Abstract
Objectives
Pseudomonas aeruginosa is a leading cause of community- and hospital-acquired infections. Successful treatment is hampered by its remarkable ability to rapidly develop resistance to antimicrobial agents, primarily through mutation. In response, WHO listed carbapenem-resistant P. aeruginosa as a Priority 1 (Critical) pathogen for research and development of new treatments. A key resource in developing effective countermeasures is access to diverse and clinically relevant strains for testing. Herein we describe a panel of 100 diverse P. aeruginosa strains to support this endeavour.
Methods
WGS was performed on 3785 P. aeruginosa isolates in our repository. Isolates were cultured from clinical samples collected from healthcare facilities around the world between 2003 and 2017. Core-genome MLST and high-resolution SNP-based phylogenetic analyses were used to select a panel of 100 strains that captured the genetic diversity of this collection. Antibiotic susceptibility testing was also performed using 14 clinically relevant antibiotics.
Results
This 100-strain diversity panel contained representative strains from 91 different STs, including genetically distinct strains from major epidemic clones ST-111, ST-235, ST-244 and ST-253. Seventy-one distinct antibiotic susceptibility profiles were identified ranging from pan-susceptible to pan-resistant. Known resistance alleles as well as the most prevalent mutations underlying the antibiotic susceptibilities were characterized for all isolates.
Conclusions
This panel provides a diverse and comprehensive set of P. aeruginosa strains for use in developing solutions to antibiotic resistance. The isolates and available metadata, including genome sequences, are available to industry, academia, federal and other laboratories at no additional cost.
Introduction
Pseudomonas aeruginosa is a ubiquitous organism whose genetic and metabolic versatility has enabled it to survive and thrive in a diverse range of environments.1 This adroit adaptability has allowed P. aeruginosa to emerge as one of the most successful opportunistic pathogens, as evidenced by its inclusion in the clinically important ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa and Enterobacter spp.) coterie of human pathogens.2 In humans, P. aeruginosa is an important source of both community- and hospital-acquired infections, ranging from skin and soft tissue infections to pneumonias and complicated bloodstream infections.3,4 In particular, P. aeruginosa is a signature pathogen of patients with cystic fibrosis and a significant source of infection in burn wounds, resulting in substantial morbidity and mortality.4,5
The population structure of P. aeruginosa is as complex as it is varied and significant overlap between environmental and clinical isolates has been observed.6 More recent studies using WGS have further refined this structure, with the species being segregated into two major clades (hereafter referred to as Group A and Group B as per the nomenclature used by Ozer et al.7) and up to three smaller clades.7–11 Group A and B are most often associated with human infections and there is evidence that recombination between the two groups is limited, suggesting infrequent ecological overlap.7 Genetically, the two clades can be distinguished to a high degree of accuracy based on the presence of one of two genes encoding the major effector proteins secreted by the P. aeruginosa type III secretion system, exoS and exoU. Specifically, in a recent study of 739 P. aeruginosa from diverse environments, 98% of isolates from Group A carried exoS but not exoU, whereas 95% of Group B isolates carried exoU but not exoS.7 While isolates from both groups cause human infections, those carrying exoU have long been associated with more severe outcomes and higher mortality rates.12–15 The smaller clades range from one to three groups depending on the study,6–8 with the largest study defining three groups: 3, 4 and 5.8 Among them the unusual PA7 clade16 was consistently identified. This clade is a taxonomic outlier that groups isolates with seemingly reduced virulence compared with Group A and B, but increased biofilm formation and antibiotic resistance.17
A major contributor to the success of P. aeruginosa has been its remarkable ability to resist the action of antibiotics.18,19 The organism can employ a wide range of intrinsic mechanisms to resist these agents, including porin loss through mutation, hyperproduction of intrinsic AmpC enzymes and efflux pumps, and modification of antibiotic targets (for a recent review, see Lopez-Causape et al.19). The challenges posed by these formidable intrinsic mechanisms have been further compounded by the emergence of MDR and XDR strains carrying a variety of transferable antibiotic resistance genes (ARGs), including those encoding ESBLs, carbapenemases and 16S rRNA methyltransferases.20–22 Contrasting with the high genetic diversity within the antibiotic susceptible population,21 MDR and XDR isolates have been closely associated with a small number of globally distributed clones (for a review see Oliver et al.21). The most prevalent of these ‘high-risk’ clones is ST-235 (the founding member of clonal complex 235), which appears to have arisen in Europe during the early 1980s, shortly after the introduction of fluoroquinolones for treating Pseudomonas infections.23 The MDR and XDR phenotypes exhibited by ST-235 and other high-risk clones, such as ST-111 and ST-175,21 are a combination of both mutation-driven resistance and acquisition of transferable ARGs, particularly ESBLs and carbapenemases.20 Of particular concern, these high-risk clones are increasingly being encountered worldwide, making treatment of these challenging pathogens even more difficult.21,24,25
Though antimicrobial drug discovery had languished from the 1980s until the early 2010s, the emerging antibiotic resistance crisis has seen renewed interest by governmental, academic and industrial organizations.26–28 In 2017, WHO listed carbapenem-resistant P. aeruginosa as a Priority 1 (Critical) pathogen for research and development of new treatments.29 In addition, the US CDC listed MDR P. aeruginosa as a serious public health threat that requires ‘prompt and sustained action’.30 Recent advances in genomic, proteomic and bioinformatic technologies have opened new avenues for developing new antimicrobial agents, including new strategies to combat P. aeruginosa.31 An important consideration when developing strategies to combat P. aeruginosa is determining the most appropriate strain to use.31 While the reference strains PA01 and PA14 are the most commonly used research strains worldwide, they are not ‘high-risk’ clones and do not provide a comprehensive representation of the species.
Here, akin to our previous effort with A. baumannii,32 the construction of a reference panel of 100 strains that captures the genetic diversity of clinical P. aeruginosa and maximizes phylogenetic distance and pan-genome diversity is described. The panel was designed based on the WGS of 3785 P. aeruginosa isolates housed at the Multi-drug resistant organism Repository and Surveillance Network (MRSN), which have been collected over the past 11 years from around the world. Though not a primary focus of this endeavour, the panel has diverse antibiotic susceptibility phenotypes, ranging from pan-susceptible to pan-resistant, due to both intrinsic and acquired resistance mechanisms. This standardized resource will prove valuable for those interested in studying this important pathogen and will aid the design and development of novel antimicrobials and diagnostics.
Methods
P. aeruginosa repository
The MRSN collects and analyses clinically relevant MDR organisms across the Military Healthcare System33 and around the world in collaboration with the US Department of Defense’s Global Emerging Infections Surveillance (GEIS) Branch. All samples are housed in a central repository that currently contains over 85 000 isolates, including 3785 P. aeruginosa that were cultured from 2078 patients between 2002 and 2020 (Figure S1, available as Supplementary data at JAC-AMR Online). The majority (84.1%) were collected from patients in the USA, including Alaska and Hawaii, though strains from South America (4.4%), the Middle East (4%), Europe (3%), Africa (2.4%) and Asia (2.1%) are also represented. The isolates were cultured from a wide diversity of clinical specimens, including respiratory (37.9%), urine (27.4%), wounds (18.7%), surveillance swabs (5%), blood (4%), sterile fluid (2%) and the environment (1%). Isolate source was unavailable for 4.7% of the strains.
Antibiotic susceptibility testing (AST)
AST was performed in the MRSN College of American Pathologists-accredited clinical lab using three commercial platforms: the BD Phoenix (Panel NMIC/ID304; BD Diagnostics, NJ, USA), the Vitek 2 (Card AST-95 and AST-XN09 cards; bioMérieux, NC, USA) and the MicroScan WalkAway (Panel NBC47; Beckman Coulter, MD, USA). The final susceptibility profile for each isolate represents the adjudicated value from all three instruments. When discrepancies were noted between instruments, the majority interpretation from the three instruments was used. Fourteen antibiotics were recorded, which were separated into seven different categories based on the P. aeruginosa antimicrobial categories outlined by Magiorakos et al.34 with the following exceptions: the phosphonic acid and polymyxin categories were replaced with a new category encompassing the β-lactam/β-lactamase inhibitor agents, ceftazidime/avibactam and ceftolozane/tazobactam. Susceptibility to these agents was used to classify the isolates as susceptible (susceptible to all 14 agents), MDR (non-susceptible to ≥1 agent in ≥3 antimicrobial categories), XDR (non-susceptible to ≥1 agent in all but ≤2 categories), pandrug-resistant (PDR, non-susceptible to all 14 agents) or non-MDR (non-susceptible to 1 or 2 categories only) based on CLSI guidelines.35
WGS and analysis
Isolates were sequenced on an Illumina MiSeq or NextSeq benchtop sequencer (Illumina Inc., San Diego, CA, USA) and analysed as previously described,32 except that in silico MLST was performed at PubMLST36 using the scheme developed by Curran and colleagues.37 Presence or absence of the exoU and exoS type III effector genes was determined as described by Ozer et al.7 Briefly, BlastN searches of the exoU (reference nucleotide locus ID PA14_51530 in strain UCBPP-PA14) and exoS (locus ID PA3841 in strain PAO1) genes were performed using default parameters against the genomic sequences of each isolate in the diversity panel.
Refinement of the P. aeruginosa repository
To reduce redundancy in the initial 3785 isolate set, successive isolates after the first from the same patient that shared the same ST were removed unless the isolates were cultured from a different body site (e.g. blood versus urine) or were cultured >6 months apart. All isolates from the same patient with different STs were retained. This refinement resulted in a final panel of 2661 isolates (from 2085 patients) available for analysis.
cgMLST analysis and initial panel selection
The draft genomes of the 2661 isolates were uploaded and analysed using SeqSphere+ software (Ridom, Germany) using the P. aeruginosa core genome MLST (cgMLST) scheme developed by Stanton and colleagues.38 To be included in the analysis, isolates had to contain 90% of the 4400 genes included in this cgMLST scheme. The resulting minimum spanning tree (MST) was then used to select 310 strains that captured the diversity of the strain collection.
Core genome SNP and accessory genome analysis
PanSeq39 was run with a fragmentation size of 500 bp to find sequences with ≥95% identity in ≥90% of the isolates to generate the core genome SNP alignment and accessory genome binary alignment for the selected diversity set of 310 isolates. The SNP-based phylogeny was built using RAxML (version 8.2.11)40 from a 534 kb variable position alignment using the GTR GAMMA model and the rapid bootstrapping option for nucleotide sequences (100 replicates). Using this approach, 100 strains were selected to represent the final diversity panel.
Prior to phylogenetic analysis, the set of 100 isolates was re-sequenced and re-analysed to confirm purity. Reads were checked for contamination at the species level with Kraken2 (version 2.0.8-beta)41 using the standard database build and at the strain level using ConFindr (version 0.4.8)42 with parameters bf = 0.05 and q = 30. ConFindr parameters were validated for P. aeruginosa using a small subset of isolates that were deemed not contaminated by ConFindr using the default settings (q = 20). For this validation set, paired reads were randomly selected using seqtk (https://github.com/lh3/seqtk) to a normalized read total of 3 million reads. These uncontaminated isolates were then mixed in silico with another isolate from a different rMLST scheme at 0%–10%, 15%, 20%, 25% and 50% of the reads. Panel isolates with greater than 1% of their reads belonging to a different species as identified by Kraken2, or those with 3 or more single nucleotide variants as identified by ConFindr, were further purified in the laboratory and re-sequenced. These purified sequences were compared to the original genomes by phylogeny to ensure identity. Finally, phylogenies were generated as described above. Specifically, the core genome alignment was 1.7 Mb long and contained 122 kb variable positions. Genome annotations were performed using NCBI Prokaryotic Genome Annotation Pipeline (version 4.8) and were made available on GenBank (BioProject: PRJNA446057).
Analysis of chromosomal mutation-associated antibiotic resistance
No comprehensive tool, with a well-curated database, currently exists to accurately predict the mutational resistome of P. aeruginosa. In an attempt to identity the most well-characterized mutations, the existing literature was examined to obtain a non-exhaustive list of most frequently observed 151 amino acid substitutions in a set of 55 P. aeruginosa chromosomal genes that have previously been shown to be involved in antibiotic resistance19,43–47. For each gene, the corresponding protein sequences from the 100 P. aeruginosa panel isolates were aligned using MUSCLE48 and the presence of known variants (including novel substitutions at known sites) was individually recorded (Figure 3 and Table S1).
Figure 3.
Characteristics of the P. aeruginosa diversity panel. Core genome SNP-based phylogenetic tree of the 100 strains in the final diversity panel. Strain ST is indicated. The assigned antimicrobial resistance (AMR) phenotype (see the text for details) is provided, and the dark red-brown squares indicate a result of non-susceptible (filled) or susceptible (open) to the tested antibiotic. The grey circles indicate the presence of a known resistance gene or mutation, with the blue-grey colour highlighting those gene families of particular interest. For each isolate, the specific effector/genotype (exoU+ or exoS+) of the type III secretion system are indicated. AMK, amikacin; GEN, gentamicin; TOB, tobramycin; ATM, aztreonam; CAZ, ceftazidime; FEP, cefepime; TZP, piperacillin/tazobactam; TIM, ticarcillin/clavulanic acid; CZA, ceftazidime/avibactam; C/T, ceftolozane/tazobactam; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; LVX, levofloxacin.
Diversity panel availability
The final diversity panel has been deposited at BEI resources (https://www.beiresources.org/) and is currently available for research purposes under catalogue # NR-51829.
Results
Strain diversity and initial isolate selection
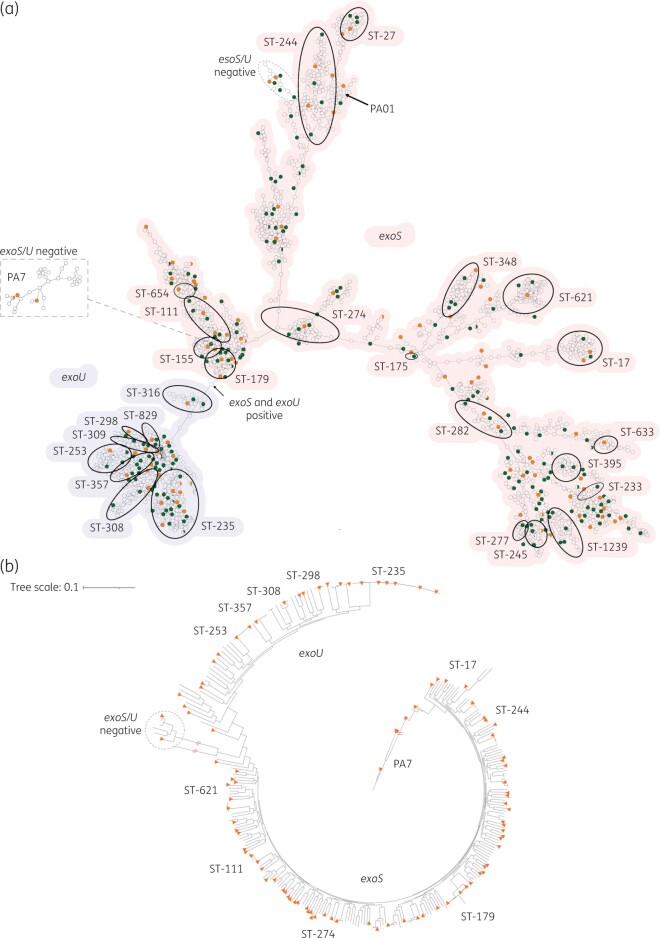
All 2661 de-duplicated P. aeruginosa passed the 90% gene threshold stipulated by cgMLST and were used to generate an MST (Figure 1a). The isolates were separated to a high degree of accuracy into two major clades by the presence of the exoS or exoU genes, consistent with the findings of Ozer et al.7 One minor clade, consisting of 40 distantly related isolates that belong to the PA7 taxonomic outlier,16 was readily apparent while a second smaller group, consisting of 15 genetically divergent isolates, was only identified using phylogenetic analysis (Figure 1b). Notably, all 40 isolates from the PA7 clade and the 15 isolates from the smallest clade lack both exoS and exoU. Finally, the location of P. aeruginosa PA01, a well-studied and frequently used strain belonging to ST-549, is indicated. This strain is exoS+/exoU− and represents the only ST-549 in our collection.
Figure 1.
Genetic diversity of P. aeruginosa in the MRSN collection. (a) cgMLST MST of the 2661 P. aeruginosa genomes used for initial panel selection. Isolates with an identical cgMLST allelic profile are represented by a single circle. For increased clarity, highly divergent isolates within the PA7 clade are displayed on a separate dendrogram wherein predicted root placement in the global population was indicated in image post-processing. The most prevalent and/or important ST (from traditional MLST typing) are indicated. The presence of exoS (red shading) or exoU (blue) is indicated for the two main clades. The absence of both alleles is noted for the PA7 and a second smaller clade. The 310 isolates initially selected to represent the breadth of diversity (see the text for details) are shown in green and orange with orange indicating the 100 strains in the final panel. (b) Core-genome SNP-based phylogenetic tree of 310 P. aeruginosa. The 100 isolates selected for the final panel are indicated with orange triangles. STs are indicated as well as the major clade designations.
Traditional MLST assigned 2304 isolates to 407 known STs and the remaining 357 isolates to 274 novel STs. Seven of the top 10 most common STs in the repository have previously been detected in at least three countries21 including ST-235 (9.7%; the most common clone), ST-244 (3.76%), ST-179 (2.86%), ST-253 (2.86%), ST-111 (2.82%), ST-17 (2.22%) and ST-274 (2.07%). Many other recognized clonal groups were also represented (Figure 1a).
Selection of a non-redundant, genetically diverse panel of P. aeruginosa
The MST was used to select a subset of 310 strains that best represented the breadth and diversity of the overall strain collection (Figure 1). This subset encompassed 193 known STs as well as 38 strains with unique and novel STs whilst retaining the temporal and geographic distribution of the full strain collection. All 310 isolates were compared at the highest resolution using a SNP-based phylogenetic tree (Figure 1b). As illustrated, the overall population structure was retained (i.e. two large exoU and exoS clades as well as smaller PA7 and exoU/S negative clades) confirming the initial selection method using cgMLST. Ultimately, this phylogeny was used to select the final panel of 100 strains, chosen to maximize genetic diversity (Figure 1b).
The selected 100 strains were cultured from a wide range of clinical samples between 2003 and 2017, with the majority (n = 97) collected from hospitals across the USA (Table S1). Genetic diversity was high, with representatives from 91 different STs, including distinct isolates from the clinically important clones ST-111, ST-235, ST-244 and ST-253. All permutations of the exoU and exoS gene were represented; 72 isolates carried exoS (including MRSN 11278, which otherwise clustered with isolates from the exoU clade in the phylogeny), 21 carried exoU, 2 carried both and 5 isolates lacked either gene (Table S1).
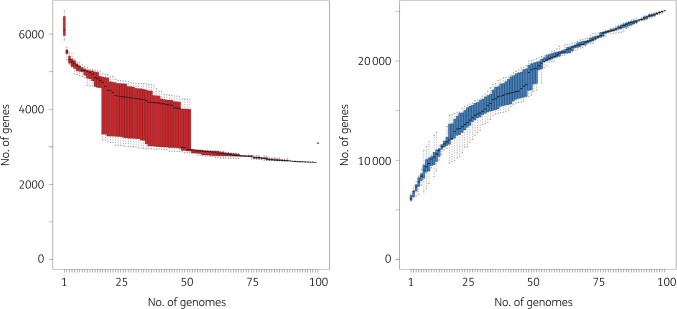
This substantial diversity is also reflected in the gene content, where just 3200 core genes are shared by all 100 isolates, while >25 000 distinct genes are represented in the pan-genome (Figure 2). Furthermore, the size of the pan-genome never plateaus, highlighting the low redundancy and large amount of gene diversity in the panel.
Figure 2.
Core and pan-genome of 100 diverse P. aeruginosa. Gene rarefaction (left, core genome) and accumulation (right, pan-genome) curves are provided with the number of genes (y-axis) as a function of the number of genomes (x-axis), from a random sample (variation indicated as boxplot) of all genomes. Core genome is defined as the number of genes found in 99% of the genomes.
Antibiotic susceptibility and antibiotic resistance mechanisms
Though the primary aim of the panel was to provide a collection of isolates with extensive genetic diversity, having comprehensive antibiotic susceptibility data for each isolate would be valuable. Phenotypically, the selected isolates composing the diversity panel encompass a diverse range of susceptibilities; 20 isolates were susceptible to all 14 agents, 23 were non-MDR, 12 were MDR, 44 were XDR and 1 isolate (MRSN 6220) was resistant to all 14 antibiotics (PDR) (Figure 3, Table S1). Overall, 71 distinct antibiotic susceptibility profiles were identified, including susceptible and non-susceptible strains across all seven categories. Notably, 64 isolates were non-susceptible to one or both carbapenems tested (imipenem and meropenem) and 19 and 12 isolates were non-susceptible to the newer β-lactam/β-lactamase combinations ceftazidime/avibactam and ceftolozane/tazobactam, respectively (Figure 3, Table S1).
Genetically, 88 ARGs were identified among the 100 isolates (Table S1), but this included 17 distinct alleles of the intrinsic blaOXA-50-like gene, 34 distinct alleles of the intrinsic blaPAO genes and 15 distinct alleles encoding aminoglycoside modifying enzymes (AMEs) (Table S1). In addition to the two intrinsic β-lactamases, all isolates carry the kanamycin resistance gene aph(3’)-IIb and the chloramphenicol resistance gene catB7, while 99 of the 100 isolates carry the fosfomycin resistance gene fosA. Remarkably, 71 isolates carry only these five intrinsic genes but exhibit antibiotic susceptibility profiles ranging from pan-susceptible to XDR indicating a vast array of acquired resistances most likely caused by spontaneous mutations. (Figure 3, Table S1).
Aminoglycoside resistance
Only six isolates were non-susceptible (i.e. resistant or intermediate) to all three aminoglycosides tested. In addition, 14 were susceptible to amikacin but non-susceptible to gentamicin and tobramycin, 12 were resistant to gentamicin alone, 2 were resistant to tobramycin alone and the remaining 66 were susceptible to all three. Overall, resistance to aminoglycosides correlated well with the presence of AME genes. For example, specific alleles of aminoglycoside N-acetyltransferases [e.g. aac(6)-Ib conferring resistance to amikacin (low-level) and tobramycin] and aminoglycoside O-nucleotidyltransferases (e.g. ant(2”)-Ia conferring resistance to gentamicin and tobramycin) were identified in 21 non-susceptible isolates (Figure 3, Table S1). Similarly, isolates susceptible to all aminoglycosides lacked these enzymes.
Thirteen isolates displayed variable aminoglycoside resistance in the absence of known AMEs, including 11 non-susceptible to gentamicin, 1 non-susceptible to tobramycin and 1 non-susceptible to all three (MRSN 20190). In the absence of acquired aminoglycoside-modifying enzymes, aminoglycoside resistance in P. aeruginosa has been linked to multiple mechanisms, but mutational overexpression of the efflux pump system MexXY-OprM is the most common.19,49 In this study, a non-exhaustive search for mutations in mexZ and/or parRS that have previously been linked to MexXY-OprM overexpression and aminoglycoside resistance was performed. A loss-of-function mutation in MexZ was identified in two gentamicin and tobramycin non-susceptible strains (MRSN 16344 and MRSN 20190), and a parS mutation was identified in gentamicin non-susceptible MRSN 12368, but the mechanism(s) underlying aminoglycoside resistance in the remaining 10 strains remains to be determined.
Carbapenem resistance
Of high clinical importance, 48 isolates were non-susceptible to both carbapenems (imipenem and meropenem) and 14 were non-susceptible to imipenem alone (Figure 3, Table S1). Notably, just three isolates carried an acquired carbapenemase (blaVIM-11 in MRSN 20176, blaVIM-6 in MRSN 6220 and blaKPC-2 in MRSN 23861) and one, MRSN 9873, carried a gene that shared 92% nucleotide identity over 98% of the gene length with blaHMB-1, a rare metallo-β-lactamase originally described in a clinical strain of P. aeruginosa in 2017.50 Besides gene acquisition, a total of 40 strains carried a loss-of-function mutation in the outer membrane protein OprD, by far the most common mechanism for carbapenem resistance in this species51 and a known modulator of colonization and virulence in a mouse model.52 A known mutation in ParS and loss-of-function mutation in MexZ were identified as the possible cause of carbapenem resistance in an additional 4 isolates while the mechanism(s) of carbapenem resistance remains undetermined for 16 isolates (Figure 3, Table S1).
Fluoroquinolone resistance
From the diversity panel, 53 strains were non-susceptible to both fluoroquinolones (FQs) (ciprofloxacin and levofloxacin), 2 were non-susceptible to just ciprofloxacin, 3 were non-susceptible to levofloxacin only and the remaining 42 were susceptible to both (Figure 3, Table S1). In just two isolates, MRSN 1906 and MRSN 8130, FQ resistance could be attributed to the acquired FQ resistance genes qnrA and aac(6)-Ib-cr5, respectively. This is consistent with the observation that high-level FQ resistance in P. aeruginosa is primarily driven by mutation, particularly mutations in the QRDRs of DNA gyrase (GyrA and GyrB) and topoisomerase IV (ParC and/or ParE) and/or overexpression of the MexEF-OprN or MexCD-OprJ efflux pumps.19,53 A non-exhaustive search for mutations known to cause FQ resistance in P. aeruginosa identified strains with mutations in gyrA (n = 29), gyrB (=3), parC (n = 12) and mexR (n = 6). However, the mechanism(s) for the observed resistance to ciprofloxacin and/or levofloxacin in an additional 22 strains remains to be determined (Figure 3, Table S1).
Discussion
WHO and the US CDC have classified MDR and carbapenem-resistant P. aeruginosa as one of the most serious antibiotic resistant threats.29,30 These classifications highlight the clinical importance of P. aeruginosa and the increasing difficulty faced by clinicians in treating infections by this organism. Fortunately, this recognition has spurred renewed interest in developing effective countermeasures, including vaccines, antimicrobial peptides, antibodies, virulence inhibitors, bacteriophages and novel antimicrobials.54,55
A key component in evaluating any new therapeutic or diagnostic is access to a diverse set of strains that capture the genetic diversity within a species. Access to a diverse source of strains can be difficult and as a result there is very little consistency in the strains used across studies. The urgency of this requirement has been recognized at the highest level of the US Government, with the US National Action Plan for Combating Antibiotic-Resistant Bacteria (CARB) indicating the need for a ‘a specimen repository to facilitate development and evaluation of diagnostic tests and treatments’. Notably, the recently released CARB 2020-2025 document directs Federal Agencies to ‘continue expanding and improving access to specimen and data repositories for research and innovation’.56
Since the publication of the first CARB national strategy and action plan in 2015, US Federal agencies have strived to develop appropriate specimen repositories. For P. aeruginosa, the US CDC and FDA have developed a useful panel of 55 P. aeruginosa isolates that were chosen to represent a diversity of AST results for drugs that are used to treat infections (https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=12). Notably, the strains carry a variety of antibiotic resistance genes, including the potent carbapenemases blaIMP, blaKPC, blaNDM and blaVIM. While this is a valuable resource for studying strains with specific antibiotic resistance patterns, there are no data on the phylogeny of the strains and how representative they are of the overall genetic diversity within this species. In 2013, De Soyza et al.57 created a panel of 43 diverse P. aeruginosa strains selected from 955 genotyped isolates by a team of clinical and research experts. Importantly, a subsequent study provided extensive phenotypic characterization of these isolates (including antimicrobial and phage susceptibilities, mucoidy, motility, biofilm formation and virulence in an insect model).58 However, no comprehensive genomic characterization was performed (e.g. no identification of resistance alleles) and the typing methodology used (ArrayTube) cannot be cross-referenced to widely used typing schemes, such as traditional MLST, to easily identify isolates from major epidemic lineages. In the intervening 8 years, newly introduced therapeutics (e.g. ceftazidime/avibactam and ceftolozane/tazobactam) and accrued knowledge on the genomic epidemiology and population structure of P. aeruginosa21 prompted the need for a comprehensive panel of isolates and genomes for research and development efforts. Finally, the PA01 strain has been used widely in studies, but our data indicate that it belongs to the very rare ST-549 and is not a good genetic representative of the species.
In an effort to address these limitations and provide a new panel that is complementary to these existing sets, the large repository of P. aeruginosa clinical isolates present in the MRSN was leveraged. These strains have been collected over the past 11 years from clinical samples around the world (Figure S1) and bolstered by historical strains stored in US Military Treatment Facility archives. Though the majority of strains were collected in the USA, it is notable that those from other continents display a similar distribution, a likely consequence of the successful spread of high-risk clones across the globe (Figure S1). In addition to providing representatives of the major high-risk clones, including ST-111, ST-235 and ST-175, the panel also includes emerging clones of concern such as ST-244, ST-357 and ST-65420 as well as sporadic clones that capture the genetic diversity across all major clades (including taxonomic outlier PA07) previously identified for this species.6–8,16
Though the panel was designed toward maximizing genetic diversity, the selected strains of P. aeruginosa strains encompass a diverse range of antibiotic susceptibility patterns from pan-susceptible to pan-resistant when tested against 14 relevant antibiotics (Table S1). Unlike the MRSN’s panel of diverse A. baumannii isolates, only a minor correlation between antibiotic susceptibility and the presence of transferable ARGs in P. aeruginosa was observed. This is a hallmark of P. aeruginosa and reflects the outsized role played by point mutations in driving antibiotic resistance in this species.53 Although non-exhaustive, literature reviews were performed to identify, within the panel isolates, an array of mutations known to contribute to aminoglycoside, carbapenem and fluoroquinolone resistance.18,19,22,43,45,47,49,51,53 As a result, only 10%–22% of the strains are phenotypically resistant due to unresolved genetic determinants. Finally, the panel includes strains with variable resistances to more contemporary agents, such as ceftazidime/avibactam and ceftolozane/tazobactam, which will be a valuable resource for investigating resistance mechanisms or developing the next generation of countermeasures. Altogether, this wide representation of resistance mechanisms (and combinations thereof) is the direct, desired consequence of the methodology used for selecting strains composing this panel.
In summary, an extensive collection of over 3500 P. aeruginosa was utilized to design a novel panel of 100 diverse strains that encompasses the genetic diversity of this species and captures both major global epidemic clones and sporadic strains. Furthermore, though not a primary aim of the endeavour, the panel also displays diverse resistance mechanisms and antibiotic susceptibility profiles when tested against a panel of 14 relevant antibiotics. The expectation is that easy access to the strains combined with high-quality draft genomes will facilitate multiple avenues of research into this critical human pathogen.
Supplementary Material
Acknowledgements
We acknowledge the participation of clinical laboratories across the Military Healthcare System, GEIS-affiliated overseas laboratories and numerous collaborators for their contributions of bacteria to the MRSN Pseudomonas aeruginosa repository.
Funding
This study was funded by the US Army Medical Command and the Defense Medical Research and Development Program.
Transparency declarations
None to declare.
Disclaimer
The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army or the Department of Defense.
Supplementary data
Figure S1 and Table S1 are available as Supplementary data at JAC-AMR Online.
References
- 1. Iglewski BH. Pseudomonas In: Baron S, ed. Medical Microbiology. University of Texas Medical Branch at Galveston, 1996. [PubMed] [Google Scholar]
- 2. Rice LB. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J Infect Dis 2008; 197: 1079–81. [DOI] [PubMed] [Google Scholar]
- 3. Driscoll JA, Brody SL, Kollef MH.. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007; 67: 351–68. [DOI] [PubMed] [Google Scholar]
- 4. Kerr KG, Snelling AM.. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect 2009; 73: 338–44. [DOI] [PubMed] [Google Scholar]
- 5. Lund-Palau H, Turnbull AR, Bush A. et al. Pseudomonas aeruginosa infection in cystic fibrosis: pathophysiological mechanisms and therapeutic approaches. Expert Rev Respir Med 2016; 10: 685–97. [DOI] [PubMed] [Google Scholar]
- 6. Pirnay JP, Bilocq F, Pot B. et al. Pseudomonas aeruginosa population structure revisited. PLoS One 2009; 4: e7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ozer EA, Nnah E, Didelot X. et al. The population structure of Pseudomonas aeruginosa is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol Evol 2019; 11: 1780–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freschi L, Vincent AT, Jeukens J. et al. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol 2019; 11: 109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hilker R, Munder A, Klockgether J. et al. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol 2015; 17: 29–46. [DOI] [PubMed] [Google Scholar]
- 10. Rutherford V, Yom K, Ozer EA. et al. Environmental reservoirs for exoS+ and exoU+ strains of Pseudomonas aeruginosa. Environ Microbiol Rep 2018; 10: 485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiehlmann L, Cramer N, Tummler B.. Habitat-associated skew of clone abundance in the Pseudomonas aeruginosa population. Environ Microbiol Rep 2015; 7: 955–60. [DOI] [PubMed] [Google Scholar]
- 12. Finck-Barbancon V, Goranson J, Zhu L. et al. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 1997; 25: 547–57. [DOI] [PubMed] [Google Scholar]
- 13. Pena C, Cabot G, Gomez-Zorrilla S. et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Dis 2015; 60: 539–48. [DOI] [PubMed] [Google Scholar]
- 14. Schulert GS, Feltman H, Rabin SD. et al. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J Infect Dis 2003; 188: 1695–706. [DOI] [PubMed] [Google Scholar]
- 15. Shaver CM, Hauser AR.. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun 2004; 72: 6969–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy PH, Tetu SG, Larouche A. et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 2010; 5: e8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Medina-Rojas M, Stribling W, Snesrud E. et al. Comparison of Pseudomonas aeruginosa strains reveals that Exolysin A toxin plays an additive role in virulence. Pathog Dis 2020; 78: ftaa010. [DOI] [PubMed] [Google Scholar]
- 18. Botelho J, Grosso F, Peixe L.. Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist Updat 2019; 44: 100640. [DOI] [PubMed] [Google Scholar]
- 19. Lopez-Causape C, Cabot G, Del Barrio-Tofino E. et al. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol 2018; 9: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Del Barrio-Tofino E, Lopez-Causape C, Oliver A.. Pseudomonas aeruginosa epidemic high-risk clones and their association with horizontally-acquired β-lactamases: 2020 update. Int J Antimicrob Agents 2020; 56: 106196. [DOI] [PubMed] [Google Scholar]
- 21. Oliver A, Mulet X, Lopez-Causape C. et al. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat 2015; 21–22: 41–59. [DOI] [PubMed] [Google Scholar]
- 22. Potron A, Poirel L, Nordmann P.. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 2015; 45: 568–85. [DOI] [PubMed] [Google Scholar]
- 23. Treepong P, Kos VN, Guyeux C. et al. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 2018; 24: 258–66. [DOI] [PubMed] [Google Scholar]
- 24. Miyoshi-Akiyama T, Tada T, Ohmagari N. et al. Emergence and spread of epidemic multidrug-resistant Pseudomonas aeruginosa. Genome Biol Evol 2017; 9: 3238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright LL, Turton JF, Livermore DM. et al. Dominance of international ‘high-risk clones’ among metallo-β-lactamase-producing Pseudomonas aeruginosa in the UK. J Antimicrob Chemother 2015; 70: 103–10. [DOI] [PubMed] [Google Scholar]
- 26. Brown ED, Wright GD.. Antibacterial drug discovery in the resistance era. Nature 2016; 529: 336–43. [DOI] [PubMed] [Google Scholar]
- 27. Hutchings MI, Truman AW, Wilkinson B.. Antibiotics: past, present and future. Curr Opin Microbiol 2019; 51: 72–80. [DOI] [PubMed] [Google Scholar]
- 28. Rossolini GM, Arena F, Pecile P. et al. Update on the antibiotic resistance crisis. Curr Opin Pharmacol 2014; 18: 56–60. [DOI] [PubMed] [Google Scholar]
- 29. WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. 2017. https://apps.who.int/iris/handle/10665/311820.
- 30. CDC, USA. Antibiotic Resistance Threats in the United States: 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- 31. Dolan SK. Current knowledge and future directions in developing strategies to combat Pseudomonas aeruginosa infection. J Mol Biol 2020; 432: 5509–28. [DOI] [PubMed] [Google Scholar]
- 32. Galac MR, Snesrud E, Lebreton F. et al. A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob Agents Chemother 2020; 64: e00840–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waterman P, Kwak Y, Clifford R. et al. A multidrug-resistance surveillance network: 1 year on. Lancet Infect Dis 2012; 12: 587–8. [DOI] [PubMed] [Google Scholar]
- 34. Magiorakos AP, Srinivasan A, Carey RB. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 35. CLSI. Performance Standards for Antimicrobial Susceptibility Testing—Thirty-First Edition: M100. 2021.
- 36. Jolley KA, Bray JE, Maiden MCJ.. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 2018; 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Curran B, Jonas D, Grundmann H. et al. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 2004; 42: 5644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stanton RA, McAllister G, Daniels JB. et al. Development and application of a core genome multilocus sequence typing scheme for the health care-associated pathogen Pseudomonas aeruginosa. J Clin Microbiol 2020; 58: e00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laing C, Buchanan C, Taboada EN. et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 2010; 11: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wood DE, Lu J, Langmead B.. Improved metagenomic analysis with Kraken 2. Genome Biol 2019; 20: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Low AJ, Koziol AG, Manninger PA. et al. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ 2019; 7: e6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cabot G, Lopez-Causape C, Ocampo-Sosa AA. et al. Deciphering the resistome of the widespread Pseudomonas aeruginosa sequence type 175 international high-risk clone through whole-genome sequencing. Antimicrob Agents Chemother 2016; 60: 7415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Domitrovic TN, Hujer AM, Perez F. et al. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis 2016; 3: ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez-Causape C, Sommer LM, Cabot G. et al. Evolution of the Pseudomonas aeruginosa mutational resistome in an international cystic fibrosis clone. Sci Rep 2017; 7: 5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sherrard LJ, Tai AS, Wee BA. et al. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One 2017; 12: e0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sobel ML, Neshat S, Poole K.. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J Bacteriol 2005; 187: 1246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poole K. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49: 479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pfennigwerth N, Lange F, Belmar Campos C. et al. Genetic and biochemical characterization of HMB-1, a novel subclass B1 metallo-β-lactamase found in a Pseudomonas aeruginosa clinical isolate. J Antimicrob Chemother 2017; 72: 1068–73. [DOI] [PubMed] [Google Scholar]
- 51. Rodriguez-Martinez JM, Poirel L, Nordmann P.. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53: 4783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Skurnik D, Roux D, Cattoir V. et al. Enhanced in vivo fitness of carbapenem-resistant oprD mutants of Pseudomonas aeruginosa revealed through high-throughput sequencing. Proc Natl Acad Sci U S A 2013; 110: 20747–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011; 2: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Burrows LL. The therapeutic pipeline for Pseudomonas aeruginosa infections. ACS Infect Dis 2018; 4: 1041–7. [DOI] [PubMed] [Google Scholar]
- 55. Merakou C, Schaefers MM, Priebe GP.. Progress toward the elusive Pseudomonas aeruginosa vaccine. Surg Infect (Larchmt) 2018; 19: 757–68. [DOI] [PubMed] [Google Scholar]
- 56. Office of Science & Data Policy. National Action Plan for Combating Antibiotic-Resistant Bacteria, 2020-2025. 2020. https://aspe.hhs.gov/reports/national-action-plan-combating-antibiotic-resistant-bacteria-2020-2025.
- 57. De Soyza A, Hall AJ, Mahenthiralingam E. et al. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2013; 2: 1010–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cullen L, Weiser R, Olszak T. et al. Phenotypic characterization of an international Pseudomonas aeruginosa reference panel: strains of cystic fibrosis (CF) origin show less in vivo virulence than non-CF strains. Microbiology (Reading) 2015; 161: 1961–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.