Abstract
Background
The aim of this review is to evaluate the global scientific literature on the utility of plasma presepsin (PSP) as a prognostic biomarker in a homogeneous group of coronavirus disease 2019 (COVID-19) positive cases.
Data retrieval
A systematic review utilizing Medline (PubMed interface), LitCovid NLM, World Health Organization (WHO)–global literature on coronavirus disease, and EBSCO CINAHL Plus was undertaken. The study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) group guidelines. The quality of individual evidence and possible risk of bias were assessed using the Quality in Prognosis Studies (QUIPS) tool. A narrative synthesis-based conclusion was compiled.
Results
A total of three articles passed through the predefined screening criteria and were included in the review. Methodological quality was evaluated to be acceptable. The aggregate study population was summed up to be 167 COVID-19 positive cases, who had undergone analysis of plasma PSP levels for the prediction of severity and mortality. Based on different PSP cutoffs utilized, a statistically significant association between PSP and COVID-19 severity was reported.
Conclusion
PSP appears as a promising prognostic biomarker of COVID-19 progression. As data are scarce on its utility, large cross-sectional studies are needed.
How to cite this article
Ahmed S, Mansoor M, Shaikh MS, Siddiqui I. Presepsin as a Predictive Biomarker of Severity in COVID-19: A Systematic Review. Indian J Crit Care Med 2021;25(9):1051–1054.
Keywords: COVID-19, Presepsin, Prognosis, Severe COVID, Systematic review
Introduction
The first case of coronavirus disease 2019 (COVID-19) was detected in Wuhan, China in 2019, which spreads at an exponential rate, and hence, it was termed as a “pandemic” by the World Health Organization (WHO) by March 2020.1 Since that time, the world faced several challenges mentally, physically, and economically as they tried to steer their way through these unchartered territories. For the medical community, it was much worse since this pneumonia of an unknown classification was difficult to diagnose and even harder to treat given the limited number of therapeutic options. These obstacles coupled with the high rate of transmission and severity of disease ranging from asymptomatic to fatal made it a pertinent topic to study and make continuous guideline improvements.2
Due to the overwhelming number of cases and mortality associated with COVID-19, studying reliable biomarkers to assess the severity and prognosis of the disease is crucial for medical practitioners and scientists to ensure timely diagnosis and appropriate management.1 When the pandemic struck, the guidelines were based on basic vital signs, such as respiratory rate, oxygen saturation, and oxygen indicator (PaO2/FiO2). These predictors are susceptible to subjective analysis.3 Hence, a multitude of biomarkers have been useful in the prediction of the severity of COVID-associated pneumonia as inflammatory reactions, cytokine storms, and coagulation cascades are involved in the pathogenesis of COVID pneumonia.1 Various biomarkers have been studied over the course of this pandemic in relation to COVID-19, ranging from the widely available procalcitonin (PCT) to ferritin, and less common angiotensin-converting enzyme assays and Krebs von den Lungen 6.1,3
CD14, a cluster of differentiation marker protein, is found on the surface of mononuclear cells, which has a propensity for lipopolysaccharide (LPS), and LPS binding protein was found on the surface of pathogens. Presepsin (PSP) is a small soluble subunit of CD14 (sCD14-ST; 64 amino acids, 13 kDa) that is found on the N-terminal sequence of CD14.4 Recent studies have shown that PSP levels rise dramatically in response to an acute infectious insult or high degree of sepsis. In the rabbit sepsis model, it was proven that phagocytosis plays a role in PSP elevation. In the lysozyme of a leukocyte, cathepsin D digests the bacteria along with CD14 creating its polypeptide subunit—PSP. This digested form of CD14 is then released into the bloodstream.4,5 In the context of COVID-19, owing to the cytokine storm syndrome, CD14 has been linked with the presentation of LPS to Toll-like receptor. Moreover, it causes elevated plasma PSP levels through intracellular signaling leading to enhanced gene expression responsible for immune-mediated response.
The normal concentration of PSP in a healthy adult is around 250 pg/mL (upper reference rate).5 The concentration of PSP rises significantly in sepsis or severe sepsis than in normal patients or patients with systemic inflammatory response syndrome (SIRS) where classification for each disease severity is based upon the definitions provided by ACCP/SCCM.6 Along with these characteristics, PSP has also shown high sensitivity and specificity for diagnosis in comparison to PCT, an otherwise frequently used biomarker.7 Coupled with these advantages, the measurement of PSP values from blood is a quick process, measured from whole blood through point-of-care (POC) devices in labs in around 15 minutes.5
In severe sepsis, PSP has shown efficacy in predicting the clinical outcome in hospitalized patients with pneumonia.8 Ugajin et al. concluded that patients who showed pneumonia severity index (PSI) class ≥4 with plasma PSP levels >470 pg/mL had a higher 30 day mortality (p = 0.005) than the milder patients with a lower PSI and PSP level, thus showing that high initial values of PSP are a good predictor for future deterioration of patients with pneumonia.8 Various studies have concluded that the use of PSP is not only a good biomarker for predicting the prognosis of pneumonia but also helpful for early diagnosis and risk stratification.9,10 The utility of PSP values in the case of COVID pneumonia has been shown to be predictive not only of its severity but also mortality in the limited research that has been conducted till now.
Given the limited amount of research done on PSP as a predictive marker for COVID pneumonia, prognosis and early diagnosis coupled with the positive results from the data available makes it a suitable biomarker to study further. With this perspective, this review was undertaken to systematically evaluate the globally available scientific studies that have assessed the prognostic utility of PSP in COVID-19.
Data Retrieval
In line with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) group, a systematic literature search was conducted. Medline (PubMed interface), LitCovid NLM, WHO–global literature on coronavirus disease, and EBSCO CINAHL Plus from the advent of COVD-19 in December 2019 till February 2021 were utilized for the search.11
The following Medical Subject Heading terms and keywords were searched: Coronavirus OR “corona virus” OR coronavirinae OR coronaviridae OR betacoronavirus OR covid19 OR “covid 19” OR nCoV OR “CoV 2” OR CoV2 OR sarscov2 OR 2019nCoV OR “novel CoV” OR “wuhan virus” OR [(wuhan OR hubei OR huanan) AND (“severe acute respiratory” OR pneumonia) AND (outbreak)] OR “Coronavirus” OR “Coronavirus Infections” OR “COVID-19” OR “severe acute respiratory syndrome coronavirus 2” OR “Betacoronavirus” AND [”presepsin protein, human” (Supplementary Concept) OR Presepsin OR PSP OR sCD14-ST], without language restrictions.
Human subject-based studies were considered only. Furthermore, the “related articles” feature in the databases was also applied to look for other potentially relevant articles, and two independent assessors from the team of authors also manually skimmed through the reference list of included articles.
In the next phase, the citations short lister were further scrutinized based on their titles and abstracts. The following inclusion criteria were applied: (1) Study population should be adult with PCR-positive COVID-19, (2) study participants must have undergone documented measurements of serum PSP, (3) study should have statistically evaluated the prognostic performance of PSP in relation to outcome, (4) outcome-based study, defining severity on the basis of the requirement of mechanical ventilation or intensive care admission or mortality, and (5) study design should be cohort or case control or case series.
Studies comprising of reviews, perspectives, opinion papers, hypothesis, viewpoints, basic sciences/nonclinical studies, and non-English language were excluded. Subsequently, full-text versions of final inclusions were assessed by two independent reviewers. Inter-rater reliability and agreement were assessed using kappa statistics.12
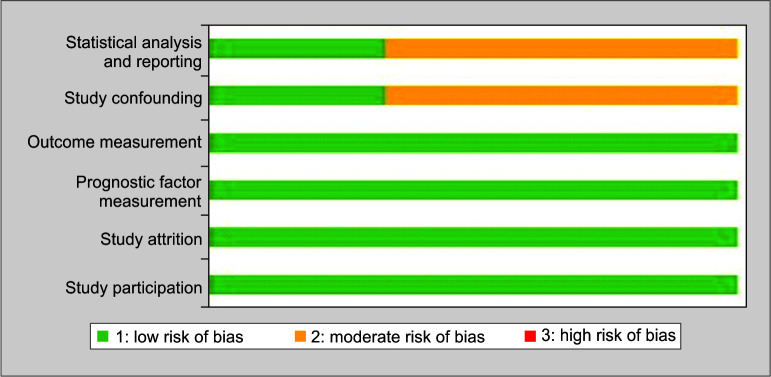
Quality in Prognosis Studies (QUIPS) tool-based risk of bias assessment was conducted, with scores ranging from low, moderate, and high risk.13 Six domains including case selection, attrition, PSP measurement, outcome assessment, confounders, statistical analysis, and reporting were assessed and stratified into 30 well-defined criteria. Attainment of low or moderate risk under all domains was rated as high and correspondingly.
Secondly, the extracted data were assembled in tabulated form, including the location from where the study is reported, eligible participants, timelines of recruitment, PSP levels, and correlation analysis with their respective p-value.
Results
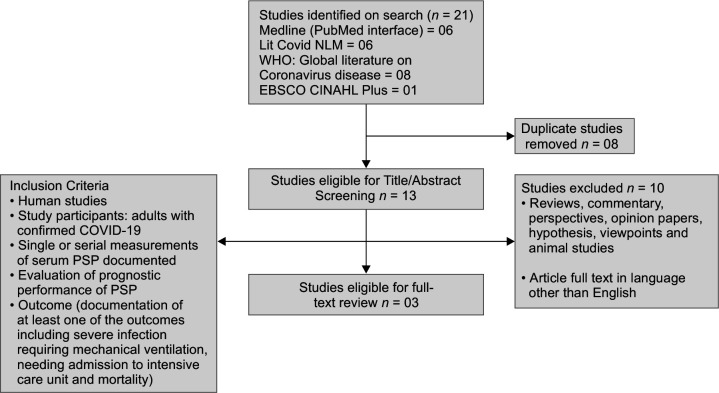
A flow diagram of the search strategy adopted and studies included is depicted in Flowchart 1. A total of 21 studies were identified, which further underwent the removal of 8 duplicate studies. The title and abstract of studies identified were further scrutinized based on the stringent exclusion criteria. Three studies fulfilled the inclusion criteria, which were included in the final analysis. The aggregate study population was summed up to be 167 COVID-19 positive cases, who had undergone analysis of plasma PSP levels for the prediction of severity and mortality. Kappa static value was yielded to be 0.90, depicting an excellent inter-investigator agreement.
Flowchart 1.
Flow diagram of selected studies for review
Summary of the literature review is provided in Table 1.3,9,14 Two studies were from Italy and one from Japan. All the included studies were published after January 2020. For the prediction of outcome, PSP cutoffs ranging from 250–2069 pg/mL were highlighted in the published literature. The QUIPS tool analysis yielded a low to moderate risk of bias for all studies as summarized domain-wise in Figure 1.
Table 1.
Summary of studies evaluated and enlisting the correlation of PSP with severity
| Author | Region | Date of recruitment | Year of recruitment initiation | Study design | Sample size (male: female) | Age (years) | PSP cutoff | PSP correlation with severity (p value) | AUC for the prediction of severity |
|---|---|---|---|---|---|---|---|---|---|
| Fukada et al.3 | Japan | February–March | 2020 | Case series | 6 | Not recorded | >250 pg/mL | p <0.05 | Not recorded |
| Schirinzi et al.14 | Italy | March–May | 2020 | Cross-sectional | 86 (58:28) | Male: 67.6 ± 12.4 Female: 65.7 ± 15.4 | 2069 pg/mL | p <0.0001 | 0.73 (95% CI: 0.684–0.786) |
| Zaninotto et al.9 | Italy | January–March | 2020 | Case series | 75 (56:19) | 67 (IQR: 56–76) | >250 pg/mL | p <0.001 | 0.72 |
Fig. 1.
Assessment of risk of bias using QUIPS tool
Discussion
Clinicians and laboratorians utilized their prior experience of biomarkers linked with respiratory viral illnesses and evaluated them widely amidst the peak pandemic months. In 2004, PSP was introduced as a new biomarker with diagnostic and prognostic utility in sepsis and SIRS.15 Since then various authors have evaluated PSP against PCT and other inflammatory biomarkers in sepsis and have reported comparable and in certain instances better performances and kinetics of PSP.16–19 The peak concentrations that are usually attained on days 1–3 of sepsis and, therefore, on admission levels can be considered to be an effective measure to predict severity. However, as shown by this review, only a few studies evaluated PSP in relation to this novel pandemic. In a recently published systematic review, PCT was found to be significantly associated with prognosis, which was further endorsed by various subsequent publications.20,21 Owing to its comparable performance with PCT, the need for the evaluation of PSP in COVID-19 was further justified.
The study population included Asians as well as Europeans, which adds strength to our findings as results can be generalized and are not specific to a certain genetic makeup subset. Additionally, the risk of bias was also acceptable as demonstrated by validation based on the QUIPS tool with excellent inter-rater agreement between the team of authors. Another distinct advantage offered by PSP is the availability of POC testing analyzer for rapid bedside testing that will aid rapid triage of patients.22
Based on a robust search, this is the first systematic review that has evaluated the utility of PSP in COVID-19, and our results revealed a significant association between PSP and severity, suggesting its further evaluation as a reliable prognostic biomarker. In addition to the limitations of less number of studies and a small aggregated sample size, a varying degree of heterogeneity was noted across the three studies. The analysis platform was also not uniform as PATHFAST Presepsin (Mitsubishi Chemical Europe GmbH, Düsseldorf, Germany) and STACIA Presepsin (LSI Medience Corporation, Tokyo, Japan) were utilized.3,9,14 The cutoffs used by the two studies were 250 pg/mL; however, Schirinzi et al. used a cutoff of 250 pg/mL and reported a much higher cutoff of 2069 pg/mL (AUC = 0.738) having sensitivity and specificity of 50 and 90%, respectively, for severe cases and a cutoff of 1179 pg/mL (AUC = 0.873) having sensitivity and specificity of 75 and 71%, respectively, for mild cases.3,9,14 Serial measurements were not undertaken for PSP; however, Schirinzi et al. showed a good correlation with human epididymis secretory protein (r = 0.621), a potential serum inflammatory biomarker having a role in natural immunity.14
Additionally, some confounding factors may influence PSP concentration, such as elderly age, renal dysfunction, superimposed bacteremia, and hemophagocytic syndrome. Therefore, PSP levels should be considered an appropriate clinical context in complementation with other laboratory parameters.
Conclusion
PSP has shown acceptable performance to predict the aggravation of severity in COVID-19, which can aid clinicians to identify high‐risk patients and determine treatment strategies at an early stage for optimal resource allocation in approximately 17 minutes by rapid POC assays. Further studies are high need of time to validate its utility with a large number of participants ideally in multinational and multicenter evaluations with serial measurement.
Authors' Contributions
S.A. conceived the idea, performed the literature search, reviewed articles, data analysis, and write-up of the work in the first draft. M.M. assisted in the writing of the first draft and table and reviewed the final draft. M.S.S. did the in-depth review of the literature included in the final analysis and assisted in the writing of the first draft and reviewed the final draft. I.S. reviewed the articles in case of conflict between S.A. and M.M. and critically revised the article for the intellectual content. All authors have reviewed the final draft and agreed upon it.
Footnotes
Source of support: Nil
Conflict of interest: None
Orcid
Sibtain Ahmed https://orcid.org/0000-0003-0316-2622
Maheen Mansoor https://orcid.org/0000-0002-8926-9318
Muhammad S Shaikh https://orcid.org/0000-0003-2938-7416
Imran Siddiqui https://orcid.org/0000-0001-6807-5429
References
- 1.Ahmed S, Jafri L, Majid H, Khan AH, Ghani F, Siddiqui I. Challenges amid COVID-19 times - review of the changing practices in a clinical chemistry laboratory from a developing country. Ann Med Surg. 2020;55:300–304. doi: 10.1016/j.amsu.2020.06.004. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu YC, Chen CS, Chan YJ. The outbreak of COVID-19: an overview. J Chin Med Assoc. 2020;83(3):217–220. doi: 10.1097/JCMA.0000000000000270. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukada A, Kitagawa Y, Matsuoka M, Sakai J, Imai K, Tarumoto N, et al. Presepsin as a predictive biomarker of severity in COVID-19: a case series. J Med Virol. 2021;93(1):99–101. doi: 10.1002/jmv.26164. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Presepsin – Mechanism. http://presepsin.com/mechanism.html. [[Accessed March 12, 2021]]. http://presepsin.com/mechanism.html Available from:
- 5.Osman AS, Awadallah MG, Tabl HA, Abed NT, Goudah ES. Presepsin as a novel diagnostic marker in neonatal septicemia. Egypt J Med Microbiol. 2015;38(3174):1–6. doi: 10.12816/0024924. DOI: [DOI] [Google Scholar]
- 6.Shozushima T, Takahashi G, Matsumoto M, Kojika M, Okamura Y, Endo S. Usefulness of presepsin (sCD14-ST) measurements as a marker for the diagnosis and severity of sepsis that satisfied diagnostic criteria of systemic inflammatory response syndrome. J Infect Chemother. 2011;17(6):764–769. doi: 10.1007/s10156-011-0254-x. DOI: [DOI] [PubMed] [Google Scholar]
- 7.Okamura Y, Yokoi H. Development of a point-of-care assay system for measurement of presepsin (sCD14-ST). Clin Chim Acta. 2011;412(23–24):2157–2161. doi: 10.1016/j.cca.2011.07.024. DOI: [DOI] [PubMed] [Google Scholar]
- 8.Ugajin M, Matsuura Y, Matsuura K, Matsuura H. Impact of initial plasma presepsin level for clinical outcome in hospitalized patients with pneumonia. J Thorac Dis. 2019;11(4):1387–1396. doi: 10.21037/jtd.2019.03.74. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaninotto M, Mion MM, Cosma C, Rinaldi D, Plebani M. Presepsin in risk stratification of SARS-CoV-2 patients. Clin Chim Acta. 2020;507:161–163. doi: 10.1016/j.cca.2020.04.020. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ham JY, Song KE. A prospective study of presepsin as an indicator of the severity of community-acquired pneumonia in emergency departments: comparison with pneumonia severity index and CURB-65 scores. Lab Med. 2019;50(4):364–369. doi: 10.1093/labmed/lmz005. DOI: [DOI] [PubMed] [Google Scholar]
- 11.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. DOI: [DOI] [PubMed] [Google Scholar]
- 12.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. doi: 10.2307/2529310. DOI: [DOI] [PubMed] [Google Scholar]
- 13.Hayden JA, Cote P, Bombardier C. Evaluation of the quality of prognosis studies in systematic reviews. Ann Intern Med. 2006;144(6):427–437. doi: 10.7326/0003-4819-144-6-200603210-00010. DOI: [DOI] [PubMed] [Google Scholar]
- 14.Schirinzi A, Cazzolla AP, Lovero R, Lo Muzio L, Testa NF, Ciavarella D, et al. New insights in laboratory testing for COVID-19 patients: looking for the role and predictive value of Human epididymis secretory protein 4 (HE4) and the innate immunity of the oral cavity and respiratory tract. Microorganisms. 2020;8(11):1718. doi: 10.3390/microorganisms8111718. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Hu L, Zhang G, Wu F, He T. Accuracy of presepsin in sepsis diagnosis: a systematic review and meta-analysis. PLoS One. 2015;10(7):e0133057. doi: 10.1371/journal.pone.0133057. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CC, Lan HM, Han ST, Chaou CH, Yeh CF, Liu SH, et al. Comparison of diagnostic accuracy in sepsis between presepsin, procalcitonin, and C-reactive protein: a systematic review and meta-analysis. Ann Intens Care. 2017;7(1):1–6. doi: 10.1186/s13613-017-0316-z. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iskandar A, Arthamin MZ, Indriana K, Anshory M, Hur M, Di Somma S. GREAT Network. Comparison between presepsin and procalcitonin in early diagnosis of neonatal sepsis. J Matern Fetal Neonatal Med. 2019;32(23):3903–3908. doi: 10.1080/14767058.2018.1475643. DOI: [DOI] [PubMed] [Google Scholar]
- 18.Abdelshafey EE, Nasa P, Elgohary AE, Khalil MF, Rashwan MA, Ghezala HB, et al. Role of presepsin for the diagnosis of sepsis and ICU mortality: a prospective controlled study. Indian J Crit Care Med. 2021;25(2):153. doi: 10.5005/jp-journals-10071-23715. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azim A. Presepsin: a promising biomarker for sepsis. Indian J Crit Care Med. 2021;25(2):117. doi: 10.5005/jp-journals-10071-23741. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed S, Jafri L, Hoodbhoy Z, Siddiqui I. Prognostic value of serum procalcitonin in COVID-19 patients: a systematic review. Indian J Crit Care Med. 2021;25(1):77. doi: 10.5005/jp-journals-10071-23706. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savio RD. Procalcitonin (in COVID-19): the incessant quest. Indian J Crit Care Medicine. 2021;25(1):1. doi: 10.5005/jp-journals-10071-23698. DOI: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chenevier-Gobeaux C, Trabattoni E, Roelens M, Borderie D, Claessens YE. Presepsin (sCD14-ST) in emergency department: the need for adapted threshold values? Clin Chim Acta. 2014;427:34–36. doi: 10.1016/j.cca.2013.09.019. DOI: [DOI] [PubMed] [Google Scholar]