Abstract
Background
Accelerated epigenetic aging using DNA methylation (DNAm)-based biomarkers has been reported in people with human immunodeficiency virus (HIV, PWH), but limited data are available among African Americans (AA), women, and older PWH.
Methods
DNAm was measured using Illumina EPIC Arrays for 107 (69 PWH and 38 HIV-seronegative controls) AA adults ≥60 years in New York City. Six DNAm-based biomarkers of aging were estimated: (1) epigenetic age acceleration (EAA), (2) extrinsic epigenetic age acceleration (EEAA), (3) intrinsic epigenetic age acceleration (IEAA), (4) GrimAge, (5) PhenoAge, and (6) DNAm-estimated telomere length (DNAm-TL). The National Institutes of Health (NIH) Toolbox Cognition Battery (domains: executive function, attention, working memory, processing speed, and language) and Montreal Cognitive Assessment (MoCA) were administered. Participants were assessed for frailty by the Fried criteria.
Results
The PWH and control groups did not differ by sex, chronological age, or ethnicity. In total, 83% of PWH had a viral load <50 copies/mL, and 94% had a recent CD4 ≥200 cells/µL. The PWH group had a higher EAA, EEAA, GrimAge, and PhenoAge, and a lower DNAm-TL compared to the controls. IEAA was not different between groups. For PWH, there were significant negative correlations between IEAA and executive function, attention, and working memory and PhenoAge and attention. No associations between biomarkers and frailty were detected.
Conclusions
Evidence of epigenetic age acceleration was observed in AA older PWH using DNAm-based biomarkers of aging. There was no evidence of age acceleration independent of cell type National Institutes of Health composition (IEAA) associated with HIV, but this measure was associated with decreased cognitive function among PWH.
Keywords: epigenetics, epigenteic age, aging, DNA methylation, cognitive function
Evidence of epigenetic age acceleration was observed in older African American adults with human immunodeficiency virus (HIV) using DNAm-based biomarkers of aging. A measure of age acceleration independent of cell type composition was associated with decreased cognitive function among the HIV group.
Antiretroviral therapy (ART) has dramatically improved life expectancy for people living with human immunodeficiency virus (HIV, PWH), leading to a growing population of PWH ≥50 years of age [1]. However, due to chronic HIV infection and long-term ART, PWH do not necessarily experience healthy aging, and have higher than expected rates of comorbidities, including geriatric syndromes (eg, frailty and cognitive impairment) compared to individuals without HIV [2–4].
These observations have led to speculation that HIV, directly or indirectly, accelerates aging processes. Although many biomarkers are sensitive to aging, such as telomere length, epigenetic clocks have emerged as sensitive aging biomarkers [5]. A number of different epigenetic clocks now exist that use DNA methylation (DNAm) patterns at specific cytosine-phosphate-guanine (CpG) sites to estimate “epigenetic age” and contrast it with chronological age to determine if there is epigenetic age acceleration [6, 7]. “First-generation” clocks include epigenetic age acceleration (EAA), extrinsic epigenetic age acceleration (EEAA), and intrinsic epigenetic age acceleration (IEAA). However, the first-generation clocks are less predictive of clinical measures and disease endpoints [8]. Two “second-generation” clocks–GrimAge and PhenoAge, were developed to overcome this limitation and predict morbidity/mortality in the elderly [9, 10]. A DNA-methylation estimator of telomere length (DNAm-TL) was also developed [11].
Accelerated epigenetic age has been reported among adult PWH, mostly male, including those on ART [12–14], as well as youth with perinatally acquired HIV [15]. However, data are lacking for women, African Americans (AAs), and older PWH (>60 years). The relationships between epigenetic age acceleration and geriatric outcomes, including cognitive function and frailty, have been examined in the general population [16–21]. Among men >50 years, EEAA was associated with greater cognitive decline [20]. In a study of twins (mean age 56 years), EEA and IEAA were found to be associated with a decline in cognition scores [21]. EEA was associated with frailty in a study of 1820 elderly adults [16]. However, less is known about these associations among PWH, particularly older AA. Accelerated epigenetic aging was observed in brain tissue samples of PWH diagnosed pre-mortem with cognitive impairment [22], and 2 studies of youth found negative correlations between epigenetic age acceleration and measures of cognitive functioning, including a study of 20–35 year-old AA PWH [15, 23]. Few studies have evaluated epigenetic age and frailty in PWH [24].
This study aimed to assess if HIV infection is associated with epigenetic age acceleration in AA adults ≥60 years by first- and second-generation epigenetic clocks and to evaluate if epigenetic age acceleration is associated with frailty and cognitive function.
METHODS
Study Participants
Participants were recruited from February 2018-March 2020 at Columbia University Irving Medical Center (CUIMC) in New York City. Recruitment was accomplished through flyers and referrals from clinical providers. Eligible participants had to identify as AA, be ≥60 years of age, and have a documented positive HIV test and be on ART for ≥5 years (PWH) or have a negative human immunodeficiency virus type 1 (HIV-1) antibody test at enrollment (control). Those with hepatitis B/C infection or any condition that, by investigator opinion, would compromise the subject’s ability to participate, were not eligible. The study was approved by the Institutional Review Board at CUIMC, and informed consent was obtained from participants.
Measurements
Demographic information was obtained by questionnaire. For PWH, ART history, nadir and recent CD4 T-cell counts, and HIV-RNA viral load were collected. Using standardized protocols, blood pressure, weight, height, and waist and iliac circumference were measured. Body mass index (BMI) (weight/height2) and waist-to-hip ratio were calculated.
The National Institutes of Health (NIH) Toolbox Cognition Battery was used to assess cognitive function across 5 domains: executive function (Dimensional Change Card Sort Test), attention (Flanker Inhibitory Control and Attention Test), working memory (List Sorting Working Memory Test), processing speed (Pattern Comparison Processing Speed Test), and language (Oral Reading Recognition Test) [25]. The battery was self-administered on an iPad. Normative scores included the age-corrected standard score (mean 100; SD 15) and fully corrected T-score (mean 50; SD 10) which compare scores to a nationally representative sample, adjusting for age, gender, race/ethnicity, and education. The Montreal Cognitive Assessment (MoCA) was also administered [26]. Total scores were used to index overall cognitive function, and a normal score was defined as ≥26. Participants were assessed for frailty [27] by the presence of ≥3 of 5 Fried criteria.
Blood samples were collected by venipuncture and DNA extracted using the Qiagen Qiamp DNA Blood Midi Kit (Qiagen, Germantown, Maryland, USA) and quantified using the Qubit dsDNA BR Assay Kit and Qubit Fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) at CUIMC. DNAm levels were measured using the Infinium MethylationEPIC BeadChip (Illumina, San Diego, California, USA) at Roswell Park Cancer Institute (Buffalo, New York, USA). Estimated DNAm age (years) was obtained from the online calculator developed by Horvath [6]. Estimated proportions of cell types (B cells, CD4 T cells, CD8 T cells, natural killer cells, granulocytes, monocytes) were calculated [28].
Six methylation-based biomarkers of aging were calculated: (1) EAA [6], (2) EEAA [29, 30], (3) IEAA (4), GrimAge [9], (5) PhenoAge [10], and (6) DNAm-TL (detailed in Supplementary Table 1). For the first 5, positive values indicate the participant’s biological age is older than expected based on chronological age, and negative values indicate the biological age is younger than expected. For DNAm-TL, a shorter telomere length is indicative of aging.
Statistical Analysis
Demographic characteristics, clinical characteristics, and cognitive function test scores for PWH and controls were summarized and compared using χ 2 or Fisher exact tests for categorical variables, and t tests for continuous measures. Each of the methylation-based biomarkers of aging were compared between groups using t tests. The association between HIV status and other characteristics with each of the biomarkers was assessed by fitting linear regression models, using linear regression models, unadjusted and adjusted for confounders that had associations of P < .10 with each of the epigenetic measures. Among PWH, we determined if viral load and absolute CD4 count were associated with each of the epigenetic age measures. Each of the biomarkers was also compared between those who were frail and not frail and those with a normal (≥26) vs abnormal MoCA score (<26) using t tests. Pearson correlations were used for correlating each of the biomarkers with cognitive function test scores for the entire population and stratified by HIV status. Statistical analyses were performed using SAS, version 9.4 (SAS Institute, Cary, North Carolina, USA).
RESULTS
The study included 107 AA participants (69 PWH, 38 controls) 60–82 years of age (Table 1). PWH and control groups did not differ by age, sex, or ethnicity. A higher proportion of controls completed some college or additional education than PWH. Current tobacco use was higher in PWH, although current alcohol use was slightly higher in the control group. PWH had a higher waist-to-hip ratio than the controls and lower BMI. Most PWH were on an integrase strand transfer inhibitor (INSTI)-based ART regimen (49.3%) or an INSTI+protease inhibitor (PI) or non-nucleoside reverse transcriptase inhibitor (NNRTI)-based regimen (21.7%). Almost all PWH had a recent viral load <1000 copies/mL and recent CD4 ≥200 cells/µL. DNAm-estimated blood cell composition differed between groups due to lower proportions of CD4 T cells (0.23 vs 0.38, P < .001) and higher CD8 T cells (0.35 vs 0.18, P < .001) in PWH compared to controls.
Table 1.
Characteristics of 69 Older African American Adults With Human Immunodeficiency Virus (HIV) and 38 HIV-Seronegative Adults (Controls)
| Characteristics | Controls (N = 38) | HIV (N = 69) | P |
|---|---|---|---|
| Sex, N (%) | |||
| Male | 22 (57.9) | 35 (50.7) | .48 |
| Female | 16 (42.1) | 34 (49.3) | |
| Age (years), range | 60–78 | 60–82 | NA |
| Age (years), mean (SD) | 66.0 (5.0) | 64.6 (4.4) | .15 |
| Ethnicity, N (%) | |||
| Hispanic or Latino | 2 (5.3) | 5 (7.3) | 1.00F |
| Not Hispanic or Latino | 36 (94.7) | 64 (92.8) | |
| Education, N (%) | |||
| Less than high school | 2 (5.3) | 20 (29.0) | .001 |
| High school completion | 4 (10.5) | 18 (26.1) | |
| Some college/associate’s | 17 (44.7) | 17 (24.6) | |
| Bachelor’s degree or advanced degree | 15 (39.5) | 14 (20.3) | |
| Tobacco use, N (%) | |||
| Never | 20 (54.1) | 9 (13.0) | <.001 |
| Past | 8 (21.6) | 33 (47.8) | |
| Current | 9 (24.3) | 27 (39.1) | |
| Missing | |||
| Alcohol use, N (%) | |||
| Never | 3 (7.9) | 7 (10.1) | .15 |
| Past | 9 (23.7) | 28 (40.6) | |
| Current | 26 (68.4) | 34 (49.3) | |
| Illicit drug use, N (%) | |||
| Never | 15 (39.5) | 19 (27.5) | .30 |
| Past | 20 (52.6) | 39 (56.5) | |
| Current- | 3 (7.9) | 11 (15.9) | |
| BP systolic, mean (SD) | 132.8 (18.6) | 130.0 (16.1) | .43 |
| Missing | 4 | 7 | |
| BP diastolic, mean (SD) | 78.0 (13.1) | 78.8 (11.1) | .73 |
| Missing | 4 | 7 | |
| Waist-hip ratio, mean (SD) | 0.89 (0.08) | 0.93 (0.09) | .023 |
| BMI (kg/m2), range | 22.0 – 53.7 | 16.8 – 45.4 | |
| BMI (kg/m2), mean (SD) | 30.9 (6.2) | 28.5 (6.4) | .063 |
| HIV characteristics | |||
| Duration of infection (years), mean (SD) | 23.9 (6.9) | ||
| ART regimen class, N (%) | |||
| INSTI | 34 (49.3) | ||
| INSTI + PI/NNRTI | 15 (21.7) | ||
| NNRTI | 9 (13.0) | ||
| PI | 6 (8.7) | ||
| Other | 1 (5.8) | ||
| Unknown | 4 (1.5) | ||
| Adherence, N (%) | |||
| 100% | 45 (75.0) | ||
| 80–100% | 13 (21.7) | ||
| <80% | 2 (3.33) | ||
| Missing | 9 | ||
| Recent VL, N (%) | - | ||
| <50 | 54 (83.1) | ||
| 50–1000 | 8 (12.3) | ||
| ≥1000 | 3 (4.6) | ||
| Missing | 4 | ||
| Recent CD4, Mean (SD) | 706 (339) | ||
| Recent CD4 (cells/µL), N (%) | |||
| <200 | 4 (6.1) | ||
| ≥200 | 62 (93.9) | ||
| Missing | 3 | ||
| Nadir CD4 missing, N (%) | - | 24 | |
| Nadir CD4, mean (SD) | 180 (145) | ||
| AIDS, N (%) | |||
| Yes | 29 (44.6) | ||
| No | 36 (55.4) | ||
| Missing | 4 | ||
| DNA methylation-estimated cell type proportions | |||
| Proportion CD4 T cells, mean (SD) | 0.383 (0.09) | 0.231 (0.13) | <.001 |
| Proportion CD8 T cells, mean (SD) | 0.178 (0.05) | 0.347 (0.10) | <.001 |
| Proportion natural killer cells, mean (SD) | 0.108 (0.05) | 0.096 (0.01) | .32 |
| Proportion B cells, mean (SD) | 0.149 (0.06) | 0.137 (0.05) | .25 |
| Proportion monocytes, mean (SD) | 0.155 (0.07)) | 0.144 (0.07) | .44 |
| Proportion granulocytes, mean (SD) | 0.010 (0.02) | 0.006 (0.02) | .15 |
Abbreviations: ART, antiretroviral therapy; BMI, body mass index; BP, blood pressure; INSTI, integrase strand transfer inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitors; PI, protease inhibitor; SD, standard deviation; VL, viral load.
Frailty status and cognitive function measurements are shown in Table 2. The proportion of participants who met criteria for frailty was similar in PWH and control groups (36.2% vs 31.6%). The individual frailty criteria were also similar in the two groups, aside from exhaustion, which was higher in PWH than controls. In cognitive ability scores measured by the NIH Toolbox Cognition Battery, Fully Corrected T-scores were lower for the PWH group than controls for all measured domains but only significantly lower for working memory and language. Mean MoCA scores were lower for the PWH group than the controls (24.2 vs 26.6, P = .001), and a larger proportion of the PWH group was categorized as having an abnormal score (<26) compared to the controls (58% vs 21%, P < .001).
Table 2.
Frailty Status and Cognitive Function Measurements Among 69 Older African American Adults With Human Immunodeficiency Virus (HIV) and 38 HIV-Seronegative Adults (Controls)
| Controls (N = 38) | HIV (N = 69) | P | |
|---|---|---|---|
| Frailty | |||
| Frailty score, median (IQR) | 1.5 (0, 3) | 2.0 (1, 3) | .34 |
| Frailty status, N (%) | |||
| Frail | 12 (31.6) | 25 (36.2) | .63 |
| Non-frail | 26 (68.4) | 44 (63.8) | |
| Frailty criteria: weight loss, N (%) | 8 (21.1) | 15 (21.7) | .93 |
| Frailty criteria: exhaustion, N (%) | 12 (31.6) | 43 (62.3) | .002 |
| Frailty Criteria: low activity, N (%) | 14 (36.8) | 24 (34.8) | .83 |
| Frailty criteria: slowness, N (%) | 12 (31.6) | 27 (39.1) | .44 |
| Frailty criteria: weakness, N (%) | 16 (42.1) | 22 (31.9) | .29 |
| NIH Toolbox cognition battery scores | |||
| Executive function, mean (SD) | |||
| Age corrected standard score | 94.0 (12.9) | 87.0 (17.2) | .03 |
| Fully corrected T-score | 46.9 (11.4) | 44.5 (12.3) | .33 |
| Attention, mean (SD) | |||
| Age corrected standard score | 83.7 (10.8) | 79.1 (10.9) | .04 |
| Fully corrected T-score | 42.8 (5.9) | 40.6 (6.5) | .09 |
| Working memory, mean (SD) | |||
| Age corrected standard score | 95.1 (14.1) | 88.8 (12.9) | .024 |
| Fully corrected T-score | 48.5 (10.4) | 44.5 (9.3) | .048 |
| Processing speed, mean (SD) | |||
| Age corrected standard score | 88.6 (19.5) | 83.2 (19.2) | .18 |
| Fully corrected T-score | 47.4 (12.9) | 43.5 (12.6) | .14 |
| Language, mean (SD) | |||
| Age corrected standard score | 103.9 (17.9) | 89.6 (17.0) | <.001 |
| Fully corrected T-score | 57.0 (10.7) | 49.8 (10.5) | .001 |
| Montreal Cognitive Assessment (MoCA) score, mean (SD) | 26.6 (2.6) | 24.2 (4.0) | .001 |
| Normal (≥26) | 30 (79.0) | 29 (42.0) | <.001 |
| Not normal (<26) | 8 (21.0) | 40 (58.0) |
Abbreviations: IQR, interquartile range; SD, standard deviation.
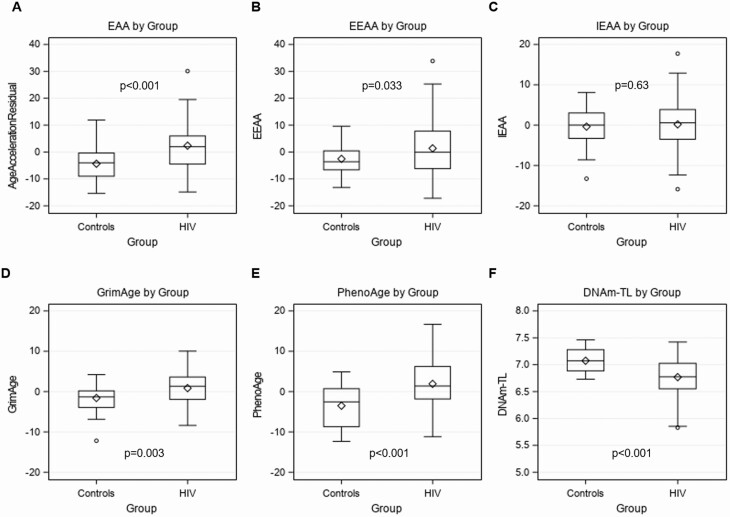
Chronological age and DNAm age were positively correlated (r = 0.36, P < .01) (Supplementary Figure 1). Correlations were similar when stratified by HIV status (controls: r = 0.52, P < .01; PWH: r = 0.41, P < .01). The PWH group had a higher EAA (2.39 ± 8.5 vs −4.34 ± 5.6, P < .001), EEAA (1.39 ± 10.4 vs −2.53 ± 5.6, P = .033), GrimAge (0.87 ± 4.3 vs −1.58 ± 3.3, P = .003), PhenoAge (1.93 ± 6.7 vs −3.50 ± 5.1, P < .001), and a lower DNAm-TL (6.77 ± 0.35 vs 7.07 ± 0.20, P < .001) compared to the controls (Figure 1A–F). IEAA was not significantly different between the PWH group and controls (−0.20 ± 6.1 vs −0.36 ± 5.0, P = .63).
Figure 1.
Box plots of (A) EAA, (B) EEAA, (C) IEAA, (D) GrimAge, (E) PhenoAge, and (F) DNAm-TL by HIV group. Abbreviations: DNAm-TL, DNAm-based telomere length; EAA, epigenetic age acceleration; EEAA, extrinsic epigenetic age acceleration; HIV, human immunodeficiency virus; IEAA, intrinsic epigenetic age acceleration.
In unadjusted linear regression models (Table 3), sex was significantly associated with GrimAge, with females having a lower GrimAge than males, but not associated with any other biomarkers. Having a high school education or less was associated with a higher EAA, GrimAge, and PhenoAge compared to having some college or additional education. Past/current tobacco use was associated with higher EAA, GrimAge, PhenoAge, and lower DNAm-TL Past/current illicit drug use was associated with a higher GrimAge. A 1 unit increase in BMI was associated with a decrease in EAA, GrimAge, and PhenoAge, and an increase in DNAm-TL. Ethnicity and alcohol use were not associated with any biomarkers.
Table 3.
Factors Associated With Methylation-Based Biomarkers of Aging in Linear Regression Models
| EAA | EEAA | IEAA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All | Est | 95% CI | P | Est | 95% CI | P | Est | 95% CI | P | |
| HIV status | HIV vs control (ref) | 6.74 | 3.70, 9.77 | <.001 | 3.92 | .33, 7.52 | .032 | 0.57 | -1.72, 2.86 | .63 |
| Sex | Female vs male (ref) | −2.14 | −5.28, 1.00 | .18 | −2.14 | −5.64, 1.36 | .23 | −0.63 | −2.82, 1.57 | .57 |
| Ethnicity | Hisp/Lat vs not (ref) | 0.37 | −6.02, 6.77 | .91 | 3.57 | −3.50, 10.6 | .32 | −1.50 | −5.93, 2.92 | .50 |
| Education | HS or less vs college+ (ref) | 4.44 | 1.34, 7.54 | .005 | 2.13 | −1.42, 5.68 | .24 | 1.95 | −0.25, 4.15 | .08 |
| Tobacco | Past/current vs never (ref) | 3.98 | .51, 7.45 | .02 | 2.41 | −1.50, 6.32 | .22 | 0.44 | −2.04, 2.92 | .73 |
| Alcohol | Past/current vs never (ref) | −2.35 | −7.77, 3.06 | .39 | −4.55 | −10.5, 1.43 | .13 | −2.49 | −6.23, 1.24 | .19 |
| Illicit drug | Past/current vs never (ref) | 0.04 | −3.61, 3.69 | .98 | 0.47 | −3.56, 4.49 | .82 | −0.71 | −3.28, 1.87 | .59 |
| BMI | −0.34 | −.58, −.10 | .006 | −0.25 | −.52, .03 | .076 | −0.10 | −.27, .07 | .24 | |
| HIV only | ||||||||||
| HIV RNA VL | ≥50 vs <50 (ref) | 2.84 | −2.78, 8.46 | .32 | 5.88 | −.84, 12.6 | .085 | −1.27 | −5.23, 2.69 | .53 |
| CD4 count | <200 vs >200 (ref) | 9.20 | .73, 17.7 | .0337 | 12.4 | 2.19, 22.6 | .018 | 0.18 | −5.97, 6.32 | .95 |
| CD4 count | −0.0003 | −.007, .006 | .92 | −0.004 | −.01, .003 | .24 | 0.005 | .001, .009 | .026 | |
| Duration of infection | 0.19 | −.11, .49 | .22 | 0.25 | −.13, .62 | .19 | −0.014 | −.233, .205 | .90 | |
| GrimAge | PhenoAge | DNAm-TL | ||||||||
| All | Est | 95% CI | P | Est | 95% CI | P | Est | 95% CI | P | |
| HIV status | HIV vs control (ref) | 2.45 | .86, 4.04 | .003 | 5.43 | 2.95, 7.91 | <.001 | −0.30 | −.43, −.18 | <.001 |
| Sex | Female vs male (ref) | −2.28 | −3.81, −.76 | .004 | −1.30 | −3.87, 1.27 | .32 | 0.09 | −.04, .22 | .19 |
| Ethnicity | Hisp/Lat vs not (ref) | −1.99 | −5.17, 1.20 | .22 | 0.90 | −4.31, 6.11 | .73 | 0.90 | −4.31, 6.11 | .73 |
| Education | HS or less vs college+ (ref) | 2.17 | .62, 3.73 | .007 | 3.63 | 1.10, 6.15 | .005 | −0.07 | −.20, .06 | .31 |
| Tobacco | Past/current vs never (ref) | 3.53 | 1.87, 5.19 | <.001 | 3.08 | .25, 5.91 | .03 | −0.16 | −.30, −.01 | .035 |
| Alcohol | Past/current vs never (ref) | 0.54 | −2.19, 3.26 | .70 | −3.75 | −8.12, .62 | .09 | 0.097 | −.13, .32 | .40 |
| Illicit drug | Past/current vs never (ref) | 2.06 | .27, 3.86 | .025 | 1.41 | −1.51, 4.33 | .34 | −0.022 | −.016, .12 | .76 |
| BMI | −0.12 | −.25, −.003 | .045 | −0.22 | −.42, −.02 | .028 | 0.012 | .002, .022 | .021 | |
| HIV only | ||||||||||
| HIV RNA VL | ≥50 vs <50 (ref) | 1.26 | −1.57, 4.09 | .38 | 2.26 | −2.20, 6.72 | .32 | −0.27 | −.50, −.04 | .024 |
| CD4 count | <200 vs >200 (ref) | 1.39 | −3.00, 5.78 | .53 | 5.89 | −.99, 12.8 | .09 | −0.43 | −0.78, −.08 | .018 |
| CD4 count | −0.003 | −.006, −.0005 | .023 | −0.004 | −.009, .001 | .13 | 0.0003 | .0001, .0006 | .007 | |
| Duration of infection | −0.05 | −.20, .10 | .49 | 0.06 | −.19, .30 | .63 | −0.004 | −.02, .008 | .51 | |
Abbreviations: BMI, body mass index; CI, confidence interval; EAA, epigenetic age acceleration; EEAA, extrinsic epigenetic age acceleration; HIV, human immunodeficiency virus; IEAA, intrinsic epigenetic age acceleration; VL, viral load.
In a multivariable model adjusted for education, tobacco use, and BMI, the PWH group had a significantly higher EAA than controls (β = 4.72, 95% confidence interval [CI]: 1.16, 8.28) and BMI was significantly associated with EAA (β = −0.26, 95% CI: −.49, −.03). HIV status was no longer significantly associated with EEAA (β = 3.44, 95% CI: −.19, 7.08), after adjusting for BMI. Similarly, HIV status was no longer associated with GrimAge, after adjusting for sex, ethnicity, education, tobacco use, illicit drug use, and BMI. The PWH group had a significantly higher PhenoAge (β = 3.29, 95% CI: .13, 6.44) compared to controls, after adjusting for education, tobacco use, alcohol use, and BMI. The PWH group had a significantly lower DNAm-TL compared to controls (β = −0.26, 95% CI: −.41, −.12), after adjusting for tobacco use and BMI.
DNAm-TL was significantly lower among PWH with a detectable HIV-RNA viral load ≥50 copies/mL compared to <50 copies/mL (β = −0.27, 95% CI: −.50, −.04), and in those with a CD4 count <200 cells/mm3 compared to >200 cells/mm3(β = −0.43, 95% CI: −.78, −.08). EAA and EEAA were also higher in older adults with HIV with a CD4 count <200 cells/mm3 compared to >200 cells/mm3 (EEA: β = 9.20, 95% CI: .73, 17.7; EEAA: β = 12.4, 95% CI: 2.19, 22.6). On a continuous scale, a higher CD4 count was associated with a lower IEAA (β = 0.005, 95% CI: .001, .009) and GrimAge (β = −0.003, 95% CI: −.006, −.0005). Duration of infection was not associated with any of the aging biomarkers.
We examined whether biomarkers were different by frailty status and those who had a normal vs abnormal MoCA score (Supplementary Table 2). Although those with frailty had a higher EAA, EEAA, IEAA, GrimAge, and PhenoAge than non-frails, none of the differences were significantly different. DNAm-TL was the same for those with and without frailty. Findings were similar when stratified by HIV status. Those with abnormal MoCA had a higher EAA (1.75 ± 9.1 vs −1.42 ± 7.1, P = .047) and PhenoAge (1.85 ± 6.9 vs −1.51 ± 6.1, P = .009), and a lower DNAm-TL (6.78 ± 0.4 vs 6.95 ± 0.3, P = .009) compared to those with a normal score. EEAA, IEAA, and GrimAge were higher in those with an abnormal score vs normal score but not significantly different. When stratified by HIV status, differences were no longer statistically significant.
Finally, we correlated each methylation-based biomarker of aging with each of the cognitive function test scores for each domain from the NIH Toolbox and the MoCA for all participants, and stratified by HIV status (Table 4). For all participants, there was a significant negative linear relationship between EAA and scores for executive function (r = −0.22, P = .02), working memory (r = −0.023, P = .02), and MoCA score (r = −0.21, P = .03). When stratified by HIV status, the negative correlations were primarily observed in PWH and significant for working memory (r = −0.26, P = .03). In a multivariable model EAA was negatively associated with executive function after adjusting for HIV status and education (P = .04). For EEAA, there was a significant negative linear relationship between EEAA and attention (r = −0.20, P = .04). This remained in the same direction but not statistically different after adjusting for HIV status and education (P = .08). For IEAA, there was a negative linear relationship between IEAA and executive function (r = −0.19, P = .053) and working memory (r = −0.22, P = .03). This was consistent after adjusting for HIV status and education. When stratified by HIV status, there was a negative linear relationship between IEAA and executive function, attention, and working memory for PWH. GrimAge was not correlated with any of the cognition scores. PhenoAge was significantly negatively corelated with executive function (r = −0.22, P = .02), attention (r = −0.27, P = .006), working memory (r = −0.26, P = .01), and MoCA score (r = −0.21, P = .03). PhenoAge remained significantly associated with executive function (P = .04) and attention (P = .04) after adjusting for HIV status and education. When stratified by HIV status, PhenoAge remained significantly negatively correlated with attention (r = −0.25, P = .04). DNAm-TL was significantly positively correlated with MoCA score for all participants (r = 0.21, P = .03). In a multivariable model adjusted for HIV status and education, this association was not significant.
Table 4.
Pearson Correlations Between Methylation-Based Biomarkers of Aging and Cognitive Function Domains Measured by the National Institutes of Health (NIH) Toolbox Cognition Battery Scores and Overall Score on the Montreal Cognitive Assessment (MoCA) for the Full Sample and Stratified by HIV Status
| NIH Toolbox Cognition Battery | |||||||
|---|---|---|---|---|---|---|---|
| Epigenetic Age | Group | Executive Function | Attention | Working Memory | Processing Speed | Language | MoCA Score |
| EAA | All | r = −0.22 | r = −0.19 | r = −0.23 | r = −0.07 | r = −0.18 | r = −0.21 |
| P = .02 | P = .05 | P = .02 | P = .48 | P = .07 | P = .03 | ||
| Controls | r = −0.19 | r = 0.13 | r = 0.01 | r = 0.13 | r = −0.03 | r = 0.01 | |
| P = .25 | P = .43 | P = .94 | P = .45 | P = .88 | P = .94 | ||
| HIV | r = −0.21 | r = −0.24 | r = −0.26 | r = −0.07 | r = −0.08 | r = −0.12 | |
| P = .09 | P = .06 | P = .03 | P = .59 | P = .54 | P = .32 | ||
| EEAA | All | r = −0.13 | r = −0.20 | r =−0.14 | r = −0.11 | r = −0.15 | r = −0.14 |
| P = .17 | P = .04 | P = .15 | P = .26 | P = .12 | P = .14 | ||
| Controls | r = −0.12 | r = −0.17 | r = −0.07 | r = −0.25 | r = 0.02 | r = 0.03 | |
| P = .47 | P = .30 | P = .68 | P = .13 | P = .91 | P = .85 | ||
| HIV | r = −0.12 | r = −0.18 | r = −0.12 | r = −0.03 | r = −0.13 | r = −0.10 | |
| P = .33 | P = .14 | P = .33 | P = .78 | P = .29 | P = .40 | ||
| IEAA | All | r = −0.19 | r = −0.13 | r = −0.22 | r = 0.07 | r = −0.03 | r = −0.11 |
| P = .053 | P = .21 | P = .03 | P = .51 | P = .74 | P = .27 | ||
| Controls | r = −0.14 | r = 0.20 | r = −0.04 | r = 0.21 | r = −0.12 | r = 0.03 | |
| P = .42 | P = .23 | P = .82 | P = .21 | P = .46 | P = .85 | ||
| HIV | r = −0.20 | r = −0.26 | r = −0.30 | r = 0.01 | r = 0.03 | r = −0.14 | |
| P = .09 | P = .03 | P = .01 | P = .93 | P = .80 | P = .27 | ||
| GrimAge | All | r = 0.14 | r = 0.002 | r = −0.003 | r = 0.03 | r =−0.08 | r = −0.06 |
| P = .15 | P = .99 | P = .97 | P = .79 | P = .40 | P = .52 | ||
| Controls | r = 0.14 | r = −0.17 | r = 0.09 | r = 0.02 | r = −0.03 | r = 0.02 | |
| P = .42 | P = .33 | P = .60 | P = .92 | P = .86 | P = .89 | ||
| HIV | r = 0.20 | r = 0.14 | r = 0.05 | r = 0.10 | r = 0.03 | r = 0.03 | |
| P = .11 | P = .25 | P = .70 | P = .43 | P = .80 | P = .82 | ||
| PhenoAge | All | r = −0.22 | r = −0.27 | r = −0.26 | r = −0.07 | r = −0.18 | r = −0.21 |
| P = .02 | P = .006 | P = .01 | P = .51 | P = .07 | P = .03 | ||
| Controls | r = −0.23 | r = −0.16 | r = −0.30 | r = 0.11 | r = −0.06 | r = 0.03 | |
| P = .18 | P = .35 | P = .07 | P = .53 | P = .74 | P = .85 | ||
| HIV | r = −0.20 | r = −0.25 | r = −0.15 | r = −0.06 | r = −0.07 | r = −0.12 | |
| P = .11 | P = .04 | P = .23 | P = .65 | P = .59 | P = .32 | ||
| DNAm-TL | All | r = 0.07 | r = 0.12 | r = 0.19 | r = 0.14 | r = 0.16 | r = 0.21 |
| P = .48 | P = .22 | P = .06 | P = .16 | P = .10 | P = .03 | ||
| Controls | r = 0.14 | r = 0.09 | r = 0.13 | r = 0.29 | r = 0.06 | r = 0.009 | |
| P = .41 | P = .59 | P = .46 | P = .09 | P = .75 | P = .96 | ||
| HIV | r = 0.002 | r = 0.05 | r = 0.11 | r = 0.02 | r = 0.02 | r = 0.11 | |
| P = .98 | P = .71 | P = .36 | P = .86 | P = .89 | P = .37 | ||
Abbreviations: DNAm-TL, DNAm-based telomere length; EAA, epigenetic age acceleration; EEAA, extrinsic epigenetic age acceleration; HIV, human immunodeficiency virus; IEAA, intrinsic epigenetic age acceleration.
DISCUSSION
Epigenetic age acceleration in blood was observed in AA PWH 60 years of age and older on ART using both first- and second-generation epigenetic clocks. The PWH group had a higher EAA, EEAA, GrimAge, and PhenoAge, and a lower DNAm-TL compared to the controls. IEAA was not significantly different between groups. After confounder adjustment, EAA and PhenoAge remained significantly higher and DNAm-TL remained significantly lower for PWH than controls.
Our findings in older AA PWH are largely consistent with other studies of both adults and youth PWH who have reported higher EAA and EEAA, using first-generation clocks [12–14]. Consistent with our study of AA young adults with and without HIV in New York City, EAA and EEAA were higher in this older population of adults with HIV compared to the controls, and IEAA, a measure of epigenetic age acceleration independent of blood cell composition, was not different between groups.
A recent longitudinal study measured EAA, EEAA, PhenoAge, and GrimAge in 15 PWH (mean age 45) pre-ART, 6–12 months post-ART, and 18–24 months post-ART and in controls, and found all 4 to be higher in pre-ART PWH compared to the controls [31]. By 18–24 months post-ART, PhenoAge and GrimAge were no longer significant between groups, although EAA and EEAA remained higher in PWH compared to controls, indicating that ART initiation may lead to a reduction in age acceleration. In contrast, although the older adults in our study were all on ART and mostly virally suppressed, we still found evidence of epigenetic age acceleration using second-generation epigenetic clocks, including PhenoAge, even after adjusting for confounders. PWH in our study were older (mean age 65) than in the aforementioned study [31]; therefore, they may have been exposed to HIV for a longer period prior to ART initiation and may also have more aging-related comorbidities that are not modifiable with ART. Duration of HIV did not appear to be associated with any of the aging biomarkers in our study. However, in a study of 378 ART-naive PWH and 34 controls without HIV, Yang et al found that duration of HIV was a risk factor for age acceleration [32].
DNAm-TL correlates negatively with age in different tissues and cell types and has been found to be associated with smoking history and other age-related conditions [11]. We found lower DNAm-TL in PWH compared to controls. To our knowledge, this is the first study to report on this epigenetic biomarker in a study of older adults with HIV. In the HIV literature, shorter telomere length has been reported in peripheral blood mononuclear cells (PBMC) isolated from ART-naive or ART-treated adults PWH compared with controls [33, 34], providing additional evidence for accelerated aging.
In our prior study of young AA PWH 20–35 years, we found a signal that IEAA may be correlated with working memory and hypothesized there may be potential associations between epigenetic aging and cognition not mediated by immune system aging [23]. Further evidence was found in the present study of older PWH ≥60 years with associations between IEAA and executive function, attention, and working memory. These findings align with longitudinal data from a study of twins, which found that those who were biologically older by IEAA were more likely to show a 2.5–3% decline in executive function and memory function. We did not, however, find associations between EEAA and cognitive measures among PWH. This was unexpected given prior findings of associations between EEAA and cognitive performance among adults without HIV [19, 20] and youth with HIV [15]. Differences in the age range of the participants in this cross-sectional sample may play a role in the inconsistent results. Of note, we did find associations between PhenoAge and executive function and attention.
There is some evidence in the general population that frail individuals may have epigenetic age acceleration [17]. Although all markers of epigenetic age acceleration were higher in frail compared to non-frail patients, there were no statistically significant differences between groups. This remained the same when stratified by HIV status. This was in contrast to a small study of 20 adults with HIV, which reported higher epigenetic age in frail compared to non-frail participants [24, 35]. This could be due to sample size or the use of the Weidner epigenetic model, rather than the 6 methylation-based biomarkers of aging in our study.
Our study had several limitations. Given the cross-sectional nature of our study, we were unable to fully explore how cognitive trajectories or unmeasured health behaviors may be related to epigenetic age acceleration. We did not have measures of mood disorders (eg, depression or anxiety), which are highly prevalent among PWH and may affect cognitive functioning. Given race differences in extrinsic and intrinsic measures of epigenetic aging, our findings may not be generalizable to other racial groups [36, 37]. Many of our participants had been treated with older antiretroviral agents and we excluded participants with history of hepatitis B or C infection, which may limit generalizability of our findings to PWH with more recent diagnoses and ART regimens and coinfections with hepatitis B or C.
In conclusion, our study demonstrates that epigenetic age acceleration is detectable by DNAm-based biomarkers of aging in AA PWH on ART. Associations between IEAA and lower cognitive function suggest that the impact of HIV on immune cells may not fully account for the cognitive impairment observed in PWH. Additional studies are warranted to examine how these biomarkers of aging change over time in relation to changes in cognition over time.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute on Aging (grant number R21 AG056175 to M. T. Y. and S. M. A.) and the National Institute of Mental Health (grant number R25 MH108389 to S. S.).
Potential conflicts of interest. S. M. A. reports grants/support from the National Institutes of Health (NIH) (5R21AG056175 Biological aging in older human immunodeficiency virus [HIV]-infected African Americans) during the conduct of the study/outside the submitted work. A. M. B. reports Technologies for white matter hyperintensity quantification (9867566) patent. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
REFERENCES
- 1. Antiretroviral Therapy Cohort Collaboration. Survival of HIV-positive patients starting antiretroviral therapy between 1996 and 2013: a collaborative analysis of cohort studies. Lancet HIV 2017; 4:e349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shiau S, Bender AA, O’Halloran JA, et al. . The current state of HIV and aging: findings presented at the 10th International Workshop on HIV and Aging. AIDS Res Hum Retroviruses 2020; 36:973–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gabuzda D, Jamieson BD, Collman RG, et al. . Pathogenesis of aging and age-related comorbidities in people with HIV: highlights from the HIV ACTION Workshop. Pathog Immun 2020; 5:143–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nguyen A, Rinaldi S, Martinez C, Perkins M, Holstad MM. HIV and aging in special populations: from the mitochondria to the metropolis—Proceedings From the 2019 Conference. J Assoc Nurses AIDS Care 2021;. 32:214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jylhävä J, Pedersen NL, Hägg S. Biological age predictors. EBioMedicine 2017; 21:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013; 14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hannum G, Guinney J, Zhao L, et al. . Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013; 49:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 2018; 19:371–84. [DOI] [PubMed] [Google Scholar]
- 9. Lu AT, Quach A, Wilson JG, et al. . DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019; 11:303–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levine ME, Lu AT, Quach A, et al. . An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018; 10:573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu AT, Seeboth A, Tsai PC, et al. . DNA methylation-based estimator of telomere length. Aging (Albany NY) 2019; 11:5895–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gross AM, Jaeger PA, Kreisberg JF, et al. . Methylome-wide analysis of chronic HIV infection reveals five-year increase in biological age and epigenetic targeting of HLA. Mol Cell 2016; 62:157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nelson KN, Hui Q, Rimland D, et al. . Identification of HIV infection-related DNA methylation sites and advanced epigenetic aging in HIV-positive, treatment-naive U.S. veterans. AIDS 2017; 31:571–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horvath S, Levine AJ. HIV-1 infection accelerates age according to the epigenetic clock. J Infect Dis 2015; 212:1563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Horvath S, Stein DJ, Phillips N, et al. . Perinatally acquired HIV infection accelerates epigenetic aging in South African adolescents. AIDS 2018; 32:1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 2016; 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gale CR, Marioni RE, Harris SE, Starr JM, Deary IJ. DNA methylation and the epigenetic clock in relation to physical frailty in older people: the Lothian Birth Cohort 1936. Clin Epigenetics 2018; 10:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bressler J, Marioni RE, Walker RM, et al. . Epigenetic age acceleration and cognitive function in African American adults in midlife: the atherosclerosis risk in communities study. J Gerontol A Biol Sci Med Sci 2020; 75:473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marioni RE, Shah S, McRae AF, et al. . The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 2015; 44:1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beydoun MA, Shaked D, Tajuddin SM, Weiss J, Evans MK, Zonderman AB. Accelerated epigenetic age and cognitive decline among urban-dwelling adults. Neurology 2020; 94:e613–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaccarino V, Huang M, Wang Z, et al. . Epigenetic age acceleration and cognitive decline: a twin study. J Gerontol A Biol Sci Med Sci 2021; glab047. doi: 10.1093/gerona/glab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levine AJ, Quach A, Moore DJ, et al. . Accelerated epigenetic aging in brain is associated with pre-mortem HIV-associated neurocognitive disorders. J Neurovirol 2016; 22:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiau S, Cantos A, Ramon CV, et al. . Epigenetic age in young African American adults with perinatally-acquired HIV. J Acquir Immune Defic Syndr 2021; 87:1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sánchez-Conde M, Rodriguez-Centeno J, Dronda F, et al. . Frailty phenotype: a clinical marker of age acceleration in the older HIV-infected population. Epigenomics 2019; 11:501–9. [DOI] [PubMed] [Google Scholar]
- 25. Gershon RC, Wagster MV, Hendrie HC, Fox NA, Cook KF, Nowinski CJ. NIH toolbox for assessment of neurological and behavioral function. Neurology 2013; 80:S2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nasreddine ZS, Phillips NA, Bédirian V, et al. . The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc 2005; 53:695–9. [DOI] [PubMed] [Google Scholar]
- 27. Fried LP, Tangen CM, Walston J, et al. ; Cardiovascular Health Study Collaborative Research Group . Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 2001; 56:M146–56. [DOI] [PubMed] [Google Scholar]
- 28. Houseman EA, Accomando WP, Koestler DC, et al. . DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012; 13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen BH, Marioni RE, Colicino E, et al. . DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016; 8:1844–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klemera P, Doubal S. A new approach to the concept and computation of biological age. Mech Ageing Dev 2006; 127:240–8. [DOI] [PubMed] [Google Scholar]
- 31. Sehl ME, Rickabaugh TM, Shih R, et al. . The effects of anti-retroviral therapy on epigenetic age acceleration observed in HIV-1-infected adults. Pathog Immun 2020; 5:291–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang CX, Schon E, Obeidat M, et al. . Accelerated epigenetic aging and methylation disruptions occur in human immunodeficiency virus infection prior to antiretroviral therapy. J Infect Dis 2021; 223:1681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lagathu C, Cossarizza A, Béréziat V, Nasi M, Capeau J, Pinti M. Basic science and pathogenesis of ageing with HIV: potential mechanisms and biomarkers. AIDS 2017; 31 Suppl 2:105–19. [DOI] [PubMed] [Google Scholar]
- 34. Pathai S, Lawn SD, Gilbert CE, et al. . Accelerated biological ageing in HIV-infected individuals in South Africa: a case-control study. AIDS 2013; 27:2375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weidner CI, Lin Q, Koch CM, et al. . Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol 2014; 15:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Horvath S, Gurven M, Levine ME, et al. . An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 2016; 17:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tajuddin SM, Hernandez DG, Chen BH, et al. . Novel age-associated DNA methylation changes and epigenetic age acceleration in middle-aged African Americans and whites. Clin Epigenetics 2019; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.