Abstract
In vitro studies have implicated nuclear respiratory factor 1 (NRF-1) in the transcriptional expression of nuclear genes required for mitochondrial respiratory function, as well as for other fundamental cellular activities. We investigated here the in vivo function of NRF-1 in mammals by disrupting the gene in mice. A portion of the NRF-1 gene that encodes the nuclear localization signal and the DNA-binding and dimerization domains was replaced through homologous recombination by a β-galactosidase–neomycin cassette. In the mutant allele, β-galactosidase expression is under the control of the NRF-1 promoter. Embryos homozygous for NRF-1 disruption die between embryonic days 3.5 and 6.5. β-Galactosidase staining was observed in growing oocytes and in 2.5- and 3.5-day-old embryos, demonstrating that the NRF-1 gene is expressed during oogenesis and during early stages of embryogenesis. Moreover, the embryonic expression of NRF-1 did not result from maternal carryover. While most isolated wild-type and NRF-1+/− blastocysts can develop further in vitro, the NRF-1−/− blastocysts lack this ability despite their normal morphology. Interestingly, a fraction of the blastocysts from heterozygous matings had reduced staining intensity with rhodamine 123 and NRF-1−/− blastocysts had markedly reduced levels of mitochondrial DNA (mtDNA). The depletion of mtDNA did not coincide with nuclear DNA fragmentation, indicating that mtDNA loss was not associated with increased apoptosis. These results are consistent with a specific requirement for NRF-1 in the maintenance of mtDNA and respiratory chain function during early embryogenesis.
The electron transport and oxidative phosphorylation system in mammalian mitochondria requires contributions from both the nuclear and the mitochondrial genetic systems. The mitochondrial DNA encodes 13 respiratory subunits, as well as the 22 tRNAs and 2 rRNAs required for their mitochondrial translation. However, most respiratory proteins and all of the gene products required for mitochondrial DNA (mtDNA) replication and transcription are nucleus encoded (reviewed in references 39 and 42). Nuclear respiratory factor 1 (NRF-1) was identified as a nuclear transcription factor that transactivates the promoters of a number of mitochondrion-related genes in vitro (7, 10, 11, 48). These include respiratory subunits, the rate-limiting heme biosynthetic enzyme, and factors involved in the replication and transcription of mtDNA (reviewed in reference 39). Among the most intriguing is TFAM, a nucleus-encoded transcription factor that acts on bidirectional promoters within the mitochondrial D-loop regulatory region (12). TFAM was recently shown to be essential for mitochondrial biogenesis during embryonic development (29) and for normal function of the heart (50). Moreover, NRF-1 is involved in the transcriptional control of mitochondrial biogenesis during adaptive thermogenesis through its interaction with the cold-inducible coactivator PGC-1 (53).
In addition to its proposed role in respiratory chain expression, NRF-1 has also been implicated in other cellular functions. Most recently, genes encoding two rate-limiting enzymes in purine nucleotide biosynthesis (8), a receptor involved in chemokine signal transduction (52), a subunit of a neural receptor (33), and the human poliovirus receptor CD155 (44) were all shown to have functional NRF-1 binding sites in their promoters. These observations are consistent with a broader role for NRF-1 in the integration of diverse cellular functions.
Although many transcription factors are members of gene families, NRF-1 is a single-copy gene in vertebrates, and no family members have yet been detected. However, two invertebrate factors, P3A2 and EWG (erect wing gene product), have high sequence similarity to the NRF-1 protein that is restricted to the DNA-binding domain (9, 48). Although P3A2 acts as a negative regulator of a cytoskeletal actin gene (54), target genes for EWG have yet to be identified (9). In contrast, the NRF-1 homologue in zebrafish, not really finished (nrf), has 91% identity to the human protein and was recently disrupted in vivo by insertional mutagenesis (3). Each of these genes has been implicated in embryonic or larval development and both erect wing and nrf have been associated with the central nervous system.
We describe here the targeted disruption of the NRF-1 gene in mice. The results establish that NRF-1 is essential for early embryogenesis in mammals, and its loss of function results in a peri-implantation lethal phenotype. In addition, NRF-1−/− blastocysts show a dramatic decrease in the amount of mtDNA. The results are in keeping with the proposed role for NRF-1 in the maintenance of the respiratory apparatus.
MATERIALS AND METHODS
Construction of the targeting vector.
The 5′ homologous region was derived from two genomic fragments. A 5-kb EcoRI-Acc65I genomic fragment contains an intron between exons 1 and 2. An adjacent 1-kb fragment contains the first 33 nucleotides (nt) of CE2 terminated at its 3′ end with a site-directed Acc65I site. Ligation of these fragments generated the 6-kb 5′ homology region of the targeting vector. The 3′ homologous region encompasses a 3.2-kb XhoI-XbaI genomic fragment beginning with the 3′-terminal 108 nt of CE4 and extending into the intron between CE4 and CE5. The β-galactosidase–neomycin cassette separates truncated coding exons 2 and 4, and the herpes simplex virus thymidine kinase selectable marker lies outside of the homologous regions as depicted in Fig. 1A.
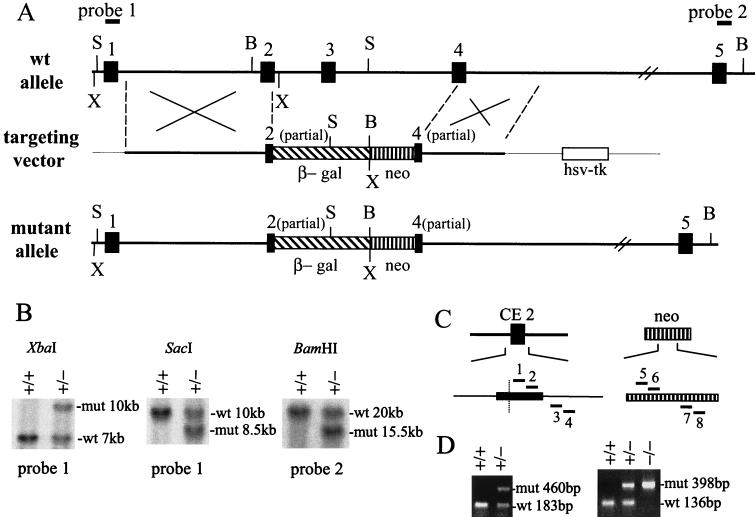
FIG. 1.
Targeted disruption of the mouse NRF-1 gene. (A) Schematic representation of the wild-type and the mutant mouse NRF-1 alleles covering CE1 to CE5 (filled boxes). The bold solid lines represent introns, and the thin solid lines indicate the plasmid backbone in the targeting vector. A promoterless β-galactosidase gene cassette (β-gal, hatched box) was inserted downstream of the first 33 bp of CE2 in the targeting vector. The neomycin cassette (neo, vertical hatched box) and the herpes simplex virus thymidine kinase cassette (hsv-tk, open box) each contain a mouse phosphoglycerate kinase promoter, and their transcriptional orientations are from right to left. The 5′ and 3′ homologous regions are 6 and 3.2 kb, respectively. Upon homologous recombination, the 5.3-kb β-gal-neo sequence replaces approximately 7 kb of endogenous NRF-1 gene sequence. Restriction enzyme cleavage sites shown above and below: B, BamHI; S, SacI; X, XbaI. The positions of probes used for genotyping by Southern blot analysis are indicated above the wild-type allele. Exons and the intron between CE4 and CE5 are not drawn to scale. (B) Southern blot analysis used to screen ES clones and genotype the progeny from heterozygous matings. The restriction enzymes and probes are shown above and below, respectively. The sizes of the hybridizing fragments in the wild-type (wt) and mutant (mut) alleles are shown on the right. (C) PCR primers for genotyping. The positions of the primers are indicated by solid bars. Primers 1 to 4 were designed to detect the wild-type allele, and primers 5 to 8 were designed to detect the mutant allele. In CE2, the sequence to the right of the dotted line is deleted in the mutant allele. Primers 1, 2, 5, and 6 are in the sense orientation, and primers 3, 4, 7, and 8 are in the antisense orientation. (D) PCR genotyping of the progeny from heterozygous matings. As described in Materials and Methods, newborn mice and 6.5- to 8.5-day-old embryos were genotyped with primers 2, 4, 5, and 7 shown in panel C, while preimplantation embryos were genotyped with primers 1 to 8 by a nested-PCR method. The sizes of the PCR products are indicated on the right of each panel.
Details of the vector construction are as follows. mouse P1 clone 11713 isolated from a 129/OLA ES cell library (23) was probed with DNA fragments containing different human NRF-1 coding exons (CE) in Southern blots. pSK-mNRF1Sa10.5 was generated by cloning a 10.5-kb SacI fragment hybridizing to CE1, -2, and -3 into pBluescript SK(+) (Stratagene). pSK-mNRF1Xh10.5 was generated by cloning a 10.5-kb XhoI fragment hybridizing to CE4 into pBluescript SK(+). From pSK-mNRF1Sa10.5, a 5-kb EcoRI-Acc65I intron fragment between CE1 and CE2 was cloned into pBluescript SK(+), resulting in pSK-mNRF1EA5. A 2-kb Acc65I-EcoRI fragment containing CE2 from pSK-mNRF1Sa10.5 was cloned into M13mp18, and an Acc65I site was created 31 bp downstream of the 5′ end of CE2 by oligonucleotide-mediated site-directed mutagenesis (27), using primer mN1cDNA366MS (5′-CTGCTGTGGTACCAGGGAAGAAAC-3′). The 1-kb Acc65I fragment containing 33 bp of the CE2 sequence was digested from the resulting construct and cloned into pBluescript SK(+) to generate pSK-mNRF1Ac1M. pSK-mNRF1Xh10.5 was digested with XbaI, and the 6.2-kb fragment hybridizing to CE4 was self-ligated to generate pSK-mNRF1XX3.2. This construct contained 3 kb of pBluscript SK(+) vector sequence and a 3.2-kb insert between the XhoI and XbaI sites of the vector. The 5-kb EcoRI-Acc65I fragment from pSK-mNRF1EA5 and the 3.5-kb Acc65I-XbaI fragment from pSV-β-Galactosidase Control Vector (Promega) were cloned in a three-way ligation into EcoRI-XbaI-digested pPNT (46), generating pPNT-5′intron-LacZ. Then pPNT-5′homol-LacZ was generated by cloning the 1-kb Acc65I fragment from pSK-mNRF1Ac1M into the Acc65I site of pPNT-5′intron-LacZ. pPNT-5′homol-LacZ was sequenced with primer mN1INTRON3S (5′-CAGTGTGTTGCTGTGTCTCTCC-3′) to ensure that the remaining CE2 sequence and the β-galactosidase sequence were in frame. The 3.2-kb insert in pSK-mNRF1XX3.2 was cut out with XhoI and NotI and cloned into pPNT-5′homol-LacZ between its XhoI and NotI sites. This resulted in the 20-kb targeting vector, named pmNRF1KO.
Generation of targeted ES cells and mutant mice.
pmNRF1KO was linearized with NotI prior to electroporation. Embryonic stem (ES) cell culture, electroporation, blastocyst injection, and generation of germ line-transmitting founder mice were carried out by the Targeted Mutagenesis Facility in the Children's Memorial Institute for Education and Research at Northwestern University.
Screening of ES clones and genotyping of newborn mice and 6.5- to 8.5-day-old embryos.
The following oligonucleotides were used in screening and genotyping, as depicted in Fig. 1C: mN1cDNA385S (primer 1), 5′-GAAACGGAAACGGCCTCATGTG-3′; mN1cDNA419S (primer 2), 5′-CCATCTATCCGAAAGAGACAGCAGAC-3′; mN1INTRON4AS (primer 3), 5′-CCTCAAGACACTGGCATGG AG-3′; mN1INTRON4AS2 (primer 4), 5′-AGGTTTAGACTTGGAATCACTCCCGT-3′; pPNT778S-neo (primer 5), 5′-TGAATGAACTGCAGGACGAGG-3′; pPNT841S-neo (primer 6), 5′-CAGCTGTGCTCGACGTTGTCA-3′; pPNT1237AS-neo (primer 7), 5′-CCACAGTCGATGAATCCAGAA-3′; and pPNT1279AS-neo (primer 8), 5′-GCCAACGCTATGTCCTGATAG-3′.
Southern blot analysis was performed to identify ES clones that had undergone homologous recombination. SacI-digested and XbaI-digested ES cell DNA was probed with probe 1, a 200-bp DraIII-PstI genomic fragment containing mouse NRF-1 coding exon 1 sequence (Fig. 1A). BamHI-digested ES cell DNA was probed with probe 2, a 570-bp SacI-HindIII mouse genomic fragment hybridizing to human NRF-1 cDNA nt 761 to 1020 (48). The sizes of the hybridizing fragments from the wild-type and mutant alleles are indicated in Fig. 1B. Tails from newborn mice and 6.5- to 8.5-day-old embryos were genotyped by PCR with PTC-100 programmable thermal controller (M. J. Research, Inc.). A 183-bp NRF-1 sequence absent in the mutant allele was amplified to identify the wild-type allele, and a 460-bp neomycin fragment was amplified from the mutant allele as shown in Fig. 1D. Tails from about 100 newborn mice from three generations were also genotyped by Southern blot as described above to verify the accuracy of the PCR method. PCR was performed using AmpliTaq polymerase (Perkin-Elmer) with 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates (dNTPs) 0.3 μM concentrations of primers pPNT778S-neo and pPNT1237AS-neo, and 0.1 μM concentrations of primers mN1cDNA419S and mN1INTRON4AS2 in a total volume of 25 μl. The cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 30 cycles.
Genotyping of preimplantation embryos.
We isolated 2.5- and 3.5-day-old embryos by flushing mouse uteri, and each was collected in 10 to 15 μl of phosphate-buffered saline (pH 7.2). DNA was isolated by incubation at 95°C for 10 min, with 20-μg proteinase K treatment at 55°C for 3 h, followed by inactivation of the proteinase at 95°C for 10 min. Half of the DNA from each embryo was used for genotyping by a nested PCR method using Platinum Taq DNA polymerase (GIBCO-BRL). The first PCR was carried out with 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 μM concentrations of primers pPNT778S-neo and pPNT1279AS-neo, and 0.1 μM concentrations of primers mN1cDNA385S and mN1INTRON4AS2 in a total volume of 25 μl. The cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 25 cycles. The second PCR was carried out with 1 μl of product mixture from the first PCR, 0.3 μM concentrations of primers pPNT841S-neo and pPNT1237AS-neo, and 0.15 μM concentrations of primers mN1cDNA419S and mN1INTRON4AS. The cycling conditions were the same as for the first PCR except that the cycle number was 20. Products of the second PCR included a 136-bp fragment from the wild-type allele and a 398-bp fragment from the mutant allele as depicted in Fig. 1D.
RNase protection assays.
To generate the mouse NRF-1 riboprobe, primers mN1cDNA671S (5′-CTGCCGCCTCTCACCATCGAT-3′) and mN1cDNA1016AS (5′-GATGAGCTATACTGTGTGTGGTG-3′) were used to PCR-amplify a 346-bp NRF-1 cDNA fragment located 3′ of the deleted region. The PCR product was cloned into pGEM-T (Promega), and a 580-bp antisense riboprobe was synthesized with Sp6 RNA polymerase after the plasmid DNA was linearized with PvuII. To generate the NRF-1–β-galactosidase riboprobe, a 192-bp fragment was amplified with genomic DNA isolated from NRF-1 heterozygous mice, using primers mN1INTRON3S and pSVbgal710AS (5′-CGGGATCGATCTCGCCATACA-3′). The product, containing 69 bp of NRF-1 intron sequence, the first 33 bp of NRF-1 CE2 sequence and 90 bp of sequence from pSV-β-Galactosidase Control Vector, was cloned into pGEM-T. The plasmid DNA was linerized with MluI, and a 292-bp antisense riboprobe for the NRF-1–β-galactosidase fusion gene in the mutant allele was generated with T7 RNA polymerase. Hybridization was performed as described previously (23), with 10 μg of total RNA from mouse tissues or yeast tRNA as a negative control.
β-Galactosidase staining of embryos.
We stained 2.5- and 3.5-day-old embryos for β-galactosidase activity as described previously (47). Briefly, freshly isolated embryos were fixed for 10 min in 1% (vol/vol) paraformaldeyde, 0.2% (vol/vol) glutaraldehyde, and 1% (vol/vol) calf serum in phosphate-buffered saline (pH 7.2). They were then rinsed in phosphate-buffered saline (pH 7.2) and transferred to a mixture containing 0.02% NP-40, 0.01% sodium deoxycholate, 5 mM K4Fe(CN)6 · 3H2O, 5 mM K3Fe(CN)6, 2 mM MgCl2, and 1 mg of 4-chloro-5-bromo-3-indolyl-β-galactoside (X-Gal) per ml in phosphate-buffered saline (pH 7.3). Positively stained embryos were scored after 20 h of incubation at 37°C.
β-Galactosidase staining of the ovary.
Ovaries were isolated freshly from sexually mature mouse females. They were fixed in phosphate-buffered saline (pH 7.2) containing 2% paraformaldehyde, 0.02% glutaraldehyde, and 2 mM MgCl2 for 90 min at 4°C and then equilibrated in 10% sucrose–2 mM MgCl2 in phosphate-buffered saline (pH 7.2) for 2 h at 4°C and in 20% sucrose–2 mM MgCl2 in phosphate-buffered saline (pH 7.2) for 2 h at 4°C. Samples were then embedded in OCT and frozen at −80°C. Cryostat sections (8 μm) were made, and the slides were stained for β-galactosidase activity as described previously (21). Briefly, the slides were first treated with 0.02% NP-40, 0.01% sodium deoxycholate, and 2 mM MgCl2 in phosphate-buffered saline (pH 7.3) for 5 min at 25°C. Samples were then fixed in phosphate-buffered saline (pH 7.2) containing 2% paraformaldehyde, 0.02% glutaraldehyde, and 2 mM MgCl2 for 5 min at 25°C and stained in 0.02% NP-40, 0.01% sodium deoxycholate, 5 mM K4Fe(CN)6 · 3H2O, 5 mM K3Fe(CN)6, 2 mM MgCl2, and 1 mg of X-Gal per ml in phosphate-buffered saline (pH 7.3) for 20 h at 37°C in 5% CO2 in a humidified incubator. The slides were rinsed in phosphate-buffered saline (pH 7.2) and H2O and stained in 0.25% eosin–85% ethanol for 1 min at room temperature for better visualization of the staining.
Embryo culture.
To culture blastocysts in vitro, Dulbecco modified Eagle medium (GIBCO-BRL 11965-092) was supplemented with 15% (vol/vol) fetal bovine serum (HyClone), 100 U of penicillin per ml, 100 μg of streptomycin per ml, 2 mM l-glutamine, 0.1 mM minimal essential medium with nonessential amino acids, 8 μg of adenosine per ml, 8.5 μg of guanosine per ml, 7.3 μg of cytidine per ml, 7.3 μg of uridine per ml, 2.4 μg of thymidine per ml, 0.1 mM β-mercaptoethanol, and 200 U of murine leukemia inhibitory factor (GIBCO BRL) per ml (1). Blastocysts were cultured individually in microdrops under mineral oil at 37°C in 5% CO2 in a humidified incubator. They were examined after 40 and 100 h in culture and were genotyped by the nested-PCR method described above.
Rhodamine 123 staining of blastocysts.
Freshly isolated blastocysts were incubated in 10 μg of rhodamine 123 per ml and 1% calf serum in phosphate-buffered saline (pH 7.2) for 30 min at 37°C (24). They were then washed in phosphate-buffered saline (pH 7.2) and viewed under a fluorescent microscope at 485 nm.
mtDNA copy number analysis in blastocysts.
For mtDNA copy number analysis, half of the DNA from each genotyped blastocyst was used to amplify a 648-bp mouse mt DNA fragment corresponding to nt 267 to 914 of the cytochrome oxidase subunit 1 gene and a 316-bp 5S ribosomal DNA (rDNA) fragment in a PCR with Platinum Taq DNA polymerase. The 5S rDNA fragment corresponded to nt 699 to 1014 in the GenBank sequence database under accession number D17317. The PCR was carried out with 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 μM concentrations of primers m5SrDNA699S (5′-ACCGTCTAGCCGTCCTCCTT-3′) and m5SrDNA1014AS (5′- CCCACTGAGGATGGATACATG-3′), and 0.15 μM concentrations of primers mCO1-267S (5′- CCCAGATATAGCATTCCCACGA-3′) and mCO1-914AS (5′-AGCAAGCTCGTGTGTCTACATC- 3′). The cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 25 cycles. To generate standards for comparison, PCRs were also performed under the same conditions, in each experiment, with a series of twofold dilutions of heart DNA isolated from an adult wild-type mouse as templates. The products from the standards and tested blastocysts were resolved on a 1.2% agarose gel. The mtDNA fragment was visualized by ethidium bromide staining. The 5S rDNA fragment was detected by autoradiography following Southern blotting.
mtDNA copy number analysis in unfertilized eggs.
Superovulation was induced as previously described (21). Female mice that were 3 to 5 weeks old and weighed between 12 and 16 g were injected intraperitoneally with 5 IU of pregnant mare's serum (Sigma G4877), followed by an intraperitoneal injection of 5 IU of human chorionic gonadotropin (Sigma C1063) 45 h later. They were sacrificed the next day, and unfertilized eggs were obtained from the oviducts. The cumulus cells surrounding the eggs were removed by incubation in 300 μg of hyaluronidase per ml in phosphate-buffered saline (pH 7.2) for 5 to 10 min. Eggs from the same female were pooled in phosphate-buffered saline (pH 7.2) at a concentration of 1 egg/μl. DNA was isolated as described above for blastocysts. To analyze the amount of mtDNA in unfertilized eggs from wild-type and heterozygous females, The total DNA amounts isolated from 2.5, 5, or 10 wild-type eggs were used as PCR templates to generate standards, and the total DNA amounts from 5 eggs of heterozygous females were used as templates for comparison. The same mtDNA fragment and 5S rDNA fragment described above for the blastocysts were amplified in a PCR with Platinum Taq DNA polymerase. The PCR was carried out with 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 μM concentrations of primers m5SrDNA699S and m5SrDNA1014AS, and 0.1 μM concentrations of primers mCO1-267S and mCO1-914AS. The cycling conditions were 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 22 cycles. The products were resolved on a 1.2% agarose gel, the mtDNA fragment was visualized by ethidium bromide staining, and the 5S rDNA fragment was detected by autoradiography following Southern blotting.
TUNEL staining of 3.5-day-old embryos.
We stained 3.5-day-old embryos for DNA strand breaks by TUNEL (terminal deoxynucleotidyl-transferase-mediated dUTP-biotin nick end labeling) method using the In Situ Cell Death Detection Kit, Fluorescein (Boehringer Mannheim). Briefly, freshly isolated embryos were fixed in 4% (vol/vol) paraformaldehyde and 1% (vol/vol) calf serum in phosphate-buffered saline (pH 7.2) for 5 min and then permeabilized in 0.1% Triton X-100, 0.1% sodium citrate, and 1% (vol/vol) calf serum in phosphate-buffered saline (pH 7.2) for 5 min on ice. They were then rinsed in phosphate-buffered saline (pH 7.2) and incubated in the TUNEL reaction mixture for 1 h at 37°C. Positive controls were treated the same except that before incubation in the reaction mixture, they were incubated in phosphate-buffered saline (pH 7.5) containing 50 U of RQ1 DNase (Promega) per ml, 10 mM Tris-HCl, 1 mM MgCl2, and 1 mg of bovine serum albumin per ml for 30 min at 37°C. Stained embryos were viewed under a fluorescent microscope at 485 nm and then collected for genotyping as described above.
RESULTS
Targeted disruption of the mouse NRF-1 gene.
To disrupt the mouse NRF-1 gene, a targeting vector was constructed in which the NRF-1 sequence encoding amino acids 86 to 166 and encompassing coding exons 2 through 4 was replaced by a β-galactosidase–neomycin cassette (Fig. 1A). This region contains the nuclear localization signals and the DNA-binding and dimerization domains, and its deletion upon homologous recombination should completely eliminate NRF-1 function (15, 16, 48). In the mutant allele, the β-galactosidase coding sequence is fused in frame with the first 85 amino acids of NRF-1, thus placing β-galactosidase expression under the control of the NRF-1 promoter and 5′-untranslated region. The targeting vector was electroporated into 129/SvJ-derived (ES) cells, and G418 and ganciclovir double-resistant cells were screened. Homologous integration into the NRF-1 locus was confirmed in approximately 20% of the selected ES clones (Fig. 1B) by Southern blot hybridization. Five positive ES clones were microinjected into C57BL/6 blastocysts, and three chimeras from one of the clones showed germline transmission. Most of the heterozygous NRF-1 mice were viable and fertile. They were interbred, and their offspring were genotyped by Southern blotting and PCR analysis (Fig. 1B to D). Among the initial 412 newborns generated by heterozygous matings, none were homozygous mutants, and the ratio of heterozygous to wild-type offspring was 1.78 (Table 1). This was indicative of an embryonic lethal phenotype associated with the NRF-1−/− genotypes.
TABLE 1.
Genotypes of progeny from heterozygous matings
| Stage (dpc) | No. of progeny (%) with genotype:
|
Total no. | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Newborna | 148 (36) | 264 (64) | 0 | 412 |
| 6.5–8.5 | 27 (38) | 45 (62) | 0 | 72 |
| 3.5 | 15 (22) | 35 (51) | 18 (26) | 68 |
DNA was isolated from 1-day-old to 3-week-old animals.
The stage of embryonic death associated with NRF-1 loss of function was investigated by determining the genotypes of embryos between 6.5 and 8.5 days postcoitus (dpc). Surprisingly, no homozygous NRF-1 embryos were identified at this stage (Table 1). Therefore, a nested-PCR strategy was developed to genotype preimplantation embryos (Fig. 1C and D). Among 68 blastocysts isolated at 3.5 dpc, 15 were wild-type, 35 were heterozygous, and 18 were homozygous mutant (Table 1). This result is consistent with the expected ratio for Mendelian inheritance. Thus, homozygous null mutations in NRF-1 result in lethality between embryonic days 3.5 and 6.5.
NRF-1 expression in preimplantation embryos.
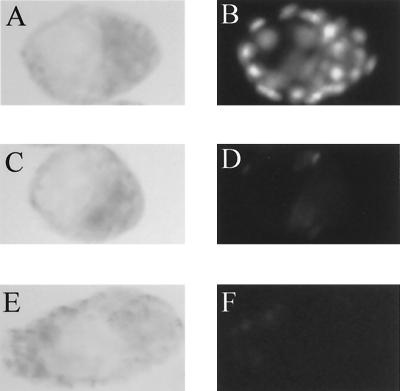
The death of NRF-1−/− embryos around the time of implantation indicated that NRF-1 function was required for development beyond this stage. It was demonstrated recently that NRF-1 expression can be detected in 7.5-dpc mouse embryos by Northern analysis (40). However, it is unknown whether NRF-1 is expressed earlier. The targeting vector was designed with the expectation that, by fusing β-galactosidase in frame with NRF-1 coding exon 2 (Fig. 1A), β-galactosidase activity could be measured as a good approximation of NRF-1 expression. To confirm this, RNase protection assays were performed to detect the expression of NRF-1–β-galactosidase fusion transcript relative to that of the endogenous NRF-1 transcript in tissues obtained from NRF-1−/− mice (Fig. 2A). Total RNA (10 μg) isolated from various tissues was hybridized simultaneously to a specific NRF-1 riboprobe and to an NRF-1–β-galactosidase fusion gene riboprobe. The results demonstrated that the relative expression of the fusion gene in various tissues closely paralleled that observed for the NRF-1 transcript itself, with the highest expression in the testis and the lowest expression in the heart and liver (lanes 1 to 5). The heterogeneity of protected fusion gene transcripts (approximately 120 nt) did not result from degradation of the NRF-1 transcript because the fusion gene riboprobe alone yielded the same heterogeneous protected bands upon hybridization to kidney RNA from NRF-1+/− animals (lane 6). The specificity of the fusion gene riboprobe was tested by hybridizing total kidney RNA from a wild-type mouse to both riboprobes. In this case, only the protected NRF-1 transcript from the wild-type allele was observed (lane 7). The results support the conclusion that the expression of the β-galactosidase gene is directed from the NRF-1 promoter and therefore reflects NRF-1 expression.
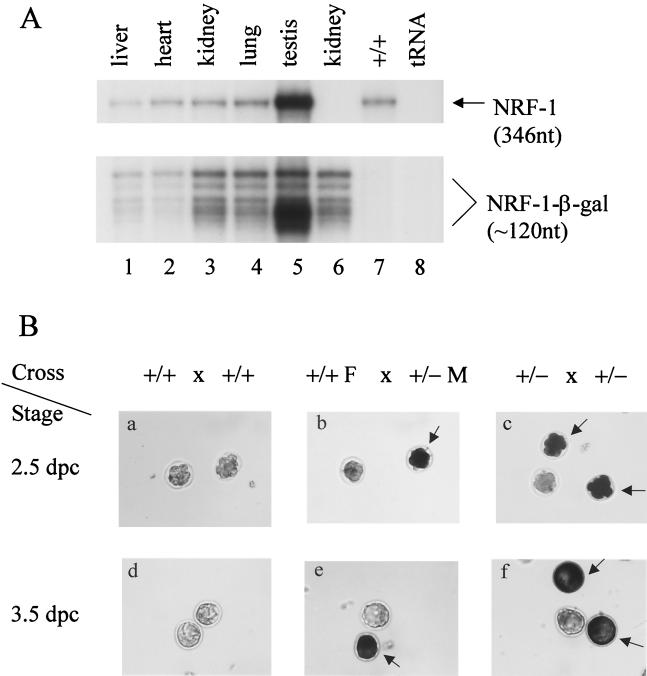
FIG. 2.
β-Galactosidase expression. (A) Expression of NRF-1 and the NRF-1–β-galactosidase fusion gene transcripts in adult mice. A total of 10 μg of RNA isolated from the indicated tissues of an NRF-1+/− mouse (lanes 1 to 6) or from kidney of a wild-type littermate (lane 7) was analyzed for the expression of NRF-1 and NRF-1–β-galactosidase transcripts by RNase protection assay. Lane 6, with the NRF-1–β-galactosidase riboprobe alone; other lanes, with both NRF-1 and NRF-1–β-galactosidase riboprobe. The sizes of the protected products are indicated on the right. (B) β-Galactosidase expression in embryos. Embryonic stages are indicated on the left, and the various crosses are shown above. F, female; M, male. Positively stained embryos are indicated by arrows. β-Galactosidase activity was readily detected in eight-cell morulae (b and c) and in both the inner cell mass and trophectoderm cells in blastocysts (e and f).
To determine whether NRF-1 is expressed in preimplantation embryos, 2.5- and 3.5-dpc embryos were isolated, and β-galactosidase staining was performed. The results showed that 17 of 23 blastocysts (74%) and 15 of 19 eight-cell morulae (79%) obtained from heterozygous matings stained positive for β-galactosidase activity (Fig. 2Bc and f; Table 2), whereas all embryos from wild-type crosses were negative (Fig. 2Ba and d). In addition, representative stained embryos were genotyped. All positively stained embryos were either NRF-1−/− or NRF-1+/−, and all negatively stained ones were wild type (not shown). This is consistent with the expected expression of β-galactosidase activity from the endogenous promoter.
TABLE 2.
β-Galactosidase staining of embryos
| Mating type | Stage (dpc) | Litter (n) | No. of embryos
|
||
|---|---|---|---|---|---|
| Stained | Not stained | Total | |||
| +/− × +/− | Blastocyst (3.5) | 3 | 17 | 6 | 23 |
| Eight-cell morula (2.5) | 2 | 15 | 4 | 19 | |
| +/+F × +/−Ma | Blastocyst (3.5) | 2 | 5 | 7 | 12 |
| Eight-cell morula (2.5) | 2 | 6 | 9 | 15 | |
F, female; M, male.
Active gene expression has been demonstrated during oocyte growth at the stage of the first meiotic prophase, and transcripts and proteins expressed during this time could be carried over to early embryos (reviewed in references 41 and 51). If the β-galactosidase activity observed in NRF-1 embryos results from the inheritance of maternal gene products, one would expect to see positive staining in all embryos from heterozygous crosses because all wild-type haploid eggs from heterozygous females should also obtain the gene products expressed before the first meiotic division. The observation that wild-type 2.5- and 3.5-dpc embryos from heterozygous matings stained negative for β-galactosidase activity thus argues against maternal inheritance of β-galactosidase. To confirm this, wild-type females were crossed to heterozygous males, and embryos were isolated for β-galactosidase staining. Since wild-type females do not express any bacterial β-galactosidase, any detected activity would thus be expressed from the mutant allele obtained from the male. The results show that 5 of 12 blastocysts (42%) and 6 of 15 eight-cell morulae (40%) from such matings stained positive for β-galactosidase activity (Fig. 2Bb and e; Table 2). Therefore, NRF-1 is expressed in preimplantation embryos at no later than the eight-cell morular stage.
NRF-1 is required for growth of blastocysts in vitro.
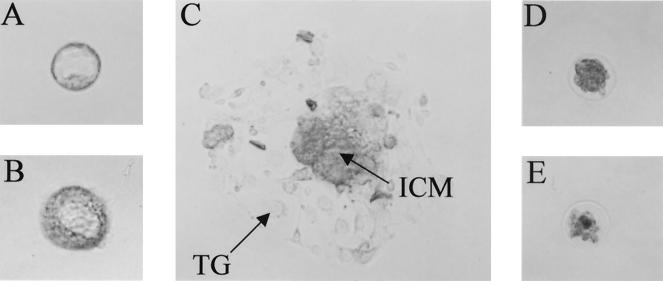
The morphology of NRF-1−/− blastocysts was indistinguishable from that of NRF-1+/− or wild-type blastocysts. Given the lack of an obvious phenotype, it was of interest to determine whether the arrest at this stage was associated with an intrinsic defect in cellular proliferation. To this end, blastocysts from heterozygous matings were isolated and cultured in vitro. Among 36 blastocysts obtained from five litters, 23 showed typical morphology in culture after 5 days, with the trophoblast spreading out on the culture dish and a proliferating inner cell mass on top (Fig. 3C; Table 3). In contrast, the other 13 blastocysts were unable to attach to the culture dishes and showed no sign of growth (Fig. 3D and E). Some of them were lost after 5 days in culture, presumably from lysis of the entire embryo. Some appeared to collapse and showed a decrease in size (Fig. 3D and E). Attempts to genotype these blastocysts by PCR were unsuccessful. The 23 embryos that grew in culture were genotyped, among which 14 were heterozygous and 9 were wild-type (Table 3). These results suggested that none of the homozygous NRF-1 blastocysts could grow normally in vitro.
FIG. 3.
Growth of blastocysts in culture. Blastocysts were cultured in vitro as described in Materials and Methods. (A) Blastocyst in culture medium immediately after isolation (day 0 in culture). (B and D) Blastocyst in culture for 40 h (day 2). (C and E) Blastocyst in culture for 100 h (day 5). Most blastocysts attached and started to grow at day 2 (B) and showed typical morphology at day 5, including the trophoblastic giant cells (TG) and the inner cell mass (ICM) (C). Putative NRF-1−/− blastocysts failed to grow (D and E). (B and C) The grown blastocyst was genotyped as NRF-1+/+. All of the images are at the same magnification.
TABLE 3.
Growth of blastocysts in culture
| Mating type | Litter (n) | No. of blastocysts with no growth | No. of blastocysts with normal growth
|
Total | ||
|---|---|---|---|---|---|---|
| Total | +/− | +/+ | ||||
| +/− × +/− | 5 | 13 | 23 | 14 | 9 | 36 |
| +/+ × +/− | 2 | 2 | 12 | NDa | ND | 14 |
| +/+ × +/+ | 2 | 0 | 16 | ND | ND | 16 |
ND, the genotypes of progeny from +/+-to-+/− matings and wild-type matings were not determined.
Wild-type and NRF-1+/− blastocysts were further tested for their ability to grow in culture. A total of 30 blastocysts (four litters) from wild-type matings or heterozygous-to-wild-type matings were cultured under the same conditions. Among them, 28 grew normally. The two that did not grow were from heterozygous-to-wild-type matings, suggesting that a small number of heterozygous blastocysts may also be defective in in vitro development (Table 3). Thus, despite their normal morphology, NRF-1−/− blastocysts displayed a generalized defect in their ability to develop further in vitro.
Homozygous disruption of NRF-1 is associated with a decrease in mitochondrial staining and a reduction in mtDNA content.
Given the proposed role for NRF-1 in respiratory gene expression, it was of interest to determine whether NRF-1−/− blastocysts had a mitochondrion-related phenotype. Rhodamine 123 staining requires a normal mitochondrial membrane potential and has been used for fluorescent staining of functional mitochondria with low background (24). Embryos from heterozygous or wild-type matings were isolated at 3.5 dpc and stained with rhodamine 123. The 17 blastocysts (three litters) obtained from wild-type matings were similar in the overall intensity of staining (Fig. 4Aa). In contrast, among 13 embryos (two litters) from heterozygous matings, five blastocysts had a weaker and more diffuse staining pattern than those of their littermates, which resembled wild-type blastocysts in staining intensity (Fig. 4Ab). Although the genotypes of the stained embryos were unknown because rhodamine 123 staining interfered with genotyping, the results were consistent with a reduction in the number or function of mitochondria associated with the loss of NRF-1.
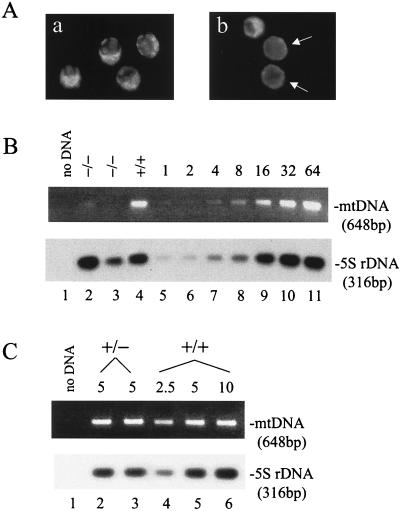
FIG. 4.
Mitochondrial staining and mtDNA copy number analyses. (A) Rhodamine 123 staining of blastocysts. Blastocysts were stained with rhodamine 123 as described in Materials and Methods. Four blastocysts from a wild-type mating (a) are compared to three blastocysts from a heterozygous mating (b). Those with weak fluorescence are indicated by the arrows. (B) mtDNA copy number in blastocysts. Half of the DNA isolated from each of the indicated embryos (lanes 2 to 4) was used to amplify a mtDNA fragment and a 5S rDNA fragment in a PCR. For standards, genomic DNA prepared from the heart of an adult wild-type mouse was diluted and subjected to the same PCR amplification (lanes 5 to 11). The amount of template was indicated in fold at the top for each standard. Products were resolved on a 1.2% agarose gel, and the mtDNA product was visualized by ethidium bromide staining. The 5S rDNA product was detected by Southern blotting. (C) mtDNA copy number in unfertilized eggs. Unfertilized eggs were isolated from wild-type or heterozygous NRF-1 females for DNA preparation as described in Materials and Methods. DNA from 5 eggs from a heterozygous animal (lanes 2 and 3) or from the equivalent of 2.5, 5, or 10 eggs from a wild-type animal (lanes 4, 5, and 6, respectively) was used as the template in PCR amplifications of the same mtDNA sequence as described in panel B. The numbers of eggs from which the template DNA was isolated are indicated above the panels.
The correlation between NRF-1 and mitochondrial function was further examined by comparing the amount of mtDNA in NRF-1−/− and wild-type blastocysts. This was accomplished by first genotyping individual blastocysts and then subjecting total DNA from NRF-1−/− and wild-type blastocysts to PCR amplifications of a 648-bp mtDNA fragment and a 316-bp 5S rDNA fragment. The latter served as an internal loading control for quantifying mtDNA levels (Fig. 4B). Standards were generated using adult heart genomic DNA (Fig. 4B, lanes 5 to 11). In comparison, NRF-1−/− blastocysts showed a reduced amount of mtDNA ranging from 30 to <5% of wild-type controls (lanes 2 to 4). This is consistent with the reduction in rhodamine staining indicative of a loss of mitochondrial function. Thus, NRF-1 is required for the maintenance of normal levels of mtDNA in blastocysts.
NRF-1 expression in the ovary.
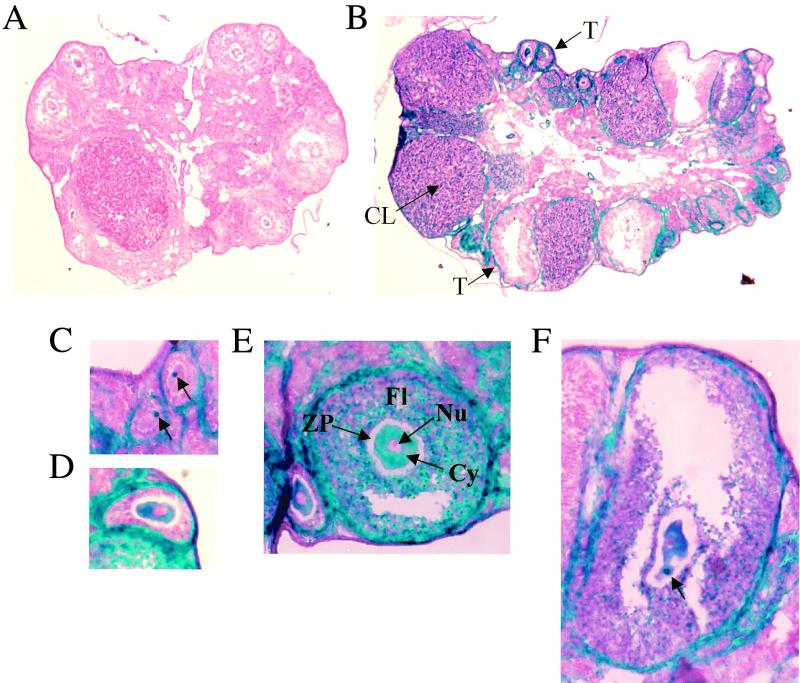
During mouse oocyte growth, the paired homologous chromosomes are fully extended and gene expression occurs. This accounts for the maternally inherited mRNAs and proteins in the early embryos. It is also well documented that mammalian mtDNA is amplified at least 100-fold during oocyte growth so that the final mtDNA copy number in the mature egg is approximately 100,000 (31, 36). To determine whether NRF-1 expression coincides with maternal gene expression and mtDNA amplification in oocytes, ovaries from sexually mature female heterozygotes were sectioned and stained for β-galactosidase activity. As shown in Fig. 5B, both the corpus luteum and the thecal cells surrounding the follicles were most darkly stained, while some of the follicular cells were lightly stained. Careful examinations of serial sections revealed that oocytes at all stages of maturation also stained positive for β-galactosidase activity (Fig. 5C to F), with medium-sized ones being the most darkly stained (panel E). The β-galactosidase activity was cytoplasmic with a small area away from the nucleus stained most intensely, resulting in the appearance of a dark blue spot in the relatively lightly stained cytoplasm (Fig. 5C, D, and F). The nucleus was unstained, presumably because a nuclear localization signal was absent in the fusion protein. Ovaries from wild-type controls stained negative (Fig. 5A), indicating that the staining resulted from the expression of the mutant NRF-1 allele. These results demonstrate that NRF-1 is expressed throughout oocyte maturation where gene expression and a massive mtDNA amplification occur.
FIG. 5.
NRF-1 expression in ovary. (A) β-Galactosidase staining of an ovary from a wild-type mouse. (B) β-Galactosidase staining of an ovary from a NRF-1+/− mouse. (C to F) β-Galactosidase staining of follicles from a NRF-1+/− mouse containing eggs at various stages of maturity. The corpus luteum (CL), the thecal cell layer surrounding the follicle (T), the follicle (Fl), the zona pellucida (ZP, unstained), the cytoplasm (Cy), and the nucleus (Nu) are indicated (B and E). The concentrated regions of β-galactosidase accumulation within the cytoplasm are indicated by arrows in panels C and F. Panels A and B are at the same magnification; panels C to F are at the same magnification.
Measurements of the amount of mtDNA in unfertilized eggs.
It is possible that the reduced mtDNA levels observed in NRF-1−/− blastocysts is caused by the effect of NRF-1 heterozygosity on mtDNA amplification during oocyte growth. If this is the case, unfertilized eggs from heterozygotes should exhibit decreased mtDNA levels. To test this possibility, unfertilized eggs were obtained from wild-type and heterozygous female mice, and the amounts of mtDNA were compared after PCR amplification of the 648-bp mtDNA fragment (Fig. 4C). Again, the 5S rDNA fragment was amplified as an internal loading control. When DNA isolated from five eggs was used as the PCR template, roughly the same amount of mtDNA product was obtained for wild-type and heterozygous females (lanes 2, 3, and 5). DNA isolated from 10 wild-type eggs did not generate proportionately more product than that from 5 eggs, suggesting that the PCR may be close to saturation (lane 6). However, DNA isolated from the equivalent of 2.5 wild-type eggs gave a linear decrease in product (lane 4), indicating that the difference in the amount of mtDNA between wild-type and heterozygous eggs is <2-fold. Thus, mtDNA amplification in oocytes does not appear to be dramatically affected by the loss of one copy of the NRF-1 gene. This indicates that the massive loss of mtDNA observed in the blastocyst occurs after fertilization.
Loss of mtDNA in NRF-1−/− blastocysts is not associated with increased apoptosis.
NRF-1 may affect apoptosis through its regulation of mitochondrial functions. One explanation for the loss of mtDNA in NRF-1−/− blastocysts is that it is associated with apoptotic cell death that occurs after fertilization as a consequence of the absence of NRF-1. It is well established that limited apoptosis does occur during early mammalian embryogenesis starting around the blastocyst stage (18), and it is possible that NRF-1 loss leads to massive cell death and the loss of the embryo. To examine this possibility, TUNEL staining was performed on both wild-type and NRF-1−/− blastocysts to detect the chromosomal DNA breakage associated with apoptosis. For comparison, a positive control was generated by treating wild-type blastocysts with DNase (Fig. 6A and B). The results demonstrate that while small numbers of apoptotic cells are detectable in both wild-type and NRF-1−/− blastocysts, there is no difference between the two in the overall levels of apoptotic cell death (Fig. 6C to F). This suggests that the loss of mtDNA is not a by-product of increased apoptosis but rather a consequence of the disruption of NRF-1-dependent pathways of mtDNA maintenance.
FIG. 6.
Detection of chromosomal DNA fragmentation in blastocysts by TUNEL staining. TUNEL staining was performed on blastocysts as described in Materials and Methods. Blastocysts with expanded blastocoel cavities were selected to ensure that they had entered the stage when limited apoptosis would normally occur. Bright-field microscopy (A, C, and E) and the corresponding fluorescent images of TUNEL-stained blastocysts (B, D, and F) are shown. (A and B) Wild-type blastocyst treated with RQ1 DNase as a positive control. (C and D) Wild-type blastocyst showing very few stained cells. (E and F) NRF-1−/− blastocyst showing essentially the same staining intensity as the wild-type embryos.
DISCUSSION
NRF-1 expression and embryonic development.
We demonstrate here that homozygous disruption of the mouse NRF-1 gene leads to embryonic death around the time of implantation. Loss of NRF-1 also coincides with the reduction of mtDNA in the blastocyst, thus providing the first evidence that NRF-1 is required for mitochondrial maintenance in vivo.
The targeting vector was designed so that in the mutant allele the first 85 amino acids of NRF-1 are fused in frame to β-galactosidase and polyadenylation signals are present at the 3′ end of the transcriptional unit. This feature, along with the opposite transcriptional orientation of the neomycin cassette, would direct the termination of the NRF-1–β-galactosidase fusion transcript at the 3′ end of the β-galactosidase gene, without any downstream NRF-1 sequence expressed. The deletion of amino acids 1 to 77 has no affect on the ability of NRF-1 to transactivate the cytochrome c promoter in transfection assays (16). β-Galactosidase is expressed from the endogenous NRF-1 promoter, and the fidelity of fusion gene expression was confirmed by the similar tissue expression patterns of NRF-1 and the fusion transcripts. β-Galactosidase activity was readily detected in growing oocytes of heterozygous females, as well as in heterozygous and homozygous eight-cell morulae and blastocysts. According to a thorough investigation of accumulation profiles of known transcripts in preimplantation embryos, most genes, once activated at the two-cell stage, continue to be transcribed at least into the blastocyst stage (reviewed in reference 25). Although β-galactosidase staining was not performed in embryos before the eight-cell stage, it is likely that NRF-1 expression starts at an earlier time and that its continuous expression throughout preimplantation development accounts for the observed β-galactosidase activity in the early embryo.
It is interesting to note that the loss of NRF-1 or its relatives results in different phenotypes in both vertebrates and invertebrates. P3A2 is a negative regulator of transcription in sea urchins that functions in the spatial expression of a cytoskeletal actin gene during development. Its loss of function in the egg affects the morphogenesis of the archenteron and results in embryonic lethality prior to gastrulation (4). In Drosophila melanogaster, the erect wing locus is required for proper neuromuscular development, and certain alleles result in late embryonic or early larval lethality presumably due to nervous system dysfunction (13, 14). Unlike the invertebrate relatives, the sequence similarity between nrf in zebra fish and the mammalian NRF-1 is not restricted to the DNA-binding domain but rather is distributed throughout the coding region. During development, expression of nrf occurs predominantly in the central nervous system and the optic tracts. A homozygous insertional disruption of the nrf locus in zebra fish results in the loss of photoreceptors in the retina and ultimately death at the larval stage (3). In each case, loss-of-function mutations led to developmental arrest. However, in mice, the early lethality of the NRF-1-null embryos suggests that, in mammals, NRF-1 expression is required to proceed beyond the small number of cell divisions leading to the blastocyst stage.
Several recently described transcription factor gene knockouts also exhibit a peri-implantation lethal phenotype. As examples, the stage of developmental arrest has been characterized in vitro for B-myb (45), GATA6 (26), vHNF1 (2), and Max (43) knockouts. Blastocysts from the vHNF1 homozygous null mutants are morphologically identical to the wild-type and exhibit normal outgrowth. In contrast, blastocysts from both B-myb and GATA6 homozygous null mutants generate trophoblastic giant cells in culture but are severely impaired in the proliferation of the inner cell mass. Blastocysts from Max homozygous null mutants are more severely impaired in that they appear normal initially but outgrowth ceases after 2 to 3 days in culture. NRF-1−/− blastocysts differ markedly from all of these cases in that morphologically normal blastocysts form in vivo, but they do not attach when placed in culture and there is no proliferative outgrowth. Interestingly, the timing of this arrest coincides approximately with the resumption of mtDNA replication that occurs around the time of implantation (30).
NRF-1 and mitochondrial function in early embryogenesis.
Although NRF-1−/− blastocysts were morphologically normal, a striking decrease was observed in their mtDNA content. This depletion of mtDNA occurred in the absence of a generalized increase in nuclear DNA fragmentation that may occur as a result of increased apoptosis. The results are consistent with a specific role for NRF-1 in maintaining mtDNA during early embryogenesis. The fact that NRF-1−/− embryos can survive to the blastocyst stage with severely diminished mtDNA levels is consistent with previous findings concerning the role of mitochondrial function during early embryogenesis. It has been demonstrated that oxygen consumption starts to rise at the eight-cell stage and increased by 3.5-fold in the blastocysts (32), coinciding with changes in the mitochondrial ultrastructure (35). Although drastic increases in mitochondrial rRNA and mRNA expression have been observed from two-cell embryos to blastocysts (37), mitochondrial genetic activities do not appear to be required for blastocyst development (35). Blastocysts that had been treated with inhibitors of mitochondrial RNA and protein synthesis underwent normal development when transplanted to the uteri of foster mothers. Thus, although mitochondrial function may be necessary for embryonic development to the blastocyst stage, normal expression of the mitochondrial genome may not be absolutely required. In the case of NRF-1−/− blastocysts, a low level of expression from mtDNA combined with transcripts and proteins carried from the oocyte may be sufficient to achieve this stage of development.
mtDNA is massively amplified during mammalian oocyte growth before the first meiotic devision (31), and no mtDNA replication occurs from the egg through the blastocyst stage (36, 37). The reduced amount of mtDNA in homozygous blastocysts could thus be due to the effect of NRF-1 disruption on the mtDNA amplification during oocyte growth, the maintenance of mtDNA during early embryogenesis, or a combination of both. The positive β-galactosidase staining in the ovary of heterozygous mice is consistent with the expression of NRF-1 in growing oocytes, therefore suggesting a possible role for NRF-1 in mtDNA amplification. However, unfertilized eggs from wild-type and heterozygous females had the same amount of mtDNA, arguing against an abnormal mtDNA amplification in heterozygous oocytes. Nonetheless, in addition to transcriptional regulation of the target genes in early embryos, NRF-1 may play a role in the normal maintenance of mtDNA in early embryogenesis by regulating the expression of genes in growing oocytes, whose products are essential when carried into the embryos.
Several NRF-1 target genes have been implicated in mitochondrial maintenance. Among them, TFAM is required for mtDNA transcription and replication (12, 34), and its expression correlates with mtDNA levels in patients (28, 38) and in an in vivo animal model (29). NRF-1 binding was demonstrated to be important for human TFAM promoter activity in vitro (49). Thus, it is plausible that the disruption of NRF-1 may cause a reduction in mtDNA through reduced expression of TFAM. However, TFAM-null mice die after embryonic day 8.5, suggesting that the earlier death of NRF-1 homozygous embryos may not be due to the downregulation of TFAM. One possibility is that the phenotype is only in part contributed by TFAM, while involving other NRF-1-regulated mitochondrial genes. Such candidates include MRP RNA, the RNA component of the endonuclease necessary for generating primer RNAs for mitochondrial transcription (6), and mtSSB, the single-stranded DNA-binding protein that binds to mtDNA D-loops and enhances the rate of the DNA polymerase reaction (17). These and other unidentified NRF-1 target genes, together with TFAM, may act in concert in mitochondrial maintenance.
NRF-1 disruption and embryonic lethality.
Could the reduced amount of mtDNA in homozygous blastocysts be the cause of embryonic death? The mouse embryo does not grow in size until the blastocyst stage and begins to grow rapidly during implantation (19). mtDNA synthesis is activated at around this time (30). Therefore, although reduced mtDNA content could provide sufficient mitochondrial expression at the blastocyst stage, the rapid growth of the embryo starting at implantation may require more efficient energy production. Hence, the reduced number of mtDNA molecules may become inadequate, leading to cell death because of low levels of oxidative phosphorylation.
In light of the broader role for NRF-1 in transcriptional regulation, it is unlikely that this simple model can adequately explain the embryonic lethal phenotype. Since NRF-1 was identified, several genes whose functions are not directly related to mitochondria have been found to be regulated by NRF-1 at the transcriptional level in vitro. Some of these gene products are involved in fundamental cellular functions, including amino acid metabolism (tyrosine aminotransferase), translation initiation (eIF-2α), chromatin structure (histone h5), and purine synthesis (GPAT and AIRC) (reviewed in reference 39). It is possible that the disruption of NRF-1 function causes their abnormal expression, and one or more of these proteins are essential for embryonic development. Conceivably, other unidentified NRF-1-dependent factors could be required for embryogenesis. Computer searches revealed potential NRF-1 binding sites in a variety of mammalian genes of diverse functions (48). Null mutations of some of these genes in the mouse have demonstrated that they are essential for embryonic development. Examples include the mouse Dad1 gene (defender against apoptotic cell death) (22), the mouse cdh5 gene (vascular endothelial cadherin) (5), and the mouse Lrp gene (low-density lipoprotein receptor-related protein) (20). Thus, the disruption of NRF-1 may lead to dysfunction of a number of cellular pathways in addition to mitochondrial deficiency via abnormal gene regulation. These effects may act in combination to cause embryonic lethality.
ACKNOWLEDGMENTS
We acknowledge the assistance of the Targeted Mutagenesis Facility in the Children's Memorial Institute for Education and Research at Northwestern University. We are grateful to Jan Reddy for his expertise and advice. We thank Kristel Vercauteren for excellent technical assistance and Ulf Andersson for critical reading of the manuscript.
This work was supported by United States Public Health Service Grant GM32525-18.
REFERENCES
- 1.Abbondanzo S J, Gadi I, Stewart C L. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225:803–855. doi: 10.1016/0076-6879(93)25052-4. [DOI] [PubMed] [Google Scholar]
- 2.Barbacci E, Reber M, Ott M-O, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- 3.Becker T S, Burgess S M, Amsterdam A H, Allende M L, Hopkins N. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor 1 and avian initiation binding repressor. Development. 1998;124:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- 4.Bogarad L D, Arnone M I, Chang C, Davidson E H. Interference with gene regulation in living sea urchin embryos: transcription factor knock out (TKO), a genetically controlled vector for blockade of specific transcription factors. Proc Natl Acad Sci USA. 1998;95:14827–14832. doi: 10.1073/pnas.95.25.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmeliet P, Lampugnani M G, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oostuyse B, Dewerchin M, Zanetti A, Angellilo A, Mattot V, Nuyens D, Lutgens E, Clotman F, de Ruiter M C, Gittenberger-de Groot A, Poelmann R, Lupu F, Herbert J M, Collen D, Dejana E. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. doi: 10.1016/s0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 6.Chang D D, Clayton D A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 7.Chau C A, Evans M J, Scarpulla R C. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2α, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992;267:6999–7006. [PubMed] [Google Scholar]
- 8.Chen S H, Nagy P L, Zalkin H. Role of NRF-1 in bidirectional transcription of the human GPAT-AIRC purine biosynthesis locus. Nucleic Acids Res. 1997;25:1809–1816. doi: 10.1093/nar/25.9.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desimone S M, White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol Cell Biol. 1993;13:3641–3949. doi: 10.1128/mcb.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans M J, Scarpulla R C. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF and intron Sp1 recognition sites. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- 11.Evans M J, Scarpulla R C. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 12.Fisher R P, Clayton D A. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 1988;8:3496–3509. doi: 10.1128/mcb.8.8.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleming R J, Desimone S M, White K. Molecular isolation and analysis of the erect wing locus in Drosophila melanogaster. Mol Cell Biol. 1989;9:719–725. doi: 10.1128/mcb.9.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleming R J, Zusman S B, White K. Developmental genetic analysis of lethal alleles at the ewg locus and their effects on muscle development in Drosophila melanogaster. Dev Genet. 1983;3:347–363. [Google Scholar]
- 15.Gugneja S, Scarpulla R C. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J Biol Chem. 1997;272:18732–18739. doi: 10.1074/jbc.272.30.18732. [DOI] [PubMed] [Google Scholar]
- 16.Gugneja S, Virbasius C A, Scarpulla R C. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S, Van Tuyle G C. The gene and processed pseudogenes of the rat mitochondrial single-strand DNA-binding protein: structure and promoter strength analyses. Gene. 1998;212:269–278. doi: 10.1016/s0378-1119(98)00148-6. [DOI] [PubMed] [Google Scholar]
- 18.Hardy K. Cell death in the mammalian blastocyst. Mol Hum Reprod. 1997;3:919–925. doi: 10.1093/molehr/3.10.919. [DOI] [PubMed] [Google Scholar]
- 19.Hensleigh H C, Weitlauf H M. Effect of delayed implantation on dry weight and lipid content of mouse blastocysts. Biol Reprod. 1974;10:315–320. doi: 10.1095/biolreprod10.3.315. [DOI] [PubMed] [Google Scholar]
- 20.Herz J, Couthier D E, Hammer R E. LDL receptor-related protein internalizes and degrades uPA-PAL-1 complexes and is essential for embryo implantation. Cell. 1993;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 21.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 22.Hong N, Flannery M, Hsieh S N, Cado D, Pedersen R, Winoto A. Mice lacking Dad1, the defender against apoptotic death-1, express abnormal N-linked glycoproteins and undergo increased embryonic apoptosis. Dev Biol. 2000;220:76–84. doi: 10.1006/dbio.2000.9615. [DOI] [PubMed] [Google Scholar]
- 23.Huo L, Scarpulla R C. Multiple 5′-untranslated exons in the nuclear respiratory factor 1 gene span 47 kb and contribute to transcript heterogeneity and translational efficiency. Gene. 1999;233:213–224. doi: 10.1016/s0378-1119(99)00135-3. [DOI] [PubMed] [Google Scholar]
- 24.Johnson L V, Walsh M L, Chen L B. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidder G M. The genetic program for preimplantation development. Dev Genet. 1992;13:319–325. doi: 10.1002/dvg.1020130502. [DOI] [PubMed] [Google Scholar]
- 26.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126:723–732. [PubMed] [Google Scholar]
- 27.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson N-G, Oldfors A, Holme E, Clayton D A. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem Biophys Res Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- 29.Larsson N G, Wang J M, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh G S, Clayton D A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 30.McLaren A. Growth from fertilization to birth in the mouse: embryogenesis in mammals. Ciba Found Symp. 1976;40:47–51. [Google Scholar]
- 31.Michaels G S, Hauswirth W W, Laipis P J. Mitochondrial DNA copy number in bovine oocytes and somatic cells. Dev Biol. 1982;94:246–251. doi: 10.1016/0012-1606(82)90088-4. [DOI] [PubMed] [Google Scholar]
- 32.Mills R M, Jr, Brinster R L. Oxygen consumption of preimplantation mouse embryos. Exp Cell Res. 1967;47:337–344. [Google Scholar]
- 33.Myers S J, Peters J, Huang Y, Comer M B, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisi M A, Clayton D A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 35.Piko L, Chase D G. Role of the mitochondrial genome during early development in mice. Effects of ethidium bromide and chloramphenicol. J Cell Sci. 1973;58:357–378. doi: 10.1083/jcb.58.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piko L, Matsumoto L. Number of mitochondria and some properties of mitochondrial DNA in the mouse egg. Dev Biol. 1976;49:1–10. doi: 10.1016/0012-1606(76)90253-0. [DOI] [PubMed] [Google Scholar]
- 37.Piko L, Taylor K D. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol. 1987;123:364–374. doi: 10.1016/0012-1606(87)90395-2. [DOI] [PubMed] [Google Scholar]
- 38.Poulton J, Morten K, Freeman-Emmerson C, Potter C, Sewry C, Dubowitz V, Kidd H, Stephenson J, Whitehouse W, Hansen F J, Parisi M, Brown G. Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum Mol Genet. 1994;3:1763–1769. doi: 10.1093/hmg/3.10.1763. [DOI] [PubMed] [Google Scholar]
- 39.Scarpulla R C. Nuclear control of respiratory chain expression in mammalian cells. J Bioenerg Biomembr. 1997;29:109–119. doi: 10.1023/a:1022681828846. [DOI] [PubMed] [Google Scholar]
- 40.Schaefer L, Engman H, Miller J B. Coding sequence, chromosomal localization, and expression pattern of Nrf-1: the mouse homolog of Drosophila erect wing. Genome. 2000;11:104–110. doi: 10.1007/s003350010021. [DOI] [PubMed] [Google Scholar]
- 41.Schultz R M. Molecular aspects of mammalian oocyte growth and maturation. In: Rossant J, Pederson R A, editors. Experimental approaches to mammalian embryonic development. Cambridge, England: Cambridge University Press; 1986. pp. 195–237. [Google Scholar]
- 42.Shadel G S, Clayton D A. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 43.Shen-Li H, Hagan R C, Jr, Hou H, Horner II J W, Lee H-W, DePinho R A. Essential role for Max in early embryonic growth and development. Genes Dev. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- 44.Solecki D, Bernhardt G, Lipp M, Wimmer E. Identification of a nuclear respiratory factor-1 binding site within the core promoter of the human polio virus receptor/CD155 gene. J Biol Chem. 2000;275:12453–12462. doi: 10.1074/jbc.275.17.12453. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka Y, Patestos N, Maekawa T, Ishii S. B-myb is required for inner cell mass formation at an early stage of development. J Biol Chem. 1999;274:28067–28070. doi: 10.1074/jbc.274.40.28067. [DOI] [PubMed] [Google Scholar]
- 46.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 47.Vernet M, Bonnerot C, Briand P, Nicolas J. Application of LacZ gene fusions to preimplantation development. Methods Enzymol. 1993;225:434–451. doi: 10.1016/0076-6879(93)25030-6. [DOI] [PubMed] [Google Scholar]
- 48.Virbasius C A, Virbasius J V, Scarpulla R C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 49.Virbasius J V, Scarpulla R C. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning J C, Kahn C R, Clayton D A, Barsh G S, Thoren P, Larsson N-G. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- 51.Wassarman P M, Kinloch R A. Gene expression during oogenesis in mice. Mutat Res. 1992;296:3–15. doi: 10.1016/0165-1110(92)90028-8. [DOI] [PubMed] [Google Scholar]
- 52.Wegner S A, Ehrenberg P K, Chang G, Dayhoff D E, Sleeker A L, Michael N M. Genomic organization and functional characterization of the chemokine receptor CXCR4, a major entry co-receptor for human immunodeficiency virus type 1. J Biol Chem. 1998;273:4754–4760. doi: 10.1074/jbc.273.8.4754. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 54.Zeller R W, Britten R J, Davidson E H. Developmental utilization of SpP3A1 and SpP3A2: two proteins which recognize the same DNA target site in several sea urchin gene regulatory regions. Dev Biol. 1995;170:75–82. doi: 10.1006/dbio.1995.1196. [DOI] [PubMed] [Google Scholar]