Abstract
Background
Diagnosing and treating patients with multiple primary lung cancers (MPLCs) bring challenges to the clinic, and the preliminary evidence has revealed unsatisfying outcomes after targeted therapy and immunotherapy. Therefore, we surveyed genomic profiles of MPLCs and their possible associations with tumor mutation burden (TMB), programmed death-ligand 1 (PD-L1), and the immune cell infiltration landscape.
Materials and methods
A total of 112 patients with MPLCs with surgically resected 294 tumors were eligible, and 255 tumors were sequenced using a 1021-gene panel. Immunohistochemistry staining was performed to evaluate the levels of PD-L1 and the density of CD3+/CD8+ tumor-infiltrating lymphocytes (TILs), and CD68+/CD163+ tumor-associated macrophages (TAMs) at the central tumor and invasive margin, and immunotypes were generated based on those variables.
Results
MPLCs often occur simultaneously in non-smoker women younger than 60 years and manifest as ground-glass opacities, adenocarcinoma, and stage I lung lesions. The most frequently mutated genes in the 255 tumors were EGFR (56%), ERBB2 (12%), TP53 (12%), BRAF (11%), RBM10 (11%), and KRAS (9%). We found 87 (77.7%) patients with diverse genomic profiles, and 61 (54.5%) who shared at least one putative driver gene between different tumors presented more aggressive tumors. The median TMB was 1.92 mutations/Mb, and high-TMB (≥3) lesions often harbored EGFRL858R/KRASG12C/RBM10/TP53/LRP1B mutations or wild-type ERBB2. Only 8.1% of patients and 3.9% of lesions were positive for PD-L1 on tumor cells, and this positivity was more frequent in LRP1B/TP53-mutant tumors. EGFRL858R/RBM10/TP53 mutations were positively associated with specific immune cells and an inflamed immunotype, but ERBB2 mutations were negatively correlated. TMB, CD3+TILs, and CD68+/CD163+ TAMs presented with significant heterogeneity among paired tumors (all kappa <0.2), but PD-L1 and CD8 +TILs were more uniformly present in tumor pairs.
Conclusion
MPLCs are driven by different molecular events and often exhibit low TMB, low PD-L1, and a heterogeneous immune infiltration landscape. Specific genomic profiles are associated with TMB and the tumor immune microenvironmental landscape in MPLCs. Our findings can help to guide MPLCs diagnoses and to identify patient populations that may benefit from immunotherapy and targeted therapy.
Keywords: lung neoplasms, genetic markers, tumor microenvironment, lymphocytes, tumor-infiltrating, macrophages
Background
Lung cancer is the most lethal malignancy in the world.1 Multiple primary lung cancers (MPLCs) are defined in patients harboring two or more primary lung cancers simultaneously or sequentially. MPLCs have been detected increasingly with the widespread utilization of CT. The three diagnostic criteria commonly used to identify MPLCs from intrapulmonary metastases2–4 do not lead to accurate diagnoses. Thus, MPLCs pose clinical diagnostic and therapeutic challenges. With the development of next-generation sequencing (NGS), the American College of Chest Physicians (ACCP) guidelines and American Joint Committee on Cancer (AJCC) Cancer Stage Manual (eighth edition)2 3 have added different molecular genetic characteristics as a diagnostic criterion, but no specific genetic characteristic was mentioned. Therefore, identifying genomic alteration patterns in MPLCs is necessary to achieve accurate diagnoses.
MPLCs are often diagnosed during early stages, and radical surgery and stereotactic body radiotherapy are the main therapies.5 6 However, some patients cannot tolerate wide range or multiple surgeries due to their limited pulmonary function. Systemic treatments are another option. The representative targeted therapy has shown a limited response in patients with resected EGFR-mutant lung cancers companied by residual ground-glass opacities (GGOs).7 However, the mechanism of the poor efficacy of EGFR tyrosine kinase inhibitors (TKIs) in MPLCs remains unclear. Immune checkpoint inhibitors (ICIs) have revolutionized the therapeutic scenario against advanced lung cancers and have been gradually applied for the treatment of early-stage lung cancers.8 Preliminary small-sample studies have revealed that most high-risk GGOs in patients with stage I or advanced non-small cell lung cancer (NSCLC) have no diameter reduction after receiving ICIs.9 10 Therefore, exploring the distribution of predictive biomarkers of immunotherapy in patients with MPLC is important to evaluate the application value of ICIs in this group of patients.
Multiple factors including genetic and immune indexes influenced the efficacy of ICIs in lung cancers, and programmed-death ligand 1 (PD-L1) is one of the most widely adopted biomarkers.11 A few small-sampled studies investigated the expression levels and heterogeneity of PD-L1 in MPLCs without reaching a consensus.12 13 Tumor mutation burden (TMB) is a predictive and prognostic biomarker in multiple solid tumors treated with ICIs,14 but the TMB expression pattern among MPLCs remains unclear. Moreover, various immune cells including lymphocytes and macrophages infiltrate the tumor parenchyma. Tumor-infiltrating lymphocytes (TILs) at baseline and after treatment have been associated with the efficacy and prognosis of patients with cancer receiving immunotherapy.11 Tumor-associated macrophages (TAMs, CD68+), divided into M1 and M2 (CD163+) subtypes, contribute to tumor progression and immune resistance.15 The polarization status of M1 and M2 TAMs exerts an influence on the efficacy of ICIs.16 Zhou et al10 proposed that synchronous GGOs with limited response to ICIs have higher levels of CD68+TAMs and lower levels of CD8+TILs than the main lesions by performing single-cell sequencing. However, there still lacks large-sample studies that investigate the expression level of PD-L1 among MPLCs, and little is known about the distribution pattern of TMB, TILs, and TAMs among patients with MPLCs.
Herein, we examined the genomic profiles of MPLCs and their correlation with TMB, PD-L1 expression, and the immune cell infiltration landscapes (CD3+/CD8 +TILs, CD68+/CD163 +TAMs) by performing 1021-gene NGS and immunohistochemistry (IHC) staining on patient samples. Our results should help clinicians with diagnoses and with the selection of therapy for patients with MPLCs (especially for the application of systematic therapies including targeted therapy and immunotherapy).
Materials and methods
Patients and samples
We enrolled 112 patients with lung cancer who had undergone radical resection of at least two primary lung cancer lesions between January 2018 and March 2020. The multifocal lung cancers in each patient were diagnosed as MPLCs based on ACCP guidelines3: different histological types, or distinct molecular genetic characteristics, or at least one lesion derived from carcinoma in situ, or identical histological types but located in different lobes without N2 or N3 lymph nodes or systemic metastases. Clinical, radiological, and histopathological information were obtained from electronic medical records. Informed consent was obtained from the patients.
NGS and alteration identification
At least two primary lung cancers from each patient were sequenced using a customized panel of 1021 cancer-related genes in the Geneplus-Beijing Institute (Beijing, China). These 1021 sequenced genes are listed in the online supplemental table S1. Detected somatic variations included single nucleotide variations, small insertions and deletions (InDels), copy number variations, and gene fusions. Detailed information on sample processing, DNA extraction, library construction, target capture, NGS, and data analysis is described in the online supplemental Methods S1-3.
jitc-2021-003773supp001.pdf (712.9KB, pdf)
Assessment of TMB
We defined TMB as the number of somatic mutations and indels per megabyte bases in coding regions detected in tumor tissues and categorized it into high-TMB and low-TMB. TMB in the top quartile (>25%) was considered high-TMB.
IHC staining
Formalin-fixed, paraffin-embedded tumor specimens were sectioned into 4 µm thick slices. All sections were dewaxed with xylene, hydrated with gradient alcohol, washed with phosphate-buffered saline·tween 20, and treated with antigen retrieval buffer (MVS-0098, MXB). The tissue samples were blocked with goat serum (SP KIT-B2, MXB) and the endogenous peroxidase activity was blocked with 3% H2O2 solution (SP KIT-A3, MXB) for 15 min at room temperature. Next, the tissue samples were incubated with rabbit/rat antihuman PD-L1 antibodies (Dako 22C3, 1:50) overnight at 4°C or with CD3 (Kit-0003, MXB), CD8 (MAB-0021, MXB), CD68 (Kit-0026, MXB), or CD163 (MAB-0206, MXB) antibodies for 2 hours at room temperature. The detailed procedure was performed according to the manufacturer’s instructions. Cells with positive staining was defined as those with a yellowish–brown staining of the cytoplasm or cell membrane. Two independent observers determined staining results and discussed them to reach a consensus in contradictory cases.
Quantification of PD-L1, CD3, CD8, CD68, and CD163
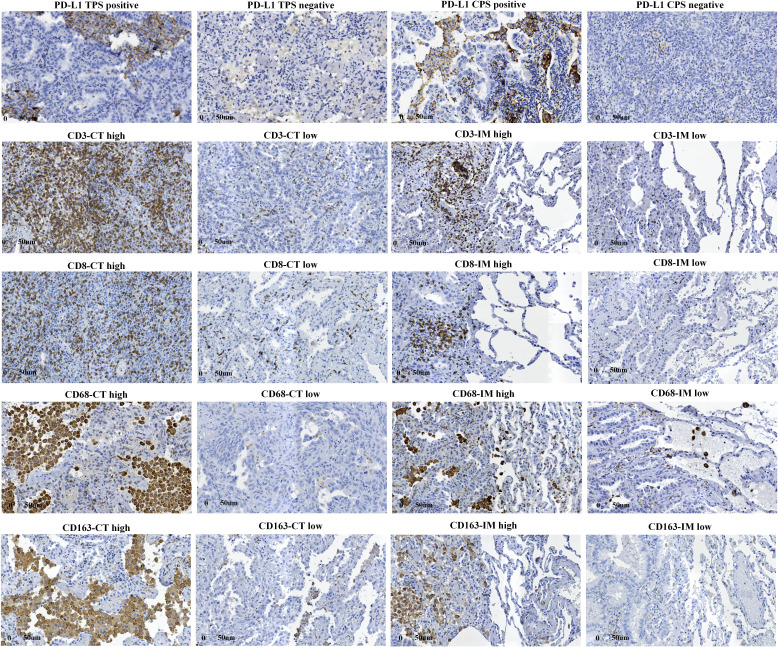
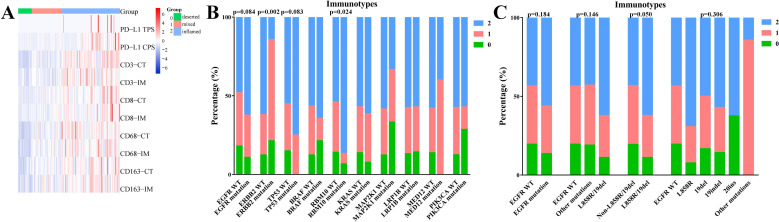
PD-L1 tumor proportion score (TPS) was estimated as the percentage of tumor cells for membranous PD-L1 staining for each section. PD-L1 combined positive score (CPS) was defined as the ratio of the total number of tumor cells, lymphocytes, and macrophages positive for PD-L1 to the number of tumor cells in the whole section. For CD3, CD8, CD68, and CD163, we selected two visual fields with the highest density at the central tumor (CT) region and invasive margin (IM) at 200X.17 18 Tumor IM was defined as a 1 mm area with the center of the boundary line between tumor tissues and pericarcinomatous tissues. The positive cut-off values of PD-L1 TPS and CPS were 1% and 1 (median), respectively. Tumors were divided into high and low-density groups based on the median number of positive immune cells per unit area (figure 1). IHC images were quantified with Image Pro Plus V.6.0 (Media Cybernetics, Washington, USA).
Figure 1.
Expression levels of immune indicators on immunohistochemistry staining. CPS, combined positive score; CT, central tumor; IM, invasive margin; PD-L1, programmed death ligand 1; TPS, tumor proportion score.
Statistical analysis
Categorical variables were described with numbers and percentages. Normally distributed measurement data were expressed as mean and SD, and non-normally distributed data were described using median and IQRs. The Pearson χ2, Fisher’'s exact, and non-parameter tests were performed to compare the distribution of TMB and immune indexes. Pearson correlation analysis and Spearman rank correlation analysis were used to analyze the correlations. We performed cluster analysis to obtain immunotypes. In addition, agreement tests including Bland-Altman analysis (numerical) and kappa test (categorical) were used to evaluate the heterogeneity of TMB and immune indexes among paired lesions in patients with MPLCs. For the patients with more than three tumors, all tumor foci were constituted tumor pairs to compare the heterogeneity. Statistical analyses were completed using IBM SPSS Statistics V.22.0 (IBM). A two-sided α of less than 0.05 was considered to be statistically significant.
Results
Characteristics of patients and tumors in MPLCs
A total of 112 patients with MPLCs and 294 primary lung cancer lesions were eligible for this study, and the characteristics of patients and tumors were listed in tables 1 and 2, respectively. Most patients were women (73.2%), non-smokers (88.4%), younger than 60 years at initial diagnosis (76.8%), and had multiple lesions in different pulmonary lobes (76.8%). All patients presented synchronous MPLCs, most underwent one-stage surgery (92.9%) and had the same histological type (97.3%), as described in previous studies.19 The highest T, N, and p.TNM stages in most of patients were T1, N0, and Ⅰ, respectively. Most tumors were in the bilateral upper lobes (58.9%), manifested as GGOs (89.8%), adenocarcinomas (99.0%), T1 stage (86.4%), N0 stage (99.0%), and stage Ⅰ (98.7%) (table 2). Squamous cell carcinoma was more common in past studies,4 20 21 but adenocarcinoma and GGO have become more common in recent studies,22 23 in accordance with an increasing trend of lung cancer in non-smoking women.24
Table 1.
Clinicopathological characteristics of 112 patients with multiple primary lung cancers
| Characteristics | Number of patients (%) | Characteristics | Number of patients (%) |
| Total | 112 (100.0) | Total | 112 (100.0) |
| Sex | Surgery types | ||
| Female | 82 (73.2) | (Bi)lobectomy | 23 (20.5) |
| Male | 30 (26.8) | Lobectomy+sublobectomy | 41 (36.6) |
| Age | (Multi)sublobectomy | 48 (42.9) | |
| <60 | 86 (76.8) | Lymphadenectomy | |
| ≥60 | 26 (23.2) | Yes | 86 (76.8) |
| Smoking | No | 26 (23.2) | |
| No | 99 (88.4) | Surgical time | |
| Yes | 13 (11.6) | Synchronous | 104 (92.9) |
| Malignancy | Metachronous | 8 (7.1) | |
| No | 108 (96.4) | Histopathology | |
| Yes | 4 (3.6) | Same | 109 (97.3) |
| Family malignancy | Different | 3 (2.8) | |
| No | 97 (86.6) | Highest T stage | |
| Yes | 15 (13.4) | T1 | 90 (80.4) |
| Tumor numbers | T2 | 22 (19.6) | |
| 2 | 68 (60.7) | Highest N stage | |
| 3 | 23 (20.5) | N0 | 109 (97.3) |
| ≥4 | 21 (18.8) | N1 | 2 (1.8) |
| Tumor locations | N2 | 1 (0.9) | |
| Same lobe | 26 (23.2) | Highest TNM stage | |
| Ipsilateral | 43 (38.4) | ⅠA | 87 (77.7) |
| Bilateral | 43 (38.4) | ⅠB | 21 (18.8) |
| Tumor presentation time | ⅡA | 0 (0.0) | |
| Synchronous | 112 (100.0) | ⅡB | 3 (2.7) |
| Metachronous | 0 (0.0) | ⅢA | 1 (0.8) |
Table 2.
Clinicopathological characteristics of 294 tumors in patients with multiple primary lung cancers
| Characteristics | Number of tumors (%) | Characteristics | Number of tumors (%) |
| Total | 294 (100.0) | Total | 294 (100.0) |
| Tumor location | Vascular invasion | ||
| RUL | 96 (32.7) | No | 290 (98.6) |
| RML | 21 (7.1) | Yes | 4 (1.4) |
| RLL | 50 (17.0) | T stage | |
| LUL | 77 (26.2) | T1a | 183 (62.2) |
| LLL | 50 (17.0) | T1b | 59 (20.1) |
| Radiology | T1c | 12 (4.2) | |
| GGO | 264 (89.8) | T2a | 38 (12.9) |
| Solid | 30 (10.2) | T2b | 1 (0.3) |
| Surgery types | T3 | 1 (0.3) | |
| Lobectomy | 108 (36.7) | N stage | |
| Wedge resection | 116 (39.5) | N0 | 291 (99.0) |
| Segmentectomy | 70 (23.8) | N1 | 2 (0.7) |
| Histopathology | N2 | 1 (0.3) | |
| AIS | 36 (12.2) | TNM stage | |
| MIA | 54 (18.4) | ⅠA1 | 182 (61.9) |
| ADC | 201 (68.4) | ⅠA2 | 59 (20.1) |
| Other | 3 (1.0) | ⅠA3 | 10 (3.4) |
| Diameter | ⅠB | 39 (13.3) | |
| ≤1 cm | 198 (67.3) | ⅡA | 0 (0.0) |
| 1<×≤2 cm | 75 (25.5) | ⅡB | 3 (1.0) |
| 2<×≤3 cm | 18 (6.1) | ⅢA | 1 (0.3) |
| >3 cm | 3 (1.1) | Gene tests | |
| Pleural invasion | Yes | 255 (86.7) | |
| No | 254 (86.4) | No | 39 (13.3) |
| Yes | 40 (13.6) |
ADC, adenocarcinoma; AIS, adenocarcinoma in situ; GGO, ground-glass opacity; LLL, left lower lobe; LUL, left upper lobe; MIA, minimally invasive adenocarcinoma; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe.
Mutational profile of MPLCs
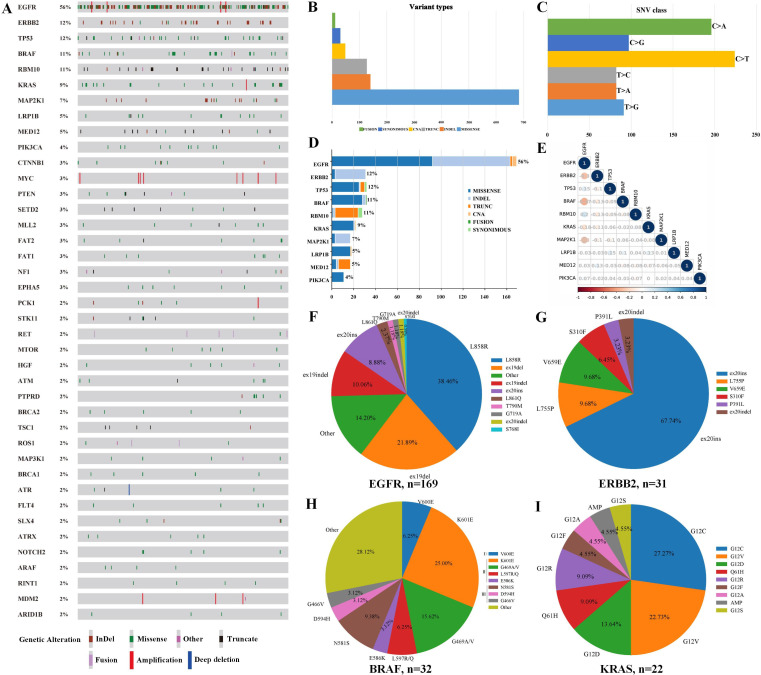
We detected 833 somatic variations in 248 lesions after performing 1021-gene panel NGS on 255 lesion samples from 112 patients with MPLC (figure 2A). Missense mutations were the most common variant and C>T substitutions the most common single nucleotide variation (figure 2B–C). In addition, the top 10 mutated genes in 255 tumors were EGFR (56%), ERBB2 (12%), TP53 (12%), BRAF (11%), RBM10 (11%), KRAS (9%), MAP2K1 (7%), LRP1B (5%), MED12 (5%), and PIK3CA (4%) (figure 2D). Correlation analysis showed that EGFR mutations were mutually exclusive with ERBB2, BRAF, KRAS, and MAP2K1 mutations but significantly co-occurred with TP53 and RBM10 mutations (all p<0.05) (figure 2E).
Figure 2.
Gene mutation spectrum in MPLCs. Gene mutation spectrum of top 40 mutated genes in MPLCs (A). Variation types (B), single nucleotide variations (C), top 10 mutated genes (D), and correlation analysis of top 10 mutated genes (E) in MPLCs. Frequency distributions of EGFR (F), ERBB2 (G), BRAF (H), and KRAS (I) mutation subtypes. MPLCs, multiple primary lung cancers.
We further analyzed the most frequent gene mutation subtypes in MPLCs (figure 2F–I). EGFR p.L858R and exon 19del comprised 38.46% and 21.89% of 169 EGFR mutations, respectively. The most common mutant subtype of ERBB2 was exon 20ins (21/31, 67.74%). BRAF mutations could be divided into three classes: class Ⅰ (V600D/E/K/R), class Ⅱ (G464V/G469X/E586K/L597X/K601X), and class Ⅲ (G466V/N581X/D594X/G596R).25 Type Ⅰ (V600E), Ⅱ, and Ⅲ mutations made up 6.25%, 50.00%, 15.63% of 32 BRAF mutations, respectively. KRAS p.G12C (27.27%) and p.G12V (22.73%) were the most common mutant subtypes. PIK3CA p.H1047R/E545Q/D and MAP2K1 exon 2del were the most common PIK3CA and MAP2K1 mutations, respectively (Supplementary figure S1). In addition, lung cancer driver genes including EML4-ALK fusions, ROS1 fusions, and KIF5B-RET fusions were detected in two, two, and five tumors, respectively.
Of the 112 patients with MPLCs, 87 (77.7%) shared no genetic mutations among their individual multiple lesions. We found 61 patients (54.5%) sharing ≥1 putative driver gene between at least two of their lesions (online supplemental figure S2 and table S2). These 61 patients had a higher proportion of the highest T stage (≥T1 c; p=0.027) and pTNM stage (≥ⅠA3; p=0.039), and their tumors had a higher percentage of T stage ≥T1 c (p=0.038) and pTNM stage≥ⅠA3 (p=0.044) than the patients sharing no putative driver gene (online supplemental table S3). These findings suggest that patients sharing one driver gene may have more aggressive cancers. We found 25 (55.6%) patients with ≥2 EGFRL858R/19del-mutant tumors among 45 patients with sensitive EGFR mutations. Besides, 25 (22.3%) patients shared driver gene alterations in ≥2 of their lesions, including EGFR p.L858R/19del/19indel/L861Q/20ins, ERBB2 exon 20ins, KRAS p.G12C/G12V/G12R, and BRAF p.K601E (online supplemental table S4).
Association of genomic landscape with TMB
The median TMB was 1.92 mutations/megabases (muts/Mb; range: 0–26.88) in 112 patients with MPLCs with 255 lesions, and the 25th and 75th percentiles of TMB were 0.96 and 3.00, respectively. Tumors were divided into high and low groups based on the 75th percentiles of TMB, and 31 patients (31/112, 27.7%) and 66 (66/255, 25.9%) tumors had high TMB. Higher TMB was observed in invasive adenocarcinoma and tumors with higher T stage and TNM stage (online supplemental figure S3).
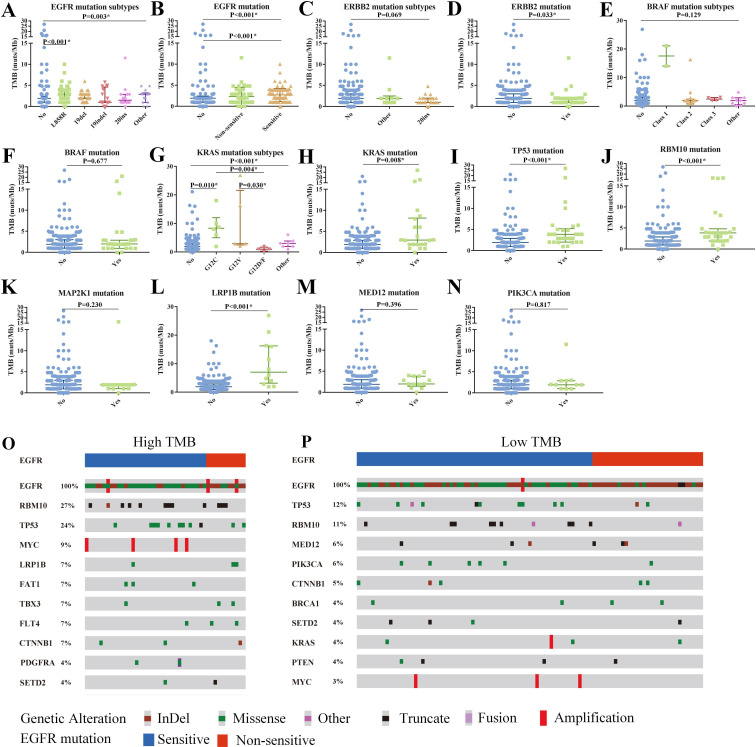
We further investigated the association of the top 10 high-frequency mutated genes with TMB. Tumors with EGFR p.L858R had higher TMB compared with wild-type tumors (2.88 vs 1.92, p=0.003) but did not differ from tumors with other EGFR subtypes (figure 3A). And tumors with sensitive EGFR mutations (L858R/19del) had significantly higher TMB than those with wild-type EGFR (2.88 vs 1.92, p<0.001, figure 3B). Tumors with ERBB2 mutations particularly exon 20ins had lower TMB than tumors with wild-type ERBB2 (exon 20ins: other mutations: wild type=1: 1.92: 1.92, p=0.069; mutations: wild type=1.00 vs 1.92, p=0.033; figure 3C and D). In addition, we found no significant difference in TMB among tumors with different BRAF subtypes, but two BRAFV600E-mutant tumors had a high median TMB (17.6muts/Mb, figure 3E and F). KRASG12C-mutant tumors had higher TMB than those with KRASG12D/F (8.32 vs 0.96, p=0.004) or wild type (8.32 vs 1.92, p=0.010), and KRAS-mutant tumors had higher TMB than those with wild type (2.94 vs 1.92, p=0.008) (figure 3G and H). In addition, tumors with TP53, RBM10, or LRP1B mutations had higher TMB levels vs those with wild type (TP53/RBM10: 3.84 vs 1.92, LRP1B: 7.00 vs 1.92, all p<0.05), this was not the case for tumors with MAP2K1, MED12, and PIK3CA mutations (figure 3I–N).
Figure 3.
Genomic profile and its associations with TMB. Association of TMB with EGFR mutation subtypes (A), EGFR mutation (B), ERBB2 mutation subtypes (C), ERBB2 mutation (D), BRAF mutation subtypes (E), BRAF mutation (F), KRAS mutation subtypes (G), KRAS mutation (H), TP53 mutation (I), RBM10 mutation (J), MAP2K1 mutation (K), LRP1B mutation (L), MED12 mutation (M), and PIK3CA mutation (N). Top 10 concomitant mutations of high-TMB (O) and low-TMB tumors (P) in EGFR-mutant patients with multiple primary lung cancers. InDel, insertions and deletions; TMB, tumor mutation burden.
To investigate the reason for higher TMB in EGFR-mutant tumors, we further depicted the gene mutation spectra in EGFR-mutant tumors. We found that high-TMB tumors with EGFR sensitive or non-sensitive mutations possessed a higher proportion of concurrent TP53/RBM10/LRP1B mutations that also predicted a high TMB (figure 3O–P). However, of 193 tumors without TP53/RBM10/LRP1B mutations, the ones with sensitive EGFR mutations (particularly L858R) still had higher TMB compared with tumors with wild-type EGFR (online supplemental figure S4).
Correlation between mutational landscape and PD-L1 and immune cells
PD-L1 TPS, PD-L1 CPS, CD3+/CD8 +TILs, and CD68+/CD163 +TAMs were detected in 75 patients with MPLCs with 157 lesions. The median PD-L1 TPS and CPS were 0 and 1, respectively. PD-L1 TPS was positive in only 6 (8.0%) patients and 6 (3.8%) tumors, and nobody had ≥2 positive lesions. Fifty-three (70.7%) patients with 83 (52.9 %) lesions had at least one lesion with positive PD-L1 CPS. The density ranges of tumor-infiltrating immune cells and broad positive correlations among them were displayed in the online supplemental figure S5.
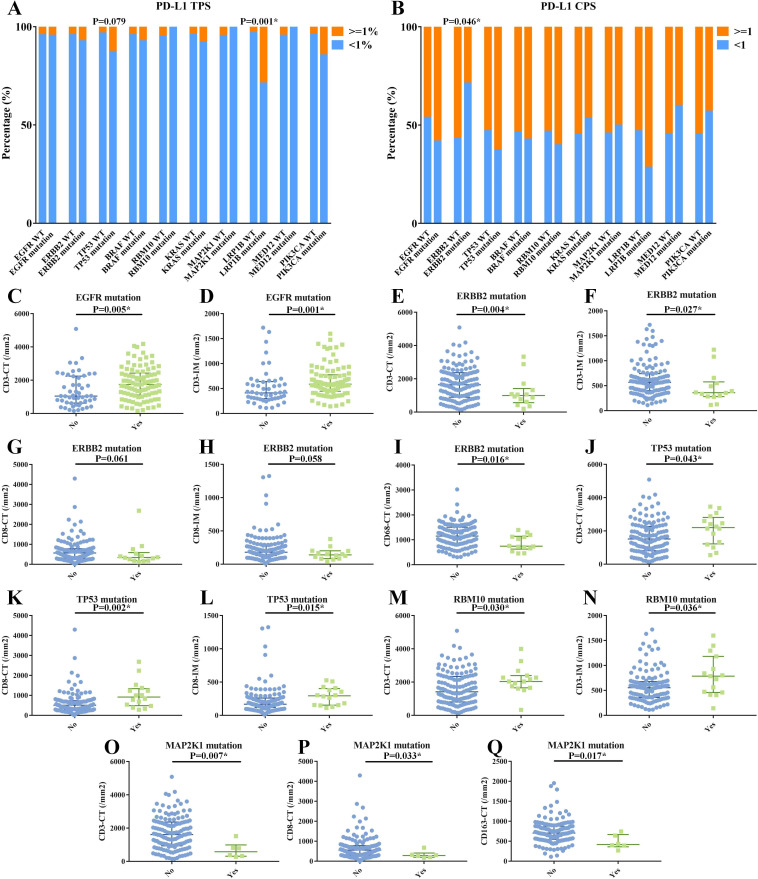
We began by analyzing the correlation between PD-L1 positivity and clinicopathological features in 157 lesions and the top 10 mutant genes in 143 lesions. Positive PD-L1 TPS was more often seen in solid tumors, invasive adenocarcinoma, tumors ≥2 cm, and higher-stage tumors (p<0.05), but PD-L1 CPS had no association with clinicopathological characteristics (online supplemental table S5). LRP1B-mutant tumors had a significantly higher percentage of positive PD-L1 TPS vs those with wild-type LRP1B (28.6% vs 2.9%; p=0.001), and TP53-mutant tumors presented with a positive trend (12.5% vs 3.1%; p=0.079) (figure 4A). And PD-L1 CPS positivity was more frequent in tumors with wild-type ERBB2 (56.6% vs 28.6%; p=0.046) (figure 4B).
Figure 4.
Correlation between the top 10 mutated genes and immune indicators. Correlation between the top 10 mutated genes and PD-L1 TPS (A) and PD-L1 CPS (B). Significant correlations between the top 10 mutated genes and immune cells (C–Q), including the associations of EGFR mutation with CD3-CT (C) and with CD3-IM (D); of ERBB2 mutation with CD3-CT (E), CD3-IM (F), CD8-CT (G), CD8-IM (H), and CD68-CT (I); of TP53 mutation with CD3-CT (J), CD8-CT (K), and CD8-IM (L); of RBM10 mutation with CD3-CT (M) and CD3-IM (N); of MAP2K1 mutation with CD3-CT (O), CD8-CT (P), and CD163-CT (Q). CPS, combined positive score; CT, central tumor; IM, invasive margin; PD-L1, programmed death ligand 1; TPS, tumor proportion score.
We then investigated the immune cell infiltration landscapes according to different clinicopathological and genomic features. We observed higher levels of CD3+/CD8+ TILs at both CT and IM and CD68+/CD163+ TAMs at CT were more often observed in patients with larger tumor sizes and higher stages (p<0.05; online supplemental tables S6,7). Moreover, we found that EGFR/TP53/RBM10 mutations positively correlated with specific immune cells, while ERBB2/MAP2K1 mutations were negatively correlated with them (figure 4C–Q, online supplemental table S8). Specifically, tumors with EGFR mutations (especially L858R) had higher levels of CD3-CT and CD3-IM (p<0.05), even in the 112 tumors without concomitant TP53/RBM10 mutations (figure 4C–D, online supplemental figure S6). ERBB2-mutant tumors presented with significantly lower levels of CD3-CT, CD3-IM, CD68-CT, and a trend toward lower CD8-CT (p=0.061) and CD8-IM (p=0.058) (figure 4E–I). TP53-mutant tumors exhibited higher levels of CD3-CT, CD8-CT, and CD8-IM (p<0.05) (figure 4J–L). RBM10-mutant tumors showed higher levels of CD3-CT and CD3-IM (p<0.05) (figure 4M and N). MAP2K1-mutant tumors presented with lower levels of CD3-CT, CD3-IM, and CD163-CT (figure 4O–Q). BRAF, KRAS, LRP1B, MED12, and PIK3CA mutations presented no statistical correlation with immune indexes (online supplemental table S8).
Association between genetic mutations and immunotypes
We further studied the association of frequent genetic mutations with immune indexes as a whole. Three immunotypes were generated based on systematic cluster analysis of PD-L1 TPS and CPS, CD3+/CD8+ TILs, and CD68+/CD163+ TAMs at CT and IM: high immune cells infiltration (‘inflamed’); low immune cells infiltration (‘deserted’); mixed type (figure 5A). We found that RBM10-mutant tumors exhibited a higher proportion of the inflamed immunotype (p=0.024), EGFR/TP53-mutant tumors had a high inflamed trend, and ERBB2-mutant tumors had a lower percentage of inflamed immunotype (p=0.002) (figure 5B). Considering the comutated pattern in EGFR and RBM10/TP53 genes, we further analyzed the association of EGFR mutations with immunotypes in tumors with wild-type RBM10/TP53. A higher percentage of inflamed immunotype was also observed in tumors with sensitive EGFR mutations (p=0.050), particularly EGFRL858R (figure 5C).
Figure 5.
Association between frequent genetic mutations and immunotypes. (A) Cluster analysis of PD-L1 TPS and CPS, CD3+/CD8 +TILs, and CD68+/CD163 +TAMs at CT and IM to obtain three immunotypes: inflamed (2); mixed type (1); deserted (0). (B) Correlation between top 10 mutated genes and immunotypes. (C) Correlation between EGFR mutation subtypes and immunotypes in tumors with wild-type RBM10/TP53. CPS, combined positive score; CT, central tumor; IM, invasive margin; PD-L1, programmed death ligand 1; TAMs, tumor-associated macrophages; TILs, tumor-infiltrating lymphocytes; TPS, tumor proportion score.
Heterogeneity of TMB, PD-L1 expression, and immune cell infiltration
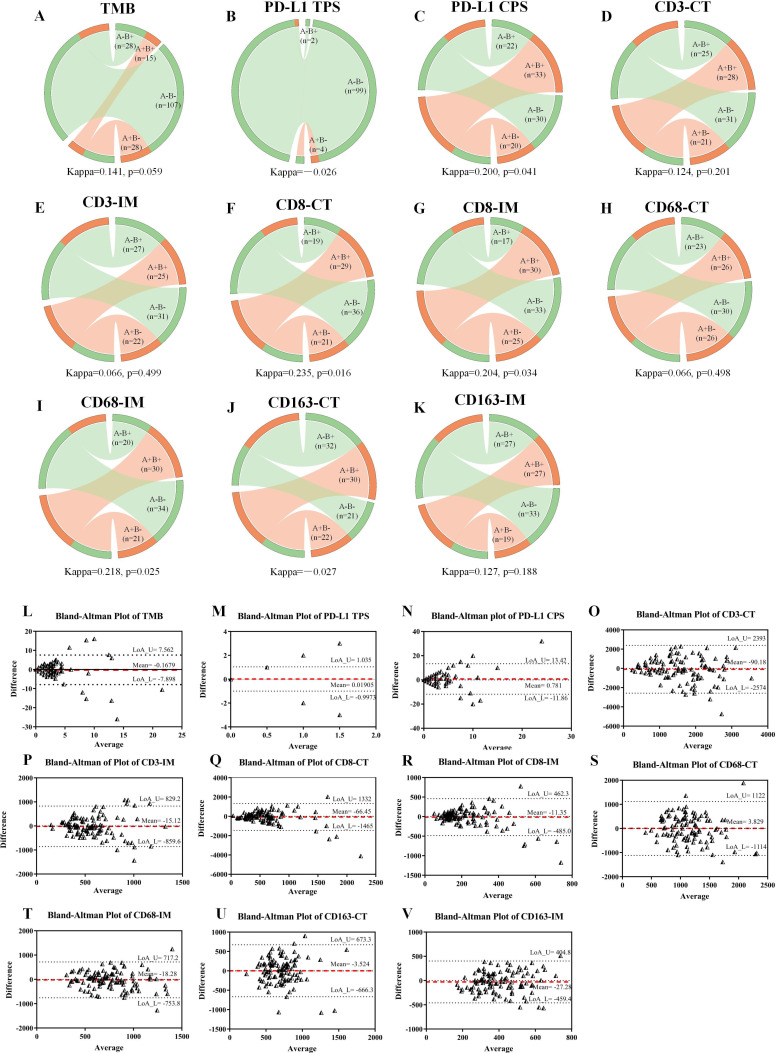
We used data from 178 tumor pairs in 112 patients with MPLCs with 255 lesions and from 105 tumor pairs in 63 patients with 146 lesions to perform agreement analyses of TMB and immune indexes, respectively. We analyzed the distributional agreement of each immune index as both categorical variables by kappa tests (figure 6A–K) and continual variables (figure 6L–V) by Bland-Altman tests among paired tumors.
Figure 6.
Heterogeneity of TMB, PD-L1, and immune cells in paired tumors. Circos diagrams display the distributional agreement of TMB (A), PD-L1 TPS (B), PD-L1 CPS (C), CD3-CT (D), CD3-IM (E), CD8-CT (F), CD8-IM (G), CD68-CT (H), CD68-IM (I), CD163-CT (J), and CD163-IM (K) in tumor pairs as categorical variables. Bland-Altman plots showed the LoA of TMB (L), PD-L1 TPS (M), PD-L1 CPS (N), CD3-CT (O), CD3-IM (P), CD8-CT (Q), CD8-IM (R), CD68-CT (S), CD68-IM (T), CD163-CT (U), and CD163-IM (V) in tumor pairs as continual variables. Two tumors from the same patient constitute a tumor pair (for instance, three tumors in an individual constitute three tumor pairs). The left side of a circos diagram represents the second lesions, and the right side represents the paired first lesions. The positive and negative tumors are marked with orange and green, respectively. Ribbons within each circos diagram connect paired tumors. Ribbons connecting the circos diagram of the same color (A+B+ and A−B−) represent agreement, otherwise they represent heterogeneity. The bigger the positive kappa value, the less significant the heterogeneity. In the Bland-Altman plots, the differences in each immune index are plotted against the mean of each immune index between paired tumors. The mean differences are displayed with red dotted lines, and 95% CIs of the LoA are displayed within the black dotted horizontal lines. The wider the 95% LoA, the more significant the heterogeneity. CPS, combined positive score; CT, central tumor; IM, invasive margin; LoA, limits of agreement; PD-L1, programmed death ligand 1; TMB, tumor mutation burden; TPS, tumor proportion score.
There was agreement of TMB levels in 122 (68.5%) paired tumors and disagreement in 56 (31.5%) paired tumors (kappa=0.141, p=0.059; figure 6A). Bland-Altman plot revealed a relatively wide 95% CI limit of agreement (LoA) (figure 6L). None of the tumor pairs were positive for PD-L1 TPS, an agreement was observed in 99 (94.3%, both negative) tumor pairs, and disagreement in 6 (5.7%) pairs (kappa=−0.026; figure 6B). The negative kappa value (coefficient of agreement) of PD-L1 TPS had no clinical significance. Sixty-three (60%) tumor pairs presented uniform PD-L1 CPS expression, and 42 (40%) were heterogeneous (kappa=0.200, p=0.041; figure 6C). Bland-Altman test revealed a narrow 95% LoA for PD-L1 TPS and CPS between tumor pairs, indicating no obvious heterogeneity (figure 6M–N).
In terms of the immune cells, kappa tests showed no significant heterogeneity of CD8 +TILs at both CT (kappa=0.235, p=0.016) or IM (kappa=0.204, p=0.034) and CD68-IM (kappa=0.218, p=0.025), and Bland-Altman tests also showed narrow 95% LoA (figure 6F–G1, Q–R and T). However, we observed significant heterogeneity and a relatively wide 95% LoA in CD3-CT (kappa=0.124, p=0.201) and CD3-IM (kappa=0.066, p=0.499), CD68-CT (kappa=0.066, p=0.498), CD163-CT (kappa=−0.027), and CD163-IM (kappa=0.127, p=0.188) (kappa=0.008, p=0.925) (figure 6D–E, H, J–K, O–P, S). Overall, TMB, CD3 +TILs, CD68-CT, and CD163 +TAMs presented with significant heterogeneity among paired tumors (kappa <0.2), while PD-L1, CD8 +TILs, and CD68-IM did not.
Discussion
MPLCs have become a new challenge even in the era of precision medicine. Therefore, we investigated genomic profiles of MPLCs and their associations with TMB, PD-L1 expression, and immune cell infiltration landscapes to facilitate precise diagnosis and therapy. In this study, we detected many known driver genes in MPLCs and depicted the mutation subtypes. Most patients had different mutation spectra, but half of the patients shared ≥1 putative driver gene among their multiple tumors. We found low TMB and PD-L1 expression with heterogeneous immune cell infiltration landscapes, but frequent gene mutations and subtypes were associated with these variables.
We portrayed the mutation subtypes of the most frequently mutated genes in MPLCs (such as EGFR, ERBB2, BRAF, and TP53). EGFR p.L858R and exon 19del were the predominant subtypes in MPLCs, but their ratios (38% vs 21%, almost 2:1) differed from another study including 2410 EGFR-mutant patients with early-stage lung cancer (almost 1:1).26 As for BRAF mutations in MPLCs, the most common mutations were class Ⅱ mutations rather than class Ⅰ V600E that was the predominant BRAF mutations in single lung cancer,27 indicating that BRAF/MEK inhibitors may not be efficacious against MPLCs. ERBB2 exon 20ins and KRAS p.G12C were the most common subtypes of ERBB2 ad KRAS mutations in MPLCs, similar to single lung cancer.28 29 To our knowledge, ours is the first elaborate description of mutational subtypes of high-frequency mutated genes in MPLCs, especially those manifesting as GGOs. Together, except for BRAF, there were similarities in the main mutation subtypes of frequent driver genes between MPLCs and single lung cancer.
In this study, the majority of patients with MPLCs had different mutational background, indicative of independent clonal origin, but about half of patients shared ≥1 putative driver gene. Ma et al30 proposed the theory of ‘convergent evolution’ in multifocal lung cancers, that is, heterogeneous driver mutations in multiple tumors from the same patient aggregate on the same signaling pathway to activate key carcinogenic pathways. The evidence from other studies31 and from our own study (where half of the patients shared ≥1 putative driver gene and presented more aggressive tumors) supports this theory. MPLCs have been shown to share hotspot driver mutations (EGFRL858R/19del/KRASG12X) that can be added into a histomolecular algorithm for diagnosing MPLCs.32 In this study, we provide detailed shared mutational repertoire that are of great importance for the accurate diagnosis of MPLCs. Moreover, more than half of the patients had ≥2 EGFRL858R/19del-mutant tumors among 45 patients with sensitive EGFR mutations, particularly patients with higher T and TNM stages, providing an opportunity for EGFR-TKI therapy. A retrospective study revealed the limitedobjective response rate (ORR, about 15%) of targeted therapy against residual GGOs in patients with resected EGFR-mutant lung cancers, of whom more than 90% received first-generation EGFR-TKI.7 Osimertinib has been approved as an adjuvant therapy for EGFR-mutant patients with stage IB-IIIA NSCLC due to significantly prolonged disease-free survival.33 This may be a novel choice for EGFR-mutant patients with resected stage IB-IIIA NSCLC accompanied by residual GGOs. Collectively, MPLCs are driven by different molecular events but may have convergent evolution. Depicting genomic profiles of MPLCs may facilitate the accurate diagnosis and the application of targeted therapy particularly EGFR-TKI.
In our study, the median TMB in MPLCs was extremely low, and high-TMB lesions often harbored EGFRL858R/RBM10mut/LRP1Bmut/TP53mut/KRASG12C mutations or wild-type ERBB2. Chang et al34 enrolled 37 patients with MPLCs who had an average TMB of 7.0 (0–41.2) muts/Mb, significantly higher than that in our study (1.92). Higher TMB was more common in invasive and advanced tumors or in smokers35; however, our research populations included mainly non-smoker women with stage Ⅰ, which may partly explain the discrepancy. Tumors with sensitive EGFR mutations (particularly L858R) had a higher TMB and a higher proportion of concomitant TP53/RBM10/LRP1B mutations in MPLCs. This finding differs from the results of studies with single advanced lung cancer in which EGFR-mutant patients had a lower TMB.36 37 Several reasons may partly explain the discrepancy. TP53/RBM10/LRP1B mutations have been associated with high TMB in NSCLC,38–41 and their high concomitant incidence in EGFR-mutant MPLCs lesions may in part account for high TMB. In addition, tumors with sensitive EGFR mutations but no TP53/RBM10/LRP1B mutation also had a higher TMB level than tumor with wild-type EGFR. Lung cancer with EGFR mutations has an advantage in growth and invasion, but advanced lung cancers with wild-type EGFR need an abundance of molecular events to drive tumor progression into advanced stage and it may lead to higher TMB in advanced cancers with wild-type EGFR. For early-stage lung cancers, tumors with EGFR mutations are more aggressive and may harbor more concomitant mutations versus those with wild-type EGFR, which may explain the higher TMB level in EGFR-mutant cancers. Therefore, concomitant mutations and distinct biological behaviors at different tumor stages may account for the higher TMB in EGFR-mutant tumors in MPLCs, but studies are needed to confirm this. Moreover, a significantly higher TMB was also observed in patients with MPLCs with KRASG12C/ERBB2wt. A study involving 4064 patients with NSCLC found that ERBB2-mutant patients had no significant difference in TMB versus patients with wild-type ERBB2, but KRAS-mutant patients had a higher TMB versus patients with wild-type KRAS,36 similar to our study. Overall, specific gene mutations and concomitant mutations had a close correlation with TMB in MPLCs, which may help identify patients who benefit from immunotherapy.
PD-L1 TPS was positive in few patients with MPLCs, but LRP1B-mutant tumors exhibited significantly higher levels of PD-L1 TPS, while TP53-mutant tumors had a positive trend. The PD-L1 TPS positive rate in locally advanced or metastatic NSCLC ranges from 24% to 60%,42 and it is approximately 47% in early-stage NSCLC.43 And 13%–48% of patients and 15%–30% of tumors showed PD-L1 positivity in MPLCs,12 13 44 higher than our study. PD-L1 positivity on tumor cells occurred more frequently in advanced cancers, but our study had a higher percentage of stage Ⅰ patients in comparison with other MPLCs studies (≥80% vs ≤60%), which may partially explain the low expression level of PD-L1 in our study. Furthermore, we found that PD-L1 TPS positivity was more often observed in LRP1B/TP53-mutant tumors in MPLCs. LRP1B and TP53 mutations were positive predictive and prognostic biomarkers of immunotherapy in multiple solid and hematological tumors.38 45 A study involving 1586 patients with NSCLC showed that TP53 mutations were associated with positive PD-L1 expression.46 Patients with advanced NSCLC with TP53 mutations had a higher ORR and better overall survival (OS) under ICIs.38 Collectively, the expression level of PD-L1 TPS in patients with MPLCs is extremely low, indicating a lack of ICI efficacy, but LRP1B and TP53 mutations may help predict PD-L1 levels and help optimize the immunotherapy strategy for patients with MPLCs.
Our study depicted the infiltration landscape of CD3+/CD8 +TILs or CD68+/CD163 +TAMs at both CT and IM. These immune cells were associated with immunotherapy resistance and prognosis in a variety of solid tumors.11 15 47 48 CD8 +TILs, especially at IM, could directly predict the efficacy of PD-1 inhibitors in melanoma.49 Prognostic analysis in 17 solid tumors including lung cancer demonstrated CD8 +TILs had the most robust association with favorable outcomes.50 TAMs mediated immunotherapy resistance by capturing PD-1 inhibitors from the surface of CD8 +T cells,15 and their repolarization status could potentiate the efficacy of ICIs.16 A meta-study involving 2572 patients with NSCLC from 20 studies found that high levels of CD68+ TAMS and M1 TAMs in the tumor parenchyma were associated with longer OS, and that high levels of CD68 +TAMs and CD163 +TAMs in the stroma were associated with shorter OS.51 Therefore, combining multiple immune cells at different sites may better predict the efficacy of immunotherapy and prognosis.
After integrating all immune indicators, we found that EGFRL858R/19del mutations (often accompanied by RBM10/TP53 mutations that predict high levels of CD3+/CD8 +TILs) were associated with higher TMB, higher CD3 +TILs, and an inflamed immunotype. Multiple retrospective studies have shown that patients with NSCLC with EGFR mutations rarely benefit from ICIs with ORR ranging from 4% to 20%.52–54 IMMUNOTARGET, one of the largest retrospective studies, enrolled 551 patients with advanced lung cancer with at least one driver mutation to evaluate the efficacy of ICIs monotherapy; 125 EGFR-mutant patients achieved an ORR of 12%.52 Neoadjuvant immunotherapy studies in early lung cancer have also shown that few EGFR-mutant patients benefit from neoadjuvant immunotherapy.55 This poor efficacy of immunotherapy in EGFR-mutant patients has been ascribed to a variety of factors, including EGFR mutation site, TMB, PD-L1 expression, tumor-infiltrating immune cells.56 Patients with EGFR-mutant lung cancer had lower TMB, PD-L1 levels, and numbers of TILs and CD8+TILs.54 57 In terms of EGFR mutation subtypes, NSCLCpatients with EGFR exon 19del had significantly lower TMB levels than those with EGFR p.L858R, and they had a significantly lower ORR to ICIs (7% vs 22%) and shorter PFS/OS than those with wild-type EGFR.58 However, the scenario is slightly different in MPLCs: of 20 patients with resected NSCLC who had at least one high-risk residual GGOs and received sintilimab, 6 patients with 12 GGOs received secondary operations, and EGFR-mutant lesions displayed fewer residual tumor cells.9 Similarly, we found that EGFRL858R/19del-mutant tumors (particularly EGFRL858R) were often accompanied by RBM10/TP53 mutations and were mainly characterized by higher TMB and inflamed immunotypes in MPLCs. Our results suggest a potential benefit from ICIs for patients with MPLCs with EGFRL858R/19del mutations (particularly EGFRL858R) and concurrent RBM10/TP53 mutations, but further studies are needed to confirm it.
ERBB2-mutant tumors in MPLCs primarily exhibited lower TMB, fewer CD3+/CD8+ TILs and CD68+ TAMs, and non-inflamed immunotypes. Limited evidence on the efficacy of immunotherapy for ERBB2-positive patients with NSCLC has revealed an ORR of approximately 7%–12%.52 59 In the IMMUNOTARGET study, 29 patients with ERBB2-positive advanced lung cancer received ICIs monotherapy and showed an ORR of only 7%.52 Another study with 122 ERBB2-positive advanced NSCLC showed that 26 of them receiving immunotherapy had an ORR of only 12%.59 Compared with a non-selective lung cancer cohort (n=3000), ERBB2-positive patients with NSCLC had a similar TMB but lower PD-L1 positivity,59 which may account for the poor efficacy of ICIs in these patients. In our study, ERBB2-mutant tumors were mainly characterized by low immunogenicity and non-inflamed immune microenvironment, suggesting a poor efficacy of ICIs in ERBB2-mutant patients with MPLCs.
In addition, we found an agreement in PD-L1 positivity among most of the paired tumors. A study on 43 patients with MPLCs with 112 tumors reported disagreement of PD-L1 in 12 (27.9%) patients.12 Another retrospective study on 23 patients with MPLCs showed PD-L1 expression disagreement in 11 and agreement in 12 patients, showing the heterogeneity as a whole.13 With larger sample size, the proportion of patients with negative PD-L1 expression in all lesions rises gradually, and the heterogeneity diminishes gradually. Moreover, TMB, CD3+ TILs, and CD68+/CD163+ TAMS but not CD8+ TILs exhibited significant heterogeneity between paired lesions in patients with MPLCs. Differentially-expressed TMB and differentially infiltrated CD3+CD8- TILs and TAMs among paired tumors may lead to heterogeneous responses to ICIs in patients with MPLCs.
Finally, we acknowledge the limitations in our study. First, this is a single-center retrospective study, so it may have selection bias. Second, we diagnosed MPLCs based on ACCP guidelines in the absence of accurate diagnostic standard, so we might not completely exclude metastatic lung cancer. Establishing comprehensive diagnostic criteria that combine clinical imaging, histopathology, and molecular genetic characteristics is important to achieve an accurate diagnosis. Third, we calculated TMB based on 1021-gene panel NGS, which may be less accurate than whole-exome sequencing (WES). Future studies performing WES may better investigate the association of genomic profile with TMB. Fourth, we selected two representative fields in the immunohistochemical sections when counting tumor-infiltrating immune cells, which may not reflect the complete immune landscape. Finally, patients with MPLCs in this study underwent surgery within 3 years; therefore, disease-free survival and OS data were not available for us to analyze prognostic factors.
Conclusion
We depicted the genomic profiles of tumors in patients with MPLCs and demonstrated that MPLCs are driven by different molecular events but may have convergent evolution. Our results suggested that MPLCs often exhibited low TMB, low PD-L1 expression, and a heterogeneous immune infiltration landscape. Some frequent gene mutations (such as EGFR/ERBB2) and concomitant mutations (such as TP53/RBM10/LRP1B) were associated with the immunogenicity and tumor immune microenvironmental features. Our findings can be helpful for the accurate diagnosis of MPLCs and the identification of patient populations that may benefit from immunotherapy and targeted therapy.
Acknowledgments
The authors sincerely thank the multidisciplinary team of thoracic oncology, the Second Xiangya Hospital, Central South University.
Footnotes
CH, LZ and WL contributed equally.
Contributors: Conception and design: LZ, FW. Administrative support: CH, FW. Provision of study materials or patients: WL, FW, SF. Collection and assembly of data: LZ, DY, YL, YP, YZ, SZ, SS, YP. Experimental operation and technique support: LZ, JL, YL, XHL, LS, SF, CC, YJ, QL, LY, YS, QH. Data analysis and interpretation: LZ, YL, FW, XLL, CD, ZQ, PL. Manuscript writing and final approval: All authors. FW acts as the guarantor for overall content.
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University (No.2021002). All patients signed consents form to participate in this study.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7–33. 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Rami-Porta RAH, Travis WD, Rusch VW. AJCC cancer staging manual AJCC cancer staging manual. 8th edn. France: Springer International Publishing, 2017: 431–56. [Google Scholar]
- 3.Kozower BD, Larner JM, Detterbeck FC, et al. Special treatment issues in non-small cell lung cancer: diagnosis and management of lung cancer, 3rd edn: American College of chest physicians evidence-based clinical practice guidelines. Chest 2013;143:e369S–99. 10.1378/chest.12-2362 [DOI] [PubMed] [Google Scholar]
- 4.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606–12. 10.1016/S0022-5223(19)40289-4 [DOI] [PubMed] [Google Scholar]
- 5.Kang X, Zhang C, Zhou H, et al. Multiple pulmonary resections for synchronous and metachronous lung cancer at two Chinese centers. Ann Thorac Surg 2020;109:856–63. 10.1016/j.athoracsur.2019.09.088 [DOI] [PubMed] [Google Scholar]
- 6.Nikitas J, DeWees T, Rehman S, et al. Stereotactic body radiotherapy for early-stage multiple primary lung cancers. Clin Lung Cancer 2019;20:107–16. 10.1016/j.cllc.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng B, Li C, Zhao Y, et al. The impact of postoperative EGFR-TKIs treatment on residual GGO lesions after resection for lung cancer. Signal Transduct Target Ther 2021;6:73. 10.1038/s41392-020-00452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mielgo-Rubio X, Calvo V, Luna J, et al. Immunotherapy moves to the early-stage setting in non-small cell lung cancer: emerging evidence and the role of biomarkers. Cancers 2020;12:3459. 10.3390/cancers12113459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Shi M, Li M, et al. An exploratory study of PD-1 inhibitor for high-risk multiple ground-glass nodules (mGGNs) in synchronous stage I non-small cell lung cancer patients. JCO 2020;38:e21068. 10.1200/JCO.2020.38.15_suppl.e21068 [DOI] [Google Scholar]
- 10.Wu F, Li W, Zhao W, et al. Synchronous ground‐glass nodules showed limited response to anti‐PD‐1/PD‐L1 therapy in patients with advanced lung adenocarcinoma. Clin Transl Med 2020;10:e149. 10.1002/ctm2.149 [DOI] [Google Scholar]
- 11.Yi M, Jiao D, Xu H, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer 2018;17:129. 10.1186/s12943-018-0864-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haratake N, Toyokawa G, Takada K, et al. Programmed death-ligand 1 expression and EGFR mutations in multifocal lung cancer. Ann Thorac Surg 2018;105:448–54. 10.1016/j.athoracsur.2017.09.025 [DOI] [PubMed] [Google Scholar]
- 13.Mansfield AS, Murphy SJ, Peikert T, et al. Heterogeneity of programmed cell death ligand 1 expression in multifocal lung cancer. Clin Cancer Res 2016;22:2177–82. 10.1158/1078-0432.CCR-15-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sha D, Jin Z, Budczies J, et al. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discov 2020;10:1808–25. 10.1158/2159-8290.CD-20-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arlauckas SP, Garris CS, Kohler RH, et al. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci Transl Med 2017;9:eaal3604. 10.1126/scitranslmed.aal3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choo YW, Kang M, Kim HY, et al. M1 macrophage-derived Nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors. ACS Nano 2018;12:8977–93. 10.1021/acsnano.8b02446 [DOI] [PubMed] [Google Scholar]
- 17.Feng W, Li Y, Shen L, et al. Clinical impact of the tumor immune microenvironment in completely resected stage IIIA(N2) non-small cell lung cancer based on an immunoscore approach. Ther Adv Med Oncol 2021;13:1758835920984975. 10.1177/1758835920984975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagès F, Kirilovsky A, Mlecnik B, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol 2009;27:5944–51. 10.1200/JCO.2008.19.6147 [DOI] [PubMed] [Google Scholar]
- 19.Finley DJ, Yoshizawa A, Travis W, et al. Predictors of outcomes after surgical treatment of synchronous primary lung cancers. J Thorac Oncol 2010;5:197–205. 10.1097/JTO.0b013e3181c814c5 [DOI] [PubMed] [Google Scholar]
- 20.Sugimura H, Watanabe S, Tsugane S, et al. Case-control study on histologically determined multiple primary lung cancer. J Natl Cancer Inst 1987;79:435–41. [PubMed] [Google Scholar]
- 21.Kono M, Fujii M, Adachi S, et al. Multiple primary lung cancers: radiographic and bronchoscopic diagnosis. J Thorac Imaging 1993;8:63–8. [PubMed] [Google Scholar]
- 22.Yang H, Sun Y, Yao F, et al. Surgical therapy for bilateral multiple primary lung cancer. Ann Thorac Surg 2016;101:1145–52. 10.1016/j.athoracsur.2015.09.028 [DOI] [PubMed] [Google Scholar]
- 23.Chang Y-L, Wu C-T, Lee Y-C. Surgical treatment of synchronous multiple primary lung cancers: experience of 92 patients. J Thorac Cardiovasc Surg 2007;134:630–7. 10.1016/j.jtcvs.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 24.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 25.Negrao MV, Raymond VM, Lanman RB, et al. Molecular landscape of BRAF-mutant NSCLC reveals an association between clonality and driver mutations and identifies targetable Non-V600 driver mutations. J Thorac Oncol 2020;15:1611–23. 10.1016/j.jtho.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suda K, Mitsudomi T, Shintani Y, et al. Clinical impacts of EGFR mutation status: analysis of 5780 surgically resected lung cancer cases. Ann Thorac Surg 2021;111:269–76. 10.1016/j.athoracsur.2020.05.041 [DOI] [PubMed] [Google Scholar]
- 27.Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307–16. 10.1016/S1470-2045(17)30679-4 [DOI] [PubMed] [Google Scholar]
- 28.Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res 2005;65:1642–6. 10.1158/0008-5472.CAN-04-4235 [DOI] [PubMed] [Google Scholar]
- 29.Scheffler M, Ihle MA, Hein R, et al. K-Ras mutation subtypes in NSCLC and associated Co-occuring mutations in other oncogenic pathways. J Thorac Oncol 2019;14:606–16. 10.1016/j.jtho.2018.12.013 [DOI] [PubMed] [Google Scholar]
- 30.Ma P, Fu Y, Cai M-C, et al. Simultaneous evolutionary expansion and constraint of genomic heterogeneity in multifocal lung cancer. Nat Commun 2017;8:823. 10.1038/s41467-017-00963-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X, Fujimoto J, Ying L, et al. Multi-region exome sequencing reveals genomic evolution from preneoplasia to lung adenocarcinoma. Nat Commun 2019;10:2978. 10.1038/s41467-019-10877-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansuet-Lupo A, Barritault M, Alifano M, et al. Proposal for a combined histomolecular algorithm to distinguish multiple primary adenocarcinomas from intrapulmonary metastasis in patients with multiple lung tumors. J Thorac Oncol 2019;14:844–56. 10.1016/j.jtho.2019.01.017 [DOI] [PubMed] [Google Scholar]
- 33.Wu Y-L, Tsuboi M, He J, et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med 2020;383:1711–23. 10.1056/NEJMoa2027071 [DOI] [PubMed] [Google Scholar]
- 34.Chang JC, Alex D, Bott M, et al. Comprehensive next-generation sequencing unambiguously distinguishes separate primary lung carcinomas from intrapulmonary metastases: comparison with standard histopathologic approach. Clin Cancer Res 2019;25:7113–25. 10.1158/1078-0432.CCR-19-1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai L, Bai H, Duan J, et al. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Immunother Cancer 2019;7:198. 10.1186/s40425-019-0660-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a Clinicogenomic database. JAMA 2019;321:1391–9. 10.1001/jama.2019.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Offin M, Rizvi H, Tenet M, et al. Tumor mutation burden and efficacy of EGFR-tyrosine kinase inhibitors in patients with EGFR-mutant lung cancers. Clin Cancer Res 2019;25:1063–9. 10.1158/1078-0432.CCR-18-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Assoun S, Theou-Anton N, Nguenang M, et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 2019;132:65–71. 10.1016/j.lungcan.2019.04.005 [DOI] [PubMed] [Google Scholar]
- 39.Ozaki Y, Muto S, Takagi H, et al. Tumor mutation burden and immunological, genomic, and clinicopathological factors as biomarkers for checkpoint inhibitor treatment of patients with non-small-cell lung cancer. Cancer Immunol Immunother 2020;69:127–34. 10.1007/s00262-019-02446-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Chong W, Wu Q, et al. Association of LRP1B mutation with tumor mutation burden and outcomes in melanoma and non-small cell lung cancer patients treated with immune check-point blockades. Front Immunol 2019;10:1113. 10.3389/fimmu.2019.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang Y, Li X, Liu W, et al. Comprehensive genomic profile of Chinese lung cancer patients and mutation characteristics of individuals resistant to icotinib/gefitinib. Sci Rep 2020;10:20243. 10.1038/s41598-020-76791-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu H, Boyle TA, Zhou C, et al. PD-L1 expression in lung cancer. J Thorac Oncol 2016;11:964–75. 10.1016/j.jtho.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Chen Z, Ballman KV, et al. Correlation of PD-L1 expression with tumor mutation burden and gene signatures for prognosis in early-stage squamous cell lung carcinoma. J Thorac Oncol 2019;14:25–36. 10.1016/j.jtho.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jia X, Zhang L, Wu W, et al. Driver mutation analysis and PD-L1 expression in synchronous double primary lung cancer. Appl Immunohistochem Mol Morphol 2018;26:246–53. 10.1097/PAI.0000000000000412 [DOI] [PubMed] [Google Scholar]
- 45.Hodkinson BP, Schaffer M, Brody JD, et al. Biomarkers of response to ibrutinib plus nivolumab in relapsed diffuse large B-cell lymphoma, follicular lymphoma, or Richter's transformation. Transl Oncol 2021;14:100977. 10.1016/j.tranon.2020.100977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoenfeld AJ, Rizvi H, Bandlamudi C, et al. Clinical and molecular correlates of PD-L1 expression in patients with lung adenocarcinomas. Ann Oncol 2020;31:599–608. 10.1016/j.annonc.2020.01.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boscolo A, Fortarezza F, Lunardi F, et al. Combined immunoscore for prognostic stratification of early stage non-small-cell lung cancer. Front Oncol 2020;10:564915. 10.3389/fonc.2020.564915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell 2015;27:462–72. 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruni D, Angell HK, Galon J. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20:662–80. 10.1038/s41568-020-0285-7 [DOI] [PubMed] [Google Scholar]
- 51.Mei J, Xiao Z, Guo C, et al. Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: a systemic review and meta-analysis. Oncotarget 2016;7:34217–28. 10.18632/oncotarget.9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321–8. 10.1093/annonc/mdz167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haratani K, Hayashi H, Tanaka T, et al. Tumor immune microenvironment and nivolumab efficacy in EGFR mutation-positive non-small-cell lung cancer based on T790M status after disease progression during EGFR-TKI treatment. Ann Oncol 2017;28:1532–9. 10.1093/annonc/mdx183 [DOI] [PubMed] [Google Scholar]
- 54.Gainor JF, Shaw AT, Sequist LV, et al. Egfr mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016;22:4585–93. 10.1158/1078-0432.CCR-15-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang J, Zhang C, Zhong W-Z. Neoadjuvant immunotherapy for non-small cell lung cancer: state of the art. Cancer Commun 2021;41:287–302. 10.1002/cac2.12153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin A, Wei T, Meng H, et al. Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Mol Cancer 2019;18:139. 10.1186/s12943-019-1062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong Z-Y, Zhang J-T, Liu S-Y, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017;6:e1356145. 10.1080/2162402X.2017.1356145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hastings K, Yu HA, Wei W, et al. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann Oncol 2019;30:1311–20. 10.1093/annonc/mdz141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.W-CV L, Feldman DL, Buonocore DJ. PD-L1 expression, tumor mutation burden and response to immune checkpoint blockade in patients with HER2-mutant lung cancers. J Clin Oncol 2018;36:9060. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003773supp001.pdf (712.9KB, pdf)
Data Availability Statement
Data are available upon reasonable request.