Neuroserpin preferentially inhibits amyloid formation; a 14-mer region in neuroserpin is implicated in binding to amyloid.
Abstract
Neuroserpin is a secreted protease inhibitor known to inhibit amyloid formation by the Alzheimer’s beta peptide (Aβ). To test whether this effect was constrained to Aβ, we used a range of in vitro assays to demonstrate that neuroserpin inhibits amyloid formation by several different proteins and protects against the associated cytotoxicity but, unlike other known chaperones, has a poor ability to inhibit amorphous protein aggregation. Collectively, these results suggest that neuroserpin has an unusual chaperone selectivity for intermediates on the amyloid-forming pathway. Bioinformatics analyses identified a highly conserved 14-residue region containing an α helix shared between neuroserpin and the thyroxine-transport protein transthyretin, and we subsequently demonstrated that transthyretin also preferentially inhibits amyloid formation. Last, we used rationally designed neuroserpin mutants to demonstrate a direct involvement of the conserved 14-mer region in its chaperone activity. Identification of this conserved region may prove useful in the future design of anti-amyloid reagents.
INTRODUCTION
Various physical and chemical stresses in the body, and mutations, can lead to the misfolding of proteins, which then expose regions of hydrophobicity to the surrounding solvent and bind via hydrophobic interactions with other nearby misfolded protein molecules. These aggregating species have the potential to form various stable structures, including soluble aggregates and insoluble deposits. Depending on the individual protein and the prevailing conditions, the protein aggregates formed may be either unstructured (amorphous) or consist of β sheet–rich fibrillar structures (amyloid). All of these protein structures have been found associated with disease states. For example, soluble oligomeric protein aggregates have been implicated as cytotoxic species in various neurodegenerative disorders, and insoluble protein deposits play an important role in the pathologies of a variety of protein deposition diseases (e.g., diabetes type 2 and amyloidoses) (1). In recent years, it has become clear that there is a growing family of abundant chaperone proteins in the extracellular fluids of metazoans that are able to bind to, and keep soluble, proteins that are misfolded as a result of mutations or stresses to inhibit their aggregation and toxicity. Furthermore, these extracellular chaperones are strongly implicated in clearing aggregating proteins from extracellular spaces and facilitating their degradation (2). Thus, the discovery and characterization of the action of new extracellular chaperones is highly relevant to understanding the mechanisms of extracellular protein homeostasis (proteostasis) and may help advance us toward the development of new therapies for serious human diseases.
Neuroserpin (NS) is a secreted, monomeric protein, approximately 46 kDa in mass, primarily expressed throughout the nervous system and secreted into the cerebrospinal fluid (CSF), where it is present at a concentration of ~5 to 10 ng/ml (3, 4). NS is well known as a serine protease inhibitor, and its three-dimensional structure includes three β sheets (A, B, and C), nine α helices, and a major reactive center loop (RCL), which is the known binding site for target proteases (5). NS preferentially interacts via its RCL with tissue plasminogen activator (tPA) (6). The expression of tPA increases in animal models of stroke and epilepsy, and administration of NS reduces epileptic seizures in animal models (7). In contrast, certain mutants of NS polymerize in vivo to form deposits associated with familial encephalopathy with NS inclusion bodies (8). Thus, while wild-type NS can exert important neuroprotective effects, destabilization of its native state can result in undesired pathologies. NS also colocalizes with amyloid β (Aβ) plaques in the brains of patients with Alzheimer’s disease and has been reported to reroute the aggregation pathway of Aβ peptide away from amyloid fibrils, an ability that was shown to protect cultured neuronal cells from Aβ toxicity (9).
Multiple nonchaperone proteins have been shown to influence the aggregation and toxicity of the hydrophobic Aβ peptide (10). We therefore asked whether the effects of NS on Aβ could result from a genuine chaperone activity capable of inhibiting the aggregation of a range of different misfolding proteins. If this were true, then it would establish NS as a member of a growing family of extracellular chaperones implicated in protecting the human body from a range of serious diseases arising from the inappropriate aggregation of proteins in body fluids (1, 2). NS was tested in a range of in vitro assays to assess its ability to inhibit the aggregation of a variety of proteins proceeding via either the amorphous or amyloid-forming pathways. In contrast to most typical molecular chaperones, the results indicated that NS can efficiently inhibit amyloid formation by a range of different proteins and is protective against the associated cytotoxicity, but it has limited ability to inhibit amorphous protein aggregation.
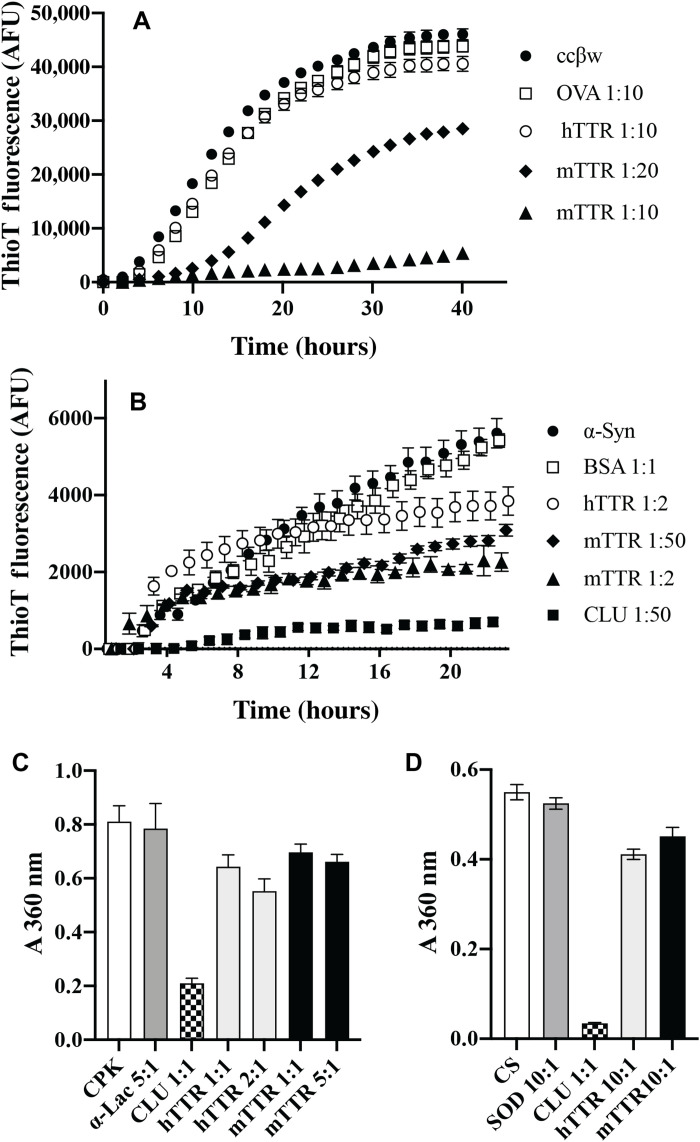
Next, to explore whether other amyloid-selective extracellular chaperones may exist and to investigate the molecular basis of the observed anti-amyloid activity of NS, we performed bioinformatics analyses to identify possible similarities with other proteins known to inhibit amyloid formation. We identified in this way a short 14-mer sequence containing an α helix that is highly conserved between NS and transthyretin (TTR). This region in TTR had earlier been proposed as a binding site for Aβ (11, 12), although this suggestion has been challenged (13, 14). TTR is a 55-kDa secreted homotetrameric protein, rich in β sheet structure and approximately 55 kDa in mass, present in plasma and CSF at concentrations of 0.2 to 0.4 mg/ml and 0.02 to 0.04 mg/ml, respectively (15). Human TTR (hTTR) exists in vivo predominantly in its native tetrameric form in equilibrium with monomeric TTR (mTTR), which, especially in the case of certain pathological TTR mutations, is suggested to assemble to form amyloid deposits in familial amyloidotic polyneuropathy (16, 17). TTR is known to play a key role in transporting thyroxine, and a complex formed between retinol and retinol binding protein, via the blood to different parts of the body (18). Previous studies reported that TTR inhibits the in vitro formation of amyloid by Aβ (14) and CsgA (a protein precursor subunit found in Enterobacteriaceae biofilms) (13) and also inhibits Aβ cytotoxicity (19), but the ability of TTR to inhibit amorphous protein aggregation was untested. We performed additional in vitro analyses to show that a genetically engineered mTTR (which cannot form tetramers) reduced amyloid formation more efficiently than tetrameric hTTR but that both were poorly effective at inhibiting amorphous protein aggregation. These results confirmed that NS and TTR share both a highly conserved 14-mer region containing an α helix and a previously unknown amyloid-selective chaperone activity.
We extended these studies to examine whether the conserved 14-mer sequence in NS might be directly involved in its interactions with amyloid. Rationally designed point mutations in this region did not alter the overall structure of NS but substantively reduced its ability to inhibit amyloid formation and to protect cells from amyloid-associated toxicity, strongly implicating this region as playing a direct role in the anti-amyloidogenic activity. Collectively, our results have identified NS and TTR as extracellular chaperones that preferentially interact with amyloid-forming proteins and identified a short region in the NS protein as a likely binding site for amyloid species.
RESULTS
Recombinant NS is an active protease inhibitor
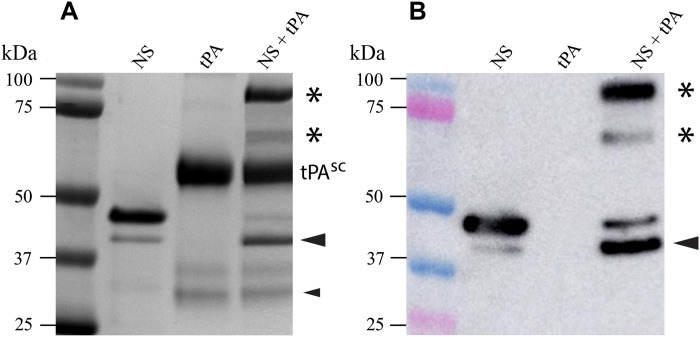
Wild-type NS of the expected size was purified by nickel affinity chromatography to near homogeneity, as judged by SDS–polyacrylamide gel electrophoresis (SDS-PAGE); a minor band of lower molecular weight, probably representing a cleavage product, was variably detected in different production batches (Fig. 1A) and has been previously reported for NS expressed in bacteria (20). The identity of NS was confirmed by immunoblotting (Fig. 1B), and its enzymatic activity was confirmed by SDS-PAGE and immunoblotting analyses of mixtures of NS and tPA, which demonstrated the expected formation of covalent NS-tPA complexes (Fig. 1, A and B). The commercial Actilyse (Boehringer-Ingelheim) used to provide tPA contains both the single-chain (sc) and the two-chain (tc) forms of tPA, although mostly the sc form. Both forms of tPA rapidly form acyl-enzyme complexes with NS in vitro; the interactions are short-lived, rapidly progressing to complete cleavage of NS and regeneration of fully active enzyme (21). Under the conditions tested, complexes formed between NS and both sc- and tc-tPA were detected, together with cleaved NS (Fig. 1).
Fig. 1. Detection of covalently linked NS-tPA complexes.
(A) Nonreducing 10% SDS-PAGE analysis of purified wt-NS (NS), tPA, and a mixture of NS and tPA. Molecular markers are indicated on the left side of the image, and the sample loaded into each well is indicated at the top of the image. The position of sc tPA (tPAsc) is indicated; the Actilyse tPA preparation also appears to contain small amounts of twin-chain tPA (tPAtc) dissociated into the A and B chains (lowest two bands migrating at ~31 and 28 kDa, respectively). When NS was incubated with tPA, two high–molecular weight, SDS-stable species formed (asterisks) and an increase in cleaved NS was also apparent (large black arrowhead). The small black arrowhead indicates the position of the B chain of tPAtc containing the serine protease domain that reacted with NS to produce the faster-migrating NS-tPA complex (lower asterisk). (B) Immunoblotting using an antibody specific for NS confirmed that the high–molecular weight complexes (asterisks) contained NS and only form when NS is incubated with tPA. The larger complex (upper asterisk) corresponds to one containing tPAsc, while the smaller complex (lower asterisk) corresponds to one formed between NS and the B chain of tPAtc.
NS inhibits amyloid formation by a variety of proteins
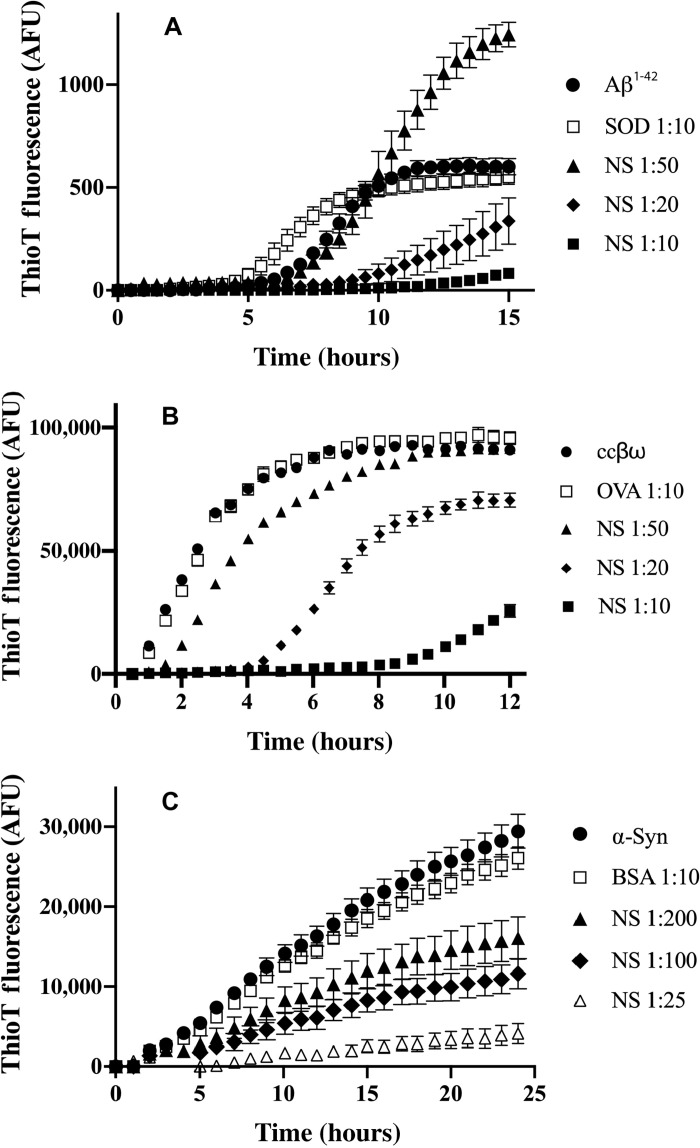
NS showed specific, dose-dependent inhibition of the in vitro formation of amyloid by Aβ1-42, coiled-coil βω peptide (ccβω) and α-synuclein (α-syn) (Fig. 2). In each case, when used at the highest molar ratio tested, the nonchaperone control proteins [superoxide dismutase (SOD), ovalbumin (OVA), or bovine serum albumin (BSA)] did not significantly decrease the endpoint level of thioflavin T (ThioT) fluorescence. Relative to Aβ1-42 alone, SOD caused a small increase in ThioT fluorescence between 5 and 9 hours, but by 10 to 15 hours, this difference was no longer significant (Fig. 2A). At the lowest molar ratio of NS:Aβ1-42 tested (1:50), relative to Aβ alone, NS increased the levels of ThioT fluorescence after about 10 hours of incubation but significantly inhibited the production of ThioT fluorescence when tested at the higher ratios of 1:20 and 1:10 (Fig. 2A). NS also showed a clear dose-dependent inhibition of the generation of ThioT fluorescence associated with amyloid formation by both ccβω and α-syn (Fig. 2, B and C). When incubated alone under any of the conditions used in these amyloid-forming assays, NS generated negligible levels of ThioT fluorescence (fig. S1).
Fig. 2. Effects of NS on the in vitro formation of amyloid by Aβ1-42, ccβω, and α-syn.
Plots of ThioT fluorescence [arbitrary fluorescence units (AFU)] as a function of time for aggregation reactions containing (A) Aβ1-42, (B) ccβω, and (C) α-syn. The aggregation reactions for Aβ1-42 and ccβω were unseeded but that for α-syn was seeded (see Materials and Methods). Nonchaperone control proteins (SOD, OVA, and BSA) and the molar ratio of these or NS to the aggregating protein are indicated in the respective keys. When incubated alone under these conditions, NS did not generate any significant ThioT fluorescence (fig. S1). Data points plotted are means ± SEM (n = 3), and each result shown is representative of two to three independent experiments.
NS has limited effects on amorphous protein aggregation
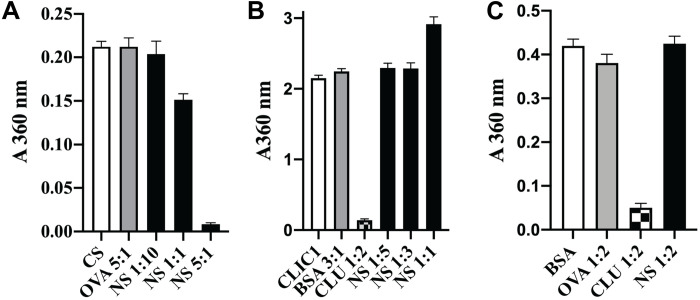
In contrast to the results obtained with amyloid-forming proteins, NS was either ineffective or of limited potency as a specific inhibitor of the amorphous aggregation of a variety of different proteins tested, including citrate synthase (CS), cysteine-less chloride intracellular channel protein (CLIC1) (22), and BSA. For CS, a fivefold molar excess of NS was required to achieve substantial inhibition of aggregation, a 1:1 ratio only achieving ~25% inhibition under the conditions tested (Fig. 3A). NS was completely unable to inhibit CLIC1 aggregation and, when present at an equimolar level to CLIC1, actually slightly increased the endpoint absorbance value (Fig. 3B). In contrast, when the extracellular chaperone clusterin (CLU) was added to give CLU:CLIC1 = 1:2, it inhibited ~95% of CLIC1 aggregation (Fig. 3B). Similarly, although CLU inhibited ~90% of the amorphous aggregation of BSA when present at a CLU:BSA = 1:2, at the same molar ratio, NS had no significant effect (Fig. 3C). As NS lacks any cysteine residues or disulfide bonds, its structure is expected to be unaffected by the reducing conditions used to induce the amorphous aggregation of BSA. Representative examples of complete protein aggregation assay curves for these results are provided in fig. S2. NS is known to undergo heat-induced polymerization (23); however, under the conditions used to induce proteins to form either amyloid or amorphous aggregates, negligible NS polymerization was detected (fig. S3).
Fig. 3. NS has limited ability to inhibit amorphous protein aggregation.
(A to C) Histograms showing the endpoint absorbance values (A360 nm), which were used to measure the amorphous aggregation of CS (A), CLIC1 (B), and BSA (C). See fig. S2 for the corresponding complete protein aggregation assay time courses. Nonchaperone control proteins (OVA, BSA, and SOD) and the molar ratio of these, CLU or NS to the aggregating protein, are indicated on the respective plots. Data points plotted are means ± SEM (n = 3), and each result shown is representative of two independent experiments.
NS contains a 14-mer region highly homologous with a proposed TTR binding site for Aβ
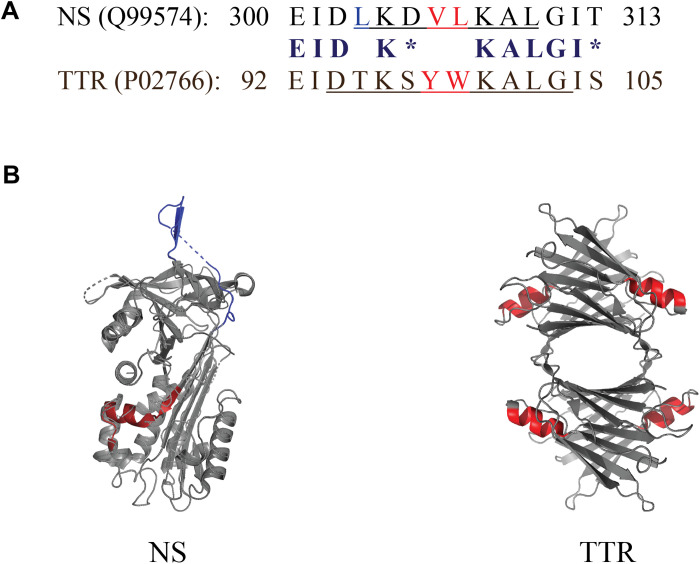
Pairwise sequence alignment of NS and TTR sequences obtained from UniProtKB/Swiss-Prot performed using the National Center for Biotechnology Information (NCBI) Align Sequences Protein BLAST tool revealed a homologous 14-residue sequence, which incorporates an α helix on both NS (residues 300 to 313) and TTR (residues 92 to 105; Fig. 4A) (24–26). The specific amino acid residues corresponding to the α helix on NS (LKDVLKAL; obtained from UniProtKB/Swiss-Prot) and TTR (DTKSYWKALG) (11) are underlined in each sequence in Fig. 4A. Each conserved region shares 9 identical and 2 similar residues; thus, in this 14-mer motif, 11 residues (~79%) are identical or similar. PyMOL was used to generate three-dimensional renderings of the structures of both NS and TTR, and the 14 residues corresponding to the homologous region are highlighted in red on each structure (Fig. 4B). The structure of these conserved regions is notably similar between the two proteins (fig. S1A). Note that in each of these regions, the three positions occupied by nonidentical/nonsimilar residues are not exposed to the solvent (fig. S1B) and are therefore unlikely to be directly involved in interactions with chaperone client proteins. This region in TTR was proposed as an Aβ binding site (11, 12), although more recent work has challenged this suggestion (13, 14). In NS, this region is located distant from the interface involved in its formation of dimers (fig. S4) (23).
Fig. 4. Short conserved region shared between NS and TTR.
(A) Comparison of the sequences of NS and TTR corresponding to the conserved 14-mer region. Identical residues are indicated in bold, chemically similar residues (blosum62 score equal to or greater than 0) are indicated by asterisks, and hydrophobic residues are shown in red. The specific residues corresponding to an α helix are underlined. Residue numbering includes the N-terminal signal peptides in both cases (16 residues for NS and 20 residues for TTR). (B) PyMOL-generated structures of NS and TTR indicating the position of the 14-residue homologous region (shown in red), which incorporates an α helix in both NS and TTR (four α helices in TTR, one per identical monomeric subunit). The PyMOL rendering of NS has two small regions (indicated by dashed lines) for which the structure was not resolved; the NS RCL is shown in dark blue at the top of the image.
TTR also has a molecular chaperone activity that preferentially inhibits amyloid formation
TTR had previously been reported to inhibit amyloid formation by both Aβ and the bacterial biofilm scaffold protein CsgA (13, 14), but its ability to inhibit amorphous protein aggregation was unknown. mTTR is a significantly more potent inhibitor of Aβ and CsgA amyloid aggregation than tetrameric hTTR (13, 14, 27, 28). We first examined whether hTTR/mTTR had the ability to inhibit amyloid formation by two other proteins, ccβω and α-syn, and then their ability to inhibit amorphous protein aggregation. mTTR showed specific dose-dependent inhibition of amyloid formation by ccβω, reducing the endpoint ThioT fluorescence by ~90% when present at a molar ratio of mTTR:ccβω = 1:10 (Fig. 5A). At the same molar ratio, relative to ccβω alone, both hTTR and OVA (a nonchaperone control protein) had little effect on the level of ThioT fluorescence (Fig. 5A). mTTR showed a less potent ability to inhibit the seeded aggregation of α-syn, reducing endpoint ThioT fluorescence by ~60% when present at a molar ratio of mTTR:ccβω = 1:2. Relative to α-syn alone, at the same molar ratio, hTTR actually increased the level of ThioT fluorescence at 3 to 8 hours but reduced endpoint ThioT fluorescence by ~30%. In contrast, a molar ratio of CLU:α-syn = 1:50 reduced endpoint ThioT fluorescence by ~87% (Fig. 5B). In the same assay, an equimolar concentration of the nonchaperone control protein BSA had no significant effect on ThioT fluorescence. As expected, in amorphous protein aggregation assays, a 1:1 molar ratio of CLU to creatine phosphokinase (CPK) or CS provided very substantial inhibition of aggregation. However, even when used at a molar excess of from 2:1 up to as high as 10:1, neither hTTR nor mTTR reduced CPK or CS aggregation by more than ~25% (Fig. 5, C and D). At the highest molar ratios tested, the nonchaperone control proteins [α-lactalbumin (α-lac) for CPK and SOD for CS] had negligible effects on protein aggregation. Complete amorphous protein aggregation assay curves corresponding to these results are provided in fig. S5.
Fig. 5. TTR inhibits amyloid formation but has limited effects on amorphous protein aggregation.
(A and B) Plots of ThioT fluorescence (AFU) as a function of time for aggregation reactions containing (A) ccβω and (B) α-syn. Nonchaperone control proteins (OVA and BSA) and the molar ratio of these or hTTR/mTTR to the aggregating protein are indicated in the respective keys. (C and D) Histogram plots showing absorbance values (A360 nm) used to quantify the amorphous aggregation of CPK (at 5 hours) (C) and CS (at 1.5 hours) (D). See fig. S5 for the corresponding complete aggregation time courses. Nonchaperone control proteins (α-lac and SOD) and the molar ratio of these, CLU or hTTR/mTTR to the aggregating protein, are indicated on the respective plots. For (A) to (D), data points plotted are means ± SEM (n = 3), and each result shown is representative of two independent experiments. In some cases, the error bars are too small to be visible.
Generation and structural characterization of rationally designed NS mutants
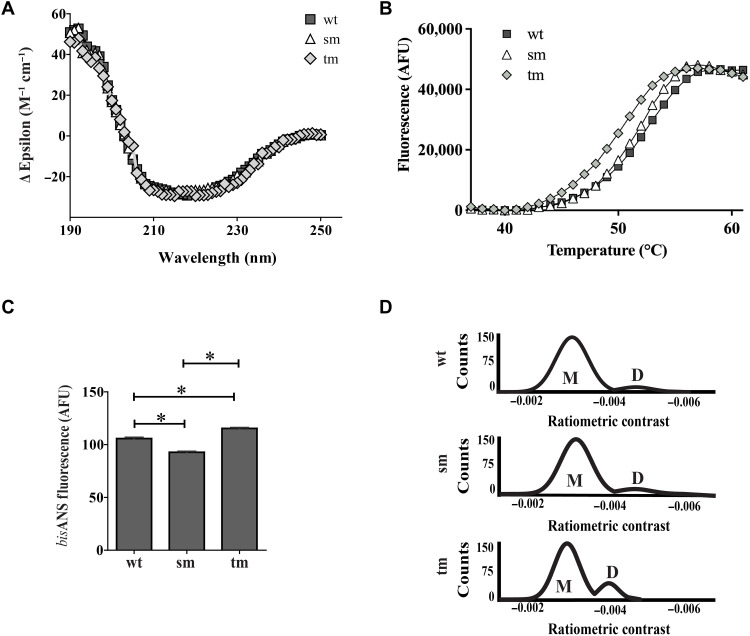
To investigate the possibility that the 14-mer conserved region might be, at least in part, responsible for the amyloid-selective chaperone action of NS, we rationally designed mutations to solvent-exposed residues in this region that were identified as least likely to affect the structure of the region while being solvent-exposed and harboring large physicochemical changes to maximally affect any activity (see fig. S6 and associated text). Two NS mutants were expressed and purified, K308E (single mutant, NSsm) and K304A, K308E, L310Q (triple mutant, NStm), and these behaved similarly to wild-type NS (NSwt) when analyzed by SDS-PAGE, native gel electrophoresis, and immunoblotting (fig. S7). These proteins were further analyzed by far ultraviolet (UV) circular dichroism (CD) spectrophotometry (to assess secondary structure content and stability at different temperatures), bisANS fluorescence assays (to measure levels of solvent-exposed hydrophobicity), and mass photometry (to measure solution size) (Fig. 6). At 37°C, for NSwt, NSsm, and NStm, the far UV CD spectra collected using the spectral measurement program (plotted as mean residue ellipticity) were virtually superimposable (Fig. 6A), indicating little difference in secondary structure content (table S1). Subsequently, to detect any differences in thermal stability, we used a thermal shift assay, which measures an increase in SPYRO Orange fluorescence when this dye binds to hydrophobic regions exposed on proteins as they unfold with increasing temperature (29). Using this approach, under the conditions tested, the calculated apparent “melting temperatures” (Tm) for NSwt, NSsm, and NStm were 51.8°, 51.3°, and 49.3°C, respectively; NStm was significantly less stable than both NSwt and NSsm (Fig. 6B and table S2). BisANS fluorescence assays indicated that although all three proteins had similar levels of solvent-exposed hydrophobicity, relative to NSwt, this level was slightly reduced for NSsm but slightly increased for NStm (both differences were statistically significant; P < 0.05, Student’s t test, pairwise comparisons; Fig. 6C). We also compared the relative abilities of NSwt and the two mutants to interact with tPA; we found that both mutants were still able to interact with tPA, albeit at a similarly lower efficiency when compared with NSwt (fig. S8). Last, mass photometry analysis suggested that the dominant species in solution for all three molecules was a monomer. However, a small population of putative dimer was also detected in all three samples analyzed; this was most abundant for NStm (~17%) compared with NSsm (~9%) and NSwt (~8%) (Fig. 6D). Ratiometric contrast is defined as the ratio of light scattered by a molecule at its interface with the measurement surface to that of light reflected at that surface and can be affected by both refractive index and mass (30). The ratiometric contrast values for the two populations of particles detected for NStm appeared slightly lower than those detected for NSwt and NSsm; this difference may result from small changes in refractive index and/or mass associated with partial destabilization of the NStm molecule. Collectively, the structural analyses suggested that the chosen mutations did not substantively change overall NS structure; however, the thermal shift assay and mass photometry results suggest that the triple mutation partially destabilized NStm and led to an increase in its formation of dimers.
Fig. 6. Biophysical characterization of wild-type NS and mutants.
(A) Far UV CD spectra of NSwt (wt), NSsm (sm), and NStm (tm), plotted as mean residue ellipticity against wavelength at 37°C. Means ± SEM (n = 10) are plotted. (B) Results of a thermal shift assay, measuring SPYRO Orange fluorescence (AFU) as a function of temperature for NSwt (wt), NSsm (sm), and NStm (tm). Means ± SEM (n = 4) are plotted. For (A) and (B), each result is representative of at least two independent experiments and the error bars are too small to be visible. (C) BisANS analysis of wt, sm, and tm NS. Mean fluorescence values (AFU) ± SEM (n = 3) are plotted; error bars are too small to be easily seen. Asterisks indicate statistically significant differences (Student’s t test, pairwise comparisons, P < 0.05). Results are representative of three independent experiments. (D) Mass photometry analyses showing the distribution of putative monomeric (M) and dimeric (D) forms of wt, sm, and tm NS. Results are representative of three independent experiments.
Mutations in the 14-mer conserved region reduced the ability of NS to inhibit amyloid aggregation and associated cytotoxicity
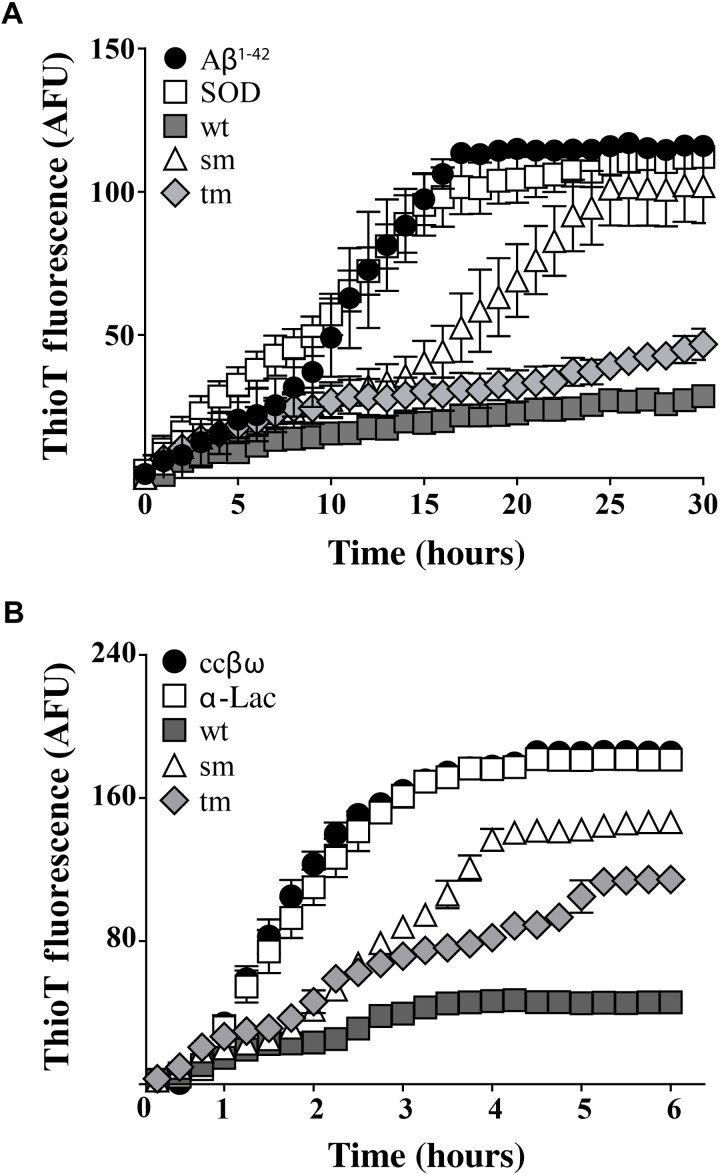
The single and triple mutations introduced into the conserved 14-mer region of NS both reduced the ability of NS to inhibit in vitro amyloid formation by Aβ1-42 and ccβω (Fig. 7). Under the conditions tested, relative to Aβ incubated with the nonchaperone control protein SOD, NSwt reduced the endpoint ThioT fluorescence by ~74%. In contrast, NSsm significantly delayed Aβ1-42 aggregation but, at the endpoint, had reduced ThioT fluorescence by only ~10% (Fig. 7A). The ability of NStm to reduce endpoint ThioT fluorescence was greater in these assays than NSsm, achieving a ~58% reduction (still significantly less than NSwt; P < 0.001, Student’s t test). Similarly, relative to ccβω incubated with the nonchaperone control protein α-lac, NSwt reduced endpoint ThioT fluorescence by ~74%, while the corresponding reductions for NSsm and NStm were ~19 and ~26%, respectively (Fig. 7B). We also tested the abilities of NSwt, NSsm, and NStm to bind to intermediates formed during Aβ1-42 aggregation, which revealed a similar pattern: The binding of NSsm to Aβ1-42 intermediates was significantly less than that of NSwt, while the reduction in binding for NStm was less pronounced (fig. S9).
Fig. 7. Effects of mutations in the conserved 14-mer region on the ability of NS to inhibit in vitro amyloid formation.
ThioT fluorescence (AFU) was used to monitor the formation of amyloid by (A) Aβ1-42 and (B) ccβω. The molar ratio of nonchaperone control proteins (SOD and α-lac) or NSwt/NSsm/NStm to the aggregating protein was, in all cases, 1:10. Data plotted are means ± SEM (n = 3). In some cases, the error bars are too small to be visible. Each result shown is representative of two independent experiments.
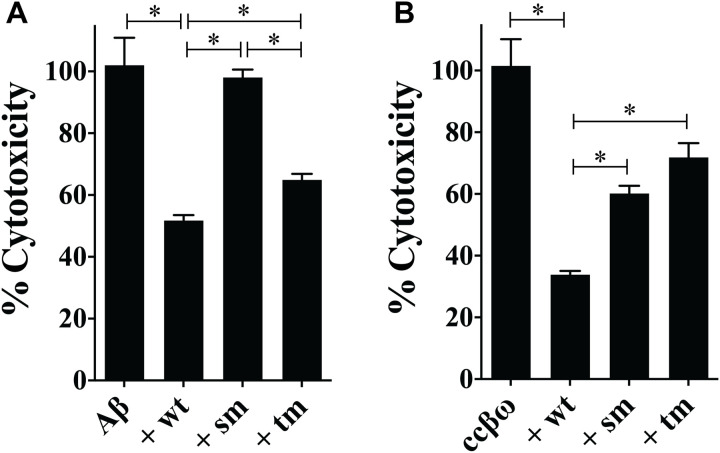
Last, the single and triple mutations were found to significantly reduce the ability of NS to protect cells against toxic species generated during the formation of amyloid by Aβ1-42 and ccβω. Under the conditions tested, NSwt reduced the cytotoxicity of Aβ1-42 by ~50% but NSsm effectively lost any protective ability (Fig. 8A). In contrast, NStm retained some ability to protect against Aβ1-42, reducing its cytotoxicity by ~37% (Fig. 8A). In the case of ccβω, NSwt reduced cytotoxicity by ~68%, and the corresponding lesser reductions for NSsm and NStm were 41 and 29%, respectively (Fig. 8B).
Fig. 8. Effects of mutations in the conserved 14-mer region on the ability of NS to inhibit amyloid cytotoxicity.
Samples taken from aggregation mixtures of Aβ1-42 and ccβω were incubated with (or without) NSwt/NSsm/NStm before adding to NSC34 cells; cell viability was later quantified using calcein-AM as described in Materials and Methods. In all cases, the molar ratio of NS:Aβ1-42 or NS:ccβω was 1:10. Results are presented as percent cytotoxicity relative to Aβ1-42 or ccβω alone. Under the conditions tested, relative to the control, the percentage of viable cells for cultures treated with Aβ1-42 or ccβω alone were 45 and 31%, respectively. Values plotted represent means ± SEM (n = 16). Statistically significant differences have been indicated by an asterisk [one-way analysis of variance (ANOVA), P < 0.05]. The results shown are each representative of two independent experiments.
DISCUSSION
We have previously reported the discovery of a growing family of chaperones present in human body fluids at μg-mg/ml levels, including CLU (31, 32), haptoglobin (33), and α2-macroglobulin (34). These extracellular chaperones are likely to be involved in binding to toxic species formed during extracellular protein aggregation to neutralize their toxicity and stabilize them in a soluble nontoxic form before mediating their safe clearance by receptor-mediated endocytosis and lysosomal degradation (2, 35). In the current study, we developed a new optimized expression system to produce enzymatically active recombinant human NS (Fig. 1) and demonstrated that at 20 μM Aβ1-42, a molar ratio of NS:Aβ1-42 = 1:10 was sufficient to nearly abolish the generation of ThioT fluorescence in Aβ1-42 aggregation assays. Furthermore, under the conditions tested, a 1:20 molar ratio of NS:Aβ1-42 reduced Aβ1-42 cytotoxicity by ~50%. Our results confirm that NS inhibits Aβ amyloid formation and cytotoxicity and further demonstrate that NS potently inhibits amyloid formation by two other proteins, ccβω and α-syn, and substantively protects neuroblastoma cells against cytotoxic ccβω species generated during amyloid formation (Figs. 2 and 8). The observation that, at the lowest molar ratio of NS:Aβ tested (1:50), NS actually increased the level of ThioT-reactive Aβ1-42 species (Fig. 2A) is consistent with a similar behavior that we reported previously for the effects of very low levels of CLU on Aβ1-42 amyloid formation (36). One speculative explanation for this effect is that when present at limiting, very low levels, these chaperones may facilitate amyloid formation by stabilizing small aggregates (amyloid nuclei) that act as seeding points for fibril formation. Collectively, the results presented suggest that NS has a broad capacity to inhibit the generation of amyloid by a range of structurally unrelated proteins and the associated cytotoxicity.
Previously described extracellular chaperones all have a broad and promiscuous ability to inhibit many different proteins from aggregating by either the amyloid or amorphous pathways (2). On the basis of the molar ratio of chaperone to client protein required to inhibit protein aggregation, many chaperones (including CLU) appear to be more efficient inhibitors of amyloid formation than amorphous aggregation. This results from the fact that during the early stages of amyloid formation, typically only a very small fraction of the protein molecules in solution (~1 to 2%) are actually misfolded (37), making the job of the chaperone less challenging than in the case of amorphous protein aggregation. As an example, CLU has been shown to inhibit amyloid formation by a variety of proteins when present at molar ratios of CLU:client protein ranging from 1:10 to 1:50 (36) and to inhibit the amorphous aggregation of different client proteins at lesser but still often substoichiometric molar ratios, for example, between ~1:2 to 1:4 (32) and ~1:1 to 1:2 (Figs. 3, B and C, and 5, C and D). We found that while NS was an effective inhibitor of in vitro amyloid formation by several different proteins (Aβ1-42, ccβω, and α-syn), it was, to an unexpected extent, much less effective at inhibiting amorphous protein aggregation. For three amorphously aggregating proteins tested (CS, CLIC1, and BSA), even when NS was added at (or near to) an equimolar concentration, it had limited effects on protein aggregation. For comparison, in two of these cases (CLIC1 and BSA), we tested the effects of adding similar levels of CLU and showed that it almost completely abolished all protein aggregation (Fig. 3). We have previously published similarly potent effects of CLU on the aggregation of CS (38). Only when NS was added at a fivefold molar excess to CS did it abolish almost all protein precipitation. Collectively, these results suggest that NS preferentially interacts with amyloid-forming proteins, in that it can efficiently inhibit amyloid formation by a variety of proteins but is generally much less effective in the context of amorphous protein aggregation.
Bioinformatic analysis identified a short 14-mer region conserved between NS and TTR, which, in both molecules, contains an α helix (Fig. 4) and, in TTR, has been proposed to act as an Aβ binding site (11, 12). This conserved region suggested to us the possibility that NS and TTR might share functional similarities. As TTR had itself earlier been reported as having the ability to inhibit amyloid formation by Aβ and CsgA, we further examined the chaperone abilities of TTR and found that, under the conditions tested, the tetrameric form of the molecule (hTTR) had limited chaperone activity but that the less abundant monomeric form (mTTR) has a potent, dose-dependent ability to inhibit ccβω amyloid formation (Fig. 5A). Previous studies have reported similar more potent effects of mTTR on amyloid formation than tetrameric hTTR and have proposed that dissociation of tetrameric TTR to a monomer enhances its chaperone activity (13, 14, 28). Although mTTR partially inhibited the seeded aggregation of α-syn, there was limited dose dependence and, even at a molar ratio of mTTR:α-syn = 1:2, endpoint ThioT fluorescence was only reduced by ~60%. In contrast, under the same conditions, a molar ratio of CLU:α-syn = 1:50 reduced endpoint ThioT fluorescence by nearly 90% (Fig. 5B). Therefore, mTTR appears less effective at inhibiting seeded α-syn aggregation than unseeded amyloid formation by ccβω. Further studies will be required to determine the specific amyloid species targeted by mTTR. The reasons why hTTR modestly increased the level of ThioT fluorescence at early time points during α-syn aggregation (hTTR:α-syn = 1:2) but reduced endpoint ThioT fluorescence by ~30% are unclear but presumably arise from limited interactions between hTTR and intermediate species formed during the α-syn aggregation process.
The apparent ability of NS and TTR to generally more effectively inhibit amyloid-forming protein aggregation than amorphous protein aggregation cannot be explained by the proposal that they preferentially interact with long-lived intermediates present only in kinetically slow protein aggregation reactions. In all cases tested, the amyloid-forming reactions required a period of many hours to reach maximum ThioT fluorescence (e.g., Aβ, 10 to 15 hours; ccβω, 5 to 10 hours; and α-syn, >20 hours; see Fig. 2). If anything, the amorphous aggregation reactions were, on average, a little faster (CS, 3 hours; CLIC1, 5 hours; CPK, 7 hours; and BSA, 18 hours; see Materials and Methods), nor were there any systematic differences in the conditions used for the amyloid-forming versus amorphous aggregation reactions. In most cases, aggregation reactions were performed in phosphate-buffered saline (PBS) or a phosphate buffer at pH 7.4 [the only exceptions being ccβω in Hepes buffer (pH 7.5) and CS in a tris buffer (pH 8.0)]. Similarly, there were no systematic differences in the incubation temperature or whether the reactions were shaken or not (see Materials and Methods). Therefore, together, our results strongly suggest that NS and TTR are extracellular chaperones that preferentially interact with intermediates on the amyloid-forming pathway regardless of their amino acid sequence. To the best of our knowledge, the only other chaperone reported to have a comparable preference for interacting with amyloid-forming species is the bacterial secreted chaperone CsgC, which is restricted in its ability to inhibit amyloid formation by the sequence of the aggregating protein (CsgC inhibits amyloid formation by α-syn but not Aβ) (39). We next examined the possibility that the short 14-mer sequence conserved between NS and TTR might be directly involved in the interactions of NS with amyloid species. We expressed and purified two rationally designed NS mutants (NSsm and NStm) and, in a variety of structural assays, did not detect any substantive changes in structure between the mutants and wild-type NS (Fig. 6A and fig. S7). However, the thermal shift assay indicated that NStm was significantly less stable than both NSwt and NSsm (Fig. 6B and table S2). Furthermore, mass photometry analyses indicated that for all three NS proteins, ~80 to 90% of species in solution were of the same size (putative monomer) but that the abundance of the only other species detected (putative dimer) was about twofold greater for NStm than either of the other two proteins (Fig. 6D). Thus, collectively, the structural analyses suggested that relative to NSwt and NSsm, the NStm may be partially destabilized and that this leads to the association of partly misfolded structures, resulting in a greater abundance of NStm dimers.
These mutants were then compared with NSwt for their relative abilities to inhibit amyloid formation and cytotoxicity. When tested for their effects on in vitro amyloid formation by Aβ1-42 and ccβω, both mutants were less effective inhibitors than NSwt (Fig. 7). It was the single point mutant NSsm (K308E) that showed the greatest reduction in chaperone efficiency, followed by the triple mutant NStm (K304A, K308E, L310Q). Similarly, relative to NSwt, both mutants showed a significant reduction in their ability to protect cells from cytotoxic species generated during amyloid formation by Aβ1-42 and ccβω (Fig. 8). Notably, the single K308E mutation completely abolished the ability of NS to protect from Aβ1-42 cytotoxicity, while the cytoprotective activity of NStm was ~26% less than NSwt (Fig. 8A). NStm was slightly less effective than NSsm in protecting cells from cytotoxic ccβω species (although this difference was not statistically significant), and both were significantly less effective than NSwt (Fig. 8B). Our initial expectation was that NStm would show greater loss of ability to inhibit amyloid formation and toxicity than NSsm; however, the converse was found to be the case. We do not currently have sufficient data to unequivocally identify the reasons behind this. One possibility, however, is that the increased level of dimers detected for NStm in solution (Fig. 6D) may, by virtue of increased avidity, bind more tightly to amyloidogenic species, thereby leading to greater inhibition of amyloid formation and toxicity than NSsm. Regardless of the reasons for the differences between NStm and NSsm, both mutational variants remain significantly less active than NSwt. In particular, as NSsm and NSwt have the same thermal stability, the results strongly support our hypothesis that the 14-mer conserved region in NS is involved in its interactions with intermediate species formed during amyloid formation.
Analysis of the 8-mer α-helical region contained within this conserved sequence in NS indicates that it is an amphipathic α helix (fig. S10), a structural feature often implicated in binding to regions of exposed hydrophobicity such as those present on misfolding proteins (40, 41). The corresponding α-helical region in TTR may also be functionally amphipathic (fig. S10). Future studies will be needed to clarify whether the short, conserved region in TTR has any involvement in its amyloid-selective chaperone activity. Most previous in vitro studies of NS, including the current study, have used recombinant NS expressed in Escherichia coli. NS is secreted from eukaryotic cells, however, as a glycoprotein bearing N-linked sugars attached at two points, including at N321 which is located ~10 residues C-terminal to the 14-mer sequence implicated in the interactions of NS with amyloid species (42). Whether the sugars conjugated to NS have any effects on its chaperone activity is a valid question to be addressed in the future. To conclude, this study represents the first to report that NS and TTR are two extracellular chaperones with an unusual ability to preferentially interact with amyloid-forming proteins and that a short 14-mer region in NS containing an amphipathic α helix is at least partly responsible for the ability of NS to inhibit amyloid formation and protect cells from amyloid toxicity.
MATERIALS AND METHODS
Materials
All chemicals were of analytical grade and prepared in Milli-Q water (Millipore; Billerica, MA, USA) unless otherwise stated. Bacto-tryptone, yeast extract, NaCl, agar, and ampicillin were from Roche (Dee Why, NSW, Australia). cOmplete EDTA-free protease inhibitor cocktail tablets and all other chemicals and reagents were purchased from Sigma-Aldrich (Australia) unless otherwise specified.
Proteins
α-Lac, alcohol dehydrogenase (ADH), BSA, CS, OVA, and CPK were purchased from Sigma-Aldrich. Purified human SOD was a gift from L. McAlary (Illawarra Health and Medical Research Institute, University of Wollongong, Australia). Aβ1-42 was purchased from China Peptides (Shanghai, China) or Australian Biosearch (Perth, Australia). ccβω, a modified version of the ccβ peptide (43) that has an additional tryptophan at its N terminus, was purchased from GenScript (Piscataway, NJ, USA). The ccβ peptide was purpose-designed to provide an experimentally convenient model system to study amyloid formation (43); the addition of the N-terminal tryptophan increases the rate at which the peptide forms amyloid and allows determination of the concentration of the peptide in solution by measurement of its absorbance at 280 nm (36). Human plasma CLU was purified by immunoaffinity chromatography of plasma prepared from human blood donated by Wollongong Hospital (Wollongong, Australia), as described before (44). Purified hTTR and a stable monomeric mutant of hTTR that carries F87M and L110M mutations and is unable to form a tetramer (mTTR) were produced as previously described (13, 14).
Plasmids
Purified plasmid containing an insert encoding wild-type human NS (NSwt) with a N-terminal 6His tag (pQE-81L-NSwt) was a gift from D. Lomas (University of Cambridge, UK). The numbering of NS amino acid residues referred to hereafter includes the 16 residue signal peptide. Site-directed mutagenesis as described in (45) was used to mutate NS residue 308 from lysine to aspartate. Briefly, Q5 polymerase (NEB, Australia) was used to polymerase chain reaction (PCR) amplify the NSwt sequence using pQE-81L-NSwt as the template, with forward primer 5′-TTAAAAGATGTTTTGGAAGCTCTTGGAATAACT-3′ and reverse primer 5′-AGTTATTCCAAGAGCTTCCAAAACATCTTTTAA-3′. Putative clones were verified using sequencing primer (5′-TCATAAAAAATTTATTTGCTTTGTGAGCGG-3′) in an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems; Rotkreuz, Switzerland) at the Sequencing Facility, School of Earth and Atmospheric Life Sciences, University of Wollongong. The resulting plasmid was designated pQE-81L-NSsm. A second expression plasmid encoding NS bearing a triple mutation (tm) in the same 14-mer predicted α-helical region (K304A, K308E, L310Q) was produced. The sequence encoding this mutant NS was synthesized as a gene fragment by Gene Universal USA and then cloned into the pDEST17 vector (Invitrogen) to produce pDEST17-NStm. pQE-81L-NSwt, pQE-81L-NSsm, and pDEST17-NStm were transformed into NiCo21 (DE3) E. coli cells for the expression of recombinant proteins; the constructs were designed such that all NS molecules expressed incorporated a 6-histidine tag at the N terminus of the protein.
Recombinant proteins
Neuroserpin
The expression protocol for wild-type NS (NSwt) was optimized at the Protein Production Unit (Monash University, Melbourne, Australia) to significantly increase the yield of recombinant product obtained compared to the previously published method (9). NiCo21(DE3) E. coli cells were transformed with plasmid and grown in LB media containing ampicillin (100 μg/ml) before inoculating into 950 ml of Overnight Express Instant TB autoinduction media (Novagen), also containing ampicillin (100 μg/ml), and incubated overnight at 28°C. Bacterial cells were then harvested by centrifugation at 6000g for 15 min at 4°C. NS was extracted from the cell pellet using BugBuster Master Mix (Novagen). Cellular debris was removed from the lysate by centrifugation at 16,000g for 20 min at 4°C and subsequent filtration through a 0.22-μm pore size membrane. The clarified lysate was then loaded at 0.5 ml/min onto a 5-ml HisTrap nickel affinity column (GE Healthcare) pre-equilibrated in loading buffer [50 mM tris, 0.3 M NaCl, and 5 mM imidazole (pH 8)] using an NGC Liquid Chromatography System (Bio-Rad) before washing the column with 5 column volumes of loading buffer. Bound recombinant NS was eluted using an imidazole gradient of 5 to 125 mM imidazole in loading buffer, and fractions were collected. Fractions were analyzed by SDS-PAGE, and those containing purified NS were pooled and dialyzed against PBS [137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8 mM Na2HPO4 (pH 7.4)] before being stored at −80°C. NSsm and NStm were expressed and purified as described above. The yields of NSwt, NSsm, and NStm were all in the culture volume range of 6 to 8 mg/liter.
CLIC1
A cDNA insert encoding mutant CLIC1 (22) cloned into a pET28a plasmid was a gift from P. Curmi (University of New South Wales, Sydney). BL21(DE3) E. coli cells transformed with this plasmid were used to express the recombinant CLIC1 protein, which was then purified by Ni2+ affinity chromatography as described in (22). Purified CLIC1 was stored at 4°C in PBS containing 0.1% (w/v) sodium azide (PBS/Az).
NS-tPA interaction
Actilyse, purchased from Boehringer-Ingelheim, was used as the source of human tPA. The well-known protease inhibitor activity of NS was assessed by measuring, in mixtures of NS and tPA, the formation of SDS-stable NS-tPA complexes and cleaved NS (20). NS and tPA were incubated together for 5 min at room temperature at a molar ratio of NS:tPA = 2:3. Samples were then diluted 1:1 with 2× SDS sample buffer [100 mM tris-Cl (pH 6.8), 4% (w/v) SDS, 0.2% (w/v) bromophenol blue, and 20% (v/v) glycerol] and heated at 95°C for 5 min before separation by nonreducing SDS-PAGE, electrophoretic transfer to nitrocellulose membrane, and immunoblot analysis.
Gel electrophoresis
SDS-PAGE
SDS-PAGE was conducted using either hand-poured gels (Hoefer SE 260 Mighty Small II gel system) composed of separate stacking and resolving gels, 5 and 10% (w/v) bis-acrylamide, respectively, or 4 to 12% bis-tris gels (Thermo Fisher Scientific, Australia). Samples were prepared under nonreducing conditions by incubating with sample buffer [50 mM tris-HCl (pH 6.8), 10% (w/v) glycerol, 2% (w/v) SDS, and 0.01% (w/v) bromophenol blue] and heated for 5 min at 95°C. Electrophoresis was performed at 120 to 150 V in SDS running buffer [0.025 M tris, 0.192 M glycine, and 0.1% (w/v) SDS (pH 8.5)] or at 150 V in 1× MES SDS running buffer (Thermo Fisher Scientific, Australia) until the dye front reached the bottom of the gel. Following electrophoresis, gels were stained with either Coomassie Brilliant Blue or InstantBlue Protein Stain (Expedeon). Precision Plus Protein Dual Color Standards (Bio-Rad) were used to estimate molecular weights.
Native gel electrophoresis
To confirm that purified wt-NS was monomeric and remained so under the conditions used in the protein aggregation assays, aliquots of wt-NS in PBS were either freshly thawed or heated to 37° and 43°C for 6 and 4 hours, respectively. NS was also incubated for 30 min at 50°C to induce polymerization. Each NS sample (20 μg) was then incubated with nondenaturing sample buffer [100 mM tris-HCl, 10% glycerol, and 0.0025% (w/v) bromophenol blue (pH 8.6)] and analyzed by native PAGE. Hand-poured gels were produced as above, except that the stacking and resolving gels contained 5 and 8% (w/v) bis-acrylamide, respectively, and lacked SDS. The samples were loaded onto the gel and electrophoresed in running buffer [25 mM tris-base and 192 mM glycine (pH 8.3)] at 90 V for ~4 hours. To compare wt- and mutant NS, samples were diluted in native PAGE sample buffer [100 mM tris-HCl, 10% glycerol, and 0.0025% (w/v) bromophenol blue (pH 8.6)] and analyzed on prepoured 4 to 20% Mini-PROTEAN TGX Stain-Free Gel (Bio-Rad, Australia). Electrophoresis was performed at 100 V and 4°C before staining the gels with InstantBlue Protein Stain (Expedeon).
Immunoblotting
After separation by SDS-PAGE, proteins were transferred at 4°C to BioTrace NT nitrocellulose membranes (Pall Life Sciences) using a Mini Trans-Blot Cell Western blotting apparatus (Bio-Rad). The transfer buffer used was 0.025 M tris, 0.192 M glycine, and 20% (v/v) methanol (pH 8.5), and the transfer was performed at 90 V for 120 min. Afterward, the membrane was blocked with PBS [2.7 mM KCl, 1.75 mM KH2PO4, 135 mM NaCl, and 10 mM Na2HPO4 (pH 7.4)] containing 5% (w/v) skim milk (PBS/SM) for 1 hour at room temperature or overnight at 4°C. To detect wt- and mutant NS, the blot was incubated overnight at 4°C with anti-6His tag monoclonal antibody (Abcam, ab137839) diluted 1:2000 in PBS/SM. The membrane was then washed three times with PBS containing 0.1% (v/v) Triton X-100 and twice with PBS. To detect bound primary antibody, the membrane was then incubated for 1 hour at room temperature with a 1:5000 dilution of goat anti-rabbit immunoglobulin G–horseradish peroxidase (Dako Agilent) in PBS/SM. Last, the membrane was washed again as described above, and reactive protein bands were detected using an enhanced chemiluminescence (ECL) SuperSignal Western Pico substrate kit (Pierce Biotechnology; Rockford, IL, USA). The ECL signal was imaged using an Amersham Imager 600 (GE Healthcare).
Bioinformatic analysis of NS and TTR sequences
A pairwise sequence alignment was performed for NS and TTR using the NCBI Align Sequences Protein BLAST tool; default settings and algorithm parameters were used for all pairwise analyses. All protein sequences were obtained from UniProtKB/Swiss-Prot reviewed entries. The TTR sequence used in all bioinformatic analyses included the 20–amino acid signal sequence. The PyMOL Molecular Graphics System (version 1.2.8; Schrodinger LLC) was used to visualize the three-dimensional structures of NS and TTR with structural data obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB); the PDB IDs for NS and TTR used in this study were 3F5N (25) and 4N85 (46), respectively.
Protein aggregation assays
Protein aggregation assays were carried out in clear, flat-bottom 96- or 384-well microplates (Greiner; Kremsmünster, Austria) using a total volume of 50 μl per well and a POLARstar OPTIMA microplate reader (BMG LABTECH; Offenburg, Germany) to record changes in absorbance or fluorescence. For amorphously aggregating client proteins (CS, CPK, CLIC1, ADH, and BSA), changes in turbidity (an indicator of protein aggregation) were monitored by measuring the absorbance at 360 nm over time. To induce aggregation, CS (1.8 μM) was incubated for 3 hours at 43°C in 50 mM tris-HCl and 5 mM Hepes (pH 8), CPK (15 μM) was incubated for 5 hours at 43°C in PBS, CLIC1 (60 μM) was incubated for 5 hours at 37°C in PBS, and BSA (10 μM) was incubated for 18 hours at 37°C in PBS containing 20 mM dithiothreitol. In all cases, samples were shaken (double orbital, 100 rpm) for 20 s before each measurement. The duration of these amorphous aggregation reactions was chosen to correspond to the minimum period of time in which the turbidity measured reached a plateau. Amyloid formation by Aβ1-42, ccβω, and α-syn was monitored by measuring changes in ThioT fluorescence. Aβ1-42 (20 μM) and ccβω (80 μM) were incubated with double orbital shaking (200 rpm) at either 30°C in PBS (Aβ) or at 37°C in 20 mM Hepes (pH 7.5) (ccβω). α-syn (25 μM) containing 1.25 μM α-syn seeds was incubated at 37°C in PBS without shaking. α-Syn seeds were prepared by first incubating 1.25 μM monomeric α-syn in PBS at 42°C in a water bath with gentle stirring for 24 hours (to initiate fibril formation), followed by 15 cycles of sonication (cycle time, 1.5 s; power set at 30%) using a Branson 250 Digital Sonifier (Branson Ultrasonics, CT, USA) to break up the newly formed fibrillar material. This process was repeated once more before storing the fibril seeds at −80°C. Aggregation mixtures were supplemented with 25 μM ThioT, and fluorescence was measured using the following band-pass filters: excitation, 440 ± 10 nm, and emission, 480 ± 10 nm. CLU was used as a known chaperone protein, and α-Lac, BSA, OVA, or SOD was used as nonchaperone control proteins. It was confirmed that, when incubated alone, neither NS nor TTR (or any of the control proteins) generated turbidity or ThioT fluorescence under any of the assay conditions used. Buffer alone was also used as a control in all assays. Samples were incubated in triplicate, and all assays were repeated at least twice.
Far UV CD spectroscopy
Far UV CD measurements were performed using a Jasco Model J-810 spectropolarimeter connected to a CDF-426S/L Peltier system (Jasco). Data were collected for NS proteins using the “spectral measurement program” (data pitch, 0.2 nm; scan speed, 100 nm/min; response, 2 s; bandwidth, 1 nm; and wavelength, 190 to 240 nm). Purified NSwt, NSsm, and NStm proteins were analyzed at 0.1 mg/ml in 5 mM phosphate buffer (pH 7.4; filtered and degassed); all samples were analyzed using a 0.1-cm cuvette set at 37°C. Output spectra were averaged from 10 scans. Data were analyzed using DichroWeb software against the SMP 180 dataset optimized for 190 to 240 nm using the CONTIN program (47, 48) and plotted as mean residue ellipticity versus wavelength.
Thermal shift assay
Thermal shift assays were performed as previously described (29). Briefly, proteins were diluted to a final concentration of 200 μg/ml in 7 mM Na2HPO4, 3 mM NaH2PO4, and 50 mM NaCl (pH 7.0), containing SPYRO Orange (diluted 1 in 625 from a 5000× stock; Molecular Probes). The same solution lacking protein was used as a no protein control to measure background fluorescence. Samples were examined in quadruplicate (20 μl per well) in Multiply μStripPro LP qPCR strips (Sarstedt) in a QuantStudio 5 Real-Time PCR instrument with firmware version 1.2.x (Applied Biosystems). Using QuantStudio Design and Analysis Software version 1.5.2 (Applied Biosystems), the protocol had an initial hold step for 2 min at 35°C. The temperature was then increased in increments of 1.0°C/min with a ramp speed of 0.017°C/min until 65°C was reached. The fluorescence was acquired at the end of a 1-min hold at each temperature; background fluorescence estimated from the no protein control was subtracted from all data points. To estimate the apparent “melting temperature” (Tm), the equation for a Boltzmann sigmoidal curve was fitted to each dataset using GraphPad Prism version 5 (GraphPad Software).
BisANS assays
NS (wt, sm, and tm) proteins at 0.1 mg/ml in PBS were incubated with 20 μM bisANS for 10 min at room temperature in a Greiner 384-well clear well plate. Samples were analyzed in triplicate. Subsequently, fluorescence was measured on a POLARstar Omega plate reader using 360/10 nm and 490/10 nm band-pass filters for excitation and emission, respectively. All readings were corrected for the fluorescence of 20 μM bisANS in PBS.
Mass photometry
Proteins were diluted to a concentration of 1 nM in PBS. Using a Refeyn One mass photometer, 10 μl of each sample was analyzed over 10 min at a rate of 600 frames/min. Sample acquisition and data analysis were done as reported previously (49).
Cytotoxicity
To prepare toxic Aβ oligomers for cytotoxicity experiments, 5 μM Aβ1-42 was aggregated as described above, except that the buffer used was a 1:1 mixture of PBS and Dulbecco’s minimum essential medium (DMEM):F12; samples were taken at 5 hours and flash-frozen using liquid nitrogen. ccβω was aggregated in 20 mM Hepes (pH 7.5), exactly as described in the “Protein aggregation assays” section above, and samples were taken at 1 hour frozen as for Aβ. The cytotoxicity of these samples was tested on NSC34 (motor neuroblastoma) cells (50). NSC34 cells were cultured at 37°C and 5% (v/v) CO2 in DMEM-F12 (Gibco) containing 10% (v/v) fetal bovine serum (Thermo Fisher Scientific). Cells were seeded in a 96-well Greiner cell culture plate (Sigma-Aldrich, Australia) with 5000 cells per well; twenty-four hours later, the culture medium was removed and the cells were gently washed three times with prewarmed PBS before adding 100 μl of DMEM-F12 to each well. Aβ1-42 (2 μM) and ccβω (80 μM) were incubated for 15 to 20 min at room temperature with or without supplementation with, respectively, 0.1 or 4 μM NS (wt, sm, or tm), and then, 100 μl of these mixtures was added to the wells containing cells and 100 μl of DMEM-F12. Thus, the final concentrations of Aβ1-42 and ccβω added to the cells were 1 and 40 μM, respectively. Control wells for the Aβ1-42 and ccβω treatments were supplemented with a matching volume of PBS or 20 mM Hepes (pH 7.5), respectively. Cells were cultured for 5 days after these additions before using calcein-AM to measure cell viability (51). Calcein-AM (1 μM) was added to cells and left to incubate for 30 min at 37°C before gently washing twice with PBS. Calcein-AM fluorescence was measured using a POLARstar plate reader using 480/20 nm and 520/10 nm band-pass filters for excitation and emission, respectively.
Statistical tests
Statistical analyses of data were performed using either one-way analysis of variance (ANOVA) and Bonferroni comparisons between paired samples or Student’s t test (pairwise comparisons); differences with P < 0.05 were considered to be statistically significant.
Acknowledgments
We thank L. McAlary for help with the generation of PyMOL images.
Funding: J.W., S.S., D.R.W., and E.-J.P. are grateful for Australian Government postgraduate scholarships. M.R.W. thanks the Australian Research Council for financial support in the form of a Discovery Project grant (DP160100011). P.S. is supported by the Royal Society through a University Research Fellowship (URF\R1\201461). M.V. and P.S. acknowledge the support of the Centre for Misfolding Diseases (UK).
Author contributions: J.W., S.S., D.R.W., M.K., N.J.G., and E.-J.P. all performed experimental work, had input into experimental design, and reviewed the manuscript. P.S., M.V., J.N.B., and M.R. contributed to experimental design and authoring of the manuscript. M.R.W. supervised experimental design and work and was primary author of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Methods to rationally design NS mutants
Methods to generate pinwheel structures
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.Yerbury J. J., Stewart E. M., Wyatt A. R., Wilson M. R., Quality control of protein folding in extracellular space. EMBO Rep. 6, 1131–1136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wyatt A. R., Yerbury J. Y., Ecroyd H., Wilson M. R., Extracellular chaperones and proteostasis. Annu. Rev. Biochem. 82, 295–322 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Nielsen H. M., Minthon L., Londos E., Blennow K., Miranda E., Perez J., Crowther D. C., Lomas D. A., Janciauskiene S. M., Plasma and CSF serpins in Alzheimer disease and dementia with Lewy bodies. Neurology 69, 1569–1579 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Miranda E., Lomas D. A., Neuroserpin: A serpin to think about. Cell. Mol. Life Sci. 63, 709–722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loebermann H., Tokuoka R., Deisenhofer J., Huber R., Human α1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J. Mol. Biol. 177, 531–557 (1984). [PubMed] [Google Scholar]
- 6.Hastings G. A., Coleman T. A., Haudenschild C. C., Stefansson S., Smith E. P., Barthlow R., Cherry S., Sandkvist M., Lawrence D. A., Neuroserpin, a brain-associated inhibitor of tissue plasminogen activator is localized primarily in neurons. Implications for the regulation of motor learning and neuronal survival. J. Biol. Chem. 272, 33062–33067 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Yepes M., Sandkvist M., Coleman T. A., Moore E., Wu J. Y., Mitola D., Bugge T. H., Lawrence D. A., Regulation of seizure spreading by neuroserpin and tissue-type plasminogen activator is plasminogen-independent. J. Clin. Invest. 109, 1571–1578 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R. L., Shrimpton A. E., Holohan P. D., Bradshaw C., Feiglin D., Collins G. H., Sonderegger P., Kinter J., Becker L. M., Lacbawan F., Krasnewich D., Muenke M., Lawrence D. A., Yerby M. S., Shaw C. M., Gooptu B., Elliott P. R., Finch J. T., Carrell R. W., Lomas D. A., Familial dementia caused by polymerization of mutant neuroserpin. Nature 401, 376–379 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Kinghorn K. J., Crowther D. C., Sharp L. K., Nerelius C., Davis R. L., Chang H. T., Green C., Gubb D. C., Johansson J., Lomas D. A., Neuroserpin binds Aβ and is a neuroprotective component of amyloid plaques in Alzheimer disease. J. Biol. Chem. 281, 29268–29277 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Luo J., Wärmländer S. K. T. S., Gräslund A., Abrahams J. P., Non-chaperone proteins can inhibit aggregation and cytotoxicity of Alzheimer amyloid ß peptide. J. Biol. Chem. 289, 27766–27775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho P. Y., Joshi G., Johnson J. A., Murphy R. M., Transthyretin-derived peptides as β-amyloid inhibitors. ACS Chem. Nerosci. 5, 542–551 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du J., Cho P. Y., Yang D. T., Murphy R. M., Identification of beta-amyloid-binding sites on transthyretin. Protein Eng. Des. Sel. 25, 337–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain N., Ådén J., Nagamatsu K., Evans M. L., Li X., McMichael B., Ivanova M. I., Almqvist F., Buxbaum J. N., Chapman M. R., Inhibition of curli assembly and Escherichia coli biofilm formation by the human systemic amyloid precursor transthyretin. Proc. Natl. Acad. Sci. U.S.A. 114, 12184–12189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., Zhang X., Ladiwala A. R. A., Du D., Yadav J. K., Tessier P. M., Wright P. E., Kelly J. W., Buxbaum J. N., Mechanisms of transthyretin inhibition of β-amyloid aggregation in vitro. J. Neurosci. 33, 19423–19433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naylor H. M., Newcomer M. E., The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry 38, 2647–2653 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Peterson S. A., Klabunde T., Lashuel H. A., Purkey H., Sacchettini J. C., Kelly J. W., Inhibiting transthyretin conformational changes that lead to amyloid fibril formation. Proc. Natl. Acad. Sci. U.S.A. 95, 12956–12960 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miroy G. J., Lai Z. H., Lashuel H. A., Peterson S. A., Strang C., Kelly J. W., Inhibiting transthyretin amyloid fibril formation via protein stabilization. Proc. Natl. Acad. Sci. U.S.A. 93, 15051–15056 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Power D. M., Elias N. P., Richardson S. J., Mendes J., Soares C. M., Santos C. R. A., Evolution of the thyroid hormone-binding protein, transthyretin. Gen. Comp. Endocrinol. 119, 241–255 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Yang D. T., Joshi G., Cho P. Y., Johnson J. A., Murphy R. M., Transthyretin as both a sensor and a scavenger of β-Amyloid oligomers. Biochemistry 52, 2849–2861 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belorgey D., Crowther D. C., Mahadeva R., Lomas D. A., Mutant neuroserpin (S49P) that causes familial encephalopathy with neuroserpin inclusion bodies is a poor proteinase inhibitor and readily forms polymers in vitro. J. Biol. Chem. 277, 17367–17373 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Barker-Carlson K., Lawrence D. A., Schwartz B. S., Acyl-enzyme complexes between tissue-type plasminogen activator and neuroserpin are short-lived in vitro. J. Biol. Chem. 277, 46852–46857 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Goodchild S. C., Angstmann C. N., Breit S. N., Curmi P. M. G., Brown L. J., Transmembrane extension and oligomerization of the CLIC1 chloride intracellular channel protein upon membrane interaction. Biochemistry 50, 10887–10897 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Noto R., Santangelo M. G., Ricagno S., Mangione M. R., Levantino M., Pezzullo M., Martorana V., Cupane A., Bolognesi M., Manno M., The tempered polymerization of human neuroserpin. PLOS ONE 7, e32444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takehara S., Onda M., Zhang J., Nishiyama M., Yang X., Mikami B., Lomas D. A., The 2.1-Å crystal structure of native neuroserpin reveals unique structural elements that contribute to conformational instability. J. Mol. Biol. 388, 11–20 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Ricagno S., Caccia S., Sorrentino G., Antonini G., Bolognesi M., Human neuroserpin: Structure and time-dependent inhibition. J. Mol. Biol. 388, 109–121 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hamilton J. A., Steinrauf L. K., Braden B. C., Liepnieks J., Benson M. D., Holmgren G., Sandgren O., Steen L., The X-ray crystal structure refinements of normal human transthyretin and the amyloidogenic Val-30 → Met variant to 1.7-Å resolution. J. Biol. Chem. 268, 2416–2424 (1993). [PubMed] [Google Scholar]
- 27.Mangrolia P., Yang D. T., Murphy R. M., Transthyretin variants with improved inhibition of β-amyloid aggregation. Protein Eng. Des. Sel. 29, 209–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Buxbaum J. N., Transthyretin and the brain re-visited: Is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol. Neurodegener. 6, 79 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huynh K., Partch C. L., Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 79, 28.9.1–28.9.14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu D., Piszczek G., Measuring the affinity of protein-protein interactions on a single-molecule level by mass photometry. Anal. Biochem. 592, 113575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson M. R., Easterbrook-Smith S. B., Clusterin is a secreted mammalian chaperone. Trends Biochem. Sci. 25, 95–98 (2000). [DOI] [PubMed] [Google Scholar]
- 32.Humphreys D. T., Carver J. A., Easterbrook-Smith S. B., Wilson M. R., Clusterin has chaperone-like activity similar to that of small heat shock proteins. J. Biol. Chem. 274, 6875–6881 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Yerbury J. J., Rybchyn M. S., Easterbrook-Smith S. B., Henriques C., Wilson M. R., The acute phase protein haptoglobin is a mammalian extracellular chaperone with an action similar to clusterin. Biochemistry 44, 10914–10925 (2005). [DOI] [PubMed] [Google Scholar]
- 34.French K., Yerbury J. J., Wilson M. R., Protease activation of α2-macroglobulin modulates a chaperone-like action with broad specificity. Biochemistry 47, 1176–1185 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Wyatt A. R., Yerbury J. J., Berghofer P., Greguric I., Katsifis A., Dobson C. M., Wilson M. R., Clusterin facilitates in vivo clearance of extracellular misfolded proteins. Cell. Mol. Life Sci. 68, 3919–3931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yerbury J. J., Poon S., Meehan S., Thompson B., Kumita J. R., Dobson C. M., Wilson M. R., The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J. 21, 2312–2322 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Narayan P., Orte A., Clarke R. W., Bolognesi B., Hook S., Ganzinger K. A., Meehan S., Wilson M. R., Dobson C. M., Klenerman D., The extracellular chaperone clusterin sequesters oligomeric forms of the amyloid-β1−40 peptide. Nat. Struct. Mol. Biol. 19, 79–83 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyatt A. R., Yerbury J. J., Wilson M. R., Structural characterization of clusterin-chaperone client protein complexes. J. Biol. Chem. 284, 21920–21927 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landreh M., Rising A., Presto J., Jörnvall H., Johansson J., Specific chaperones and regulatory domains in control of amyloid formation. J. Biol. Chem. 290, 26430–26436 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaari A., Eliezer D., Ladjimi M., The C-terminal α-helices of mammalian Hsc70 play a critical role in the stabilization of α-synuclein binding and inhibition of aggregation. Int. J. Biol. Macromol. 83, 433–441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Härndahl U., Kokke B. P. A., Gustavsson N., Linse S., Berggren K., Tjerneld F., Boelens W. C., Sundby C., The chaperone-like activity of a small heat shock protein is lost after sulfoxidation of conserved methionines in a surface-exposed amphipathic α-helix. Biochim. Biophys. Acta 1545, 227–237 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Visentin C., Broggini L., Sala B. M., Russo R., Barbiroli A., Santambrogio C., Nonnis S., Dubnovitsky A., Bolognesi M., Miranda E., Achour A., Ricagno S., Glycosylation tunes neuroserpin physiological and pathological properties. Int. J. Mol. Sci. 21, 3235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kammerer R. A., Kostrewa D., Zurdo J., Detken A., Garcia-Echeverria C., Green J. D., Muller S. A., Meier B. H., Winkler F. K., Dobson C. M., Steinmetz M. O., Exploring amyloid formation by a de novo design. Proc. Natl. Acad. Sci. U.S.A. 101, 4435–4440 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson M. R., Easterbrook-Smith S. B., Clusterin binds by a multivalent mechanism to the Fc and Fab regions of IgG. Biochim. Biophys. Acta 1159, 319–326 (1992). [DOI] [PubMed] [Google Scholar]
- 45.Liang X., Peng L., Li K., Peterson T., Katzen F., A method for multi-site-directed mutagenesis based on homologous recombination. Anal. Biochem. 427, 99–101 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Yokoyama T., Kosaka Y., Mizuguchi M., Crystal structures of human transthyretin complexed with glabridin. J. Med. Chem. 57, 1090–1096 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Whitmore L., Wallace B. A., Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 89, 392–400 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Whitmore L., Wallace B. A., DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole D., Young G., Weigel A., Sebesta A., Kukura P., Label-free single-molecule imaging with numerical-aperture-shaped interferometric scattering microscopy. ACS Photonics 4, 211–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cashman N. R., Durham H. D., Blusztajn J. K., Oda K., Tabira T., Shaw I. T., Dahrouge S., Antel J. P., Neuroblastoma × spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 194, 209–221 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Bratosin D., Mitrofan L., Palii C., Estaquier J., Montreuil J., Novel fluorescence assay using calcein-AM for the determination of human erythrocyte viability and aging. Cytometry A 66, 78–84 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Zhou J., Grigoryan G., Rapid search for tertiary fragments reveals protein sequence-structure relationships. Protein Sci. 24, 508–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Methods to rationally design NS mutants
Methods to generate pinwheel structures
Tables S1 and S2
References