Supplemental Digital Content is available in the text.
Keywords: Antibiotic resistance, Heavy metals, Lead, C. diff, Infectious disease epidemiology, Environmental epidemiology
Abstract
Background:
Infection by antibiotic resistant bacteria (ARB) is a global health crisis and asymptomatic colonization increases risk of infection. Nonhuman studies have linked heavy metal exposure to the selection of ARB; however, few epidemiologic studies have examined this relationship. This study analyzes the association between urinary lead level and colonization by ARB in a nonclinical human population.
Methods:
Data came from the Survey of the Health of Wisconsin 2016–2017, and its ancillary Wisconsin Microbiome Study. Urinary lead levels, adjusted for creatinine, were used to assess exposure. ARB included methicillin resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), fluoroquinolone resistant Gram-negative bacilli (RGNB), and Clostridium difficile (C. diff), from skin, nose, and mouth swabs, and saliva and stool samples. Logistic regression, adjusted for covariates, was used to evaluate associations between Pb and ARB. Secondary analysis investigated Pb resistance from ARB isolates.
Results:
Among 695 participants, 239 (34%) tested positive for ARB. Geometric mean urinary Pb (unadjusted) was 0.286 µg/L (95% confidence intervals [CI] = 0.263, 0.312) for ARB negative participants and 0.323 µg/L (95% CI = 0.287, 0.363) for ARB positive participants. Models adjusted for demographics, diet, and antibiotic use showed elevated odds of positive colonization for those in the 95th percentile (vs. below) of Pb exposure (odds ratio [OR] = 2.05, 95% CI = 0.95, 4.44), and associations were highest in urban residents (OR = 2.85, 95% CI = 1.07, 7.59). RGNB isolates were most resistant to Pb.
Discussion:
These novel results suggest that Pb exposure is associated with increased colonization by ARB, and that RGNB are particularly resistant to Pb.
What This Study Adds
Antibiotic resistance, a global public health challenge, threatens the efficacy of some of the world’s only treatments for bacterial infections. Environmental toxicants contribute to the presence of antibiotic resistance genes in the environment. Translation of this knowledge to community settings is limited. This study examines general population-based levels of urinary lead, a biomarker of chronic exposure, and the prevalence of antibiotic resistant organisms. It adds new epidemiologic links between lead exposure and colonization by multiple antibiotic resistant bacteria at multiple body sites. Results highlight additional interdisciplinary environmental epidemiology and infectious disease collaborations are needed to tackle antibiotic resistance.
Introduction
Heavy metals are among the top 10 list of priority pollutants contributing to morbidity and mortality globally.1 Despite significant progress in environmental regulation in the United States, data from the National Health and Nutrition Exam Survey (NHANES) show heavy metal exposures in the general US population, including lead (Pb), remain prevalent.2,3 In 2017, the global burden of disease reports found Pb exposure accounted for 1.06 million deaths and 24.4 million years of healthy life lost (disability-adjusted life years [DALYs] 2017), considering only cardiovascular and neurological health effects.4 However, human exposure to Pb has also been shown to affect almost every aspect of immune function, even at low levels of Pb exposure.5 These immune effects may also lead to changes in human microbiome composition and reduce the body’s ability to resist harmful bacteria, and may lead to an abundance of antimicrobial resistance (AMR) within the host. However, the role of environmental pollution and its intersection with AMR has been largely understudied.
Infection by antibiotic resistant bacteria (ARB) remains a global health crisis6 and the World Health Organization (WHO) has declared AMR one of the top 10 global public health threats.7,8 Some pathogens of particular concern are methicillin resistant Staphylococcus aureus (MRSA), vancomycin resistant enterococci (VRE), antibiotic resistant Gram-negative bacilli (RGNB), and Clostridium difficile (C. diff.). Infection by these bacteria can lead to increased medical care usage and costs, and can result in severe morbidity and mortality.9 In the United States, infections by ARB cause more than 23,000 deaths annually.6 Exposure to these bacteria is common in health care settings, but community acquisition is also on the rise.10–12 As bacterial resistance to antibiotics continues to evolve and spread, treatment options become increasingly sparse.13 Although overuse or misuse of antibiotics and other antimicrobials is a main driver, AMR has also been associated with lack of clean water and sanitation, and increasingly, environmental pollutants including heavy metals.14,15
There is increasing recognition that environmental conditions and presence of pollutants can alter the bacterial composition within environmental ecosystems, as well as within the human microbiome, contributing in part to growing risk of AMR bacteria in the environment and in hosts, including humans.16–18 The interaction between pollutant exposures, gut microbiome composition and immune function, including host susceptibility is an emerging area of environmental health research.19 To date, few studies have examined how pollutant exposure may alter the prevalence of ARB in the human microbiota, and how continued efforts to address pollutant exposure may also lead to improved control of infectious diseases.
Centuries before the development of antibiotics, heavy metals were considered key therapeutics in the fight against infectious diseases20; however, they have also been shown to select for antibiotic resistance.16,21 The genes that code for heavy metal resistance are often physically or transcriptionally linked to antibiotic resistance genes, and in some instances, the same set of genes can provide resistance to both heavy metals and antibiotics.22 Often, these genes reside on mobile genetic units called plasmids that can be vertically transferred during replication or horizontally transferred to other bacteria. Bacterial plasmids require selective pressure to be maintained within a given bacterial population; otherwise they are easy targets for removal during replication when energy and resources are limited.21 When antibiotic and metal resistance genes are encoded on these plasmids and either xenobiotic enters the environment, the selective advantage of maintaining, rather than eliminating the plasmid, is greatly increased and proliferation of these genes is amplified.21 Pb, in particular, increases the prevalence of ARB in soil and water, increasing prevalence of ARB in runoff and other environmental media.23,24
Another mechanism to consider in the association between heavy metal exposures and antibiotic resistance is through altered gut microbial composition. The gut microbiome plays an important role as a first line of defense in xenobiotic metabolism.25 Many bacteria are susceptible to the toxic effects of heavy metals such as Pb, and introduction of metals into a microbial community can alter the metabolic activity and overall abundance of different bacteria within the microbiome.26 Previous studies in both animal models and humans have shown Pb and heavy metal exposure is associated with altered microbial diversity within the gut microbiome,27,28 which can alter the presence of healthy and commensal bacteria that would normally out compete pathogens in the gut.29 Taken together, the potential of Pb to select for antibiotic resistance, reduce competitive inhibition by altering the gut microbiota, and impede immune function, make it highly plausible that human Pb exposure would be associated with colonization by ARB.
Epidemiologic evidence of this association is in its infancy. Our group was among the first to find an association between increased blood Pb level and increased odds of MRSA colonization in NHANES.30 NHANES is a cross-sectional study representative of the general US population that includes biomarkers of environmental chemicals.30 The number of ARB in NHANES was limited to nasal MRSA concentrations. The aim of this study was to identify the association between urinary Pb concentration and colonization by MRSA and other ARB in a nonclinical, population-based sample of adults from urban, and rural communities using multiple biospecimens. As a secondary analysis, we use in vitro methods to test isolated ARB found in human samples for resistance to Pb as a means to increase understanding of these complex associations between heavy metal exposures and ARB in the population.
Methods
Data source
The study sample includes 695 adults age 18 years or older who participated between 2016 and 2017 in the Survey of the Health of Wisconsin (SHOW) and its ancillary Wisconsin Microbiome Study (WMS).19 Details of both studies have been previously described.31,32 In brief, SHOW was initiated in 2008 as an annual cross-sectional health examination survey including randomly selected participants, representative of Wisconsin, a geographically diverse population of both urban and rural residents. SHOW collects interviews, physical exams, and biospecimens. Questionnaires cover a broad range of social determinants of health including health history, behaviors, diet, neighborhood and housing characteristics, current health status, and biologic measurements and samples. The WMS was designed to assess the relationship between dietary fiber intake, gut microbial composition, and ARB colonization. The WMS added more detailed dietary history and 24-hour diet-recall survey questions on factors known to influence the gut microbiota and risk of ARB colonization. Oral, nasal and skin swabs, along with saliva and stool samples were also collected in additional to spot urines. Study protocols were reviewed and approved by the University of Wisconsin Institutional Review Board, and all participants provided informed consent to study participation.
Primary exposure variable
The main exposure variable is creatinine adjusted urinary Pb concentration as an estimate of Pb exposure. Urinary lead represents body burden of exposure and is correlated with blood levels in a subsample of this population.27,33 Creatinine adjustment accounts for heterogeneity in urinary concentrations and kidney function. Urinary lead was measured using inductively coupled plasma mass spectrometry. Any value with a result below the limit of detection (LOD) was replaced with the LOD/√2. As lead levels in this study population are reflective of a general population, we analyzed those in the 95th percentile of Pb exposure versus below to determine associations with high Pb exposure. Although currently, US regulatory agencies consider no exposure to be safe, the 95th percentile levels in this study were selected because they correspond to urinary levels previously associated with immune-toxicity, all cause and cancer mortality in humans.34,35 As a sensitivity analyses, models using continuous log Pb and quartile estimates were also examined.
Primary outcome variable
The primary outcome was ARB colonization as defined by presence of at least one of four different ARB: MRSA, VRE, RGNB, and C. diff. Presence of ARB colonization was pooled across the multiple microbial samples collected as part of the WMS. MRSA, VRE, and RGNB were isolated from stool and saliva samples, and nasal, oral, and skin swabs. C. diff was isolated from a subset of stool samples, as previously described.32 MRSA and RGNB were tested for resistance to cefoxitin, and ciprofloxacin, respectively, using Kirby-Bauer disc diffusion methods and cut points established by the Clinical Laboratory Standards Institute.36,37 Vancomycin resistance in VRE was determined using the E-test (Bio-Merieux, Marcy l’Etoile, France) of minimum inhibitory concentration (MIC). C. diff isolates were tested for toxin genes using PCR. Results from each test were pooled into a single ARB variable, which was dichotomized as either positive, for any participant with a positive or intermediate resistance result across any sample or ARB type, or negative, for any participant with no positive or intermediate results across any sample or ARB type. In a subanalysis, the most Pb resistant ARB was considered individually.
Confirmatory analysis using in vitro assay
Confirmatory in vitro analysis was also conducted on ARB isolates (except C. diff.) from stool samples to determine Pb resistance by MIC analysis using Pb (II) Acetate solution in a microtiter plate. The testing method is based on a protocol reported by Kafilzadeh et al.38 Data are reported as MIC for MRSA, VRE, and RGNB by participant.
Covariates
Several additional variables were used to control for potential confounding factors, as determined through the use of a DAG (directed acyclic graph). These included the self-reported demographic variables of age, gender, race/ethnicity, education, income, smoking status, and length of residence in the current home. Urbanicity was also included as a covariate, defined using geocoded participant addresses and cross referencing them with the 2010 Rural Urban Commuting Area (RUCA) codes.39,40 Urbanicity and length of residence were considered potential confounders because they indicate many components of environmental exposures, and the general length of those exposures. Additional potential confounding variables considered were self-reported antibiotic use in the last year (yes/no), ownership of an indoor pet (yes/no), and dietary nutrients including Iron (mg), Vitamin C (mg), Calcium (mg), and Fiber (g). Dietary values were calculated based on self-reported usual diet over the last year as queried by the National Cancer Institute’s DHQ-II.41 Although antibiotic use is not necessarily associated with Pb exposure, making it a true confounder, it was included as a covariate in our analysis because it has been previously shown to be associated with antibiotic resistance and the human microbiome, and eliminating those who have taken antibiotics in the last year would greatly reduce our sample.
Statistical analysis
Frequency tables were used to evaluate the distribution of demographics and confounding variables by ARB colonization. P values were calculated by χ2 for categorical variables, and a P for trend was calculated for continuous variables to test for significant differences by colonization. Primary analyses included use of adjusted logistic regression models to estimate odds of ARB positive versus negative associated with creatinine adjusted urinary Pb in the 95th percentile versus below. Sensitivity analysis included the same models with continuous Log Pb, and Pb in quartiles (creatinine adjusted). Logistic regression models were adjusted for age, gender, antibiotic use, race/ethnicity, education, urbanicity, length of residence, and dietary fiber and Vitamin C. Income, smoking, body mass index, indoor pet ownership, and dietary iron and calcium were also considered as covariates; however, they were not included in the final models, as they did not empirically confound the relationship. Interaction models were also tested for statistical significance. Subanalysis also examined the most Pb-resistant ARB (RGNB) positive versus negative (not pooled with other ARBs). Confirmatory-analysis of Pb resistance was performed using linear regression of MIC by isolate type and of MIC by urinary Pb in the 95th percentile for the most Pb resistant ARB (RGNB). Statistical analysis was performed in SAS v. 9.4 (Carry, North Carolina, USA).
Results
Study population
Among 695 participants, 239 (34%) tested positive for ARB colonization: 22 (3.2%) tested positive for MRSA, 135 (19.4%) tested positive for RGNB, 96 (13.8%) tested positive for VRE, and of 276 tested positive for C. diff., 21 (7.6%) were positive for toxigenic C. diff., and 2 (0.7%) were positive for nontoxigenic. The distribution of ARB colonization by sample site and ARB type can be found in eTable 1; http://links.lww.com/EE/A157. One participant had a Pb value below the limit of detection. Among participants testing negative for ARB, geometric mean urinary lead was 0.29 µg/L, ranging from 0.01 to 2.47 µg/L. The geometric mean was 0.32 µg/L, ranging from 0.03 to 3.72 µg/L for positive participants. Of the variables examined in univariate analysis (Table 1), only race/ethnicity was significantly associated with ARB colonization (P = 0.014), with Non-Hispanic Black participants having the highest prevalence of ARB colonization. As previously reported, urinary Pb concentration (adjusted for creatinine) was highest in those age 70 and above, women, non-Hispanic Whites, those in the low income group, former smokers, and those who do not own an indoor pet.27
Table 1.
Distribution of demographics and potential covariates by ARB colonization status (±), from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017
| Exposure (continuous) | TotalN | ARB–GM (SE) | ARB+GM (SE) | P for trend | Health (categorical) | TotalN | ARB–n (%) | ARB+n (%) | χ2 P value |
|---|---|---|---|---|---|---|---|---|---|
| Urinary Pb (µg/L) | 695 | 0.27 (1.0) | 0.32 (1.1) | 0.240a | Smoking | 684 | 0.899 | ||
| Demographics (categorical) | N | n (%) | n (%) | χ2 P value | Current | 93 | 61 (65.6) | 32 (34.4) | |
| Age | 695 | 0.219 | Former | 205 | 138 (67.3) | 67 (32.7) | |||
| 18–29 | 58 | 42 (72.4) | 16 (27.6) | Never | 386 | 253 (65.5) | 133 (34.5) | ||
| 30–49 | 172 | 118 (68.6) | 54 (31.4) | Antibiotic Use | 652 | 0.749 | |||
| 50–69 | 338 | 221 (65.4) | 117 (34.6) | Yes | 227 | 154 (67.8) | 73 (32.2) | ||
| ≥70 | 127 | 75 (59.1) | 52 (40.9) | No | 425 | 283 (66.6) | 142 (33.4) | ||
| Gender | 695 | 0.734 | BMI | 689 | 0.518 | ||||
| Female | 398 | 259 (65.1) | 139 (34.9) | Underweight/normal | 165 | 112 (67.9) | 53 (32.1) | ||
| Male | 297 | 197 (66.3) | 100 (33.7) | Overweight/obese | 524 | 341 (65.1) | 183 (34.9) | ||
| Race/ethnicity | 694 | 0.014 | Residence (categorical) | N | n (%) | n (%) | χ2 P value | ||
| Non-Hispanic White | 576 | 386 (67.0) | 190 (33.0) | Indoor pet | 692 | 0.109 | |||
| Non-Hispanic Black | 70 | 34 (48.6) | 36 (51.4) | Yes | 378 | 258 (68.3) | 120 (31.7) | ||
| Hispanic | 24 | 18 (75.0) | 6 (25.0) | No | 314 | 196 (62.4) | 118 (37.6) | ||
| Non-Hispanic Other | 24 | 17 (70.8) | 7 (29.2) | Urbanicity | 694 | 0.847 | |||
| Family income | 695 | 0.866 | Urban | 459 | 299 (65.1) | 160 (34.9) | |||
| Low income | 204 | 133 (65.2) | 71 (34.8) | Suburban | 76 | 52 (68.4) | 24 (31.6) | ||
| Middle income | 220 | 142 (64.5) | 78 (35.5) | Rural | 159 | 105 (66.0) | 54 (34.0) | ||
| High income | 271 | 181 (66.8) | 90 (33.2) | Length of residence (years) | 687 | 0.769 | |||
| Education | 694 | 0.640 | <1 | 62 | 44 (71.0) | 18 (29.0) | |||
| ≤High school | 187 | 122 (65.2) | 65 (34.8) | 1–3 | 105 | 68 (64.8) | 37 (35.2) | ||
| Some college | 246 | 167 (67.9) | 79 (32.1) | 3–10 | 145 | 97 (66.9) | 48 (33.1) | ||
| ≥Bachelor’s degree | 261 | 167 (64.0) | 94 (36.0) | >10 | 375 | 242 (64.5) | 133 (35.5) | ||
| Diet (continuous) | N | Mean ± SE | Mean ± SE | P for trend | |||||
| Dietary iron (mg/1,000 kcal) | 614 | 7.5 ± 0.1 | 7.7 ± 0.2 | 0.351 | |||||
| Dietary calcium (mg/1,000 kcal) | 614 | 753.8 ± 23.9 | 758.1 ± 34.9 | 0.918 | |||||
| Dietary fiber (g/1,000 kcal) | 614 | 11.0 ± 0.2 | 10.8 ± 0.3 | 0.626 | |||||
| Dietary vitamin C (mg/1,000 kcal) | 614 | 58.4 ± 1.9 | 64.2 ± 2.8 | 0.068 | |||||
Categorical distribution statistics calculated using frequency tables adjusted for household clustering, including P values from the χ2 statistic. Continuous covariate statistics calculated including adjustment for household clustering, including P for trend.
aP for trend calculated using Log Pb, adjusted for creatinine.
GM, geometric mean; N, n, number of observations; Pb, lead; SE, standard error.
Urinary Pb and ARB colonization
The primary predictor in our regression analysis was creatinine-adjusted-urine Pb above (n = 34) versus below (n = 661) the 95th percentile, with geometric mean Pb (not creatinine adjusted) of 0.80 µg/L (95% CI = 0.57, 1.10) and 0.28 µg/L (95% CI = 0.26, 0.30), respectively. In our logistic regression analysis, Pb was associated with a significant increase (P = 0.020) in ARB colonization in the unadjusted model (Table 2). In a model adjusted for age, gender, antibiotic use, race, education level, fiber, Vitamin C, urbanicity and length of residence in current home, Pb in the 95% was associated with 2 times increased odds (P = 0.069) of ARB colonization. Dietary fiber intake was associated with a slight but significant decrease in odds of ARB colonization, while increasing age and Vitamin C intake were associated with slight but significantly increased odds of ARB colonization.
Table 2.
Results of logistic regression of ARB colonization, unadjusted and adjusted for covariates, from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017
| Unadjusted | Adjusted | |||
|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |
| 95%ile Pba (yes) | 2.24 | 1.14, 4.41 | 2.05 | 0.95, 4.44 |
| Age | 1.01 | 1.00, 1.03 | ||
| Gender (female vs. male) | 1.04 | 0.71, 1.53 | ||
| Antibiotic use (yes vs. no) | 0.92 | 0.62, 1.37 | ||
| Race/ethnicity (Non-White vs. Non-Hispanic White) | 1.47 | 0.91, 2.39 | ||
| Education | ||||
| ≤High-school diploma | 0.66 | 0.41, 1.08 | ||
| Some college | 0.77 | 0.5, 1.18 | ||
| ≥Bachelor degree | Ref | – | ||
| Dietary fiberb | 0.95 | 0.91, 1.00 | ||
| Dietary vitamin Cb | 1.01 | 1.00, 1.01 | ||
| Urban (vs. rural) | 0.94 | 0.64, 1.39 | ||
| Length of residence (years) | ||||
| 0–1 | 0.73 | 0.36, 1.45 | ||
| 1–3 | 1.06 | 0.61, 1.86 | ||
| 3–10 | 0.87 | 0.53, 1.44 | ||
| >10 | Ref | – | ||
Bold values are considered statistically significant.
aUrinary measurement, creatinine-adjusted.
bkcal/1000 kcal.
Pb, lead; Ref, reference level.
Urban and rural comparisons of urinary Pb and ARB
Urban residents in our study population (n = 459) had a geometric mean urinary Pb level of 0.31 µg/L (95% CI = 0.28, 0.34), although nonurban (suburban and rural) residents had a geometric mean urinary Pb level of 0.28 µg/L (95% CI = 0.24, 0.31). Stratified regression analysis by urbanicity (Table 3) showed that for urban residents, high urinary Pb levels were associated with nearly 3 times increased odds of ARB colonization (P = 0.037) compared with low lead. In the same stratified model of urban residents, non-White participant had 76% increased odds of ARB colonization (P = 0.045).
Table 3.
Results of logistic regression of ARB colonization, stratified by urbanicity, adjusted for covariates, from the microbiome study sample of the Survey of the Health of Wisconsin 2016–2017
| Urban | Suburban/rural | |||
|---|---|---|---|---|
| OR | 95% CI | OR | 95% CI | |
| 95 %ile Pba (yes) | 2.85 | 1.07, 7.59 | 1.07 | 0.28, 4.05 |
| Age | 1.01 | 0.99, 1.03 | 1.01 | 0.99, 1.04 |
| Gender (female vs. male) | 1.30 | 0.82, 2.07 | 0.60 | 0.30, 1.23 |
| Antibiotic use (yes vs. no) | 0.96 | 0.60, 1.54 | 0.82 | 0.38, 1.77 |
| Race/ethnicity (Non-White) | 1.76 | 1.01, 3.05 | 0.46 | 0.10, 2.16 |
| Education | ||||
| ≤High-school diploma | 0.83 | 0.45, 1.53 | 0.49 | 0.20, 1.19 |
| Some college | 0.87 | 0.51, 1.48 | 0.55 | 0.26, 1.14 |
| ≥Bachelor degree | Ref | – | Ref | – |
| Dietary fiberb | 0.98 | 0.93, 1.03 | 0.90 | 0.80, 1.01 |
| Dietary vitamin Cb | 1.01 | 1.00, 1.01 | 1.01 | 1.00, 1.02 |
| Length of residence (years) | ||||
| 0–1 | 0.74 | 0.32, 1.69 | 0.46 | 0.10, 2.00 |
| 1–3 | 1.30 | 0.67, 2.53 | 0.40 | 0.12, 1.33 |
| 3–10 | 0.84 | 0.45, 1.60 | 0.94 | 0.38, 2.29 |
| >10 | Ref | – | Ref | – |
Bold values are considered statistically significant.
aUrinary measurement, creatinine-adjusted.
bkcal/1000kcal.
Pb, lead; Ref, reference level.
Continuous log Pb and ARB colonization
In sensitivity analysis of continuous log Pb, estimates from unadjusted and adjusted analyses showed smaller odds ratios, but consistent direction as the primary analysis (eTable 2; http://links.lww.com/EE/A158 and eTable 3; http://links.lww.com/EE/A159). When stratified by urbanicity, log Pb was not associated with ARB colonization in either subgroup. Analysis comparing quartiles of Pb exposure had similar results (eTable 4; http://links.lww.com/EE/A160 and eTable 5; http://links.lww.com/EE/A161). These results indicate that the association between Pb exposure and ARB colonization is strongest in those most highly exposed within this study population.
In vitro Pb and RGNB colonization
Because RGNB had the highest average Pb MIC (see results of confirmatory analysis), we conducted a subanalysis of only RGNB colonization by 95th percentile Pb level. Results were of similar effect size and direction to the main analysis of any ARB colonization but were not significant (eTable 6; http://links.lww.com/EE/A162).
Confirmatory analysis: Pb resistance
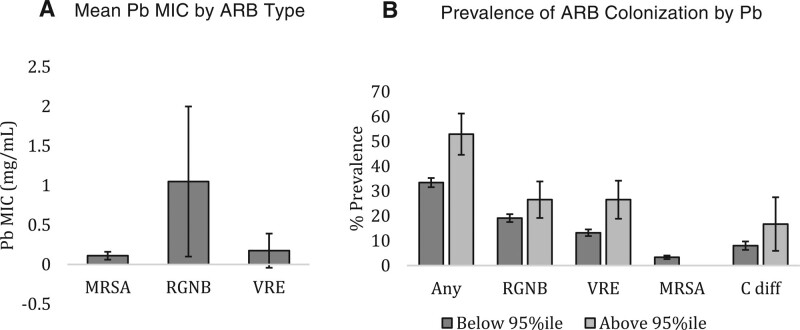
Pb MIC was significantly higher in isolates of RGNB from stool samples than MRSA and VRE (P < 0.001) (Figure 1A). Pb MIC was not tested in C. diff. isolates. Pb MIC level within the RGNB isolates was not associated with Pb exposure in the 95th percentile (data not shown). Of participants in the 95th percentile of Pb, over 50% are colonized by at least one ARB, with VRE and RGNB having the highest prevalence (26.5% percent each) (Figure 1B).
Figure 1.
A, Mean Pb MIC by ARB isolate type. B, Prevalence of ARB colonization overall and by type, in groups of creatinine adjusted urinary Pb level above and below the 95th percentile. C. diff. indicates Clostridium difficile; MIC, maximum inhibitory concentration; MRSA, methicillin-resistant Staphylococcus aureus; Pb, lead; RGNB, resistant Gram-negative bacilli; VRE, vancomycin-resistant enterococci.
Discussion
This cross-sectional investigation adds to a growing, but very limited epidemiologic literature regarding environmental exposures and their impacts on antibiotic resistance within the human microbiome. Specifically, we found an association between high levels of Pb exposure and the presence of ARB in a general population based sample. We found individuals with the highest Pb exposure (geometric mean 0.80 µg/L for the 95th percentile) had significantly higher ARB colonization, and that the effects were strongest among urban residents. However, among this study population with average urinary Pb levels, ARB colonization was not statistically significantly associated with increasing log Pb when modeled as a continuous variable.
In vitro testing of Pb resistance in ARB positive isolates from stool showed that, on average, RGNB positive isolates have higher tolerance to the presence of Pb, but increasing Pb levels in humans was not associated with increased Pb tolerance in their colonizing RGNB. Findings are consistent with previous animal and experimental evidence that suggests a complex relationship between heavy metal exposure and the human microbiome, and in particular, antibiotic resistance.
Heavy metal exposures including Pb continue to be a significant public health threat globally, and a paucity of research examines the immunological impacts of lead exposure particularly as it relates to the global crisis of antibiotic microbial resistance. A recent analysis found the population attributable fraction of Pb with mortality in the US population was 18%, equivalent to approximately 412,000 annual deaths.42 Impacts on mortality were found in blood lead levels ranging from the 10th to the 90th percentile of lead exposures among NHANES,42 including levels of exposure found in this study population. Results from this investigation suggest reducing heavy metal exposures may play an important role in tackling AMR.
Study findings also reveal built environment, namely urbanicity, can shape AMR susceptibility. Geometric mean urinary Pb was higher in urban vs. nonurban residents, and stratified regression analysis indicating a stronger association between Pb and ARB for urban residents. These findings are consistent with environmental studies of ARB in urban settings. Urban aquatic environments in particular contain many selective pressures and potential reservoirs for the proliferation of antibiotic resistance genes.43–45 Urban wastewater contains higher amounts of hospital effluents and contamination from community antibiotic use.43 The wastewater treatment cycle and drinking water distribution infrastructure also provide opportunities for antibiotic resistance selection and proliferation through biofilms, and mixed contamination by ARB of human and animal origin, metals, parabens, and pharmaceuticals including antibiotics.43,44 It has been hypothesized that bacteria can move through different sectors of the urban water cycle and convey antibiotic resistance genes to the human microbiome.46 RGNB have been shown to be particularly problematic in moving antibiotic resistance from hospital wastewater into the urban wastewater treatment system47 and were shown to be the most Pb resistant in this study. When our epidemiologic findings are considered in the context of these environmental studies, they suggest continued investigation of ARB colonization, infection, and transmission pathways in urban environments.
Among the various ARB, molecular mechanisms driving RGNB may offer clues to the complex relationship between heavy metal exposure and antibiotic resistance. Several RGNB have been identified as urgent threats to public health including Escherichia coli, Salmonella spp., Campylobacter spp., Helicobacter pylori, Pseudomonas aeruginosa, and Vibrio cholera.6 RGNB often have intrinsic antibiotic resistance due to their outer membrane, which serves as a permeability barrier that is not present in Gram-positive bacteria. However, RGNB resistance genes can also be carried on plasmids with large concentrated multiresistance regions that confer resistance to multiple classes of antibiotics.48 Typical mechanisms for bacterial resistance to heavy metals that may be plasmid-derived include binding of metals to intra- or extracellular compounds, or precipitating into salt limiting the ability of metals to interfere with cellular mechanisms. Other approaches include cellular transformation of metals into nonvolatile forms, or effluxing from the cell.49,50 It is not clear from our analysis whether the RGNB resistance to Pb and ciprofloxacin in this population is plasmid-derived or intrinsic. However, environmental heavy metal contamination may play a role in selection for plasmid-derived antibiotic resistance within the natural reservoirs for RGNB including water, soil, sewage, and the gut of animals, including humans.51
The finding that increased Pb exposure in subjects is not associated with increased Pb tolerance in colonizing RGNB can be explained in several ways. If the Pb resistance measured in the isolated RGNB is intrinsic, Pb exposure would not add any selective pressure, thus we would not expect to see an association. For the other ARB included in this analysis, the relationship between Pb exposure and ARB colonization in the most highly exposed of this population may not be due to in vivo selection for antibiotic resistance, but are likely due to the immune effects of Pb, and the shifts in gut microbial composition that reduce competitive inhibition. Moreover, the geometric mean urinary Pb in this population was lower than the national average, thus Pb exposure in this population may not have been high enough for antibiotic resistance selection, but would likely be high enough to affect immune function.5,27,52 The use of a pooled ARB indicator variable across multiple body sites also captures more systemic effects of circulating Pb than selection for resistance via direct exposure. It may also be that increased exposure to Pb is associated with increased abundance of Pb resistant (and likely antibiotic resistant) bacteria within individuals, which was not assessed in this study. Such were the findings of Nisanian et al., who fed increasing levels of Pb to leghorn chickens, and saw a dose response effect between the amount of Pb consumed, and the number of ARB present in each individual.53 If this were the case in our study population, the null relationship between Pb exposure and Pb MIC would not be inconsistent.
Despite offering some novel insights regarding human Pb levels and ARB prevalence, this study is not without limitations and offers several opportunities for future inquiry. The cross-sectional nature of this investigation limits conclusions regarding causal relationships, and potential for reverse causality exists. The use of urine Pb level as a marker of chronic Pb exposure, although highly correlated with blood Pb, is perhaps not the best measure of Pb exposure to the human microbiome. Only 10% of ingested Pb is absorbed into the blood stream, with even less filtered into urine.33 Thus, the level of Pb to which gut microbes are exposed is likely much higher than what is excreted in urine. Moreover, some of the circulating Pb stored in bones and blood cells can end up in urine without interacting with bacteria in the human microbiome.33 Because the gut microbiota likely plays a role in metabolizing the Pb that enters the body,54 any association found between urine Pb level and presence of ARB may be due to reverse causality. For instance, if Pb resistant (and antibiotic resistant) bacteria use a naturally thick cell wall for resistance, or efflux Pb out of their cells,50 it is more likely to be absorbed into the blood stream and ultimately leave the body in urine. This would result in higher Pb measurement in the urine because of the presence of Pb resistant bacteria.
The use of ARB colonization as an endpoint instead of infection is also somewhat problematic. Not all bacteria that are resistant to antibiotics are also pathogenic, meaning that not every colonizing organism will lead to an infection. Antibiotic resistant isolates identified in this study are not tested for the presence of pathogenicity genes. Colonization by an ARB is, however, a strong risk factor for subsequent infection.55 Although ARB colonization is an imperfect surrogate measure of infection, it is useful in this population-based sample where prevalence of infection by ARB is likely to be low.
In conclusion, despite limitations, this study is among the first observational studies to assess the associations between heavy metals and ARB colonization in real-world settings and thus lays an important foundation for future validation and translational research. Additional laboratory studies are needed to fully establish a causal association of Pb and ARB and to tease out the complex temporal relationships and potential for reverse causality. Further, this study provides a good model to be replicated in other populations with potentially higher Pb exposure and in areas with endemic ARB presence. Additional work in populations across the spectrum of heavy metal exposures, and with mixtures are also needed. Examining combined exposure levels to multiple heavy metals would also add to our understanding of the link between heavy metals and ARB colonization in humans.
Acknowledgments
The authors would like to thank the study participants, and staff at the Survey of the Health of Wisconsin, the University of Wisconsin Survey Center, Dr. Garret Suen in the Department of Bacteriology at the University of Wisconsin, Dr. Safdar’s Infectious Disease Research Laboratory, especially Megan Duster for her work on the Pb resistance testing protocol, and Noel Stanton at the Wisconsin State Laboratory of Hygiene for his support of urinary lead analyses.
Supplementary Material
Footnotes
Published online 3 November 2021
Funding for this work came from the University of Wisconsin School of Medicine and Public Health’s (UWSMPH) Wisconsin Partnership Program and UWSMPH’s Department of Medicine Pilot Award Program. Funding for SE’s time comes from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (T32 HD049311). A.K. was supported by a National Library of Medicine training grant to the Computation and Informatics in Biology and Medicine Training Program, USA (T15 LM007359). Authors also acknowledge support from the National Institutes of Health core grant to the Center for Demography and Ecology at the University of Wisconsin-Madison (P2C HD047873). K.M.C.M. is also a member of the Center for Demography and Aging at the University of Wisconsin Madison (P30 AG017266) and supported by related NIH grant (R21 Al142481).
The data and code used in this analysis are available upon reasonable request to the corresponding author (kmalecki@wisc.edu).
Supplemental digital content is available through direct URL citations in the HTML and PDF versions of this article (www.environepidem.com).
The authors declare that they have no conflicts of interest with regard to the content of this report.
References
- 1.Substance Priority List. ATSDR. 2020. Available at: https://www.atsdr.cdc.gov/spl/index.html. Accessed August 19, 2021.
- 2.Miao H, Liu Y, Tsai TC, Schwartz J, Ji JS. Association between blood lead level and uncontrolled hypertension in the US population (NHANES 1999-2016). J Am Heart Assoc. 2020;9:e015533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettinger AS, Egan KB, Homa DM, Brown MJ. Blood lead levels in U.S. Women of Childbearing Age, 1976-2016. Environ Health Perspect. 2020;128:17012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GBD Compare. IHME Viz Hub. Available at: http://vizhub.healthdata.org/gbd-compare. Accessed November 17, 2020.
- 5.Dietert RR, Piepenbrink MS. Lead and immune function. Crit Rev Toxicol. 2006;36:359–385. [DOI] [PubMed] [Google Scholar]
- 6.Center for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2013. U.S. Department of Health and Human Services; 2013. [Google Scholar]
- 7.Jee Y, Carlson J, Rafai E, et al. Antimicrobial resistance: a threat to global health. Lancet Infect Dis. 2018;18:939–940. [DOI] [PubMed] [Google Scholar]
- 8.WHO. Global action plan on AMR. WHO. Available at: http://www.who.int/antimicrobial-resistance/global-action-plan/en/. Accessed November 17, 2020. [Google Scholar]
- 9.Nelson RE, Hatfield KM, Wolford H, et al. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the United States. Clin Infect Dis. 2021;72(suppl 1):S17–S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H, Flynn NM, King JH, Monchaud C, Morita M, Cohen SH. Comparisons of community-associated methicillin-resistant Staphylococcus aureus (MRSA) and hospital-associated MSRA infections in Sacramento, California. J Clin Microbiol. 2006;44:2423–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vysakh PR, Jeya M. A comparative analysis of community acquired and hospital acquired methicillin resistant staphylococcus aureus. J Clin Diagn Res JCDR. 2013;7:1339–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosas Auguet O, Betley JR, Stabler RA, et al. Evidence for community transmission of community-associated but not Health-Care-Associated Methicillin-Resistant Staphylococcus Aureus Strains Linked to Social and Material Deprivation: Spatial Analysis of Cross-sectional Data. PLoS Med. 2016;13:e1001944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol. 2014;18:56–60. [DOI] [PubMed] [Google Scholar]
- 14.Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. [DOI] [PubMed] [Google Scholar]
- 15.Alonso A, Sánchez P, Martínez JL. Environmental selection of antibiotic resistance genes. Environ Microbiol. 2001;3:1–9. [DOI] [PubMed] [Google Scholar]
- 16.Seiler C, Berendonk TU. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front Microbiol. 2012;3:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu K, Mahbub R, Fox JG. Xenobiotics: interaction with the intestinal microflora. ILAR J. 2015;56:218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Sun Y, Huang J, Wang H, Tang D. Effects of soil heavy metal pollution on microbial activities and community diversity in different land use types in mining areas. Environ Sci Pollut Res Int. 2020;27:20215–20226. [DOI] [PubMed] [Google Scholar]
- 19.Strategic Plan 2018-2023. National Institute of Environmental Health Sciences. Available at: https://www.niehs.nih.gov/about/strategicplan/index.cfm. Accessed January 7, 2020.
- 20.Gould K. Antibiotics: from prehistory to the present day. J Antimicrob Chemother. 2016;71:572–575. [DOI] [PubMed] [Google Scholar]
- 21.Gullberg E, Albrecht LM, Karlsson C, Sandegren L, Andersson DI. Selection of a multidrug resistance plasmid by sublethal levels of antibiotics and heavy metals. mBio. 2014;5:e01918–e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemme CL, Green SJ, Rishishwar L, et al. Lateral gene transfer in a heavy metal-contaminated-groundwater microbial community. mBio. 2016;7:e02234–e02215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauthier PT, Norwood WP, Prepas EE, Pyle GG. Metal-PAH mixtures in the aquatic environment: a review of co-toxic mechanisms leading to more-than-additive outcomes. Aquat Toxicol. 2014;154:253–269. [DOI] [PubMed] [Google Scholar]
- 24.Imran M, Das KR, Naik MM. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere. 2019;215:846–857. [DOI] [PubMed] [Google Scholar]
- 25.Claus SP, Guillou H, Ellero-Simatos S. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes. 2016;2:16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elizabeth George S, Wan Y. Advances in characterizing microbial community change and resistance upon exposure to lead contamination: implications for ecological risk assessment. Crit Rev Environ Sci Technol. 2019;50:2223–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eggers S, Safdar N, Sethi AK, et al. Urinary lead concentration and composition of the adult gut microbiota in a cross-sectional population-based sample. Environ Int. 2019;133(Pt A):105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao B, Chi L, Mahbub R, et al. Multi-Omics reveals that lead exposure disturbs gut microbiome development, key metabolites, and metabolic pathways. Chem Res Toxicol. 2017;30:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. [DOI] [PubMed] [Google Scholar]
- 30.Eggers S, Safdar N, Malecki KM. Heavy metal exposure and nasal Staphylococcus aureus colonization: analysis of the National Health and Nutrition Examination Survey (NHANES). Environ Health. 2018;17:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieto FJ, Peppard PE, Engelman CD, et al. The Survey of the Health of Wisconsin (SHOW), a novel infrastructure for population health research: rationale and methods. BMC Public Health. 2010;10:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggers S, Malecki KM, Peppard P, et al. Wisconsin microbiome study, a cross-sectional investigation of dietary fibre, microbiome composition and antibiotic-resistant organisms: rationale and methods. BMJ Open. 2018;8:e019450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai T. Biomarkers of lead exposure. Ind Health. 2000;38:127–142. [DOI] [PubMed] [Google Scholar]
- 34.Fenga C, Gangemi S, Di Salvatore V, Falzone L, Libra M. Immunological effects of occupational exposure to lead (Review). Mol Med Rep. 2017;15:3355–3360. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Wang J, Zhang B, et al. Urinary lead concentration is an independent predictor of cancer mortality in the U.S. General Population. Front Oncol. 2018;8:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 10th ed. Clinical and Laboratory Standards Institute; 2006. [Google Scholar]
- 37.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; 26th Informational Supplement. Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 38.Kafilzadeh F, Afrough R, Johari H, Tahery Y. Range determination for resistance/tolerance and growth kinetic of indigenous bacteria isolated from lead contaminated soils near gas stations (Iran). Eur J Exp Biol. 2012;2:62–69. [Google Scholar]
- 39.U.S. Department of Agriculture Economic Research Service, John Cromartie. USDA ERS - Rural-Urban Commuting Area Codes. 2021. Available at: https://www.ers.usda.gov/data-products/rural-urban-commuting-area-codes/. Accessed September 17, 2021.
- 40.Morrill R, Cromartie J, Hart G. UW RHRC Rural Urban Commuting Area Codes - RUCA. Available at: http://depts.washington.edu/uwruca/. Accessed September 17, 2021.
- 41.Background on Diet History Questionnaire II (DHQ II). EGRP/DCCPS/NCI/NIH. 2019. Available at: https://epi.grants.cancer.gov/dhq2/about/. Accessed August 12, 2019.
- 42.Lanphear BP, Rauch S, Auinger P, Allen RW, Hornung RW. Low-level lead exposure and mortality in US adults: a population-based cohort study. Lancet Public Health. 2018;3:e177–e184. [DOI] [PubMed] [Google Scholar]
- 43.Manaia CM, Macedo G, Fatta-Kassinos D, Nunes OC. Antibiotic resistance in urban aquatic environments: can it be controlled? Appl Microbiol Biotechnol. 2016;100:1543–1557. [DOI] [PubMed] [Google Scholar]
- 44.Auguet O, Pijuan M, Borrego CM, et al. Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ. 2017;605-606:1047–1054. [DOI] [PubMed] [Google Scholar]
- 45.Almakki A, Jumas-Bilak E, Marchandin H, Licznar-Fajardo P. Antibiotic resistance in urban runoff. Sci Total Environ. 2019;667:64–76. [DOI] [PubMed] [Google Scholar]
- 46.Vaz-Moreira I, Nunes OC, Manaia CM. Bacterial diversity and antibiotic resistance in water habitats: searching the links with the human microbiome. FEMS Microbiol Rev. 2014;38:761–778. [DOI] [PubMed] [Google Scholar]
- 47.Zagui GS, de Andrade LN, Moreira NC, et al. Gram-negative bacteria carrying β-lactamase encoding genes in hospital and urban wastewater in Brazil. Environ Monit Assess. 2020;192:376. [DOI] [PubMed] [Google Scholar]
- 48.Partridge SR. Analysis of antibiotic resistance regions in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:820–855. [DOI] [PubMed] [Google Scholar]
- 49.Gadd GM, Griffiths AJ. Microorganisms and heavy metal toxicity. Microb Ecol. 1977;4:303–317. [DOI] [PubMed] [Google Scholar]
- 50.Jarosławiecka A, Piotrowska-Seget Z. Lead resistance in micro-organisms. Microbiology (Reading). 2014;160(Pt 1):12–25. [DOI] [PubMed] [Google Scholar]
- 51.Exner M, Bhattacharya S, Christiansen B, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:Doc05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buser MC, Ingber SZ, Raines N, Fowler DA, Scinicariello F. Urinary and blood cadmium and lead and kidney function: NHANES 2007-2012. Int J Hyg Environ Health. 2016;219:261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nisanian M, Holladay SD, Karpuzoglu E, et al. Exposure of juvenile Leghorn chickens to lead acetate enhances antibiotic resistance in enteric bacterial flora. Poult Sci. 2014;93:891–897. [DOI] [PubMed] [Google Scholar]
- 54.Breton J, Massart S, Vandamme P, De Brandt E, Pot B, Foligné B. Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacol Toxicol. 2013;14:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Safdar N, Bradley EA. The risk of infection after nasal colonization with Staphylococcus aureus. Am J Med. 2008;121:310–315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.