Abstract
As the practice of medicine becomes more reliant on imaging and laboratory tests, medical decisions will be increasingly based on numbers. Accordingly, following the introduction of solid-phase testing to the HLA testing repertoire, laboratory directors and physicians have employed preset mean fluorescence intensity (MFI) thresholds as the basis for decisions in the management of transplant patients. However, what do MFI values mean? The literature is rife with reports detailing numerous factors that influence antibody assessment including (but not limited to) sensitization history of the patient, level of mismatch between donor and recipient, presence of interfering substances in the serum, whether the antigen on multiplex beads is native or denatured, day-to-day and technologist variability, and the historical performance of an assay in a given institution. How are these variables incorporated into the interpretation of MFI values? Herein, the pitfalls and complexities of single antigen bead (SAB) testing and interpretation are discussed with specific attention to what can and cannot be inferred by MFI.
Keywords: HLA, SAB, MFI, Solid phase single antigen, C1q
1. Introduction
Physicians rely heavily on numbers when screening for illness, making a diagnosis, adjusting medications, and when monitoring a patient’s response to treatment. Consequently, more attention is placed on laboratory testing with diminishing focus on physical examination skills [1]. In fact, the role of the clinical laboratory in medical decision-making is now vital with an estimated 70% of all medical decisions based on laboratory values [2]. Since the late 1960s, the histocompatibility laboratory has been integral to transplantation due in no small part to the pioneering work of Dr. Paul I. Terasaki and his collaborators. Not only did they highlight the risk of hyperacute allograft rejection in the face of a positive crossmatch [3], but they forged the path for the development of modern-day clinical histocompatibility testing. Histocompatibility testing has evolved from the early days of cell-based tests to modern-day solid-phase antibody assays and molecular-based HLA typing. In particular, single antigen bead (SAB) platforms permit the detection of individual HLA specificities. The reporting of HLA antibody activity as mean fluorescence intensity (MFI) values has led to an incorrect perception that those values represent the “strength” of the antibodies, including donor specific antibodies (DSA). Clinicians use preset MFI thresholds as decision points pre-and post-transplant to determine the necessity of pre-or post-transplant desensitization, to predict crossmatch results and to help diagnose and monitor antibody-mediated rejection (AMR) [4]. Similar to physicians comparing a patient’s hemoglobin and electrolyte values from one time point to the next, it is easy to understand a transplant clinician’s temptation to compare MFI values from one sample to the next. Should clinical decisions be based on these MFI values? What do these numbers actually mean? Do they truly reflect the level and/or strength of an HLA antibody? Is it valid to assume that a high MFI number is indicative of a stronger and clinically relevant antibody while a low MFI value is reflective of a weak and clinically irrelevant antibody? Following is a detailed description of the pitfalls and complexities of SAB testing and interpretation, and questions of what can and cannot be inferred by MFI are addressed.
2. MFI: near misses
Though solid-phase SABs have led to advances including calculated PRA (cPRA) and the virtual crossmatch (vXM) [5], they are not without their limitations. In clinical practice, the vast majority of investigators would consider MFI values of 100 as negative. Nonetheless, antibodies with MFI values as low as 100 have been associated with poor AMR free graft survival [6]. The following case is an example in which the MFI alone failed to properly identify potential clinically relevant antibodies.
Case:
Class II solid-phase SAB assay (LS2A01, Lot#11) demonstrated low-level DR17 (DRB1*03:01), DR18 (DRB1*03:02), and DR52 (DRB3) reactivity with MFI values ranging from ~500 to ~1200 (Fig. 1A). If MFI levels alone were used to assign the presence/absence of antibody, the above antibodies would be considered negative, as the MFI values were <2000, the threshold in our laboratory. However, the corresponding FlowPRA® (One Lambda, Inc.) screening assay was positive with a definitive peak (Fig. 1B). Analysis with the FlowPRA® Specific bead assay revealed positive reactivity with DR17 (DRB1*03:01), DR18 (DRB1*03:02), and DR52 (DRB3) (Fig. 1C), substantiating the reactivity seen on the SAB. Further evidence that the antibodies were present is based upon utilizing the Epitope Registry. The software revealed a shared epitope (74R) among the specificities suggesting that the observed reactions were due to a single antibody targeting a shared epitope. If only SAB analysis had been performed and a pre-set threshold applied, the cPRA would have inaccurately been reported as 0%. From a laboratory perspective, the difference would be deeming a candidate unsensitized vs. sensitized.
Fig. 1.
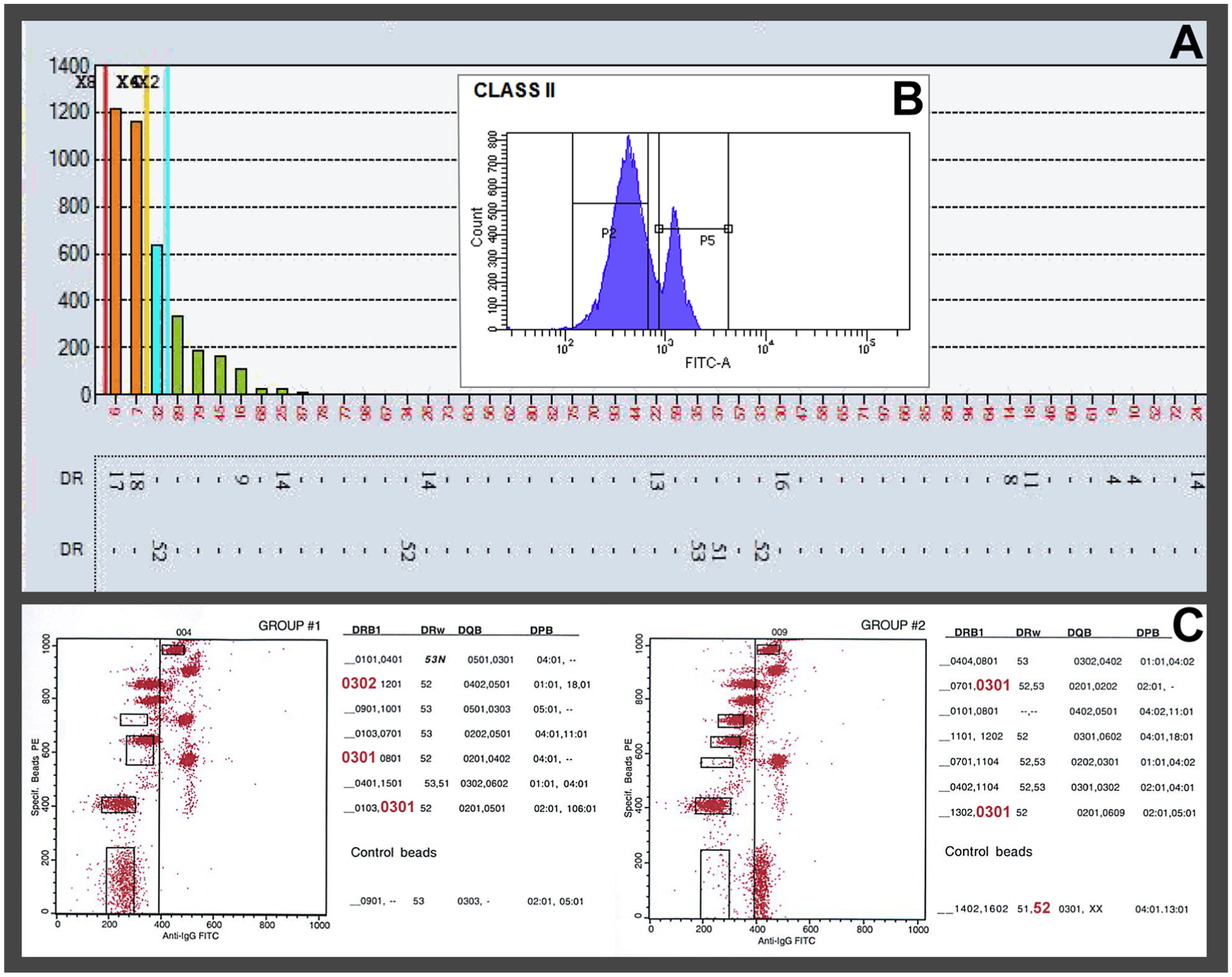
(A) LabScreen® single antigen bead assay: Patient serum demonstrating low-level DR17 (DRB1*03:01), DR18 (DRB1*03:02), and DR52 (DRB3) reactivity with MFI values below the laboratory’s cutoff of 2000 MFI. (B) Corresponding cytometric histogram of Class II FlowPRA®: Patient serum showing a definitive positive peak (approx. 24%). (C) Dot plot of Class II FlowPRA® Specific bead assay: Patient serum with specificity beads showing reactivity with DR17- (DRB1*03:01), DR18- (DRB1*03:02), and DR52-(DRB3) bearing beads, corroborating the presence of specificities demonstrated on the LabScreen® single antigen bead assay.
Granted such low MFI values on SAB would likely result in a negative physical crossmatch with lymphocytes bearing one of the corresponding antigens. However, does this correspond to a low-risk transplant? Post-transplant, an anamnestic response could lead to acute/accelerated AMR and/or graft loss. Additionally, DSA appearing post-transplant and considered to be de novo may actually be pre-existing antibodies that were missed because the MFI cutoffs pre-transplant were set too high [7]. This case illustrates how a single HLA assay may not capture the entire picture. Though the cost of additional assays is not trivial, the additional information garnered can be critical in the assessment of a patient’s alloimmune status and in alerting the clinicians to the possibility of increased risk.
3. MFI threshold: Mostly a Fluid Idea?
In biology, the term threshold is used to define a point at which a reaction occurs; e.g., neuronal action potential. In HLA testing (specifically antibody assessment) threshold was originally defined as the MFI value at which flow crossmatches would be positive. However, the MFI does not always predict positive crossmatches: In fact, sera with MFIs >10,000 can still result in a negative crossmatch [8]. More recently the MFI threshold has evolved into risk for AMR. But how rejection is defined is dependent on transplant program philosophy, the types of patients transplanted, whether desensitization will be considered and/or whether transplanting across “weak” DSA is acceptable. Thus, as far as determining the MFI threshold for clinical transplantation, there is no clear-cut consensus. In fact, the literature is fraught with studies that reveal a wide range of MFI cutoff values: 100 [6], 300 [9,10], 500 [11–14], 600 [15], 1000 [16–22], and 2000 [23]. Additionally, thresholds can also be locus specific; for example, in our laboratory, antibodies against C-locus specificities have a higher threshold (5000 MFI) compared to A and B loci (2000 MFI), as antigen expression for the C-locus is lower than other HLA loci [24]. The multitude of cutoffs provides a snapshot of the variation that exists among HLA laboratories and transplant centers. Hence, a universal MFI threshold is an unreasonable expectation, at least for now. The “appropriate” threshold should be a programmatic decision involving the HLA laboratory and members of the transplant program.
The value of a cutoff lies in its reliability to act as a decision point, e.g., its ability to predict a positive crossmatch and/or undesirable clinical outcome (e.g., AMR, graft survival, etc.). Regrettably, as just detailed, establishing a single threshold is not straightforward. For one thing, not all solid-phase tests are created equal; there are differences in antigen type (native vs. recombinant), antigen display (phenotype vs. single antigen), ancillary reagents (EDTA, DTT), vendors (One Lambda vs. Immucor) and how the test is technically performed [25]. For example, certain platforms contain beads that express a complete class I or II HLA phenotype (e.g., FlowPRA, Flow Specificity Beads) while others have beads that each express a single HLA allele (SAB) with the former expressing less antigen per bead than the latter. As a result, certain sera may display what has been termed the “peanut butter” phenomenon [26], wherein the antibody to a shared epitope is limiting, spread over multiple beads that each express the epitope resulting in a signal too weak to be detected. Interestingly, phenotypic beads yield a positive result with the same serum sample as the antibody concentrates onto fewer beads containing the shared epitope [4]. Conversely, when antibody is ample and antigen is limited (such as on phenotype beads), SABs can yield a positive result when phenotypic beads are negative, as the SAB contains a higher density per bead of the given antigen [4]. Of note, even within the same assay, variability in MFI has been reported between lots of antigen beads [27]. Furthermore, day-to-day and technologist variability within a laboratory add additional layers of technical complexities in MFI interpretation [28]. Thus, even if an antibody level increases from 2000 to 4000 MFI in a month, this upturn in MFI does not necessarily mean that the level of antibody has doubled. One approach to resolve the question of whether MFI values have actually increased or remained the same would be to run both samples (current and historical) simultaneously. Though costly, this approach reduces variability in testing conditions and could help in deciphering relevance of acute fluctuations in MFI. The cost of additional testing would ultimately be less expensive than desensitizing the patient unnecessarily.
4. MFI confounders: interfering substances and denatured antigens
In addition to technical concerns, solid-phase SAB testing is susceptible to so-called “interfering factors” that mask antibody detection. Most often seen in highly sensitized patients, this phenomenon is referred to as the “prozone effect,” which by strict definition, is a misnomer. Prozone refers to inhibition of agglutination or precipitation in a fluid phase assay, not to antibody binding in solid-phase assays such as SAB. Nevertheless, the term prozone is used to describe the observation of bead reactivity increasing with serum dilutions indicating the presence of an interfering substance. In the SAB test, the inhibition has been attributed to blocking of the secondary PE-conjugated antibody by activated complement components [16,29]. Briefly, antibody-antigen binding activates complement resulting in C1qrs and C3d deposition on solid-phase beads [16,30]. These large complexes inhibit binding of a PE-conjugated anti-Ig secondary antibody (Fig. 2). Addition of EDTA, DTT, heat inactivation or serum dilution can overcome complement-mediated inhibition of antibody binding [31,32]. Hypotonic dialysis has also been reported to eliminate/reduce effects of interfering substances [33]. IgM HLA antibodies present in patient sera have also been reported to block HLA antibody from binding the beads [34]. In these cases, DTT pretreatment increases IgG binding. Each of the above approaches requires pre-test manipulation, which translates to added technologist time, increased expense and opportunities for technical errors to occur.
Fig. 2.
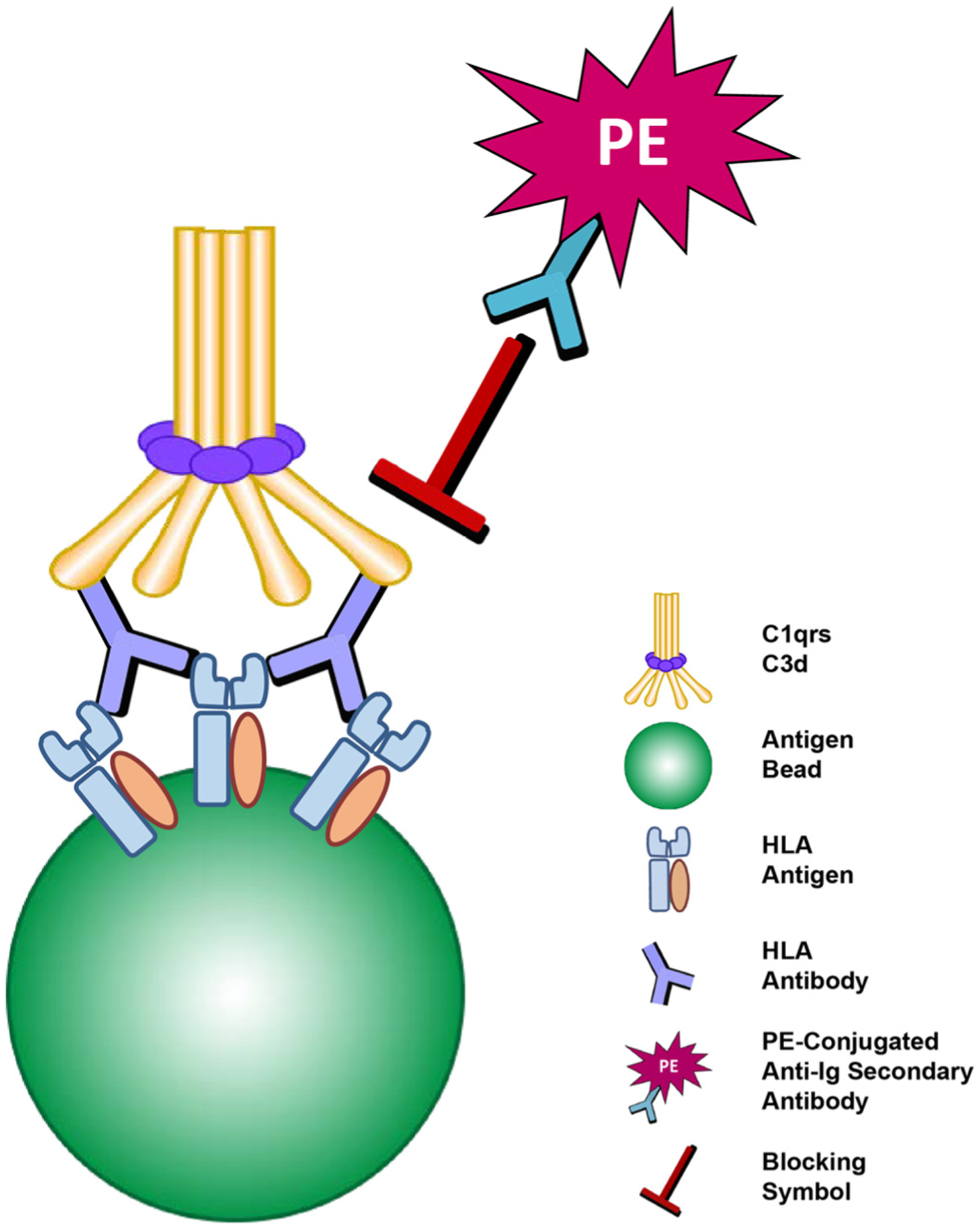
Complement-mediated interference (a.k.a. “prozone” effect). The large C1qrs and C3d complex, bound to primary HLA antibody and residing on the surface of the bead, are thought to sterically block the binding of PE-conjugated anti-Ig secondary antibody.
To avoid pre-test manipulation of patient sera, we developed and implemented an enhanced detection method to identify antibodies masked by EDTA-sensitive interfering factors [29]. Initially, we identified sera displaying the “prozone effect” and incubated them with HLA class I or II single antigen beads in the presence/absence of EDTA. We assessed the sera using a standard LABScreen® Single Antigen assay employing a secondary antibody directly conjugated to phycoerythrin (PE) (STD-SAB). In comparison, a modified indirect assay was run. In that situation, the secondary antibody was conjugated to biotin and subsequently probed with PE-conjugated streptavidin (BIO-SAB). As expected, the MFI of antibodies in the STD-SAB assay was higher with EDTA treatment (Fig. 3A). Importantly, the “prozone effect” did not occur with any of the sera in the BIO-SAB assay. In fact, MFIs resulting from the BIO-SAB method were generally higher than in the STD-SAB, EDTA-treated samples (Fig. 3A) [35]. Mechanistically, we hypothesize that the biotinylated secondary is not hindered in its binding to IgG as biotin is a low molecular weight molecule (325d). Additionally, the biotin uses a long spacer between the anti-IgG and streptavidin (https://www.jacksonimmuno.com/technical/products/conjugate-selection/biotin), which allows the anti-IgG to bind to HLA antibody despite the presence of activated complement components (Fig. 3B).
Fig. 3.
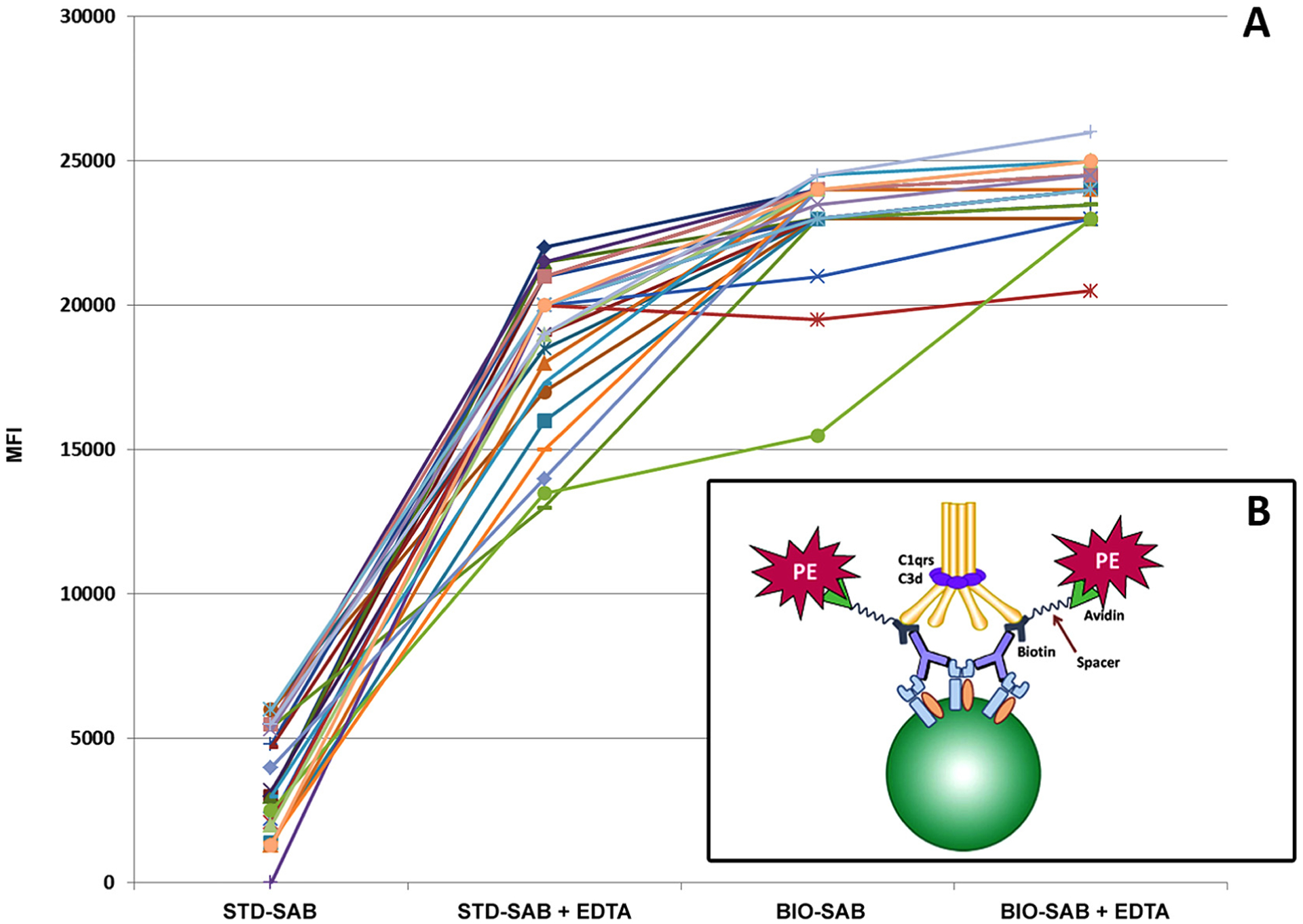
(A) Graphical representation of LabScreen® single antigen bead assay: Patient sample demonstrating higher reactivity with BIO-SAB assay compared to standard method without biotin. The MFI values of the standard method increased further with the addition of EDTA; however, the addition EDTA did not greatly affect the MFI values of the BIO-SAB method. (B) BIO-SAB assay with biotin/streptavidin detection system surmounts complement-mediated inhibition in standard method. Mechanistically, the biotinylated anti-IgG may minimize steric hindrance due to the presence of a long spacer biotin molecule that extends away from the IgG allowing binding of the streptavidin-PE.
Another obstacle uncovered in SAB testing was the existence of what are now referred to as denatured antigens, i.e., misfolded or degraded HLA proteins on the surface of single antigen beads. Denatured antigens were first reported in sera obtained from normal, healthy, and (self-reported) non-alloimmunized males that displayed HLA antibodies. The antibodies were mostly to rare HLA specificities, making prior exposure to alloantigen unlikely [36]. Subsequently, these unexpected alloantibodies were reported to target cryptic epitopes on HLA molecules not exposed in native configuration [37]; The current interpretation is that due to steps in manufacturing recombinant HLA antigens, these otherwise unexposed epitopes become immunologically accessible. Not surprisingly, such antibodies presented a clinical challenge. Should the corresponding HLA antigens be considered as clinically unacceptable or should they be ignored? Frequently, antibodies to denatured antigens that equated to positive virtual crossmatches resulted in negative flow cytometric crossmatches [38,39]. Nonetheless, as stated earlier, such data did not address whether the antibodies were clinically relevant.
Subsequently, studies showed that recipients possessing DSAs to denatured antigens did not experience the poor graft outcomes that occurred in recipients with DSA to native, intact antigens [40]. Some studies reported graft survival was comparable to recipients who had no DSA [41]. Consequently, many centers have considered antibodies to denatured antigens as clinically irrelevant. In fact, some laboratories ignored these antibodies and reported them as negative. This practice has been called into question by recent data generated by Visentin et al. who demonstrated that sera containing antibodies against denatured antigen could also bind to native, intact HLA. Furthermore, they also demonstrated that serum containing antibodies to native antigen could bind to denatured antigen on acid-treated Luminex single antigen beads [42]. Collectively, these data suggest that antibodies that appear to be directed to denatured antigens cannot necessarily be ignored. Accordingly, every patient should be carefully considered by examining patterns of reactivity amongst the positive specificities and examining whether reactivity can be explained by antibodies to shared epitopes. As HLA antigens are composed of multiple and overlapping epitopes, a single antibody to a shared amino acid sequence can react with multiple antigens (Fig. 4). It is here where knowledge of serology and cross-reactive groups (CREGs) can assist in the interpretation of modern assays. Online tools that assist with the identification of shared epitopes include HLA MatchMaker (http://www.epitopes.net) and the HLA Epitope Registry (http://www.epregistry.com.br/terms/index). The latter allows relatively quick identification of shared epitopes amongst different Luminex bead antigens. Though this approach is not yet commonplace and needs thorough validation (e.g., applying adsorption and elution techniques to explain the positive reactivity of multiple beads), it substantiates the presence of a biologically plausible antibody to a shared epitope. This is particularly useful when there is low level reactivity amongst seemingly unrelated beads.
Fig. 4.
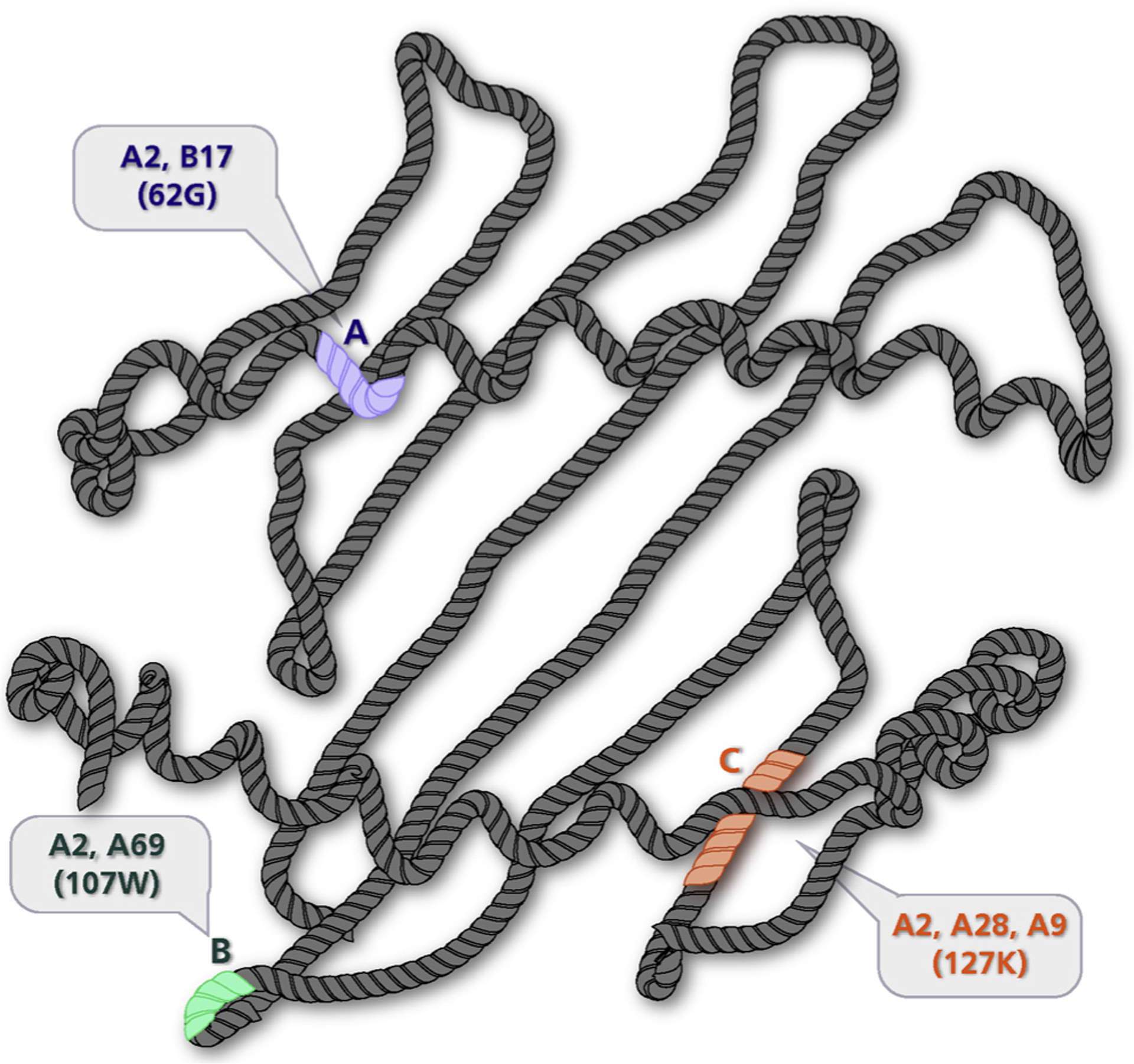
Shared epitopes found with HLA-A2. Structural representation of HLA-A2 molecule highlighting different antibody-defined epitopes (shaded). Each epitope provides the target for an antibody with reciprocal specificity (paratope). The shaded epitopes represent amino acid sequences shared with other antigens (i.e., Public Epitopes). A: 62G is shared between HLA-A2 and HLA-B17. B: 107W is shared between HLA-A2 and HLA-A69. C: 127K is shared among HLA-A2, HLA-A28, and HLA-A9. Antibodies targeting these shared determinants can help explain reactivity amongst different beads. Figure adapted from Fuller et al. Human Immunology, 1990 [86]) G – Glycine, W – Tryptophan, K – Lysine.
5. Beyond MFI
MFI values are at best a semi-quantitative surrogate of antibody level. As we have discussed, MFI level alone does not necessarily equate with a deleterious outcome. Thus, the question becomes whether alternative assays could be utilized to better characterize an antibody’s pathogenic potential. To address this question, modifications to solid-phase SAB and other techniques have been developed with the aim of defining clinically relevant antibody. These tests seek to predict which antibodies will lead to unfavorable outcomes (i.e., AMR and/or graft failure) based on different physical characteristics.
5.1. Complement fixation
Given that complement fixation is the original mechanism by which antibody was thought to contribute to organ injury in vivo [43,44], efforts to determine in vitro complement fixing ability of antibody and thereby infer pathogenic potential were reasonable. Complement activation has long been used in assays to determine antibodies’ potential to engage the complement system. Terasaki and Patel used the complement-dependent cytotoxicity (CDC) assay to demonstrate the association between a positive crossmatch and allograft failure [3]. Current studies focus on assessment of complement fixation via solid-phase platforms. The test garnering the most attention is the C1qScreen assay (One Lambda, Inc), which spikes serum with C1q reagent prior to single bead analysis. As C1q binding is the first step in the complement cascade, leading to the membrane attack complex and subsequent cell lysis, antibodies that bind C1q in vitro are postulated to correlate with in vivo pathogenicity.
The C1q assay attracted considerable attention after Loupy et al. reported that complement-binding DSA lead to significantly lower graft survival than non-complement binding DSA and non-DSA antibody in a cohort of 1016 renal transplant patients [13]. Smaller studies also demonstrated C1q-binding as a predictive factor of graft loss [45]. An important caveat is that these studies only found clinical relevance of C1q-binding capacity in the post-transplant setting. As such, the results cannot be extrapolated to predict the significance of preformed complement-binding DSA. In fact, in the pre-transplant setting, studies have not found a strong correlation between C1q binding and AMR or graft loss [21,23,46]. In our opinion, there appears to be little, if any, value to use the C1q assay as a pre-screening tool. Interestingly, Yell et al. diluted C1q binding DSA to levels at which the MFI of neat serum correlated to C1q negative DSA, and converted those C1q binding DSA to C1q-negative DSA. Conversely, by concentrating serum, they converted C1q-negative DSA to C1q binding DSA [15]. Thus, simply performing a C1q assay and determining that DSA is not complement fixing can be misleading, as it appears to be dependent on additional factors such as antibody concentration.
Though C1q binding is the first step in the complement cascade, its binding does not assure the cascade will go to completion. Downstream components in the cascade theoretically provide a better approximation of an antibody’s potential to induce inflammation and organ damage through a complement-dependent process. As such, immunohistochemical staining for C4d, a downstream complement degradation product, has been used as part of the Banff criteria for routine renal biopsy evaluation [47]; and some have demonstrated C4d detection in FlowPRA assays correlate well with immunohistochemical C4d staining [48]. Subsequently, complement binding SAB assays incorporating detection of complement degradation products, mainly C3d and C4d-binding, were developed to assess complement-binding ability of HLA antibodies. In terms of the C3d assays, the presence of C3d-positive DSAs has been associated with higher risk of graft loss [49] and lower 10-year graft survival [20]. Again, as with C1q assays, this association has only been demonstrated in the post-transplant setting. Similarly, assessments of C4d-binding in SAB testing have not lead to definitive consensus on its clinical utility. While some have noted correlation with graft survival [50,51] and AMR [9], others have not been able to find an increased association between C4d-positive DSA and AMR [52,53] or graft loss [52]. In addition, it is now well recognized that C4d-negative AMR exists and is associated with poor outcomes [54]. Hence, with conflicting clinical results, the verdict is still out on the clinical advantage of complement-binding assays.
Of note, none of the complement-binding assays addresses the issue of complement-independent AMR. Though antibody-mediated complement activation has long been viewed as the main mechanism by which graft injury occurs, C4d deposition is frequently not present on renal biopsies showing histologic evidence of transplant glomerulopathy; some studies demonstrate C4d staining in approximately just one third of cases [55,56]. Recent data suggests that antibody can induce organ injury independent of complement. For example, FcγR-dependent antibody interaction with effector NK cells [57,58], macrophages and monocytes [59,60] has been shown to facilitate injury through antibody-dependent cell cytotoxicity [43]. HLA antibody has also been implicated in endothelial cell [44,61] and smooth muscle activation [44,62] as well as in chemoattraction of chemokines and proinflammatory mediators [44]. The concept of complement-independent AMR has now been incorporated into the Banff classification for rejection [47], which acknowledges the need to address instances in which DSA is present despite negative C4d immunohistochemical staining. Hence, complement activation assays would miss cases in which complement does not directly mediate graft injury despite the presence of DSA. For these reasons, considering an antibody to be clinically innocuous because of its inability to fix complement does not seem reasonable.
5.2. IgG subclass analysis
As the classic complement pathway cannot proceed in the absence of antibody-antigen binding, focus has been placed on IgG subclass analysis of HLA antibody. Analysis of complement fixing (CF) antibodies (IgG1/IgG3) and non-complement fixing (NCF) antibodies (IgG2/IgG4) has been studied for years in the context of older methods of antibody detection including ELISA based methods [63,64] and traditional flow cytometry [65,66]. Not surprisingly, with the advent of newer methods, investigators have now applied IgG subclass analysis to SABs in attempts to classify antibody pathogenicity.
Teasing out the significance between CF and NCF HLA antibodies has proven to be somewhat difficult due to the preponderance of CF antibodies, alone or in conjunction with NCF. The high prevalence of CF antibodies is likely due to the natural progression of immunoglobulin maturation. After class switching from IgM, IgG is first composed of IgG3 and IgG1, which can then expand or class-switch to IgG2 and IgG4 [67]. As a result, most studies to date have demonstrated that HLA antibodies are composed of predominantly CF or a mixture of CF and NCF antibodies with only a small percentage containing NCF alone (1–4%) [12,22,68]. With such small numbers of NCF-only cases, studies are merely able to compare cases of CF-only antibodies to those with a mixture of CF and NCF antibodies [22,68]. Notably, compared to CF antibodies alone, cases in which there are both CF and NCF antibodies do not seem to have differing outcomes [22,68], indicating that pathogenicity is more reliant on the presence of CF antibodies. One study did suggest that NCF may be less pathologic with a better AMR-free post-transplant course, but this observation was based on only 3 patients who had >50% IgG2/IgG4 DSA [69].
Notwithstanding some of the limitations in studying IgG subclasses, some studies have demonstrated a correlation between certain IgG subclasses and clinical outcomes. In particular, IgG3 DSA has been reported to be significantly associated with renal allograft failure [12] and increased risk of chronic rejection and graft loss in liver transplantation [70]. As in the case of complement-binding assessment, these IgG subclass studies only found clinical significance post-transplantation. Pretransplantation, IgG subclasses have not seemed to add predictive value to outcomes [65,66]. Interestingly, Schaub et al. found that 1840 of 1974 (93%) cases with C1q-negative SAB were positive for CF antibodies [71]. This finding questions the utility of in vitro subclass analysis to predict an antibody’s potential for causing cytotoxicity if they do not actually engage C1q. Moreover, IgG1 is the predominant HLA IgG subtype identified in the studies presented [18,68,70,71], yet it is also present in patients exhibiting normal graft function despite its strong complement fixing ability [70]. The lack of overt pathology in the presence CF DSA would denote that factors, other than CF, are necessary to effect complement-mediated injury. Weighing the evidence, we agree with previous opinions that, at this time, IgG subclass analysis does not appear to provide significant clinical information for the characterization of HLA antibody [72].
5.3. Titers
In a recent review of SAB interpretation, it was astutely observed that neat MFI values provide the relative concentration of antibody bound to a bead but not necessarily the amount of antibody found in the serum [7]. By definition, titer is the extent to which an antibody can be diluted before losing its ability to react with a specific antigen. In the same review, examples of two sera with comparable neat MFI values were shown. Upon dilution, the MFI of the first serum sample remained stable, indicating that antibody concentration was in excess of antigen load. In contrast, the MFI of the second serum sample decreased substantially, signifying that there was less initial antibody compared to the first serum. This example highlights that the neat MFI does not always accurately reveal the true serum concentration of an antibody. Recently, performing antibody titration has been proposed as a method to characterize the relative strength of HLA antibody in SAB assays [7,17,32]. However, such titrations may only be helpful when the antibody level is at or above saturation (ie; the point at which a bead can no longer bind additional antibody).
In comparing titers to neat MFI and C1q assay values, Tambur et al. highlighted that a single serum sample could contain antibodies of multiple specificities, some of which are preferentially affected by the so-called “prozone effect” [32]. In these cases, the MFI values of some antibodies unexpectedly increased with serial dilutions of the neat serum while the values of other antibodies decreased. Increasing MFI values with serial dilution indicates the presence of interfering substances, such as activated complement components or IgM. In this situation, the interfering entity is preferentially diluted out, allowing for better antibody detection. Although they noted a good correlation between C1q assay and tittered peak MFI values, the authors concluded that titers were a better indicator of antibody strength, as they were more sensitive and allowed the detection of the highest MFI values via serial dilution. This conclusion presumes again that a higher MFI value is a surrogate for pathogenicity.
Papers presented here only examine titers in relation to other methods; studies examining HLA/DSA antibody titers pre- or post-transplant as predictive indicators of AMR or graft loss are lacking. One group did report de novo DSA with higher titers post-transplant correlated with graft loss, but this association disappeared after adjusting for clinical phenotype and non-adherence to immunosuppressive regimen [73]. Another study looked at antibody titers in the clinical context of antibody removal therapies in cases of AMR and pre-transplant desensitization [17]. Comparing pre- to post-antibody removal values, the authors reported that delta-reduction values of antibody titrations were more uniform compared to other methods, including C1q and SAB testing on neat serum, and concluded that antibody titers could better evaluate responsiveness to antibody removal therapies and monitor their efficacy. More studies like these are needed and could provide insight on whether titers may be worth the extra time and monetary cost.
Though antibody titers do provide a relative assessment of antibody concentration compared to other methods, titration studies are not without their drawbacks. For one, there are no uniform or standardized methods to measure titers. This variability can make comparisons of inter- and intra-laboratory comparisons difficult. As a study by Reed et al. demonstrated, inter-laboratory MFI variation using solid-phase SAB arrays can be as high as 62% [28], and that is without the added variability of serial dilutions. Even with standardization, the question would remain which titer would be used as the clinical threshold. Would it be the titer at which the MFI is the highest value? Or the titer at which the MFI reaches an accepted significant value? Furthermore, though titers are informative as to the amount or concentration of antibody present in serum, they are not necessarily indicative of an antibody’s affinity and/or avidity for cognate antigen, and therefore, may not necessarily predict an antibody’s pathogenicity.
5.4. Role of conformation
The issue of affinity and avidity underlines the notion of conformational compatibility between antibody and antigen. That is, how does the conformation of the antigen affect the ability of antibody to bind? Clearly, if antigen is physically inaccessible to its corresponding antibody, then binding and subsequent complement engagement cannot occur despite antibody specificity. Our group, in part, previously described a case that highlights the potential importance of conformation [74].
Case:
The α-helix of HLA-B*49:01, B*50:01, and B*50:02 share amino acid sequences at residues 152–156 that are believed to confer the B21 serological reactivity [74,75]. Yet, there are antibodies targeting this shared sequence that bind B*49:01 and B*50:01 but not B*50:02. B*50:02 has an additional polymorphism at residue 167 (W→S), which confers a serological reactivity of B45 [76]. One possible explanation for the lack of B21 sero-reactivity would be that the B*50:02 has some inherent structural or conformational difference, imparted by the polymorphism at residue 167, that prevents the antibody from binding despite the presence of the corresponding linear sequence. Where B*49:01 and B*50:01 do differ from B*50:02 at residue 167, B*49:01 and B*50:01 have a tryptophan while B*50:02 has a serine. As serine (89.0 A3) is smaller than tryptophan (227.8 A3), it stands to reason that this difference could alter the protein confirmation by rotating the α-helix, making the region inaccessible to the antibody. Clinically, knowledge of this conformational difference between the antigens is critical; a patient with antibody identified against B*49:01 and B*50:01 may not be considered acceptable for a donor organ possessing a B*50:02 if the assumption of crossreactivity, based on linear sequence alone, is made [74].
The relevance of conformation has been emphasized with a recent focus on eplets, noncontiguous fragments of a protein sequence which, due to folding, can come together to form a functional epitope that can be recognized by an antibody [77]. So, the notion that conformation impacts antibody-antigen engagement is only beginning to emerge, giving birth to new and innovative research.
6. MFI: only part of the elephant
When ordering a laboratory test for clinical purposes, it is imperative to know how the resulting data will be applied. If treatment decisions are to be made based on the data, a thorough understanding of the test results is necessary. Similarly, in assessing solid-phase SAB results, the question should be considered carefully. Most experts would agree that an antibody to A34 (A*34:01) on SAB (Fig. 5A) with an MFI ~10,000 would pose a potential immunological risk. However, looking at a concurrent FlowPRA, though, two distinct peaks are observed for Class I (Fig. 5B). This is an example of gene dosage in which the first peak represents a bead that is heterozygous for A*34:01 while the second peak represents a bead that is homozygous for A*34:01. As stated before, the amount of antigen, or target, on the bead affects the perceived strength of the antibody (when antibody is in excess). Likewise, a crossmatch with donor cells that are heterozygous for a given antigen will react weaker than with cells that are homozygous for that antigen (when antibody is in excess). Therefore, an MFI of 10,000 or even >20,000 represents but one description of an antibody and can only tell so much about a possible outcome, as the amount and accessibility of the target are unknown but clearly crucial pieces of the puzzle.
Fig. 5.
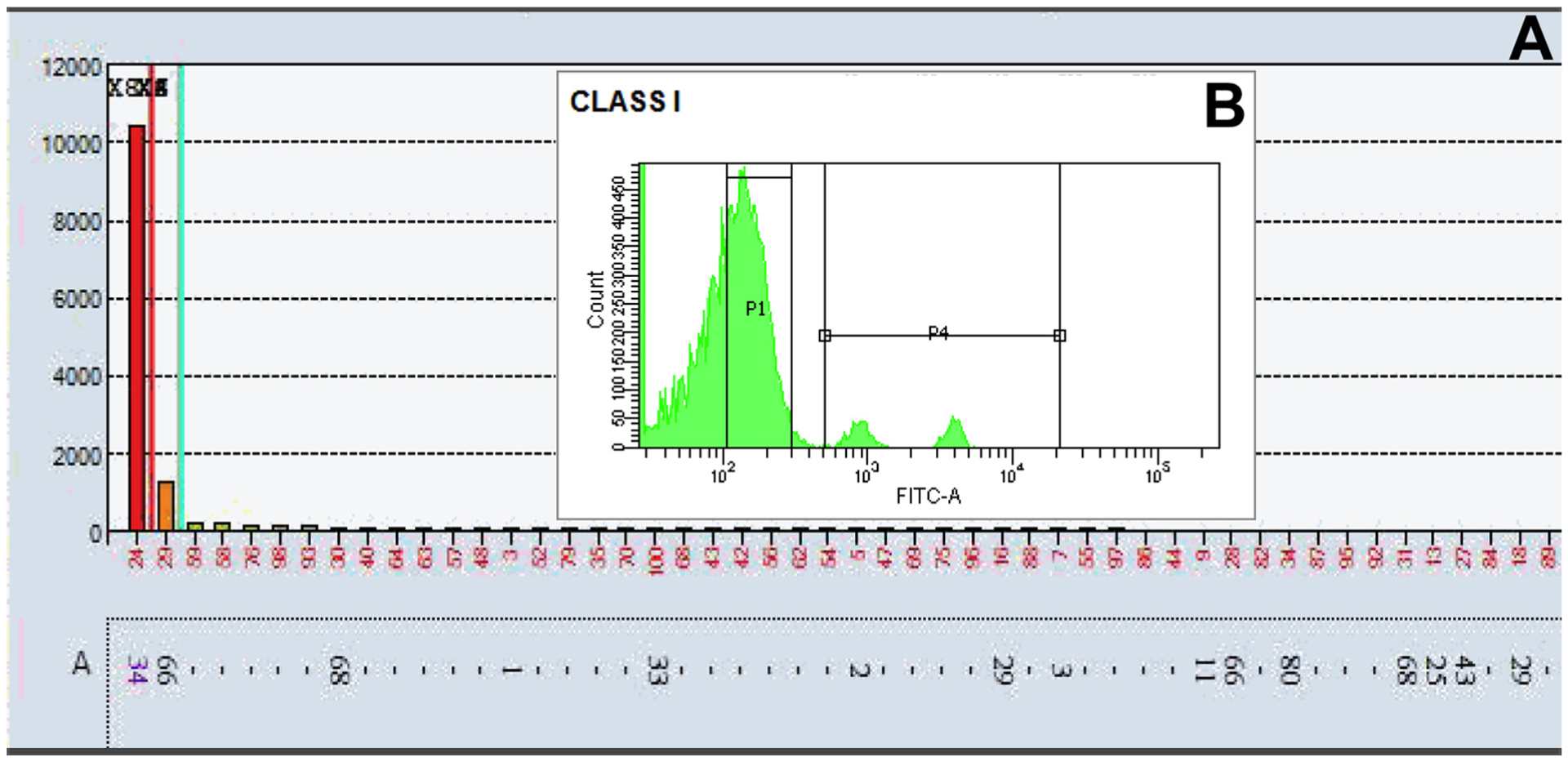
(A) LabScreen® single antigen bead assay: Patient serum demonstrating an antibody to A34 (A*34:01) with an MFI ~10,000 (B) Corresponding cytometric histogram of Class I FlowPRA®: Patient serum shows two distinct peaks, which likely represent an example of the gene-dosage effect with the first peak representing a bead that is heterozygous for A*34:01 and the second peak representing a bead that is homozygous for A*34:01.
Defining the strength of an antibody is not trivial. There are no definite reference ranges and no clear definition of what “strength” means. The value ascribed to the MFI is simply a number representing the relative amount of antibody adhering to the solid-phase bead under specified test conditions. While common practice is to interpret the MFI as a semi-quantitative number, it can be misleading. Although solid-phase SAB testing offers improved sensitivity and specificity compared to other testing modalities, after all is said and done, it is an in vitro test that attempts to predict how an antibody will react in vivo. Allegorically, looking at MFI in isolation is like three blind men touching an elephant for the first time: The first man touches the elephant’s leg, the second holds its tail, and the third feels a tusk. Each man is experiencing only one part of the whole picture.
Another piece of the puzzle that has profound relevance in all aspects of transplant medicine is the immense complexity of human biology and the individual immune response. Patient-centric factors, including but certainly not limited to, disease type, drug therapy, immunosuppressive regimens [7], and overall immuno-competence of an individual can all influence the allogeneic response. There is also a growing body of literature, albeit controversial, implicating non-HLA antibodies, such as anti-endothelial cell and anti-angiotensin type-1 receptor antibodies, as risk factors for rejection [78,79]; therefore, there may be more pieces to the puzzle than HLA antibody and its associated MFI. Moreover, the breadth of the immune response to alloantigen varies greatly among individuals. While some people will respond to an immunizing event, others do not [80,81]. For instance, only 5%–28% of patients exposed to foreign HLA antigen through platelet transfusion develop antibodies [80], however, up to 50% of women develop HLA alloantibodies as a result of pregnancy [82]. Accordingly, just because a recipient receives an HLA mismatched organ does not necessarily mean they will become sensitized. The reason why some patients respond and others do not is unknown, but for certain the answer does not lie in an MFI value alone. Undoubtedly, a better understanding of the responder and non-responder phenotype could potentially lead to mitigating strategies to prevent alloantibody production.
7. MFI: Where do we go from here?
What to do with so many variables? Though neat MFI may not be optimal, it may be the most practical option with adequate reliability, at least in certain clinical settings. The detection of DSA assessed by MFI alone on solid phase SAB has repeatedly been shown to be predictive of poor outcomes such as AMR [14,19,83] and graft survival [11,83]. Even low level, pre-formed DSA detected by solid phase in the face of a negative CDC and flow cytometric crossmatch (FCXM) appears to confer some increased risk of AMR and graft failure [10,84], with higher MFI values correlating with increased risk of rejection [85]. Though new assays (e.g., C1q, IgG subclass, etc.) and techniques have been developed in recent years, the literature is conflicting in terms of their utility. Notably, many studies indirectly advocate the use of MFI values. Correlations between MFI and complement binding assays support the use of MFI values as a surrogate for C1q assays [21,23] and C3d-binding assays [20]. In some cases, MFI has been found to be more diagnostically sensitive than C1q assays [15]. It should be recognized, though, that if a transplant center adopts a more conservative practice, the more sensitive the assay, the more likely that donors and recipients will be deemed incompatible even though some donor organs may have functioned well despite the presence of low-level DSA in their prospective recipients.
So, how to proceed? At least for now, a practical and pragmatic approach is to utilize the MFI values in conjunction with patient HLA type, patient alloimmunization history, epitope specificities of the antibodies all in the context of an assay’s performance history [7]. As mentioned above, utilizing supplementary assays (e.g., FlowPRA Screening and Specificity) can be extremely helpful in certain circumstances to resolve questionable and incomplete results. Additionally, a close working relationship and constant communication with the transplant surgeons/clinicians is important, lending clarity and constancy when reporting antibody specificities with MFI values. Practically speaking, MFI values should be viewed as a guiding reference, not an absolute determinant, of whether a patient should receive or be denied a specific allograft. In other words, transplant decisions should be based on evidence and experience, not just a number. Remember the words of Plato: “A good decision is based on knowledge and not on numbers.”
Acknowledgement
The authors thank Christina Dean, MD and Donna Martin of the Emory University Department of Pathology and Laboratory Medicine for their contributions and excellent work.
Abbreviations:
- MFI
mean fluorescence intensity
- HLA
human leukocyte antigen
- SAB
single antigen bead
- DSA
donor-specific antibody
- AMR
antibody mediated rejection
- cPRA
calculated PRA
- EDTA
Ethylenediaminetetraacetic acid
- DTT
Dithiothreitol
- PE
phycoerythrin
- CDC
complement-dependent cytotoxicity
- CF
complement fixing
- NCF
non-complement fixing
- FCXM
flow cytometric crossmatch
Footnotes
Disclosures
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- [1].Cook C, The lost art of the clinical examination: an overemphasis on clinical special tests, J. Man. Manip. Ther 18 (2010) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].70% of Medical Decisions Are Based on Lab Results: Solutions to Optimize Operations, Improve Quality and Lower Costs at Hospital Clinical, Quest Diagnostics Incorporated, 2015, QuestDiagnostics.com.
- [3].Patel R, Terasaki PI, Significance of the positive crossmatch test in kidney transplantation, N. Engl. J. Med 280 (1969) 735. [DOI] [PubMed] [Google Scholar]
- [4].Gebel HM, Bray RA, HLA antibody detection with solid phase assays: great expectations or expectations too great?, Am J. Transplant 14 (2014) 1964. [DOI] [PubMed] [Google Scholar]
- [5].Cecka JM, Kucheryavaya AY, Reinsmoen NL, Leffell MS, Calculated PRA: initial results show benefits for sensitized patients and a reduction in positive crossmatches, Am. J. Transplant 11 (2011) 719. [DOI] [PubMed] [Google Scholar]
- [6].Singh N, Djamali A, Lorentzen D, Pirsch JD, Leverson G, Neidlinger N, et al. , Pretransplant donor-specific antibodies detected by single-antigen bead flow cytometry are associated with inferior kidney transplant outcomes, Transplantation 90 (2010) 1079. [DOI] [PubMed] [Google Scholar]
- [7].Ellis TM, Interpretation of HLA single antigen bead assays, Transplant Rev. (Orlando) 27 (2013) 108. [DOI] [PubMed] [Google Scholar]
- [8].Phanish MK, Immunological risk assessment and human leukocyte antigen antibody testing in kidney transplantation, Indian J. Nephrol 26 (2016) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lawrence C, Willicombe M, Brookes PA, Santos-Nunez E, Bajaj R, Cook T, et al. , Preformed complement-activating low-level donor-specific antibody predicts early antibody-mediated rejection in renal allografts, Transplantation 95 (2013) 341. [DOI] [PubMed] [Google Scholar]
- [10].Lefaucheur C, Loupy A, Hill GS, Andrade J, Nochy D, Antoine C, et al. , Preexisting donor-specific HLA antibodies predict outcome in kidney transplantation, J. Am. Soc. Nephrol 21 (2010) 1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wu P, Jin J, Everly MJ, Lin C, Terasaki PI, Chen J, Impact of alloantibody strength in crossmatch negative DSA positive kidney transplantation, Clin. Biochem 46 (2013) 1389. [DOI] [PubMed] [Google Scholar]
- [12].Lefaucheur C, Viglietti D, Bentlejewski C, Duong van Huyen JP, Vernerey D, Aubert O, et al. , IgG donor-specific anti-human HLA antibody subclasses and kidney allograft antibody-mediated injury, J. Am. Soc. Nephrol 27 (2016) 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Loupy A, Lefaucheur C, Vernerey D, Prugger C, Duong van Huyen JP, Mooney N, et al. , Complement-binding anti-HLA antibodies and kidney-allograft survival, N. Engl. J. Med 369 (2013) 1215. [DOI] [PubMed] [Google Scholar]
- [14].Amico P, Honger G, Mayr M, Steiger J, Hopfer H, Schaub S, Clinical relevance of pretransplant donor-specific HLA antibodies detected by single-antigen flow-beads, Transplantation 87 (2009) 1681. [DOI] [PubMed] [Google Scholar]
- [15].Yell M, Muth BL, Kaufman DB, Djamali A, Ellis TM, C1q binding activity of De Novo donor-specific HLA antibodies in renal transplant recipients with and without antibody-mediated rejection, Transplantation 99 (2015) 1151. [DOI] [PubMed] [Google Scholar]
- [16].Guidicelli G, Anies G, Bachelet T, Dubois V, Moreau JF, Merville P, et al. , The complement interference phenomenon as a cause for sharp fluctuations of serum anti-HLA antibody strength in kidney transplant patients, Transpl. Immunol 29 (2013) 17. [DOI] [PubMed] [Google Scholar]
- [17].Tambur AR, Glotz D, Herrera ND, Chatroop EN, Roitberg T, Friedewald JJ, et al. , Can solid phase assays be better utilized to measure efficacy of antibody removal therapies?, Hum Immunol. (2016). [DOI] [PubMed] [Google Scholar]
- [18].Kannabhiran D, Everly MJ, Walker-McDermott JK, Tiongko S, Friedlander R, Putheti P, et al. , Changes in IgG subclasses of donor specific anti-HLA antibodies following bortezomib-based therapy for antibody mediated rejection, Clin. Transpl 229 (2012). [PubMed] [Google Scholar]
- [19].Malheiro J, Tafulo S, Dias L, Martins LS, Fonseca I, Beirao I, et al. , Analysis of preformed donor-specific anti-HLA antibodies characteristics for prediction of antibody-mediated rejection in kidney transplantation, Transpl. Immunol 32 (2015) 66. [DOI] [PubMed] [Google Scholar]
- [20].Comoli P, Cioni M, Tagliamacco A, Quartuccio G, Innocente A, Fontana I, et al. , Acquisition of C3d-binding activity by de novo donor-specific HLA antibodies correlates with graft loss in nonsensitized pediatric kidney recipients, Am. J. Transplant 16 (2016) 2106. [DOI] [PubMed] [Google Scholar]
- [21].Thammanichanond D, Wiwattanathum P, Mongkolsuk T, Kantachuvesiri S, Worawichawong S, Vallipakorn SA, et al. , Role of pretransplant complement-fixing donor-specific antibodies identified by C1q assay in kidney transplantation, Transplant Proc. 48 (2016) 756. [DOI] [PubMed] [Google Scholar]
- [22].Arnold ML, Ntokou IS, Doxiadis II, Spriewald BM, Boletis JN, Iniotaki AG, Donor-specific HLA antibodies: evaluating the risk for graft loss in renal transplant recipients with isotype switch from complement fixing IgG1/IgG3 to noncomplement fixing IgG2/IgG4 anti-HLA alloantibodies, Transpl. Int 27 (2014) 253. [DOI] [PubMed] [Google Scholar]
- [23].Crespo M, Torio A, Mas V, Redondo D, Perez-Saez MJ, Mir M, et al. , Clinical relevance of pretransplant anti-HLA donor-specific antibodies: does C1q-fixation matter?, Transpl Immunol. 29 (2013) 28. [DOI] [PubMed] [Google Scholar]
- [24].McCutcheon JA, Gumperz J, Smith KD, Lutz CT, Parham P, Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA, J. Exp. Med 181 (1995) 2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gebel HM, Liwski RS, Bray RA, Technical aspects of HLA antibody testing, Curr. Opin. Organ Transplant 18 (2013) 455. [DOI] [PubMed] [Google Scholar]
- [26].Roberts-Wilson TKTG, Gebel HM, Bray RA, Disconnects with solid-phase HLA antibody assays: reconnecting the dots, ASHI Quarterly 37 (2013) 3. [Google Scholar]
- [27].Friedlander R, Putheti P, Diaz E, Menon A, Ponce B, Muthukumar T, et al. , On the detection of anti-HLA antibodies using single antigen bead Luminex assay: lot-to-lot variations in MFI, Transplantation 96 (2013) e24. [DOI] [PubMed] [Google Scholar]
- [28].Reed EF, Rao P, Zhang Z, Gebel H, Bray RA, Guleria I, et al. , Comprehensive assessment and standardization of solid phase multiplex-bead arrays for the detection of antibodies to HLA, Am. J. Transplant 13 (2013) 1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Weinstock C, Schnaidt M, The complement-mediated prozone effect in the Luminex single-antigen bead assay and its impact on HLA antibody determination in patient sera, Int. J. Immunogenet 40 (2013) 171. [DOI] [PubMed] [Google Scholar]
- [30].Schwaiger E, Wahrmann M, Bond G, Eskandary F, Bohmig GA, Complement component C3 activation: the leading cause of the prozone phenomenon affecting HLA antibody detection on single-antigen beads, Transplantation 97 (2014) 1279. [DOI] [PubMed] [Google Scholar]
- [31].Schnaidt M, Weinstock C, Jurisic M, Schmid-Horch B, Ender A, Wernet D, HLA antibody specification using single-antigen beads–a technical solution for the prozone effect, Transplantation 92 (2011) 510. [DOI] [PubMed] [Google Scholar]
- [32].Tambur AR, Herrera ND, Haarberg KM, Cusick MF, Gordon RA, Leventhal JR, et al. , Assessing antibody strength: comparison of MFI, C1q, and titer information, Am. J. Transplant 15 (2015) 2421. [DOI] [PubMed] [Google Scholar]
- [33].Zachary AA, Lucas DP, Detrick B, Leffell MS, Naturally occurring interference in Luminex assays for HLA-specific antibodies: characteristics and resolution, Hum. Immunol 70 (2009) 496. [DOI] [PubMed] [Google Scholar]
- [34].Kosmoliaptsis V, Bradley JA, Peacock S, Chaudhry AN, Taylor CJ, Detection of immunoglobulin G human leukocyte antigen-specific alloantibodies in renal transplant patients using single-antigen-beads is compromised by the presence of immunoglobulin M human leukocyte antigen-specific alloantibodies, Transplantation 87 (2009) 813. [DOI] [PubMed] [Google Scholar]
- [35].Brannon PHD, Marchman C, Waslaske S, Saw CL, Gebel HM, Bray RA, Improved detection of HLA antibodies by LabScreen single antigen using a Biotin-Streptavidin complex system, Hum. Immunol 68 (2007) S21 (Poster Presentation at 33rd Annual Meeting of the American Society for Histocompatibility and Immunogenetics (ASHI)). [Google Scholar]
- [36].Morales-Buenrostro LE, Terasaki PI, Marino-Vazquez LA, Lee JH, El-Awar N, Alberu J, “Natural” human leukocyte antigen antibodies found in nonalloimmunized healthy males, Transplantation 86 (2008) 1111. [DOI] [PubMed] [Google Scholar]
- [37].El-Awar N, Terasaki PI, Nguyen A, Sasaki N, Morales-Buenrostro LE, Saji H, et al. , Epitopes of HLA antibodies found in sera of normal healthy males and cord blood, Clin. Transpl 199 (2008). [PubMed] [Google Scholar]
- [38].Jacob EK, De Goey SR, Gandhi MJ, Positive virtual crossmatch with negative flow crossmatch results in two cases, Transpl. Immunol 25 (2011) 77. [DOI] [PubMed] [Google Scholar]
- [39].Pereira S, Perkins S, Lee JH, Shumway W, LeFor W, Lopez-Cepero M, et al. , Donor-specific antibody against denatured HLA-A1: clinically nonsignificant?, Hum Immunol. 72 (2011) 492. [DOI] [PubMed] [Google Scholar]
- [40].Otten HG, Verhaar MC, Borst HP, van Eck M, van Ginkel WG, Hene RJ, et al. , The significance of pretransplant donor-specific antibodies reactive with intact or denatured human leucocyte antigen in kidney transplantation, Clin. Exp. Immunol 173 (2013) 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Visentin J, Marroc M, Guidicelli G, Bachelet T, Nong T, Moreau JF, et al. , Clinical impact of preformed donor-specific denatured class I HLA antibodies after kidney transplantation, Clin. Transplant 29 (2015) 393. [DOI] [PubMed] [Google Scholar]
- [42].Visentin J, Guidicelli G, Moreau JF, Lee JH, Taupin JL, Deciphering allogeneic antibody response against native and denatured HLA epitopes in organ transplantation, Eur. J. Immunol 45 (2015) 2111. [DOI] [PubMed] [Google Scholar]
- [43].Thomas KA, Valenzuela NM, Reed EF, The perfect storm: HLA antibodies, complement, FcgammaRs, and endothelium in transplant rejection, Trends Mol. Med 21 (2015) 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Valenzuela NM, McNamara JT, Reed EF, Antibody-mediated graft injury: complement-dependent and complement-independent mechanisms, Curr. Opin. Organ Transplant 19 (2014) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sutherland SM, Chen G, Sequeira FA, Lou CD, Alexander SR, Tyan DB, Complement-fixing donor-specific antibodies identified by a novel C1q assay are associated with allograft loss, Pediatr. Transplant 16 (2012) 12. [DOI] [PubMed] [Google Scholar]
- [46].Otten HG, Verhaar MC, Borst HP, Hene RJ, van Zuilen AD, Pretransplant donor-specific HLA class-I and -II antibodies are associated with an increased risk for kidney graft failure, Am. J. Transplant 12 (2012) 1618. [DOI] [PubMed] [Google Scholar]
- [47].Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, et al. , Banff 2013 meeting report: inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions, Am. J. Transplant 14 (2014) 272. [DOI] [PubMed] [Google Scholar]
- [48].Bartel G, Wahrmann M, Exner M, Regele H, Huttary N, Schillinger M, et al. , In vitro detection of C4d-fixing HLA alloantibodies: associations with capillary C4d deposition in kidney allografts, Am. J. Transplant 8 (2008) 41. [DOI] [PubMed] [Google Scholar]
- [49].Sicard A, Ducreux S, Rabeyrin M, Couzi L, McGregor B, Badet L, et al. , Detection of C3d-binding donor-specific anti-HLA antibodies at diagnosis of humoral rejection predicts renal graft loss, J. Am. Soc. Nephrol 26 (2015) 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bartel G, Wahrmann M, Schwaiger E, Kikic Z, Winzer C, Horl WH, et al. , Solid phase detection of C4d-fixing HLA antibodies to predict rejection in high immunological risk kidney transplant recipients, Transpl. Int 26 (2013) 121. [DOI] [PubMed] [Google Scholar]
- [51].Smith JD, Hamour IM, Banner NR, Rose ML, C4d fixing, luminex binding antibodies – a new tool for prediction of graft failure after heart transplantation, Am. J. Transplant 7 (2007) 2809. [DOI] [PubMed] [Google Scholar]
- [52].Wahrmann M, Bartel G, Exner M, Regele H, Kormoczi GF, Fischer GF, et al. , Clinical relevance of preformed C4d-fixing and non-C4d-fixing HLA single antigen reactivity in renal allograft recipients, Transpl. Int 22 (2009) 982. [DOI] [PubMed] [Google Scholar]
- [53].Honger G, Wahrmann M, Amico P, Hopfer H, Bohmig GA, Schaub S, C4d-fixing capability of low-level donor-specific HLA antibodies is not predictive for early antibody-mediated rejection, Transplantation 89 (2010) 1471. [DOI] [PubMed] [Google Scholar]
- [54].Orandi BJ, Alachkar N, Kraus ES, Naqvi F, Lonze BE, Lees L, et al. , Presentation and outcomes of C4d-negative antibody-mediated rejection after kidney transplantation, Am. J. Transplant 16 (2016) 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sis B, Campbell PM, Mueller T, Hunter C, Cockfield SM, Cruz J, et al. , Transplant glomerulopathy, late antibody-mediated rejection and the ABCD tetrad in kidney allograft biopsies for cause, Am. J. Transplant 7 (2007) 1743. [DOI] [PubMed] [Google Scholar]
- [56].Regele H, Bohmig GA, Habicht A, Gollowitzer D, Schillinger M, Rockenschaub S, et al. , Capillary deposition of complement split product C4d in renal allografts is associated with basement membrane injury in peritubular and glomerular capillaries: a contribution of humoral immunity to chronic allograft rejection, J. Am. Soc. Nephrol 13 (2002) 2371. [DOI] [PubMed] [Google Scholar]
- [57].Hidalgo LG, Sis B, Sellares J, Campbell PM, Mengel M, Einecke G, et al. , NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection, Am. J. Transplant 10 (2010) 1812. [DOI] [PubMed] [Google Scholar]
- [58].Akiyoshi T, Hirohashi T, Alessandrini A, Chase CM, Farkash EA, Neal Smith R, et al. , Role of complement and NK cells in antibody mediated rejection, Hum. Immunol 73 (2012) 1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Christen T, Nahrendorf M, Wildgruber M, Swirski FK, Aikawa E, Waterman P, et al. , Molecular imaging of innate immune cell function in transplant rejection, Circulation 119 (2009) 1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tinckam KJ, Djurdjev O, Magil AB, Glomerular monocytes predict worse outcomes after acute renal allograft rejection independent of C4d status, Kidney Int. 68 (2005) 1866. [DOI] [PubMed] [Google Scholar]
- [61].Zhang X, Valenzuela NM, Reed EF, HLA class I antibody-mediated endothelial and smooth muscle cell activation, Curr. Opin. Organ Transplant 17 (2012) 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Galvani S, Trayssac M, Auge N, Thiers JC, Calise D, Krell HW, et al. , A key role for matrix metalloproteinases and neutral sphingomyelinase-2 in transplant vasculopathy triggered by anti-HLA antibody, Circulation 124 (2011) 2725. [DOI] [PubMed] [Google Scholar]
- [63].Regan J, Monteiro F, Speiser D, Kalil J, Pouletty P, Buelow R, Pretransplant rejection risk assessment through enzyme-linked immunosorbent assay analysis of anti-HLA class I antibodies, Am. J. Kidney Dis 28 (1996) 92. [DOI] [PubMed] [Google Scholar]
- [64].Monteiro F, Mineiro C, Rodrigues H, de Paula FJ, Kalil J, Pretransplant and posttransplant monitoring of anti-HLA class I IgG1 antibodies by ELISA identifies patients at high risk of graft loss, Transplant Proc. 29 (1997) 1433. [DOI] [PubMed] [Google Scholar]
- [65].Gao ZH, McAlister VC, Wright JR Jr., McAlister CC, Peltekian K, MacDonald AS, Immunoglobulin-G subclass antidonor reactivity in transplant recipients, Liver Transpl. 10 (2004) 1055. [DOI] [PubMed] [Google Scholar]
- [66].Karuppan SS, Ohlman S, Moller E, The occurrence of cytotoxic and non-complement-fixing antibodies in the crossmatch serum of patients with early acute rejection episodes, Transplantation 54 (1992) 839. [DOI] [PubMed] [Google Scholar]
- [67].Schaub S, Honger G, Amico P, The complexity of the humoral immune response against HLA antigens, Transpl. Int 27 (2014) 249. [DOI] [PubMed] [Google Scholar]
- [68].Honger G, Hopfer H, Arnold ML, Spriewald BM, Schaub S, Amico P, Pretransplant IgG subclasses of donor-specific human leukocyte antigen antibodies and development of antibody-mediated rejection, Transplantation 92 (2011) 41. [DOI] [PubMed] [Google Scholar]
- [69].Lobashevsky A, Rosner K, Goggins W, Higgins N, Subtypes of immunoglobulin (Ig)-G antibodies against donor class II HLA and crossmatch results in three kidney transplant candidates, Transpl. Immunol 23 (2010) 81. [DOI] [PubMed] [Google Scholar]
- [70].Kaneku H, O’Leary JG, Taniguchi M, Susskind BM, Terasaki PI, Klintmalm GB, Donor-specific human leukocyte antigen antibodies of the immunoglobulin G3 subclass are associated with chronic rejection and graft loss after liver transplantation, Liver Transpl. 18 (2012) 984. [DOI] [PubMed] [Google Scholar]
- [71].Schaub S, Honger G, Koller MT, Liwski R, Amico P, Determinants of C1q binding in the single antigen bead assay, Transplantation 98 (2014) 387. [DOI] [PubMed] [Google Scholar]
- [72].Filippone EJ, Farber JL, Humoral immunity in renal transplantation: epitopes, Cw and DP, and complement-activating capability–an update, Clin. Transplant 29 (2015) 279. [DOI] [PubMed] [Google Scholar]
- [73].Wiebe C, Gareau AJ, Pochinco D, Gibson IW, Ho J, Birk PE, et al. , Evaluation of C1q status and titer of De Novo donor specific antibodies as predictors of allograft survival, Am. J. Transplant (2016). [DOI] [PubMed] [Google Scholar]
- [74].Bray RA, Nickerson PW, Kerman RH, Gebel HM, Evolution of HLA antibody detection: technology emulating biology, Immunol. Res 29 (2004) 41. [DOI] [PubMed] [Google Scholar]
- [75].Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, et al. , IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex, Nucleic Acids Res. 31 (2003) 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Darke C, Guttridge MG, Thompson J, Street J, Thomas M, Molecular, serological and population studies on a novel HLA-B allele–HLA-B*5002, Tissue Antigens 51 (1998) 666. [DOI] [PubMed] [Google Scholar]
- [77].Duquesnoy RJ, Update of the HLA class I eplet database in the website based registry of antibody-defined HLA epitopes, Tissue Antigens 83 (2014) 382. [DOI] [PubMed] [Google Scholar]
- [78].Zhang X, Reinsmoen NL, Impact of non-human leukocyte antigen-specific antibodies in kidney and heart transplantation, Front. Immunol 8 (2017) 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pinelli DF, Friedewald JJ, Haarberg KMK, Radhakrishnan SL, Zitzner JR, Hanshew WE, et al. , Assessing the potential of angiotensin II type 1 receptor and donor specific anti-endothelial cell antibodies to predict long-term kidney graft outcome, Hum. Immunol 78 (2017) 421. [DOI] [PubMed] [Google Scholar]
- [80].Hess JR, Trachtenberg FL, Assmann SF, Triulzi DJ, Kaufman RM, Strauss RG, et al. , Clinical and laboratory correlates of platelet alloimmunization and refractoriness in the PLADO trial, Vox Sang. (2016). [DOI] [PubMed] [Google Scholar]
- [81].Higgins JM, Sloan SR, Stochastic modeling of human RBC alloimmunization: evidence for a distinct population of immunologic responders, Blood 112 (2008) 2546. [DOI] [PubMed] [Google Scholar]
- [82].Picascia A, Grimaldi V, Sabia C, Napoli C, Comprehensive assessment of sensitizing events and anti-HLA antibody development in women awaiting kidney transplantation, Transpl. Immunol 36 (2016) 14. [DOI] [PubMed] [Google Scholar]
- [83].Kannabhiran D, Lee J, Schwartz JE, Friedlander R, Aull M, Muthukumar T, et al. , Characteristics of circulating donor human leukocyte antigen-specific immunoglobulin G antibodies predictive of acute antibody-mediated rejection and kidney allograft failure, Transplantation 99 (2015) 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mohan S, Palanisamy A, Tsapepas D, Tanriover B, Crew RJ, Dube G, et al. , Donor-specific antibodies adversely affect kidney allograft outcomes, J. Am. Soc. Nephrol 23 (2012) 2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Adebiyi OO, Gralla J, Klem P, Freed B, Davis S, Wiseman A, et al. , Clinical significance of pre-transplant donor specific antibodies in the setting of negative cell-based flow cytometry crossmatching in kidney transplant recipients, Am. J. Transplant (2016). [DOI] [PubMed] [Google Scholar]
- [86].Fuller AA, Trevithick JE, Rodey GE, Parham P, Fuller TC, Topographic map of the HLA-A2 CREG epitopes using human alloantibody probes, Hum. Immunol 28 (1990) 284. [DOI] [PubMed] [Google Scholar]


