Abstract
Background:
Stickleback fish are widely used to study the genetic and ecological basis of phenotypic evolution. Although several major loci have now been identified that contribute to evolutionary differences between wild populations, further study of the phenotypes associated with particular genes and mutations has been limited by the difficulty of generating targeted mutations at precise locations in the stickleback genome.
Approach and aims:
We compared different methods of expressing single-guide RNAs (sgRNAs) and Cas9 activity in fertilized stickleback eggs. We used an easily scored pigmentation gene (SLC24A5) to screen for molecular lesions, phenotypic effects, and possible germline transmission of newly induced alleles. We then used the optimized CRISPR methods to target two major evolutionary loci in sticklebacks, KITLG and EDA. We hypothesized that coding region mutations in the KITLG gene would alter body pigmentation and possibly sex determination, and that mutations in the EDA gene would disrupt the formation of most armor plates, fin rays, spines, teeth, and gill rakers.
Results:
Targeted deletions were successfully induced at each target locus by co-injecting one-cell stage stickleback embryos with either Cas9 mRNA or Cas9 protein, together with sgRNAs designed to protein-coding exons. Founder animals were typically mosaic for multiple mutations, which they transmitted through the germline at overall rates of 21 to 100%. We found that the copy of KITLG on the X chromosome (KITLGX) has diverged from the KITLG on the Y chromosome (KITLGY). Predicted loss-of-function mutations in the KITLGX gene dramatically altered pigmentation in both external skin and internal organ, but the same was not true for KITLGY mutations. Predicted loss-of-function mutations in either the KITLGX or KITLGY genes did not lead to sex reversal or prevent fertility. Homozygous loss-of-function mutations in the EDA gene led to complete loss of armor plates, severe reduction or loss of most soft rays in the dorsal, anal, and caudal fins, and severe reductions in tooth and gill raker number. In contrast, long dorsal and pelvic spines remained intact in EDA mutant animals, suggesting that common co-segregation of plate loss and spine reduction in wild populations is unlikely to be due to pleiotropic effects of EDA mutations.
Conclusion:
CRISPR-Cas9 approaches can be used to induce germline mutations in key evolutionary loci in sticklebacks. Targeted coding region mutations confirm an important role for KITLG and EDA in skin pigmentation and armor plate reduction, respectively. They also provide new information about the functions of these genes in other body structures.
Keywords: genome editing, CRISPR-Cas9, stickleback, SLC24A5, KITLG, EDA
INTRODUCTION
Sticklebacks have become a powerful system for combining molecular and ecological studies of vertebrate evolution (Gibson, 2005). More than a century of detailed research has identified dramatic morphological and physiological differences that have evolved repeatedly among abundant marine, lake, and stream populations (Bell and Foster, 1994). More recently, the development of genetic mapping approaches has made it possible to identify some of the major chromosome regions that underlie classical skeletal, pigmentation, physiological, and behavioral differences (Kingsley and Peichel, 2007; Peichel and Marques, 2017). Unfortunately, the candidate regions from genetic mapping studies typically contain many different genes (Miller et al., 2014). It can thus be challenging to determine which particular genes in a quantitative trait locus (QTL) interval are responsible for a phenotype of interest. In addition, multiple different phenotypes have often been mapped to the same or overlapping chromosome regions (Miller et al., 2014; Peichel and Marques, 2017). It is usually not clear whether co-mapping of phenotypes is due to pleiotropic effects of single genes or to separate effects of multiple linked genes in a given genetic interval.
Transgenic methods have previously been used to study the regulatory or phenotypic effects of single genes in sticklebacks. However, conventional microinjection of foreign DNA sequences typically results in random integration, generating substantial variability between individuals and lines (Hosemann et al., 2004; Colosimo et al., 2005; Chan et al., 2010; Mills et al., 2014; O’Brown et al., 2015; Erickson et al., 2015; Erickson, Ellis, et al., 2016; Indjeian et al., 2016; Howes et al., 2017; Cleves et al., 2018; Thompson et al., 2018).
Targeted editing of endogenous loci in sticklebacks would provide a powerful additional tool for examining the function of engineered DNA sequence changes within a normal chromosomal context. Multiple methods have been developed in other organisms for targeting DNA nucleases to particular DNA sequences in the genome. For example, TALEN nucleases can be engineered to recognize and cut particular DNA sites (Miller et al., 2011; Dahlem et al., 2012), and have previously been used successfully to generate targeted germline deletions in sticklebacks (Erickson et al., 2015, 2018; Erickson, Ellis, et al., 2016; Cleves et al., 2018). However substantial effort is required to generate TALEN nucleases with the desired specificity.
Recently, CRISPR-Cas9 has emerged as a simple and efficient system for genome editing. To create targeted mutations, two components are required: 1) the Cas9 protein, which cuts DNA, and 2) a single-guide RNA (sgRNA), which targets Cas9 protein to a specific DNA region. The complex formed by Cas9 and sgRNA generates double-stranded breaks at the site specified by the sgRNA (Jinek et al., 2012). Repair machinery in cells will usually attempt to repair introduced breaks. However, this process is imperfect and mutations are often generated as a result. Induced mutations can range from single base pair substitutions to deletions of hundreds of base pairs. It is simple to design sgRNAs to target specified locations in the genome, and CRISPR-Cas9 targeting has been successfully implemented in many different model systems ranging from yeast (DiCarlo et al., 2013) to cell lines (Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013) to mice (Yang et al., 2013). Promising initial CRISPR studies have also been reported in sticklebacks (Hart and Miller, 2017; Xie et al., 2019). Here, we test additional ways of introducing Cas9 activity into stickleback embryos and use optimized protocols to achieve targeting and germline transmission of engineered mutations in three different stickleback genes of evolutionary interest: SLC24A5, KITLG, and EDA.
SLC2 4A5
To optimize and test whether we could efficiently induce stickleback mutations using CRISPR-Cas9, we initially targeted the SLC24A5 gene. SLC24A5, also known as the golden locus in zebrafish, is required for normal pigmentation in zebrafish, and has also played an important evolutionary role in lightening skin pigmentation in humans at Northern latitudes (Lamason et al., 2005). Golden mutant phenotypes also provide an easy way to quickly and visually test the efficacy of different CRISPR-Cas9 conditions because loss of SLC24A5 results in loss of eye and body pigmentation early in development at 5 days post fertilization (Swarup, 1958). These phenotypes are typically recessive requiring biallelic loss-of-function in order to see a phenotype in a given cell, so CRISPR-Cas9 must highly efficiently introduce loss-of-function mutations to produce a visible phenotype (Jao et al., 2013).
KIT LG
KITLG encodes a ligand to the c-KIT receptor, also known as Steel (Sl) and mast cell growth factor (MGF) in mice (Copeland et al., 1990; Williams et al., 1990). The KITLGX locus has previously been linked to repeated evolution of pigmentation changes in both sticklebacks and humans (Miller et al., 2007; Guenther et al., 2014) and is known to be important for gametogenesis and the survival, proliferation, and differentiation of the primordial germ cells (PGC) in mice (Silvers, 1979; Brannan et al., 1992; Bedell et al., 1995; Gu et al., 2009, 2011). Given its location on LG19 (Miller et al. 2007), which is known to be the master sex determining chromosome in stickleback (Peichel et al., 2004), we hypothesized that the KITLG genes on the diverging sex chromosomes (KITLGX and KITLGY) might also play a role in sex determination. Teleost fish use a variety of different sex determining mechanisms. Many characterized systems, such as those in medaka or mice, involve a master sex determining gene, but no such gene has yet been found in sticklebacks (Gubbay et al., 1990; Koopman et al., 1991; Matsuda et al., 2002). However, previous studies have found that an increase in the number of primordial germ cells may drive the unspecified gonad to an ovarian fate (Lewis et al., 2008). Given their location in the genome and known roles in PGC migration and proliferation, KITLGX and KITLGY are promising candidates for genes that could determine gonad sex determination by altering the number of PGCs that arrive or proliferate in the unspecified gonad. In addition, generating loss-of-function mutations in the KITLGX gene can provide a critical genetic test of its previously postulated role in ecotypic color differences in sticklebacks (Miller et al., 2007).
E DA
EDA has been linked to the evolution of several traits in stickleback, including repeated armor plate reduction, changes in lateral line neuromast patterns, and changes in schooling behavior (Colosimo et al., 2005; Mills et al., 2014; Greenwood et al., 2016). Quantitative trait locus (QTL) mapping has also identified peaks near EDA for a number of additional skeletal traits, including pelvic reduction; the length of dorsal spines one, two, and three; tooth number; and gill raker number (Shapiro et al., 2004, 2009; Rogers et al., 2012; Miller et al., 2014; Erickson, Glazer, et al., 2016; Howes et al., 2017). Loss-of-function EDA mutants in other species, such as zebrafish, lack all of their fin rays, including their pelvic fins. They are also missing scales, pharyngeal teeth, and gill rakers (Harris et al., 2008). Given the mapping results and phenotypes in other species, we wanted to determine the range of skeletal phenotypes caused by introducing mutations into the stickleback EDA gene.
By conducting the current experiments with three different genes (SLC24A5, KITLG, and EDA), we examine whether CRISPR-Cas9 targeting provides a general approach for probing the functions of interesting evolutionary loci in sticklebacks.
MATERIALS AND METHODS
mRNA synthesis of Cas9
nls-zCas9-nls mRNA was transcribed from PT3TS-nCas9n from the Chen lab, obtained via Addgene (46757) (Jao et al., 2013). The plasmid was linearized using XbaI (New England BioLabs, R0145S). The linear plasmid was purified (QIAquick PCR Purification kit), and 1 μg was used for an in vitro transcription reaction (T3 mMessage mMachine, Life Technologies, AM1348M). The reaction was incubated at 37°C in a thermocycler for 1 hour, and the mRNA was purified by phenol-chloroform and isopropanol precipitation as detailed in the mMessage mMachine manual. A nanodrop spectrophotometer was used to check the concentration and purity of the Cas9 mRNA. 1 μl of the product was run on a 1% agarose gel to check the integrity of the transcript.
Cas9 protein
Cas9-nls protein was purchased from the QB3 MacroLab Facility at University of California, Berkeley.
Characterization of X- and Y-linked Kitlga genes
The stickleback ortholog of the zebrafish kitlga gene was previously reported (Miller et al., 2007). This gene maps to Linkage Group 19 (LG19), the evolving sex chromosome pair in sticklebacks (Peichel et al., 2004). To avoid possible confusion between the divergent loci on the evolving female and male sex chromosomes, we here refer to the X-linked locus as KITLGX, and the Y-linked locus as KITLGY. Expressed sequence tags (ESTs) derived from mixed adult organs of the Conner Creek (CC), Washington, freshwater population show partial homology to KITLGX, but also have regions of unique sequence (see Genbank accession numbers DN681761.1 and DN681762.1, produced from the ends of cDNA clone CGX20-B06). We completely sequenced the corresponding CGX20-B06 clone, which revealed a divergent KITLG predicted open reading frame. Predicted KITLGX and KITLGY protein sequence alignment was made with ClustalW, and alignment displayed using the output from the BOXSHADE server (https://embnet.vital-it.ch/software/BOX_form.html). We mapped this gene to the Y chromosome in a marine x freshwater F2 cross (Shapiro et al., 2004; Miller et al., 2007) using primers TTCTCCTTCGGCCTACTCCT and TGTGCGAAAGCCTATTTCAA, which amplify a ~200 base pair (bp) fragment off the Y chromosome and named this gene KITLGY. We screened all BAC filters from the stickleback CHORI 213 and 215 BAC libraries (Kingsley et al., 2004), using two pairs of overgoes for each gene, designed to distinguish these two genes. Overgo screening was performed as described at www.chori.org/bacpac/overgohyb.htm. The KITLGX overgoes (all overgo and primers sequences are 5’ to 3’) were CAACGACGGACCTCCCCTAGTTAA and CGTTTTAGAGCCATCATTAACTAG, and GAACCTGAAACCCTGTCTGGGGTG and GAAGGCTTTGCTCCAACACCCCAG. The KITLGY overgoes were TGACACTGTCTCTGCAACCACCAA and GAGTCTGGCCATCGCCTTGGTGGT, and GAAACGAGAGCCCTGTCTGCGGTG and GAAAGCTTTGCTCCACCACCGCAG. KITLGX positives were screened by PCR with CTGTCGGCCTGATCAGTTTC and TGCAGTGTATCCTCCATGAATC, and TGCTGCTTTGAATAACTCACAT and ATTTAGGTCTGGACGGCATT. From this screening, two 213 and two 215 KITLGX BACs were found to overlap the KITLGX locus (213–42J11, 213–167N05, 215–250I22, and 215–264A13). These pairs of 213 and 215 BACs span ~360 kb and ~300 kb respectively in the reference assembly (Jones et al., 2012), centered on the KITLGX gene.
KITLGY positives were screened by PCR with CACCACTCAGGCTTTGGCTG and CAGCTGCTAGAGGATTTGCT, and TTCCTGATTCACAGGTGACG and GTGGTCCACACTTTGCCTCT, (designed to the SP6 and T7 ends of BAC 213–21C23, respectively, accession numbers CL641946.1 and CL641947.1) since 213–21C23 was a KITLGY positive found here. From this screening, one 213 and one 215 BAC was found to contain portions of the KITLGY gene (213–21C23 and 215–203C15). The 21C23 BAC lacks the 3’ end of the KITLGY gene, while the 215–203C15 BAC spans the entire gene. These BACs were fully sequenced by the Stanford Human Genome Center as part of a larger sex chromosome sequencing project to be reported elsewhere.
We confirmed that KITLGX and KITLGY are both expressed in stickleback tissues by BLASTN (Altschul et al., 1990) of the exon sequences of both against large RNA-sequencing datasets generated from testes and liver of the Bear Paw Lake (BEPA), Alaska, freshwater population (NCBI Sequence Read Archive (SRA): SRX146597 and SRX146599). Recovered sequence reads were assigned to either KITLGX or KITLGY when they covered at least three different sequence positions that differ between KITLGX and KITLGY, and did not differ from both of the KITLGX and KITLGY reference sequences at more than one position. This analysis showed 131 and 22 reads that could be assigned to KITLGX and KITLGY respectively in the testes dataset; and 23 and 47 reads that could be assigned to KITLGX and KITLGY respectively in the liver dataset. Both datasets contained reads that supported the KITLGY exon-exon junctions at the skipped sixth exon (9 reads in testes, 6 in liver) and novel ninth exon (3 reads in testes, 2 in liver).
Design and synthesis of single-guide RNAs (sgRNAs)
sgRNAs were designed using CHOPCHOP (http://chopchop.cbu.uib.no/index.php) based on the G. aculeatus genome assembly (BROADS1) (Montague et al., 2014; Labun et al., 2016). To target SLC24A5, the target sequence 3’-GAAGCCGTCGGGGAACTCGGAGG-5’ was chosen for three reasons. First, the target sequence starts with a G allowing for better transcription using T7 RNA polymerase than other possible bases (Chamberlin and Ring, 1973). Second, the closest possible off-target sequence, identified by a BLASTN search (Altschul et al., 1990) of the G. aculeatus genome assembly (BROADS1), differed by eight bases and lacked the required NGG PAM site (shown in bold above), therefore minimizing potential off-target effects. Third, this sequence was highly ranked in CHOPCHOP’s computational predictions of efficiency, with a score of 0.70. To target KITLG, four sgRNAs were used in combination because it is located on the stickleback sex chromosome (LG19), and there are coding differences (see Table 1) not only between the X and Y linked copies of the gene, but also between the reference genome (Bear Paw Lake, Alaska) and the population used for injection (Matadero Creek (MATA), California).
Table 1.
sgRNA target sequences for KITLG.
| sgRNA1 designed in reference | GAACTACATTCCGAGGGAAGAGG |
| KITLGX in MATA | GAACTACATTCCAAGGGAAGAGG |
| KITLGY in MATA | ACATTACGTTCTGAGGGAAAAGG |
| sgRNA2 designed in reference | AGGGAAGAGGTAAGACATGGTGG |
| KITLGX in MATA | AGGGAAGAGGTAAGACATGGTGG |
| KITLGY in MATA | AGGGAAAAGGTAAGATGTGGTGG |
| sgRNA3 designed in reference | AGTGCCACTACAGGGAAGAGAGG |
| KITLGX in MATA | AGTGCCACTACAGGGAAGAGAGG |
| KITLGY in MATA | AGTGCCACTACAGGCAAGAGAGG |
| sgRNA4 designed in reference | GGTGTTGTGGGACAGGGAGGAGG |
| KITLGX in MATA | GGTCTTGTGGGACAGGGAGGAGG |
| KITLGY in MATA | GGTGTTGTGGGACAGGGAGGAGG |
The sgRNAs were made with a two oligo-PCR method that uses one gene-specific primer and one scaffold primer as described by (Shah et al., 2015). The gene-specific guide contained a T7 RNA polymerase promoter sequence, the 20 bp targeting sequence in the gene of interest (N20; the target site excluding the NGG PAM sequence), and 20bp of homology to the scaffold oligo. The sequence was 5’- AATTAATACGACTCACTATA(N20)GTTTTAGAGCTAGAAATAGC-3’. The scaffold oligo sequence was 5’-GATCCGCACCGACTCGGTGCCACTTTTTCAAGTTGATA ACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’. The scaffold oligo was HPLC purified (synthesized by IDT), but the gene specific oligo was not.
To make the sgRNA template for transcription, 2x Phusion Master Mix (New England BioLabs, M0531L) was used for a 20 μl reaction with 5 μl of 10 μM gene-specific oligo and 5 μl of 10 μM scaffold oligo. PCR program was 95°C for 30 s; 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 10 s; and 72°C for 5 min. The PCR product was run on a 2% agarose gel, and the correct size band (121bp) was cut out and purified (QIAquick Gel Extraction kit) or purified if there was no double band (QIAquick PCR purification) and eluted in 20μl of water. For transcription, 7 μl of this gel-purified guide template was used in a T7 in vitro transcription (MEGAscript T7, Life Technologies, AM1334M). The transcription was incubated at 37°C in a thermocycler for 16 hours and cleaned up using LiCl as detailed in the MEGAscript manual. sgRNAs were resuspended in nuclease-free water from the kit and stored at a concentration of 1 μg/μl at −80°C.
SLC24A5 injection and assessment of efficiency
Injections with sgRNAs targeting SLC24A5 were done with both Cas9 protein and Cas9 mRNA. For SLC24A5 Cas9 mRNA injections, the mix contained 300 ng/μl of Cas9 mRNA and 300ng/μl of the sgRNA. For SLC24A5 Cas9 protein injections, the mix contained 1μg/μl of Cas9 protein and 300 ng/μl of the sgRNA. A final concentration of 0.05% Phenol red was used, and the volume was adjusted with 10mM Tris pH 7.5. Two clutches of fertilized stickleback eggs were pooled and split in approximately 3 equal portions. One third was injected with Cas9 mRNA, another third was injected with Cas9 protein, and the remaining third was kept as uninjected controls. At 5 days post fertilization, a total of 26 controls, 38 mRNA injected, and 31 protein injected embryos were screened for pigmentation defects. To assess the efficiency of the mutagenesis, five embryos were sacrificed (two injected with Cas9 mRNA, two injected with Cas9 protein, and one control). To extract DNA, embryos were placed in lysis buffer (10mM Tris pH 8, 100mM NaCl, 10 mM EDTA, 0.5% SDS) with Proteinase K (333μg/mL) at 55ºC overnight, and then phenol chloroform extracted. The target SLC24A5 genomic region was amplified with the forward primer 5’-AAACCCATTCCACCAGAGTG −3’ and the reverse primer 5’-TACCACAGCATTCCAGACCA −3’. The PCR product was cloned into pCR Blunt-II TOPO using the Zero Blunt TOPO PCR Cloning Kit. DNA was isolated from approximately 30 colonies from each individual (except for 12 for the control individual) by miniprep and sequenced to determine the lesion.
To measure germline transmission of targeted mutations, mosaic founder fish were grown to sexual maturity (~seven months). A total of 34 Cas9 mRNA injected fish and 22 Cas9 protein injected fish successfully hatched and were raised to adulthood. Sperm was isolated from the males and used to fertilize eggs of wild-type females from the same stickleback population (Matadero Creek, CA). Because no obvious pigmentation defect was seen in adult F0 animals, males were chosen for breeding based only on reproductive condition. Offspring from these crosses were sacrificed and screened for transmitted DNA lesions by amplifying the SLC24A5 target region as above and directly sequencing the PCR product.
KITLGX and KITLGY injection and assessment of efficiency
For KITLGX and KITLGY injections, the mix contained 300 ng/μl Cas9 mRNA and 125 ng/μl total of all four sgRNAs (each sgRNA was at a concentration of 31 ng/μl). One clutch of eggs was injected, and eleven of the fish were raised to sexual maturity. To determine the transmission of the mutations and identify associated phenotypes, F0 fish were crossed either to each other (to create potential homozygotes) or to wild-type fish (to create potential heterozygotes). KITLGX was amplified using forward 5’-GCACTTTGCATGTTAGCCCC-3’ and reverse 5’-ACAAACAAGCTCTCCCCGAG-3’. KITLGY was amplified using 5’-AGAAGACCCAGCGGAAAAGG-3’ and 5’-TGAGGAACATCAGTGCACCC-3’primers. These products contained all four sgRNA sites. Because environment and age affect pigmentation, age-matched fish of each genotypic class were raised together for eight months before comparing their pigmentation patterns.
EDA Mutants
One clutch from fully-plated anadromous fish (Little Campbell River, British Columbia) was injected with Cas9 protein and an sgRNA (N20: 5’-CGCTCTTCACAGGGAAAAAG-3’) targeting the fourth exon of EDA using the same procedures described above for SLC24A5. Fish were raised to sexual maturity and then F0 founders were crossed with each other. Fish skeletons were stained with Alizarin Red as described in (Peichel et al., 2001; Miller et al., 2007) except that fish were first fixed in 70% ethanol. Branchial skeletons were dissected and flat-mounted as described in (Ellis and Miller, 2016), with an additional 10 day clearing step in 1% potassium hydroxide after dissection. To examine DNA lesions, a 1394 bp region was amplified using 5’- ACACACGCAAACACACATGG-3’ and 5’- GAAGCCCCACCTTCTCTCAC-3’ primers. The resulting product was cloned into pCR Blunt-II TOPO and sequenced. Differences in lesions and linked SNPs were used to verify that both alleles were detected.
RESULTS
Generation of SLC24A5 mutants
To optimize the efficiency of CRISPR-Cas9 mutations, we first tested two ways to introduce Cas9 activity and sgRNAs into stickleback embryos. Cas9 can be introduced either as a premade protein or as mRNA that will be translated by the machinery of the cell. The advantage of using Cas9 protein is that it can begin cutting almost immediately when injected into the cell, whereas if Cas9 mRNA is injected, it must first be translated, and the injected message may also compete with endogenous transcripts for translation. While in theory Cas9 protein should be more efficient, previous studies have not always seen increased efficiency using Cas9 protein (Gagnon et al., 2014). We chose to test both by introducing either Cas9 mRNA or Cas9 protein with a single sgRNA into single cell stage stickleback embryos by microinjection, using the SLC24A5 locus as a test because of its easy to score phenotypes.
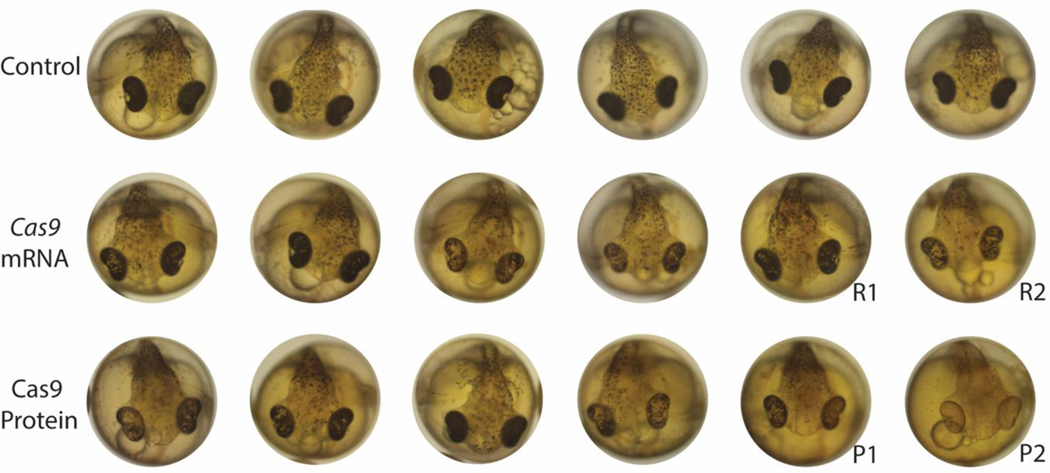
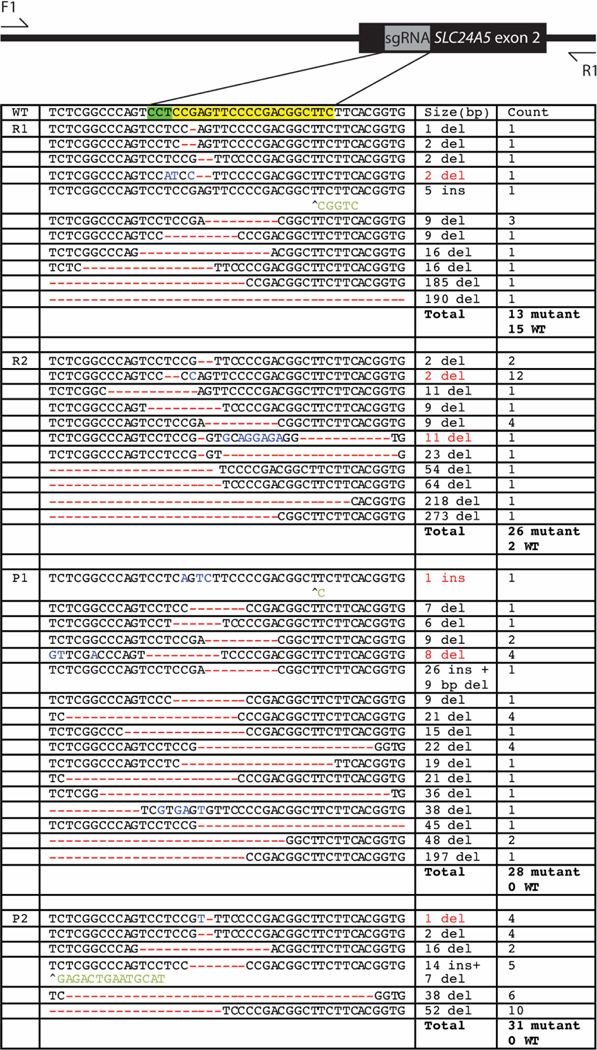
Sticklebacks generated by injecting fertilized eggs with SLC24A5 CRISPR-Cas9 showed reduced melanization in the eye and body of the fish at five days post fertilization (Figure 1). These defects were no longer visible by seven days post fertilization. No obvious pigmentation defects persisted into adulthood, but more subtle differences may have been missed due to natural variation in stickleback pigmentation. Severely reduced melanin pigmentation was only seen when fish were injected with Cas9 protein, but the amount of pigment that remained varied widely within both protein- and mRNA-injected embryos (Figure 1). To study the types of molecular lesions introduced by SLC24A5 CRISPR targeting, we amplified and cloned a genomic region around the targeting site and then sequenced multiple sub-clones from two founder (F0) individuals injected with Cas9 mRNA, and two F0 individuals injected with Cas9 protein. All fish were mosaic for multiple lesions, which ranged in size from single base pair deletions to a 273 bp deletion around the sgRNA target (Figure 2).
Figure 1. Pigmentation phenotypes in SLC24A5 targeted F0 mosaic stickleback embryos.
Six representative embryos are shown for each condition: uninjected controls, fish injected with Cas9 mRNA and sgRNA, and fish injected with Cas9 protein and sgRNA. Images were chosen to represent the range of phenotypes seen at five days post fertilization. DNA lesions in R1 and R2 fish (injected with mRNA Cas9) and P1 and P2 fish (injected with Cas9 protein) were sequenced and are shown in figure 2.
Figure 2. Diversity of molecular lesions induced around SLC24A5 targeting site.
A region of the SLC24A5 locus was amplified and subcloned from two mosaic F0 embryos injected with Cas9 mRNA (R1 and R2) and two F0 embryos injected with Cas9 protein (P1 and P2). Red indicates deletions, blue indicates substitutions, and green indicates insertions. If both a deletion and an insertion were seen, the size indicated in red denotes only the size of the deletion.
To determine the efficiency of male germline transmission, three male F0 fish injected with Cas9 mRNA and three injected with Cas9 protein were crossed to wild-type fish from the same population. Sequencing of ~30 offspring from each cross demonstrated that the transmission rate was between 33 and 41% for fish injected with Cas9 mRNA (Table 2). The transmission rate for fish injected with Cas9 protein was more variable between 21 and 100% (Table 2). All of these efficiencies are probably underestimates because the assay would not detect lesions that also remove one or both of the primer binding sites for PCR amplification.
Table 2.
Transmission efficiency from SLC24A5 F0 mosaic fish
| Cas9 type F0 Mosaic Individual % of F1 offspring with mutations | ||
|---|---|---|
| mRNA | Male #1 | 39% (11/28) |
| Male #2 | 41% (12/29) | |
| Male #3 | 33% (10/30) | |
| protein | Male #4 | 26% (07/27) |
| Male #5 | 100% (27/27) | |
| Male #6 | 21% (06/28) | |
Once we had had successfully targeted the SLC24A5 gene, we chose to target two major evolutionary loci in sticklebacks: KITLGX on Linkage Group 19 (LG19) (the sex chromosome) and EDA on LG4.
Effects of KITLGX and KITLGY on pigmentation
CRISPR targeted F0 KITLGX and KITLGY fish injected with Cas9 mRNA and four sgRNAs had melanized pigmentation defects that were visible starting at five days post fertilization. These defects persisted into adulthood and were visible in both the males and females (Figure 3). Viable eggs and sperm were obtained from both female and male F0 fish respectively. Amplification, cloning, and sequencing of induced mutations revealed a range of lesions around all the sgRNA sites in KITLGX (sgRNA 1 and 2 could not be distinguished because they are so close). In contrast, KITLGY only had mutations at the site of sgRNA 4, probably due to many sequence differences between KITLGX and KITLGY (Table 1).
Figure 3. External pigmentation phenotypes in F0 mosaic sticklebacks following KITLGX and KITLGY targeting.
A. Schematic showing exon 3–5 of KITLGX and KITLGY and the location of the four sgRNA sites that were targeted. B. WT male C. F0 mosaic KITLGX and KITLGY CRISPR male D. WT female E. F0 mosaic KITLGX and KITLGY CRISPR female. Scale 5mm.
KITLGX and KITLGY contain numerous sequence differences and produce different transcripts. KITLGY skips the sixth exon and has a long divergent C-terminus (Figure 4). To determine if KITLGX and KITLGY have different roles in pigmentation, F0 males and females were crossed to wild type and each other to generate all possible mutant genotypic classes following germline transmission. The survival rate among putative homozygous fish was low. Only 30% (n=27/88) of the fish from two crosses that survived until 15 days post fertilization lived to two months.
Figure 4. KITLGX and KITLGY predicted amino acid sequence divergence.
LC = Little Campbell anadromous marine (see Miller et al. 2007), FTC = Fish Trap Creek freshwater (see Miller et al. 2007), CC = Conner Creek freshwater (cDNA clone, see Methods).
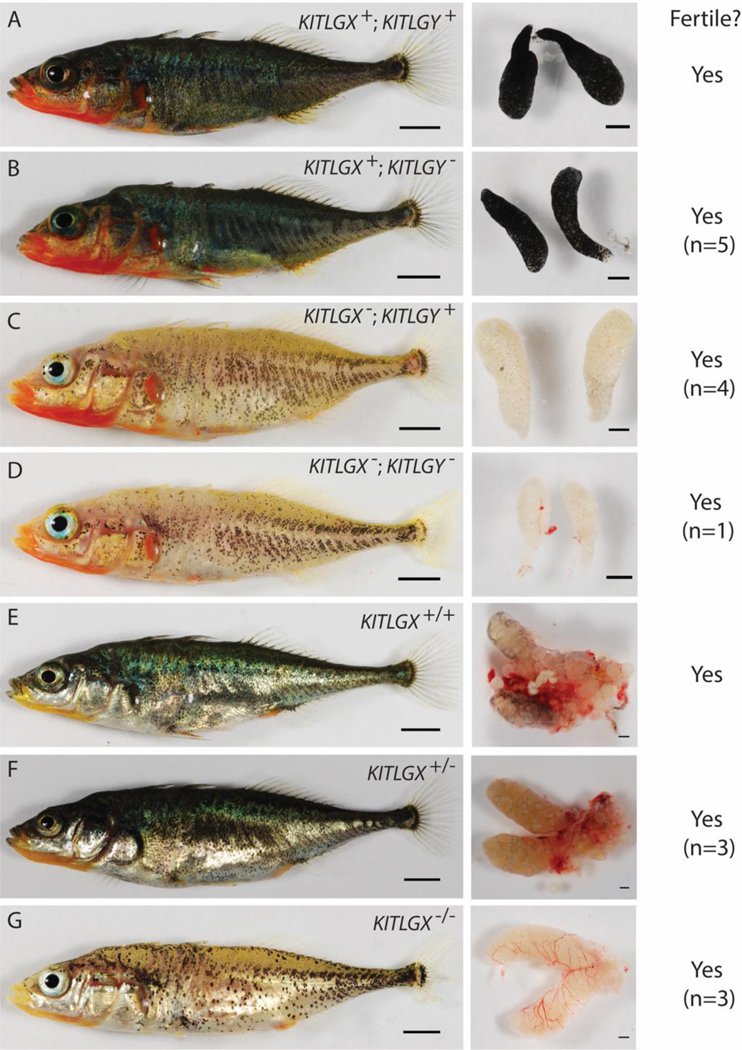
Males with mutations in KITLGX (total n=4; KITLGX allele 1 n=3; KITLGX allele 2 n=1), KITLGY (total n=5; Y allele 1 n=3; Y allele 2 n=1; Y allele 3 n=3), or both KITLGX and KITLGY (KITLGX allele 2 and KITLGY allele 1 n=1) all produced viable sperm (Figure 5A–D; Table 3). The males only carrying frameshift mutations in KITLGY did not show dramatic pigmentation differences in the skin or testes compared to wild type, though sometimes appeared lighter in ventral regions (Fig. 5B). The males only carrying frameshift mutations in KITLGX lost much of their normal dark external melanophore pigmentation and had testes devoid of obvious melanized pigmentation (Fig. 5C). While the testes were consistently white, the extent of remaining external body pigmentation was somewhat variable even among fish of the same age, genotype, and tank. Nevertheless, KITLGX mutant genotypically male fish could be clearly distinguished from wild type and carriers of mutant KITLGY alleles. Only one fish was recovered with frameshift mutations in both KITLGX and KITLGY (Fig. 5D), and appeared similar to its genotypically male siblings that only had KITLGX mutations (Fig. 5C).
Figure 5. Altered pigmentation but normal sex determination in F1 fish carrying different induced KITLGX and KITLGY alleles.
A. Wild-type male and testes. B. Body and testes from male carrying frameshift KITLGY allele 1 (see table 3). Sperm from males carrying frameshift mutations in KITLGY (n=5) successfully fertilized eggs. C. Body and testes from male fish with frameshift KITLGX allele 1. Sperm from F1 males with frameshift in KITLGX (n=4) also successfully fertilized eggs. D. Body and testes from male carrying both frameshift KITLGX allele 2 and KITLGY allele 1. Sperm from male with frameshift mutations in both alleles successfully fertilized eggs (n=1). E. Wild-type female and ovaries. F. Body and ovaries from female carrying frameshift KITLGX allele 1. Eggs were produced and successfully fertilized (n=3). G. Body and ovaries from female fish carrying both frameshift KITLGX alleles 1 and 8. Eggs were produced and successfully fertilized (n=3). Scale 5mm (left column) and 1mm (right column).
Table 3. Transmitted frameshift alleles found in F1 KITLGX and KITLGY mutants.
Identified mutations near different sgRNA target sites are shown (underlining indicates the sgRNA sequence, bold type denotes the PAM sequence, and italic type is used to indicate intron sequences of KITLGX and KITLGY).
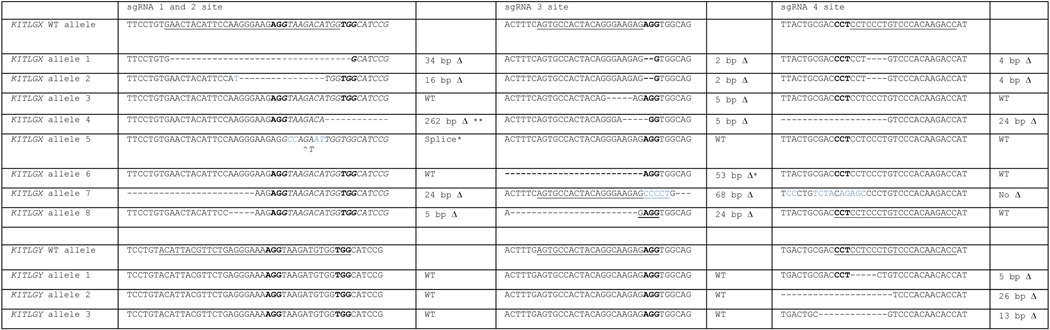
|
indicates the deletion of a base.
indicates the deletion or mutation of splice donor or acceptor sequence.
indicates the deletion of the fourth exon. Blue bases are substitutions.
Both the females heterozygous for frameshift mutations (total n=3; KITLGX allele 3 n=1; KITLGX allele 4 n=1; KITLGX allele 5 n=1) and the females homozygous for frameshift mutations (total n=3; KITLGX alleles 4 and 3 n=1; KITLGX alleles 6 and 7 n=1; KITLGX alleles 2 and 8 n=1) produced viable eggs (Figure 5E–G; Table 3). While the heterozygous female mutants did not show obvious external pigmentation defects, the homozygous female KITLGX mutants lacked most of the melanized pigment on the dorsal and ventral sides of the fish (Fig. 5F and G). The heterozygous females also showed less pigmentation of the dissected ovaries compared to the wild-type controls, and the homozygous females lacked all ovarian melanin pigmentation (Fig. 5F and G). KITLG is linked to pigmentation in the gills (Miller et al., 2007); however, the Matadero Creek population in which the mutants were made already has little pigmentation in the gills, so the effect of KITLG loss on this trait could not be determined.
EDA mutants
We targeted the EDA protein coding region with an sgRNA, and subsequently intercrossed founder fish to transmit lesions through the germline. One F0 x F0 intercross produced 24 F1 siblings that were successfully raised to seven months of age and >30 mm in length. Of these surviving progeny, one was homozygous wild-type (Figure 6A), and two were homozygous for mutations predicted to reduce or eliminate EDA function (Figure 6B–C). The fish shown in Figure 6B inherited two alleles with the same two-bp deletion resulting in an early stop codon in the fifth of eight exons. One allele had an additional point mutation eight bases upstream that falls in the intron and probably does not affect the transcript. The fish shown in Figure 6C had the same two-bp deletion allele as well as a second allele with 75-bp deletion that removed the splice acceptor of the fourth exon and the first 60 bp of the exon.
Figure 6. Skeletal phenotypes in sticklebacks homozygous for induced EDA CRISPR mutations.
A. Lateral, dorsal and ventral views of a wild-type sibling, stained with Alizarin Red to reveal calcified bones. B. Fish carrying two mutant EDA alleles. Allele 1 is a two-bp deletion. Allele 2 is a two-bp deletion with an additional substitution in an intron. C. Second fish carrying two mutant EDA alleles. Allele 1 is the same two-bp deletion as in B. Allele 2 is a 75-bp deletion that removes the last 60 bp of exon 4 and the first 15 bp of the following intron. The wild type control fish shown in row A had long pectoral, dorsal, anal and caudal fins, but the right pectoral fin was removed for DNA preparation. The two EDA mutant fish in rows B and C developed with no armor plates and largely missing pectoral, dorsal, and anal fins. Portions of the reduced caudal fins have been removed for DNA preparation prior to photography. DS1 Dorsal Spine 1. DS2 Dorsal Spine 2. DS3 Dorsal Spine 3. DF Dorsal Fin. CF Caudal Fin. AF Anal Fin. AS Anal Spine, Pel Pelvis, Pec Pectoral Fin. AP Armor Plates. PS Pelvic Spine. PR Pelvic Ray. Scale 5mm (left and center column) and 1mm (right column).
External skeletal armor:
While the wild-type control sibling had a full assemblage of fins, spines, and armor plates, the homozygous mutants had no armor plates and severely deformed or absent caudal, anal, dorsal, and pectoral fins (Figure 6 B–C). The first and second dorsal spines were long and prominent, but the third dorsal spine and anal spines were absent in the mutants even though the underlying pterygiophores were present. The homozygous mutants also had a full assemblage of pelvic spines and processes, but the pelvic ray did not form.
Gill rakers and teeth:
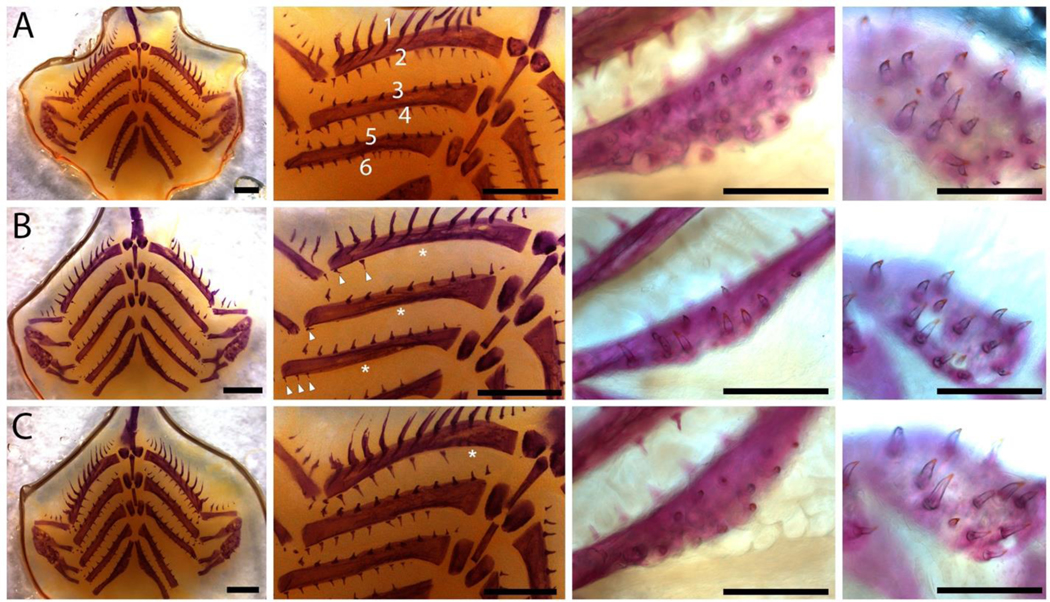
EDA homozygous mutants displayed reductions in gill raker and pharyngeal tooth number in the branchial skeleton. A two-bp/two-bp deletion homozygote displayed severe reductions in posterior facing gill rakers, while anterior facing gill rakers appeared normal (Figure 7A–B). A two-bp/75-bp deletion mutant displayed mild reductions in gill raker number, again specifically affecting posterior facing gill rakers (Figure 7A, C). Relative to wild type, both mutants displayed severely reduced dorsal and ventral pharyngeal dentitions, with smaller tooth fields but also reduced tooth number within each tooth field (Figure 7A–C).
Figure 7. Branchial skeleton phenotypes in sticklebacks homozygous for induced EDA CRISPR mutations.
A. (left to right) flat-mounted branchial skeleton, zoomed view of gill rakers, ventral pharyngeal tooth plate, and large dorsal pharyngeal tooth plate in wild-type sibling. In the two left-most columns, anterior is the top; in the two right-most columns, anterior is to the left. Wild-type fish have both anterior facing gill rakers (odd rows) and posterior facing gill rakers (even rows) B. Fish homozygous for two-bp deletion display severe reductions in posterior facing gill rakers (white asterisks), as well as reduced tooth plate size and tooth number C. Fish trans-heterozygous for a two-bp and a 75-bp deletion display more moderate reductions in posterior facing gill rakers (e.g. missing medial row 2 gill rakers labeled with white asterisk), as well as reduced tooth plate size and tooth numbers. Scale bars: left two columns 1 mm, right two columns 0.5 mm.
DISCUSSION
Our studies show that CRISPR-Cas9 is a powerful tool to induce mutations at defined loci in sticklebacks and to study the biology of major evolutionary loci.
CRISPR-Cas9
Both Cas9 mRNA and Cas9 protein could be used to efficiently create deletions and insertions in prechosen regions of the stickleback genome. In targeting SLC24A5, severe loss of pigmentation was only seen when fish were injected with Cas9 protein, but both mRNA and protein injections resulted in a range of phenotypes. This result is consistent with other studies which show a general trend of increased efficiency using injections with Cas9 protein, but in some cases, there is no significant difference between protein and mRNA (Gagnon et al., 2014).
Even though targeting is highly efficient, injected founders are clearly mosaic for a variety of different alleles. Pigmentation traits (e.g. in SLC24A5 and KITLG mutants) could easily be scored in F0 founder animals because even somatic patches of mutant cells produce visible phenotypes. In contrast, EDA founders had to be bred to homozygosity to reveal easily visible phenotypes. Germline transmission efficiencies were high, and founders typically transmitted more than one type of allele to various progeny in the next generation.
The sgRNAs used to target KITLG highlight that the sgRNA does not need to be an exact sequence match to cut the genome. As has been seen in other studies, mismatches that occur in the sgRNA target sequence furthest from the PAM site are tolerated and may not even reduce the cutting efficiency, whereas those close to the PAM significantly reduce the cutting efficiency (Hsu et al., 2013; Hwang et al., 2013). These are important considerations in sticklebacks due to numerous SNPs between populations. The possibility of polymorphisms should always be considered when designing sgRNAs based on the reference genome and doing experiments in other populations. At the same time, population polymorphisms might be used as an advantage to generate allele-specific CRISPR, targeting only the allele of one population but not another.
KIT LG
Frameshift mutations in KITLGX and KITLGY demonstrate that the two different sex-chromosome versions of KITLG play different roles in the pigmentation of sticklebacks. Frameshift mutations in KITLGY in males did not result in dramatic external or testes pigmentation defects compared to the wild type, whereas frameshift mutations in KITLGX resulted in a loss of both external and testes pigment. In females, one copy of KITLGX appears sufficient for the wild-type external pigmentation of the fish, but less pigment is seen in the ovaries suggesting that one copy is not fully sufficient for a wild-type phenotype. KITLGX’s role in pigmentation in sticklebacks was originally identified based on X-linked gill and ventral skin pigmentation differences in crosses between marine and freshwater fish (Miller et al., 2007), but the likely severe loss-of-function mutations in KITLGX clearly affect both dorsal and ventral pigmentation patterning.
Although the KITLGX mutants confirm that this gene plays an important role in the pigmentation of sticklebacks, the sterility phenotype seen in mice was not seen in sticklebacks. While we cannot rule out that KITLGX and KITLGY mutant stickleback have reduced fertility (reduced sperm or egg production), we did not see the total sterility seen in some mouse mutants.
The fertility of the stickleback KITLGX and KITLGY mutants is consistent with what has been seen in zebrafish. Mutants have been made for both kitlg and its receptor c-kit in zebrafish. The ligand mutant, kitlga or sparse-like, and the receptor mutant, kita or sparse, have a reduced melanophore number (Kelsh et al., 1996; Parichy et al., 1999; Dooley et al., 2013). In contrast to the mouse mutants, hematopoiesis and primordial germ cell development appeared normal, even though kita is expressed in the cells that give rise to the hematopoietic precursors. Although zebrafish kitlga mutants do not have documented reductions in viability, we do see a reduction in the viability of the fish homozygous for stickleback KITLG mutations, indicating that the stickleback gene likely plays important roles in addition to its contribution to pigmentation.
Although KITLGX and KITLGY map to the sex chromosomes in sticklebacks, our experiments indicate KITLG is unlikely to be the sex-determining locus. Genotypically female KITLGX heterozygous and homozygous mutants produced viable eggs and did not develop testes or produce sperm. Additionally, it does not appear that KITLGY is required for the production of sperm, as genotypic males with mutations in KITLGY still produced functional sperm. It is possible KITLGY has become a pseudogene on the young evolving stickleback Y chromosome. However, the gene is expressed and spliced based on a cDNA clone and RNA sequences from testes and liver, and these sequences predict a large encoded protein with intact KIT binding domains (Fig. 4). We only recovered a single viable stickleback that was simultaneously carrying induced mutant alleles at both KITLGX and KITLGY, suggesting that fish that inherit mutations in both genes may have reduced viability (Figure 5). We thus think it is likely that KITLGY has some functions in development that are separate from pigmentation or sex determination, and this possibility can be tested in the future by additional phenotypic studies of fish carrying KITLGY mutations.
ED A
Many freshwater stickleback populations have prominent armor reduction, including common reduction in armor plates, shortening or loss of dorsal spines, and rarer loss of the pelvic hindfin. All of these phenotypes have been linked to the EDA region of LG4 in some genetic crosses (Shapiro et al., 2004, 2009; Rogers et al., 2012; Miller et al., 2014; Erickson, Glazer, et al., 2016; Howes et al., 2017), raising the possibility that armor, spine, and pelvic reduction may be due to pleiotropic effects of the EDA gene in one or more populations. Our homozygous EDA mutants confirm that EDA is critical for armor plate formation, as no armor plates formed in the homozygous mutants. However, the homozygous EDA fish do not support an obvious requirement for EDA in formation of the most prominent dorsal spines in sticklebacks, or in formation of the pelvis and pelvic spine, which remain robust in mutant fish even when armor plates and most fins were lost. It is possible that other types of mutations in the EDA gene might generate a different range of phenotypes, including dorsal spine and pelvic defects. However, it is striking that the spines and pelvis appear relatively unaffected in fish with drastic reductions in other skeletal structures, suggesting that the dorsal and pelvic spines of sticklebacks are less sensitive to changes in EDA activity than other armor structures. The only part of the pelvis that was clearly affected was the pelvic ray. Threespine sticklebacks generally have one pelvic ray on each side (Bell and Foster, 1994). The loss of the pelvic ray is consistent with the fact that most of the other soft fin rays are lost in the stickleback EDA mutants, and that the pelvic rays are lost in zebrafish EDA mutants (Harris et al., 2008).
The severe reductions in pharyngeal tooth number support the hypothesis that natural variant EDA alleles modify tooth number, as previously suggested by a large-effect tooth number QTL detected in a large marine by freshwater F2 cross that mapped to LG4 and had a peak marker at EDA (Miller et al., 2014). Freshwater alleles of this tooth number QTL reduced tooth number in this cross, a direction opposite to the overall pattern of increased tooth numbers observed in the grandparental freshwater fish (Cleves et al., 2014; Miller et al., 2014). However, the tooth reductions seen in the current experiments match the predicted direction for EDA alleles in freshwater populations being partial loss-of-function (Colosimo et al., 2005; O’Brown et al., 2015). In contrast, the posteriorly biased gill raker phenotypes do not resemble the phenotypic effects of the large effect gill raker number QTL previously mapped to LG4, which have strong effects on anterior facing gill rakers as well (Glazer et al., 2014; Miller et al., 2014). However, this EDA mutant phenotype strongly resembles the gill raker phenotype described in EDAR hypomorphic mutant zebrafish (Harris et al., 2008), suggesting posterior gill rakers are most sensitive to reductions in EDA/EDAR function in teleosts. These posterior-biased reductions in gill rakers seen in EDA mutants also resemble the natural phenotypes seen in some fish species (e.g. herrings) (Gibson, 1988).
The two homozygous mutant fish recovered in this study had somewhat different phenotypes, possibly related to the nature of the different mutant alleles and their predicted effects on EDA transcript and/or protein. The EDA ligand is synthesized as a transmembrane protein with a transmembrane, collagen, and tumor necrosis factor domain. In the two-bp deletion allele, a premature stop codon occurs before the tumor necrosis factor (TNF) domain. Premature stop codons often lead to nonsense-mediated decay of the corresponding transcript (He and Jacobson, 2015), which could reduce transcript levels and EDA activity. In addition, the TNF domain is crucial for receptor binding, therefore any protein encoded by the mutant transcript would be unlikely to bind the receptor. In addition, the 75-bp deletion allele removes 60 bp at the start of the fourth exon and 15 bp in the intron and presumably prevents the fourth exon from being normally spliced. However, the mutation does not produce an obligate frameshift mutation that would disrupt the TNF domain. Instead, the mutation alters the collagen domain which is thought to be required for the oligomerization of the ligand. Cell culture experiments suggest that similar mutations may decrease the activity of EDA, but not eliminate it entirely (Schneider et al., 2001; Swee et al., 2009).
The diversity of alleles generated by CRISPR targeting can be seen as both an advantage and disadvantage of current methods. The diversity clearly requires careful screening of fish to identify the types of mutations present. These mutations can range from single base pair substitutions, insertion and deletions on the order of tens or hundreds of bases (such as those detected in this study), to deletions larger than 1,000 bases and rearrangements that have been reported in other studies (Kosicki et al., 2018). However, having multiple induced alleles at a locus may also be helpful for revealing the range of phenotypic effects at a locus, or for recovering viable fish carrying mutations in powerful developmental control genes. The complexity of alleles present in founder fish becomes greatly simplified upon germline transmission of individual alleles, allowing the investigator to then choose which mutations to further propagate and study. In the future, it may be possible to further enrich for particular types of edited genome changes during targeting, by including oligo donor nucleotides or other DNA sequences containing homology arms to direct the repair process surrounding Cas9-induced lesions (Irion et al., 2014; Albadri et al., 2017; Zhang et al., 2018). We have also recently used injections with multiple sgRNAs to recapitulate larger non-coding deletions upstream of the PITX1 locus, which mimic the type of regulatory alleles and phenotypes that repeatedly occur in wild stickleback populations (Xie et al., 2019).
Conclusion
Our studies show that CRISPR-Cas9 can be used to efficiently generate targeted mutations to study the roles of key evolutionary loci in sticklebacks. We show that both Cas9 protein and Cas9 mRNA can be used to efficiently induce mutations, although Cas9 protein may be slightly more efficient. KITLG mutants have pigmentation defects that differ depending on whether the X- or Y-linked copy is mutated, but they are fertile. Mutations in EDA cause a host of phenotypic effects, including the loss of armor plates and soft fin rays and severe reductions in tooth and gill raker number, while leaving the large dorsal spines and pelvis intact. The ability to edit defined locations in the genome will add a powerful new tool to the stickleback model system, making it possible to dissect the roles of individual genes and regulatory elements, rather than large linked chromosome regions, in a range of phenotypes.
ACKNOWLEDGMENTS
We thank Jane Grimwood, Jeremy Schmutz, and Richard Myers for BAC sequences generated during studies of the stickleback sex chromosomes; Ana Sousa for CRISPR-Cas9 advice; Abbey Thompson and Garrett Kingman for help collecting and maintaining Matadero Creek fish; Tim Howes and Felicity Jones for collecting Little Campbell River fish; and Semiahmoo First Nation for permission to collect sticklebacks from the Little Campbell River. This work was supported in part by NIH graduate training grant 2T32GM007790 (J.I.W.), and by NIH DE021475 and NSF-IOS 1645170 (C.T.M.). David Kingsley is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- Albadri S, Del Bene F. and Revenu C. 2017. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods, 121–122: 77–85. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW and Lipman DJ 1990. Basic local alignment search tool. J. Mol. Biol, 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bedell MA, Brannan CI, Evans EP, Copeland NG, Jenkins NA and Donovan PJ 1995. DNA rearrangements located over 100 kb 5′ of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev, 9: 455–470. [DOI] [PubMed] [Google Scholar]
- Bell M. and Foster SA 1994. The evolutionary biology of the threespine stickleback. Oxford: Oxford University Press. [Google Scholar]
- Brannan CI, Bedell MA, Resnick JL, Eppig JJ, Handel MA, Williams DE, et al. 1992. Developmental abnormalities in Steel17H mice result from a splicing defect in the steel factor cytoplasmic tail. Genes Dev, 6: 1832–1842. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. and Ring J. 1973. Characterization of T7-specific ribonucleic acid polymerase. J. Biol. Chem, 248: 2235–2244. [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, Brady SD, et al. 2010. supp. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science, 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, et al. 2014. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc. Natl. Acad. Sci, 111: 13912–13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Hart JC, Agoglia RM, Jimenez MT, Erickson PA, Gai L, et al. 2018. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet, 14: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal GJ, Dickson M, Grimwood J, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science, 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science, 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Gilbert DJ, Cho BC, Donovan PJ, Jenkins NA, Cosman D, et al. 1990. Mast cell growth factor maps near the steel locus on mouse chromosome 10 and is deleted in a number of steel alleles. Cell, 63: 175–183. [DOI] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, et al. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet, 8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J. and Church GM 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res, 41: 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley CM, Mongera A, Walderich B. and Nusslein-Volhard C. 2013. On the embryonic origin of adult melanophores: the role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development, 140: 1003–1013. [DOI] [PubMed] [Google Scholar]
- Ellis NA and Miller CT 2016. Dissection and flat-mounting of the threespine stickleback branchial skeleton. J. Vis. Exp, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Baek J, Hart JC, Cleves PA and Miller CT 2018. Genetic dissection of a supergene implicates Tfap2a in craniofacial evolution of threespine sticklebacks. Genetics, 209: 591–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Cleves PA, Ellis NA, Schwalbach KT, Hart JC and Miller CT 2015. A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev. Biol, 401: 310–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Ellis NA and Miller CT 2016. Microinjection for transgenesis and genome editing in threespine sticklebacks. J. Vis. Exp, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Glazer AM, Killingbeck EE, Agoglia RM, Baek J, Carsanaro SM, et al. 2016. Partially repeatable genetic basis of benthic adaptation in threespine sticklebacks. Evolution., 70: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, et al. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One, 9: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. 2005. The synthesis and evolution of a supermodel. Science, 307: 1890–1891. [DOI] [PubMed] [Google Scholar]
- Gibson RN 1988. Development, morphometry and particle retention capability of the gill rakers in the herring, Clupea harengus L. J. Fish Biol, 32: 949–962. [Google Scholar]
- Glazer AM, Cleves PA, Erickson PA, Lam AY and Miller CT 2014. Parallel developmental genetic features underlie stickleback gill raker evolution. Evodevo, 5: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AK, Mills MG, Wark AR, Archambeault SL and Peichel CL 2016. Evolution of schooling behavior in threespine sticklebacks is shaped by the Eda gene. Genetics, 203: 677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Runyan C, Shoemaker A, Surani A. and Wylie C. 2009. Steel factor controls primordial germ cell survival and motility from the time of their specification in the allantois, and provides a continuous niche throughout their migration. Development, 136: 1295–1303. [DOI] [PubMed] [Google Scholar]
- Gu Y, Runyan C, Shoemaker A, Surani MA and Wylie C. 2011. Membrane-bound steel factor maintains a high local concentration for mouse primordial germ cell motility, and defines the region of their migration. PLoS One, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Münsterberg A, et al. 1990. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature, 346: 245–250. [DOI] [PubMed] [Google Scholar]
- Guenther CA, Tasic B, Luo L, Bedell MA and Kingsley DM 2014. A molecular basis for classic blond hair color in Europeans. Nat. Genet, 46: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P. and Nüsslein-Volhard C. 2008. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet., 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JC and Miller CT 2017. Sequence-based mapping and genome editing reveal mutations in stickleback Hps5 cause oculocutaneous albinism and the casper phenotype. G3 Genes, Genomes, Genet., 7: 3123–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F. and Jacobson A. 2015. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu. Rev. Genet, 49: 339–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosemann KE, Colosimo PF, Summers BR and Kingsley DM 2004. A simple and efficient microinjection protocol for making transgenic sticklebacks. Behaviour, 141: 1345–1355. [Google Scholar]
- Howes TR, Summers BR and Kingsley DM 2017. Dorsal spine evolution in threespine sticklebacks via a splicing change in MSX2A. BMC Biol., 15: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol, 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Shengdar Q, Sander JD, et al. 2013. Efficient in vivo genome editing using RNA-guided nucleases. Nat. Biotechnol, 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Kingman GA, Jones FC, Guenther CA, Grimwood J, Schmutz J, et al. 2016. Evolving new skeletal traits by cis-regulatory changes in bone morphogenetic proteins. Cell, 164: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Krauss J. and Nusslein-Volhard C. 2014. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development, 141: 4827–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L-E, Wente SR and Chen W. 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci, 110: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA and Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science, 337: 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E. and Doudna J. 2013. RNA-programmed genome editing in human cells. Elife, 2013: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature, 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, Brand M, Jiang YJ, Heisenberg CP, Lin S, Haffter P, et al. 1996. Zebrafish pigmentation mutations and the processes of neural crest development. Development, 123: 369–389. [DOI] [PubMed] [Google Scholar]
- Kingsley DM and Peichel CL 2007. The molecular genetics of evolutionary change in sticklebacks. In: Biology of the three-spined stickleback (Östlund-Nilsson S, Mayer I, and Huntingford F, eds), pp. 41–81. Boca Raton, FL: CRC Press. [Google Scholar]
- Kingsley DM, Zhu B, Osoegawa K, Jong PJ De, Marra M, Peichel C., et al. 2004. New genomic tools for molecular studies of evolutionary change in threespine sticklebacks. Behaviour, 141: 1331–1344. [Google Scholar]
- Koopman P, Gubbay J, Vivian Ni, Goodfellow P. and Lovell-Badge R. 1991. Male development of chromosomally female mice transgenic for Sry. Nature, 351: 117–121. [DOI] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K. and Bradley A. 2018. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol, 36: 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB and Valen E. 2016. CHOPCHOP v2 : a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res, 44: 272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MPK, Mest JR, Wong AC, Norton HL, Aros MC, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science, 310: 1782–1787. [DOI] [PubMed] [Google Scholar]
- Lewis ZR, McClellan MC, Postlethwait JH, Cresko WA and Kaplan RH 2008. Female-specific increase in primordial germ cells marks sex differentiation in threespine stickleback (Gasterosteus aculeatus). J. Morphol, 269: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. 2013. RNA-guided human genome engineering via Cas9. Science, 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A. and Sato T. 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature, 399: 559–563. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, et al. 2007. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell, 131: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Glazer AM, Summers BR, Blackman BK, Norman AR, Shapiro MD, et al. 2014. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics, 197: 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. 2011. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol, 29: 143–150. [DOI] [PubMed] [Google Scholar]
- Mills MG, Greenwood AK and Peichel CL 2014. Pleiotropic effects of a single gene on skeletal development and sensory system patterning in sticklebacks. Evodevo, 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz M, Gagnon JA, Church GM and Valen E. 2014. CHOPCHOP : a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res, 42: 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brown NM, Summers BR, Jones FC, Brady SD and Kingsley DM 2015. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. Elife, 4: e05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Rawls JF, Pratt SJ, Whitfield TT and Johnson SL 1999. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ cell development. Development, 126: 3425–3436. [DOI] [PubMed] [Google Scholar]
- Peichel CL and Marques DA 2017. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Philos. Trans. R. Soc. Lond. B Biol. Sci, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, et al. 2001. The genetic architecture of divergence between threespine stickleback species. Nature, 414: 901–905. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, et al. 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol, 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Rogers SM, Tamkee P, Summers B, Balabahadra S, Marks M, Kingsley DM, et al. 2012. Genetic signature of adaptive peak shift in threespine stickleback. Evolution., 66: 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P, Street SL, Gaide O, Hertig S, Tardivel A, Tschopp J, et al. 2001. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member Ectodysplasin-A. J. Biol. Chem, 276: 18819–18827. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC and Moens CB 2015. Rapid reverse genetic screening using CRISPR in zebrafish. Nat. Methods, 12: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jónsson B, et al. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature, 428: 717–723. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Summers BR, Balabhadra S, Aldenhoven JT, Miller AL, Cunningham CB, et al. 2009. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr. Biol, 19: 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers WK 1979. The coat colors of mice: a model for mammalian gene action and interaction. Springer-Verlag New York. [Google Scholar]
- Swarup H. 1958. Stages in the Development of the Stickleback Gasterosteus aculeatus (L.). Development, 6: 373–383. [PubMed] [Google Scholar]
- Swee LK, Ingold-Salamin K, Tardivel A, Willen L, Gaide O. and Favre M. 2009. Biological activity of Ectodysplasin A is conditioned by its collagen and heparan sulfate proteoglycan-binding domains. J. Biol. Chem, 284: 27567–27576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AC, Capellini TD, Guenther CA, Chan YF, Infante CR, Menke DB, et al. 2018. A novel enhancer near the Pitx1 gene influences development and evolution of pelvic appendages in vertebrates. Elife, e38555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DE, Eisenman J, Baird A, Rauch C, Van Ness K, March CJ, et al. 1990. Identification of a ligand for the c-kit proto-oncogene. Cell, 63: 167–174. [DOI] [PubMed] [Google Scholar]
- Xie KT, Wang G, Thompson AC, Wucherpfennig JI, Reimchen TE, Maccoll ADC, et al. 2019. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science, 363: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L. and Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas mediated genome engineering. Cell, 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Z. and Ge W. 2018. An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by CRISPR/Cas9-mediated gene editing. J. Biol. Chem, 293: 6611–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]