Abstract
We initially identified c-myc promoter-binding protein 1 (MBP-1) from a human cervical carcinoma cell expression library which negatively regulates c-myc promoter activity. A recent study demonstrated that MBP-1 acts as a general transcriptional repressor (A. K. Ghosh, R. Steele, and R. B. Ray, Mol. Cell. Biol. 19:2880–2886, 1999). In order to identify the cellular protein(s) interacting with MBP-1 for transcriptional regulation, a HeLa cell cDNA expression library was screened using a yeast two-hybrid system. An MBP-1-interacting cDNA encoding a polypeptide of 140 amino acid residues with an approximate molecular mass of 16 kDa was identified and named MBP-1 interacting protein-2A (MIP-2A). MIP-2A has a sequence similarity with an unknown mRNA and SEDL. Mutations in the SEDL gene, located at human chromosome Xp22, has recently been implicated with an X-linked genetic disease, although the function of SEDL gene product was not determined (A. K. Gedeon et al., Nat. Genet. 22:400–404, 1999). However, our results suggested the localization of MIP-2A at human chromosome 19. The specificity of interaction between MBP-1 and MIP-2A was verified by an in vitro glutathione S-transferase pulldown experiment, a mammalian two-hybrid analysis, and in vivo coimmunoprecipitation assays. Further analysis revealed that the amino-terminal domain of MBP-1 (amino acids 1 to 95) interacts with MIP-2A. Immunofluorescent staining suggested colocalization of MIP-2A and MBP-1 primarily in the perinuclear membrane of cells. Functional analysis demonstrated that MIP-2A relieves MBP-1 mediated transcriptional repression on c-myc promoter. Additionally, MIP-2A antagonizes cell growth regulatory role of MBP-1. Taken together, these results suggest the functional interaction of MIP-2A and MBP-1 in cell growth regulation.
We initially identified c-myc promoter-binding protein 1 (MBP-1) from a human cervical carcinoma (HeLa) cell expression library (25). MBP-1 is ubiquitously expressed in different human tissues (26) and is located at human chromosome 1p35-pter (31). This protein binds to the TATA box sequences of the c-myc P2 promoter. In vitro transient-transfection assay suggested that MBP-1 negatively regulates both human and mouse c-myc promoter activity (24, 25) through the N-terminal half (27, 30). Further studies have shown that MBP-1 and TATA-binding protein (TBP) bind simultaneously in the minor groove of the c-myc P2 promoter (4). Ectopic expression of MBP-1 induces cell death and the reduction of c-myc expression (24) and regresses tumor growth (28). However, Bcl2, a cell survival gene (1, 29), protects against MBP-1-mediated apoptotic cell death, suggesting that, besides c-myc regulation, MBP-1 exerts a regulatory role on cell growth through other unknown mechanisms (24).
A recent study indicated that MBP-1, when brought to the promoter by a Gal4 DNA-binding domain, can significantly repress transcriptional activity (7). Structure-function analysis of MBP-1 mutants in the context of the Gal4 DNA-binding domain revealed that MBP-1 transcriptional repressor domains are located in the N terminus (amino acids 1 to 47) and C terminus (amino acids 232 to 338). This further suggests that MBP-1 can modulate cellular gene transcription through an alternative mechanism, besides blocking the c-myc transcription. To understand the regulatory role of MBP-1 in cell growth, it is important to characterize the interacting cellular protein(s) and to evaluate molecular mechanisms involved in MBP-1-mediated transcriptional repression. We undertook a search for cellular proteins that interact with MBP-1 using a two-hybrid interaction cloning strategy in yeast. An MBP-1 interacting protein (MIP-2A) was identified which relieves the transcriptional repression of MBP-1 and antagonizes MBP-1-mediated cell death. Thus, MBP-1 may regulate transcriptional modulation by forming a complex with MIP-2A and also exerts its effect for cell growth regulation.
MATERIALS AND METHODS
Yeast two-hybrid screening.
The coding region of MBP-1 was cloned in frame with the LexA DNA-binding domain into the pLexA plasmid vector (Clontech) at BamHI and XhoI restriction sites. The yeast strain EGY48 carrying both the Leu and LacZ reporter genes was transformed with Lex-MBP-1 plasmid DNA and selected positive clones on a synthetic dropout (sd) His− Ura− agar plate. The positive yeast colonies were tested for MBP-1 expression by Western blot analysis using anti-LexA monoclonal antibody (Clontech). Lex–MBP-1 positive yeast cells were grown in appropriate liquid medium lacking histidine and uracil and transformed with human cervical carcinoma (HeLa) cell cDNA library (kindly provided by Alain Nepveu, McGill University, Montreal, Quebec, Canada) constructed in pB42AD plasmid vector under the control of an inducible promoter Gal1 which drives the expression of library-encoded proteins fused to the B42 activation domain. Yeast colonies bearing library proteins were primarily selected on agar plates lacking histidine, tryptophan, and uracil over a period of 7 days. Positive colonies were scraped and replated on galactose-raffinose-containing agar medium lacking histidine, leucine, tryptophan, and uracil following a 4-h growth period at 30°C in galactose-raffinose-containing liquid medium lacking the above-mentioned amino acids. Colonies appeared on galactose-raffinose agar medium were transferred onto fresh medium and grown, and then we performed a β-galactosidase (β-Gal) assay. Positive interaction was determined by the appearance of blue colonies. The β-Gal-positive candidate colonies were picked up and grown in selective medium for library plasmid isolation. Isolated plasmids were then transformed into Escherichia coli KC8 strain and selected on M9-Trp− agar plates. The putative MBP-1-interacting cDNA inserts were retransformed into EGY48 yeast strain bearing the Lex-MBP-1 fusion gene, followed by growth on selective medium and a β-Gal assay. Candidate clones were subsequently sequenced and analyzed.
Chromosomal localization of human MIP-2A.
Genomic DNAs from the NIGMS Hybrid Mapping Panel was obtained from the NIGMS Genetic Mutant Cell Repository (Coriell Cell Institute for Medical Research, Camden, N.J.). Mapping panel 2 consisted of DNA isolated from 24 human-rodent cell hybrids retaining one or more chromosomes. All but two hybrids retained a single intact human chromosome and therefore mapping panel 2 is referred to as a nonredundant mapping panel. Approximately 5 μg of DNA from hamster, human, and mouse DNA were digested with BamHI, HindIII, and PstI to find a suitable restriction fragment length polymorphism (RFLP) or unique genomic fragment for use of mapping. Subsequently, genomic DNAs from mapping panel 2 were digested with HindIII. Southern blots were probed with a human 0.75-kb MIP-2A cDNA as previously described (31). Hybrids were scored for the appropriate human-specific restriction endonuclease fragment on autoradiographs.
Plasmid constructs for mammalian two-hybrid analysis.
The full-length coding sequence of putative candidate MBP-1-interacting clone (MIP-2A) was PCR amplified using a sense primer (5′-ATATATTGAATTCATGTCTGG-3′) and an antisense primer (5′-GGAATTTTCTCGAGTCAGCTT-3′) and cloned in frame with the DNA-binding domain of Gal4 under the control of the simian virus 40 promoter (pM1MIP-2A). A hybrid polypeptide containing the transactivation domain of herpesvirus VP16 fused to full-length MBP-1 was also constructed (VPMBP-1) using the mammalian expression vector VPFlag (34). Similarly, a series of expression plasmids were constructed in VPFlag that encode hybrid polypeptides containing the VP16 transactivation domain fused to MBP(1–234), MBP(1–155), MBP(1–95), and MBP(190–338) deletion mutants.
In vitro pulldown experiment.
A glutathione S-transferase (GST)–MBP-1 fusion construct was expressed in bacteria and immobilized onto GST-beads (8). The beads were incubated with [35S]methionine-labeled full-length MIP-2A in vitro-translated product, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography as previously described (8).
Coimmunoprecipitation assay.
For coimmunoprecipitation, 3 μg of the VPMBP-1 and pM1MIP-2A or pM1 expression plasmid DNAs was cotransfected into subconfluent NIH 3T3 cells. Cells were lysed 48 h after transfection in low-stringency lysis buffer (10 mM HEPES, pH 7.5; 150 mM NaCl; 5 mM EDTA; 0.1% NP-40) containing a cocktail of protease inhibitors and sonicated briefly. Cleared lysates were immunoprecipitated with monoclonal antibodies to Flag, followed by the addition of protein A-Sepharose beads. Precipitates were washed five times with lysis buffer and separated by SDS-PAGE, followed by immunoblotting with anti-Gal4 rabbit polyclonal antibody for detection of MIP-2A fusion protein. A secondary antibody conjugate (anti-rabbit immunoglobulin G–horseradish peroxidase; Amersham) was used for detection of the peroxidase signal by enhanced chemiluminescence.
Immunofluorescence study.
HeLa cells were transfected with GalMBP-1 and/or CMVHA–MIP-2A (MIP-2A cDNA cloned in frame with the hemagglutinin [HA] tag in pcDNA3 vector) using Lipofectamine for immunofluorescence studies. Cells were washed 48 h after transfection and fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 30 min. After they were fixed, cells were washed twice with PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Cells were incubated with anti-Gal4 rabbit polyclonal antibody, anti-HA mouse monoclonal antibody, or both for 1 h at room temperature. Cells were washed and incubated with fluorochrome-conjugated secondary antibodies for 30 min at room temperature. Finally, washed cells were mounted for confocal microscopy using a Bio-Rad 1024 confocal microscope as described previously (9). Fluorescence images were superimposed digitally to allow fine comparison. Colocalization of green (fluorescein isothiocyanate [FITC]) and red (tetramethyl rhodamine isocyanate [TRITC]) signals in a single pixel produces yellow color, while separated signals remain green or red. There was no detectable staining when normal control sera were used.
Cell transfection and chloramphenicol acetyltransferase (CAT) assay.
Full-length MIP-2A was cloned into pcDNA3 vector at EcoRI/XhoI restriction sites under the control of the cytomegalovirus (CMV) promoter. GalMBP-1 fusion protein constructed in CMVGal4 plasmid vector was used in this study (7). Subconfluent NIH 3T3 cells were transfected with 1.0 μg of Gal4TKCAT reporter plasmid, 2.0 μg GalMBP-1, and/or various amounts of CMVMIP-2A using Lipofectamine. For the mammalian two-hybrid assay, cells were cotransfected with 1.0 μg of Gal4-responsive reporter gene (G5E1bCAT), 2.0 μg of pM1MIP-2A and 2.0 μg of VPMBP-1 or its deletion hybrid effector plasmids. At 48 h after transfection, cell extracts were prepared, and a CAT assay was performed as previously described (9). In all of the transfection experiments, β-Gal gene was included to normalize the transfection efficiency.
Transformation assay.
Full-length MBP-1 cDNA (pSV2MBP-1) cloned into pSV2neo vector (25) was used in this study. Mouse embryo fibroblasts (NIH 3T3) were cotransfected with pSV2MBP-1 and CMVMIP-2A or pcDNA3 in a ratio of 5:1, with appropriate controls (pSV2MBP-1 or CMVMIP-2A alone), using Lipofectamine. Transfected cells were split 48 h after transfection at a ratio of 1:9 and treated with 500 μg of G418 per ml for selection of drug-resistant colonies. At 3 weeks following G418 treatment, cells were fixed and stained with crystal violet, and then we counted the transformed foci.
Nucleotide sequence accession number.
The GenBank accession number is AF291676.
RESULTS
Yeast two-hybrid screening for MBP-1-interacting proteins.
We undertook a search for cellular proteins that interact with MBP-1 using yeast two-hybrid interaction cloning. EGY48 yeast cells expressing MBP-1 were transformed with HeLa cell cDNA library plasmid DNAs, and we selected the candidate interacting colonies on the basis of their ability to grow in appropriate selection medium and turn on the LacZ reporter gene. We picked up 50 clones which were grown in leucine-deficient selective medium and exhibited β-Gal activity. Plasmid DNA was isolated from 21 clones and retransformed into EGY48 yeast cells expressing Lex–MBP-1 gene for the confirmation of positive interaction. All 21 clones exhibited positive growth on selective medium and β-Gal activity following retransformation. The putative interacting clones were sequenced and analyzed by the BLAST program. Sequence analysis revealed that the majority of these isolates represent cDNA clones of “unknown mRNA” (accession no. AF058918), and we named this novel protein MIP-2A. However, while our work was in progress, a computer search using the BLAST program matched MIP-2A with SEDL (accession no. NM-014563) gene (6). Interestingly, the SEDL coding sequence (sedlin) has perfect homology with MIP-2A; however, the function of sedlin has not yet been determined.
Chromosomal localization of human MIP-2A.
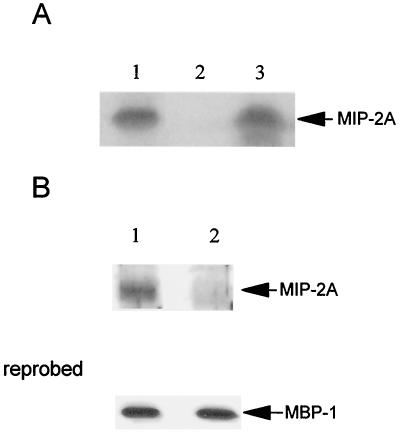
The coding sequences of MIP-2A and SEDL genes are identical; however, there are significant differences in their untranslated regions. Therefore, we wanted to determine the chromosomal localization of MIP-2A. For this purpose, we performed rodent-human somatic hybridization (31). Hamster, human, and mouse DNAs were digested with BamHI, HindIII, and PstI to identify a specific RFLP pattern for MIP-2A gene in each species. Southern blot analysis was performed using the entire MIP-2A cDNA, including the untranslated regions as a probe. A human-specific 1.0-kb fragment was found on Southern blots of HindIII-digested genomic DNAs from the parental cell lines (hamster, human, and mouse) (Fig. 1). DNAs from the parental and the somatic cell hybrid cell lines were digested with HindIII, Southern blotted, and probed. An analysis of mapping panel 2 indicated that the human-specific HindIII pattern was observed in cell line 19, which contains human chromosome 19. Results from this experiment indicated that MIP-2A is localized only at human chromosome 19.
FIG. 1.
Mapping of MIP-2A in humans to chromosome 19 using a monochromosomal mapping panel. HindIII-digested genomic DNAs from hamster (h), human (H), and mouse (M) sources, as well as 24 human-rodent somatic cell hybrids (lanes 1 to 22, X, and Y) probed with entire MIP-2A cDNA. The human-specific fragment is indicated with an arrow and can be seen in the human control and in lane 19, which contain hamster-human cell hybrid DNA.
Association of MBP-1 with MIP-2A in mammalian cells.
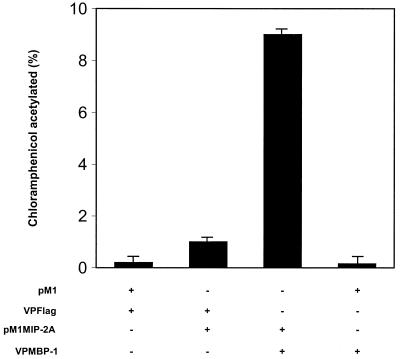
The mammalian version of the conventional yeast two-hybrid assay was used to ascertain whether the MIP-2A associates with MBP-1 in mammalian cells (34). For this purpose, we constructed the mammalian expression plasmid vectors that encode VPMBP-1 fusion protein and Gal4 DNA-binding protein fused to MIP-2A (pM1MIP-2A). The mammalian two-hybrid assay was then performed by transfecting NIH 3T3 cells with a Gal4-responsive reporter gene (G5E1bCAT) and pairwise combinations of the appropriate expression vectors. Reporter gene expression was determined by measuring CAT activity in cell lysates from each transfected culture. A significant increase in CAT activity was observed following coexpression of the VPMBP-1 and pM1MIP-2A hybrid (Fig. 2). However, CAT activity was not enhanced by coexpression of the VPMBP-1 and empty vector (pM1). CAT activity was also detected in pM1MIP-2A and VPFlag vector-transfected cells, although the level of CAT expression was much lower compared to the hybrid. These results indicated that MIP-2A forms a complex with MBP-1 in mammalian cells. Specificity of this interaction was similarly examined using a number of other cellular proteins (MAD, ZEB, and CtBP) as Gal4 fusion constructs for association with MBP-1 or MIP-2A. Results from these studies suggested that MBP-1 or MIP-2A did not physically associate with these cellular proteins in a mammalian two-hybrid assay (data not shown).
FIG. 2.
Interaction of MBP-1 with MIP-2A in a mammalian two-hybrid system. NIH 3T3 cells were transfected with 1 μg of G5E1bCAT reporter gene, 2 μg of VPMBP-1, and 2 μg of pM1MIP-2A. A CAT assay was performed at 48 h posttransfection. The amount of DNA was kept constant in each transfection by adding the empty vector DNA. The MBP-1–MIP-2A hybrid shows a high level of CAT activity compared to pM1MIP-2A or VPMBP-1 alone.
MBP-1 interacts with MIP-2A in vitro.
An in vitro binding assay was employed to determine MBP-1–MIP-2A interactions. GST–MBP-1 was expressed in bacteria, immobilized onto beads, and incubated with 35S-labeled full-length MIP-2A generated by in vitro translation. The binding mixture was then subjected to SDS-PAGE, followed by autoradiography. The results showed specific binding between MBP-1 and MIP-2A (Fig. 3A). However, the GST-beads did not bind with the in vitro-translated MIP-2A. The molecular mass of MIP-2A is about 16 kDa as determined from the relative electrophoretic mobility of the in vitro-translated product with respect to the protein molecular mass standards on the same SDS-polyacrylamide gel. Similarly, histidine-tagged MIP-2A immobilized onto Ni-beads or unrelated histidine-tagged protein beads (used as a negative control) were incubated with 35S-labeled in vitro-translated full-length MBP-1. Results from this set of experiment also showed a specific physical association between MBP-1 and MIP-2A (data not shown). However, an unrelated viral protein used as negative control failed to show binding toward the in vitro-translated MBP-1 product.
FIG. 3.
MBP-1 physically associates with MIP-2A. (A) In vitro-translated [35S]methionine labeled MIP-2A was subjected to a GST pulldown analysis with GST–MBP-1 fusion protein immobilized on beads (lane 3) or on GST-beads (lane 2). Ten percent of the in vitro-translated MIP-2A (lane 1) was loaded in gel for electrophoresis. (B) In vivo coimmunoprecipitation of MBP-1 with MIP-2A. NIH 3T3 cells were cotransfected with Flag-tagged MBP-1 and Gal4-tagged MIP-2A or empty vector and lysed after 48 h. MIP-2A-transfected (lane 1) and vector-transfected (lane 2) cell lysates were immunoprecipitated with a monoclonal antibody to Flag. Immunoprecipitates were separated by SDS–12% PAGE and immunoblotted with a rabbit antibody to Gal4 DNA-binding domain. The molecular weight of the MIP-2A fusion protein band was ascertained from the migration of standard protein molecular weight markers. The blot was reprobed with a monoclonal antibody to Flag for the detection of MBP-1. A similar level of MBP-1 expression was detected in each immunoprecipitate.
Detection of MBP-1–MIP-2A complexes in vivo.
A coimmunoprecipitation assay was also performed using lysates of NIH 3T3 cells cotransfected with VPMBP-1 and pM1MIP-2A or empty vector as a negative control to verify the in vivo association of MBP-1 with MIP-2A. Cell lysates were immunoprecipitated with a monoclonal antibody to Flag and separated by SDS-PAGE. The presence of Gal4–MIP-2A (pM1MIP-2A) fusion protein was then detected by immunoblotting with antibodies to Gal4. MIP-2A was coimmunoprecipitated as Gal4 fusion protein (lane 1) from cells expressing Flag-tagged MBP-1 (Fig. 3B). However, the empty vector (pM1) alone expressing Gal4 DNA-binding domain, used as a negative control, did not exhibit binding to MBP-1 (lane 2) under similar experimental conditions. Therefore, the specific association of MBP-1 and MIP-2A in mammalian cells was demonstrated by two independent procedures: the two-hybrid assay and coimmunoprecipitation analysis.
Colocalization of MBP-1 with MIP-2A.
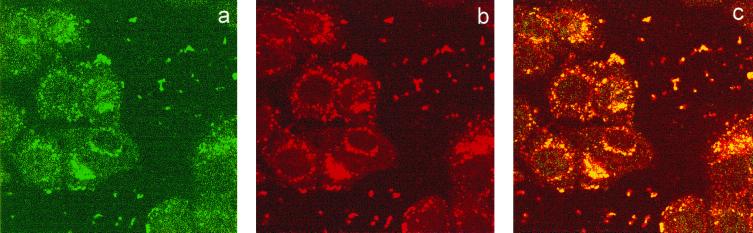
To determine the biological significance of interaction between MBP-1 and MIP-2A, we investigated whether these two cellular proteins colocalize in the cells. We first examined the localization of MBP-1 and MIP-2A in GalMBP-1 or CMVHA–MIP-2A transfected HeLa cells by indirect immunofluorescence. A predominant perinuclear localization of MIP-2A and a nucleocytoplasmic localization of MBP-1 was observed (data not shown). When cells were cotransfected with both the plasmids expressing the MBP-1 and MIP-2A, immunofluorescent staining exhibited a predominant colocalization of these two proteins in the perinuclear region of cells (Fig. 4).
FIG. 4.
Colocalization of transfected GalMBP-1 and CMVHA–MIP-2A in HeLa cells. Immunofluorescent staining was performed following cotransfection of MBP-1 and MIP-2A into HeLa cells using a rabbit polyclonal antibody against Gal4 DNA-binding domain for MBP-1 (a) and a monoclonal antibody to HA for MIP-2A (b). (c) Fluorescence images of panels a and b were superimposed digitally for fine comparison.
Identification of MIP-2A binding domain of MBP-1.
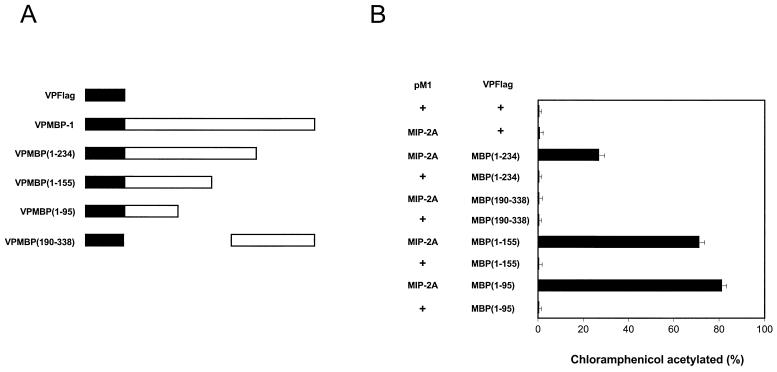
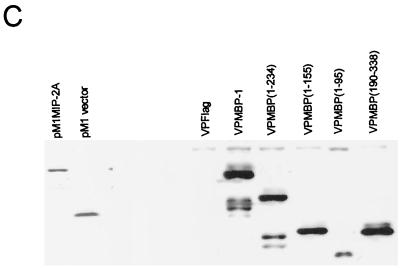
A library of MBP-1 deletion mutants were constructed by cloning in frame downstream of the VP16 acidic transactivation domain into the vector VPFlag (Fig. 5A) to identify the MIP-2A binding region of MBP-1 using the mammalian two-hybrid assay. For this purpose, NIH 3T3 cells were cotransfected with pM1MIP-2A and these deletion mutants of MBP-1, along with the reporter gene G5E1bCAT as described earlier. A significant increase in CAT activity was noted when MBP-1 N-terminal deletion mutants [VPMBP(1–234), VPMBP(1–155), or VPMBP(1–95)] and pM1MIP-2A were coexpressed (Fig. 5B). However, CAT activity was not altered by the coexpression of MBP-1 deletion mutants and empty Gal4 vector (pM1). No significant increase in CAT activity was detected in VPMBP(190–338) and pM1MIP-2A coexpressed cell lysates compared to pM1MIP-2A and VP16 vector coexpression. These results suggested that the MIP-2A interacting domain of MBP-1 is localized within the first 95 amino acid residues. Protein expression from pM1MIP-2A and MBP-1 deletion mutants was examined by Western blot analysis (Fig. 5C). Multiple polypeptides appearing from some of the MBP-1 deletion constructs may represent proteolytically cleaved products, as was observed earlier (7).
FIG. 5.
Analysis of MIP-2A interacting domain of MBP-1. (A) Schematic representation of VPMBP-1 deletion mutants used in the analysis for the MIP-2A interacting domain. The filled box represents the VP16 transactivation domain, and the open box represents the MBP-1 sequences. The numbers following MBP are amino acid positions. (B) Mammalian two-hybrid assay for the MIP-2A interacting domain of MBP-1. NIH 3T3 cells were transfected with G5E1bCAT reporter gene and the two indicated expression vectors. A CAT assay was performed at 48 h posttransfection. The amount of DNA was kept constant in each transfection by adding the empty vector DNA. MIP-2A cloned into pM1 expression vector was used as indicated under pM1. MBP-1 deletion mutants were cloned into VPFlag expression vector and used as indicated under VPFlag. The use of empty vector is indicated by “+” sign in the figure. (C) Expression of pM1MIP-2A and deletion mutants of MBP-1 by Western blot analysis using monoclonal antibodies to the Gal4 DNA-binding domain and the Flag epitope, respectively.
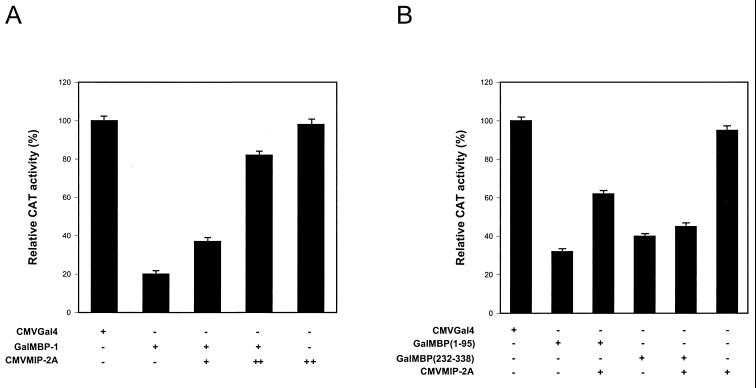
MIP-2A relieves transcriptional repression by MBP-1.
To examine the functional effect of MIP-2A on MBP-1-mediated transcriptional repression of a Gal4-dependent promoter in the reporter plasmid Gal4TKCAT, the MIP-2A cDNA fragment was cloned into pcDNA3 expression vector under the control of CMV early promoter (CMVMIP-2A). GalMBP-1 repressed thymidine kinase (TK) promoter activity approximately 70% in NIH 3T3 cells. However, cotransfection of NIH 3T3 cells with GalMBP-1 and increasing amounts of CMVMIP-2A restores promoter activity in a dose-dependent manner (Fig. 6A). Expression of CMVMIP-2A alone had no significant effect on TK promoter activity under similar experimental conditions. To further examine the correlation between MIP-2A binding domain of MBP-1 (amino acid residues 1 to 95) and the ability of MIP-2A to relieve MBP-1-mediated transcriptional repression, the amino- and carboxy-terminal deletion mutants of MBP-1 encompassing the repressor domains [GalMBP(1–95) and GalMBP(232–338)] were used for functional analysis. GalMBP(1–95) or GalMBP(232–338) was cotransfected with CMVMIP-2A and Gal4TKCAT for the CAT assay. The results suggested that MIP-2A relieves transcriptional repression only from the GalMBP(1–95) construct (Fig. 6B). To further verify the functional significance of MBP-1–MIP-2A interaction, two other transcriptional repressor proteins (ZEB and MAD) were used in the in vitro reporter assay. The results exhibited that MIP-2A does not alter the repressor activity of ZEB and MAD (data not shown). These studies suggested a specific association between MBP-1 and MIP-2A for functional activity.
FIG. 6.
MBP-1-mediated transcriptional repression is relieved by MIP-2A. (A) Effector plasmids (GalMBP-1 and/or CMVMIP-2A) were cotransfected with Gal4TKCAT as the reporter plasmid in NIH 3T3 cells. The total amount of plasmid DNA was kept constant by the addition of empty vector in each transfection. Cell extracts were prepared 48 h posttransfection, and a CAT assay was performed. (B) Deletion mutants of MBP-1 [GalMBP(1–95) or GalMBP(232–338)] were cotransfected with CMVMIP-2A and Gal4TKCAT in NIH 3T3 cells for CAT assay to determine the functional significance of MBP-1 and MIP-2A interaction. In each set of experiments, triplicate transfections were performed and the relative CAT activities are presented.
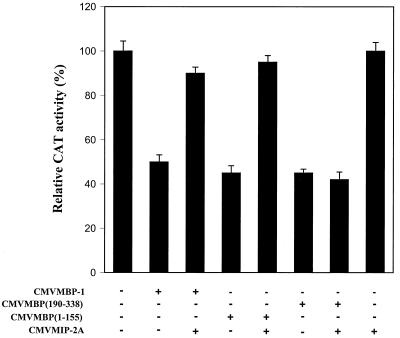
MIP-2A relieves MBP-1-mediated transcriptional repression on the c-myc promoter.
In order to investigate whether the interaction of MBP-1 with MIP-2A has an effect on c-myc promoter activity, an in vitro transient reporter assay was performed. CV1 cells were cotransfected with a c-myc–CAT reporter gene (25) and MBP-1 with or without CMVMIP-2A. MBP-1 alone repressed the c-myc promoter activity approximately 50%. However, c-myc promoter activity was significantly restored following coexpression of MIP-2A with MBP-1 (Fig. 7). On the other hand, expression of MIP-2A alone had no detectable effect on the c-myc promoter activity. Similar experiment was done using p53 promoter as a control, where neither MBP-1 nor MIP-2A had a trans-regulatory effect (data not shown). Functional association of MIP-2A with MBP-1 for trans-repression activity was further ascertained using deletion mutants of MBP-1. An in vitro transient reporter assay using c-myc–CAT, MIP-2A, and MBP-1 deletion mutants was performed (Fig. 7). Results exhibited that MIP-2A can relieve the transcriptional repression of c-myc promoter mediated by the MBP-1 N-terminal half (MBP1–155) but not its C-terminal half, MBP(190–338). Together, these results suggest that MIP-2A modulates MBP-1-mediated transcriptional repression on the c-myc promoter through protein-protein interaction.
FIG. 7.
Association of MIP-2A with MBP-1 relieves repressor activity on the c-myc promoter. CV1 cells were cotransfected with the c-myc–CAT reporter gene and the indicated expression constructs. The total amount of plasmid DNA was kept constant by the addition of empty vector in each transfection. Cell extracts were prepared at 48 h posttransfection, and a CAT assay was performed. In each set of experiments, triplicate transfections were performed and the relative CAT activities are presented.
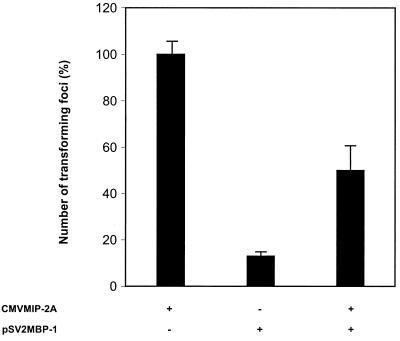
Effects of MIP-2A on MBP-1-mediated cell growth suppression.
Previous studies have shown that the overexpression of full-length MBP-1 induces cell death in murine fibroblasts (24). To examine the effect of MIP-2A, NIH 3T3 cells were transfected with pSV2MBP-1 or CMVMIP-2A or cotransfected with pSV2MBP-1 and CMVMIP-2A at a ratio of 5:1, and the G418-resistant colonies were selected over a period of 3 weeks. Following G418 selection, the colonies were stained and the numbers of resistant colonies were counted. The numbers of colonies obtained from three independent experiments of transfection with MBP-1 alone were markedly reduced (∼87%) compared to CMVMIP-2A-transfected colonies (Fig. 8). However, coexpression of MBP-1 and MIP-2A significantly increased the number of colonies. These results suggested that MIP-2A antagonizes MBP-1-mediated cell growth regulation. Since the C-terminal deletion mutant of MBP-1 [MBP(190–338)] significantly reduced the cell growth (27), we further examined its antiproliferative activity in the presence of MIP-2A. While the expression of MBP(190–338) in NIH 3T3 cells leads to growth arrest, coexpression with MIP-2A did not significantly alter the colony numbers. This raises the question regarding how MIP-2A antagonizes MBP-1-mediated cell growth suppression. A plausible explanation comes from the relief of MBP-1-mediated transcriptional repression of c-myc by MIP-2A, which plays a role in cell proliferation. However, the mechanism of cell growth regulation by MIP-2A–MBP-1 interaction remains to be elucidated.
FIG. 8.
MIP-2A antagonizes MBP-1-mediated cell death in murine fibroblasts. NIH 3T3 cells were cotransfected with pSV2MBP-1 and CMVMIP-2A at a ratio of 5:1 with appropriate controls (pSV2MBP-1 or CMVMIP-2A alone). Transfected cells were split after 48 h and treated with 500 μg of G418 per ml for the selection of drug-resistant colonies. At 3 weeks following treatment, cells were fixed and stained with crystal violet to count the foci. Results are presented as the means from three independent experiments.
DISCUSSION
Previous studies have shown that MBP-1 transcriptionally downregulates c-myc promoter activity by direct binding to its P2 promoter sequences (25). MBP-1 also acts as a general transcriptional repressor when brought to the promoter using the Gal4 DNA-binding domain (7), although the molecular basis of this repressor activity has not been well understood. Recruitment of histone deacetylase by MBP-1 is one of the mechanisms by which repression could occur; however, other factors may also be involved for transcriptional regulation by MBP-1 (8). Eukaryotic repressors are typically modular, consisting of a single polypeptide with functionally distinct activities distributed among the domains (reviewed in reference (13)). These domains can target different components of the transcription machinery to affect distinct steps in initiation. Thus, multiple mechanisms built within a single repressor ensure that a gene can be silenced in an efficient manner (20).
In search of the potential partner(s) of MBP-1, a yeast two-hybrid interacting screening was employed. Our results demonstrated that MIP-2A physically associates with MBP-1 both in vitro and in vivo. Functional analysis indicates that MIP-2A relieves the transcriptional repression of MBP-1 and antagonizes the MBP-1-mediated cell death in fibroblasts. The specificity and functional significance of this interaction were examined using a number of cellular proteins as a control. Structure-function analysis suggested that MIP-2A binds to the amino-terminal repressor domain of MBP-1 and relieves the repressor activity. The association of MIP-2A also relieves transcriptional repression of MBP-1 on the natural c-myc promoter. Analysis of predicted amino acids suggests that MIP-2A possesses two potential CKII phosphorylation sites. MIP-2A is a phosphoprotein (R.B.R. and A.K.G., unpublished observation); however, the biological significance of phosphorylation remains to be elucidated. Additionally, the absence of a DNA-binding domain in MIP-2A raises interesting possibilities for serving as a coactivator or an adaptor-like molecule in MBP-1-mediated transcriptional repression. Although MIP-2A exhibited identity with the SEDL coding sequence, this gene is localized at human chromosome 19 and does not appear to be a pseudogene (6). Future characterization of MIP-2A genome should clarify the relationship with SEDL in the context of its functional relevance.
Protein-protein interactions play a key role in transcriptional regulation. Repressors interact with their target to transmit signals to the transcription machinery. However, interaction with another protein can also impair the ability of a transcriptional repressor to downregulate mRNA synthesis. There are a number of examples to this phenomenon. The γ5 subunit of a heterotrimeric G protein has been shown to bind specifically to the adipocyte enhancer-binding protein 1 to attenuate its transcriptional repression (23). On the other hand, the adenoviral protein E1A is known to exert several biological activities, including the transformation of cells and the activation or repression of cellular and viral genes (reviewed in reference (22)). E1A blocks the ability of the CREB-binding protein (CBP) to function as a coactivator for a number of transcription factors and binds to three sites within CBP. Recent studies have demonstrated that CBP interacts with SRCAP and influences its ability to activate transcription (17). Different promoters which utilize CBP to activate transcription have diverse requirements for coactivators (18). In contrary, Mad member interacting protein 1 (Mmip1) can suppress the antiproliferative actions of Mad family of proteins and indirectly upregulates the transcriptional activity of c-myc (11). Another newly identified Mad-interacting protein (Mmip2) can reverse the suppressive effect of Mad proteins on c-myc-responsive target genes and on c-myc–c-ras-mediated focus formation in fibroblasts (33). A protein complex may prevent transcription factor from binding to a specific DNA sequence or from interacting with basal or other transcription factors (reviewed in reference (5)). Formation of myc-Max heterocomplexes activate transcription (2, 12, 19) and Mad-Max complexes repress transcription (3, 14, 32). Likewise, Mnt or Rox interacts with Max in vivo and functions as a transcriptional repressor (15, 21). Mga, a recently identified Max-interacting protein, suppresses transcriptional activation by c-myc and inhibits c-myc-dependent cell transformation, thereby regulating myc-Max target genes in vivo (16). Several studies have demonstrated an inverse correlation between the expression of c-myc and Mad family proteins (10). Whether a similar correlation occurs between MBP-1 and MIP-2A is not known at this time. However, MBP-1, being a negative regulator for c-myc, is concurrently expressed with c-myc gene in the human breast carcinoma cells (26). The results presented here further exemplify a complex regulatory role of MBP-1 in cellular machineries through the identification of its cellular partner MIP-2A as a negative regulator of MBP-1 functions.
ACKNOWLEDGMENTS
We thank G. Chinnadurai and Michael M. C. Lai for valuable suggestions on the yeast two-hybrid system, Alain Nepveu for the HeLa cell cDNA library, Richard Baer for providing plasmid constructs of the mammalian two-hybrid system, Douglas Dean and Bob Eisenman for providing the Gal4 expression vectors, and Jane McHowat and Ranjit Ray for helpful discussions.
This research was supported by PHS grant CA52799 from the National Cancer Institute.
REFERENCES
- 1.Adams J M, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Dalton S, Brooks M W, Littlewood T D, Evan G I, Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992;359:423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- 3.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–223. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary D, Miller D M. The c-myc promoter binding protein (MBP-1) and TBP bind simultaneously in the minor groove of the c-myc P2 promoter. Biochemistry. 1995;34:3438–3445. doi: 10.1021/bi00010a036. [DOI] [PubMed] [Google Scholar]
- 5.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 6.Gedeon A K, Colley A, Jamieson R, Thompson E M, Rogers J, Sillence D, Tiller G E, Mulley J C, Gecz J. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet. 1999;22:400–404. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh A K, Steele R, Ray R B. Functional domains of c-myc promoter binding protein 1 involved in transcriptional repression and cell growth regulation. Mol Cell Biol. 1999;19:2880–2886. doi: 10.1128/mcb.19.4.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh A K, Steele R, Ray R B. MBP-1 physically associates with histone deacetylase for transcriptional repression. Biochem Biophys Res Commun. 1999;260:405–409. doi: 10.1006/bbrc.1999.0921. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh A K, Majumder M, Steele R, Yaciuk P, Chrivia J, Ray R, Ray R B. Hepatitis C virus NS5A protein modulates transcription through a novel transcription factor SRCAP. J Biol Chem. 2000;275:7184–7188. doi: 10.1074/jbc.275.10.7184. [DOI] [PubMed] [Google Scholar]
- 10.Grandori C, Eisenman R N. Myc target genes. Trends Biochem Sci. 1997;22:177–181. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 11.Gupta K, Anand G, Yin X-Y, Grove L, Prochownik E V. Mmip1: a novel leucine zipper protein that reverses the suppressive effects of Mad family members on c-myc. Oncogene. 1998;16:1149–1159. doi: 10.1038/sj.onc.1201634. [DOI] [PubMed] [Google Scholar]
- 12.Gu W, Cechova K, Tassi V, Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci USA. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 14.Hurlin P J, Queva C, Koskinen P J, Steingrimsson E, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 16.Hurlin P, Steingrimsson E, Copeland N G, Jenkins N A, Eisenman R N. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–1028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston H, Kneer J, Chackalaparapil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- 18.Korzus E, Torchia J, Rose D, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–706. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 19.Kretzner L, Blackwood E M, Eisenman R N. Transcriptional activities of the Myc and Max proteins in mammalian cells. Curr Top Microbiol Immunol. 1992;182:435–443. doi: 10.1007/978-3-642-77633-5_55. [DOI] [PubMed] [Google Scholar]
- 20.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 21.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro C L, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran E. Cell growth control mechanisms reflected through protein interactions with adenovirus E1A gene products. Semin Virol. 1994;5:327–340. [Google Scholar]
- 23.Park J-G, Muise A, He G-P, Kim S-W, Ro H-S. Transcriptional regulation by the gamma5 subunit of a heterotrimeric G protein during adipogenesis. EMBO J. 1999;18:4004–4012. doi: 10.1093/emboj/18.14.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray R. Induction of cell death in murine fibroblasts by a c-myc promoter binding protein. Cell Growth Differ. 1995;6:1089–1096. [PubMed] [Google Scholar]
- 25.Ray R, Miller D M. Cloning and characterization of a human c-myc promoter-binding protein. Mol Cell Biol. 1991;11:2154–2161. doi: 10.1128/mcb.11.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray R B, Sheikh M S, Fontana J F, Miller D M. Human breast carcinoma cells show correlation in expression of c-myc oncogene and the c-myc binding protein (MBP-1) Int J Oncol. 1994;5:1433–1436. doi: 10.3892/ijo.5.6.1433. [DOI] [PubMed] [Google Scholar]
- 27.Ray R B, Steele R. Separate domains of MBP-1 involved in c-myc promoter binding and growth suppressive activity. Gene. 1997;186:175–180. doi: 10.1016/s0378-1119(96)00693-2. [DOI] [PubMed] [Google Scholar]
- 28.Ray R B, Steele R, Seftor E, Hendrix M. Human breast carcinoma cells transfected with the gene encoding c-myc promoter binding protein (MBP-1) shows tumor suppression in nude mice. Cancer Res. 1995;55:3747–3751. [PubMed] [Google Scholar]
- 29.Reed J C. Bcl-2 and the regulation of programmed cell death. J Cell Biol. 1994;124:1–6. doi: 10.1083/jcb.124.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Miller D M. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene. J Biol Chem. 2000;275:5958–5965. doi: 10.1074/jbc.275.8.5958. [DOI] [PubMed] [Google Scholar]
- 31.White R A, Adkinson L R, Dowler L L, Ray R B. Chromosomal localization of the human gene encoding c-myc promoter-binding protein (CMBP1) at chromosome 1p35-pter. Genomics. 1997;39:406–408. doi: 10.1006/geno.1996.4499. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, Pena A, Korcz A, Soprano D R, Soprano K J. Overexpression of Mxi1 inhibits the induction of the human ornithine decarboxylase gene by the Myc/Max protein complex. Oncogene. 1996;12:621–629. [PubMed] [Google Scholar]
- 33.Yin X-Y, Gupta K, Han W P, Levitan E S, Prochownik E V. Mmip-2, a novel RING finger protein that interacts with mad members of the Myc oncoprotein network. Oncogene. 1999;18:6621–6634. doi: 10.1038/sj.onc.1203097. [DOI] [PubMed] [Google Scholar]
- 34.Yu X, Wu L C, Bowcock A M, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]