Abstract
Despite recent declines in incidence, Pneumocystis carinii pneumonia (PCP) remains the most commonly occurring opportunistic illness among persons with AIDS in the United States. While P. carinii DNA has been detected in patient respiratory specimens and in air samples collected from various indoor environments housing PCP patients, the viability of these organisms is unknown. For this reason, we have developed and evaluated a molecular viability assay for P. carinii. This method is based upon the detection of P. carinii mRNA by a reverse transcription-PCR that employs specific primers from a member of the heat shock protein 70 family. Under optimal assay conditions, these primers were capable of detecting as few as 100 viable trophozoites as determined by ethidium bromide staining, while no signal was obtained from 106 trophozoites killed by heat, desiccation, or UV radiation. This assay was also capable of distinguishing P. carinii from other common fungi present in the air. Therefore, this molecular viability assay may be useful in conjunction with standard bioaerosol collection devices and procedures for the detection of viable P. carinii collected from various indoor environments. It may also be useful in confirming the presence of viable trophozoites in respiratory specimens collected by noninvasive techniques from putatively infected individuals.
Despite the recent declines in incidence due to widespread chemoprophylaxis and antiretroviral therapy in immunosuppressed human immunodeficiency virus (HIV)-infected individuals, Pneumocystis carinii pneumonia (PCP) is still the most common opportunistic infection during the course of AIDS in the United States (7). However, for those persons who cannot tolerate antipneumocystis therapy, or who are unaware of their HIV status, the risk of PCP remains high (41).
Basic knowledge of P. carinii ecology and epidemiology is still lacking. The once widely accepted theory of PCP reactivation in persons with severe immunosuppression has come into question with recent studies providing evidence to support the hypothesis that most episodes of PCP result from a de novo acquisition. Support for this new hypothesis includes epidemiological studies of hospital outbreaks (8, 13, 16), animal studies of airborne transmission and limited persistence after primary infections (9, 17, 34, 38, 39), the absence of P. carinii in respiratory samples collected from healthy individuals as well as asymptomatic HIV-infected individuals (28, 30, 31), and the demonstration that strain types of P. carinii isolated from patients during subsequent episodes of infection differ approximately 50% of the time (20, 21, 36).
While airborne transmission of P. carinii appears likely, the source of infective organisms remains largely unproven. It has been hypothesized that an important source of P. carinii is patients with patent PCP, who act as amplifiers and then shed large numbers of viable organisms into their immediate environment. This shedding would result in local concentrations of P. carinii that considerably exceed the levels found in uncontaminated environments. This conclusion is suggested from the facts that P. carinii sp. f. hominis DNA has been frequently detected in low-volume air samples collected from the environments of PCP patients and that it has rarely been detected in low-volume air samples collected from indoor environments not recently housing a PCP patient or from the outdoors (3, 5, 29). Furthermore, P. carinii sp. f. hominis strain types detected in air samples collected from PCP patient hospital rooms matched the strain types isolated from the same PCP patients in 12 of 14 instances (3).
It has also been hypothesized that P. carinii is ubiquitous, occurring at low levels in the general environment. Evidence for this hypothesis comes from studies in which both human and rat P. carinii DNAs have been detected in large-volume air samples collected from the outdoor air (40) and in which the seroprevalence of antibodies against P. carinii found in young children has been high (32).
The method of choice for the diagnosis of PCP involves collection of specimens by either induction of sputum or bronchoalveolar lavage (BAL), followed by microscopic visualization of P. carinii trophozoites and/or cysts. The sensitivity of microscopic examination of induced sputum has been shown to be variable. For this reason, it has been recommended that BAL be performed on patients presenting with symptoms of PCP but who exhibit a negative induced-sputum sample (41). On the other hand, because of the intrusiveness of these techniques, many clinicians empirically treat suspected PCP cases with antipneumocystis drugs, although this has been demonstrated to be a highly nonspecific approach (6). Recent studies have demonstrated that P. carinii DNA can be detected in oral wash samples by PCR (2, 15, 37). This technique represents a less invasive method for diagnosing the presence of P. carinii organisms in presumptively infected individuals, providing a strong incentive to demonstrate organism viability in these samples.
Although it is possible to detect P. carinii DNA by PCR in environmental and clinical samples, DNA detection gives no indication of organism viability. It is therefore unclear what the above-mentioned results mean in terms of potential risk for susceptible individuals, potential exposure control measures, or the ecology of P. carinii. In order to evaluate the viability of P. carinii organisms collected from environmental and clinical sources, we have developed a molecular viability assay to be used in conjunction with standard bioaerosol collection techniques as well as noninvasive clinical specimen collection methods.
MATERIALS AND METHODS
P. carinii sp. f. carinii culture.
P. carinii sp. f. carinii trophozoites and cysts were harvested from preparations derived from spinner-flask cultures prepared as previously described (22). Briefly, mixed cultures of P. carinii sp. f. carinii trophozoites and cysts isolated from infected rat lungs were grown in a spinner-flask culture containing human embryonic cells sheeted to Cytodex microcarrier beads. An approximate fivefold increase in the number of organisms is typically seen in 7 days, as determined by organism count, antigen detection, and DNA quantification. These cultures consist primarily of trophozoites (95 to 99%), with a small proportion of cysts (1 to 5%).
P. carinii sp. f. carinii isolation and cyst enrichment.
P. carinii sp. f. carinii cysts and trophozoites were isolated from the infected lungs of immunosuppressed Sprague-Dawley rats as previously described (26). A cyst enrichment protocol was then performed on the isolated organisms. Isolated organisms were resuspended in 1 ml of phosphate-buffered saline (PBS; 1.5 mM KH2PO4, 8.1 mM Na2HPO4 [pH 7.4], 1.5 mM KCl, 137 mM NaCl; Life Technologies) and subjected to one freeze-thaw cycle, followed by centrifugation at 5,000 × g for 10 min at 4°C. The pelleted organisms were then resuspended in a 0.1× PBS solution containing 0.1% sodium dodecyl sulfate (SDS) for 10 min at 25°C. The remaining cysts were pelleted at 5,000 × g for 10 min at 4°C and then washed five times in PBS to remove cell debris and free nucleic acids. This protocol typically yielded approximately 59% of the original cyst population while removing most host cells and trophozoites.
Microscopic organism enumeration and viability assessment.
Before RNA extraction from either a cyst-enriched preparation or a spinner-flask harvest, organism density and viability were determined microscopically. Organism density was assessed as described by Lee et al. (22). Viability was assessed essentially as described by Kaneshiro et al. (18). Briefly, the fluorescent dye calcein-acetoxy methyl ester (Molecular Probes, Inc., Eugene, Oreg.) was used in conjunction with the nucleic acid stain ethidium bromide (EB). An aliquot from the P. carinii sp. f. carinii preparation was placed in 200 μl of PBS, to which calcein-acetoxy methyl ester was added for a final concentration of 13 μM and EB was added for a final concentration of 25 μM. The organisms were placed in an incubator at 37°C for 30 min, after which time they were examined by epifluorescence microscopy. Organisms that fluoresced green were considered viable, and those that fluoresced red were considered nonviable.
RNA extraction.
Total RNA was extracted from P. carinii sp. f. carinii trophozoites and cysts derived from spinner-flask cultures by employing a commercially available RNA isolation kit (RNeasy; Qiagen Inc., Valencia, Calif.), according to the manufacturer's instructions. Total RNA was extracted from the enriched-cyst preparation derived from rat lung as well as three species of fungi: Aspergillus niger, Saccharomyces cerevisiae, and Penicillium chrysogenum. A. niger and Penicillium sp. were examined because they are soil fungi that are commonly present in indoor air (14, 23), while S. cerevisiae was chosen because it is commonly present in indoor environments and because it has been shown to be genetically very similar to P. carinii (12). The cysts and fungal harvests were incubated in a solution containing 100 mM NaCl, 40 mM EDTA, 0.2% SDS, and 200 μg of proteinase K per ml for 6 h at 37°C. A commercially available guanidinium isothiocyanate solution (RNAzol B; Teltest, Inc., Friendswood, Tex.) was then added directly to the mixture, and total RNA isolation was carried out according to the manufacturer's instructions. The mRNA species were then purified from the total RNA preparation by adsorption to oligo(dT) beads (Oligotex; Qiagen Inc.) by following the manufacturer's instructions.
RT-PCR assay.
mRNA-specific primers from a member of the P. carinii sp. f. carinii heat shock protein 70 (HSP70) multigene family (PcSA1; GenBank accession number U80967 [35]) were designed for use in a reverse transcription-PCR (RT-PCR) assay to assess organism viability. The coding primer (5′-TTGAGAAAGCAATTGGTATT-3′) was designed to cross the second intron splice site found in the PcSA1 gene. The sequence of the noncoding primer was 5′-CTGCTGCAGTAGGCTCATTG-3′. RNA preparations prepared as described above were initially annealed in a solution containing a 4 μM concentration of the noncoding primer by heating to 70°C for 15 min and cooling to 0°C for 5 min. The solution containing the annealed complex was brought to a final volume of 10 μl in a solution containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, a 0.5 mM concentration (each) of dATP, dCTP, dGTP, and dTTP, 10 U of RNasin RNase inhibitor (Promega Inc., Madison, Wis.), and 100 U of Superscript II RNase H− reverse transcriptase (BRL Life Technologies, Gaithersburg, Md.). RT reaction mixtures were incubated at 45°C for 50 min and then at 70°C for 10 min. After the completion of the cDNA first-strand synthesis, the reaction mixture was diluted to 50 μl for PCR amplification. The final PCR solution contained 60 mM Tris-HCl (pH 8.5), 15 mM (NH4)2SO4, 3.5 mM MgCl2, a 0.5 μM concentration of each primer, a 100 μM concentration (each) of dATP, dGTP, dCTP, and dTTP, and 2.5 U of Amplitaq (Applied Biosystems Inc., Branchburg, N.J.). The reaction mixture was then overlaid with 50 μl of mineral oil and subjected to 40 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min, followed by an incubation at 72°C for 7 min. RT-PCR products were electrophoresed on 1.5% agarose gel and detected after being stained with EB.
RT-PCR products derived from P. carinii were gel purified and cloned into the pCR2.1 vector (Invitrogen Inc., La Jolla, Calif.). The clones were then manually sequenced, and their identities were confirmed by comparison with the PcSA1 gene sequence.
Southern blot hybridization.
A Southern blot was prepared by capillary blotting an agarose gel containing the PcSA1 PCR products onto Hybond nylon membrane (Amersham Life Sciences Inc., Little Chalfont, Buckinghamshire, England). The blot was hybridized in the formamide buffer recommended by the manufacturer at 42°C using radiolabeled PCR products derived from a pure culture of P. carinii sp. f. carinii as a probe. Following hybridization, the membrane was washed twice at 50°C in a solution of 15 mM NaCl, 1.5 mM sodium citrate, and 0.1% SDS.
RESULTS
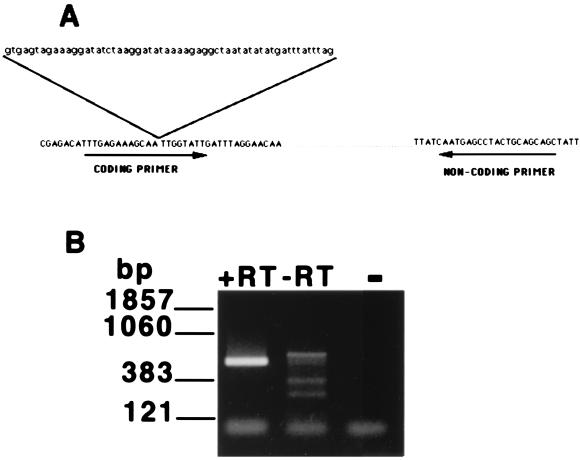
mRNA-specific primers from a member of the P. carinii sp. f. carinii HSP70 multigene family (PcSA1) were designed for use in an RT-PCR assay to assess organism viability. While the majority of the HSP70 genes in P. carinii are constitutively expressed, expression of PcSA1 is upregulated by brief exposure to moderate heat (35). The 5′ (coding orientation) primer was designed from the cDNA sequence so that the sequence of the primer included the second intron splice site in the PcSA1 sequence (GenBank accession number U80967) (Fig. 1A). The 3′ (noncoding) primer was derived from the third exon (Fig. 1A). Use of these primers in an RT-PCR assay employing total RNA extracted from roughly 106 trophozoites isolated from a spinner-flask culture resulted in the production of a 530-bp PCR product, equivalent in size to that predicted from analysis of the mature mRNA (Fig. 1B). DNA sequence analysis of this product confirmed that it was derived from the mature PcSA1 transcript. Furthermore, the 530-bp product was not obtained from mRNA extracted from uninfected rat lungs, providing additional evidence that this product was derived from P. carinii and not host cell mRNA (data not shown). As expected, the 530-bp product was also absent from reaction mixtures lacking reverse transcriptase, as the design of the coding primer across the intron splice site prevented the amplification of this product from contaminating P. carinii genomic DNA (Fig. 1B).
FIG. 1.
Specific amplification of PcSA1 sequences from P. carinii mRNA. (A) Design of primers for the PcSA1 RT-PCR assay. The 5′ (coding orientation) primer crosses a 58-bp intron to prevent the amplification of contaminating genomic DNA derived from P. carinii. The exon sequences are shown in uppercase letters, and the intron sequence is shown in lowercase letters. (B) Results from the PcSA1 RT-PCR assay. Lane 1, RT-PCR carried out on total RNA extracted from 106 P. carinii organisms derived from a spinner-flask culture; lane 2, control RT-PCR lacking reverse transcriptase carried out on total RNA extracted from 106 P. carinii organisms derived from a spinner-flask culture; lane 3, PCR negative control. +RT, with reverse transcriptase; −RT, without reverse transcriptase.
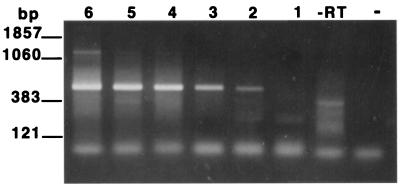
In order to determine the sensitivity of the assay, RNAs extracted from serial dilutions of spinner-flask-cultured organisms were employed as templates in the RT-PCR assay. The assay was found to detect as few as 102 trophozoites as determined by EB staining (Fig. 2). To examine the specificity of the RT-PCR for the detection of viable organisms (i.e., the ability of the assay to distinguish viable from nonviable organisms), four aliquots, each containing 106 viable trophozoites, were prepared. Total RNA was extracted from an untreated dilution series prepared from one aliquot (104 to 102 trophozoites), while the other three aliquots of trophozoites were subjected either to heat (autoclave), desiccation (left on bench top for 1 week), or UV light exposure (left under a germicidal lamp for 24 h). Loss of viability in these treated samples was confirmed by microscopy, as described in Materials and Methods. Total RNA was then extracted from each of the treated samples in the same manner as for the untreated sample. A positive signal was obtained from the aliquot containing 102 viable trophozoites, while no signal was obtained from any of the treated samples (Fig. 3). Thus, the assay was capable of distinguishing 102 viable P. carinii trophozoites from 106 nonviable P. carinii trophozoites.
FIG. 2.
Sensitivity of the PcSA1 RT-PCR. RT-PCRs were carried out on total RNA preparations prepared from log-unit dilutions of viable P. carinii. Lane 6, 106 P. carinii organisms; lane 5, 105 P. carinii organisms; lane 4, 104 P. carinii organisms; lane 3, 103 P. carinii organisms; lane 2, 102 P. carinii organisms; lane 1, 101 P. carinii organisms; lane −RT, without reverse transcriptase (RNA from 106 organisms); lane −, PCR negative control.
FIG. 3.
Discrimination of viable from nonviable organisms by the PcSA1 RT-PCR. RT-PCR assays were carried out as described in Materials and Methods by employing RNAs extracted from treated cultures of P. carinii as templates. Lane 1, RNA derived from 104 untreated P. carinii organisms; lane 2, RNA derived from 103 untreated organisms; lane 3, RNA derived from 102 untreated organisms; lane 4, RNA derived from 106 P. carinii organisms killed by desiccation; lane 5, RNA derived from 106 organisms killed by heat; lane 6, RNA derived from 106 organisms killed by UV light; lane 7, no RNA; lane 8, PCR negative control.
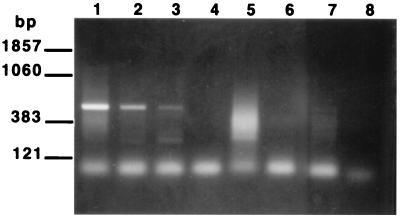
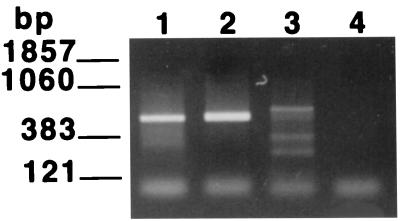
One major application of a molecular viability assay for P. carinii will be to detect the presence of viable organisms in the environment. It was therefore of interest to explore the level of species specificity of the RT-PCR. To accomplish this, the P. carinii primers were employed to attempt to amplify a related sequence from RNAs prepared from three different species of ascomycetous fungi as described in Materials and Methods. The PcSA1 primers amplified a small fragment from Penicillium chrysogenum and a fragment derived from S. cerevisiae of approximately the same size as that amplified from P. carinii (Fig. 4A). However, the band derived from S. cerevisiae was easily distinguished from that of P. carinii on the basis of hybridization with the bona fide P. carinii PCR product (Fig. 4B).
FIG. 4.
Species specificity of the PcSA1 RT-PCR. RT-PCRs were carried out on fungal RNA preparations prepared as described in Materials and Methods. (A) EB-stained gel of PCR products; (B) Southern blot of the gel shown in panel A probed with the labeled PcSA1 PCR product. In each panel, lane 1 contains RNA extracted from P. carinii, lane 2 contains RNA extracted from S. cerevisiae, lane 3 contains RNA extracted from Penicillium chrysogenum, and lane 4 contains RNA extracted from A. niger.
Since spinner-flask cultures consisted primarily of trophozoites (95 to 99%), it was unlikely that the small number of cysts in the harvest preparations contributed much to the results obtained from the spinner-flask cultures. Therefore, it was necessary to determine if the heat shock RT-PCR assay was capable of detecting a preparation primarily composed of cysts. To accomplish this, P. carinii organisms were isolated from rat lungs with overt P. carinii infection. The mixed-life cycle preparation was then treated in order to isolate the cysts from the trophozoites, as described in Materials and Methods. Bright-field microscopic examination of the cyst-enriched preparation revealed no intact trophozoites in 20 fields, while the cyst concentration was estimated to be at least 107/ml. Together, these data suggested that the enriched-cyst preparation contained less than 0.1% trophozoite contamination. Epifluorescence microscopic examination of the preparation suggested that greater that 80% of the cysts in the enriched preparation were viable.
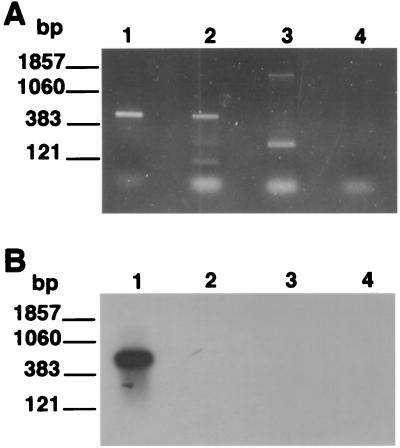
Total RNA was isolated from the enriched-cyst preparation as described in Materials and Methods, and poly(A)+ RNAs purified from the total RNA preparations were used as templates in the RT-PCR. The additional purification step [poly(A)+ RNA isolation] was found to be necessary to eliminate inhibitors of the RT reaction found to be present in the total RNA preparations prepared from the enriched cysts (data not shown). Following RT-PCR, the poly(A)+ RNA preparation from the enriched-cyst preparation resulted in material that supported the amplification of the PcSA1 product (Fig. 5).
FIG. 5.
Detection of P. carinii cysts by the PcSA1 RT-PCR. Lane 1, poly(A)+ mRNA extracted from a cyst-enriched preparation; lane 2, RNA derived from a spinner-flask culture; lane 3, cyst-enriched preparation of RNA amplified in the absence of reverse transcriptase; lane 4, PCR negative control.
DISCUSSION
The rationale for the viability assay described above was that mRNA molecules, as opposed to DNA or rRNA, are usually unstable following the death of an organism. By designing primers that span an intron splice site in the PcSA1 sequence, we anticipated that only intact mRNA molecules would serve as a template in the RT-PCR and that these molecules would be labile following the death of the organism. The data described above suggest that this was the case. The RT-PCR assay was capable of detecting as few as 102 viable trophozoites as determined by EB staining. In contrast, 106 nonviable trophozoites killed by either high heat, desiccation, or UV exposure produced no signal in the RT-PCR assay. This is significant, as the last two methods of killing represent the two most important mechanisms resulting in the loss of viability of airborne microorganisms (10). Apart from its ability to distinguish viable and nonviable P. carinii, this RT-PCR assay can distinguish between viable P. carinii and other fungi that commonly occur in the indoor environment. Both Aspergillus and Penicillium sp. are commonly present in the indoor environment (14), a finding confirmed with our own air sampling of an urban indoor residence, in which moderate levels of viable A. niger, Aspergillus fumigatus, and Penicillium were found (data not shown).
Although most of the experiments carried out in the work described above involved trophozoites obtained from spinner-flask-cultured organisms, the assay was also capable of detecting the PcSA1 transcript in a cyst-enriched preparation obtained directly from an infected rat lung. Although this preparation was highly enriched for cysts, we cannot rule out the possibility that the positive signal obtained from this preparation resulted from contamination of the cyst preparation, either with intact trophozoites or with residual mRNA released from the lysed trophozoites.
While the life cycle stage involved in airborne transmission is unknown, the likely candidate would be the P. carinii cyst due the structure of its cell wall, which possibly allows for protection against environmental stressors such as desiccation. Thus, determining how well this RT-PCR assay assesses cyst viability is key to its use with environmental samples. There are precedents for use of the RT-PCR in assessing the viability of both Giardia cysts (1, 19) and Cryptosporidium oocysts collected from environmental samples (19). These assays all exploited the heat shock responses of the two organisms, distinguishing viable from nonviable cysts by detecting the heat shock transcript by RT-PCR after heat treatment.
Culturing of air samples collected from the environment is standard procedure in the field of aerobiology when the presence of viable biological agents is assessed (11). Thus, one possible approach for viable P. carinii cyst detection might involve short-term (<24 h) culture of cysts collected from the environment by bioaerosol collection methods such as filtration or liquid impingement. This would allow for maturation of the eight intracystic bodies to trophozoites. RNAs extracted from the cultures containing the newly emerged trophozoites could then be used as templates in the RT-PCR assay.
When bacteria are selectively sampled, a fungicide is added to the medium, and when fungal spores are selectively sampled, antibiotics are added to the medium. Unlike any other known species of fungus, P. carinii lacks the steroid ergosterol in its cell wall. Thus, it is not susceptible to the class of fungicidal drugs that inhibit ergosterol biosynthesis (4). Therefore, antibiotics and antifungals such as the imidazole drugs can be added to a short-term culture of P. carinii, allowing for its growth while preventing the growth of other fungi and bacteria collected in the sample.
One additional application of this viability assay may be in clinical diagnosis of PCP. It is possible to envision a two-step system to utilize the RT-PCR in combination with the DNA-based PCR for this purpose. The DNA-based PCR might first be used to detect P. carinii DNA in patient oral wash samples. If evidence for the presence of P. carinii DNA is obtained, positive individuals can be retested by employing the RT-PCR assay to confirm the presence of viable trophozoites, which are the most prevalent life cycle stage found in an infected host (24). This strategy might represent a sensitive and specific diagnostic protocol that is much less invasive than BAL or induction of sputum. This approach might also prove useful in detecting low levels of viable organisms in the face of highly active therapies where organism burdens may be lower than in untreated disease.
Additional work on this assay will be needed before it can used as a reliable tool in a field study of the occurrence of viable P. carinii sp. f. hominis in the environment. Issues such as organism recovery efficiency from bioaerosol collection devices and potential environmental interference must be worked out. Finally, the system must be adapted for use with the human-specific variant of P. carinii. With the recent successes in the development of a human P. carinii culture system (27), this should be relatively straightforward.
Determination of organism viability does not prove infectiousness. However, determining where, when, and under what circumstances viable P. carinii occurs in the environment will help to narrow down the list of potential sources of infection. If the theory of PCP patients contaminating indoor environments with infectious organisms holds true, then there are many infection control approaches that can be used to limit exposure of at-risk individuals to the organism. Given the potential for antibiotic resistance (25) and the toxicity associated with P. carinii prophylaxis among HIV patients (33), exposure prevention may be preferable if person-to-person transmission can be confirmed as an important mode of infection in studies employing the molecular viability assay described herein.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant R01 (AI54586 01 A1).
REFERENCES
- 1.Abbaszadegan M H, Huber M S, Gerba C P, Pepper I L. Detection of viable Giardia cysts by amplification of heat shock-induced mRNA. Appl Environ Microbiol. 1997;63:324–328. doi: 10.1128/aem.63.1.324-328.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atzori C, Agostini F, Gubertini G, Cargneal A. Diagnosis of PCP by ITS nested PCR on noninvasive oropharyngeal samples. J Eukaryot Microbiol. 1996;43(5):44. doi: 10.1111/j.1550-7408.1996.tb04977.x. Suppl. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett M S, Lu J-J, Lee C-H, Durant P J, Queener S F, Smith J W. Types of Pneumocystis carinii detected in air samples. J Eukararyot Microbiol. 1996;43(5):44. doi: 10.1111/j.1550-7408.1996.tb04980.x. Suppl. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett M S, Queener S F, Shaw M M, Richardson J D, Smith J W. Pneumocystis carinii is resistant to imidazole antifungal agents. Antimicrob Agents Chemother. 1994;38:1859–1861. doi: 10.1128/aac.38.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett M S, Vermund S H, Jacobs R R, Durant P, Shaw M M, Smith J W, Tang X, Lu J-J, Li B-H, Jin S, Lee C-H. Detection of Pneumocystis carinii DNA in air samples: likely environmental risk to susceptible persons. J Clin Microbiol. 1997;35:2511–2513. doi: 10.1128/jcm.35.10.2511-2513.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baughman R P, Dohn M N, Frame P T. The continuing utility of bronchoalveolar lavage to diagnose opportunistic infection in AIDS patients. Am J Med. 1994;97:515–522. doi: 10.1016/0002-9343(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. HIV/AIDS surveillance report 9 2. Atlanta, Ga: Centers for Disease Control and Prevention; 1997. [Google Scholar]
- 8.Chave J P, Wauters J P, Van Melle G, Francioli P. Transmission of Pneumocystis carinii from AIDS patients to other immunosuppressed patients: a cluster of Pneumocystis carinii pneumonia in renal transplant recipients. AIDS. 1991;5:927–932. doi: 10.1097/00002030-199108000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Gigliotti F, Harmsen A G. Latency is not an inevitable outcome of infection with Pneumocystis carinii. Infect Immun. 1993;61:237–242. doi: 10.1128/iai.61.12.5406-5409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox C S. Stability of airborne microbes and allergens. In: Cox C S, Wathes C M, editors. Bioaerosols handbook. Boca Raton, Fla: CRC Press; 1995. pp. 77–98. [Google Scholar]
- 11.Dillon H K, Heinsohn P A, Miller J D. AIHA: field guide for the determination of biological contaminants in the environment. Fairfax, Va: American Industrial Hygiene Association; 1996. [Google Scholar]
- 12.Edman J C, Kovacs J A, Masur H, Santi D V, Elwood H J, Sogun M L. Ribosomal RNA sequences show Pneumocystis to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 13.Goesch T R, Gotz G, Stellbrunk K H, Albrecht H, Weh H J, Hossfield D K. Possible transfer of Pneumocystis carinii between immunodeficient patients. Lancet. 1990;336:627. doi: 10.1016/0140-6736(90)93420-t. [DOI] [PubMed] [Google Scholar]
- 14.Hay R J, Clayton Y M, Goodley J M. Fungal aerobiology: how, when and where? J Hosp Infect. 1995;30(Suppl.):352–357. doi: 10.1016/0195-6701(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 15.Helweglarsen J, Jensen J S, Benfield T, Svendsen U G, Lundgren J D, Lundgren B. Diagnostic use of PCR for detection of Pneumocystis carinii in oral wash samples. J Clin Microbiol. 1998;36:2068–2072. doi: 10.1128/jcm.36.7.2068-2072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennequin C, Page B, Roux P, Legendre C, Kreis H. Outbreak of Pneumocystis carinii pneumonia in a renal transplant unit. Eur J Clin Microbiol Infect Dis. 1995;14:122–126. doi: 10.1007/BF02111870. [DOI] [PubMed] [Google Scholar]
- 17.Hughes W T. Natural mode of acquisition for de novo infection with Pneumocystis carinii. J Infect Dis. 1982;145:842–848. doi: 10.1093/infdis/145.6.842. [DOI] [PubMed] [Google Scholar]
- 18.Kaneshiro E S, Wider M A, Wu Y-P, Cushion M T. Reliability of calcein acetoxy methyl ester and ethidium homodimer or propidium iodide for viability assessment of microbes. J Microbiol Methods. 1993;17:1–16. [Google Scholar]
- 19.Kaucner C, Stinear T. Sensitive and rapid detection of viable Giardia cysts and Crytosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription-PCR. Appl Environ Microbiol. 1998;64:1743–1749. doi: 10.1128/aem.64.5.1743-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keely S P, Stringer J R. Sequences of Pneumocystis carinii f. sp. hominis strains associated with recurrent pneumonia vary at multiple loci. J Clin Microbiol. 1997;35:2745–2747. doi: 10.1128/jcm.35.11.2745-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keely S P, Stringer J R, Baughman R P, Linke M J, Walzer P D, Smulian A G. Genetic variation among Pneumocystis carinii hominis isolates in recurrent Pneumocystis. J Infect Dis. 1995;172:595–598. doi: 10.1093/infdis/172.2.595. [DOI] [PubMed] [Google Scholar]
- 22.Lee C-H, Bauer N L, Shaw M M, Durkin M M, Bartlett M S, Queener S F, Smith J W. Proliferation of rat Pneumocystis carinii on cells sheeted on microcarrier beads in spinner flasks. J Clin Microbiol. 1993;31:1659–1662. doi: 10.1128/jcm.31.6.1659-1662.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levetin E. Fungi. In: Burge H, editor. Bioaerosols. Ann Arbor, Mich: Lewis Publishers; 1995. pp. 104–107. [Google Scholar]
- 24.Levine S J. Pneumocystis carinii. Clin Chest Med. 1996;17:665–695. doi: 10.1016/s0272-5231(05)70339-7. . (Review.) [DOI] [PubMed] [Google Scholar]
- 25.Mei Q, Gurunathan S, Masur H, Kovacs J A. Failure of co-trimoxazole in Pneumocystis carinii infection and mutations in dihydropteroate synthase gene. Lancet. 1998;351:1631–1632. doi: 10.1016/S0140-6736(05)77687-X. [DOI] [PubMed] [Google Scholar]
- 26.Merali S, Clarkson A B., Jr Polyamine content of Pneumocystis carinii and response to the ornithine decarboxylase inhibitor dl-α-difluoromethylornithine. Antimicrob Agents Chemother. 1996;40:973–978. doi: 10.1128/aac.40.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merali S, Frevert U, Williams J H, Chin K, Bryan R, Allen J, Clarkson B. Continuous axenic cultivation of Pneumocystis carinii. Proc Natl Acad Sci USA. 1999;96:2402–2407. doi: 10.1073/pnas.96.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millard P R, Heryet A R. Observation favouring Pneumocystis carinii as a primary infection: a monoclonal antibody study on paraffin sections. J Pathol. 1988;154:365–368. doi: 10.1002/path.1711540413. [DOI] [PubMed] [Google Scholar]
- 29.Olsson M, Lidman C, Latouche S, Bjorkman A, Roux P, Linder E, Wahlgren M. Identification of Pneumocystis carinii f. sp. hominis gene sequences in filtered air in hospital environments. J Clin Microbiol. 1998;36:1737–1740. doi: 10.1128/jcm.36.6.1737-1740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ongibene F P, Masur J, Rogers P, Travis A F, Suffredini L, Feuerstein I, Gill V J, Baird B F, Carrasquillo J S, Parrillo J E, Lane H C, Shelmamer J H. Nonspecific interstitial pneumonitis without evidence of Pneumocystis carinii in asymptomatic patients infected with human immunodeficiency virus (HIV) Ann Intern Med. 1988;109:874–878. doi: 10.7326/0003-4819-109-11-874. [DOI] [PubMed] [Google Scholar]
- 31.Peters S E, Wakefield A E, Sinclair K, Millard P R, Hopkins J M. A search for Pneumocystis carinii in post mortum lungs by DNA amplification. J Pathol. 1992;166:195–198. doi: 10.1002/path.1711660217. [DOI] [PubMed] [Google Scholar]
- 32.Pifer L L, Hughes W T, Stagno S, Woods D. Pneumocystis carinii infection: evidence for high prevalence in normal and immunosuppressed children. Pediatrics. 1978;61:35–41. [PubMed] [Google Scholar]
- 33.Ryan C, Madalon M, Wortham D W, Graziano F M. Sulfa hypersensitivity in patients with HIV infection: onset, treatment, critical review of the literature. Wis Med J. 1998;97(5):23–27. [PubMed] [Google Scholar]
- 34.Sepkowitz K, Schulager N, Godwin T, Armstrong C, Bucula R. DNA amplification in experimental pneumocystosis: characterization of serum Pneumocystis carinii DNA and potential P. carinii carrier states. J Infect Dis. 1993;168:421–426. doi: 10.1093/infdis/168.2.421. [DOI] [PubMed] [Google Scholar]
- 35.Stedman T T, Butler D R, Buck G A. The HSP70 gene family in Pneumocystis carinii: molecular and phylogenetic characterization of cytoplasmic members. J Eukaryot Microbiol. 1998;45:589–599. doi: 10.1111/j.1550-7408.1998.tb04554.x. [DOI] [PubMed] [Google Scholar]
- 36.Tsolaki A G, Miller R F, Underwood A P, Bajeri S, Wakefield A E. Genetic diversity at the internal transcribed spacer regions of the rRNA operon among isolates of Pneumocystis carinii from AIDS patients with recurrent pneumonia. J Infect Dis. 1996;174:141–156. doi: 10.1093/infdis/174.1.141. [DOI] [PubMed] [Google Scholar]
- 37.Tsolaki A G, Miller R F, Wakefield A E. Oropharyngeal samples for genotyping and monitoring response to treatment in AIDS patients with Pneumocystis carinii pneumonia. J Med Microbiol. 1999;48:897–905. doi: 10.1099/00222615-48-10-897. [DOI] [PubMed] [Google Scholar]
- 38.Vargas S L, Hughes W T, Wakefield A E, Oz H S. Limited persistence and subsequent elimination of Pneumocystis carinii from the lungs after P. carinii pneumonia. J Infect Dis. 1995;172:506–510. doi: 10.1093/infdis/172.2.506. [DOI] [PubMed] [Google Scholar]
- 39.Vogel P, Miller C J, Lowenstine L L, Lackner A A. Evidence of horizontal transmission of Pneumocystis carinii pneumonia in simian immunodeficiency virus-infected rhesus monkeys. J Infect Dis. 1993;168:836–843. doi: 10.1093/infdis/168.4.836. [DOI] [PubMed] [Google Scholar]
- 40.Wakefield A E. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilken A, Feinberg J. Pneumocystis carinii pneumonia: a clinical review. Am Family Phys. 1999;60:1699–1708. [PubMed] [Google Scholar]