Abstract
Background
A new coronavirus disease named COVID-19, caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is rapidly spreading worldwide. However, there is currently no effective drug to fight COVID-19.
Methods
In this study, we developed a Virus-Drug Association (VDA) identification framework (VDA-RWLRLS) combining unbalanced bi-Random Walk, Laplacian Regularized Least Squares, molecular docking, and molecular dynamics simulation to find clues for the treatment of COVID-19. First, virus similarity and drug similarity are computed based on genomic sequences, chemical structures, and Gaussian association profiles. Second, an unbalanced bi-random walk is implemented on the virus network and the drug network, respectively. Third, the results of the random walks are taken as the input of Laplacian regularized least squares to compute the association score for each virus-drug pair. Fourth, the final associations are characterized by integrating the predictions from the virus network and the drug network. Finally, molecular docking and molecular dynamics simulation are implemented to measure the potential of screened anti-COVID-19 drugs and further validate the predicted results.
Results
In comparison with six state-of-the-art association prediction models (NGRHMDA, SMiR-NBI, LRLSHMDA, VDA-KATZ, VDA-RWR, and VDA-BiRW), VDA-RWLRLS demonstrates superior VDA prediction performance. It obtains the best AUCs of 0.885 8, 0.835 5, and 0.862 5 on the three VDA datasets. Molecular docking and dynamics simulations demonstrated that remdesivir and ribavirin may be potential anti-COVID-19 drugs.
Conclusions
Integrating unbalanced bi-random walks, Laplacian regularized least squares, molecular docking, and molecular dynamics simulation, this work initially screened a few anti-SARS-CoV-2 drugs and may contribute to preventing COVID-19 transmission.
Keywords: SARS-CoV-2, Antiviral drug, Virus-drug association, Unbalanced bi-random walk, Laplacian regularized least squares, Molecular docking, Molecular dynamics simulation
1. Introduction
1.1. Background
In December 2019, a new coronavirus disease named COVID-19 by the World Health Organization broke out in Wuhan, China. COVID-19 is caused by an unknown coronavirus called severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). As of May 13, 2021, COVID-19 had a cumulative total of 159,949,065 confirmed cases with over 3,322,439 deaths [1]. With the continuing threat of SARS-CoV-2 to global health, it is urgent to develop effective prevention and treatment strategies for SARS-CoV-2 transmission [[2], [3], [4], [5], [6], [7]].
Although pharmaceutical companies have invested enormous money and time in drug research and development over the past two decades, the number of new chemical entity drugs approved by the FDA is still relatively limited every year. Drug repositioning, applied to reposition an existing active pharmaceutical ingredient for a new indication, helps to reduce the attrition experienced in the process of new drug discovery [8,9]. Therefore, drug repositioning has been a research hotspot in drug discovery. It is unrealistic to find therapeutic options for COVID-19 patients by experimental methods under such an urgent situation. Subsequently, systematic identification of anti-SARS-CoV-2 drugs by repositioning FDA-approved drugs offers an effective way to find possible pharmaceutical ingredients for COVID-19 [10,11].
However, due to a lack of knowledge of complete drug-target interactions, designing promising and affordable methods for the effective treatment of COVID-19 is challenging. SARS-COV-2 is a new single-stranded RNA virus [12] and has high homology with severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) [13,14]. In addition, the spike (S) protein and host angiotensin-converting enzyme 2 (ACE2) have been recognized as two target proteins of COVID-19 in clinical trials [15]. The above knowledge provides clues for repositioning available drugs for COVID-19 [[15], [16], [17]].
1.2. Related work
1.2.1. Initial screening of potential antifungal drugs for COVID-19
Potential drugs against COVID-19 can be roughly divided into three main categories: antifungal drugs, antibacterial drugs, and antiviral drugs [18]. Roudbary et al. [19] and salehi et al. [20] observed that patients with COVID-19 infection are still extremely susceptible to fungal or bacterial infections. Chen et al. [21] found fungals related to co-infections in COVID-19, such as Aspergillus spp., Candida albicans, Candida dubliniensis, Candida glabrata, Candida krusei, Candida tropicalis, and Candida parapsilosis sensu stricto. Roudbary et al. [19] reported that fungals including aspergillosis, candidemia, Histoplasma spp., Rhizopus spp., Mucor spp., Cryptococcus spp. have been increasingly associated with COVID-19 co-infections caused by opportunistic fungal diseases. Therefore, new antifungal drug screening is currently urgently needed among COVID-19 patients [22,23]. Song et al. [24] provided a clinical diagram to help clinicians and laboratory experts manage aspergillosis, candidiasis, cryptococcosis, and mucormycosis in COVID-19 patients. Mohamed et al. [25] revealed that itraconazole, an antifungal drug, is one of the best-documented multi-target drugs.
1.2.2. Initial screening of potential antibacterial drugs for COVID-19
A bacterium plays an important role in various environments [26]. In COVID-19, bacterial co-pathogens are observed to be co-infectious and drastically increase morbidity and mortality rates [27]. Rawson et al. [28] reported that a few COVID-19 patients have been co-infected by bacteria, such as Acinetobacter baumannii, Escherichia coli, Haemophilus influenzae, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Biosurfactants generated by bacterial species have wide antimicrobial activity, biocompatibility, biodegradability, low toxicity and better interfacial activity than traditionally produced chemical surfactants [26,29]. Hassan Mahmoudi [30] obtained blood cultures and endotracheal aspirates from COVID-19 patients based on a cross-sectional study and confirmed the bacterial isolates. They identified bacteria isolated through blood culture and endotracheal aspirate, for example, Enterobacter species 5, Escherichia coli 7, Klebsiella species 11, methicillin-resistant Staphylococcus aureus 6, methicillin-sensitive Staphylococcus aureus 9, Pseudomonas aeruginosa 4, and Streptococcus pneumoniae 1. Li et al. [31] revealed that secondary bacterial infection is one of the main complications in COVID-19 patients with high mortality. They found that Gram-negative bacteria (especially A. baumannii and K. pneumoniae) are the main bacteria. Thus, precise application of antibacterial drugs constitutes a promising alternative to COVID-19 patients with secondary bacterial infection [29,31,32]. Several works are dedicated to finding antimicrobial compounds produced by bacterial species to prevent the continuing transmission of COVID-19. Narendrakumar et al. [33] presented that doxycycline can inhibit bacterial protein synthesis and shows antibacterial activity in COVID-19 [33]. Pan et al. [34] reported that Shuanghuanglian, an antiviral and antibacterial compound, has been applied to COVID-19 treatment in China.
1.2.3. Initial screening of potential antiviral drugs for COVID-19
To accelerate drug discovery of COVID-19, multiple computational models were presented to assist in finding antiviral drugs by drug repositioning. Currently, computational drug-repurposing methods for initially screening anti-COVID-19 drugs can be roughly classified into structure-based approaches, machine learning-based approaches, and network-based approaches. Structure-based approaches [[35], [36], [37], [38]] aim to understand the binding mechanism of compounds with the targets of COVID-19 mainly by molecular docking, Molecular Dynamic Simulations (MDS) and binding energy computation.
Machine learning-based methods mainly use machine learning models, especially deep learning models, to fight COVID-19 effects. BenevolentAI [39] extracted numerous structured medical information from recent reports by machine learning and predicted that baricitinib may be therapeutic strategy for COVID-19. Ge et al. [5] developed a data-driven drug repurposing framework integrating machine learning models and statistical analysis approaches to find possible drugs against SARS-CoV-2. Ray et al. [40] used a variational graph autoencoder to fight COVID-19. Ke et al. [41] developed a deep learning method to search available compounds effective against coronaviruses. Beck et al. [42] proposed a previously trained deep learning model called molecule transformer-drug-target interactions to find commercially available antiviral drugs against COVID-19. Although machine learning researchers have been very active in their efforts to prevent COVID-19 infections, few studies have concerned drug repositioning. More importantly, only few studies have been accepted for publication or are available online in a journal although their applications against COVID-19 have become publicly available [43].
Network-based methods computed biological similarity between drugs, viruses, targets and proposed network propagation algorithms to find therapeutic strategies for COVID-19. Gysi et al. [44] developed a graph neural network-based method and identified 81 potential repositioning candidates associated with SARS-CoV-2. Peng et al. [45] constructed a single-stranded RNA Virus-Drug Association (VDA) dataset and developed a bipartite local model to find potential treatments for COVID-19. Zhou et al. [46] designed a KATZ measurement for predicting therapeutic agents of COVID-19. Following by the above methods, Peng et al. [47] constructed three VDA datasets and developed a random walk with restart to discover potential antiviral drugs. The above methods effectively combined network-based drug repositioning, molecular docking, and document retrieval and are state-of-the-art virus-drug association prediction methods.
1.2.4. Molecular docking and dynamics simulation
To design novel inhibitors for various diseases, computational methods such molecular docking and MDS have been broadly carried out [48,49]. Molecular docking is a powerful technique widely applied to model the interactions between ligands and target proteins at the atomic level [50]. We can characterize the behavior of ligands binding to target proteins and elucidate fundamental biochemical processes through molecular docking. Molecular docking contains two basic steps: prediction of the ligand conformation and its position within binding sites and computation of the ligand-protein binding affinity. A higher binding energy shows more stable ligand-protein binding capability. Similarly, the binding affinity between chemical agents and the S protein/human ACE2 can be computed by molecular docking [51]. For example, Thillainayagam et al. [52] revealed the anti-malarial feature of quinolinyl chalcone derivatives by molecular docking, three-dimensional QSAR, and comparative molecular field analysis.
MDS has been used to confirm ligand-protein stable interactions [53]. For example, by molecular docking and MDS, Thillainayagam et al. [48] studied the inhibitory action of epoxyazadiradione on malaria. Ragunathan [54] et al. reported that FtsA is a cidal target of Staphylococcus aureus. Ragunathan et al. [55] found that quercetin may be a stable inhibitor of Murb. More importantly, Basu et al. [53] identified a new cyclohexanone compound, ZINC07333416, that has crucial associations with most active-site residues of the SARS-CoV-2 main protease by molecular docking and confirmed these associations through MDS.
1.3. Study contributions
In this study, we developed a VDA prediction model (VDA-RWLRLS) based on biological information of viruses and drugs, unbalanced bi-Random Walks, Laplacian Regularized Least Squares (LRLS), molecular docking, and MDS. This study has four main contributions:
-
●
The unbalanced bi-random walk algorithm is designed to globally exploit the weighted circular bigraphs in the virus network and the drug network.
-
●
LRLS is presented to iteratively score each virus-drug pair.
-
●
Viral sequences, drug chemical structures, and VDA information are utilized to integrate the biological features of the viruses and drugs.
-
●
Molecular docking and MDS are applied to validate the inferred anti-SARS-CoV-2 drugs.
2. Materials and methods
2.1. Experimental data
2.1.1 drug. Virus- association
In this study, we use three VDA datasets provided by Peng et al. [47]. Dataset 1 was provided by Zhou et al. [46] and contains 96 confirmed VDAs between 12 human RNA viruses, including SARS-CoV-2, and 78 small molecules. Dataset 2 was obtained by manually retrieving recent documents based on the text mining technique and contains 770 confirmed VDAs between 69 viruses and 128 small molecules. Dataset 3 was constructed by preprocessing VDA data from the Drug Virus.info database [56] and consists of 407 known VDAs from 34 viruses and 203 chemical agents. The details are shown in Table 1 .
Table 1.
The description of virus-drug association datasets.
| Dataset | Virus | Drugs | VDAs |
|---|---|---|---|
| Dataset 1 | 12 | 78 | 96 |
| Dataset 2 | 69 | 128 | 770 |
| Dataset 3 | 34 | 203 | 407 |
A VDA matrix is represented as X (n×m) (n and m are the number of viruses and drugs, respectively), where x ij = 1 if the i-th virus v i associates with the j-th drug d j and x ij = 0 otherwise. The adjacency matrix of a VDA network is represented as Y (n×m) with the element y ij by Eq. (1):
| (1) |
2.1.2. Drug chemical structure similarity
Based on the assumption that two drugs sharing more chemical substructures are more similar [57], drug chemical structure similarity can be calculated. Chemical substructures were first downloaded from the DrugBank database [58]. An open-source cheminformatics software, RDKit [59], is then applied to compute extended connectivity fingerprints [60] of drugs based on their chemical substructures. Finally the drug chemical structure similarity matrix S d was calculated based on the extended connectivity fingerprints.
2.1.3. Virus sequence similarity
The complete genomic sequences of viruses were obtained from the NCBI database [61], and the virus sequence similarity matrix S v was computed based on MAFFT [62], a multiple sequence alignment software.
2.2. Methods
2.2.1. Problem description
A heterogeneous virus-drug network is comprised of a virus similarity network, a drug similarity network and VDA data. Let V (n×n), D (m×m) and Y (n×m) denote the affinity matrices of the virus similarity network, drug similarity network, and VDA network, respectively. The virus similarity network is constructed by using viruses as vertices, virus-virus similarities as edges, and similarity between two viruses as the weight of their linked edge. Similarly, a drug similarity network is built. The VDA network is the constructed VDA matrix. Our objective is to find missing VDAs on the heterogeneous network by reconstructing an association matrix P (n×m).
2.2.2. Overview of VDA-RWLRLS
In this study, we developed a VDA prediction framework, VDA-RWLRLS, based on various biological information, unbalanced bi-random walk, LRLS, molecular docking, and MDS. Fig. 1 shows the flowchart of VDA-RWLRLS. As shown in Fig. 1, the VDA-RWLRLS framework contains six main steps. (1) Similarity computation. Viral similarities are fused based on genomic sequences and Gaussian association profiles. Drug similarities are integrated based on chemical structures and Gaussian association profiles. (2) Label propagation. An unbalanced bi-random walk is designed to propagate association labels based on the left random walk on the virus network and the right random walk on the drug network. (3) Virus-drug association probability computation. LRLS is applied to compute association probabilities for unknown virus-drug pairs based on the results obtained from unbalanced bi-random walks. The prediction scores from the virus network and the drug network are averaged as the results at the current iteration. (4) Iteratively updating. Steps (2) and (3) are iteratively updated t times and the final predicted association scores (P t) are computed. (5) Molecular docking. Molecular docking was employed to measure the binding abilities between the predicted top anti-SARS-CoV-2 drugs and the crystal structure of the S protein-binding domain bound to human ACE2. (6) MDS. MDS was used to evaluate the interaction-dynamics of the predicted possible anti-COVID-19 drug-target proteins of the COVID-19 complex.
Fig. 1.
The flowchart of VDA prediction based on an unbalanced bi-random walk, LRLS, molecular docking, and MDS (VDA-RWLRLS).
2.2.3. Gaussian association profile kernel similarity
Considering that any two viruses associated with more common drugs could tend to share higher similarity, we utilize Gaussian Association Profile Kernels (GAPK) [63] to compute virus and drug GAPK similarity matrices. The association profile of a virus v i, denoted as IP(v i) (the i-th row of Y), describes the relationship between virus v i and all observed drugs. For two viruses v i and v j, their GAPK similarity K v is computed by Eq. (2).
| (2) |
where is the bandwidth parameter. Similarly, drug GAPK similarity can be defined by Eq. (3).
| (3) |
where is the bandwidth parameter. IP(d j) is the j-th column of Y and represents the correlation between drug d j and all observed viruses. Sequence similarity and GAPK similarity are integrated to measure virus similarity by Eq. (4).
| (4) |
Similarly, the fused drug similarity can be calculated by Eq. (5).
| (5) |
2.2.4. Balanced random walk
Suppose that a circular bigraph is represented as a subgraph comprised of a virus path and a drug path with two ends connected by two VDAs and . The circular bigraph denotes a vicinity relationship between two VDAs and by generalizing the similarity between v 1 and v n, and one between d 1 and d m. The smallest circular bigraph can be represented based on the “guilt-by-association” principle:
-
●
The triangle with one virus and two drugs follows the assumption that similar drugs may be applied to the same disease.
-
●
The rectangle with two viruses and two drugs follows the assumption that virus v i tends to associate with drug d m when virus v j associates with drug d n, v i is similar to v j and d m is similar to d n.
-
●
The triangle with two viruses and one drug follows the assumption that diseases caused by similar viruses may be treated by the same drug.
A Balanced Bi-Random Walk method (VDA-BiRW) is designed to identify potential VDAs by investigating circular bigraphs with high frequency in a VDA network. By multiplying V on the left and D on the right, VDA-BiRW iteratively extends the virus path and the drug path to identify the circular bigraphs by Eq. (6).
| (6) |
In Eq. (6), both Y and P are normalized to make the sum of all elements 1. Initially, P 0 is equal to Y. The known VDA matrix Y is integrated to balance the bi-random walk and the prior knowledge. Parameter has two functions: (I) Decreasing the importance of a circular bigraph when the path is getting longer. (II) Balancing potential VDAs and known VDAs. Successive matrix multiplication is used to implement mimic jumps on the virus network, the drug network, and the VDA network. In the first walking, the -th element in the matrix denotes the number of circular bigraphs obtained by linking the i-th target virus with the path length of 1 to the j-th candidate drug with the path length of 1. When α = 1, after t steps of successive multiplication , we can find circular bigraphs with a path length of t.
2.2.5. Unbalanced Bi-random walk
Theoretically, the above balanced random walk model can converge to a unique solution. However, circular bigraphs with small path lengths are more informative for VDA identification. In particular, circular bigraphs with long path lengths could result in false positives. More importantly, the virus similarity network and the drug similarity network contain different topological structures, and thus the optimal walking step size could be different in the process of random walk on the two networks. Therefore, we develop an unbalanced bi-random walk-based VDA prediction method by introducing two parameters as the maximal iteration numbers on the left and right random walks on the two networks, respectively:
| (7) |
| (8) |
where Eqs. (7), (8) denote the left random walk with the step size of l on the virus network, and the right random walk with the step size of r on the drug network, respectively. At each step, virus similarity is integrated to the left random walk by multiplying V on the left of Eq. (7). Similarly, drug similarity is integrated by Eq. (8).
2.2.6. Laplacian Regularized Least Squares
In the unbalanced bi-random walk algorithm, the jump condition depends on the computed similarity matrices and known VDAs. For a given node in a VDA network, if there exist two nodes that have the same similarity with the given node, the two nodes will equally contribute to the direction of the jump. However, the node with relatively smaller similarities with the other nodes may have more contribution. Therefore, LRLS [64] is introduced to solve the above problem. First, the virus Laplacian matrix L V and drug Laplacian matrix L D are normalized to measure the jump for each node by Eqs. (9), (10).
| (9) |
| (10) |
where M V and M D are the diagonal matrices of viruses and drugs whose elements M V(i, i) and M D(j, j) represent the summation of the i-th row of V and the j-th row of D, respectively.
Second, the cost function in the virus space and the drug space can be defined to describe the minimum optimization problem based on Laplacian matrices L V and L D by Eqs. (11) and (12), respectively:
| (11) |
| (12) |
where ‖ ⋅‖F represents the Frobenius norm, (⋅)T denotes the transpose, and η v and η d are trade-off parameters. The above two models (11) and (12) can be solved by Eqs. (13), (14)), respectively:
| (13) |
| (14) |
To comprehensively consider the effect of unbalanced random walk on the prediction performance, we replace Y with association scores obtained from unbalanced random walks. Suppose that Eqs. (15) and (16) be defined as follows:
| (15) |
| (16) |
At the t-th walking, the corresponding models integrating unbalanced random walk and LRLS can be defined by Eqs. (17), (18), respectively.
| (17) |
| (18) |
Finally, VDA-RWLRLS computes the association probability for each virus-drug pair by integrating the interaction scores from the virus network and the drug network by Eq. (19):
| (19) |
2.2.7. Molecular docking and dynamics simulation
Molecular docking has been widely applied to measure the intermolecular binding affinity between one ligand-receptor pair. We implemented molecular docking to evaluate the binding abilities between the predicted anti-SARS-CoV-2 drugs and the S protein/human ACE2. Drug chemical structures can be obtained from the DrugBank database [58] and are represented based on the PDB format files. AutoDockTool is used to convert the PDB files into the pdbqt format files required by AutoDock 4. The structure of the S protein-binding domain bound to human ACE2 (PDB ID: 6M0J) was downloaded from the RCSB Protein Data Bank [65]. The predicted anti-SARS-CoV-2 compounds are used as ligands, and the binding domain of the S protein bound to human ACE2 is used as a receptor to implement molecular docking.
We used PyMOL [66] to remove solvent and organic compounds. Receptor atoms are set as the AD4 type and Gasteiger charges are computed before docking. Binding pockets are defined using AutoGrid 4. The grid size is set as 126 × 126 × 126 with a spacing of , and the grid center is placed at the area of the S protein-binding domain bounding with ACE2. AutoDock [67] is finally applied to molecular docking where the Lamarckian genetic approach with default parameters [68] is used as the search algorithm.
Basu et al. [53] employed MDS and evaluated the interaction dynamics of one ligand-protein complex for COVID-19. Inspired by MDS proposed by Basu et al. [53], we carried out MDS to analyze our predicted anti-SARS-CoV-2 drugs after molecular docking. We employ GROMACS 2021-2 for MDS with a Charmm36 force field [69]. First, we obtained topological files of ligands through CGENFF [63]. Second, we generated topological files of receptors by ignoring H atoms and using the tip 3-point for the water model based on gromacs. Third, the ligand-protein complex is placed in a dodecahedral box filled with simple point charge water, distance between the solvent and the box is set to 1 nm, and ions including Na or Cl are added to the solvation system to balance the charge. Fourth, the energy is minimized to the initial structure before simulation by the steepest descent minimization method. In the process, the iteration stop condition is set as the maximum force with less than 1000 kJ/mol nm−1 and the maximum number of iterations is set as 50 000. Five, the solvents and ions around the complex are balanced. The process is divided into two stages: NVT (constant number of particles, constant volume, and constant temperature) and NPT (constant number of particles, constant pressure P, and constant temperature). In NVT stage, we set the target temperature to 300 K and perform 100 ps simulation where unit time is set to 2 fs and the maximum number of iterations is set to 50 000. In the NPT stage, we use the same 100 ps balance, and the reference pressure is set to 1 bar. After balancing the density of the system, we conduct an MDS of 50 ns.
3. Results
3.1. Experimental setting
We perform 5-fold Cross Validations (CVs) to investigate the performance of our proposed VDA-RWLRLS method. 80% of the VDAs of Y were randomly extracted as the training set, and the remaining VDAs as testing set. Sensitivity, specificity, accuracy, F1 score, and AUC were used as evaluation metrics. Sensitivity and specificity denote the ability of a VDA identification model to correctly predict positive VDAs and negative VDAs, respectively. Accuracy denotes the ratio of the correctly predicted positive and negative VDAs to all known positive VDAs and screened negative VDAs. The F1 score is a harmonic mean between precision and recall. Precision is the ratio of the correctly predicted positive VDAs to all predicted positive VDAs. Recall is the same for sensitivity. Let TP, FP, TN, and FN indicate true positive, false positive, true negative, and false negative, respectively. The definitions of sensitivity, specificity, accuracy, and F1 score are by Eqs. (20), (21), (22), (23), (24), (25):
| (20) |
| (21) |
| (22) |
| (23) |
where
| (24) |
| (25) |
AUC is widely applied to evaluate the performance of a binary classifier. It denotes the area under the Receiver Operating Characteristic (ROC) curve [70,71]. The ROC curve is a plot that characterizes the trade-off between sensitivity and (1-specificity) or between the true positive rate and false positive rate across a few cut-off points. Higher sensitivity, specificity, accuracy, F1 score and AUC indicate better performance for VDA prediction models. In addition, the AUC is a more important measurement than the other four evaluation metrics. The experiments are repeated 20 times, and the final performance is achieved by averaging the results from 20 experiments.
3.2. Parameter settings
There are seven parameters used in VDA-RWLRLS: α, l, r, λ v, λ d, and γ d/γ v. The parameter α is used to control the importance between the predicted VDAs and known VDAs during random walks. l and r control the iteration steps on the left and right random walks, respectively. λ v and λ d measure the importance of Gaussian association profile similarity and virus sequence similarity/drug chemical structure similarity. γ d and γ v are bandwidth parameters. η v and η d are set as 0.01. We use grid search to obtain the optimal parameter combination. Table 2 shows the optimal parameter settings where VDA-RWLRLS computes the best performance.
Table 2.
The parameter settings of VDA-RWLRLS.
| Dataset | α | l | r | λv | λd | γd/γv |
|---|---|---|---|---|---|---|
| dataset1 | 0.3 | 11 | 11 | 0.1 | 0.1 | 2.5 |
| dataset2 | 0.001 | 31 | 1 | 0.5 | 0.5 | 2.5 |
| dataset3 | 0.001 | 11 | 1 | 0.1 | 0.1 | 2.5 |
3.3. Comparison with six state-of-the-art methods
The proposed VDA-RWLRLS method is compared with six VDA prediction methods (NGRHMDA [72], SMiR-NBI [73], LRLSHMDA [64], VDA-KATZ [46], VDA-RWR [47], and VDA-BiRW) on three VDA datasets. NGRHMDA is a recommendation-based microbe-disease association prediction approach. SMiR-NBI is a network inference-based method applied to identify biomarkers of cancers. LRLSHMDA is an LRLS classifier with Gaussian interaction profile kernel similarity. The three methods computed good accuracy in the corresponding application area. VDA-KATZ and VDA-RWR are the two newest VDA prediction methods and obtained better performance. VDA-BiRW is a balanced random walk-based VDA identification model. Table 3 illustrates the comparison results of seven VDA prediction models on three VDA datasets. The experimental results show that VDA-RWLRLS obtains better AUCs on the three datasets. We predicted the top 10 drugs associated SARS-CoV-2 and found that two small molecules, remdesivir and ribavirin, came together in three datasets, and three small molecules, nitazoxanide, favipiravir, and niclosamide, came together in two datasets. Molecular docking and MDS were implemented between the five drugs and the crystal structure of the S protein-binding domain bound with ACE2. The results suggest that remdesivir, ribavirin, nitazoxanide, and niclosamide have higher binding abilities with the crystal structure.
Table 3.
The performance of seven VDA prediction methods on three datasets.
| Datasets | Methods | Sensitivity | Specificity | F1 score | Accuracy | AUC |
|---|---|---|---|---|---|---|
| Dataset 1 | NGRHMDA | 0.578 3 | 0.556 7 | 0.061 5 | 0.557 2 | 0.645 9 |
| SMiR-NBI | 0.833 1 | 0.193 6 | 0.038 5 | 0.207 9 | 0.572 3 | |
| LRLSHMDA | 0.803 4 | 0.581 3 | 0.111 9 | 0.586 3 | 0.840 3 | |
| VDA-KATZ | 0.697 6 | 0.668 4 | 0.151 7 | 0.669 1 | 0.880 3 | |
| VDA-RWR | 0.482 4 | 0.783 1 | 0.115 3 | 0.827 8 | 0.858 2 | |
| VDA-BiRW | 0.832 3 | 0.636 8 | 0.133 2 | 0.641 1 | 0.876 5 | |
| VDA-RWLRLS | 0.562 6 | 0.838 0 | 0.225 9 | 0.831 9 | 0.885 8 | |
| Dataset 2 | NGRHMDA | 0.454 4 | 0.356 2 | 0.021 8 | 0.358 1 | 0.301 1 |
| SMiR-NBI | 0.834 9 | 0.094 2 | 0.033 6 | 0.108 1 | 0.415 6 | |
| LRLSHMDA | 0.783 8 | 0.484 0 | 0.073 3 | 0.489 6 | 0.824 8 | |
| VDA-KATZ | 0.551 2 | 0.757 4 | 0.080 5 | 0.753 5 | 0.829 6 | |
| VDA-RWR | 0.502 2 | 0.664 3 | 0.057 4 | 0.661 3 | 0.667 5 | |
| VDA-BiRW | 0.557 4 | 0.752 4 | 0.110 5 | 0.748 7 | 0.832 2 | |
| VDA-RWLRLS | 0.513 3 | 0.826 4 | 0.123 2 | 0.820 5 | 0.835 5 | |
| Dataset 3 | NGRHMDA | 0.358 2 | 0.408 1 | 0.011 9 | 0.407 4 | 0.255 4 |
| SMiR-NBI | 0.923 0 | 0.042 7 | 0.023 0 | 0.053 6 | 0.436 5 | |
| LRLSHMDA | 0.812 9 | 0.523 9 | 0.055 2 | 0.527 5 | 0.816 9 | |
| VDA-KATZ | 0.711 6 | 0.566 6 | 0.062 6 | 0.568 4 | 0.847 8 | |
| VDA-RWR | 0.505 3 | 0.705 7 | 0.055 6 | 0.703 2 | 0.712 3 | |
| VDA-BiRW | 0.707 8 | 0.574 1 | 0.072 6 | 0.575 8 | 0.851 1 | |
| VDA-RWLRLS | 0.519 8 | 0.843 8 | 0.118 9 | 0.844 6 | 0.862 5 |
VDA-RWLRLS obtains better specificity, F1 score, accuracy, and AUC than the other six methods on the three datasets. On dataset 1, for example, the AUCs obtained from VDA-RWLRLS are 27.08%, 35.39%, 5.14%, 0.62%, 3.12%, and 1.05% better than NGRHMDA, SMiR-NBI, LRLSHMDA, VDA-KATZ, VDA-RWR, and VDA-BiRW, respectively. On dataset 2, the AUCs computed by VDA-RWLRLS are 63.96%, 50.26%, 1.28%, 0.71%, 20.11%, and 0.39% better than those of six methods, respectively. On dataset 3, the AUC calculated by VDA-RWLRLS increases 70.39%, 49.39%, 5.29%, 1.70%, 17.41%, and 1.32%, respectively. Fig. 2 illustrates the AUC values achieved from the seven VDA prediction methods on the three datasets. The AUC and F1 score can better evaluate the performance of the association prediction methods compared to the other three measurements. VDA-RWLRLS computes the best F1 scores and AUCs on the three VDA datasets. Therefore, VDA-RWLRLS can be effectively applied to rank unknown virus-drug pairs at a more accurate rate based on known VDAs.
Fig. 2.
The AUC values predicted by seven VDA prediction methods on three VDA datasets.
More importantly, VDA-KATZ and VDA-RWR are two of the newest VDA prediction models and obtained good prediction performance. VDA-KATZ used the KATZ measurement, and VDA-RWR applied the random walk with restart on the constructed heterogeneous virus-drug network. Compared to the two state-of-the-art methods, VDA-RWLRLS boosts the prediction performance under the majority of conditions on the three datasets. Therefore, VDA-RWLRLS improves the VDA prediction accuracy.
VDA-RWR, VDA-BiRW and VDA-RWLRLS use bi-random walk for VDA prediction. The three methods use random walk with restart, balanced bi-random walk, and unbalanced bi-random walk. VDA-RWLRLS significantly outperforms VDA-RWR and VDA-BiRW. The results demonstrate the contributions of LRLS to VDA classification ability.
LRLSHMDA is an LRLS-based association prediction method. VDA-RWLRLS obtains better performance than LRLSHMDA on the three datasets. The results may be due to the left random walk on the virus network and the right random walks on the drug network: thus, considering the different maximal random walking steps based on two sides of the networks may boost the VDA prediction performance. In summary, integrating unbalanced bi-random walk and LRLS helps to mine underlying VDAs.
In addition, all unknown drug-virus pairs were chosen as negative samples. Sensitivity was defined as TP/(TP + FN). In datasets 1, 2, and 3, there were 96, 770, and 407 known VDAs, respectively, and 840, 8062, and 6495 unknown virus-drug pairs, respectively. The ratios of known VDAs to unknown virus-drug pairs in the three VDA datasets were 11.43%, 9.55%, and 6.27%, respectively. Thus, FN is much greater than TP because of the unbalanced characteristics between negative VDAs and positive VDAs. Therefore, the sensitivity and F1 score are relatively lower for VDA prediction algorithms. In addition, on dataset 1, NGRHMDA and SMiR-NBI calculated AUCs larger than 0.5. However, they computed AUCs smaller than 0.5 on datasets 2 and 3. In contrast, if we re-evaluate the logical direction of the cut-off points, NGRHMDA and SMiR-NBI will obtain AUCs larger than 0.5 on datasets 2 and 3: however, AUCs from the two methods will be smaller than 0.5 on dataset 1. This may be caused by the poor generalization ability of NGRHMDA and SMiR-NBI.
3.4. Case study
We aimed to identify possible clues of treatment for COVID-19 after confirming the performance of VDA-RWLRLS. Table 4, Table 5, Table 6 list the top 10 predicted drugs against SARS-CoV-2 in the three datasets. The majority of the top 10 predicted antiviral drugs have been supported by recent literature. The powerful results indicate the prediction confidence of VDA-RWLRLS. Among the inferred antiviral drugs, there are two small molecules coming in three VDA datasets, that is, remdesivir and ribavirin. In addition, three chemical agents were combined into two VDA datasets: nitazoxanide, favipiravir, and niclosamide.
Table 4.
The predicted top 10 drugs against SARS-CoV-2 on dataset 1.
| Rank | Drug | Evidence |
|---|---|---|
| 1 | Remdesivir | PMID: 32 020 029, 31 996 494, 32 022 370, 31 971 553, 32 035 018, 32 035 533, 32 036 774, 32 194 944, 32 275 812, 32 145 386, 32 838 064 |
| 2 | Oseltamivir | PMID: 32 034 637, 32 127 666 |
| 3 | Zanamivir | PMID: 32 511 320 |
| 4 | Ribavirin | PMID: 32 034 637, 32 127 666, 32 227 493, 26 492 219,32 771 797 |
| 5 | Presatovir | PMID: 32 147 628 |
| 6 | Elvitegravir | PMID: 32 147 628 |
| 7 | Zidovudine | PMID: 32 568 013 |
| 8 | Emtricitabine | PMID: 32 488 835 |
| 9 | Laninamivir | https://arxiv.org/abs/2009.10333 |
| 10 | Peramivir | unconfirmed |
Table 5.
The predicted top 10 drugs against SARS-CoV-2 on dataset 2.
| Rank | Drug | Evidence |
|---|---|---|
| 1 | Favipiravir | PMID: 32 346 491, 32 967 849, 32 972 430 |
| 2 | Niclosamide | PMID: 32 125 140, 32 221 153 |
| 3 | Remdesivir | PMID: 32 020 029, 31 996 494, 32 022 370, 31 971 553, 32 035 018, 32 035 533, 32 036 774, 32 194 944, 32 275 812, 32 145 386, 32 838 064 |
| 4 | Cyclosporine | PMID: 32 777 170, 32 505 466 |
| 5 | Nitazoxanide | PMID: 32 127 666, 32 568 620, 32 448 490 |
| 6 | Mycophenolic acid | PMID: 32 579 258 |
| 7 | BCX4430 (Galidesivir) | PMID: 32 711 596 |
| 8 | Emetine | PMID: 32 251 767,32 278 693,32 340 120 |
| 9 | Amodiaquine | PMID: 32 246 834, 32 834 612, 32 631 083, 32 317 408 |
| 10 | Ribavirin | PMID: 32 034 637, 32 127 666, 32 227 493, 26 492 219,32 771 797 |
Table 6.
The predicted top 10 drugs against SARS-CoV-2 on dataset 3.
| Rank | Drug | Evidence |
|---|---|---|
| 1 | Ribavirin | PMID: 32 034 637, 32 127 666, 32 227 493, 26 492 219, 32 771 797 |
| 2 | Nitazoxanide | PMID: 32 127 666, 32 568 620, 32 448 490 |
| 3 | Chloroquine | PMID: 32 020 029, 32 145 363, 32 074 550, 32 236 562 |
| 4 | Camostat | PMID: 32 347 443 |
| 5 | Umifenovir | PMID: 32 941 741 |
| 6 | Favipiravir | PMID: 32 346 491, 32 967 849, 32 972 430 |
| 7 | Amantadine | PMID: 32 361 028 |
| 8 | Niclosamide | PMID: 32 125 140, 32 221 153 |
| 9 | Remdesivir | PMID: 32 020 029, 31 996 494, 32 022 370, 31 971 553, 32 035 018, 32 035 533, 32 036 774, 32 194 944, 32 275 812, 32 145 386, 32 838 064 |
| 10 | Berberine | PMID: 33 670 363 |
Remdesivir is a triphosphate analog first reported as a potential therapeutic agent against Ebola. The drug has broad antiviral activity against the Arenaviridae, Coronaviridae, Filoviridae, Flaviviridae, Paramyxoviridae, and Pneumoviridae viral families. It has been confirmed as a non-obligate chain terminator of RNA-dependent RNA polymerase from SARS-CoV-2 and investigated in many COVID-19 clinical trials. On November 19, 2020, it was authorized for the treatment of COVID-19 in combination with baricitinib [58].
Ribavirin is an antiviral drug. This drug can inhibit viral RNA synthesis and mRNA capping. Thus it has broad-spectrum activity against a few RNA and DNA viruses. Currently, dual therapy with ribavirin and peginterferon alfa-2a/peginterferon alfa-2b is recommended as the first-generation and standard antiviral treatment [58].
3.5. Molecular docking
We used AutoDock 4.2, a molecular docking software, to conduct molecular docking between the predicted five antiviral drugs and the crystal structure of the S protein-binding domain bound with ACE2. The chemical agents are taken as ligands and the crystal structure is taken as the receptor. Molecular binding energy was used to measure the binding stabilities between the predicted five antiviral drugs and the crystal structure.
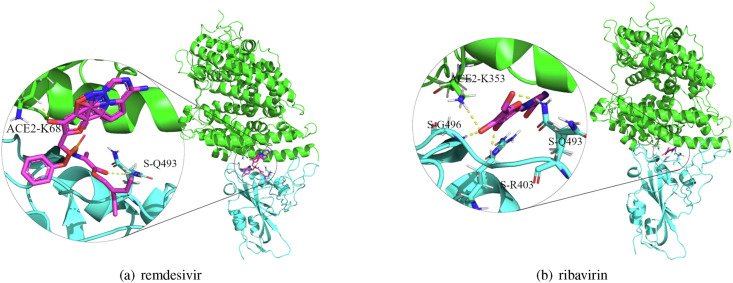
Fig. 3 shows the molecular docking between remdesivir and ribavirin and the crystal structure. The results show that remdesivir has a binding energy of −7.00 kcal/mol with the crystal structure and that binding sites are K68 with ACE2 and Q493 with the S protein. Ribavirin has a binding energy of −6.59 kcal/mol, and its binding sites are K353 with ACE2 and G496, Q493, and R403 with the S protein. Molecular docking results for the other three drugs, nitazoxanide, favipiravir, and niclosamide and the crystal structure are illustrated in Figs. 1–3 in Supplementary Materials 1. The corresponding binding energies and sites are shown in Table 1 in Supplementary Materials 1.
Fig. 3.
Molecular docking between remdesivir and ribavirin and the crystal structure of the S protein-ACE2 binding domain.
3.6. Molecular dynamics simulation
After performing molecular docking, we continued to conduct MDAs between the predicted possible drugs and the S protein, human ACE2, and the crystal structure of the S protein-binding domain bound with ACE2. Fig. 4 illustrates Root Mean Square Deviation (RMSD) trajectories of ACE2 and remdesivir, Radius of gyration (Rg) pattern of the remdesivir-ACE2 complex, Root Mean Square Fluctuation (RMSF) pattern of ACE2, and H-bond during MDS, and Solvent Accessible Surface Area (SASA) of the remdesivir-ACE2 complex backbone. From Fig. 4, we can observe that the RMSD trajectory of ACE2 (Fig. 4 (a)) obtains equilibrium beyond 10 ns with an average value of approximately 0.25 nm. The RMSD of remdesivir (Fig. 4 (b)) is approximately stable during MDS, with an average value of approximately 0.6 nm. The two average values indicate that the relative change in ligand position is less than that of human ACE2, thereby demonstrating the stability of the remdesivir-human ACE2 pose.
Fig. 4.
MDS between remdesivir and human ACE2 protein during 50ns.
The lower mean value (2.5 nm) and stable trajectory of Rg (Fig. 4(c)) depict the compactness of the remdesivir-ACE2 complex during the simulation. In Fig. 4(d), the SASA of the complex is 0.1–0.5 nm2, indicating that the hydrophobic core is compact and that the conformational geometry of the complex is stable during MDS. In Fig. 4(e), remdesivir and ACE2 interact with 1–5 hydrogen bonds; however, only two bonds are observed to be consistent throughout MDA. In Fig. 4(f), RMSF is applied to measure the number of positional fluctuations of each residue in the remdesivir-ACE2 backbone during MDS. The RMSF value in the backbone is in the range of 0.05–0.4 nm with an average of approximately 0.15 nm and minimum fluctuations of the key active-site residues. The other MDSs between the predicted ligands (remdesivir, ribavirin, nitazoxanide, favipiravir, and niclosamide) and three receptors (the S protein, human ACE2, and the crystal structure of the S protein-ACE2 binding domain) under 50 ns and 10 ns are illustrated in Figs. 1–25 in Supplementary Materials 2.
4. Discussion and conclusion
The key to preventing the transmission of SARS-CoV-2 lies in a deep understanding of associations among viruses, target proteins and targeting drugs. Laboratory techniques may not be realistic under such an emergent situation. Drug repositioning is a more powerful approach to find possible clues for treatment of COVID-19. Identification of potential associations between viruses including SARS-CoV-2 and post-marked drugs through drug repositioning, provides an effective way to prioritize chemical agents related to SARS-CoV-2. Toward this goal, we first analyzed the pattern of associations between viruses and drugs and observed that the majority of known VDAs are circular bigraphs with short path lengths. This result provides the foundation for new VDA prediction based on known linkages in a VDA network. An Unbalanced bi-random walk is specifically developed to mine circular bigraphs and thus reconstructs associations between viruses and chemical agents. In addition, during an unbalanced bi-random walk, the jumps of nodes severely depend on virus similarities, drug similarities and known VDAs. To reduce the limitations, LRLS is integrated into the unbalanced bi-random walk model. VDA-RWLRLS is compared to six classical VDA prediction models. The experimental results show that VDA-RWLRLS outperforms the six association prediction methods.
The novelty of VDA-RWLRLS comes from the combination of unbalanced bi-random walk, LRLS, molecular docking, and MDS. Our proposed VDA-RWLRLS algorithm performs consistently better on three constructed datasets. It may be contributed to by the following four features. First, VDA-RWLRLS uses the unbalanced bi-random walk method and takes advantage of different combinations in left and right random walks. Second, VDA-RWLRLS globally explores the weighted circular bigraphs by bi-random walk on the virus network and the drug network. The number of circular bigraphs affects the probability of a virus associated with a drug. Third, LRLS is utilized to decrease the drawback of unbalanced bi-random walk. Finally, VDA-RWLRLS computes the virus similarity and drug similarity by fusing biological information and association information in a VDA network.
In addition, morbidity and mortality rates in the COVID-19 pandemic are sharply increasing because of bacterial and fungal co-infections. However, there is currently a lack of efficient clinical trials to screen antibacterial and antifungal compounds in COVID-19 patients [74]. Therefore, it is urgent to boost the knowledge about antibacterial and antifungal drugs to optimize the prevention, diagnosis, and therapeutic strategies in the COVID-19 pandemic. In the future, we will further mine drug banks and pharmacological resources and build two datasets, that is, datasets involved in associations between antibacterial drugs and single stranded RNA virus-caused diseases and datasets involved in associations between antifungal drugs and single stranded RNA virus-caused diseases, to initially screen small molecules for COVID-19 treatment.
Author contributions
LS, L-HP, F-XL, and L-QZ proposed computational models, L-HP and LS wrote the paper, L-HP and L-QZ revised original draft, LS run the computational models, LS conducted molecular docking, LS, HL and G-YL performed MDS, LS, L-HP, and L-QZ discussed the computational models and gave conclusion. All authors read and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant 61803151, 62072172, 62172158), the Natural Science Foundation of Hunan province (Grant 2021JJ30219), scientific research project of Hunan Provincial Department of Education (20C0636), scientific research and innovation Foundation of Hunan University of Technology (Grant CX2031).
Data availability
Source code and dataset are freely downloadable at https://github.com/plhhnu/VDA-RWLRLS/. PDB ID of ACE2: 6M0J, DrugBank ID of Remdesivir: DB14761, Ribavirin: DB00811, Nitazoxanide: DB00507, Favipiravir: DB12466, Niclosamide: DB06803.
Declaration of competing interest
All authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We really appreciate four anonymous reviewers for their valuable comments. We would like to thank all authors of the cited references.
Biographies

Ling Shen: He is a postgraduate student in the School of Computer, Hunan University of Technology, China. His research interests include machine learning and bioinformatics.

Fuxing Liu: He is a postgraduate student in the School of Computer, Hunan University of Technology, China. His research interests include machine learning and bioinformatics.

Li Huang: He is a PhD candidate in the Future Laboratory, Tsinghua University, China. His research interests include bioinformatics and visual analytics.

Guangyi Liu: He is an undergraduate student in the School of Computer, Hunan University of Technology, China. His research interests include machine learning and bioinformatics.

Liqian Zhou: He is working in Hunan University of Technology as a professor. He received a PhD in Xiangtan University, China. His research interests include Machine Learning, Data Mining and Bioinformatics.

Lihong Peng: She is working in Hunan University of Technology as an associate professor. She received a PhD in the College of Information Science and Engineering, Hunan University, China. Her research interests include Machine Learning, Data Mining and Bioinformatics.
Footnotes
This research was funded by the National Natural Science Foundation of China (Grant 61803151, 62072172, 62172158), the Natural Science Foundation of Hunan province (Grant 2021JJ30219), Scientific Research Project of Hunan Provincial Department of Education (20C0636), Scientific Research and Innovation Foundation of Hunan University of Technology (Grant CX2031).
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.105119.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.WHO, Who coronavirus (covid-19) overview, Website, Access on May 13, 2021. URL: https://covid19.who.int.
- 2.L. Riva, S. Yuan, X. Yin, L. Martin-Sancho, N. Matsunaga, S. Burgstaller-Muehlbacher, L. Pache, P. P. D. Jesus, M. V. Hull, M. Chang, J. F.-W. Chan, J. Cao, V. K.-M. Poon, K. Herbert, T.-T. Nguyen, Y. Pu, C. Nguyen, A. Rubanov, L. Martinez-Sobrido, W.-C. Liu, L. Miorin, K. M. White, J. R. Johnson, C. Benner, R. Sun, P. G. Schultz, A. Su, A. Garcia-Sastre, A. K. Chatterjee, K.-Y. Yuen, S. K. Chanda, A Large-Scale Drug Repositioning Survey for SARS-CoV-2 Antivirals, bioRxiv (2020) 2020.04.16.044016. Publisher: Cold Spring Harbor Laboratory Section: New Results.
- 3.Serafin M.B., Bottega A., Foletto V.S., da Rosa T.F., Hörner A., Hörner R. Drug repositioning is an alternative for the treatment of coronavirus COVID-19. Int. J. Antimicrob. Agents. 2020;55:105969. doi: 10.1016/j.ijantimicag.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zohner Y.E., Morris J.S. Covid-track: world and USA sars-cov-2 testing and covid-19 tracking. BioData Min. 2021;14:1–15. doi: 10.1186/s13040-021-00233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Y. Ge, T. Tian, S. Huang, F. Wan, J. Li, S. Li, H. Yang, L. Hong, N. Wu, E. Yuan, L. Cheng, Y. Lei, H. Shu, X. Feng, Z. Jiang, Y. Chi, X. Guo, L. Cui, L. Xiao, Z. Li, C. Yang, Z. Miao, H. Tang, L. Chen, H. Zeng, D. Zhao, F. Zhu, X. Shen, J. Zeng, A Data-Driven Drug Repositioning Framework Discovered a Potential Therapeutic Agent Targeting COVID-19, bioRxiv (2020) 2020.03.11.986836. Publisher: Cold Spring Harbor Laboratory Section: New Results.
- 6.Gao K., Song Y.-P., Song A. Exploring active ingredients and function mechanisms of ephedra-bitter almond for prevention and treatment of corona virus disease 2019 (covid-19) based on network pharmacology. BioData Min. 2020;13:1–20. doi: 10.1186/s13040-020-00229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anbarasu A., Ramaiah S., Livingstone P. Vaccine repurposing approach for preventing COVID 19: can MMR vaccines reduce morbidity and mortality? Hum. Vaccines Immunother. 2020;16:2217–2218. doi: 10.1080/21645515.2020.1773141. Publisher: Taylor & Francis _eprint: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jourdan J.-P., Bureau R., Rochais C., Dallemagne P. Drug repositioning: a brief overview. J. Pharm. Pharmacol. 2020;72:1145–1151. doi: 10.1111/jphp.13273. https://onlinelibrary.wiley.com/doi/pdf/10.1111/jphp.13273 _eprint: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu Y., Shan X., Chen T., Jiang M., Wang Y., Wang Q., Salahub D.R., Xiong Y., Wei D.-Q. DTI-MLCD: predicting drug-target interactions using multi-label learning with community detection method. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbaa205. [DOI] [PubMed] [Google Scholar]
- 10.Khan M.T., Irfan M., Ahsan H., Ahmed A., Kaushik A.C., Khan A.S., Chinnasamy S., Ali A., Wei D.-Q. vol. 64. Publisher: Karger Publishers; 2021. pp. 55–68. (Structures of SARS-CoV-2 RNA-Binding Proteins and Therapeutic Targets, Intervirology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rian K., Esteban-Medina M., Hidalgo M.R., Çubuk C., Falco M.M., Loucera C., Gunyel D., Ostaszewski M., Peña-Chilet M., Dopazo J. Mechanistic modeling of the sars-cov-2 disease map. BioData Min. 2021;14:1–8. doi: 10.1186/s13040-021-00234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.In A., Au E., Jo M., Sa F., Ib O., Is O., C O.-G., Ao S., J N. Exploring the genetics, ecology of SARS-COV-2 and climatic factors as possible control strategies against COVID-19. Infezioni Med. Le. 2020;28:166–173. [PubMed] [Google Scholar]
- 15.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. https://onlinelibrary.wiley.com/doi/pdf/10.1002/jmv.25726 _eprint: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., Mchugh T.D., Memish Z.A., Drosten C., Zumla A., Petersen E. vol. 91. Publisher: Elsevier; 2020. pp. 264–266. (The Continuing 2019-nCoV Epidemic Threat of Novel Coronaviruses to Global Health — the Latest 2019 Novel Coronavirus Outbreak in Wuhan, China, International Journal of Infectious Diseases). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng G., Zheng K.I., Yan Q.-Q., Rios R.S., Targher G., Byrne C.D., Van Poucke S., Liu W.-Y., Zheng M.-H. Covid-19 and liver dysfunction: current insights and emergent therapeutic strategies. J. Clin. Transl. Hepatol. 2020;8:18. doi: 10.14218/JCTH.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roudbary M., Kumar S., Kumar A., Černáková L., Nikoomanesh F., Rodrigues C.F. Overview on the prevalence of fungal infections, immune response, and microbiome role in covid-19 patients. J. Fungi. 2021;7:720. doi: 10.3390/jof7090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salehi M., Ahmadikia K., Mahmoudi S., Kalantari S., Jamalimoghadamsiahkali S., Izadi A., Kord M., Dehghan Manshadi S.A., Seifi A., Ghiasvand F., et al. Oropharyngeal candidiasis in hospitalised covid-19 patients from Iran: species identification and antifungal susceptibility pattern. Mycoses. 2020;63:771–778. doi: 10.1111/myc.13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Liao B., Cheng L., Peng X., Xu X., Li Y., Hu T., Li J., Zhou X., Ren B. The microbial coinfection in covid-19. Appl. Microbiol. Biotechnol. 2020:1–9. doi: 10.1007/s00253-020-10814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arastehfar A., Carvalho A., van de Veerdonk F.L., Jenks J.D., Koehler P., Krause R., Cornely O.A., Perlin D.S., Lass-Flörl C., Hoenigl M. Covid-19 associated pulmonary aspergillosis (capa)—from immunology to treatment. J. Fungi. 2020;6:91. doi: 10.3390/jof6020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao X., Wu C., Wang S., Tong S., Wang G., Wu G., Zhou R. The importance of overweight in covid-19: a retrospective analysis in a single center of wuhan, China. Medicine. 2020;99 doi: 10.1097/MD.0000000000022766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song G., Liang G., Liu W. Fungal co-infections associated with global covid-19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020:1–8. doi: 10.1007/s11046-020-00462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamed K., Yazdanpanah N., Saghazadeh A., Rezaei N. Computational drug discovery and repurposing for the treatment of covid-19: a systematic review. Bioorg. Chem. 2021;106:104490. doi: 10.1016/j.bioorg.2020.104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nayarisseri A. Most promising compounds for treating covid-19 and recent trends in antimicrobial & antifungal agents. Curr. Top. Med. Chem. 2020;20:2119–2125. doi: 10.2174/156802662023201001094634. [DOI] [PubMed] [Google Scholar]
- 27.Mazumder P., Kalamdhad A., Chaminda G.T., Kumar M. Coalescence of co-infection and antimicrobial resistance with sars-cov-2 infection: the blues of post-covid-19 world. Case Stud. Chem. Environ. Eng. 2021;3:100093. doi: 10.1016/j.cscee.2021.100093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawson T.M., Moore L.S., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support covid-19 antimicrobial prescribing. Clin. Infect. Dis. 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano-Aroca Á., Takayama K., Tuñón-Molina A., Seyran M., Hassan S.S., Pal Choudhury P., Uversky V.N., Lundstrom K., Adadi P., Palù G., et al. Carbon-based nanomaterials: promising antiviral agents to combat covid-19 in the microbial-resistant era. ACS Nano. 2021;15:8069–8086. doi: 10.1021/acsnano.1c00629. [DOI] [PubMed] [Google Scholar]
- 30.Mahmoudi H. Bacterial co-infections and antibiotic resistance in patients with covid-19. GMS Hygiene Infect. Contr. 2020;15 doi: 10.3205/dgkh000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J., Wang J., Yang Y., Cai P., Cao J., Cai X., Zhang Y. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with covid-19 in wuhan, China: a retrospective analysis. Antimicrob. Resist. Infect. Control. 2020;9:1–7. doi: 10.1186/s13756-020-00819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahoney A.R., Safaee M.M., Wuest W.M., Furst A.L. The silent pandemic: emergent antibiotic resistances following the global response to sars-cov-2. Iscience. 2021:102304. doi: 10.1016/j.isci.2021.102304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narendrakumar L., Joseph I., Thomas S. Potential effectiveness and adverse implications of repurposing doxycycline in covid-19 treatment. Expert Rev. Anti-infect. Ther. 2021;19:1001–1008. doi: 10.1080/14787210.2021.1865803. [DOI] [PubMed] [Google Scholar]
- 34.Pan X., Dong L., Yang L., Chen D., Peng C. Potential drugs for the treatment of the novel coronavirus pneumonia (covid-19) in China. Virus Res. 2020;286:198057. doi: 10.1016/j.virusres.2020.198057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hijikata A., Shionyu-Mitsuyama C., Nakae S., Shionyu M., Ota M., Kanaya S., Shirai T. Knowledge-based structural models of SARS-CoV-2 proteins and their complexes with potential drugs. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2020;594:1960–1973. doi: 10.1002/1873-3468.13806. https://febs.onlinelibrary.wiley.com/doi/pdf/10.1002/1873-3468.13806 _eprint: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farag A., Wang P., Ahmed M., Sadek H. Publisher: ChemRxiv; 2020. Identification of FDA Approved Drugs Targeting COVID-19 Virus by Structure-Based Drug Repositioning. [Google Scholar]
- 37.Singh N., Decroly E., Khatib A.-M., Villoutreix B.O. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur. J. Pharmaceut. Sci. 2020;153:105495. doi: 10.1016/j.ejps.2020.105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan A., Tahir Khan M., Saleem S., Junaid M., Ali A., Shujait Ali S., Khan M., Wei D.-Q. Structural insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput. Struct. Biotechnol. J. 2020;18:2174–2184. doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet (London, England) 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray S., Lall S., Mukhopadhyay A., Bandyopadhyay S., Schönhuth A. 2020. Predicting Potential Drug Targets and Repurposable Drugs for COVID-19 via a Deep Generative Model for Graphs. arXiv:2007.02338 [q-bio. ArXiv: 2007.02338. [Google Scholar]
- 41.Ke Y.-Y., Peng T.-T., Yeh T.-K., Huang W.-Z., Chang S.-E., Wu S.-H., Hung H.-C., Hsu T.-A., Lee S.-J., Song J.-S., Lin W.-H., Chiang T.-J., Lin J.-H., Sytwu H.-K., Chen C.-T. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020;43:355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dotolo S., Marabotti A., Facchiano A., Tagliaferri R. A review on drug repurposing applicable to COVID-19. Briefings Bioinf. 2021;22:726–741. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gysi D.M., Valle Í.d., Zitnik M., Ameli A., Gan X., Varol O., Ghiassian S.D., Patten J.J., Davey R.A., Loscalzo J., Barabási A.-L. Proceedings of the National Academy of Sciences 118. Publisher: National Academy of Sciences Section: Biological Sciences; 2021. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng L., Tian X., Shen L., Kuang M., Li T., Tian G., Yang J., Zhou L. Identifying effective antiviral drugs against SARS-CoV-2 by drug repositioning through virus-drug association prediction. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.577387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou L., Wang J., Liu G., Lu Q., Dong R., Tian G., Yang J., Peng L. Probing antiviral drugs against SARS-CoV-2 through virus-drug association prediction based on the KATZ method. Genomics. 2020;112:4427–4434. doi: 10.1016/j.ygeno.2020.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng L., Shen L., Xu J., Tian X., Liu F., Wang J., Tian G., Yang J., Zhou L. vol. 11. Publisher: Nature Publishing Group; 2021. p. 6248. (Prioritizing Antiviral Drugs against SARS-CoV-2 by Integrating Viral Complete Genome Sequences and Drug Chemical Structures, Scientific Reports). Number: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thillainayagam M., Malathi K., Anbarasu A., Singh H., Bahadur R., Ramaiah S. Insights on inhibition of Plasmodium falciparum plasmepsin I by novel epoxyazadiradione derivatives – molecular docking and comparative molecular field analysis. J. Biomol. Struct. Dyn. 2019;37:1–32. doi: 10.1080/07391102.2018.1510342. (Publisher: Taylor & Francis) [DOI] [PubMed] [Google Scholar]
- 49.Thillainayagam M., Ramaiah S., Anbarasu A. Molecular docking and dynamics studies on novel benzene sulfonamide substituted pyrazole-pyrazoline analogues as potent inhibitors of Plasmodium falciparum Histo aspartic protease. J. Biomol. Struct. Dyn. 2020;38:3235–3245. doi: 10.1080/07391102.2019.1654923. Publisher: Taylor & Francis _eprint: [DOI] [PubMed] [Google Scholar]
- 50.Khan S., Hussain Z., Safdar M., Khan A., Wei D.-Q. Targeting the N-terminal domain of the RNA-binding protein of the SARS-CoV-2 with high affinity natural compounds to abrogate the protein-RNA interaction: a molecular dynamics study. J. Biomol. Struct. Dyn. 2021:1–9. doi: 10.1080/07391102.2021.1882337. 0. Publisher: Taylor & Francis _eprint: [DOI] [PubMed] [Google Scholar]
- 51.Meng X.-Y., Zhang H.-X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thillainayagam M., Pandian L., Murugan K.K., Vijayaparthasarathi V., Sundaramoorthy S., Anbarasu A., Ramaiah S. In silico analysis reveals the anti-malarial potential of quinolinyl chalcone derivatives. J. Biomol. Struct. Dyn. 2015;33:961–977. doi: 10.1080/07391102.2014.920277. Publisher: Taylor & Francis _eprint: [DOI] [PubMed] [Google Scholar]
- 53.Basu S., Veeraraghavan B., Ramaiah S., Anbarasu A. Novel cyclohexanone compound as a potential ligand against SARS-CoV-2 main-protease. Microb. Pathog. 2020;149:104546. doi: 10.1016/j.micpath.2020.104546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ragunathan A., Malathi K., Ramaiah S., Anbarasu A. FtsA as a cidal target for Staphylococcus aureus: molecular docking and dynamics studies. J. Cell. Biochem. 2019;120:7751–7758. doi: 10.1002/jcb.28049. https://onlinelibrary.wiley.com/doi/pdf/10.1002/jcb.28049 _eprint: [DOI] [PubMed] [Google Scholar]
- 55.Ragunathan A., Malathi K., Anbarasu A. MurB as a target in an alternative approach to tackle the Vibrio cholerae resistance using molecular docking and simulation study. J. Cell. Biochem. 2018;119:1726–1732. doi: 10.1002/jcb.26333. https://onlinelibrary.wiley.com/doi/pdf/10.1002/jcb.26333 _eprint: [DOI] [PubMed] [Google Scholar]
- 56.Andersen P.I., Ianevski A., Lysvand H., Vitkauskiene A., Oksenych V., Bjørås M., Telling K., Lutsar I., Dumpis U., Irie Y., et al. vol. 93. Publisher: Elsevier; 2020. pp. 268–276. (Discovery and Development of Safe-In-Man Broad-Spectrum Antiviral Agents, International Journal of Infectious Diseases). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horton D.A., Bourne G.T., Smythe M.L. vol. 103. Publisher: American Chemical Society; 2003. pp. 893–930. (The Combinatorial Synthesis of Bicyclic Privileged Structures or Privileged Substructures, Chemical Reviews). [DOI] [PubMed] [Google Scholar]
- 58.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landrum G., et al. 2016. Rdkit: Open-source Cheminformatics Software, 2016.http://www.rdkit.orghttps://github.com/rdkit/rdkit [Google Scholar]
- 60.Rogers D., Hahn M. vol. 50. Publisher: American Chemical Society; 2010. pp. 742–754. (Extended-Connectivity Fingerprints, Journal of Chemical Information and Modeling). [DOI] [PubMed] [Google Scholar]
- 61.Brister J.R., Ako-adjei D., Bao Y., Blinkova O. NCBI viral genomes resource. Nucleic Acids Res. 2015;43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Laarhoven T., Nabuurs S.B., Marchiori E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics. 2011;27:3036–3043. doi: 10.1093/bioinformatics/btr500. [DOI] [PubMed] [Google Scholar]
- 64.Wang F., Huang Z.-A., Chen X., Zhu Z., Wen Z., Zhao J., Yan G.-Y. vol. 7. Publisher: Nature Publishing Group; 2017. p. 7601. (LRLSHMDA: Laplacian Regularized Least Squares for Human Microbe–Disease Association Prediction, Scientific Reports). Number: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rose P.W., Beran B., Bi C., Bluhm W.F., Dimitropoulos D., Goodsell D.S., Prlić A., Quesada M., Quinn G.B., Westbrook J.D., Young J., Yukich B., Zardecki C., Berman H.M., Bourne P.E. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002;40:82–92. (Publisher: Citeseer) [Google Scholar]
- 67.Forli W., Halliday S., Belew R., Olson A.J. AutoDock version 4.2. J. Med. Chem. 2012;55:623–638. doi: 10.1021/jm2005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. https://onlinelibrary.wiley.com/doi/pdf/10.1002/%28SICI%291096-987X%2819981115%2919%3A14%3C1639%3A%3AAID-JCC10%3E3.0.CO%3B2-B _eprint: [Google Scholar]
- 69.Yu W., He X., Vanommeslaeghe K., MacKerell A.D., Jr. Extension of the charmm general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar R., Indrayan A. Receiver operating characteristic (roc) curve for medical researchers. Indian Pediatr. 2011;48:277–287. doi: 10.1007/s13312-011-0055-4. [DOI] [PubMed] [Google Scholar]
- 71.Pegoraro J.A., Lavault S., Wattiez N., Similowski T., Gonzalez-Bermejo J., Birmelé E. Machine-learning based feature selection for a non-invasive breathing change detection. BioData Min. 2021;14:1–16. doi: 10.1186/s13040-021-00265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y.-A., You Z.-H., Chen X., Huang Z.-A., Zhang S., Yan G.-Y. Prediction of microbe–disease association from the integration of neighbor and graph with collaborative recommendation model. J. Transl. Med. 2017;15:209. doi: 10.1186/s12967-017-1304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J., Lei K., Wu Z., Li W., Liu G., Liu J., Cheng F., Tang Y. Network-based identification of microRNAs as potential pharmacogenomic biomarkers for anticancer drugs. Oncotarget. 2016;7:45584–45596. doi: 10.18632/oncotarget.10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alhazzani W., Møller M.H., Arabi Y.M., Loeb M., Gong M.N., Fan E., Oczkowski S., Levy M.M., Derde L., Dzierba A., et al. Surviving sepsis campaign: guidelines on the management of critically ill adults with coronavirus disease 2019 (covid-19) Intensive Care Med. 2020;46:854–887. doi: 10.1007/s00134-020-06022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code and dataset are freely downloadable at https://github.com/plhhnu/VDA-RWLRLS/. PDB ID of ACE2: 6M0J, DrugBank ID of Remdesivir: DB14761, Ribavirin: DB00811, Nitazoxanide: DB00507, Favipiravir: DB12466, Niclosamide: DB06803.