Abstract
Background
At the start of the 2019–2020 influenza season, concern arose that circulating B/Victoria viruses of the globally emerging clade V1A.3 were antigenically drifted from the strain included in the vaccine. Intense B/Victoria activity was followed by circulation of genetically diverse A(H1N1)pdm09 viruses that were also antigenically drifted. We measured vaccine effectiveness (VE) in the United States against illness from these emerging viruses.
Methods
We enrolled outpatients aged ≥6 months with acute respiratory illness at 5 sites. Respiratory specimens were tested for influenza by reverse-transcriptase polymerase chain reaction (RT-PCR). Using the test-negative design, we determined influenza VE by virus subtype/lineage and genetic subclades by comparing odds of vaccination in influenza cases versus test-negative controls.
Results
Among 8845 enrollees, 2722 (31%) tested positive for influenza, including 1209 (44%) for B/Victoria and 1405 (51%) for A(H1N1)pdm09. Effectiveness against any influenza illness was 39% (95% confidence interval [CI]: 32–44), 45% (95% CI: 37–52) against B/Victoria and 30% (95% CI: 21–39) against A(H1N1)pdm09-associated illness. Vaccination offered no protection against A(H1N1)pdm09 viruses with antigenically drifted clade 6B.1A 183P-5A+156K HA genes (VE 7%; 95% CI: –14 to 23%) which predominated after January.
Conclusions
Vaccination provided protection against influenza illness, mainly due to infections from B/Victoria viruses. Vaccine protection against illness from A(H1N1)pdm09 was lower than historically observed effectiveness of 40%–60%, due to late-season vaccine mismatch following emergence of antigenically drifted viruses. The effect of drift on vaccine protection is not easy to predict and, even in drifted years, significant protection can be observed.
Keywords: influenza, vaccine effectiveness, vaccination, antigenic drift, test-negative
Influenza vaccination provided 39% protection against medically attended influenza during the 2019–2020 season. Effectiveness against influenza A(H1N1)pdm09 viruses was 30% overall but reduced against an emergent viral subclade that predominated late in the season.
(See the Major Article by Izurieta et al on pages e4251–9.)
Circulating influenza viruses change constantly and effectiveness of seasonal influenza vaccines is influenced by how well the vaccine matches circulating influenza viruses. The 2019–2020 influenza season was notable for unusually intense early circulation of influenza B viruses, which were of the B/Victoria lineage [1, 2]. Whereas influenza B viruses have constituted approximately 10%–30% of annual cases in the US over the past decade [3, 4], in the 2019–2020 season B/Victoria viruses represented close to half of all influenza cases. Children experienced a high incidence of severe influenza and the number of influenza-related pediatric deaths was the highest observed since the severe 2017–2018 season, with a majority of these deaths attributed to B/Victoria viruses [5]. Nearly all circulating B/Victoria viruses belonged to a different genetic clade, V1A.3, compared to the B/Colorado/06/2017-like (Victoria lineage V1A.1) reference virus in the 2019–2020 Northern Hemisphere vaccine [1]. From the start of the season there was speculation that vaccine effectiveness (VE) would be poor against B/Victoria [6].
The 2019–2020 season was also the first time since the 2009 influenza pandemic with high H1N1 activity in 2 consecutive seasons. Hemagglutinin (HA) of co-circulating A(H1N1)pdm09 viruses belonged to various 6B.1A 183P subclades [7]. Viruses within the genetically diverse 183P-5A subclade predominated and have been co-circulating with other clades since 2018. During the season, the majority of viruses belonged to 1 of 2 183P-5A subclades with acquired amino acid substitutions. The apparent mismatch of the B/Victoria viruses and the diversity of influenza A(H1N1)pdm09 viruses gave us the ability to study the effects of antigenic drift on VE using data collected by the US Influenza Vaccine Effectiveness (Flu VE) Network.
METHODS
Study Population
The US Flu VE Network has been described previously [8, 9]. In brief, the research network enrolls participants aged 6 months or older with an acute respiratory illness (ARI) including cough of ≤7 days duration at sites in Michigan, Pittsburgh, Texas, Washington, and Wisconsin. Enrollment begins at each site based on local evidence of increasing influenza activity. Patients who received influenza antivirals in the past 7 days, with illness of more than 7 days duration, and those born after March 1, 2019 were ineligible. Following consent, research staff interviewed participants or their proxies to obtain demographic and clinical information, self-rated general health status, and details about receipt of current season influenza vaccination. High-risk medical conditions were defined as medical record documentation within the previous year of International Classification of Diseases (ICD-10) codes for high-risk conditions according to the Advisory Committee on Immunization Practices (ACIP) [10]. Enrollment ended at all study sites by March 26, 2020 due to increasing community circulation of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in the US, despite ongoing influenza activity.
Influenza Vaccination History
For the 2019–2020 season, reference strains for Northern Hemisphere influenza vaccines included A/Brisbane/02/2018 (H1N1)pdm09 (clade 6B.1A), A/Kansas/14/2017 (H3N2), and B/Colorado/06/2017 (Victoria lineage, clade V1A.1) [11]. Quadrivalent vaccines also included a B/Phuket/3073/2013-like virus (Yamagata lineage). Participant vaccination status for the 2019–2020 season was obtained using electronic medical and immunization records, state immunization registries, health insurance records, and employee health records. Adult enrollees (but not children) were also classified as vaccinated by plausible self-report, in which patients reported date and location of vaccination without documentation of vaccination [12]. For this analysis, all patients, including children aged <9 years, were classified as vaccinated if they had received one or more doses of any licensed vaccine product after July 1, 2019 [11].
Laboratory Methods
Following enrollment, nasal and oropharyngeal specimens were collected from participants (nasal specimens only in children <2 years of age). Specimens were tested at study sites using a CDC-developed real-time RT-PCR assay, with additional determination of influenza A subtype and B lineage. Influenza-positive specimens with an RT-PCR cycle threshold value ≤30, indicative of a higher viral load, were sent to the CDC for further genetic characterization, including whole-genome sequencing [4]. Hemagglutinin (HA) genetic clades and subclades were determined based on phylogenetic analysis [13].
Statistical Analysis
We described demographic and clinical characteristics of influenza cases and test-negative controls (ie, with negative RT-PCR testing for influenza) using medians for continuous variables and numbers and proportions for categorical variables. Characteristics of influenza cases and controls were compared using χ2 statistics. We estimated the effectiveness of influenza vaccination against medically attended influenza A(H1N1)pdm09 and B/Victoria lineage viruses using the test-negative study design [14], which compares the odds ratio (OR) of current-season influenza vaccination in influenza cases versus test-negative controls. VE was calculated as 100% × (1-OR) using logistic regression models, adjusting for potential confounders. Network site, age in natural cubic splines, presence of any high-risk conditions versus none, and calendar time in 2-week enrollment intervals were included in final adjusted models. Sex, race/ethnicity, self-rated general health status, and interval in days from symptoms onset to enrollment did not change OR by >5%, used as a pre-specified threshold for inclusion [15], and were therefore excluded. Influenza A(H3N2) and B/Yamagata VE were not assessed due to limited circulation during the season [5]. We stratified VE estimates by age (6 months to 8 years, 9–17 years, 18–49 years, 50–64 years, and ≥65 years of age) and within genetic subclades for genetically characterized A(H1N1)pdm09 and B/Victoria viruses. We excluded patients who received influenza vaccination less than 14 days before illness onset, with indeterminant RT-PCR results, and those who tested negative for influenza before local circulation of influenza viruses. Significance levels were set at P < .05 and analyses were conducted using SAS Version 9.4 (Cary, North Carolina, USA). This study was approved by institutional review boards at the Centers for Disease Control and Prevention and all participating sites.
RESULTS
Participant Characteristics
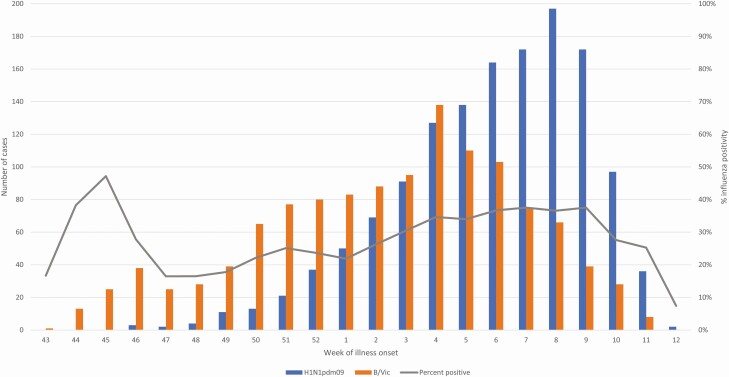
From October 29, 2019 through March 26, 2020, 9171 patients with an ARI were enrolled (9011 unique participants, with 160 enrolled more than once). We excluded from the analysis 113 (1%) test-negative patients with illness onset before local influenza circulation, 73 (1%) with indeterminant RT-PCR test results, and 140 (1%) who received influenza vaccination 0–13 days prior to illness onset. Among 8845 included participants, 2722 (31%) tested positive for influenza by RT-PCR and 6123 (69%) tested negative. Of cases, 52% (1405/2722) tested positive for influenza A(H1N1)pdm09, 44% (1209/2722) B/Victoria, 1% (29/2722) influenza A(H3N2), <1% (6/2722) B/Yamagata viruses, and 3% (83/2722) did not have subtype/lineage results. Ten tested positive for more than one virus; these were included in subtype/lineage estimates. From October 29—December 31, 2019, influenza B/Victoria viruses accounted for 77% (369/482) of influenza cases; from January 1—March 26, 2020, 58% (1307/2240) were influenza A(H1N1)pdm09 cases (Figure 1). Comparing influenza cases and test-negative controls, case patients were younger (median age 26 years versus 32 years) and more likely to be male (45% versus 40%) but similar with respect to race/ethnicity (Table 1). Overall, 51% (4528/8845) of participants received influenza vaccination, 42% (1140/2722) among influenza cases, and 55% (3388/6123) among test-negative patients.
Figure 1.
Number of patients enrolled with an acute respiratory illness testing positive for influenza B/Victoria and A(H1N1)pdm09 and percent of enrollees testing positive for influenza by epidemiologic week during the 2019–2020 season. Includes patients with medically attended acute respiratory illness enrolled in the US Influenza Vaccine Effectiveness network between October 29, 2019 through March 26, 2020. Dates on x-axis reflect the reported date of illness onset.
Table 1.
Characteristics of Participants Enrolled in the US Influenza Vaccine Effectiveness Network for the 2019–2020 Influenza Seasona
| Test result status | |||||
|---|---|---|---|---|---|
| Influenza-positive | Influenza-negative | ||||
| Characteristic | No. | (Column %) | No. | (Column %) | P-value |
| Overall | 2722 | 6123 | |||
| Study site | <.01 | ||||
| Michigan | 346 | 13 | 620 | 10 | |
| Pennsylvania | 536 | 20 | 977 | 16 | |
| Texas | 459 | 17 | 1612 | 26 | |
| Washington | 456 | 17 | 1410 | 23 | |
| Wisconsin | 925 | 34 | 1504 | 25 | |
| Sex b | <.01 | ||||
| Female | 1509 | 55 | 3660 | 60 | |
| Male | 1212 | 45 | 2463 | 40 | |
| Age group (years) | <.01 | ||||
| 6 months–8 years | 646 | 24 | 1365 | 22 | |
| 9–17 | 471 | 17 | 722 | 12 | |
| 18–49 | 1056 | 39 | 2202 | 36 | |
| 50–64 | 350 | 13 | 998 | 16 | |
| ≥65 | 199 | 7 | 836 | 14 | |
| Race/ethnicity c | .29 | ||||
| White, non-Hispanic | 1929 | 71 | 4365 | 71 | |
| Black, non-Hispanic | 252 | 9 | 549 | 9 | |
| Other, non-Hispanic | 278 | 10 | 556 | 9 | |
| Hispanic | 258 | 9 | 636 | 10 | |
| ≥1 high-risk condition d | 1014 | 37 | 2902 | 47 | <.01 |
| Vaccinated | 1140 | 42 | 3388 | 55 | <.01 |
a Included patients with medically attended acute respiratory illness enrolled in the US Influenza Vaccine Effectiveness network between October 29, 2019 through March 26, 2020.
b Sex was missing for 1 influenza-positive case.
c Race/ethnicity was missing for 22, including 5 influenza-positive cases and 17 influenza-negative controls.
d High-risk conditions included chronic cardiac diseases and circulatory diseases, chronic pulmonary diseases, diabetes mellitus, chronic renal disease, hemoglobinopathies, immunosuppressive disorders, malignancy, metabolic diseases (excluding diabetes mellitus), liver diseases, neurological/musculoskeletal conditions, cerebrovascular disease, morbid obesity, and endocrine disorders.
Genetic Characterization
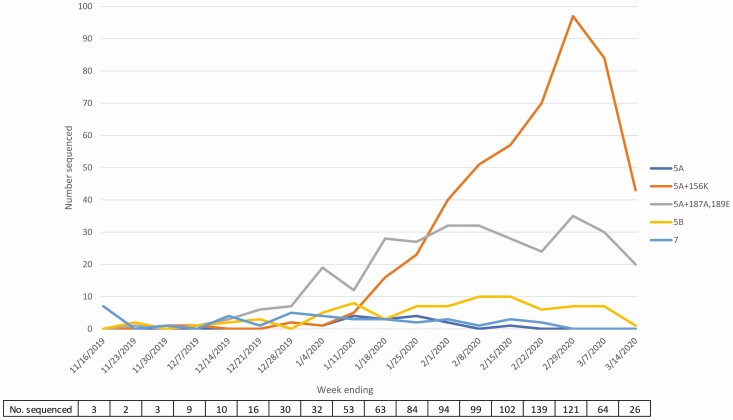
Of 999 A(H1N1)pdm09 specimens sent to CDC for sequencing, 950 (95%) successfully underwent characterization by whole-genome sequencing. Within influenza A(H1N1)pdm09 HA clade 6B.1A, which includes the vaccine strain, further genetic diversification was observed. Early in the season, 6B.1A subclade 5A viruses with additional amino acid substitutions D187A and Q189E in the HA protein (referred to here as 5A+187A,189E viruses) predominated (Figure 2). Subclade 5A viruses with additional amino acid changes K130N, N156K, L161I, V250A, and E506D (referred to here as 5A+156K viruses) increased markedly midseason, becoming the predominant subclade of sequenced viruses by February 2020. Over the entire season, 509 (54%) sequenced A(H1N1)pdm09 viruses belonged to the subclade 5A+156K group and 313 (33%) to the 5A+187A,189E group. Among B/Victoria viruses, 926 (98%) of 945 sequenced viruses belonged to clade V1A.3, which contains a 3-amino acid deletion (162–164) in the HA protein and differs antigenically from the vaccine component (V1A.1), which has a 2-amino acid deletion (162–163) in the HA protein.
Figure 2.
Number of sequenced influenza A(H1N1)pdm09 viruses and percentage within genetic subclades by epidemiologic week during the 2019–2020 season. Influenza A(H1N1)pdm09 viruses with cycle threshold values ≤30 from the US Influenza Vaccine Effectiveness network were sent to CDC for whole-genome sequencing. Virus names are abbreviated names for phylogenetic subclades: 5A=A(H1N1)pdm09 183P-5A; 5A+156K=A(H1N1)pdm09 183P-5A+156K; 5A+187A,189E=A(H1N1)pdm09 183P-5A+187A,189E; 5B=A(H1N1)pdm09 183P-5B; 7=A(H1N1)pdm09 183P-7. Dates on the x-axis reflect the reported date of illness onset.
Vaccine Effectiveness
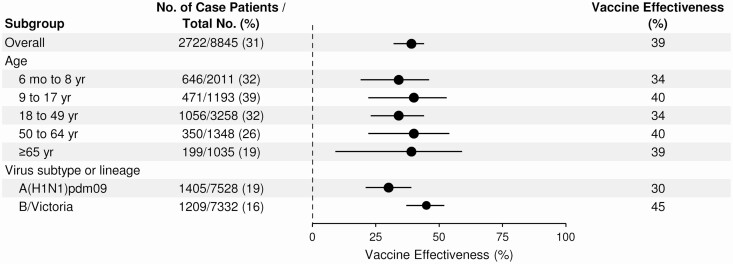
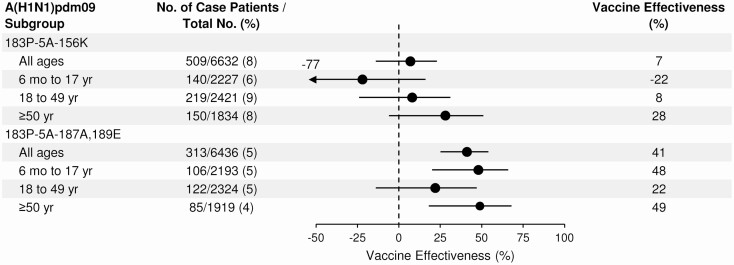
From October 29, 2019 through March 26, 2020, the overall adjusted VE against medically attended influenza A and B viruses was 39% (95% confidence interval [CI]: 32%–44%) (Figure 3), which was similar across age groups (Supplementary Table 1). Estimated VE against B/Victoria was 45% (95% CI: 37%–52%) and against influenza A(H1N1)pdm09 30% (95% CI: 21%–39%). VE against A(H1N1)pdm09 ranged from 23% to 42% across age groups; it was lowest and not statistically significant within pediatric groups (6 months to 17 years) and highest in adults ≥65 years of age [Supplementary Table 1]. No vaccine effectiveness was observed against the 5A+156K group of influenza A(H1N1)pdm09 viruses that were predominant later in the season (7%, 95% CI: –14% to 23%), whereas VE against 5A+187A,189E viruses was 41% (95% CI: 25%–54%) [Figure 4 and Supplementary Table 2]. Moderate vaccine effectiveness was observed against less common A(H1N1)pdm09 groups, but with limited estimate precision (Supplementary Table 2).
Figure 3.
Adjusted estimates of influenza vaccine effectiveness, overall and stratified by age group and virus subtype/lineage, for participants enrolled in the US Influenza Vaccine Effectiveness Network for the 2019–2020 season. Vaccine effectiveness was calculated by comparing the odds of current season influenza vaccination in participants who tested positive for influenza and participants who tested negative for influenza, or (1-OR) × 100. Logistic regression models were adjusted for network site, age, presence of ≥1 high-risk conditions, and calendar time in 2-week enrollment intervals.
Figure 4.
Adjusted estimates of influenza vaccine effectiveness within influenza A(H1N1)pdm09 subclades, overall and stratified by age group, for participants enrolled in the US Influenza Vaccine Effectiveness Network for the 2019–2020 season. Only genetically characterized influenza A(H1N1)pdm09 viruses were included in this analysis. Vaccine effectiveness was calculated by comparing the odds of current season influenza vaccination in participants who tested positive for influenza and participants who tested negative for influenza, or (1-OR) × 100. Logistic regression models were adjusted for network site, age, presence of ≥1 high-risk conditions, and time in 2-week enrollment intervals.
DISCUSSION
Seasonal influenza viruses frequently change to evade immunity, resulting in a need to regularly update vaccines. The World Health Organization (WHO) convenes an annual consultation in February to determine the composition of Northern Hemisphere influenza vaccines for the upcoming influenza season [7]. Selection of new vaccine strains relies on timely identification of emerging circulating strains. The 2019–2020 influenza vaccine provided protection against medically attended outpatient influenza, with an overall influenza vaccine effectiveness ~40%. Antigenically drifted B/Victoria viruses circulated throughout the season whereas antigenically drifted influenza A(H1N1)pdm09 viruses emerged over the course of the season [7]. Despite differences of the circulating strains with the vaccine-like strain, the 2019–2020 vaccine provided better than expected effectiveness against B/Victoria viruses at 45%. Conversely, the vaccine provided less protection against influenza A(H1N1)pdm09 viruses (30%) compared to average effectiveness of 40%–60% since the 2009 influenza pandemic [16, 17]. Genetic diversification in circulating A(H1N1)pdm09 viruses may have contributed to reduced vaccine effectiveness overall as well as within some age groups, specifically for viruses expressing 6B.1A 5A+156K hemagglutinins [7].
Influenza vaccine components are selected, among other factors, based on genetic and antigenic characterization of circulating influenza viruses, their global distribution and prevalence, and virus inhibition activity of candidate vaccine viruses. Predominant A(H1N1)pdm09 viruses during the 2019–2020 season belonged to phylogenetic subclade 6B.1A 183P-5A [7]. At the time of vaccine strain selection for the 2020–2021 influenza season, the majority of viruses in subclade 5A contained amino acid substitutions D187A and Q189E in HA antigenic site Sb, including reference strains A/Guangdong-Maonan/SWL1536/2019 selected for egg-based, and A/Hawaii/70/2019 selected for cell- or recombinant-based vaccines for the 2020–2021 season [7]. Antigenically distinct 5A viruses containing antigenic site Sb substitution N156K predominated for the rest of the season after vaccine strain selection. Recent antigenic characterization data indicates reduced inhibition of these 183P-5A+156K viruses by post-infection ferret antisera to 183P-5A+187A,189E, suggesting antigenic difference between these 2 phylogenetic subclades [7]. Effectiveness of 2020–2021 Northern Hemisphere influenza vaccines against A(H1N1)pdm09 viruses may depend on subclade circulation.
Influenza B/Victoria clade V1A.3 caused a majority of influenza cases for the first half of the 2019–2020 influenza season in the US. Prior to 2017–2018, the B/Brisbane/60/2008 (Victoria lineage clade V1A) had been the B/Victoria vaccine reference virus since 2009–2010, indicating limited antigenic drift over 8 influenza seasons. Following emergence of antigenically distinct B/Victoria viruses in 2017 within HA clade V1A.1 (containing 2 amino acid deletions at 162–163), B/Colorado/06/2017 (clade V1A.1) was selected as the 2018–2019 vaccine reference virus. B/Victoria viruses from HA clade V1A.3, with 3 amino acid deletions at 162–164, represented a small proportion of viruses in early 2019 but this group rapidly increased and displaced V1A.1 viruses. During the 2019–2020 influenza season, nearly all sequenced B/Victoria viruses belonged to clade V1A.3. Despite initial concerns for possible vaccine mismatch, VE against V1A.3 viruses was similar to estimates for B/Victoria viruses in previous seasons [16]. Antigenically characterized B/Victoria viruses during 2019–2020 showed reduced reactivity by ferret antisera to the vaccine component [18], indicating antigenic difference between the V1A.1 and V1A.3 subclades. However, human sera from vaccinated individuals suggested some cross-reactivity against circulating V1A.3 viruses [18]. This cross-reactivity and immune memory may have contributed to the observed protection against B/Victoria-associated illness. In the 2019–2020 season, over half (56%) of influenza B illness occurred in children <18 years of age, with a significant burden of severe hospitalized illness and deaths in this group [5], whereas B virus infections were uncommon in adults ≥50 years of age.
This study is subject to several limitations. While a large number of influenza viruses from Flu VE Network participants were genetically characterized, the distribution of influenza viruses at study sites may not reflect their distribution throughout the US. If distribution of viruses was not representative, overall vaccine effectiveness estimates may differ. In a minority of patients (10% of all vaccinated), we classified vaccination status based on plausible self-report rather than documentation and some misclassification may have occurred. Analysis stratified by virus subclades in this study precluded further analysis by prior season vaccination or vaccine type (eg, egg-based versus cell-culture or recombinant vaccines) based on limited sample sizes. Waning of vaccine effectiveness may also have contributed in some part to a lack of vaccine effectiveness observed for 183P-5A+156K viruses that predominated later in the season [19].
In summary, influenza vaccination nearly halved the risk of outpatient influenza illness due to antigenically distinct B/Victoria viruses but did not provide protection against the antigenically distinct A(H1N1)pdm09 viruses that predominated in the US later during the 2019–2020 influenza season. This highlights the challenges in predicting vaccine protection against drifted influenza viruses. Even in seasons when circulating viruses are antigenically drifted from the vaccine virus, vaccination can provide protection. Preventing ~40% of any influenza-associated illness through vaccination in 2019–2020 influenza season was particularly encouraging in the context of influenza virus infections causing an estimated 4.3–21 million outpatient visits, 140 000–810 000 hospitalizations, and 12 000–61 000 deaths annually in the US [20]. Strategies to prevent and control influenza are especially critical during the COVID-19 pandemic to reduce overburdening of US healthcare systems should influenza viruses and SARS-CoV-2 co-circulate. In addition to vaccination, early diagnosis through testing, and antiviral treatment including prophylaxis strategies in outbreaks among high-risk populations could be crucial in curbing influenza disease [21, 22].
Nonstandard Abbreviations: ACIP, Advisory Committee on Immunization Practices; ARI, acute respiratory illness; CI, confidence intervals; US Flu VE, US Influenza Vaccine Effectiveness; HA, hemagglutinin; ICD-10, International Classification of Diseases, Tenth Revision; OR, odds ratio; RT-PCR, reverse-transcriptase polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VE, vaccine effectiveness; WHO, World Health Organization
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We would like to acknowledge the contributions of Sarah Bauer, Kim Beney, Caroline K. Cheng, Marco Ciavaglia, Sarah Davenport, Rebecca Fong, Amy Getz, Kendra Goforth, Michelle Groesbeck, Emileigh Johnson, Asad Kamal, Anne Kaniclides, Sanaa Khechen, Seung Jun Kim, Armanda Kimberly, Lois E. Lamerato, Ryan E. Malosh, E.J. McSpadden, Joshua G. Petrie, Rachel Phillips, Hannah Segaloff, Ava Selke, Stephanie Taylor, Rachel Truscon, Miranda Viars, Regina Lehmann-Wandell, Micah Wildes, University of Michigan, Ann Arbor, and Henry Ford Health System, Detroit, Michigan; G.K. Balasubramani, Todd M. Bear, Klancie Dauer, David Figucia, Heather Eng, Stacey Engster, Edward Garofolo, Robert Hickey, Philip Iozzi, Monika Johnson, Stephanie Kirk, Jason A. Lyons, Donald B. Middleton, Jonathan M. Raviotta, Theresa Sax, Joe Suyama, Marian Vanek, Alexandra Weissman, John V. Williams, University of Pittsburgh Schools of the Health Sciences and University of Pittsburgh Medical Center, Pittsburgh, Pennsylvania; Alejandro Arroliga, Madhava Beeram, Kelsey Bounds, Lydia Clipper, Kayan Dunnigan, Jason Ettlinger, Amanda Karl, Mary Kylberg, Kempapura Murthy, Teresa O’Quinn, Deborah Price, Chandni Raiyani, Manohar Mutnal, Jeremy Ray, Michael Reis, Natalie Settele, Courtney Shaver, Michael Smith, Jennifer Thomas, Marcus Volz, Kimberley Walker, Jamie Walkowiak, Martha Zayed, Tnelda Zunie, patients and staff members from all participating clinics, Baylor Scott & White Health and Texas A&M University College of Medicine, Temple, Texas; Rachael P. Burganowski, Erika Kiniry, Matt Nguyen, Suzie Park, C. Hallie Phillips, Stacie Wellwood, Brianna M. Wickersham, Kaiser Permanente Washington Health Research Institute, Seattle, Washington; Elizabeth Armagost, Samantha Braun, Deanna Cole, Tom Dalcher, Erin Donnerbauer, Joseph Eddy, Hope Florence, Terry Foss, Sandy Freeman, Wayne Frome, Tammy Gault, Hannah Gourdoux, Gregg Greenwald, Sherri Guzinski, Kayla Hanson, Ellice Harris, Linda Heeren, Lynn Ivacic, Julie Karl, Heather Kawleski, Jennifer King, Tamara Kronenwetter Koepel, Diane Kohnhorst, Laura Konrardy, Erik Kronholm, Stacey Kyle, Megan Maronde, Carrie Marcis, Karen McGreevey, Jennifer Meece, Nidhi Mehta, Vicki Moon, Madalyn Palmquist, Cory Pike, Rebecca Pilsner, Maria Platta, DeeAnn Polacek, Martha Presson, Carla Rottscheit, Jacklyn Salzwedel, Julian Savu, Rachel Schoone, Charity Schug, Kristin Seyfert, Elisha Stefanski, Patrick Stockwell, Sandy Strey, Arin Thompson, Chelsey Thompson, Melissa Wendt, Suellyn Wojcik, Anna Zachow, Marshfield Clinic Research Institute, Marshfield, Wisconsin.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Financial support. This work was supported by the Centers for Disease Control and Prevention [cooperative agreements U01IP001034-U01IP001039]. At Pittsburgh, the project was also supported by the National Institutes of Health through grant UL1TR001857.
Potential conflicts of interest. E. A. B. reports grants from CDC during the conduct of the study. M. G. reports grants from CDC during the conduct of the study and grants from Janssen/Johnson & Johnson and CDC-Abt Associates outside the submitted work. L. A. J. reports grants from CDC during the conduct of the study and an organization research contract with Pfizer to fund clinical trials and studies outside the submitted work, and grants from Novavax outside the submitted work. M. L. J. reports grants from CDC during the conduct of the study and grants from Sanofi Pasteur outside the submitted work. E. T. M. reports personal fees from Pfizer and grants from Merck outside the submitted work. H. Q. M. reports grants from CDC during the conduct of the study and grants from Seqirus outside the submitted work. R. K. Z. reports grants from CDC during the conduct of the study and grants from Sanofi Pasteur outside the submitted work. A. S. M. reports personal fees from Sanofi and Seqirus, outside the submitted work. M. P. N. reports grants from CDC during the conduct of the study and research funding from Merck & Co, outside the submitted work. All other authors report nothing to disclose.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Dawood FS, Chung JR, Kim SS, et al. Interim estimates of 2019-20 seasonal influenza vaccine effectiveness–United States, February 2020. MMWR Morb Mortal Wkly Rep 2020; 69:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Owusu D, Hand J, Tenforde MW, et al. Early season pediatric influenza B/Victoria virus infections associated with a recently emerged virus subclade–Louisiana, 2019. MMWR Morb Mortal Wkly Rep 2020; 69:40–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaglani M, Vasudevan A, Raiyani C, et al. Effectiveness of trivalent and quadrivalent inactivated vaccines against influenza B in the United States, 2011–2012 to 2016–2017. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flannery B, Kondor RJG, Chung JR, et al. Spread of Antigenically Drifted Influenza A(H3N2) viruses and vaccine effectiveness in the United States during the 2018–2019 season. J Infect Dis 2020; 221:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention. Weekly U.S. influenza surveillance report. Available at: https://www.cdc.gov/flu/weekly/index.htm. Accessed 21 July 20.
- 6. WebMD. 2020 flu shot no match for “B” strain, season rages on. Available at: https://www.webmd.com/cold-and-flu/news/20200116/flu-shot-no-match-for-b-strain-season-rages-on#1. Accessed 3 Auguest 2020.
- 7. World Health Organization. WHO consultation and information meeting on the composition of influenza virus vaccines for use in the 2020–21 Northern Hemisphere influenza season. Available at: https://www.who.int/influenza/vaccines/virus/recommendations/consultation202002/en/. Accessed 21 July 2020.
- 8. Flannery B, Chung JR, Monto AS, et al. Influenza vaccine effectiveness in the United States during the 2016–2017 season. Clin Infect Dis 2019; 68:1798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson ML, Chung JR, Jackson LA, et al. Influenza vaccine effectiveness in the United States during the 2015–2016 season. N Engl J Med 2017; 377:534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fiore AE, Shay DK, Broder K, et al. ; Centers for Disease Control and Prevention . Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm Rep 2009; 58:1–52. [PubMed] [Google Scholar]
- 11. Grohskopf LA, Alyanak E, Broder KR, Walter EB, Fry AM, Jernigan DB. Prevention and control of seasonal influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices–United States, 2019–20 influenza season. MMWR Recomm Rep 2019; 68:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irving SA, Donahue JG, Shay DK, Ellis-Coyle TL, Belongia EA. Evaluation of self-reported and registry-based influenza vaccination status in a Wisconsin cohort. Vaccine 2009; 27:6546–9. [DOI] [PubMed] [Google Scholar]
- 13. Centers for Disease Control and Prevention. Types of influenza viruses. Available at: https://www.cdc.gov/flu/about/viruses/types.htm. Accessed 21 July 2020.
- 14. Foppa IM, Haber M, Ferdinands JM, Shay DK. The case test-negative design for studies of the effectiveness of influenza vaccine. Vaccine 2013; 31:3104–9. [DOI] [PubMed] [Google Scholar]
- 15. Zimmerman RK, Nowalk MP, Chung J, et al. 2014–2015 influenza vaccine effectiveness in the United States by vaccine type. Clin Infect Dis 2016; 63:1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belongia EA, Simpson MD, King JP, et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 2016; 16:942–51. [DOI] [PubMed] [Google Scholar]
- 17. Tenforde MW, Chung J, Smith ER, et al. Influenza vaccine effectiveness in inpatient and outpatient settings in the United States, 2015–2018. Clin Infect Dis 2020. doi: 10.1093/cid/ciaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Epperson S, Davis CT, Brammer L, et al. Update: influenza activity–United States and worldwide, May 19-September 28, 2019, and composition of the 2020 Southern hemisphere influenza vaccine. MMWR Morb Mortal Wkly Rep 2019; 68:880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferdinands JM, Fry AM, Reynolds S, et al. Intraseason waning of influenza vaccine protection: evidence from the US influenza vaccine effectiveness network, 2011-12 through 2014-15. Clin Infect Dis 2017; 64:544–50. [DOI] [PubMed] [Google Scholar]
- 20. Centers for Disease Control and Prevention. Past seasons estimated influenza disease burden. Available at: https://www.cdc.gov/flu/about/burden/past-seasons.html. Accessed 3 Auguest 2020.
- 21. Ikematsu H, Hayden FG, Kawaguchi K, et al. Baloxavir marboxil for prophylaxis against influenza in household contacts. N Engl J Med 2020; 383:309–20. [DOI] [PubMed] [Google Scholar]
- 22. Centers for Disease Control and Prevention. Influenza antiviral medications: Summary for clinicians. Available at: https://www.cdc.gov/flu/professionals/antivirals/summary-clinicians.htm. Accessed 26 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.