Summary
Fundamental aspects of brain function, including development, plasticity, learning, and memory, can take place over time scales of days to years. Chronic in vivo imaging of neural activity with cellular resolution is a powerful method for tracking the long-term activity of neural circuits. We review recent advances in our understanding of neural circuit function from diverse brain regions that have been enabled by chronic in vivo cellular imaging. Insight into the neural basis of learning and decision-making, in particular, benefit from the ability to acquire longitudinal data from genetically identified neuronal populations, deep brain areas, and subcellular structures. We propose that combining chronic imaging with further experimental and computational innovations will advance our understanding of the neural circuit mechanisms of brain function.
Introduction
A central goal of neuroscience is to achieve a mechanistic understanding of the relationship between neural circuits and behavior, including how the brain changes adaptively during learning. Long-term, repeated optical measurement of cellular activity in the living brain (in vivo), referred to here as chronic imaging, has become an important method for investigating the neural basis of behavior in animal models because of its ability to track the activity of identified types of neurons and glia in various brain areas with cellular resolution (Lütcke et al. 2013; Margolis et al. 2014; Crowe and Ellis-Davies 2014; Hamel et al. 2015; Clopath et al. 2017). Although other imaging methods exist to measure brain activation longitudinally, including fMRI and emerging technologies such as optoacoustic imaging, these methods currently lack the ability to resolve individual neurons, which is crucial for identifying the cellular and circuit mechanisms of learning-related neural changes (Jonckers et al. 2015; Ovsepian et al. 2017). In recent years, technical advances in neural activity sensors and optical microscopy have converged with novel behavioral assays and data analysis methods to make chronic in vivo imaging of neural activity an essential technique for the study of the neural circuitry underlying learning and memory formation. Since the initial applications that tracked changes in the fine-scale structure of neuronal dendritic spines in the mouse cerebral cortex (Grutzendler et al. 2002; Trachtenberg et al. 2002; Chen and Nedivi 2010), chronic in vivo imaging has been used to investigate a number of important problems in neuroscience, including synaptic formation (Holtmaat et al. 2006), cell fate in neurogenesis (Pilz et al. 2018), genetic activity markers (Wang et al. 2006), changes in disease, aging, or injury models (Hill et al. 2017, 2018; Akassoglou et al. 2017; Eyo et al. 2018; Real et al. 2018). Exciting advances in chronic imaging have also been made in a number of species from flies to non-human primates (Sadakane et al. 2015; Huang et al. 2018). In this review, we focus on novel applications of chronic in vivo imaging of neuronal activity in mice, where it is possible to leverage genetic accessibility, optical accessibility, and behavioral assays, in order to investigate the neural basis of mammalian behavior. One key feature of chronic optical imaging is the ability to resolve both active and sparsely active or “silent” neurons (Shoham et al. 2006; Margolis et al. 2012; Ovsepian 2019), which is critical for identifying how neuronal population activity changes with learning. We first cover the experimental methods that allow optical access to brain areas and cellular populations of interest, followed by advances in our understanding of the neural basis of learning-related activity changes enabled by chronic optical imaging, as well as emerging computational methods for analyzing the large resulting datasets.
Methods: imaging wide and deep
The last decade has seen a dramatic increase in the area of tool development for neuroscience, or neurotechnology (Jorgenson et al. 2015; Alivisatos et al. 2015), including advances in both electrophysiological and imaging methods for chronic recording of neural activity (Lütcke et al. 2013; Fu et al. 2016; Steinmetz et al. 2018; Piatkevich et al., 2019). Improved fluorescent sensors for the optical detection of neural activity, along with new microscopy techniques, are the primary factors that have driven the field forward by increasing the capacity for sensitive in vivo measurements of large populations of neurons.
Genetically encoded calcium indicators (GECIs) are the most commonly used class of fluorescent protein-based neural activity sensors because of the large relative change in intracellular calcium concentration that occurs with action potential firing and the resulting high signal-to-noise of the detected signals (Grienberger and Konnerth 2012). GECIs with improved sensitivity, brightness, kinetics, and expression properties have revolutionized the field, allowing many new applications for measuring neural activity in vivo (Kerr and Denk 2008; Hires et al. 2008; Margolis et al. 2011; Tian et al. 2012; Lin and Schnitzer 2016; Yang and Yuste 2017). The GCaMP family of GECIs, based on green fluorescent protein (GFP), has been widely adopted because of the large calcium-dependent fluorescence changes, photostability and capacity for imaging calcium signals in subcellular structures such as axons and dendrites (Chen et al. 2013b; Dana et al. 2019). Red-shifted GECIs have also undergone major improvements and now rival green GECIs in their ability to detect neuronal activity (Dana et al. 2016). This is important because the longer wavelength excitation and emission light used for red GECIs scatters less in tissue, allowing cellular imaging deeper in the intact brain. Calcium indicators have two main caveats. First, calcium signals are a relatively slow readout of neuronal activity because calcium entry during a single action potential lasts approximately 100 ms, with GECI fluorescence emission typically taking hundreds of ms to return to baseline, limiting the measurement of detailed temporal activity patterns during trains of action potentials. Second, calcium indicators buffer intracellular calcium, potentially influencing natural intracellular calcium dynamics (Steinmetz et al. 2017; Bootman et al. 2018; McMahon and Jackson 2018). In spite of these limitations, GECIs have become an essential tool for in vivo cellular imaging, and currently remain the most popular neural activity sensors among a growing number of promising voltage and neurotransmitter indicators (discussed further in the Outlook section, below).
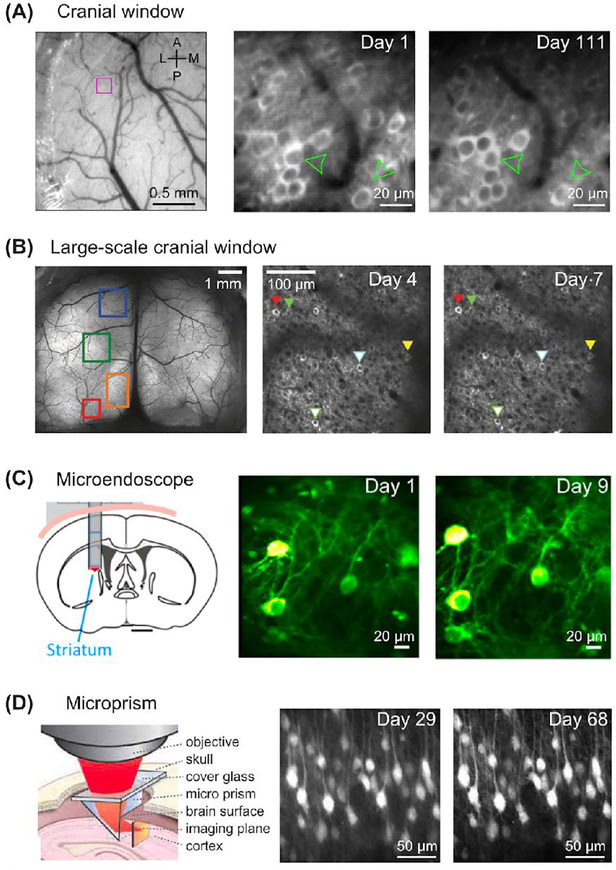
While the first chronic imaging studies using GECIs in visual, motor, and somatosensory cortex tracked population activity over an impressively long time period (Figure 1A) (Mank et al. 2008; Margolis et al. 2012), it has remained an important goal to increase the number of imaged neurons in order to understand the nature of neuronal population activity across widespread brain areas. To achieve this, advanced versions of two-photon microscopes have been developed with larger or multiple fields of view laterally (across the brain surface) (Lecoq et al. 2014; Chen et al. 2016; Sofroniew et al. 2016; Stirman et al. 2016), or rapid scanning of volumes axially (through the depth of the brain) (Song et al. 2017a; Lu et al. 2017; Nöbauer et al. 2017). In addition, newly developed optical implants have enabled hugely expanded views of neural tissue, rendering up to ~36 mm2 of the cortical surface optically accessible compared to ~1 mm2 in original glass coverslip-based windows (Figure 1B). Removable or penetrable windows have also been developed that allow access for drug application or introduction of recording electrodes (Goldey et al. 2014; Roome and Kuhn 2014). The combined advances in microscopy and optical implants have allowed an astonishing increase in the number of neurons possible to image in a single mouse, from a few in pioneering studies (Mank et al. 2008; Andermann et al. 2013) to an estimated one million with the recently developed Crystal Skull and See-Shell methods (Kim et al. 2016; Ghanbari et al. 2019).
Figure 1.
Chronic imaging approaches for tracking neuronal activity using two-photon imaging of genetically encoded calcium indicators (GECIs). A. Left: Top view of the cerebral cortex through a cranial window implanted above the dura mater. Right: layer 2/3 cortical neurons expressing YC3.60 followed over 111 days. Adapted from Margolis, Lütcke et al. (2012) (Margolis et al 2012). B. Left: Top view of cortex through a large-scale “Crystal Skull” cranial window. Right: Chronic imaging of one of many optically accessible cortical neuronal populations expressing GCaMP6s. Adapted from Kim et al. (2016) (Kim et al. 2016). C. Left: Deep brain structures can be imaged through an implanted GRIN lens. Right: Neurons in striatum expressing GCaMP6s. Note that neurons were followed for a longer time period than shown here. Adapted from Bocarsly, Jiang et al. (2015) (Bocarsly et al. 2015). D. Left: Axially oriented brain structures can be imaged through a prism implanted facing the side of the area of interest. A microprism can also be used for deeper brain structures. Right: Neurons in neocortex expressing GCaMP3 imaged over multiple weeks. Apical dendrites are visible in the x-z plane. Note that the original image was cropped for display. Adapted from Andermann et al. (2013) (Andermann et al. 2013).
Neurons within deep brain structures have been essentially hidden from view since the fluorescence excitation light needed to reach them is heavily scattered within brain tissue, especially at shorter wavelengths. Typical two-photon imaging (820–980 nm wavelength) is limited to less than 1 mm below the brain surface (Denk and Svoboda 1997; Helmchen and Denk 2005). Red fluorescent probes improve imaging depth by using longer wavelength excitation (1000–1100 nm) (Dana et al. 2016). Recently established three-photon excitation (1300 nm wavelength) (Ouzounov et al. 2017) can visualize neurons through all layers of mouse cortex to the hippocampus, more than 1 mm below the brain surface. However, even with these developments, optical access to many regions of the intact mammalian brain cannot be achieved. The introduction of chronically implanted optical relay lenses (e.g., GRIN lenses) and optical chambers (created from a glass coverslip fused to a guide tube or cannula) has been a major advance for deep brain imaging (Barretto et al. 2009; Dombeck et al. 2010). Although invasive, requiring removal or displacement of the overlying neural tissue, such implants are currently the only available method to optically access deep brain areas beyond the light penetration depth of multiphoton excitation. Optical chambers have better optical quality than GRIN lenses and have provided high-resolution data from hippocampus and striatum (Sato et al. 2016; Bloem et al. 2017), but are also more invasive because of their typically three times larger diameter. Transcortical GRIN lenses have been successfully used to perform chronic optical imaging of neurons within structures such as hypothalamus, hippocampus, and amygdala (Ziv et al. 2013; Bocarsly et al. 2015; Grewe et al. 2017), several mm below the brain surface (Figure 1C), and can be used with either two-photon or one-photon fluorescence excitation (Jung et al. 2004). Microprisms can also be used for specialized applications (Figure 1D), allowing an en bloc view of neuronal populations across different cortical layers, vertically oriented apical dendrites, or hard to reach locations such as insular or entorhinal cortex (Andermann et al. 2013; Low et al. 2014; Livneh et al. 2017). Together, the parallel improvements in fluorescent activity indicators, microscopes, and optical implants represent major advances for the study of in vivo neural function using chronic optical imaging techniques.
Stability and plasticity
How has chronic in vivo imaging of cellular activity been used to investigate unanswered questions in neuroscience? One fundamental question is the degree of stability of single neuron versus population activity over extended time periods. The balance between stability and plasticity could relate to the capacity of neural circuits to faithfully encode and store information, yet also adapt to a changing environment and learn new information (Lütcke et al. 2013; Margolis et al. 2014; Clopath et al. 2017). A number of longitudinal studies have provided insight into the nature of stability of neural function on many levels, from synapses (Ziv and Brenner 2018) to whole-brain representations (Kolasinski et al. 2016).
Chronic imaging of GECIs examines this question on the neuronal population level. One emerging view from studies of visual, somatosensory, motor, and association areas of the cerebral cortex is that neuronal population activity is largely stable over days even though the activity of individual neurons that comprise these populations is variable (Mank et al. 2008; Margolis et al. 2012; Mayrhofer et al. 2015; Clopath et al. 2017) (but see counterexamples, below). For example, the overall proportion of neurons active in response to touch in primary sensory cortex (S1) of behaving mice is stable, but the identities of the individual neurons that are active can change from day to day (Peron et al. 2015; Chen et al. 2015a). Similar results have been found in visual and motor cortex (Huber et al. 2012; Peters et al. 2014; Poort et al. 2015). The same is true of neurons in posterior parietal cortex during performance of a visually-guided virtual reality navigation task, even after learning, when behavioral performance had stabilized and the neural representation had become more refined (Driscoll et al. 2017). These data suggest that the activity of individual neurons is more variable or flexible over time than the population as a whole, implying that unknown population-level homeostatic mechanisms normalize overall population activity (LeMessurier and Feldman 2018). There are also open questions about the distribution of activity within neuronal populations. In superficial layers of sensory cortex, for example, a small fraction of neurons responds to stimuli reliably and robustly, while most other neurons are more variable and less excitable (Shoham et al. 2006; Barth and Poulet 2012; Margolis et al. 2012; Ovsepian 2019). In general, the role of individual neurons versus populations in neural signaling is an important question for understanding both local circuit computations and how information is transmitted between brain areas.
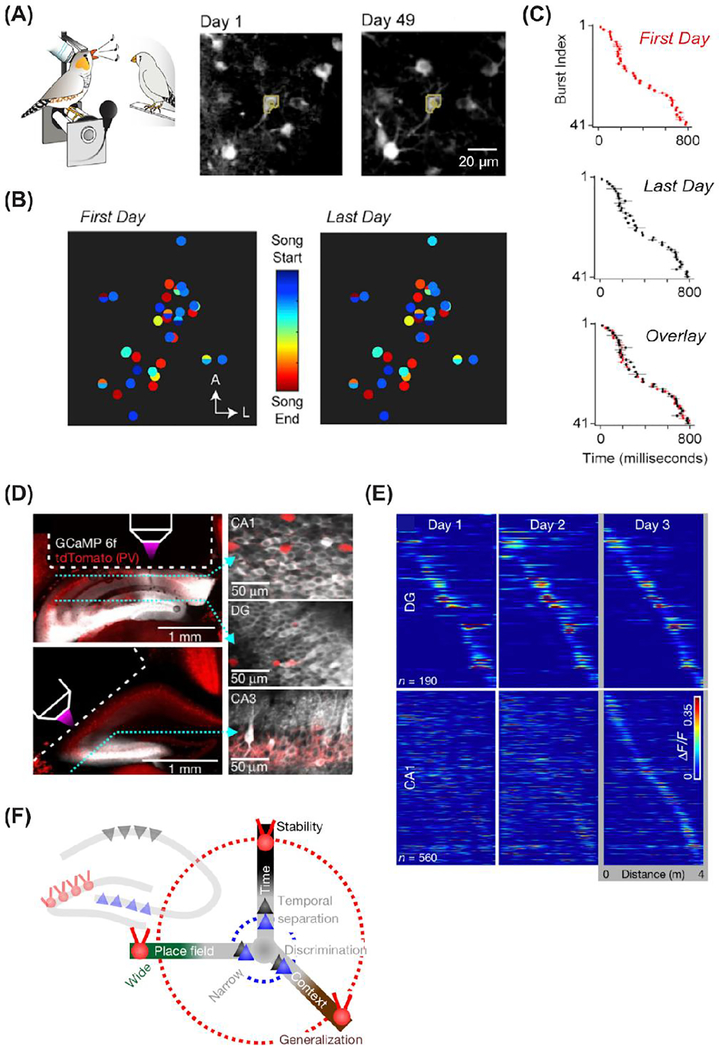
There are also striking examples of long-term stability of neuronal activity, indicating that not all single neurons are variable. Chronic two-photon imaging data from neurons in the singing-related area HVC of the songbird show maintenance of the precise sequence of population activity across several weeks (Katlowitz et al. 2018) (Figure 2A–C). A high degree of stability was also found in chronic electrophysiological recordings of cortical and subcortical neurons in rats performing natural behaviors and motor tasks (Dhawale et al. 2017). It will be important to determine whether certain subsets of neurons are more stable than others by using high density electrical recordings (Steinmetz et al. 2018) or large field-of-view neuronal population imaging (Lecoq et al. 2014; Chen et al. 2016; Sofroniew et al. 2016; Stirman et al. 2016), and to gain information from various brain regions during both learning and stable behavioral performance.
Figure 2.
Stability and flexibility of behavior-related neuronal population activity measured with chronic imaging. A. Example of stability in songbird experiment. Two-photon images of neurons expressing GCaMP6f tracked over 49 days. B. Comparison of song timing maps for a population of neurons. C. Stability of burst index measures. A-C adapted from Katlowitz et al. (2018) (Katlowitz et al. 2018). D. Subregion-specific stability in hippocampus. Chronic imaging of CA1, DG, and CA3 sub-regions. E. Heat maps of neuronal calcium signals across three days showing higher stability for DG than CA1. F. Schematic of differences in stability, width, and generalization for CA1, DG, and CA3. D-E adapted from Hainmueller and Bartos (2018) (Hainmueller and Bartos 2018).
The determinants of neuronal stability and flexibility are still unknown. One possibility is that the inherent dynamics of a given neural circuit, and its modulation during behavior, determines its degree of stability. A chronic calcium imaging study of hippocampus in behaving mice addresses whether different hippocampal subregions in the same animal show different degrees of stability (Hainmueller and Bartos 2018). The authors used an optical cannula to access three regions of hippocampus, CA1, DG and CA3 and imaged neuronal activity during performance of a visually guided task in virtual reality. Remarkably, DG was the only one of the 3 hippocampal subregions to show stability of spatial coding across 3 days (Figure 2D–F), while CA1 and CA3 were more variable (similar to (Ziv et al. 2013)). These results indicate that even closely connected sub-regions of a larger interconnected neural structure can show varying degrees of stability. Whether the differences in flexibility relate to the role of these sub-circuits in cognitive flexibility or the capacity for learning remains to be determined. It is reasonable to posit that stability and flexibility both play important roles for brain function.
Neural circuit changes during learning
How the brain changes with learning is another fundamental question that chronic optical imaging is uniquely suited to investigate. As behavioral tasks are learned and refined, multiple brain areas participate in adaptive synaptic and population-level changes in neuronal function (Romo and de Lafuente 2013; Le Merre et al. 2018; Crochet et al. 2018). Many learning paradigms in mouse models, especially those combined with imaging, involve presentation of a sensory stimulus followed by a response generated by the subject that produces a reward (usually food or water). This broad categorization of sensorimotor learning can be conceptualized as three phases of learning: perceptual, sensorimotor association, and skill learning (Makino et al. 2016). By tracking changes in functional properties of population activity over time, chronic imaging experiments can investigate the progression of neural changes within targeted brain regions through these distinct phases of learned behavior. Recent chronic imaging studies in mice have begun to make insights into longstanding issues in neuroscience, including the extent of plasticity that takes place in primary sensory areas of the cerebral cortex, how information is routed from one brain area to downstream target areas during learning, and how subsets of neurons undergo learning-related changes in selectivity for sensory, behavioral, and cognitive or reward-related features of task performance. For further consideration of data from in both non-human primates and mice, we refer the reader to (Makino et al. 2016).
Major efforts have been made to optimize experimental approaches in head-restrained mice that combine cellular resolution optical imaging with behavioral paradigms implemented around a stationary microscope, allowing high-resolution imaging during locomotion and sensory-guided decision-making tasks (Komiyama et al. 2010; Guo et al. 2014; Dombeck and Tank 2014). In a complementary approach, miniaturized head-mounted microscopes allow chronic imaging of population activity during more naturalistic behaviors (Flusberg et al. 2008; Cai et al. 2016; Jacob et al. 2018), albeit with lower optical resolution, and therefore more challenging cell identification across days (Ziv et al. 2013). The following examples illustrate key features of changes in cortical and subcortical neuronal population activity during learning, measured using either head-restrained or miniature head-mounted chronic cellular imaging techniques.
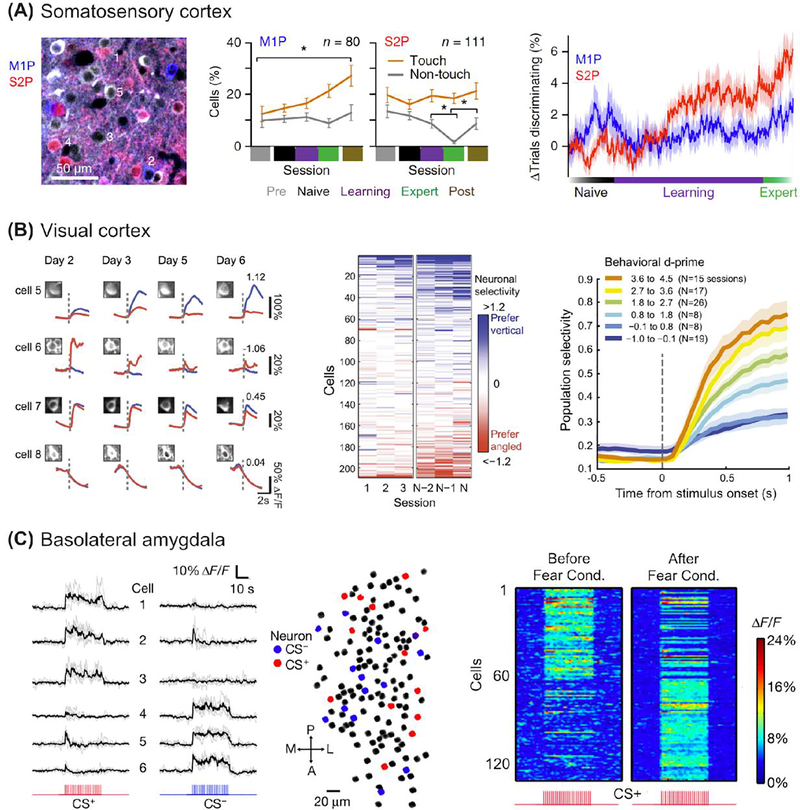
Early sensory areas of the cerebral cortex send axonal projections to many downstream target areas, and also receive feedback inputs from “higher order” areas, but it has remained unclear if certain pathways are particularly important for learning-related neural processing. In the primary somatosensory cortex (S1) of mice, intermingled populations of layer 2/3 neurons project to either S2 or M1, and can be distinguished using retrograde tracers (Chen et al. 2013a; Yamashita et al. 2013; Chen et al. 2015a; Tervo et al. 2016; Chatterjee et al. 2018). When mice learn to discriminate between textures with their whiskers, S2- and M1-projecting neurons undergo different types of functional changes: the fraction of M1-projecting neurons sensitive to touch increases, while the S2-projecting neurons remain the same in number but become more discriminative between stimuli (Chen et al. 2015a) (Fig. 3A). Strikingly, learning-related activity changes persist outside of the behavioral task for M1-projecting neurons, while the activity of S2-projecting neurons is discriminative only during task performance, highlighting the specificity of behavior-related neural dynamics for distinct populations of projection neurons within S1. These findings indicate that learning-related plasticity can occur within specific types of projection neurons, even at primary cortical stages of the sensory pathway. However, the relative contribution of the encoding of sensory stimuli versus changes in the learned behavioral strategy (i.e., stronger or more efficient whisker movements toward target objects) to S1 plasticity remains to be determined (Peron et al. 2015; Chen et al. 2015a).
Figure 3.
Learning-related changes in neuronal population activity measured with chronic cellular imaging. A. Projection neurons in primary somatosensory cortex (S1) show distinct learning-related activity during performance of a tactile discrimination task. Left: Identification of M1-projecting (M1P) and S2-projecting (S2P) neurons via retrograde tracers. Middle: The fraction of neurons classified as touch or non-touch as a function of Naive, Learning, and Expert behavioral phases. Right: The change in discrimination accuracy of M1P and S2P neurons for Go and NoGo tactile stimuli (P100 vs. P1200 textures) through learning. Adapted from (Chen et al. 2015a). B. Neurons in primary visual cortex (V1) show diverse learning-related activity during performance of a visual discrimination task. Left: Example calcium signals from four neurons across four imaging sessions in response to a vertical, rewarded stimulus (blue) or an angled, non-rewarded stimulus (red). Middle: Selectivity of neurons across the first three and last three training sessions. Right: Neuronal population selectivity as a function of learning. Each curve depicts the time course of selectivity at a range of behavioral d’. Adapted from (Poort et al. 2015). C. Neurons in basolateral amygdala change selectivity with auditory fear conditioning. Calcium signals of cells responsive to two different auditory tones (CS+ or CS−) before pairing the conditioned stimulus (CS+) with a foot shock using a fear conditioning paradigm. Right, Cell population responses in an example mouse to the CS+ tone before and after fear conditioning. A, anterior; L, lateral; M, medial; P, posterior. Adapted from (Grewe et al. 2017).
A striking example of learning-related changes in early sensory encoding comes from work in mouse primary visual cortex (V1). When mice are trained to discriminate visual stimuli of different orientation for water rewards, individual V1 neurons show prominent changes in orientation selectivity with learning (Fig. 3B) (Poort et al. 2015). Subsets of neurons become selective for either the rewarded or non-rewarded visual stimuli (with many still visually responsive but non-selective). Neuronal populations as a whole show progressive improvement in neural discriminability that parallel the increased behavioral performance with learning (Fig. 3B, right). These results, and those of studies in other sensory modalities (Shuler and Bear 2006; Hui et al. 2009; Gdalyahu et al. 2012; Kato et al. 2012; Lacefield et al. 2019), support the concept that neuronal selectivity for sensory stimuli can change dramatically depending on the stimuli’s behavioral salience (rewarded or unrewarded).
Such results indicate that cognitive aspects such as reward or salience can strongly influence neural plasticity that takes place during learning. Indeed, reward may even be represented by dedicated neuronal subpopulations, as recently found with in vivo imaging in hippocampus (Gauthier et al. 2018). In a noteworthy example from mouse visual association cortex, (Ramesh et al. 2018) found that neurons recruited to become active during learning were part of reward-related (value-coding) rather than stimulus-related neuronal subpopulations (ensembles). Imaging studies have also revealed extensive learning-related changes in neuronal population activity in subcortical brain areas with key roles in valence encoding, such as the amygdala. Here, pairing an auditory tone with an aversive foot shock led to a near-total switch in auditory-responsive neurons, with neurons initially activated by the conditioned tone losing responsiveness, and previously inactive neurons becoming active (Grewe et al. 2017) (Fig. 3C). These results emphasize the importance of behavioral salience for learning-related neural signaling and population-level functional plasticity.
While we have largely focused on functional studies of neural activity, structural plasticity of neurons, especially on the level of dendritic spines, is another type of experience-dependent and learning-related plasticity that has been extensively studied using chronic two-photon imaging (Trachtenberg et al. 2002; Holtmaat and Svoboda 2009; Berry and Nedivi 2017). Motor learning affects spine formation and maintenance in motor cortex but not in other cortical areas, while spines in sensory cortex undergo plasticity after sensory discrimination training (Yang et al. 2009; Fu et al. 2012; Kuhlman et al. 2014). Auditory fear conditioning leads to increased spine formation in primary auditory cortex (A1) and extinction leads to elimination of the newly formed spines (Lai et al. 2018). Notably, the opposite effects are seen in frontal association cortex, where extinction of learned associations causes spine formation (Lai et al. 2012). Learning-related changes in spines have been shown to be influenced by inhibitory interneuron activity in motor cortex, suggesting an interplay between excitatory and inhibitory neurotransmission in dendritic spine plasticity (Chen et al. 2015b). However, while it is likely that structural spine dynamics relate to neural activity during learning, the exact relationship has yet to be determined. Fewer in vivo studies have investigated the functional properties of dendritic spines because of the high level of mechanical stability needed to image these micron-scale structures. Thus, most data on spine function has been acquired in anesthetized animals or in ex vivo preparations (Yasuda et al. 2004; Chen et al. 2013b; Berry and Nedivi 2017). However, newer head fixation methods such as using an air supported platform has achieved stability necessary to resolve spines in awake behaving animals (Pryazhnikov et al. 2018), suggesting that functional imaging of dendritic spines will be an active area of future research.
Chronic optical imaging of axonal calcium signals in behaving mice has been performed in several studies (Glickfeld et al. 2013; Broussard et al. 2018; Dana et al. 2019), which has revealed progressive, circuit-specific changes during learning (Burgess et al. 2016; Kupferschmidt et al. 2017). In the future, it may be possible to combine cellular resolution presynaptic axonal and postsynaptic dendritic imaging (Takahashi et al. 2016; Lacefield et al. 2019) to define changes in the learning-related input-output properties of neural circuits. This could enable novel investigations of many outstanding questions related to how brain areas interact during learning and decision-making, including the influence of feedback signals from higher-order cortical areas, or subcortical structures such as thalamus and amygdala, and the effects of neuromodulator signals on neuronal population activity.
It is important to mention that in vivo imaging, in particular studies of dendritic spine structural dynamics, have provided valuable information on potential mechanisms of neural dysfunction. Alterations in dendritic spines have been associated with neurodevelopmental, neurodegenerative, and neuropsychiatric disorders, as reviewed elsewhere (Knobloch and Mansuy 2008; Glausier and Lewis 2013; Martínez-Cerdeño 2017). In Alzheimer’s disease, for example, there is a decrease in spine density that becomes more pronounced near plaques that are characteristic of the disease (Spires et al. 2005). Furthermore, whisker stimulation-related plasticity is impaired in aged compared to young mice, suggesting impairments in spine dynamics during normal aging as well (Voglewede et al. 2019). Further experiments designed to simultaneously acquire in vivo functional and structural data, in addition to further chronic neuronal population imaging studies in mouse disease models, would provide important information on the relationship between structural and functional neural circuit plasticity in both the healthy and diseased brain.
Analysis of chronic imaging data for understanding neural coding and behavior
Chronic cellular imaging experiments can produce massive datasets, including hours of movies from thousands of neurons measured repeatedly in multiple imaging sessions. Thus, optimizing data analysis is critical to avoid major bottlenecks for such experiments (Paninski and Cunningham 2018). Beyond measuring first-order features, such as the fraction of active neurons or their tuning properties, sophisticated analysis methods are essential for inferring the dynamic properties of neuronal population activity from calcium imaging data. A key first step is to estimate the underlying neural activity from the fluorescence signal, which is not trivial since calcium fluorescence signals are relatively slow and noisy compared to action potential firing, and optical signals from individual cells can be obscured by their neighbors. A number of computational methods based on template matching, deconvolution, approximate Bayesian inference, and matrix factorization have been developed to infer spiking activity of neurons from calcium imaging data. For further information, the reader is referred to recent reviews on the topic of calcium imaging analysis methods and pipelines (Stringer and Pachitariu 2019; Pnevmatikakis 2019). One popular analysis framework (Pnevmatikakis et al. 2016) uses a constrained nonnegative matrix factorization approach to identify the locations of neurons and demix the ones that are spatially overlapping, while simultaneously deconvolving their spiking activity from the spatiotemporal structure of the population calcium recordings. A limitation of these methods is the requirement of user intervention (e.g., setting parameters), and certain assumptions imposed on the model of calcium signal generation, or on the dynamics of fluorescence measurements. Such limitations can present particular challenges for analysis of chronic cellular imaging data, because the same cells must be correctly identified and tracked, and calcium signals extracted, across multiple days in order to provide meaningful measurements of long-term stability or plasticity. Newly developed analysis frameworks have started to take these specific issues related to chronic imaging into account (Giovannucci et al. 2019), and will undoubtedly be a topic of further development.
Data-driven methods, supervised learning techniques, and machine learning have great potential for improving analysis of chronic cellular imaging data by minimizing user intervention and increasing the scalability and flexibility of the frameworks (Sasaki et al. 2008; Patel et al. 2015; Speiser et al. 2017; Theis et al. 2016). One study that compared the performance of various generative and supervised spike inference algorithms (including deep neural networks) applied to specific datasets, concluded that many algorithms yield similar performance for inferring spike rates, but that each offers unique advantages and disadvantages in terms of speed and generalizability (Berens et al. 2018). Other recent work has argued that spike inference using simple non-negative deconvolution meets the performance of supervised methods, and recommended this as the preferred choice due to its simplicity and efficiency (Pachitariu et al. 2018). Regardless of the specific algorithm used, a key issue for chronic cellular imaging datasets is the accuracy and robustness of the analysis of the same cells across multiple imaging sessions. One study found that an algorithm first trained on simultaneous recordings of spikes and calcium data performed well on inferring spike rate when applied to new datasets (Theis et al. 2016). The same approach could be used to analyze data from the same neuronal populations from one day to the next, enhancing the efficiency and power of chronic imaging data throughput, thereby speeding insights into mechanisms of neural plasticity.
Calcium imaging can also be performed across large-scale, spatially separated brain areas. Spatiotemporal features of such widefield calcium signals are generally analyzed directly (without spike inference). Network analysis methods originally developed for human neuroimaging experiments (Bassett and Sporns 2017; Khambhati et al. 2018) have been applied to imaging data from various optical sensors in mice, including calcium indicators, to make insights into dynamic interactions between brain regions on the mesoscale level (Xie et al. 2016; McVea et al. 2016). For example, spectral analysis of resting-state cortical networks, constructed based on wide-field calcium imaging data, found frequency-dependent activity clusters in specific cortical regions (Vanni et al. 2017). Other studies applied visibility graph (Lacasa et al. 2008) in combination with machine learning techniques to investigate the temporal characteristics of wide-field cortical calcium dynamics related to behavioral state (Zhu et al. 2018). An emerging view from these and other studies (McGinley et al. 2015; Musall et al., 2019; Stringer et al. 2019) is the importance of global modulatory influences driven by arousal and movement on behavior-related neural dynamics. However, little is still known about how large-scale networks change during learning (Makino et al. 2017), or how such large-scale changes are related to changes in individual neurons or tractable neuronal populations. Artificial intelligence (AI) and deep learning techniques capable of automatically learning patterns and representations in data may be particularly well-suited for investigating neural mechanisms of behavior across the multiple spatial and temporal scales available in chronic imaging experiments, perhaps by generating models that are predictive of learning rate. To reach this stage, however, machine-learning approaches will have to establish their robustness for the analysis of neural data (Vogt 2018). It remains an active goal to bridge gaps in our understanding of how neural circuits operate on multiple levels of brain function, on the temporal scale between short-term (milliseconds or seconds scale) and long-term (days or weeks scale) changes in neuronal activity, and on the spatial scale between brain regions and local neuronal populations. Chronic imaging, combined with experimental and computational advances, has great potential for further mechanistic discovery of neural circuit function.
Outlook: multimodal interrogation of neural circuits
Rapid progress in the development of additional neural sensors has been an exciting area of advance. High-performance genetically encoded voltage indicators (GEVIs) have been a long-sought goal as a more direct and faster readout of neuronal activity (Knöpfel et al. 2003). Recent improvements in GEVIs have been dramatic, including the capacity for in vivo imaging, and their sensitivity is beginning to rival that of GECIs for detecting single action potentials (Hochbaum et al. 2014; Gong et al. 2015; Song et al. 2017b; Piatkevich et al. 2019). Imaging the millisecond kinetics of GEVI signals from neuronal populations requires fast frame rates generally beyond the capacity of two-photon laser-scanning microscopy, and the high light levels needed for fluorescence excitation can lead to photobleaching and photodamage (Yang and St-Pierre 2016; Xu et al. 2017; Bando et al. 2019). Despite these challenges, chronic cellular resolution imaging of GEVIs has the potential to provide unprecedented information on plasticity- or learning-related changes in the temporal patterns of neuronal firing. GEVIs, like the most sensitive GECIs, are also capable of detecting slower subthreshold voltage changes, although most population imaging studies have focused on the faster signal associated with action potential firing.
Additionally, new classes of genetically encoded indicators for detecting neurotransmitters (e.g., glutamate, GABA (Marvin et al. 2013, 2019)) and neuromodulators (e.g., dopamine, acetylcholine, or norepinephrine (Patriarchi et al. 2018; Jing et al. 2018; Feng et al. 2019)) represent major advances with potential for further circuit discovery. In future chronic imaging experiments, it may be feasible to investigate multiple aspects of neural circuit activity simultaneously by imaging different color indicators expressed in distinct cell types or cellular compartments. For example, simultaneous imaging of axons and dendrites or cell bodies could be performed using green and red GECIs, respectively, to distinguish pre- and postsynaptic sites of plasticity. Neuromodulation of neuronal population activity could be investigated with co-expression of GECIs or GEVIs and neuromodulator sensors in neurons (for example, GCaMP6 and dLight1 (Patriarchi et al. 2018)). Novel applications of viral vectors (Bedbrook et al. 2018) and a large repertoire of transgenic reporter mice (Dana et al. 2014, 2018; Madisen et al. 2015; Wekselblatt et al. 2016; Daigle et al. 2018) allow flexibility in restricting expression of genetically encoded indicators to specific cell types, subcellular structures, and brain regions (Broussard et al. 2018; Dana et al. 2019). Employed together with sophisticated behavioral paradigms in both head-fixed and freely moving mice, the growing toolkit of genetically encoded indicators holds great potential for extending our knowledge of neural circuit function and plasticity both on a fast time scale of single behavioral trials and over longer time scales during learning and memory formation. Many previously intractable questions in neuroscience are now addressable, from the functional architecture of memory, neural circuit mechanisms of injury or disease, and even undiscovered functional cell populations and projections, which will in turn lead to new insights into the functioning of the brain and open up new areas of inquiry.
All-optical circuit interrogation using optogenetics to manipulate neuronal activity combined with functional imaging to simultaneously monitor activity has become an achievable goal (Prakash et al. 2012; Carrillo-Reid et al. 2019; Marshel et al. 2019). Optogenetics can be used to selectively modulate the activity of neurons embedded within neural circuits (Deisseroth 2015). Combining optogenetics with two-photon imaging has recently been used to recapitulate activity in neuronal ensembles that can mimic visual input (Carrillo-Reid et al. 2019; Marshel et al. 2019). Future experiments combining optogenetics with GEVIs will certainly lead to additional understanding of how neural networks encode stimuli and integrate synaptic input.
Ongoing and future experiments aim to achieve an expanded view of large-scale neuronal population activity with cellular resolution, and a fine-scale view of activity within subcellular compartments, such as axons and dendrites. Combined imaging of voltage, calcium, neurotransmitters, and neuromodulator indicators could be performed on both the cellular and subcellular levels during innate and learned behaviors. Together, neurotechnology developments for chronic optical imaging, including new indicators and next-generation optical systems, have great promise for providing novel insight into neural circuit function and plasticity.
Acknowledgements
The authors are supported by grants from the US National Institutes of Health (R01NS094450, DJM), US National Science Foundation (CBET-1605646, LN and DJM; IOS-1845355, DJM), and New Jersey Commission on Brain Injury Research (CBIR16IRG032, DJM and LN).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
References
- Akassoglou K, Merlini M, Rafalski VA, et al. (2017) Imaging of CNS Injury and Disease. J Neurosci Off J Soc Neurosci 37:10808–10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos AP, Chun M, Church GM, et al. (2015) A National Network of Neurotechnology Centers for the BRAIN Initiative. Neuron 88:445–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann ML, Gilfoy NB, Goldey GJ, et al. (2013) Chronic cellular imaging of entire cortical columns in awake mice using microprisms. Neuron 80:900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando Y, Sakamoto M, Kim S, et al. (2019) Comparative Evaluation of Genetically Encoded Voltage Indicators. Cell reports 26:802–813.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto RPJ, Messerschmidt B, Schnitzer MJ (2009) In vivo fluorescence imaging with high-resolution microlenses. Nat methods 6:511–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Poulet JFA (2012) Experimental evidence for sparse firing in the neocortex. Trends Neurosci 35:345–355. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Sporns O (2017) Network neuroscience. Nat Neurosci 20:353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook CN, Deverman BE, Gradinaru V (2018) Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annu Rev Neurosci 41:323–348. [DOI] [PubMed] [Google Scholar]
- Berens P, Freeman J, Deneux T, et al. (2018) Community-based benchmarking improves spike rate inference from two-photon calcium imaging data. PLoS Comput Biol 14:e1006157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KP, Nedivi E (2017) Spine Dynamics: Are They All the Same? Neuron 96:43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloem B, Huda R, Sur M, Graybiel AM (2017) Two-photon imaging in mice shows striosomes and matrix have overlapping but differential reinforcement-related responses. [DOI] [PMC free article] [PubMed]
- Bocarsly ME, Jiang W-C, Wang C, et al. (2015) Minimally invasive microendoscopy system for in vivo functional imaging of deep nuclei in the mouse brain. Biomed Opt Express 6:4546–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman MD, Allman S, Rietdorf K, Bultynck G (2018) Deleterious effects of calcium indicators within cells; an inconvenient truth. Cell Calcium 73:82–87. [DOI] [PubMed] [Google Scholar]
- Broussard GJ, Liang Y, Fridman M, et al. (2018) In vivo measurement of afferent activity with axon-specific calcium imaging. Nat Neurosci 21:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess CR, Ramesh RN, Sugden AU, et al. (2016) Hunger-Dependent Enhancement of Food Cue Responses in Mouse Postrhinal Cortex and Lateral Amygdala. Neuron 91:1154–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, et al. (2016) A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534:115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reid L, Han S, Yang W, et al. (2019) Controlling Visually Guided Behavior by Holographic Recalling of Cortical Ensembles. Cell 178:447–457.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Sullivan HA, MacLennan BJ, et al. (2018) Nontoxic, double-deletion-mutant rabies viral vectors for retrograde targeting of projection neurons. Nat Neurosci 21:638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Carta S, Soldado-Magraner J, et al. (2013a) Behaviour-dependent recruitment of long-range projection neurons in somatosensory cortex. Nature 499:336–340. [DOI] [PubMed] [Google Scholar]
- Chen JL, Margolis DJ, Stankov A, et al. (2015a) Pathway-specific reorganization of projection neurons in somatosensory cortex during learning. Nat Neurosci 18:1101–1108. [DOI] [PubMed] [Google Scholar]
- Chen JL, Nedivi E (2010) Neuronal structural remodeling: is it all about access? Curr Opin Neurobiol 20:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Voigt FF, Javadzadeh M, et al. (2016) Long-range population dynamics of anatomically defined neocortical networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Kim AN, Peters AJ, Komiyama T (2015b) Subtype-specific plasticity of inhibitory circuits in motor cortex during motor learning. Nat Neurosci 18:1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, et al. (2013b) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clopath C, Bonhoeffer T, Hübener M, Rose T (2017) Variance and invariance of neuronal long-term representations. [DOI] [PMC free article] [PubMed]
- Crochet S, Lee S-H, Petersen CCH (2018) Neural Circuits for Goal-Directed Sensorimotor Transformations. [DOI] [PubMed]
- Crowe SE, Ellis-Davies GCR (2014) Longitudinal in vivo two-photon fluorescence imaging. J Comp Neurol 522:1708–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, et al. (2018) A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174:465–480.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Chen T-W, Hu A, et al. (2014) Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PloS one 9:e108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, et al. (2016) Sensitive red protein calcium indicators for imaging neural activity. [DOI] [PMC free article] [PubMed]
- Dana H, Novak O, Guardado-Montesino M, et al. (2018) Thy1 transgenic mice expressing the red fluorescent calcium indicator jRGECO1a for neuronal population imaging in vivo. PloS one 13:e0205444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, et al. (2019) High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat methods 16:649–657. [DOI] [PubMed] [Google Scholar]
- Deisseroth K (2015) Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci 18:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Svoboda K (1997) Photon upmanship: why multiphoton imaging is more than a gimmick. Neuron 18:351–357. [DOI] [PubMed] [Google Scholar]
- Dhawale AK, Poddar R, Wolff SB, et al. (2017) Automated long-term recording and analysis of neural activity in behaving animals. [DOI] [PMC free article] [PubMed]
- Dombeck D, Tank D (2014) Two-photon imaging of neural activity in awake mobile mice. Cold Spring Harb Protoc 2014:726–736. [DOI] [PubMed] [Google Scholar]
- Dombeck DA, Harvey CD, Tian L, et al. (2010) Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci 13:1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll LN, Pettit NL, Minderer M, et al. (2017) Dynamic Reorganization of Neuronal Activity Patterns in Parietal Cortex. Cell 170:986–999.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Mo M, Yi M-H, et al. (2018) P2Y12R-Dependent Translocation Mechanisms Gate the Changing Microglial Landscape. Cell reports 23:959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhang C, Lischinsky JE, et al. (2019) A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102:745–761.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, et al. (2008) High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat methods 5:935–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Huang S, Zhou M, et al. (2018) Hotspots of dendritic spine turnover facilitate clustered spine addition and learning and memory. Nat Commun 9:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Yu X, Lu J, Zuo Y (2012) Repetitive motor learning induces coordinated formation of clustered dendritic spines in vivo. Nature 483:92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T-M, Hong G, Zhou T, et al. (2016) Stable long-term chronic brain mapping at the single-neuron level. Nat methods 13:875–882. [DOI] [PubMed] [Google Scholar]
- Gauthier JL, Tank DW (2018) A Dedicated Population for Reward Coding in the Hippocampus. Neuron 99:179–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdalyahu A, Tring E, Polack P-O, et al. (2012) Associative fear learning enhances sparse network coding in primary sensory cortex. Neuron 75:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbari L, Carter RE, Rynes ML, et al. (2019) Cortex-wide neural interfacing via transparent polymer skulls. Nat Commun 10:1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci A, Friedrich J, Gunn P, et al. (2019) CaImAn an open source tool for scalable calcium imaging data analysis. [DOI] [PMC free article] [PubMed]
- Glausier JR, Lewis DA (2013) Dendritic spine pathology in schizophrenia. Neuroscience 251:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickfeld LL, Andermann ML, Bonin V, Reid RC (2013) Cortico-cortical projections in mouse visual cortex are functionally target specific. Nat Neurosci 16:219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldey GJ, Roumis DK, Glickfeld LL, et al. (2014) Removable cranial windows for long-term imaging in awake mice. Nat Protoc 9:2515–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, et al. (2015) High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Sci 350:1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Gründemann J, Kitch LJ, et al. (2017) Neural ensemble dynamics underlying a long-term associative memory. Nature 543:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C, Konnerth A (2012) Imaging calcium in neurons. Neuron 73:862–885. [DOI] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan W-B (2002) Long-term dendritic spine stability in the adult cortex. Nature 420:812–816. [DOI] [PubMed] [Google Scholar]
- Guo ZV, Li N, Huber D, et al. (2014) Flow of cortical activity underlying a tactile decision in mice. Neuron 81:179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainmueller T, Bartos M (2018) Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature 558:292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel EJO, Grewe BF, Parker JG, Schnitzer MJ (2015) Cellular level brain imaging in behaving mammals: an engineering approach. Neuron 86:140–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W (2005) Deep tissue two-photon microscopy. Nat methods 2:932–940. [DOI] [PubMed] [Google Scholar]
- Hill RA, Damisah EC, Chen F, et al. (2017) Targeted two-photon chemical apoptotic ablation of defined cell types in vivo. Nat Commun 8:15837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Li AM, Grutzendler J (2018) Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nat Neurosci 21:683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hires SA, Tian L, Looger LL (2008) Reporting neural activity with genetically encoded calcium indicators. Brain cell Biol 36:69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, et al. (2014) All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat methods 11:825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K (2009) Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci 10:647–658. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Wilbrecht L, Knott GW, et al. (2006) Experience-dependent and cell-type-specific spine growth in the neocortex. Nature 441:979–983. [DOI] [PubMed] [Google Scholar]
- Huang C, Maxey JR, Sinha S, et al. (2018) Long-term optical brain imaging in live adult fruit flies. Nat Commun 9:872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D, Gutnisky DA, Peron S, et al. (2012) Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature 484:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui GK, Wong KL, Chavez CM, et al. (2009) Conditioned tone control of brain reward behavior produces highly specific representational gain in the primary auditory cortex. Neurobiol Learn Mem 92:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AD, Ramsaran AI, Mocle AJ, et al. (2018) A Compact Head-Mounted Endoscope for In Vivo Calcium Imaging in Freely Behaving Mice. Curr Protoc Neurosci 84:e51. [DOI] [PubMed] [Google Scholar]
- Jing M, Zhang P, Wang G, et al. (2018) A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat Biotechnol 36:726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckers E, Shah D, Hamaide J, et al. (2015) The power of using functional fMRI on small rodents to study brain pharmacology and disease. Front Pharmacol 6:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson LA, Newsome WT, Anderson DJ, et al. (2015) The BRAIN Initiative: developing technology to catalyse neuroscience discovery. [DOI] [PMC free article] [PubMed]
- Jung JC, Mehta AD, Aksay E, et al. (2004) In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy. J Neurophysiol 92:3121–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katlowitz KA, Picardo MA, Long MA (2018) Stable Sequential Activity Underlying the Maintenance of a Precisely Executed Skilled Behavior. Neuron 98:1133–1140.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato HK, Chu MW, Isaacson JS, Komiyama T (2012) Dynamic sensory representations in the olfactory bulb: modulation by wakefulness and experience. Neuron 76:962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JND, Denk W (2008) Imaging in vivo: watching the brain in action. Nat Rev Neurosci 9:195–205. [DOI] [PubMed] [Google Scholar]
- Khambhati AN, Sizemore AE, Betzel RF, Bassett DS (2018) Modeling and interpreting mesoscale network dynamics. NeuroImage 180:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Zhang Y, Lecoq J, et al. (2016) Long-Term Optical Access to an Estimated One Million Neurons in the Live Mouse Cortex. Cell reports 17:3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch M, Mansuy IM (2008) Dendritic spine loss and synaptic alterations in Alzheimer’s disease. Mol Neurobiol 37:73–82. [DOI] [PubMed] [Google Scholar]
- Knöpfel T, Tomita K, Shimazaki R, Sakai R (2003) Optical recordings of membrane potential using genetically targeted voltage-sensitive fluorescent proteins. Methods 30:42–48. [DOI] [PubMed] [Google Scholar]
- Kolasinski J, Makin TR, Jbabdi S, et al. (2016) Investigating the Stability of Fine-Grain Digit Somatotopy in Individual Human Participants. J Neurosci Off J Soc Neurosci 36:1113–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama T, Sato TR, O’Connor DH, et al. (2010) Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature 464:1182–1186. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, O’Connor DH, Fox K, Svoboda K (2014) Structural plasticity within the barrel cortex during initial phases of whisker-dependent learning. J Neurosci Off J Soc Neurosci 34:6078–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt DA, Juczewski K, Cui G, et al. (2017) Parallel, but Dissociable, Processing in Discrete Corticostriatal Inputs Encodes Skill Learning. Neuron 96:476–489.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacasa L, Luque B, Ballesteros F, et al. (2008) From time series to complex networks: the visibility graph. Proc Natl Acad Sci United States Am 105:4972–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield CO, Pnevmatikakis EA, Paninski L, Bruno RM (2019) Reinforcement Learning Recruits Somata and Apical Dendrites across Layers of Primary Sensory Cortex. Cell reports 26:2000–2008.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CSW, Adler A, Gan W-B (2018) Fear extinction reverses dendritic spine formation induced by fear conditioning in the mouse auditory cortex. Proc Natl Acad Sci United States Am 115:9306–9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CSW, Franke TF, Gan W-B (2012) Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature 483:87–91. [DOI] [PubMed] [Google Scholar]
- Le Merre P, Esmaeili V, Charrière E, et al. (2018) Reward-Based Learning Drives Rapid Sensory Signals in Medial Prefrontal Cortex and Dorsal Hippocampus Necessary for Goal-Directed Behavior. Neuron 97:83–91.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecoq J, Savall J, Vučinić D, et al. (2014) Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat Neurosci 17:1825–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMessurier AM, Feldman DE (2018) Plasticity of population coding in primary sensory cortex. Curr Opin Neurobiol 53:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, Schnitzer MJ (2016) Genetically encoded indicators of neuronal activity. Nat Neurosci 19:1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh Y, Ramesh RN, Burgess CR, et al. (2017) Homeostatic circuits selectively gate food cue responses in insular cortex. Nature 546:611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low RJ, Gu Y, Tank DW (2014) Cellular resolution optical access to brain regions in fissures: imaging medial prefrontal cortex and grid cells in entorhinal cortex. Proc Natl Acad Sci United States Am 111:18739–18744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Sun W, Liang Y, et al. (2017) Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat Neurosci 20:620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H, Margolis DJ, Helmchen F (2013) Steady or changing? Long-term monitoring of neuronal population activity. Trends Neurosci 36:375–384. [DOI] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, et al. (2015) Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85:942–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Hwang EJ, Hedrick NG, Komiyama T (2016) Circuit Mechanisms of Sensorimotor Learning. Neuron 92:705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino H, Ren C, Liu H, et al. (2017) Transformation of Cortex-wide Emergent Properties during Motor Learning. Neuron 94:880–890.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank M, Santos AF, Direnberger S, et al. (2008) A genetically encoded calcium indicator for chronic in vivo two-photon imaging. Nat methods 5:805–811. [DOI] [PubMed] [Google Scholar]
- Margolis DJ, Lütcke H, Helmchen F (2014) Microcircuit dynamics of map plasticity in barrel cortex. Curr Opin Neurobiol 24:76–81. [DOI] [PubMed] [Google Scholar]
- Margolis DJ, Lütcke H, Helmchen F, et al. (2011) Chronic Two-Photon Imaging of Neural Activity in the Anesthetized and Awake Behaving Rodent. In: Optical Imaging of Neocortical Dynamics. Humana Press, pp 151–173 [Google Scholar]
- Margolis DJ, Lütcke H, Schulz K, et al. (2012) Reorganization of cortical population activity imaged throughout long-term sensory deprivation. Nat Neurosci 15:1539–1546. [DOI] [PubMed] [Google Scholar]
- Marshel JH, Kim YS, Machado TA, et al. (2019) Cortical layer-specific critical dynamics triggering perception. [DOI] [PMC free article] [PubMed]
- Martínez-Cerdeño V (2017) Dendrite and spine modifications in autism and related neurodevelopmental disorders in patients and animal models. Dev Neurobiol 77:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L, et al. (2013) An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat methods 10:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, et al. (2019) A genetically encoded fluorescent sensor for in vivo imaging of GABA. [DOI] [PubMed]
- Mayrhofer JM, Haiss F, Helmchen F, Weber B (2015) Sparse, reliable, and long-term stable representation of periodic whisker deflections in the mouse barrel cortex. NeuroImage 115:52–63. [DOI] [PubMed] [Google Scholar]
- McGinley MJ, Vinck M, Reimer J, et al. (2015) Waking State: Rapid Variations Modulate Neural and Behavioral Responses. Neuron 87:1143–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SM, Jackson MB (2018) An Inconvenient Truth: Calcium Sensors Are Calcium Buffers. Trends Neurosci 41:880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVea DA, Murphy TH, Mohajerani MH (2016) Large Scale Cortical Functional Networks Associated with Slow-Wave and Spindle-Burst-Related Spontaneous Activity. Front Neural circuits 10:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musall S, Kaufman MT, Juavinett AL, Gluf S, Churchland AK (2019) Single-trial neural dynamics are dominated by richly varied movements. Nat neurosci 22:1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöbauer T, Skocek O, Pernía-Andrade AJ, et al. (2017) Video rate volumetric Ca imaging across cortex using seeded iterative demixing (SID) microscopy. Nat methods 14:811–818. [DOI] [PubMed] [Google Scholar]
- Ouzounov DG, Wang T, Wang M, et al. (2017) In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat methods 14:388–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovsepian SV (2019) The dark matter of the brain. Brain Struct & Funct 224:973–983. [DOI] [PubMed] [Google Scholar]
- Ovsepian SV, Olefir I, Westmeyer G, et al. (2017) Pushing the Boundaries of Neuroimaging with Optoacoustics. Neuron 96:966–988. [DOI] [PubMed] [Google Scholar]
- Pachitariu M, Stringer C, Harris KD (2018) Robustness of Spike Deconvolution for Neuronal Calcium Imaging. J Neurosci Off J Soc Neurosci 38:7976–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L, Cunningham JP (2018) Neural data science: accelerating the experiment-analysis-theory cycle in large-scale neuroscience. Curr Opin Neurobiol 50:232–241. [DOI] [PubMed] [Google Scholar]
- Patel TP, Man K, Firestein BL, Meaney DF (2015) Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. J Neurosci methods 243:26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, et al. (2018) Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. [DOI] [PMC free article] [PubMed]
- Peron SP, Freeman J, Iyer V, et al. (2015) A Cellular Resolution Map of Barrel Cortex Activity during Tactile Behavior. Neuron 86:783–799. [DOI] [PubMed] [Google Scholar]
- Peters AJ, Chen SX, Komiyama T (2014) Emergence of reproducible spatiotemporal activity during motor learning. Nature 510:263–267. [DOI] [PubMed] [Google Scholar]
- Piatkevich KD, Bensussen S, Tseng HA, Shroff SN, Lopez-Huerta VG, Park D, Jung EE, Shemesh OA, Straub C, Gritton HJ, Romano MF, Costa E, Sabatini BL, Fu Z, Boyden ES, Han X (2019) Population imaging of neural activity in awake behaving mice. Nature In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz G-A, Bottes S, Betizeau M, et al. (2018) Live imaging of neurogenesis in the adult mouse hippocampus. Sci 359:658–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA (2019) Analysis pipelines for calcium imaging data. Curr Opin Neurobiol 55:15–21. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, et al. (2016) Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89:285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poort J, Khan AG, Pachitariu M, et al. (2015) Learning Enhances Sensory and Multiple Non-sensory Representations in Primary Visual Cortex. Neuron 86:1478–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, et al. (2012) Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat methods 9:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryazhnikov E, Mugantseva E, Casarotto P, et al. (2018) Longitudinal two-photon imaging in somatosensory cortex of behaving mice reveals dendritic spine formation enhancement by subchronic administration of low-dose ketamine. Sci reports 8:6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh RN, Burgess CR, Sugden AU, et al. (2018) Intermingled Ensembles in Visual Association Cortex Encode Stimulus Identity or Predicted Outcome. Neuron 100:900–915.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real R, Peter M, Trabalza A, et al. (2018) In vivo modeling of human neuron dynamics and Down syndrome. [DOI] [PMC free article] [PubMed]
- Romo R, de Lafuente V (2013) Conversion of sensory signals into perceptual decisions. Prog Neurobiol 103:41–75. [DOI] [PubMed] [Google Scholar]
- Roome CJ, Kuhn B (2014) Chronic cranial window with access port for repeated cellular manipulations, drug application, and electrophysiology. Front Cell Neurosci 8:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadakane O, Masamizu Y, Watakabe A, et al. (2015) Long-Term Two-Photon Calcium Imaging of Neuronal Populations with Subcellular Resolution in Adult Non-human Primates. Cell reports 13:1989–1999. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Takahashi N, Matsuki N, Ikegaya Y (2008) Fast and accurate detection of action potentials from somatic calcium fluctuations. J Neurophysiol 100:1668–1676. [DOI] [PubMed] [Google Scholar]
- Sato M, Kawano M, Yanagawa Y, Hayashi Y (2016) In vivo two-photon imaging of striatal neuronal circuits in mice. Neurobiol Learn Mem 135:146–151. [DOI] [PubMed] [Google Scholar]
- Shoham S, O’Connor DH, Segev R (2006) How silent is the brain: is there a “dark matter” problem in neuroscience? J Comp Physiol Neuroethol sensory, neural, Behav Physiol 192:777–784. [DOI] [PubMed] [Google Scholar]
- Shuler MG, Bear MF (2006) Reward timing in the primary visual cortex. Sci 311:1606–1609. [DOI] [PubMed] [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, Svoboda K (2016) A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. [DOI] [PMC free article] [PubMed]
- Song A, Charles AS, Koay SA, et al. (2017a) Volumetric two-photon imaging of neurons using stereoscopy (vTwINS). Nat methods 14:420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Barnes S, Knöpfel T (2017b) Mammalian cortical voltage imaging using genetically encoded voltage indicators: a review honoring professor Amiram Grinvald. Neurophotonics 4:031214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speiser A, Yan J, Archer EW, Buesing L, Turaga SC, Macke JH (2017) Fast amortized inference of neural activity from calcium imaging data with variational autoencoders. Advances in Neural Information Processing Systems 4024–4034. [Google Scholar]
- Spires TL, Meyer-Luehmann M, Stern EA, et al. (2005) Dendritic spine abnormalities in amyloid precursor protein transgenic mice demonstrated by gene transfer and intravital multiphoton microscopy. J Neurosci Off J Soc Neurosci 25:7278–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Buetfering C, Lecoq J, et al. (2017) Aberrant Cortical Activity in Multiple GCaMP6-Expressing Transgenic Mouse Lines. [DOI] [PMC free article] [PubMed]
- Steinmetz NA, Koch C, Harris KD, Carandini M (2018) Challenges and opportunities for large-scale electrophysiology with Neuropixels probes. Curr Opin Neurobiol 50:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman JN, Smith IT, Kudenov MW, Smith SL (2016) Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat Biotechnol 34:857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Pachitariu M (2019) Computational processing of neural recordings from calcium imaging data. Curr Opin Neurobiol 55:22–31. [DOI] [PubMed] [Google Scholar]
- Stringer C, Pachitariu M, Steinmetz N, et al. (2019) Spontaneous behaviors drive multidimensional, brainwide activity. Sci 364:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Oertner TG, Hegemann P, Larkum ME (2016) Active cortical dendrites modulate perception. Sci 354:1587–1590. [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang B-Y, Viswanathan S, et al. (2016) A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis L, Berens P, Froudarakis E, et al. (2016) Benchmarking Spike Rate Inference in Population Calcium Imaging. Neuron 90:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Looger LL (2012) Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb Protoc 2012:647–656. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, et al. (2002) Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420:788–794. [DOI] [PubMed] [Google Scholar]
- Vanni MP, Chan AW, Balbi M, et al. (2017) Mesoscale Mapping of Mouse Cortex Reveals Frequency-Dependent Cycling between Distinct Macroscale Functional Modules. J Neurosci Off J Soc Neurosci 37:7513–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglewede RL, Vandemark KM, Davidson AM, et al. (2019) Reduced sensory-evoked structural plasticity in the aging barrel cortex. Neurobiol Aging 81:222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N (2018) Machine learning in neuroscience. Nat methods 15:33. [Google Scholar]
- Wang KH, Majewska A, Schummers J, et al. (2006) In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell 126:389–402. [DOI] [PubMed] [Google Scholar]
- Wekselblatt JB, Flister ED, Piscopo DM, Niell CM (2016) Large-scale imaging of cortical dynamics during sensory perception and behavior. J Neurophysiol 115:2852–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Chan AW, McGirr A, et al. (2016) Resolution of High-Frequency Mesoscale Intracortical Maps Using the Genetically Encoded Glutamate Sensor iGluSnFR. J Neurosci Off J Soc Neurosci 36:1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zou P, Cohen AE (2017) Voltage imaging with genetically encoded indicators. Curr Opin Chem Biol 39:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Pala A, Pedrido L, et al. (2013) Membrane potential dynamics of neocortical projection neurons driving target-specific signals. Neuron 80:1477–1490. [DOI] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan W-B (2009) Stably maintained dendritic spines are associated with lifelong memories. Nature 462:920–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HH, St-Pierre F (2016) Genetically Encoded Voltage Indicators: Opportunities and Challenges. J Neurosci Off J Soc Neurosci 36:9977–9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Yuste R (2017) In vivo imaging of neural activity. Nat methods 14:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Nimchinsky EA, Scheuss V, et al. (2004) Imaging calcium concentration dynamics in small neuronal compartments. Sci STKE: Signal Transduct Knowl Environ 2004:pl5. [DOI] [PubMed] [Google Scholar]
- Zhu L, Lee CR, Margolis DJ, Najafizadeh L (2018) Decoding cortical brain states from widefield calcium imaging data using visibility graph. Biomed Opt Express 9:3017–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Brenner N (2018) Synaptic Tenacity or Lack Thereof: Spontaneous Remodeling of Synapses. Trends Neurosci 41:89–99. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Burns LD, Cocker ED, et al. (2013) Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16:264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]