E, Gabbianelli R. In vivo and in silico studies to identify mechanisms associated with Nurr1 modulation following early life exposure to permethrin in rats. Neuroscience. 2017;340:411–423. doi: 10.1016/j.neuroscience.2016.10.071. [DOI] [PubMed] [Google Scholar]
- 38.Bordoni L, Nasuti C, Fedeli D, Galeazzi R, Laudadio E, Massaccesi L, López-Rodas G, Gabbianelli R. Early impairment of epigenetic pattern in neurodegeneration: additional mechanisms behind pyrethroid toxicity. Exp Gerontol. 2019;124:110629. doi: 10.1016/j.exger.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Bordoni L, Nasuti C, Mirto M, Caradonna F, Gabbianelli R. Intergenerational effect of early life exposure to permethrin: changes in global DNA methylation and in nurr1 gene expression. Toxics. 2015;3(4):451–461. doi: 10.3390/toxics3040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bordoni L, Nasuti C, Di Stefano A, Marinelli L, Gabbianelli R. Epigenetic memory of early-life parental perturbation: dopamine decrease and dna methylation changes in offspring. Oxidative Med Cell Longev. 2019;2019:1472623–1472611. doi: 10.1155/2019/1472623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song C, Kanthasamy A, Jin H, Anantharam V, Kanthasamy AG. Paraquat induces epigenetic changes by promoting histone acetylation in cell culture models of dopaminergic degeneration. Neurotoxicology. 2011;32(5):586–595. doi: 10.1016/j.neuro.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M, Lou D, Cai Q, Chang X, Wang X, Zhou Z. Characterization of paraquat-induced miRNA profiling response in hNPCs undergoing proliferation. Int J Mol Sci. 2014;15(10):18422–18436. doi: 10.3390/ijms151018422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan M, Dou T, Lv W, Wang X, Zhao L, Chang X, Zhou Z. Integrated analysis of paraquat-induced microRNAs-mRNAs changes in human neural progenitor cells. Toxicol in Vitro. 2017;44:196–205. doi: 10.1016/j.tiv.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Huang M, Lou D, Wang YP, Cai Q, Li HH. Paraquat inhibited differentiation in human neural progenitor cells (hNPCs) and down regulated miR-200a expression by targeting CTNNB1. Environ Toxicol Pharmacol. 2016;42:205–211. doi: 10.1016/j.etap.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 45.Cai Z, Zheng F, Ding Y, Zhan Y, Gong R, Li J, Aschner M, Zhang Q, Wu S, Li H. Nrf2-regulated miR-380-3p blocks the translation of Sp3 protein and its mediation of paraquat-induced toxicity in mouse neuroblastoma N2a cells. Toxicol Sci. 2019;171(2):515–529. doi: 10.1093/toxsci/kfz162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narasimhan M, Riar AK, Rathinam ML, Vedpathak D, Henderson G, Mahimainathan L. Hydrogen peroxide responsive miR153 targets Nrf2/ARE cytoprotection in paraquat induced dopaminergic neurotoxicity. Toxicol Lett. 2014;228(3):179–191. doi: 10.1016/j.toxlet.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q, Zhan Y, Ren N, Wang Z, Zhang Q, Wu S, Li H. Paraquat and MPTP alter microRNA expression profiles, and downregulated expression of miR-17-5p contributes to PQ-induced dopaminergic neurodegeneration. J Appl Toxicol. 2018;38(5):665–677. doi: 10.1002/jat.3571. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Yang H, Wang Q, Zhang Q, Wang Z, Zhang Q, Wu S, Li H. Paraquat and MPTP induce alteration in the expression profile of long noncoding RNAs in the substantia nigra of mice: role of the transcription factor Nrf2. Toxicol Lett. 2018;291:11–28. doi: 10.1016/j.toxlet.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Shao W, Wu J, Huang S, Yang H, Luo Z, Zheng F, Wang YL, Cai P, Guo Z, Wu S, Li H. Inflammatory lncRNA AK039862 regulates paraquat-inhibited proliferation and migration of microglial and neuronal cells through the Pafah1b1/Foxa1 pathway in co-culture environments. Ecotoxicol Environ Saf. 2021;208:111424. doi: 10.1016/j.ecoenv.2020.111424. [DOI] [PubMed] [Google Scholar]
- 50.Yang H, Lin Q, Chen N, Luo Z, Zheng C, Li J, Zheng F, Guo Z, Cai P, Wu S, Wang YL, Li H. LncRNA NR_030777 alleviates Paraquat-induced neurotoxicity by regulating Zfp326 and Cpne5. Toxicol Sci. 2020;178(1):173–188. doi: 10.1093/toxsci/kfaa121. [DOI] [PubMed] [Google Scholar]
- 51.Chen N, Tang J, Su Q, Chou WC, Zheng F, Guo Z, Yu G, Shao W, Li H, Wu S. Paraquat-induced oxidative stress regulates N6-methyladenosine (m (6) a) modification of circular RNAs. Environ Pollut. 2021;290:117816. doi: 10.1016/j.envpol.2021.117816. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y, Chen LJ, Zhang ZW, Yao HD, Liu C, Li S, Xu SW. Global DNA hypomethylation: a potential mechanism in king pigeon nerve tissue damage induced by avermectin. Chem Biol Interact. 2014;219:113–122. doi: 10.1016/j.cbi.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Qian Y, Ji C, Yue S, Zhao M. Exposure of low-dose fipronil enantioselectively induced anxiety-like behavior associated with DNA methylation changes in embryonic and larval zebrafish. Environ Pollut. 2019;249:362–371. doi: 10.1016/j.envpol.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 54.Xing H, Wang C, Wu H, Chen D, Li S, Xu S. Effects of atrazine and chlorpyrifos on DNA methylation in the brain and gonad of the common carp. Comp Biochem Physiol C Toxicol Pharmacol. 2015;168:11–19. doi: 10.1016/j.cbpc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 55.Vester AI, Hermetz K, Burt A, Everson T, Marsit CJ, Caudle WM. Combined neurodevelopmental exposure to deltamethrin and corticosterone is associated with Nr3c1 hypermethylation in the midbrain of male mice. Neurotoxicol Teratol. 2020;80:106887. doi: 10.1016/j.ntt.2020.106887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding X, Li Y, Lü J, Zhao Q, Guo Y, Lu Z, Ma W, Liu P, Pestell RG, Liang C, Yu Z. piRNA-823 Is involved in cancer stem cell regulation through altering DNA methylation in association with luminal breast cancer. Front Cell Dev Biol. 2021;9:641052. doi: 10.3389/fcell.2021.641052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu TH, Liu M, Zhou XE, Liang G, Zhao G, Xu HE, Melcher K, Jones PA. Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B. Nature. 2020;586(7827):151–155. doi: 10.1038/s41586-020-2747-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onodera A, González-Avalos E, Lio CJ, Georges RO, Bellacosa A, Nakayama T, Rao A. Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells. Genome Biol. 2021;22(1):186. doi: 10.1186/s13059-021-02384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin EM, Fry RC. Environmental influences on the epigenome: exposure- associated DNA methylation in human populations. Annu Rev Public Health. 2018;39(1):309–333. doi: 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- 61.Yang N, Wei Y, Wang T, Guo J, Sun Q, Hu Y, Yan X, Zhu X, Tang B, Xu Q. Genome-wide analysis of DNA methylation during antagonism of DMOG to MnCl2-induced cytotoxicity in the mouse substantia nigra. Sci Rep. 2016;6(1):28933. doi: 10.1038/srep28933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider JS, Kidd SK, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2013;217(1):75–81. doi: 10.1016/j.toxlet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cohn WE, Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature. 1951;167(4247):483–484. doi: 10.1038/167483a0. [DOI] [Google Scholar]
- 64.Chen K, Zhao BS, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol. 2016;23(1):74–85. doi: 10.1016/j.chembiol.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duechler M, Leszczyńska G, Sochacka E, Nawrot B. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol Life Sci. 2016;73(16):3075–3095. doi: 10.1007/s00018-016-2217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cayir A, Barrow TM, Guo L, Byun HM. Exposure to environmental toxicants reduces global N6-methyladenosine RNA methylation and alters expression of RNA methylation modulator genes. Environ Res. 2019;175:228–234. doi: 10.1016/j.envres.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 67.Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71(10):3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reichel M, Köster T, Staiger D. Marking RNA: m6A writers, readers, and functions in Arabidopsis. J Mol Cell Biol 2019; 11 (10): 899–910. doi:10.1093/jmcb/mjz085. [DOI] [PMC free article] [PubMed]
- 69.Liu Q, Gregory RI. RNAmod: an integrated system for the annotation of mRNA modifications. Nucleic Acids Res. 2019;47(W1):W548–w555. doi: 10.1093/nar/gkz479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063. doi: 10.1093/nar/gkx141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riaz S, Niaz Z, Khan S, Liu Y, Sui Z. Detection, characterization and expression dynamics of histone proteins in the dinoflagellate Alexandrium pacificum during growth regulation. Harmful Algae. 2019;87:101630. doi: 10.1016/j.hal.2019.101630. [DOI] [PubMed] [Google Scholar]
- 72.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58(7):439–445. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 73.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 74.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. doi: 10.1016/j.tig.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 75.Ma L, Cao J, Liu L, Du Q, Li Z, Zou D, Bajic VB, Zhang Z. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019;47(D1):D128–d134. doi: 10.1093/nar/gky960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wallace DR, Taalab YM, Heinze S, Tariba Lovaković B, Pizent A, Renieri E, Tsatsakis A, Farooqi AA, Javorac D, Andjelkovic M, Bulat Z, Antonijević B, Buha DA. Toxic-metal-induced alteration in miRNA expression profile as a proposed mechanism for disease development. Cells. 2020;9(4):901. doi: 10.3390/cells9040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Go RCP, Corley MJ, Ross GW, Petrovitch H, Masaki KH, Maunakea AK, He Q, Tiirikainen MI. Genome-wide epigenetic analyses in Japanese immigrant plantation workers with Parkinson's disease and exposure to organochlorines reveal possible involvement of glial genes and pathways involved in neurotoxicity. BMC Neurosci. 2020;21(1):31. doi: 10.1186/s12868-020-00582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weldon BA, Shubin SP, Smith MN, Workman T, Artemenko A, Griffith WC, Thompson B, Faustman EM. Urinary microRNAs as potential biomarkers of pesticide exposure. Toxicol Appl Pharmacol. 2016;312:19–25. doi: 10.1016/j.taap.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo J, Zhang J, Wu C, Lv S, Lu D, Qi X, Jiang S, Feng C, Yu H, Liang W, Chang X, Zhang Y, Xu H, Cao Y, Wang G, Zhou Z. Associations of prenatal and childhood chlorpyrifos exposure with neurodevelopment of 3-year-old children. Environ Pollut. 2019;251:538–546. doi: 10.1016/j.envpol.2019.05.040. [DOI] [PubMed] [Google Scholar]
- 80.Chiu KC, Sisca F, Ying JH, Tsai WJ, Hsieh WS, Chen PC, Liu CY. Prenatal chlorpyrifos exposure in association with PPARγ H3K4me3 and DNA methylation levels and child development. Environ Pollut. 2021;274:116511. doi: 10.1016/j.envpol.2021.116511. [DOI] [PubMed] [Google Scholar]
- 81.Lind PM, Salihovic S, Lind L. High plasma organochlorine pesticide levels are related to increased biological age as calculated by DNA methylation analysis. Environ Int. 2018;113:109–113. doi: 10.1016/j.envint.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 82.Yu X, Zhao B, Su Y, Zhang Y, Chen J, Wu W, Cheng Q, Guo X, Zhao Z, Ke X, Danzeng W, Zhao B, Ma Q. Association of prenatal organochlorine pesticide-dichlorodiphenyltrichloroethane exposure with fetal genome-wide DNA methylation. Life Sci. 2018;200:81–86. doi: 10.1016/j.lfs.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 83.Kim S, Cho YH, Lee I, Kim W, Won S, Ku JL, Moon HB, Park J, Kim S, Choi G, Choi K. Prenatal exposure to persistent organic pollutants and methylation of LINE-1 and imprinted genes in placenta: a CHECK cohort study. Environ Int. 2018;119:398–406. doi: 10.1016/j.envint.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 84.Allen EM, Florang VR, Davenport LL, Jinsmaa Y, Doorn JA. Cellular localization of dieldrin and structure-activity relationship of dieldrin analogues in dopaminergic cells. Chem Res Toxicol. 2013;26(7):1043–1054. doi: 10.1021/tx300458b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanthasamy AG, Kitazawa M, Kanthasamy A, Anantharam V. Dieldrin-induced neurotoxicity: relevance to Parkinson's disease pathogenesis. Neurotoxicology. 2005;26(4):701–719. doi: 10.1016/j.neuro.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 86.Moretto A, Colosio C. Biochemical and toxicological evidence of neurological effects of pesticides: the example of Parkinson's disease. Neurotoxicology. 2011;32(4):383–391. doi: 10.1016/j.neuro.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 87.Matsuo N. Discovery and development of pyrethroid insecticides. Proc Jpn Acad Ser B Phys Biol Sci. 2019;95(7):378–400. doi: 10.2183/pjab.95.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barr DB, Olsson AO, Wong LY, Udunka S, Baker SE, Whitehead RD, Magsumbol MS, Williams BL, Needham LL. Urinary concentrations of metabolites of pyrethroid insecticides in the general U.S. population: National Health and nutrition examination survey 1999-2002. Environ Health Perspect. 2010;118(6):742–748. doi: 10.1289/ehp.0901275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Furlong MA, Paul KC, Cockburn M, Bronstein J, Keener A, Rosario ID, Folle AD, Ritz B. Ambient pyrethroid pesticide exposures in adult life and depression in older residents of california's central valley. Environ Epidemiol. 2020;4(6):e123. doi: 10.1097/ee9.0000000000000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li HY, Zhong YF, Wu SY, Shi N. NF-E2 related factor 2 activation and heme oxygenase-1 induction by tert-butylhydroquinone protect against deltamethrin-mediated oxidative stress in PC12 cells. Chem Res Toxicol. 2007;20(9):1242–1251. doi: 10.1021/tx700076q. [DOI] [PubMed] [Google Scholar]
- 91.Li HY, Wu SY, Shi N. Transcription factor Nrf2 activation by deltamethrin in PC12 cells: involvement of ROS. Toxicol Lett. 2007;171(1–2):87–98. doi: 10.1016/j.toxlet.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 92.Li HY, Wu SY, Ma Q, Shi N. The pesticide deltamethrin increases free radical production and promotes nuclear translocation of the stress response transcription factor Nrf2 in rat brain. Toxicol Ind Health. 2011;27(7):579–590. doi: 10.1177/0748233710393400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Carloni M, Nasuti C, Fedeli D, Montani M, Vadhana MS, Amici A, Gabbianelli R. Early life permethrin exposure induces long-term brain changes in Nurr1, NF-kB and Nrf-2. Brain Res. 2013;1515:19–28. doi: 10.1016/j.brainres.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 94.Du Y, Nomura Y, Satar G, Hu Z, Nauen R, He SY, Zhorov BS, Dong K. Molecular evidence for dual pyrethroid-receptor sites on a mosquito sodium channel. Proc Natl Acad Sci U S A. 2013;110(29):11785–11790. doi: 10.1073/pnas.1305118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kubik TD, Snell TK, Saavedra-Rodriguez K, Wilusz J, Anderson JR, Lozano-Fuentes S, Black WC, Campbell CL. Aedes aegypti miRNA-33 modulates permethrin induced toxicity by regulating VGSC transcripts. Sci Rep. 2021;11(1):7301. doi: 10.1038/s41598-021-86665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gandy MN, Corral MG, Mylne JS, Stubbs KA. An interactive database to explore herbicide physicochemical properties. Org Biomol Chem. 2015;13(20):5586–5590. doi: 10.1039/c5ob00469a. [DOI] [PubMed] [Google Scholar]
- 97.Richardson JR, Fitsanakis V, Westerink RHS, Kanthasamy AG. Neurotoxicity of pesticides. Acta Neuropathol. 2019;138(3):343–362. doi: 10.1007/s00401-019-02033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tanner CM, Kamel F, Ross GW, Hoppin JA, Goldman SM, Korell M, Marras C, Bhudhikanok GS, Kasten M, Chade AR, Comyns K, Richards MB, Meng C, Priestley B, Fernandez HH, Cambi F, Umbach DM, Blair A, Sandler DP, Langston JW. Rotenone, paraquat, and Parkinson's disease. Environ Health Perspect. 2011;119(6):866–872. doi: 10.1289/ehp.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bastías-Candia S, Zolezzi JM, Inestrosa NC. Revisiting the paraquat-induced sporadic Parkinson's disease-like model. Mol Neurobiol. 2019;56(2):1044–1055. doi: 10.1007/s12035-018-1148-z. [DOI] [PubMed] [Google Scholar]
- 100.Li H, Wu S, Wang Z, Lin W, Zhang C, Huang B. Neuroprotective effects of tert-butylhydroquinone on paraquat-induced dopaminergic cell degeneration in C57BL/6 mice and in PC12 cells. Arch Toxicol. 2012;86(11):1729–1740. doi: 10.1007/s00204-012-0935-y. [DOI] [PubMed] [Google Scholar]
- 101.Kong M, Ba M, Liang H, Ma L, Yu Q, Yu T, Wang Y. 5′-Aza-dC sensitizes paraquat toxic effects on PC12 cell. Neurosci Lett. 2012;524(1):35–39. doi: 10.1016/j.neulet.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 102.Wang X, Zhou S, Ding X, Zhu G, Guo J. Effect of triazophos, fipronil and their mixture on miRNA expression in adult zebrafish. J Environ Sci Health B. 2010;45(7):648–657. doi: 10.1080/03601234.2010.502435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.