Noncoding genetic variants cause blood disease.
Abstract
Human genetic variants are classified on the basis of potential pathogenicity to guide clinical decisions. However, mechanistic uncertainties often preclude definitive categorization. Germline coding and enhancer variants within the hematopoietic regulator GATA2 create a bone marrow failure and leukemia predisposition. The conserved murine enhancer promotes hematopoietic stem cell (HSC) genesis, and a single-nucleotide human variant in an Ets motif attenuates chemotherapy-induced hematopoietic regeneration. We describe “conditionally pathogenic” (CP) enhancer motif variants that differentially affect hematopoietic development and regeneration. The Ets motif variant functioned autonomously in hematopoietic cells to disrupt hematopoiesis. Because an epigenetically silenced normal allele can exacerbate phenotypes of a pathogenic heterozygous variant, we engineered a bone marrow failure model harboring the Ets motif variant and a severe enhancer mutation on the second allele. Despite normal developmental hematopoiesis, regeneration in response to chemotherapy, inflammation, and a therapeutic HSC mobilizer was compromised. The CP paradigm informs mechanisms underlying phenotypic plasticity and clinical genetics.
INTRODUCTION
Abundant DNA motifs within the noncoding sector of the genome mediate protein binding to control genome function. Because chromatin renders many motifs inaccessible, which underlies cellular phenotypes, organismal complexity, and interindividual variation, massive efforts have mapped chromatin accessibility and histone modifications genome wide (1, 2). Germline or acquired genetic variants beyond coding regions can disrupt or create DNA motifs, derailing genetic programs. Enhancer motif variants may abrogate, incrementally reduce, increase, or have no effect on transcription.
Discriminating between pathogenic and nonpathogenetic variants guides clinical decision-making. Variants predisposing to bone marrow failure and leukemia may justify patient surveillance or bone marrow transplantation (3–8). Because the ramifications of a motif alteration are often unclear, deeming variants pathogenic, potentially pathogenic, potentially benign, benign, or of undetermined significance is an imperfect science (9). A crux of this problem rests on cell type–specific chromatin context, as a variant may be inconsequential in most, but not all, cell types. Furthermore, multiple motifs within an enhancer may be of variable importance. Genome engineering enables rigorous testing of whether a motif is essential, modulatory, or nonessential in a genome, yet the effort may be incompatible with clinical decision-making. Chromatin accessibility and histone modification data can generate inferences of motif importance without yielding accurate predictions. Although familial genetic data are powerful, it is often unavailable. Cell type–specific mechanistic analyses of motif functions and variation will yield principles to inform the accurate curation of variants within pathogenesis-linked genes.
Cell type–specific enhancers regulate hematopoietic stem/progenitor cell (HSPC) differentiation into diverse blood cell types (10). These include enhancers regulating expression of the transcription factor GATA2, which promotes the generation and function of hematopoietic stem cells (HSCs) (11–13), myelo-erythroid progenitor cells (14–16), and erythroid precursor cells (15) and is expressed in select nonhematopoietic cell types (17). Two essential enhancers regulate GATA2 levels to promote HSPC emergence and function in the mouse and suppress the development of human GATA2 deficiency syndrome characterized by variably penetrant immunodeficiency, bone marrow failure, and acute myeloid leukemia (AML) (7, 10). Variation in a conserved GATA2 intronic enhancer 9.8 kb downstream of the transcriptional start site (+9.5 in the mouse) or coding sequences causes this syndrome. This enhancer contains E-box, GATA, and Ets motifs (18, 19). Patient mutations within this region include a c.1017+512del28 deletion (20) that eliminates E-box and GATA motifs, a c.1017+532T>A transversion (21) that disrupts the E-box, and, most frequently, a C>T transition within a 3′ Ets motif (c.1017+572C>T) (21–27). +9.5 enhancer activity requires E-box, GATA, and Ets motifs in cell- and transgenic mouse–based reporter assays (18, 23, 28), although analyses of nonessential Gata2 enhancers yielded similar results (18, 19, 29–31). Deletion of E-box and GATA motifs within the murine +9.5 enhancer (+9.5−/−; Table 1) or E-box and Ets motifs [+9.5(E-box;Ets)−/−] is lethal at about embryonic day 14 (~E14) (20, 32), in contrast to Gata2-null homozygous embryos that die at ~E10.5 (11). HSC emergence from the aorta-gonad-mesonephros (AGM) and vascular integrity is impaired in +9.5−/− and +9.5(E-box;Ets)−/− embryos (20, 32, 33). A compound heterozygous (CH) Gata2 variant lacking +9.5 on one allele and the hematopoietic progenitor regulatory −77 Gata2 enhancer (14) on the other allele was lethal at ~E14 (15). HSC emergence from the AGM was unaltered, but fetal liver HSCs decreased to that of +9.5+/− embryos, and megakaryocyte erythrocyte progenitors (MEPs) and burst-forming unit–erythroid (BFU-E) were depleted (15). Although +9.5 promotes Gata2 transcription in HSPCs during development (34), its activities, and those of its individual motifs, in adult steady-state and regenerative hematopoiesis are incompletely defined.
Table 1. Strains with variants in the Gata2 +9.5 enhancer.
| Strain | +9.5 motif mutations | |
| Allele 1 | Allele 2 | |
| +9.5−/− | E-box-GATA deletion | E-box-GATA deletion |
| +9.5(E-box;Ets)−/− | E-box disruption, Ets C>T |
E-box disruption, Ets C>T |
| +9.5(E-box)−/− | E-box disruption | E-box disruption |
| +9.5(Ets)−/− | Ets C>T | Ets C>T |
| CH | E-box-GATA deletion | Ets C>T |
In contrast to +9.5−/− E-box;GATA motif variant mice, a +9.5 Ets motif homozygous variant has little impact on embryogenesis and developmental hematopoiesis; mice are born and survive at normal Mendelian ratios (32). However, +9.5(Ets)−/− HSPCs are defective in responding to 5-fluorouracil (5-FU)–induced myeloablation and fail to efficiently reconstitute bone marrow upon transplantation (32). As human disease mutations alter different +9.5 motifs (7), motifs within an enhancer may have variable importance, and diverse hematopoietic and nonhematopoietic cells express GATA2 (17), we established systems to analyze the functional consequences of enhancer motif genetic variation in cell type–specific contexts in vivo. Our studies with a new bone marrow failure model revealed a +9.5 motif variant that dysregulates hematopoietic regeneration through a hematopoietic cell–intrinsic mechanism and identified “conditionally pathogenic” (CP) motif variants that preclude simplistic binary designations of motif variant pathogenicity status.
RESULTS
Enhancer motif variant dysregulates hematopoietic regeneration through a hematopoietic cell–intrinsic mechanism
Previously, we demonstrated that a +9.5 Ets motif variant [9.5(Ets)−/−], which generates a predisposition for human bone marrow failure and AML in human patients, severely disrupts the transcriptome of regenerating hematopoietic progenitor cells [Lin−Sca1+Kit+ (LSK)] from mice treated with the myeloablative drug 5-FU. The variant decreases immunophenotypic HSCs 12-fold (32), and the remaining HSCs have 3-fold less activity in a competitive transplant assay (32). Besides HSPCs, GATA2 is expressed in bone marrow niche nonhematopoietic cells, including endothelial cells and neurons (17), raising the question as to whether phenotypes resulting from the whole organism Ets motif knockout involve only HSC-intrinsic mechanisms or whether HSC-extrinsic mechanisms requiring nonhematopoietic microenvironment cells are also implicated.
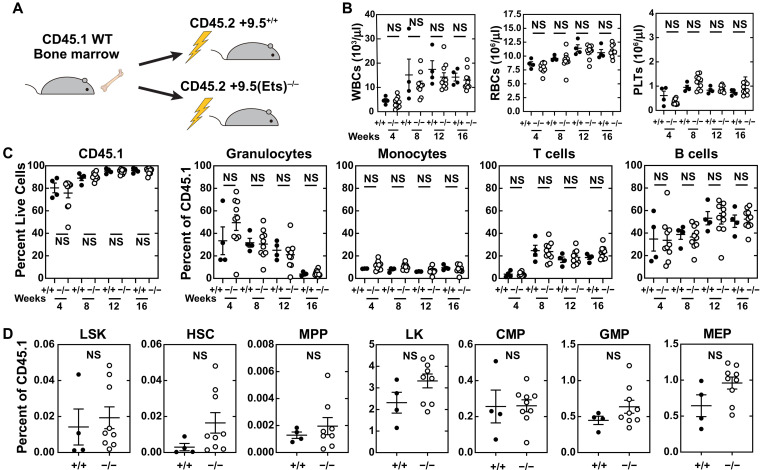
To test for potential hematopoietic cell–extrinsic phenotypes, we transplanted wild-type (WT) CD45.1 bone marrow cells (2 million) into lethally irradiated CD45.2 WT or +9.5(Ets)−/− mice (Fig. 1A). Reconstitution of differentiated cells was monitored by peripheral blood analyses 4, 8, 12, and 16 weeks after transplant. Complete blood counts (CBCs) of WT and variant recipients were indistinguishable throughout (Fig. 1B). As expected from lethally irradiated recipients, leukocytes were 76% [+9.5(Ets)−/− recipients] and 80% (WT recipients) donor-derived at week 4 and increased to 92% [+9.5(Ets)−/− recipients] and 96% (WT recipients) at week 16 (Fig. 1C). The contribution did not differ significantly between groups at any time. After 16 weeks, HSPC populations in variant recipients were indistinguishable from WT controls (Fig. 1D). Thus, the +9.5 Ets motif in nonhematopoietic cells is dispensable for regenerating the WT hematopoietic system.
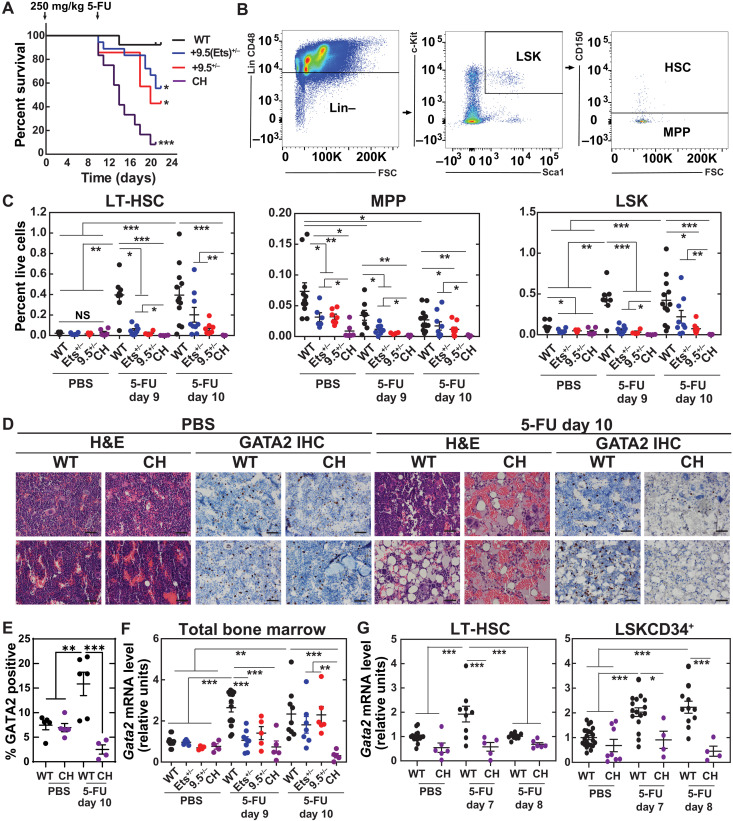
Fig. 1. Gata2 Ets motif variant dysregulates hematopoiesis through a hematopoietic cell–autonomous mechanism.
(A) Reciprocal transplantation of WT cells into WT and +9.5(Ets)−/− recipients. (B) CBCs after transplant. NS, not significant; WBCs, white blood cells; RBCs, red blood cells; PLTs, platelets. (C) Multilineage repopulating activity 16 weeks after competitive transplant (n = 4 to 10 per genotype). (D) Analysis of donor-derived LSK (Lin−CD48−Sca1+Kit+), HSC (Lin−CD48−Sca1+Kit+CD150+), MPP (Lin−CD48−Sca1+Kit+CD150−), LS−K (Lin−Sca1−Kit+), MEP (Lin−Sca1−Kit+FcγR−CD34−), CMP (Lin−Sca1−Kit+FcγR−CD34+), and GMP (Lin−Sca1−Kit+FcγR+CD34+) within bone marrow 16 weeks after transplant (n = 4 to 9 per genotype) (two-tailed unpaired Student’s t test).
Gata2 enhancer Ets motif mitigates adverse consequences of chronic inflammation
Viral or mycobacterial infections occur in ~60% of GATA2 deficiency syndrome patients (35). While the Gata2 +9.5 Ets motif promotes hematopoietic regeneration following myeloablation or transplantation (32), its role in the bone marrow response to inflammatory stress, e.g., during infection, is unknown. We tested whether the hematopoietic system requires the Ets motif to respond to the viral mimetic polyinosinic:polycytidylic acid (polyI:C or pI:C), which induces a type I interferon response and transition of quiescent to proliferative long-term (LT)–HSCs (36), and whether the motif variant compromises cell cycle entry. A single dose of polyI:C (10 mg/kg) for 24 hours increased HSCs 4.6- and 3.1-fold for WT and +9.5(Ets)−/− cells, respectively, while Gata2 expression in fluorescence-activated cell sorting (FACS)–purified bone marrow LT-HSCs was unaltered by the Ets motif variant (fig. S1).
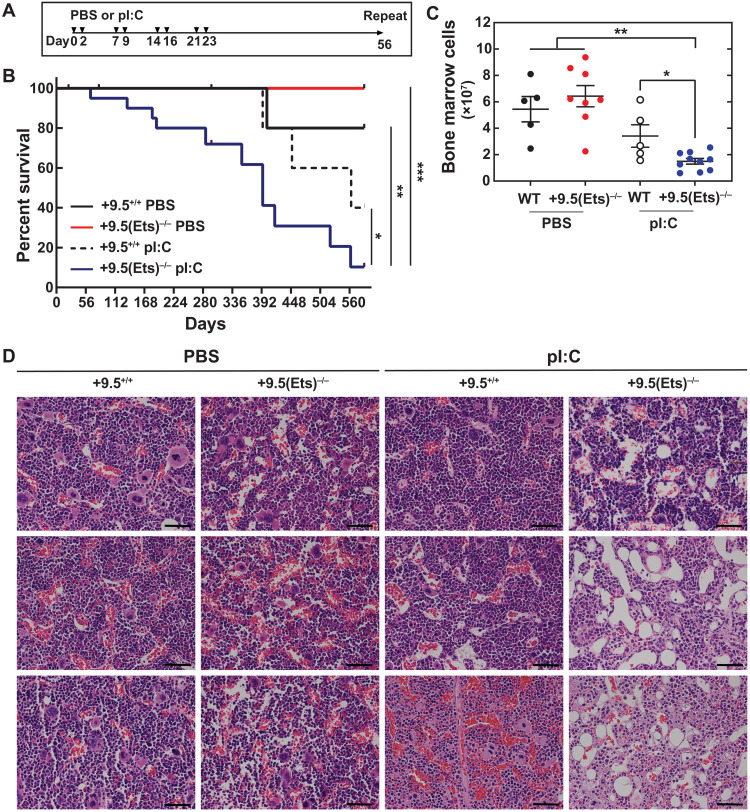
As the single-dose pI:C treatment used does not elicit chronic inflammation, we adapted the treatment scheme of Walter et al. (37) (Fig. 2A), which sustains HSC activation, decreases HSC activity, and promotes bone marrow failure in mice with defective DNA damage repair machinery (37). As a proxy for hematopoietic activity, peripheral blood parameters were quantified at the beginning of each 56-day treatment cycle. This analysis did not reveal pancytopenia in any condition (fig. S2). +9.5(Ets)−/− mice treated with polyI:C exhibited significantly increased mortality, with a median survival of 394 days (Fig. 2B). By contrast, polyI:C-treated WT mice exhibited a median survival of 563 days, with six of seven animals surviving beyond 394 days. After 10 treatment cycles (588 days), all surviving mice were euthanized. While overall bone marrow cellularity was indistinguishable between vehicle-treated WT and +9.5(Ets)−/− mice, polyI:C treatment reduced bone marrow cell number 2.3-fold in +9.5(Ets)−/− mice relative to WT, yielding hypocellular marrows (Fig. 2, C and D). Thus, the Gata2 +9.5(Ets) motif promotes hematopoietic regeneration and establishes a defense against chronic inflammation.
Fig. 2. +9.5 Ets motif mitigates lethality induced by chronic stress.
(A) Mice were given multiple doses of pI:C (10 mg/kg) or vehicle control as per Walter et al. (37). Treatment cycles were repeated every 56 days. (B) Survival [PBS: WT (n = 10), +9.5(Ets)−/− (n = 14); pI:C: WT (n = 14), +9.5(Ets)−/− (n = 20) from two experiments] (log-rank test). (C) Number of cells per single hindlimb. n = 5 to 10 per condition, Tukey’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001. (D) Representative hematoxylin and eosin (H&E) staining of femurs. Each panel represents a single mouse. Scale bars, 50 μm.
Gata2 +9.5 E-box variant delays hematopoietic regeneration kinetics
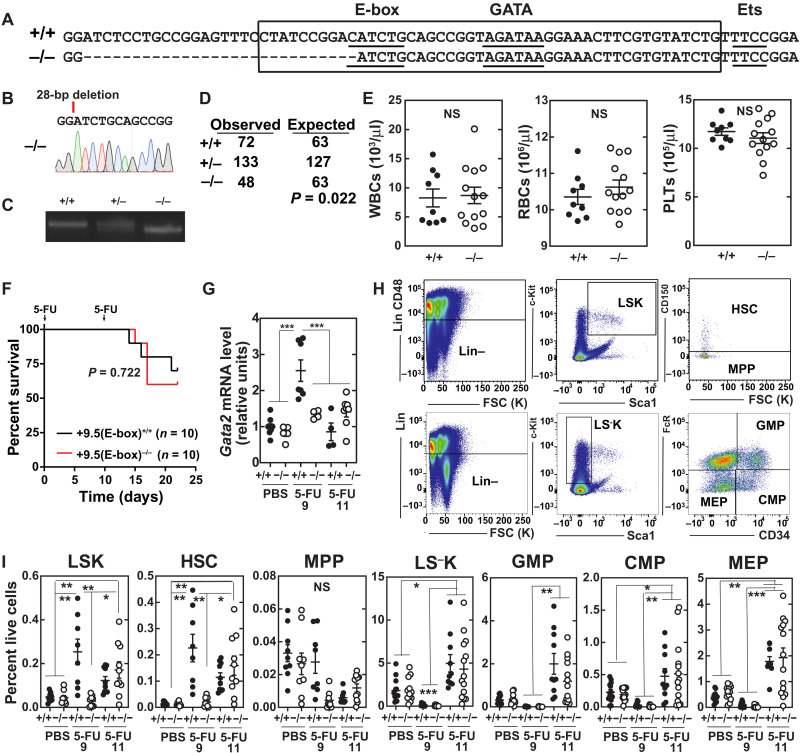
Although our studies revealed a hematopoietic cell–autonomous function of the +9.5 Ets motif variant to dysregulate hematopoiesis in adult mice subjected to hematopoietic stress, it remained unclear whether other +9.5 motifs are essential determinants of developmental, adult, and/or regenerative hematopoiesis. The embryonic lethality of combined E-box;GATA and E-box;Ets mutations precludes analysis of these alleles in adult regeneration (32). We constructed a 28–base pair (bp) deletion to eliminate a critical nucleotide of the E-box (CATCTG to GATCTG) and delete upstream sequences while retaining the spacer, GATA, and Ets motifs [+9.5(E-box)−/−; Fig. 3, A to C]. This allele shares the E-box and upstream sequence alteration that characterizes our embryonic lethal +9.5(E-box;Ets)−/− model (32). We determined whether this variant recapitulates developmental or regenerative phenotypes of other +9.5 variants or confers unique phenotypes.
Fig. 3. +9.5 E-box accelerates regeneration kinetics.
(A) Sequence of WT (+9.5+/+) and E-box variant [+9.5(E-box)−/−] mice. (B) Sequence of mutant mice. (C) Polymerase chain reaction (PCR) genotyping. (D) Genotyping results at weaning from +9.5(E-box)+/− × +9.5(E-box)+/− matings (chi-square test) (E) Steady-state blood parameters [+9.5+/+ (n = 9) and +9.5(E-box)−/− (n = 12)]. (F) Kaplan-Meier survival curve of mice following two 5-FU doses (250 mg/kg; days 0 and 11; n = 10 per genotype). P = 0.0167, log-rank test. (G) mRNA quantification from bone marrow after vehicle (PBS), 9-day 5-FU, or 11-day 5-FU (250 mg/kg) treatment (n = 5 to 11 per condition from five experiments). (H) Flow cytometry scheme for quantification of LSK (Lin−CD48−Sca1+Kit+), HSC (Lin−CD48−Sca1+Kit+CD150+), MPP (Lin−CD48−Sca1+Kit+CD150−), LS−K (Lin−Sca1−Kit+), MEP (Lin−Sca1−Kit+FcγR−CD34−), CMP (Lin−Sca1−Kit+FcγR−CD34+), and GMP (Lin−Sca1−Kit+FcγR+CD34+). (I) Quantification of HSC, MPP, and LSKs from total bone marrow after vehicle (PBS), 9-day 5-FU, or 11-day 5-FU (250 mg/kg) treatment (percentage of live bone marrow cells) [PBS: WT (n = 9), +9.5(E-box)−/− (n = 9); day 9: WT (n = 8), +9.5(E-box)−/− (n = 10); day 11: WT (n = 10), +9.5(E-box)−/− (n = 10)]. *P < 0.05, **P < 0.01, ***P < 0.001, Tukey’s multiple comparisons test.
Unlike dual E-box and GATA or Ets motif variants, +9.5(E-box)−/− mice survived through weaning, albeit at lower numbers than predicted from Mendelian ratios (Fig. 3D). Minor developmental defects included reduced numbers of emerging hematopoietic cells at E10.5, vascular integrity defects, and decreased granulocyte-macrophage progenitors (GMPs) (figs. S3 and S4). Although the +9.5 Ets motif variant decreased immunophenotypic HSCs in the embryo, in steady-state adult bone marrow, HSC numbers were indistinguishable from those of WT mice. However, the +9.5(Ets)−/− HSCs were not competent to expand and differentiate in response to 5-FU or transplantation (32). We tested whether the E-box variant exhibited overlapping or distinct adult phenotypes. White blood cell, red blood cell, and platelet numbers were indistinguishable between +9.5(E-box)−/− and WT mice (Fig. 3E). To test whether the E-box is critical for the response to myeloablation-induced stress, 5-FU was used to ablate cycling cells and promote HSC expansion and differentiation to regenerate the hematopoietic system (38). Two 5-FU doses (250 mg/kg) were administered with an 11-day interval to maximally expand HSPCs (39). Survival was unaffected, with 70% of WT and 60% of +9.5(E-box)−/− mice alive at day 22 (Fig. 3F).
During regeneration after 5-FU myeloablation, Gata2 transcripts are induced in bone marrow LSK cells (32, 39), and the +9.5 Ets motif was required for this induction. To test whether the +9.5 E-box is also required for this induction, Gata2 mRNA was quantified 9 and 11 days after 5-FU administration. Gata2 expression was comparable in untreated +9.5(E-box)−/− and WT mice. Nine days after 5-FU treatment, Gata2 expression increased 2.6-fold in WT bone marrow (Fig. 3G) and was restored to steady-state levels by day 11. By contrast, no significant induction was detected in +9.5(E-box)−/− mice at either time. To assess whether reduced cell numbers characterize +9.5(E-box)−/− bone marrow during regeneration, we quantified immunophenotypic HSPCs in untreated and treated (9 or 11 days after 5-FU) mice. In untreated mice, the populations tested did not differ significantly (Fig. 3, H and I, and fig. S5). At days 9 and 11, HSCs increased 16- and 8-fold, respectively, in WT mice. Although +9.5(E-box)−/− HSCs failed to expand at day 9, by day 11, they increased 10-fold to levels equivalent to WT. The delayed expansion did not decrease myelo-erythroid progenitor regeneration at day 11. Thus, while the +9.5 E-box variant delays HSPC regeneration kinetics after myeloablation, it does not abrogate regeneration.
Combinatorial Gata2 enhancer variant bone marrow failure model reveals hematopoietic regeneration mechanism
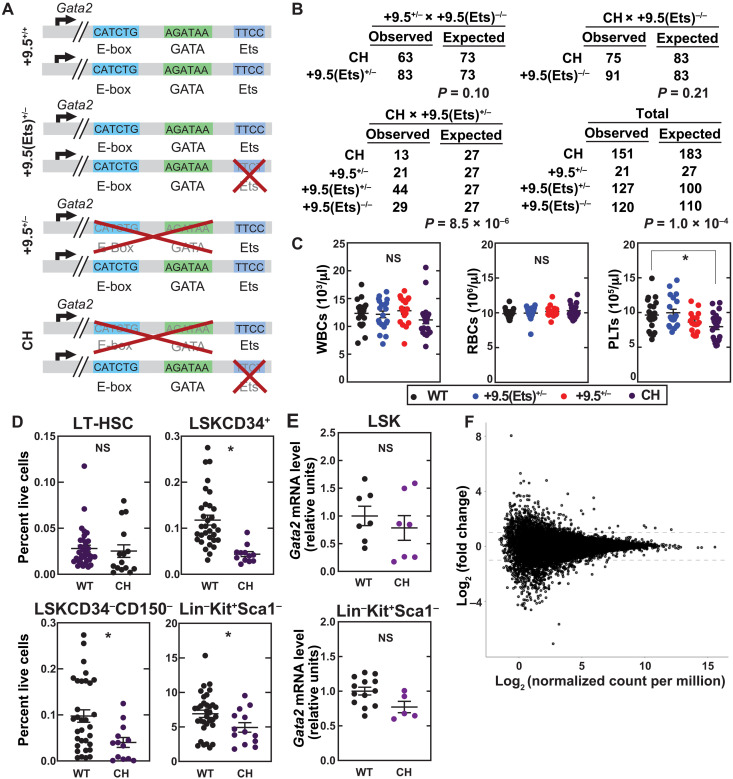
In human GATA2 deficiency syndrome, epigenetic silencing of the normal GATA2 allele correlates with disease phenotype severity (40). Thus, a single transcriptionally active WT allele appears to suppress disease phenotypes. A mechanistic explanation may involve GATA2-mediated autoregulation of GATA2 transcription (41) when the remaining WT GATA2 allele is silenced and/or GATA2 disease mutations reduce GATA2 levels/activity below that required for autoregulation (42). These results support our model that GATA2 levels/activity are critical to sustain HSPC generation and function, GATA2-dependent processes are differentially sensitive to alterations in GATA2 levels, and greater reductions in GATA2 levels/activity translate into more severe disease phenotypes (4, 7, 43). Heterozygous Gata2 mutations that modestly reduce Gata2 levels/activity in the mouse, e.g., +9.5 Ets motif mutations, fail to elicit major phenotypes, and a homozygous Ets motif variant yields relatively normal adult mice in the steady state, with a regenerative hematopoiesis defect only detected under myeloablation conditions (32). By contrast, a Gata2 heterozygous mutation with concomitant epigenetic silencing of the normal allele has a greater impact on GATA2 levels/activities and more severe phenotypes (40). In principle, it might be possible to model this scenario by combining a heterozygous Gata2 mutation with a defect in a chromatin modifying or remodeling protein that opposes Gata2 silencing. However, such proteins regulate many genes, and pleiotropic defects may affect GATA2 levels/activity via complex direct and indirect mechanisms. We therefore combined a heterozygous Ets motif variant with a more severe Gata2 variant on the second allele to mimic an epigenetically silenced allele. Loss of +9.5 E-box-spacer-GATA reduced local chromatin accessibility and transcription of the mutant allele (34). Thus, the 9.5 E-box-spacer-GATA deletion may mimic the impact of an epigenetic silencing mechanism on the locus. With a heterozygous Ets variant and an E-box-spacer-GATA deletion on the other allele (Fig. 4A), the majority of CH mice survived to adulthood; pups were born at Mendelian ratios with two of the three breeding schemes used (Fig. 4B).
Fig. 4. Enhancer-engineered bone marrow failure mouse model.
(A) Model depicting the motif configurations within the Gata2 +9.5 in WT, +9.5(Ets)+/−, +9.5+/−, and CH mice. (B) Genotypes from breeding schemes used to generate CH mice. Probability was calculated using chi-square tests. (C) Steady-state peripheral blood parameters. Data are represented as mean ± SEM; *P < 0.05 (Tukey’s multiple comparisons test). (D) Flow cytometric quantification of LT-HSC (Lin−Sca1+Kit+CD150+CD48−), LSKCD34+, LSKCD34−CD150−, and Lin−Kit+Sca1−. (E) qPCR of purified cells. (F) RNA-seq of LT-HSCs (Lin−Sca1+Kit+CD150+CD34−).
Because several hundred days of chronic inflammation were required to reveal a deleterious interaction between inflammation and the Ets motif variant, we used the CH model to analyze how acute stressors, including inflammation, interface with Gata2 variants. We conducted experiments to determine whether +9.5 E-box-spacer-GATA and Ets are both required for enhancer activity and hematopoietic regenerative functions, E-box-spacer-GATA is not required, or E-box-spacer-GATA exerts unique functions, and CH phenotypes cannot be predicted from individual heterozygous variant phenotypes.
Although patients with GATA2 deficiency syndrome exhibit monocytopenia, lymphopenia, and dendritic cell depletion (22, 25, 44, 45), in the steady state, Gata2+/− (13) and +9.5(Ets)−/− mice (32) lack major peripheral blood cell alterations. We determined whether dysregulated Gata2 expression in CH mice alters steady-state hematopoiesis. CH mice exhibited a 20% decrease (P = 0.02) in peripheral blood platelets (Fig. 4C), which has been reported in a subset of GATA2 deficiency syndrome patients (39), as well as reduced levels of bone marrow LSKCD34+, LSKCD34−CD150−, and Lin−Kit+Sca1+ progenitors (2.7-, 2.4-, and 1.4-fold, respectively; Fig. 4D). These decreases are not accompanied by significantly reduced Gata2 expression in these populations (Fig. 4E). We tested whether these changes reflected reduced Gata2 expression in LSKCD34−CD150+ LT-HSCs. While 1377 genes exhibited expression changes >2-fold, these changes were insignificant (adjusted P > 0.9995) (Fig. 4F). Thus, in the steady state, CH mutant mice have only minor hematopoietic defects, and CH LT-HSCs have a largely intact transcriptional profile.
If the +9.5 E-box-spacer-GATA motif shares Ets motif pro-regenerative activity, then CH regenerative hematopoiesis should be severely impaired. Alternatively, if the +9.5 E-box-spacer-GATA motif is committed to other mechanisms, CH mice may phenocopy Ets+/− mice. We administered two doses of 5-FU (250 mg/kg) with an 11-day interval to deplete cycling cells and expand HSPCs (39). WT mice had a 93% survival rate (12 of 13) at day 22 (Fig. 5A), whereas 66% (10 of 13) and 43% (3 of 7) of +9.5(Ets)+/− and 9.5+/− mice survived, respectively. Survival of CH mice was severely compromised, with only 8% (1 of 12) of mice surviving to day 22 and with a median survival of 14 days. As CH and +9.5(Ets)−/− regenerative capacities after myeloablation were compromised, the +9.5 E-box-spacer-GATA motif is critical for +9.5 pro-regenerative activity.
Fig. 5. Gata2 +9.5 E-box;GATA and Ets motif pro-regenerative functions.
(A) Kaplan-Meier survival curve of mice following two 5-FU doses (250 mg/kg; days 0 and 11) [WT (n = 13), +9.5(Ets)+/− (n = 18), +9.5−/− (n = 7), and CH (n = 12) from two experiments]. Significance was determined using the log-rank test. (B) Flow cytometry scheme for quantification of LSK (Lin−CD48−Sca1+Kit+), HSC (Lin−CD48−Sca1+Kit+CD150+), and MPP (Lin−CD48−Sca1+Kit+CD150−). Bone marrow was harvested 9 or 10 days after vehicle (PBS) or 5-FU (250 mg/kg) treatment. (C) Quantification of HSC, MPP, and LSK from total bone marrow (percentage of live bone marrow cells) [PBS: WT (n = 11), +9.5(Ets)+/− (n = 6), +9.5−/− (n = 7), and CH (n = 6); day 9: WT (n = 12), +9.5(Ets)+/− (n = 8), +9.5−/− (n = 8), and CH (n = 4); day 10: WT (n = 12), +9.5(Ets)+/− (n = 9), +9.5−/− (n = 8), and CH (n = 5)]. False discovery rate–adjusted *P < 0.05, **P < 0.01, ***P < 0.001, Welch’s unequal variance t tests with Benjamini, Krieger, and Yekutieli multiple-test correction. (D) Representative H&E and anti-GATA2 staining of femurs. Each panel represents a single mouse. Scale bars, 50 μm. IHC, immunohistochemistry. (E) Quantification of GATA2+ mononuclear cells from (D) (n = 4 to 6 per condition from three experiments). (F) mRNA quantification from total bone marrow after vehicle (PBS), 9-day 5-FU, or 10-day 5-FU (250 mg/kg) treatment (n = 4 to 12 per condition from five experiments). (G) mRNA quantification from sorted HSC (Lin−Sca1+Kit+CD150+CD34−) or LSKCD34+ MPP (Lin−Sca1+Kit+CD34+) after vehicle (PBS), 7-day 5-FU, or 8-day 5-FU (250 mg/kg) treatment (n = 4 to 22 per condition from eight experiments). *P < 0.05, **P < 0.01, ***P < 0.001, Tukey’s multiple comparisons test.
To establish how the CH variant regulates HSPCs to affect hematopoietic regeneration, we quantified immunophenotypic HSPC populations from bone marrow 9 and 10 days after a single administration of vehicle or 5-FU. In the steady state, LT-HSC (Lin−Sca1+Kit+CD48−CD150+) levels in CH mice were no different from WT controls (Fig. 5, B and C, and fig. S6). At 9 days after 5-FU treatment, WT HSCs increased 17-fold relative to vehicle-treated mice, while both +9.5 heterozygous groups were delayed in mounting a regenerative response. HSCs in +9.5(Ets)+/− and +9.5+/− mice increased 3.1- and 1.5-fold, respectively. At day 10, WT HSCs expanded 18-fold, while HSCs expanded 12- and 4.5-fold in +9.5(Ets)+/− and +9.5+/− mice, respectively. CH HSCs remained at or below the steady-state levels of vehicle-treated mice.
Analysis of steady-state multipotent progenitors (MPPs) (Lin−Sca1+Kit+CD48−CD150−) revealed fewer MPPs in heterozygotes [+9.5(Ets)+/−, 2.3-fold; +9.5+/−, 2.3-fold] and CH mice (8.3-fold). These defects persisted after 5-FU treatment, with the exception of +9.5(Ets)+/− 10 days after treatment. Steady-state common myeloid progenitor (CMP) (Lin−Sca1−Kit+FcγR−CD34+) levels decreased 2.6-fold in CH mice (fig. S7). While regeneration of +9.5(Ets)+/− and +9.5+/− CMP and MEP (Lin−Sca1−Kit+FcγR−CD34−) populations occurred by day 10, and regeneration of +9.5(Ets)−/− populations was detected at day 11 (32), CH progenitors failed to increase. Further supporting this conclusion, histological analysis of bone marrow from mice 10 days after treatment with 5-FU or phosphate-buffered saline (PBS) revealed that 5-FU–treated CH mice had hypocellular marrow, and immunohistochemical analysis with an affinity-purified, highly specific anti-GATA2 antibody revealed fewer GATA2-positive cells (Fig. 5, D and E).
Gata2 expression increases in regenerating bone marrow, with WT levels 2.6- and 2.3-fold higher at days 9 and 10, respectively, after 5-FU treatment (Fig. 5F). To determine whether this was intrinsic to HSPCs, we quantified Gata2 expression in LT-HSCs and LSKCD34+ MPPs. Within LT-HSCs, Gata2 increased 1.9-fold 7 days after 5-FU treatment, while no induction was detected in CH LT-HSCs at day 7 or 8 (Fig. 5G). Similar results were obtained with LSKCD34+ cells, with only WT cells showing 2- and 2.2-fold increases in Gata2 expression at days 7 and 8, respectively. Thus, single-allele +9.5 heterozygous mutations impaired hematopoietic regeneration, and biallelic mutations yielded substantially more severe defects.
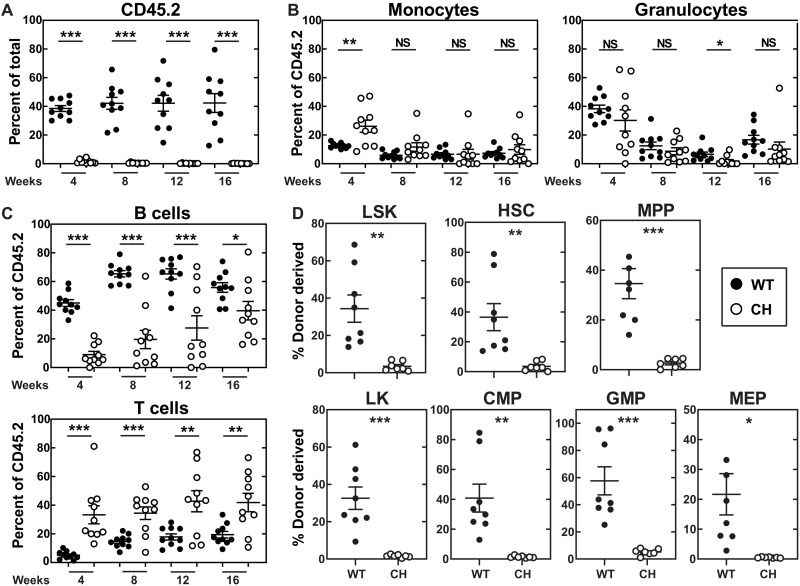
+9.5−/− fetal liver (20) and AGM (33) lack LT repopulating HSCs, and HSCs are ~3-fold lower in +9.5+/− fetal liver and AGM (20, 33). Bone marrow HSCs from adult +9.5(Ets)−/− mice exhibit a threefold decrease in LT repopulating activity (32). As CH mice are severely defective in hematopoietic regeneration after myeloablation, we determined whether the CH mutation compromises HSC activity in a competitive transplant assay. Four weeks after transplant, the number of CH progeny was 40-fold lower than progeny from WT HSPCs (Fig. 6A). CH progeny also showed a skewed lineage distribution with increased monocytes (26 versus 13%) and T cells (33 versus 5.1%) and reduced B cells (9.0 versus 45%) relative to WT donors (Fig. 6, B and C). At 8 weeks after transplant, the CH contribution was 90-fold lower than WT, and the skewed lineage distribution of B cells (20 versus 65%) and T cells (34 versus 15%) persisted. At 12 and 16 weeks after transplant, the CH contribution was 266- and 280-fold lower, respectively, with persistent lymphoid alterations sustained. Notably, B cell lymphopenia is one of the most penetrant phenotypes in human GATA2 deficiency syndrome patients (35, 44). After 16 weeks, donor-derived bone marrow HSPCs were quantified. In contrast to the 36% of donor-derived WT HSCs, CH donors only generated 3.5%. All HSPC populations were impaired, with CH contributions 11- to 40-fold lower than WT. The repopulating defect was sustained after secondary transplantation and not restored by passage through a WT microenvironment (fig. S8). Although CH and +9.5(Ets)−/− mice share phenotypes, CH mice had more severe LT repopulating and differentiated cell output phenotypes.
Fig. 6. Combinatorial E-box;GATA and Ets motif mutation abrogates LT-HSC repopulating activity.
(A) Repopulating activity in peripheral blood 16 weeks after competitive transplant of total bone marrow (n = 9 to 10 per genotype) (two-tailed unpaired Student’s t test). (B) Percentage of donor-derived myeloid cells. (C) Percentage of donor-derived lymphoid cells. (D) Donor-derived hematopoiesis of LSK, HSC, and MPP within bone marrow 16 weeks after competitive transplant (n = 7 to 8 per genotype) (two-tailed unpaired Student’s t test). Donor-derived hematopoiesis of LS−K, CMP, GMP, and MEP within bone marrow 16 weeks after competitive transplant (n = 7 to 8 per genotype) (two-tailed unpaired Student’s t test). *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired Student’s t test.
Linking enhancer variants to therapeutic stem cell mobilization efficacy
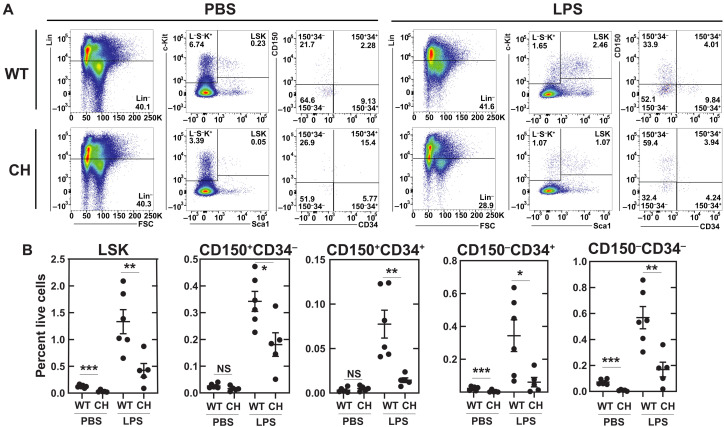
Given the sensitivity of the +9.5(Ets)−/− mutant to chronic inflammation, we determined whether CH mutant mice were also hypersensitive to acute inflammatory stress. As a component of gram-negative bacterial membranes, lipopolysaccharide (LPS) activates the membrane receptor TLR4 (Toll-like receptor 4) to promote the release of pro-inflammatory cytokines (46). LPS acts on HSCs to induce inflammatory signaling and proliferation (47, 48). Twenty-four hours after LPS (0.5 mg/kg) administration, CH LSK levels were 3.1-fold lower than WT. Considering that MPPs are reduced in steady-state CH mice (Fig. 4), and LPS induces Sca1 on progenitor cells (36), we determined whether this decrease reflects lower LT-HSCs and/or decreased progenitor cell mobilization. Immunophenotypic LT-HSCs (LSKCD150+CD34−) were 1.9-fold lower in treated CH versus WT mice. LPS reduced all LSK subpopulations; CD34−CD150−, CD34+CD150−, and CD34+CD150+ were 3.4-, 5.7-, and 5.3-fold lower, respectively (Fig. 7, A and B). Thus, the CH mutation attenuated acute inflammation-induced HSPC expansion.
Fig. 7. E-box;GATA and Ets motif variants attenuate hematopoietic regenerative response to inflammation.
(A) Flow cytometry scheme for quantification of LSK (Lin−Sca1+Kit+), HSC (Lin−Sca1+Kit+CD150+CD34−), CD150+CD34+ (Lin−Sca1+Kit+CD150+CD34+), CD150−CD34+ (Lin−Sca1+Kit+CD150−CD34+), and CD150−CD34− (Lin−Sca1+Kit+CD150−CD34−). Bone marrow was harvested 24 hours after a single dose of LPS (0.5 mg/kg) or vehicle control. (B) Quantification of HSPC populations identified in (A) (percentage of live bone marrow cells). N = 4 to 6 per condition from three experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed unpaired Student’s t test.
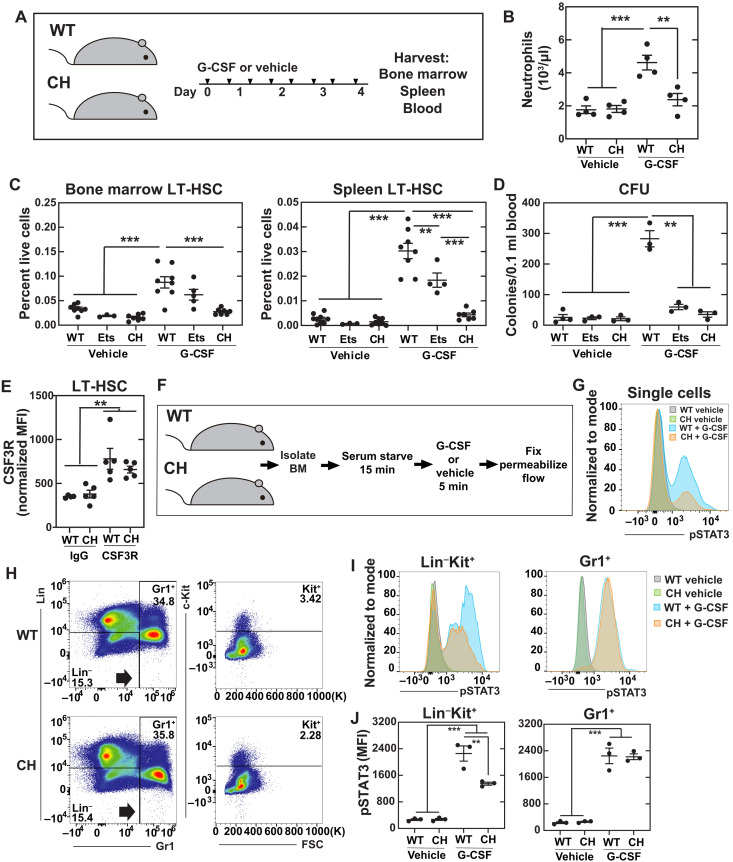
Granulocyte colony-stimulating factor (G-CSF) is among the cytokines/chemokines induced by infection (49), in part, via TLR4 signaling (50). Clinically, G-CSF is used to mobilize bone marrow HSCs to the blood for collection before transplantation. We tested whether the +9.5 mutation prevents HSPC expansion, mobilization, and/or function in response to diverse stimuli or whether these activities are +9.5 mutation insensitive. After eight doses of G-CSF (Fig. 8A), circulating neutrophil numbers increased 2.6-fold in WT, whereas neutrophils did not increase in the CH mice (Fig. 8B). G-CSF treatment increased bone marrow HSCs 2.6- and 3.3-fold in WT and +9.5(Ets)−/− mice, respectively, whereas the CH mutation abrogated this response (Fig. 8C). As a metric of mobilization, splenic HSCs were also quantified. After G-CSF treatment, WT and +9.5(Ets)−/− splenic HSCs expanded 11- to 25-fold, while CH HSCs were unchanged. To functionally analyze mobilized HSPCs, we conducted colony-forming unit (CFU) assays with peripheral blood. While CFU increased 11-fold in WT peripheral blood after treatment, neither +9.5(Ets)−/− nor CH showed increased CFU. Although +9.5(Ets)−/− HSCs expanded and mobilized, the cells lacked colony-forming activity.
Fig. 8. Enhancer variants disrupt clinically relevant stem cell mobilization.
(A) Mice were given eight doses of rhG-CSF (125 μg/kg) over 5 days. (B) Quantification of peripheral blood neutrophils. N = 4 per condition from two experiments. (C) Flow cytometric quantification of HSC (Lin−Sca1+Kit+CD150+CD48−) from bone marrow (BM) or spleen. N = 3 to 9 per condition from six experiments. (D) CFUs from 0.1 ml of peripheral blood. N = 3 to 4 per condition from two experiments. *P < 0.05, **P < 0.01, ***P < 0.001, Tukey’s multiple comparisons test. (E) Quantitation of CSF3R on LT-HSCs (Lin−CD48−Sca1+Kit+CD150+) by flow cytometry. (F) Experimental layout for ex vivo cytokine stimulation. (G) Representative phospho-flow cytometry quantitating pSTAT3 levels in all cells from WT (n = 3) and CH (n = 3) after a 5-min G-CSF stimulation. (H) Flow cytometric scheme for isolation of populations. (I) Representative phospho-flow plots of pSTAT3 levels in Gr1+ and Lin−Gr1−Kit+ populations. (J) Quantitation of (H). **P < 0.01, ***P < 0.001, Tukey’s multiple comparisons test.
We determined whether defective CH HSC mobilization resulted from decreased levels of the G-CSF receptor CSF3R. CSF3R levels were unchanged in all populations analyzed (Fig. 8E and fig. S9). To test whether G-CSF signaling capacity is affected by the CH mutation, we determined whether G-CSF treatment of bone marrow ex vivo induced signal transducer and activator of transcription 3 (STAT3) phosphorylation using a phospho-flow cytometry assay (Fig. 8F). The CH mutation attenuated G-CSF–mediated induction of phospho-STAT3 (pSTAT3) (Fig. 8G). While G-CSF induced pSTAT3 8.5-fold in WT Lin−Kit+ cells, the induction decreased significantly to 4.9-fold in CH (Fig. 8, H to J). Signaling in Gr1+ granulocytes was indistinguishable between genotypes. Thus, the CH mutation attenuated G-CSF–induced signaling and HSC mobilization, abrogated HSPC expansion, and compromised HSPC function.
These results support a model in which an enhancer can harbor motifs with shared or distinct (quantitatively or qualitatively) activities, the coordination of which endows the hematopoietic system with functional plasticity to accommodate demands imposed by development, physiology, and stress. As this plasticity includes HSC capacity to respond efficiently to stress and to a therapeutically important stem cell–mobilizing drug, these results establish a critical link between noncoding genome variation and stress- and therapy-induced stem cell mobilization.
DISCUSSION
Interindividual differences in genome sequence and structure underlie developmental, physiological, and pathogenic phenotypes. Although interpreting the phenotypic consequences of protein coding variants can often be straightforward, deducing the consequences of variants identified within noncoding sequences of the genome is, by comparison, fraught with challenges. In principle, variation within enhancer motifs reduces or increases enhancer activity and, contingent upon concordance or discordance between transcriptional and posttranscriptional mechanisms, may skew the protein concentration to alter cell and organismal phenotypes. The clustered motifs that constitute enhancers can be essential, modulatory, or nonfunctional in chromatin or functional only in select cell/tissue types and/or contexts. Thus, the rules to interpret functional ramifications of enhancer variants are not established. A variant might only manifest activity in specific physiological or pathological contexts, and functional analyses in cultured cells or mice with reporter systems may or may not unveil salient activities. Germline mutations introduce the added complexity of dysregulating processes in hematopoietic cells nonautonomously, and therefore, functional consequences may only be manifested in heterocellular populations. Rigorous rules to interpret the functional consequences of enhancer variants are not yet developed.
It is instructive to consider this enhancer variant classification problem in the context of the GATA2 Ets motif variant that creates bone marrow failure predisposition as a common prelude to acute leukemia. Nonhematopoietic cells express multiple Ets transcription factor family members (51), GATA2 is expressed in select nonhematopoietic cells (17), and nonhematopoietic bone marrow microenvironment cells are critical for hematopoiesis (52–54). Endothelial-specific deletions of Jag1, Ptn, and Egf or impairment of Vegfr, VE-cadherin, E-selectin, or Kitl impairs HSC regenerative activity (55). GATA2 establishes/maintains endothelial cell transcriptional networks (56, 57), and Gata2 +9.5 enhancer variant and Gata2 conditional knockout embryos exhibit defective vascular integrity (20, 58). We tested whether the Ets motif requirement for HSC activity involves hematopoietic cell–autonomous or nonautonomous mechanisms. Transplantation of WT bone marrow cells into WT or Ets motif−/− recipients revealed that WT cells operate normally in the Ets motif variant microenvironment. Thus, a hematopoietic cell–autonomous function of the Ets motif promotes hematopoiesis. Similarly, while HSCs deficient in the Ets factor Etv2 are defective in reconstituting hematopoiesis, a vascular endothelial cadherin (VEC)–Cre;Etv2 conditional knockout does not affect HSC activity (39).
The hematopoietic cell–autonomous Ets motif contribution to +9.5 enhancer activity is highly context dependent and not evident under steady-state conditions, despite the +9.5 activity to promote GATA2 expression and developmental hematopoiesis in the embryo (15, 20, 33). In the context of the swift hematopoietic regeneration that occurs after myeloablation, the Ets motif variant had defective regeneration capacity. Thus, the variant mice had a markedly elevated mortality in response to LT inflammation.
Because the deleterious impact of the Ets motif variant is restricted to stress-dependent processes, this variant might not seamlessly conform to the clinical genetic designations “pathogenic,” “potentially pathogenic,” “likely benign,” “benign,” or “variant of undetermined significance” (9, 59). The Ets motif mutation has been deemed pathogenic by ClinVar (NM_001145661.2) (9) and falls within Tier Ib of the clinical practice guidelines from the Association for Molecular Pathology (AMP) (60). However, hematologic disease linked to this variant is incompletely penetrant. In addition to variable age of onset and disease presentation, adult carriers can remain asymptomatic at the time of analysis (21, 35). We therefore deemed that the Ets motif variant CP in that pathogenicity is revealed only in circumscribed contexts; before these circumstances, the variant appears to be benign. Thus, the CP categorization provides an additional dimension to the functional inferences afforded by existing clinical genetic designations and more accurately defines the consequences of this type of variant. Penetrance of pathogenicity would be dictated by intraindividual parameters that dynamically change with time, including additional acquired mutations and environmentally induced epigenetic aberrations that silence the WT allele. Epigenetic silencing of the WT allele in such contexts has been described (40), and CP mechanisms may underlie the enigmatic variable penetrance of GATA2 deficiency syndrome. GATA2 coding mutations, which also cause GATA2 deficiency syndrome, can be loss of function, yielding reduced protein levels, or mutant proteins in which select activities are lost or retained as measured ex vivo (7, 43). The CP paradigm may therefore be applicable to a broader spectrum of variants that alter GATA2 levels/activities, although clinical analyses of larger patient cohorts may unveil similarities and differences between clinical attributes of patients with distinct GATA2 mutations. The context-dependent impact of a CP variant combined with interpatient genetic and environmental differences may render genotype-phenotype relationships patient specific and make broad extrapolations untenable.
On the basis of the insights articulated above, it is of considerable interest to elucidate how CP variants are triggered to yield overt pathogenesis. While myeloablation or chronic inflammation imposed upon the homozygous Ets motif variant severely disrupted regeneration, several hundred days of inflammation were required to manifest phenotypes. The Gata2 CH bone marrow failure model is characterized by relatively normal developmental hematopoiesis and adult steady-state hematopoiesis, yet the mice are markedly more vulnerable to stressors in comparison to other Ets motif variants. As the CH mice were hypersensitive to myeloablation and acute inflammation and exhibited an attenuated response to a clinical HSC-mobilizing agent, GATA2-driven regenerative hematopoiesis mitigates damage resulting from chemotherapy and inflammation and promotes G-CSF HSC-mobilizing activity. As the enhancer variant controlled HSC responsiveness to a therapeutically important stem cell–mobilizing drug, these results also link noncoding genome variation with therapeutic stem cell mobilization efficacy. On the basis of the prominent GATA2 activities described here (Fig. 9), it will be instructive to consider how the full ensemble of macromolecular components constituting the GATA2 regulatory network, including proteins, lipids, and metabolites, generates a vital protective system conferring resilience to the hematopoietic system under diverse stressors, and how genetic variants in the genes encoding the components of these pathways resemble or differ from those of GATA2 variants.
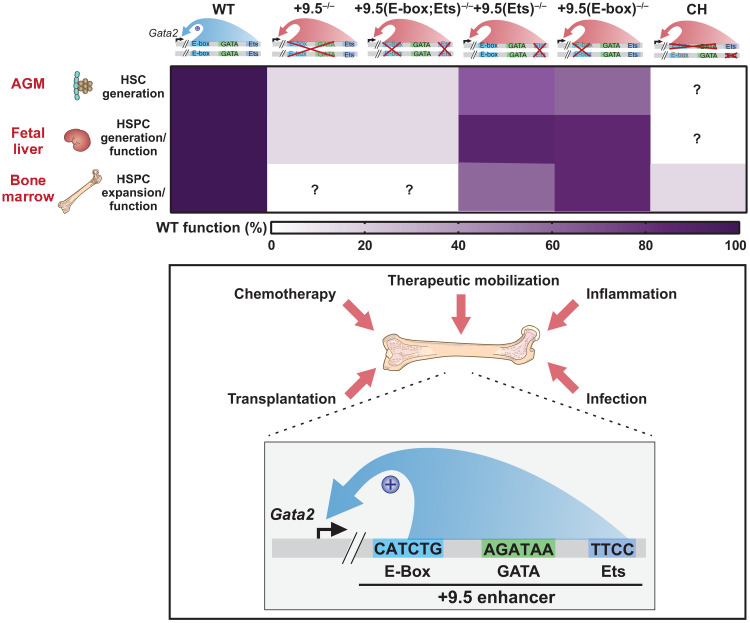
Fig. 9. Biological and pathological consequences of CP Gata2 enhancer variants.
Top: The diagram illustrates the murine Gata2 +9.5 alleles analyzed here and their capacity to support hematopoiesis in the AGM, fetal liver, and bone marrow. The heat map depicts their relative activities as a percent of the WT activity. Bottom: The model illustrates the vital +9.5 enhancer activity to mediate adult hematopoiesis in response to stressors, including chemotherapy, inflammation, infection, and therapeutic HSC mobilization with G-CSF. As the motifs constituting the +9.5 enhancer are differentially important in distinct physiological and pathological contexts, genetic variants within these motifs differentially affect GATA2-dependent processes. Thus, we term these variants “conditionally pathogenic.”
METHODS
Statistical analysis
The results are presented as mean ± SEM. Multiple independent cohorts were used in each experiment. Statistical comparisons were performed using Welch’s unequal variance t tests (significance cutoff of P < 0.05) (GraphPad Prism) with correction of statistical overrepresentation of functions calculated using Benjamini, Krieger, and Yekutieli multiple-test correction procedure; Student’s t tests with correction of statistical overrepresentation of functions calculated using Benjamini-Hochberg multiple-test correction procedure; or Tukey’s multiple comparisons test (GraphPad Prism). Outliers were detected before analysis with two-sided Grubbs’ test. A log-rank test was performed on the Kaplan-Meier survival curve (GraphPad Prism).
Generation of mice containing Gata2 variants
Pronuclear injection was conducted in the C57BL/6J background at the zygote (one-cell) stage at the University of Wisconsin–Madison Biotechnology Center using recombinant Cas9, guide RNA (gRNA) (5′-TCTCCTGCCGGAGTTTCCTATCCGG). The E-box corruption, without additional mutations, was detected in 1 of 18 live pups by DNA sequencing at and surrounding the mutation site. Further genotyping was performed using primers Gata2 −78F CACACAGCGGCCACCAA and Gata2 +120R AAAGCGGGTGAACGATTTAAAC.
+9.5+/−:+9.5(Ets)+/− CH mice were generated through three mating schemes: (i) +9.5+/− (20) and +9.5(Ets)−/− (32) (expected 50% of progeny), (ii) CH and +9.5(Ets)−/− (expected 50% of progeny), and (iii) CH and +9.5(Ets)+/− (expected 25% of progeny).
Quantitative real-time RT-PCR
Following procedure previously described in (32), total RNA was purified with TRIzol (Invitrogen). Deoxyribonuclease (Invitrogen) treatment was performed on 0.1 to 1 μg of RNA at 25°C for 15 min, followed by addition of 2.5 mM EDTA at 65°C for 10 min. Complementary DNA (cDNA) was prepared by annealing with 250 ng of a 1:5 mixture of random hexamer and oligo (dT) primers; incubated with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) with 10 mM dithiothreitol, RNasin (Promega), and 0.5 mM deoxyribonucleotide triphosphates (dNTPs) at 42°C for 1 hour; and heat-inactivated at 95°C for 5 min. cDNA was analyzed in reactions (20 μl) containing 2 μl of cDNA, primers, and 10 μl of Power SYBR Green (Applied Biosystems) by real-time reverse transcription polymerase chain reaction (RT-PCR) with a ViiA 7 real-time RT-PCR cycler (Applied Biosystems). Standard curves of serial 1:5 dilutions of cDNAs were prepared from control cDNA with the highest predicted gene expression. Values were normalized to the standard curve and 18S control.
Drug and chemical treatments
To induce in vivo cycling of HSCs, mice were injected with polyI:C (10 mg/kg) intraperitoneally (tlrl-pic; InvivoGen) as per Walter et al. (37). Mice were harvested either when moribund or after 10 cycles of treatment.
Myeloablation was induced by 5-FU (F6627; Sigma-Aldrich) administered intraperitoneally with a single dose (250 mg/kg) or two doses (250 mg/kg) at 0 and 11 days. Blood samples were collected by retro-orbital bleeding (~30 μl per collection), and hematologic parameters were quantified on a HEMAVET CBC instrument. Mice were euthanized with CO2, and bone marrow from femurs and tibias of 8- to 14-week-old mice was collected by removing epiphyses and flushing the bone marrow with a 25-gauge needle and syringe containing Iscove’s modified Dulbecco’s medium (IMDM) + 10% fetal bovine serum (FBS) (32).
A single dose of LPS (L2630; Sigma-Aldrich) was administered intraperitoneally at a dose of 0.5 mg/kg. Mice were harvested 24 hours following the dose.
Recombinant human G-CSF (rhG-CSF) (300-23; PeproTech) was administered subcutaneously at a dose of 125 μg/kg twice a day (eight divided doses) beginning in the evening of the first day, and mice were harvested 3 hours following the final dose. Stimulation of cells to detect intracellular signaling was performed essentially as described (61). Serum starvation was performed for 15 min. Cells were stimulated for 5 min using rhG-CSF (125 ng/μl).
Flow analysis
Bone marrow was dissociated and resuspended in IMDM with 10% FBS and passed through 25-μm cell strainers to obtain single-cell suspensions before antibody staining. All antibodies were purchased from eBioscience/Thermo Fisher Scientific unless stated. Lineage markers for the LSK populations were stained with fluorescein isothiocyanate (FITC)–conjugated antibodies B220 (11-0452), CD3 (11-0031), CD4 (11-0041), CD5 (11-0051), CD8 (11-0081), CD48 (11-0481), Gr-1 (11-5931), and TER-119 (11-5921). Other surface proteins were detected with phycoerythrin (PE)–conjugated CD150 (115904; BioLegend), peridinin chlorophyll protein (PerCP)–Cy5.5–conjugated Sca1 (45-5981), and allophycocyanin (APC)–conjugated c-Kit (2B8, 17-1171) antibodies. Analysis of myeloid progenitors was conducted as described by Johnson et al. (14). Lineage markers were stained with FITC-conjugated B220, CD3, CD4, CD5, CD8, CD19 (11-0193), immunoglobulin M (IgM) (11-5890), Il7Ra (11-1271), AA4.1 (11-5892), and TER-119 antibodies. Other surface proteins were detected with PE-conjugated FcγR (12-0161), eFluor 660–conjugated CD34 (50-0341), PerCP-Cy5.5–conjugated Sca1 (45-5981), PE-Cy7–conjugated c-Kit (BioLegend, 105814), or APC-CD114 (CSF3R; R&D Systems; FAB6039A) and APC-IgG (R&D Systems; IC016A). After staining, cells were washed with PBS and resuspended in IMDM + 10% FBS + 4′,6-diamidino-2-phenylindole (DAPI) and analyzed on an LSR II flow cytometer (BD Biosciences) (32).
For intracellular antigens, methanol-fixed cells were stained with rabbit antibodies against phospho(Tyr705)–STAT3 (pSTAT3) (9145; Cell Signaling Technology) for 30 min and then incubated in APC-conjugated goat anti-rabbit (1:200) (711-136-152; Jackson ImmunoResearch) with antibodies listed above for 30 min at room temperature. Samples were analyzed using a Thermo Fisher Scientific Attune NxT flow cytometer. Values for pSTAT3 levels were calculated by median fluorescence intensities (MFIs) using FlowJo version 10.6.2 (BD Life Sciences) and normalized to the maximum overall value within each experiment (relative MFI).
RNA sequencing
LT-HSCs (500 to 2000) (Lin−Sca1+Kit+CD150+CD34−) were FACS-isolated and RNA-extracted with the QIAGEN RNeasy Micro Kit (74004) as per the manufacturer’s directions. cDNA libraries were prepared using Takara SMARTer Stranded Total RNA-Seq v2 Pico as described (32). RNA-sequencing (RNA-seq) fragments were aligned by STAR (version 2.5.2b) to the mouse genome (mm10) with basic GENCODE gene annotations (version M22). Gene expression levels were quantified by RSEM (version 1.3.0), and differential expression was analyzed by edgeR (version 3.30.3). A significantly differentially expressed gene was required to have at least twofold changes and an adjusted P value of <0.05. Data have been deposited in Gene Expression Omnibus (GEO) (GSE179790).
Transplantation
Adult C57BL/6 recipient mice (CD45.1+, 6 to 8 weeks old; stock no. 002014, The Jackson Laboratory) were lethally irradiated using an X-RAD 320 irradiator for a single dose of 8.5 Gy. Bone marrow cells were harvested from individual 8-week-old mice (CD45.2+). A total of 106 bone marrow or E15.5 fetal liver cells were mixed with the same number of CD45.1+ bone marrow cells and injected into individual irradiated CD45.1+ recipients (10 per group). For reciprocal transplantation, 2 × 106 WT (CD45.1) bone marrow cells were transplanted into lethally irradiated CD45.2 [WT and +9.5(Ets)−/−] recipients. The transplanted recipient mice were maintained on Irradiated Uniprim Diet (Envigo, catalog no. TD.06596) for 2 weeks. Blood obtained from the retro-orbital venous sinus or bone marrow was isolated after transplantation and analyzed using flow cytometry for donor-derived hematopoiesis (32).
Immunohistochemistry
Femurs were fixed with 10% neutral buffered formalin (DOT Scientific; DSF10800-500) and embedded in paraffin using standard procedures. Sections (10 μm) were stained with hematoxylin and eosin (H&E). Automated immunohistochemistry was performed on Discovery Ultra (Roche-Ventana, Tucson, AZ) using Ultra protocol no. 206 and an affinity-purified rabbit anti-GATA2 antibody (62). Deparaffinization was performed on the instrument, and heat-induced epitope retrieval was completed in CC1 buffer (tris, pH 8.5) for 32 min at 95°C. One hundred microliters of antibody diluted 1:20 in Da Vinci Green (Biocare Medical, Pacheco, CA) was applied for 1 hour at 37°C. After rinsing, Discovery OmniMap anti-rabbit horseradish peroxidase (Roche, Basel, Switzerland) was applied for 8 min at 37°C, followed by rinsing. A Discovery ChromoMap DAB detection kit (Roche) was applied for the preset time, followed by rinsing and off-line counterstaining with diluted Harris modified hematoxylin. Semiquantitative analysis of GATA2 expression was performed by enumerating anti-GATA2 nuclear staining with at least 500 mononuclear cells.
Mouse embryo isolation
Embryos were isolated as per (32). Embryos were obtained from timed matings between heterozygous males and females, with the day of vaginal plug detection considered as day 0.5. Pregnant females were euthanized with CO2, and fresh embryos were transferred into ice-cold PBS for dissection.
Whole-embryo confocal microscopy
E10.5 embryos were fixed, stained, and analyzed as described (33, 63). Embryos were stained with biotinylated anti–PECAM-1 (BD Biosciences, 553371) and anti–c-Kit (BD Biosciences, 553352) antibodies. Samples were mounted in a 1:2 mix of benzyl alcohol (Sigma-Aldrich, 402834) and benzyl benzoate (Acros Organics, 105862500) to increase tissue transparency and visualized with a Nikon A1RS confocal microscope. Three-dimensional reconstructions were generated from Z-stacks (80 to 200 optical sections) using Fiji software.
Study approval
All animal protocols were approved by the University of Wisconsin–Madison Institutional Animal Care and Use Committee in accord with Association for Assessment and Accreditation of Laboratory Animal Care International regulations.
Acknowledgments
Funding: The work was supported by NIH DK68634 (E.H.B.), EvansMDS Foundation (E.H.B.), ASH Scholar Award (A.A.S.), Leukemia & Lymphoma Society Career Development Program (A.A.S.), NHLBI T32 HL07899 Training Grant in Hematology (A.A.S. and D.R.M.), Carbone Cancer Center P30CA014520, and NIH shared instrumentation grant 1S10RR025483-01.
Author contributions: A.A.S. and E.H.B. conceived and designed the research. A.A.S., D.R.M., and K.D.J. conducted experiments. P.L. conducted data analysis. A.A.S. and E.H.B. wrote the manuscript. A.A.S. and E.H.B. edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been deposited in GEO (GSE179790).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S9
REFERENCES AND NOTES
- 1.The ENCODE Project Consortium, Moore J. E., Purcaro M. J., Pratt H. E., Epstein C. B., Shoresh N., Adrian J., Kawli T., Davis C. A., Dobin A., Kaul R., Halow J., Van Nostrand E. L., Freese P., Gorkin D. U., Shen Y., He Y., Mackiewicz M., Pauli-Behn F., Williams B. A., Mortazavi A., Keller C. A., Zhang X.-O., Elhajjajy S. I., Huey J., Dickel D. E., Snetkova V., Wei X., Wang X., Rivera-Mulia J. C., Rozowsky J., Zhang J., Chhetri S. B., Zhang J., Victorsen A., White K. P., Visel A., Yeo G. W., Burge C. B., Lécuyer E., Gilbert D. M., Dekker J., Rinn J., Mendenhall E. M., Ecker J. R., Kellis M., Klein R. J., Noble W. S., Kundaje A., Guigó R., Farnham P. J., Cherry J. M., Myers R. M., Ren B., Graveley B. R., Gerstein M. B., Pennacchio L. A., Snyder M. P., Bernstein B. E., Wold B., Hardison R. C., Gingeras T. R., Stamatoyannopoulos J. A., Weng Z., Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The ENCODE Project Consortium, Snyder M. P., Gingeras T. R., Moore J. E., Weng Z., Gerstein M. B., Ren B., Hardison R. C., Stamatoyannopoulos J. A., Graveley B. R., Feingold E. A., Pazin M. J., Pagan M., Gilchrist D. A., Hitz B. C., Cherry J. M., Bernstein B. E., Mendenhall E. M., Zerbino D. R., Frankish A., Flicek P., Myers R. M., Perspectives on ENCODE. Nature 583, 693–698 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kennedy A. L., Shimamura A., Genetic predisposition to MDS: Clinical features and clonal evolution. Blood 133, 1071–1085 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Churpek J. E., Bresnick E. H., Transcription factor mutations as a cause of familial myeloid neoplasms. J. Clin. Invest. 129, 476–488 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godley L. A., Germline mutations in MDS/AML predisposition disorders. Curr. Opin. Hematol. 28, 86–93 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Klco J. M., Mullighan C. G., Advances in germline predisposition to acute leukaemias and myeloid neoplasms. Nat. Rev. Cancer 21, 122–137 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soukup A. A., Bresnick E. H., GATA2 +9.5 enhancer: From principles of hematopoiesis to genetic diagnosis in precision medicine. Curr. Opin. Hematol. 27, 163–171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X., Feurstein S., Mohan S., Porter C. C., Jackson S. A., Keel S., Chicka M., Brown A. L., Kesserwan C., Agarwal A., Luo M., Li Z., Ross J. E., Baliakas P., Pineda-Alvarez D., DiNardo C. D., Bertuch A. A., Mehta N., Vulliamy T., Wang Y., Nichols K. E., Malcovati L., Walsh M. F., Rawlings L. H., McWeeney S. K., Soulier J., Raimbault A., Routbort M. J., Zhang L., Ryan G., Speck N. A., Plon S. E., Wu D., Godley L. A., ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv. 3, 2962–2979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W. W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H. L.; ACMG Laboratory Quality Assurance Committee , Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bresnick E. H., Johnson K. D., Blood disease-causing and -suppressing transcriptional enhancers: General principles and GATA2 mechanisms. Blood Adv. 3, 2045–2056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., Alt F. W., Orkin S. H., An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Ling K.-W., Ottersbach K., van Hamburg J. P., Oziemlak A., Tsai F.-Y., Orkin S. H., Ploemacher R., Hendriks R. W., Dzierzak E., GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J. Exp. Med. 200, 871–882 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues N. P., Janzen V., Forkert R., Dombkowski D. M., Boyd A. S., Orkin S. H., Enver T., Vyas P., Scadden D. T., Haploinsufficiency of GATA-2 perturbs adult hematopoietic stem-cell homeostasis. Blood 106, 477–484 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Johnson K. D., Kong G., Gao X., Chang Y.-I., Hewitt K. J., Sanalkumar R., Prathibha R., Ranheim E. A., Dewey C. N., Zhang J., Bresnick E. H., Cis-regulatory mechanisms governing stem and progenitor cell transitions. Sci. Adv. 1, e1500503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehta C., Johnson K. D., Gao X., Ong I. M., Katsumura K. R., McIver S. C., Ranheim E. A., Bresnick E. H., Integrating enhancer mechanisms to establish a hierarchical blood development program. Cell Rep. 20, 2966–2979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson K. D., Conn D. J., Shishkova E., Katsumura K. R., Liu P., Shen S., Ranheim E. A., Kraus S. G., Wang W., Calvo K. R., Hsu A. P., Holland S. M., Coon J. J., Keles S., Bresnick E. H., Constructing and deconstructing GATA2-regulated cell fate programs to establish developmental trajectories. J. Exp. Med. 217, e20191526 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsumura K. R., Bresnick E. H.; GATA Factor Mechanisms Group , The GATA factor revolution in hematology. Blood 129, 2092–2102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wozniak R. J., Boyer M. E., Grass J. A., Lee Y., Bresnick E. H., Context-dependent GATA factor function: Combinatorial requirements for transcriptional control in hematopoietic and endothelial cells. J. Biol. Chem. 282, 14665–14674 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Wozniak R. J., Keles S., Lugus J. J., Young K. H., Boyer M. E., Tran T. M., Choi K., Bresnick E. H., Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol. Cell. Biol. 28, 6681–6694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson K. D., Hsu A. P., Ryu M.-J., Wang J., Gao X., Boyer M. E., Liu Y., Lee Y., Calvo K. R., Keles S., Zhang J., Holland S. M., Bresnick E. H., Cis-element mutated in GATA2-dependent immunodeficiency governs hematopoiesis and vascular integrity. J. Clin. Invest. 122, 3692–3704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wlodarski M. W., Hirabayashi S., Pastor V., Starý J., Hasle H., Masetti R., Dworzak M., Schmugge M., van den Heuvel-Eibrink M., Ussowicz M., De Moerloose B., Catala A., Smith O. P., Sedlacek P., Lankester A. C., Zecca M., Bordon V., Matthes-Martin S., Abrahamsson J., Kühl J. S., Sykora K.-W., Albert M. H., Przychodzien B., Maciejewski J. P., Schwarz S., Göhring G., Schlegelberger B., Cseh A., Noellke P., Yoshimi A., Locatelli F., Baumann I., Strahm B., Niemeyer C. M., Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood 127, 1387–1397 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Ganapathi K. A., Townsley D. M., Hsu A. P., Arthur D. C., Zerbe C. S., Cuellar-Rodriguez J., Hickstein D. D., Rosenzweig S. D., Braylan R. C., Young N. S., Holland S. M., Calvo K. R., GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood 125, 56–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu A. P., Johnson K. D., Falcone E. L., Sanalkumar R., Sanchez L., Hickstein D. D., Cuellar-Rodriguez J., Lemieux J. E., Zerbe C. S., Bresnick E. H., Holland S. M., GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood 121, 3830–3837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koegel A. K., Hofmann I., Moffitt K., Degar B., Duncan C., Tubman V. N., Acute lymphoblastic leukemia in a patient with MonoMAC syndrome/GATA2 haploinsufficiency. Pediatr. Blood Cancer 63, 1844–1847 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Donadieu J., Lamant M., Fieschi C., de Fontbrune F. S., Caye A., Ouachee M., Beaupain B., Bustamante J., Poirel H. A., Isidor B., van den Neste E., Neel A., Nimubona S., Toutain F., Barlogis V., Schleinitz N., Leblanc T., Rohrlich P., Suarez F., Ranta D., Chahla W. A., Bruno B., Terriou L., Francois S., Lioure B., Ahle G., Bachelerie F., Preudhomme C., Delabesse E., Cave H., Bellanné-Chantelot C., Pasquet M.; French GATA2 study group , Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica 103, 1278–1287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McReynolds L. J., Yang Y., Yuen Wong H., Tang J., Zhang Y., Mulé M. P., Daub J., Palmer C., Foruraghi L., Liu Q., Zhu J., Wang W., West R. R., Yohe M. E., Hsu A. P., Hickstein D. D., Townsley D. M., Holland S. M., Calvo K. R., Hourigan C. S., MDS-associated mutations in germline GATA2 mutated patients with hematologic manifestations. Leuk. Res. 76, 70–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z., Gao S., Diamond C., Kajigaya S., Chen J., Shi R., Palmer C., Hsu A. P., Calvo K. R., Hickstein D. D., Holland S. M., Young N. S., Sequencing of RNA in single cells reveals a distinct transcriptome signature of hematopoiesis in GATA2 deficiency. Blood Adv. 4, 2656–2670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khandekar M., Brandt W., Zhou Y., Dagenais S., Glover T. W., Suzuki N., Shimizu R., Yamamoto M., Lim K.-C., Engel J. D., A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development 134, 1703–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Snow J. W., Trowbridge J. J., Fujiwara T., Emambokus N. E., Grass J. A., Orkin S. H., Bresnick E. H., A single cis element maintains repression of the key developmental regulator Gata2. PLOS Genet. 6, e1001103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snow J. W., Trowbridge J. J., Johnson K. D., Fujiwara T., Emambokus N. E., Grass J. A., Orkin S. H., Bresnick E. H., Context-dependent function of “GATA switch” sites in vivo. Blood 117, 4769–4772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martowicz M. L., Grass J. A., Boyer M. E., Guend H., Bresnick E. H., Dynamic GATA factor interplay at a multi-component regulatory region of the GATA-2 locus. J. Biol. Chem. 280, 1724–1732 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Soukup A. A., Zheng Y., Mehta C., Wu J., Liu P., Cao M., Hofmann I., Zhou Y., Zhang J., Johnson K. D., Choi K., Keles S., Bresnick E. H., Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J. Clin. Invest. 129, 1180–1192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao X., Johnson K. D., Chang Y. I., Boyer M. E., Dewey C. N., Zhang J., Bresnick E. H., Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J. Exp. Med. 210, 2833–2842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanalkumar R., Johnson K. D., Gao X., Boyer M. E., Chang Y.-I., Hewitt K. J., Zhang J., Bresnick E. H., Mechanism governing a stem cell-generating cis-regulatory element. Proc. Natl. Acad. Sci. U.S.A. 111, E1091–E1100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spinner M. A., Sanchez L. A., Hsu A. P., Shaw P. A., Zerbe C. S., Calvo K. R., Arthur D. C., Gu W., Gould C. M., Brewer C. C., Cowen E. W., Freeman A. F., Olivier K. N., Uzel G., Zelazny A. M., Daub J. R., Spalding C. D., Claypool R. J., Giri N. K., Alter B. P., Mace E. M., Orange J. S., Cuellar-Rodriguez J., Hickstein D. D., Holland S. M., GATA2 deficiency: A protean disorder of hematopoiesis, lymphatics, and immunity. Blood 123, 809–821 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Essers M. A. G., Offner S., Blanco-Bose W. E., Waibler Z., Kalinke U., Duchosal M. A., Trumpp A., IFNα activates dormant haematopoietic stem cells in vivo. Nature 458, 904–908 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Walter D., Lier A., Geiselhart A., Thalheimer F. B., Huntscha S., Sobotta M. C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., Muedder K., Klein C., Jauch A., Schroeder T., Geiger H., Dick T. P., Holland-Letz T., Schmezer P., Lane S. W., Rieger M. A., Essers M. A. G., Williams D. A., Trumpp A., Milsom M. D., Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 520, 549–552 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Lerner C., Harrison D. E., 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol. 18, 114–118 (1990). [PubMed] [Google Scholar]
- 39.Xu C. X., Lee T.-J., Sakurai N., Krchma K., Liu F., Li D., Wang T., Choi K., ETV2/ER71 regulates hematopoietic regeneration by promoting hematopoietic stem cell proliferation. J. Exp. Med. 214, 1643–1653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Seraihi A. F., Rio-Machin A., Tawana K., Bödör C., Wang J., Nagano A., Heward J. A., Iqbal S., Best S., Lea N., McLornan D., Kozyra E. J., Wlodarski M. W., Niemeyer C. M., Scott H., Hahn C., Ellison A., Tummala H., Cardoso S. R., Vulliamy T., Dokal I., Butler T., Smith M., Cavenagh J., Fitzgibbon J., GATA2 monoallelic expression underlies reduced penetrance in inherited GATA2-mutated MDS/AML. Leukemia 32, 2502–2507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H., GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cavalcante de Andrade Silva M., Katsumura K. R., Mehta C., Velloso E. D. R. P., Bresnick E. H., Godley L. A., Breaking the spatial constraint between neighboring zinc fingers: A new germline mutation in GATA2 deficiency syndrome. Leukemia 35, 264–268 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Bresnick E. H., Jung M. M., Katsumura K. R., Human GATA2 mutations and hematologic disease: How many paths to pathogenesis? Blood Adv. 4, 4584–4592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickinson R. E., Milne P., Jardine L., Zandi S., Swierczek S. I., McGovern N., Cookson S., Ferozepurwalla Z., Langridge A., Pagan S., Gennery A., Heiskanen-Kosma T., Hämäläinen S., Seppänen M., Helbert M., Tholouli E., Gambineri E., Reykdal S., Gottfreðsson M., Thaventhiran J. E., Morris E., Hirschfield G., Richter A. G., Jolles S., Bacon C. M., Hambleton S., Haniffa M., Bryceson Y., Allen C., Prchal J. T., Dick J. E., Bigley V., Collin M., The evolution of cellular deficiency in GATA2 mutation. Blood 123, 863–874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinh D. C., Patel S. Y., Uzel G., Anderson V. L., Freeman A. F., Olivier K. N., Spalding C., Hughes S., Pittaluga S., Raffeld M., Sorbara L. R., Elloumi H. Z., Kuhns D. B., Turner M. L., Cowen E. W., Fink D., Long-Priel D., Hsu A. P., Ding L., Paulson M. L., Whitney A. R., Sampaio E. P., Frucht D. M., DeLeo F. R., Holland S. M., Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 115, 1519–1529 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ngkelo A., Meja K., Yeadon M., Adcock I., Kirkham P. A., LPS induced inflammatory responses in human peripheral blood mononuclear cells is mediated through NOX4 and Giα dependent PI-3kinase signalling. J. Inflamm. 9, 1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takizawa H., Fritsch K., Kovtonyuk L. V., Saito Y., Yakkala C., Jacobs K., Ahuja A. K., Lopes M., Hausmann A., Hardt W.-D., Gomariz Á., Nombela-Arrieta C., Manz M. G., Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 21, 225–240.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Caiado F., Pietras E. M., Manz M. G., Inflammation as a regulator of hematopoietic stem cell function in disease, aging, and clonal selection. J. Exp. Med. 218, e20201541 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinton L. J., Nelson S., Boé D. M., Zhang P., Zhong Q., Kolls J. K., Bagby G. J., The granulocyte colony-stimulating factor response after intrapulmonary and systemic bacterial challenges. J. Infect. Dis. 185, 1476–1482 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Schuettpelz L. G., Borgerding J. N., Christopher M. J., Gopalan P. K., Romine M. P., Herman A. C., Woloszynek J. R., Greenbaum A. M., Link D. C., G-CSF regulates hematopoietic stem cell activity, in part, through activation of Toll-like receptor signaling. Leukemia 28, 1851–1860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hollenhorst P. C., McIntosh L. P., Graves B. J., Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Comazzetto S., Shen B., Morrison S. J., Niches that regulate stem cells and hematopoiesis in adult bone marrow. Dev. Cell 56, 1848–1860 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu V. W. C., Scadden D. T., Heterogeneity of the bone marrow niche. Curr. Opin. Hematol. 23, 331–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao X., Xu C., Asada N., Frenette P. S., The hematopoietic stem cell niche: From embryo to adult. Development 145, dev139691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramalingam P., Poulos M. G., Butler J. M., Regulation of the hematopoietic stem cell lifecycle by the endothelial niche. Curr. Opin. Hematol. 24, 289–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linnemann A. K., O’Geen H., Keles S., Farnham P. J., Bresnick E. H., Genetic framework for GATA factor function in vascular biology. Proc. Natl. Acad. Sci. U.S.A. 108, 13641–13646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanki Y., Kohro T., Jiang S., Tsutsumi S., Mimura I., Suehiro J.-i., Wada Y., Ohta Y., Ihara S., Iwanari H., Naito M., Hamakubo T., Aburatani H., Kodama T., Minami T., Epigenetically coordinated GATA2 binding is necessary for endothelium-specific endomucin expression. EMBO J. 30, 2582–2595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim K.-C., Hosoya T., Brandt W., Ku C.-J., Hosoya-Ohmura S., Camper S. A., Yamamoto M., Engel J. D., Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J. Clin. Invest. 122, 3705–3717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landrum M. J., Lee J. M., Benson M., Brown G. R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., Karapetyan K., Katz K., Liu C., Maddipatla Z., Malheiro A., McDaniel K., Ovetsky M., Riley G., Zhou G., Holmes J. B., Kattman B. L., Maglott D. R., ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M. M., Datto M., Duncavage E. J., Kulkarni S., Lindeman N. I., Roy S., Tsimberidou A. M., Vnencak-Jones C. L., Wolff D. J., Younes A., Nikiforova M. N., Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 19, 4–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McIver S. C., Hewitt K. J., Gao X., Mehta C., Zhang J., Bresnick E. H., Dissecting regulatory mechanisms using mouse fetal liver-derived erythroid cells. Methods Mol. Biol. 1698, 67–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Im H., Grass J. A., Johnson K. D., Kim S.-I., Boyer M. E., Imbalzano A. N., Bieker J. J., Bresnick E. H., Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. U.S.A. 102, 17065–17070 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yokomizo T., Yamada-Inagawa T., Yzaguirre A. D., Chen M. J., Speck N. A., Dzierzak E., Whole-mount three-dimensional imaging of internally localized immunostained cells within mouse embryos. Nat. Protoc. 7, 421–431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S9