Abstract
PRDM9 is a DNA-binding histone methyltransferase that designates and activates recombination hotspots in mammals by locally trimethylating lysines 4 and 36 of histone H3. In mice, we recently reported two independently produced point mutations at the same residue, Glu360Pro (Prdm9EP) and Glu360Lys (Prdm9EK), which severely reduce its H3K4 and H3K36 methyltransferase activities in vivo. Prdm9EP is slightly less hypomorphic than Prdm9EK, but both mutations reduce both the number and amplitude of PRDM9-dependent H3K4me3 and H3K36me3 peaks in spermatocytes. While both mutations cause infertility with complete meiotic arrest in males, Prdm9EP, but not Prdm9EK, is compatible with some female fertility. When we tested the effects of these mutations in vitro, both Prdm9EP and Prdm9EK abolished H3K4 and H3K36 methyltransferase activity in full-length PRDM9. However, in the isolated PRDM9 PR/SET domain, these mutations selectively compromised H3K36 methyltransferase activity, while leaving H3K4 methyltransferase activity intact. The difference in these effects on the PR/SET domain vs the full-length protein shows that PRDM9 is not an intrinsically modular enzyme; its catalytic domain is influenced by its tertiary structure and possibly by its interactions with DNA and other proteins in vivo. These two informative mutations illuminate the enzymatic chemistry of PRDM9, and potentially of PR/SET domains in general, reveal the minimal threshold of PRDM9-dependent catalytic activity for female fertility, and potentially have some practical utility for genetic mapping and genomics.
Keywords: PRDM9, PR/SET domain, point mutations, enzyme activity, infertility phenotype
Introduction
Genetic recombination, the process by which genetic information is exchanged between homologous chromosomes, is both an important source of genetic diversity and a necessary step for successful meiosis in most organisms. In many taxa, recombination events are restricted to specific sites in the genome known as recombination hotspots. Many mammalian species, including mice and humans, utilize the specialized histone methyltransferase (HMT) PRDM9 to determine the locations of hotspots and to activate them for the recombination process (Paigen and Petkov 2018). In the PRDM9-mediated system, hotspots are restricted to the cognate binding sites of PRDM9 and subsequently marked by locally trimethylating lysines 4 and 36 of histone H3 (Hayashi et al. 2005; Baudat et al. 2010; Berg et al. 2010; Myers et al. 2010; Parvanov et al. 2010; Baker et al. 2014; Powers et al. 2016). This double histone mark creates a unique epigenetic signature that differentiates hotspots from H3K4me3-enriched promoters and regulatory elements, and from the H3K36me3-enriched bodies of actively transcribed genes.
PRDM9 was previously thought to be necessary for successful meiosis in mammals, largely because C57BL/6J (B6) Prdm9 knock-out mice are infertile with meiotic arrest (Hayashi et al. 2005), and also because PRDM9 mutations have been associated with azoospermia in humans (Miyamoto et al. 2008; Irie et al. 2009). The catalytic activity of PRDM9 was likewise considered necessary for fertility, as B6 mice homozygous for a methyltransferase-dead Prdm9 allele are also infertile (Diagouraga et al. 2018). However, exceptions to this rule have accumulated since the discovery of Prdm9. Male mice of the Mus musculus musculus-derived inbred strain PWD/Ph produce small amounts of mature sperm in the absence of Prdm9 (Mihola et al. 2019). Canines manage to carry out meiosis without impediment, despite pseudogenization of Prdm9 in that lineage (Munoz-Fuentes et al. 2011; Auton et al. 2013). Both male and female Norway rats remain fertile in the absence of Prdm9, though their fertility is reduced relative to wild-type (Mihola et al. 2021). And notably, there is at least one case of a fertile PRDM9-null human female (Narasimhan et al. 2016).
The structure of PRDM9 appears to be modular: it has three well-defined and well-separated functional domains—KRAB, PR/SET, and a tandem zinc finger (ZnF) array. The KRAB domain is involved in protein–protein interactions (Imai et al. 2017; Parvanov et al. 2017; Diagouraga et al. 2018). The PR/SET domain uniquely trimethylates histone H3 at both lysines 4 and 36 (Hayashi et al. 2005; Baker et al. 2014; Powers et al. 2016). The ZnF domain binds DNA in a sequence-specific manner (Brick et al. 2012; Billings et al. 2013; Walker et al. 2015; Smagulova et al. 2016). Consistent with its apparently modular structure, the PRDM9 catalytic PR/SET domain and its DNA-binding ZnF domain retain their functions when isolated from the rest of the protein in vitro, suggesting that these domains are autonomous (Billings et al. 2013; Wu et al. 2013; Eram et al. 2014; Koh-Stenta et al. 2014; Walker et al. 2015; Powers et al. 2016). Inactivating mutations in either domain cause sterility in mutant mice, emphasizing the importance of each domain for the overall function of PRDM9 (Parvanov et al. 2017; Diagouraga et al. 2018).
In mice, we recently reported sexual dimorphism in the requirement for PRDM9 for successful meiosis (Powers et al. 2020). While both male and female B6 and C3H/HeJ (C3H) mice are infertile in the absence of Prdm9, female Prdm9-null CAST/EiJ (CAST) mice are fully fertile and produce healthy offspring, although male Prdm9-null CAST mice are infertile. We also reported two catalytic mutations—Prdm9EP and Prdm9EK—that straddle the threshold for female fertility in B6 mice (Bhattacharyya et al. 2019; Powers et al. 2020). These mutations substitute glutamate 360, a residue situated within the PR/SET domain substrate-binding groove, with proline and lysine, respectively. In vivo, both mutations result in a catalytically hypomorphic PRDM9 protein that is aptly described as “methyltransferase-comatose”; both H3K4 and H3K36 methyltransferase activities are severely reduced, but not completely abolished. Prdm9EK encodes a slightly less active protein than Prdm9EP, and this results in a sex-specific fertility phenotype. Males homozygous for either mutation exhibit a phenocopy of the Prdm9-null and Prdm9-methyltransferase-dead conditions: infertility with complete meiotic arrest in the pachytene stage of prophase I. Homozygous female Prdm9EK mice are also infertile (Bhattacharyya et al. 2019). In contrast, homozygous female Prdm9EP mice are able to produce variable numbers of grossly healthy, normal offspring via natural matings (Powers et al. 2020). These observations show that oogenesis is robust to less PRDM9-dependent catalytic activity than spermatogenesis, even on the B6 genetic background.
In this paper, we characterize and contrast Prdm9EP and Prdm9EK, and present experimental data demonstrating the disparate effects of these mutations on full-length PRDM9 vs its isolated PR/SET domain. We show that both mutations severely reduce or abolish the catalytic activity of full-length PRDM9 in vivo and in vitro. However, the two mutations show separation-of-function in the context of the PR/SET domain alone in vitro. Recombinant Prdm9EP and Prdm9EK PR/SET domains show largely uncompromised H3K4 trimethylation activity, while H3K36 trimethylation activity is reduced by the Prdm9EK mutation and abolished by the Prdm9EP mutation. This observation shows that PRDM9 is not truly modular, as Prdm9EP and Prdm9EK have different effects on the full-length protein vs the isolated PR/SET domain. We discuss the implications of these mutations for different aspects of PRDM9 function—enzyme chemistry, the differential importance for female vs male meiosis, and its evolution.
Materials and methods
Ethics statement
The animal care rules used by The Jackson Laboratory are compatible with the regulations and standards of the U.S. Department of Agriculture and the National Institutes of Health. The protocols used in this study were approved by the Animal Care and Use Committee of The Jackson Laboratory (summary number 04008). Euthanasia was carried out by cervical dislocation.
Mouse strains
Mice used in this study were acquired from The Jackson Laboratory. The following strains were used: C57BL/6J (stock number 000664), C57BL/6J-Prdm9<em2Kpgn>/Kpgn (Prdm9EP), and C57BL/6J-Prdm9<em2Kpgn>/Kpgn (Prdm9EK). The latter two were generated by introducing a point mutation (Glu360Pro) via CRISPR-Cas9 gene editing, onto the C57BL/6J genetic background, as previously described (Bhattacharyya et al. 2019; Powers et al. 2020). Gene editing was performed by the Genetic Engineering Technology core at The Jackson Laboratory.
Fertility tests
Each Prdm9EK/EK female (5 weeks old) was housed with fertile, sexually mature (>8 weeks old) wild-type B6 male for a period of at least 3 and up to 7 months. A female was considered fertile if she gave live birth at least once. Litter size was determined by counting pups on the day of birth.
PRDM9 PR/SET domain synthesis and cloning
The genetic sequence encoding amino acids 187–372 of the mouse reference sequence of PRDM9 was synthesized and cloned into the pBAD-HisB expression vector, using the GenScript commercial service. In the case of Prdm9EP and Prdm9EK, the codon for glutamate 360 was altered to encode proline and lysine, respectively. Sanger sequencing confirmed that the sequences were correct, and that there were no off-target mutations.
Site-directed mutagenesis and cloning of full-length PRDM9
To induce the Prdm9EP and Prdm9EK mutations in full-length PRDM9, we used site-directed mutagenesis with a wild-type cDNA version of human PRDM9 (allele C) that we had previously cloned into pBAD-HisB. We performed site-directed mutagenesis using the QuikChange II kit (Agilent), as per the manufacturer’s instructions. Sanger sequencing confirmed that the sequences were correct, and that there were no off-target mutations.
Protein expression
To facilitate expression and purification, the sequences encoding both the PR/SET domains and full-length PRDM9 were transferred into the pMAL-c6T (New England Biolabs number N0378) vector, which contains a maltose-binding (MBP) tag. The constructs were transformed into NEB stable cells (NEB C3040I). Plasmid DNA was obtained from individual colonies and sequenced to find the correct clone. Plasmids were then transformed into Rosetta II cells (Sigma -71402-4) and grown on agar plates with 100 mg/ml ampicillin overnight at 37°C.
Single colonies of each clone were used for overnight expression culture in 100 ml in LB with 100 mg/ml ampicillin at 30°C. No inducer was added because expression is leaky and allows production of enough protein for further analysis.
Purification and HMT assays
Cell lysis, protein isolation, and HMT assays were done as previously described (Powers et al. 2016). Briefly, cells were lysed in 1× cell breakage buffer, consisting of 500 mM Tris–HCl pH 7.5, 0.15 M sucrose, 0.1% NP-40, 100 mM KCl, and Roche EDTA-free complete protein inhibitors (cat. number 04693132001) and sonicated for 1–2 min on ice. The supernatant was cleared by centrifugation at 12,000×g for 20 min. The expressed proteins were partially purified on Q sepharose (Sigma q1126) and the fractions containing the protein of interest were collected. Western blotting was done using an iBlot dry transfer system, as per the manufacturer’s instructions. Primary antibodies are identified and used in concentrations as follows: α-H3K4me3: 1:5000, EMD Millipore cat. number 07-473; α-H3K36me3: 1:5000, Active Motif cat. number 61101; α-Histone H3: 1:5000, Active Motif cat. number 61277. The secondary antibody used with all three primary antibodies was HRP-conjugated goat antirabbit (Bio-Rad cat. number 1662408EDU). Blots were imaged on a SynGene gel imaging system after a 1 min exposure to SuperSignal Femto West Maximum Sensitivity Substrate (ThermoFisher), and band density was quantified using GeneTools software. Three replicate HMT assays were done for each PR/SET domain and full-length PRDM9 protein; their means and standard deviations are plotted in Figures 1 and 2.
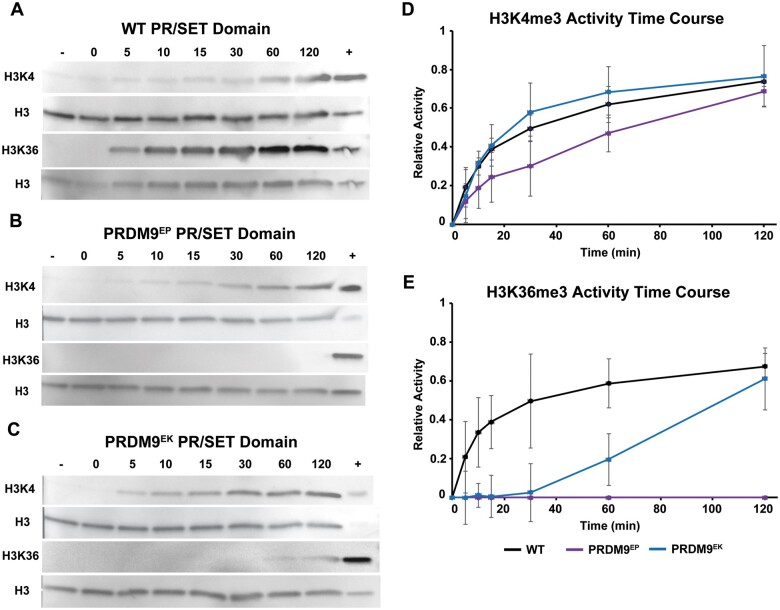
Figure 1.
HMT activities of wild-type, PRDM9EP, and PRDM9EK PR/SET domains. (A–C) Representative western blots showing the results of HMT assays. Blots were probed with α-H3K4me3 and α-H3K36me3 antibodies to show the levels of these modified histones at the denoted time points (minutes) as the assay proceeded. Negative control lanes (−) represent reactions run without s-adenosyl methionine (SAM), while positive control lanes (+) represent purified H3K4me3 or H3K36me3, as appropriate. Blots were stripped and reprobed with α-histone H3 antibody as loading controls; these are shown below their respective counterparts. Time course plots summarizing HMT activities for H3K4me3 (D) and H3K36me3 (E), quantified by band densitometry. Data points represent the mean of three replicates; error bars represent the standard deviations.
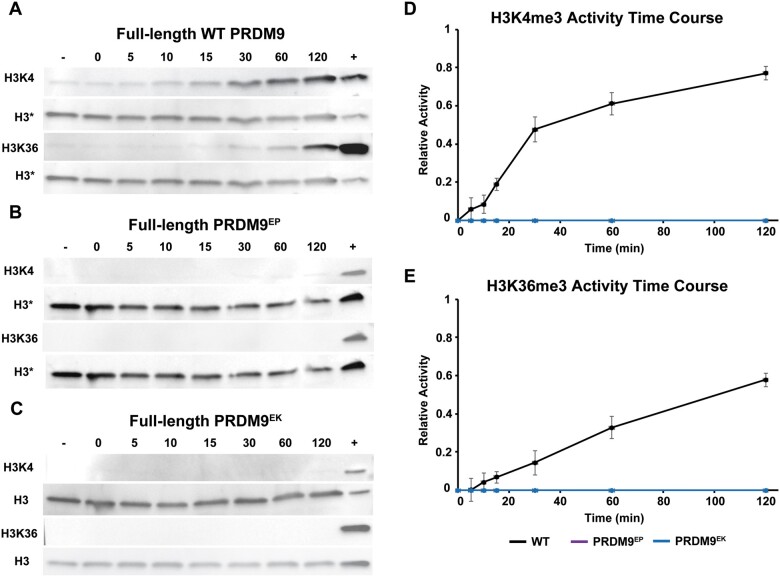
Figure 2.
HMT activities of full-length wild-type, PRDM9EP, and PRDM9EK. (A–C) Western blots showing the results of HMT assays with full-length proteins. Blots were probed with α-H3K4me3 and α-H3K36me3 antibodies to show the levels of these modified histones at the denoted time points (minutes) as the assay proceeded. Negative control lanes (−) represent reactions run without s-adenosyl methionine (SAM), while positive control lanes (+) represent purified H3K4me3 or H3K36me3, as appropriate. Blots were stripped and reprobed with α-histone H3 antibody as loading controls; these are shown below their respective counterparts. In the case of WT and PRDM9EP, the same blot was probed consecutively for H3K36me3, H3K4me3, and H3. The positive control lane on these blots contains a mixture of both H3K4- and H3K36-trimethylated histone H3. Time course plots summarizing HMT activities for H3K4me3 (D) and H3K36me3 (E), quantified by band densitometry. Data points represent the mean of three replicates; error bars represent the standard deviations.
PRDM9-dependent hotspot definition
For this study, we characterize “hotspots” as known PRDM9-dependent H3K4me3 peaks in the B6 genetic background. This peak set was ascertained by previous work from our laboratory. Baker et al. (2014) directly compared H3K4me3 ChIP-seq peaks between two strains of mice: B6 and B6-Prdm9CAST-KI. The latter is a knock-in B6 mouse with the exon encoding the DNA-binding ZnF array of Prdm9Dom2 (the B6 allele) replaced with that of Prdm9CAST (the highly divergent allele found in the CAST/EiJ strain). The Prdm9CAST knock-in binds a completely different set of hotspots relative to Prdm9Dom2, due to replacement of its DNA-binding domain. Baker et al. (2014) extracted a set of H3K4me3 peaks present in B6 spermatocytes and not B6-Prdm9CAST-KI spermatocytes, characterizing them as PRDM9Dom2-dependent H3K4me3 peaks in the B6 genetic background. The original peak set was mapped to mm9; when we remapped it to mm10, it yielded 18,838 peaks. H3K4me3 peaks outside this peak set were considered PRDM9-independent.
H3K4me3 ChIP-seq
ChIP-seq for H3K4me3 was performed using spermatocytes isolated from 14-day-old males, as described previously (Baker et al. 2014). For B6 and Prdm9EP/EP spermatocytes, ChIP-seq data our laboratory previously reported (Baker et al. 2014; Powers et al. 2020) was used. In both cases, two biological replicates were merged for analysis, with a total of 39,973,027 aligned reads for B6 and 45,931,625 aligned reads for Prdm9EP/EP spermatocytes, as in Powers et al. (2020). For Prdm9EK/EK spermatocytes, one replicate was previously reported (Bhattacharyya et al. 2019), and an additional biological replicate was performed for this study, for a total of two replicates per genotype. This additional replicate was sequenced on the Illumina NextSeq 500 platform, with 75-bp reads, and trimmed for quality using trimmomatic. Sequence data were aligned to the mouse mm10 genome using BWA (Burrows–Wheeler aligner) v0.7.9a, and duplicate reads and reads which failed to align to unique positions in the genome were discarded. For the additional Prdm9EK/EK replicate, this resulted in 56,096,669 aligned reads. The raw fastq file of this replicate was combined with that of the previous replicate for analysis, and the merged file was aligned to mm10 via the procedure above for a total of 72,730,785 aligned reads.
H3K4me3 peaks were called using MACS v1.4, default parameters, with a P value cutoff of 0.01, using treatment and control samples. The same procedure and samples that we used in our previous study (Powers et al. 2020) were used, with the addition of the merged Prdm9EK/EK sample, as well as the set of PRDM9-dependent H3K4me3 peaks described in that study. To determine overlap between known PRDM9-dependent H3K4me3 peaks and ChIP-seq peak sets, we used BedTools (v2.27.0) intersect with default parameters, except for a requirement for 20% overlap (f = 0.20).
H3K36me3 ChIP-seq
ChIP-seq for H3K36me3 was performed using spermatocytes isolated from 14-day-old males, as described previously (Powers et al. 2016). One biological replicate per genotype was performed. DNA samples were sequenced on an Illumina HiSeq 2500, with 100 bp reads, and trimmed for quality using trimmomatic. Sequence data were aligned to the mouse mm10 genome using BWA (Burrows–Wheeler aligner) v0.7.9a, and duplicate reads and reads which failed to align to unique positions in the genome were discarded. This resulted in a total of 42,288,943 aligned reads for Prdm9EP/EP spermatocytes, and 40,263,687 aligned reads for Prdm9EK/EK spermatocytes. For the B6 control, we used two previously reported biological replicates (Powers et al. 2016), merging the raw fastq files and aligning to the mm10 genome for analysis. This resulted in 95,375,802 aligned reads.
To assess detectable H3K36me3 peaks, we also used MACS v1.4, default parameters, with a P value cutoff of 0.01, but in this case with treatment samples only so that enrichment for each peak was determined relative to the surrounding background, rather than relative to a control sample. We chose this method because H3K36me3 is broadly enriched in transcriptionally active genes, and this broad enrichment can confound peak calling relative to a control sample. To determine overlap between known PRDM9-dependent H3K4me3 peaks and ChIP-seq peak sets, we used BedTools (v2.27.0) intersect with default parameters, except for a requirement for 20% overlap (f = 0.20).
Aggregation plots
Aggregation plots were done using the Aggregation and Correlation Toolbox (ACT, http://act.gersteinlab.org/), with the following parameters: nbins = 500, mbins = 0, radius = 1500.
Results
Prdm9 EP and Prdm9EK mutations compromise the H3K36 trimethylation activity of the isolated PRDM9 PR/SET domain in vitro
In wild-type mice, double-stranded breaks (DSBs) that initiate recombination occur at PRDM9-activated sites and their numbers are tightly controlled. Only about 5% of PRDM9-bound sites undergo DSB formation (Kauppi et al. 2013). In Prdm9-null B6 mice, DSBs are made in normal numbers, but at ectopic sites where H3K4me3 is enriched, such as promoters. These ectopic DSBs are not repaired efficiently and may be the principal cause of the meiotic arrest followed by apoptosis due to loss of Prdm9 (Brick et al. 2012). Presumably, this specific meiotic effect could be ascribed to the unique activity of the PR/SET domain of PRDM9, which combines two methyltransferase activities—trimethylation of histone H3 at both lysine 4 and lysine 36 (Powers et al. 2016). To further study the nature of this activity and its effect on DSB initiation and repair, we created two mutants, Prdm9EP and Prdm9EK.
We studied the effects of these mutations on PR/SET domain methyltransferase activities by performing HMT time-course assays using wild-type, Prdm9EP and Prdm9EK PR/SET domains, with three replicates of each time course. Figure 1, A–C shows representative blots, and Figure 1, D and E shows time course plots with the average and standard deviation of the three replicates. The H3K4 trimethylation curves of Prdm9EP and Prdm9EK PR/SET domains were indistinguishable from the one of wild-type PR/SET domain (Figure 1D). In contrast, the H3K36 methyltransferase activities of the mutant PR/SET domains were substantially different from the wild-type domain (Figure 1E). The initial rate of the H3K36 trimethylation activity of Prdm9EK PR/SET domain was substantially reduced, though the average did reach wild-type levels of H3K36 trimethylation at the 120-min endpoint. For the Prdm9EP PR/SET domain, H3K36 trimethylation activity was completely abolished, indicating full separation-of-function.
Prdm9 EP and Prdm9EK mutations abolish the H3K4 and H3K36 trimethylation activities of full-length PRDM9 in vitro
To characterize the effects of Prdm9EP and Prdm9EK on full-length PRDM9 in vitro, we introduced these mutations into a cDNA clone encoding human PRDM9. We then expressed and partially purified full-length wild-type PRDM9, PRDM9EP, and PRDM9EK, and repeated the HMT assays described above. In stark contrast to the isolated PR/SET domains, both mutations completely abolished the H3K4 and H3K36 methyltransferase activities of full-length PRDM9 in vitro (Figure 2).
Prdm9 EP and Prdm9EK confer severe catalytic hypomorphism in vivo
We used CRISPR/Cas9 gene editing to introduce the Prdm9EP and Prdm9EK mutations in mice on the B6 genetic background. Some of the in vivo effects of these mutations were reported elsewhere (Bhattacharyya et al. 2019; Powers et al. 2020), and here we use those data for direct comparison between the two. After breeding these mutations to homozygosity, we performed ChIP-seq for H3K4me3 and H3K36me3 in spermatocytes isolated from homozygous 14-day-old males. We then assessed overlap with known PRDM9-dependent H3K4me3 peaks in B6 spermatocytes based on previous work from our laboratory (Baker et al. 2014); “see Materials and Methods” for details. The resulting ChIP-seq data show severe reductions in both number and intensity of PRDM9-dependent H3K4me3 peaks (Table 1; Figure 3, A–F); however, in contrast to the effects of these mutations on the full-length protein in vitro (Figure 2), HMT functions are neither separated nor abolished by either mutation. There is no effect on PRDM9-independent peaks (Figure 3, A and G). Prdm9EP and Prdm9EK show 4.98- and 5.57-fold reductions in peak number, and 2.84- and 3.56-fold reductions in peak intensity at detectable H3K4me3 peaks, respectively (Table 1). The effect of PRDM9EK on H3K4me3 peaks is slightly more severe in vivo than that of PRDM9EP (Table 1; Figure 3, C–F). Although the H3K36me3 ChIP-seq experiments were done in single replicates, they matched the H3K4me3 results. We have previously shown that our H3K36me3 ChIP-seq protocol produces high-confidence results (Powers et al. 2016). We were only able to do a single replicate for H3K36me3 ChIP per mutant due to limited animal resources, and while these data are consistent with the expected pattern for H3K36me3 (enrichment in actively transcribed genes), single-replicate ChIP-seq experiments should be interpreted with caution.
Table 1.
Effect of Prdm9EP and Prdm9EK homozygosity on H3K4me3 and H3K36me3 activity at known PRDM9-dependent H3K4me3 peaks in spermatocytes
| H3K4me3 | Detectable PRDM9-dependent peaks | Fold reduction vs wild-type B6 | Mean read count in detectable peaks (RPM) | Fold reduction vs wild-type B6 |
|---|---|---|---|---|
| B6 | 18,427 | – | 3.67 | – |
| Prdm9 EP | 3,703 | 4.98 | 1.29 | 2.84 |
| Prdm9 EK | 3,307 | 5.57 | 1.03 | 3.56 |
| H3K4me3 + H3K36me3 | ||||
| B6 | 6,894 | – | 1.69 | – |
| Prdm9 EP | 412 | 16.7 | 1.22 | 1.39 |
| Prdm9 EK | 243 | 28.4 | 0.995 | 1.70 |
This table compares the number and intensity of detectable H3K4me3 and H3K36me3 ChIP-seq peaks at known PRDM9-dependent H3K4me3 peaks (n = 18,838), in wild-type B6 spermatocytes and spermatocytes homozygous for Prdm9EP and Prdm9EK. To eliminate false positives due to the nature of H3K36me3 ChIP-seq data, detectable H3K36me3 peaks were restricted to those which also had a detectable H3K4me3 peak.
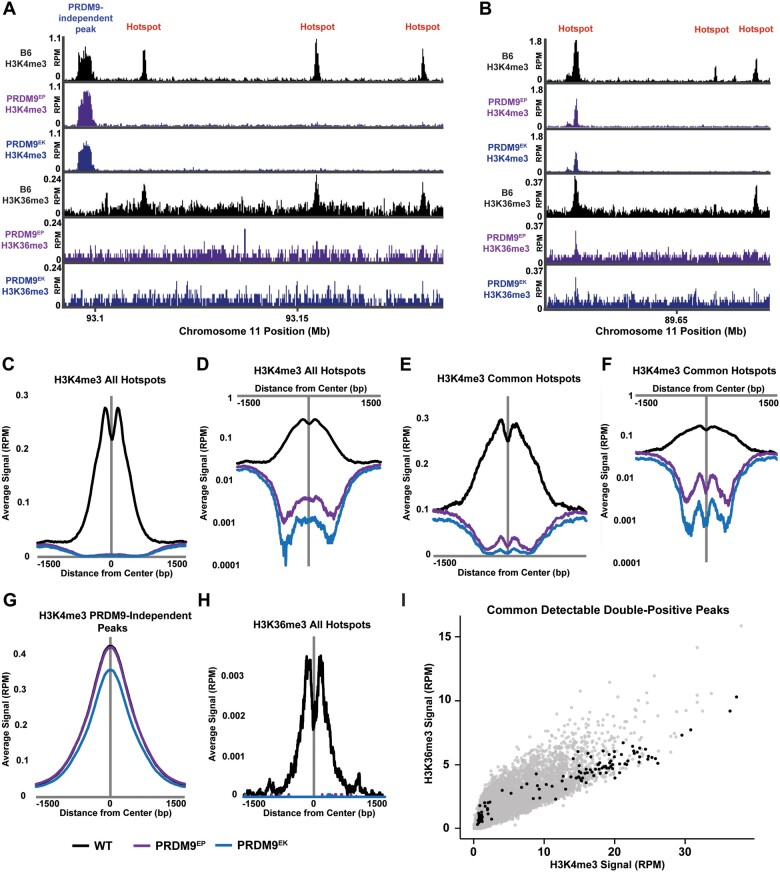
Figure 3.
Effects of PRDM9EP and PRDM9EK homozygosity in vivo. (A, B) Genome browser snapshot of H3K4me3 and H3K36me3 in wild-type B6, PRDM9EP, and PRDM9EK spermatocytes. Note the loss of both modifications at almost all hotspots, and their residual presence at one hotspot in (B). Aggregation plots of H3K4me3 at all known PRDM9-dependent H3K4me3 peaks (n = 18,838), on a linear (C) and a logarithmic (D) scale. Aggregation plots of H3K4me3 at common detectable PRDM9-dependent H3K4me3 peaks between wild-type, PRDM9EP, and PRDM9EK (n = 2632), on a linear (E) and a logarithmic (F) scale. (G) Aggregation plot of H3K4me3 at common PRDM9-independent H3K4me3 peaks between wild-type, PRDM9EP, and PRDM9EK (n = 76,995). (H) Aggregation plot of H3K36me3 at all known PRDM9-dependent H3K4me3 peaks (n = 18,838). (I) H3K4me3 signal vs H3K36me3 signal in wild-type spermatocytes at all known PRDM9-dependent H3K4me3 peaks (n = 18,838). Sites with common detectable H3K4me3 and H3K36me3 peaks between all three genotypes (n = 118) are highlighted in black. Note the enrichment of highly active wild-type peaks in this subset.
In distinct contrast to the PRDM9-dependent H3K4me3 peaks, PRDM9-dependent H3K36me3 peaks are barely detectable in both mutants (Table 1; Figure 3H), but still observable (Figure 3B). Indeed, the number of detectable H3K4me3/H3K36me3 double-positive peaks is reduced 16.7-fold in Prdm9EP homozygotes and 28.4-fold in Prdm9EK homozygotes. The intensities of the H3K36me3 peaks that do remain, however, are not as markedly reduced as those of H3K4me3 peaks—1.39- and 1.7-fold in Prdm9EP and Prdm9EK, respectively (Table 1). As shown in Figure 3I, in wild-type spermatocytes PRDM9-dependent H3K4 and H3K36 trimethylation signals are highly correlated—the most active sites tend to be high in both. These strong wild-type peaks are enriched for detectable double-positive peaks in Prdm9EP and Prdm9EK (Figure 3I). This is consistent with the expected effect if Prdm9EP and Prdm9EK confer lower catalytic activity in vivo.
Prdm9 EP and Prdm9EK straddle the threshold for female fertility in the B6 genetic background
In males, homozygosity for either Prdm9EP or Prdm9EK causes infertility with complete meiotic arrest, phenocopying the Prdm9-null condition (Bhattacharyya et al. 2019; Powers et al. 2020). Assessment of female fertility, however, yielded an unexpected result. As we reported previously, female Prdm9EP homozygotes are subfertile, producing variable numbers of litters in natural matings (data reproduced in Table 2A and B) (Powers et al. 2020). In contrast, none of the female Prdm9EK homozygotes we tested produced any pups (Table 2C). Thus, the slight reduction in PRDM9 catalytic activity exhibited by Prdm9EK relative to Prdm9EPin vivo is sufficient to abolish female fertility, in contrast to that seen in Prdm9EP homozygotes.
Table 2.
Prdm9 EP/EP and Prdm9EK/EK female fertility tests
| Genetic background | Duration (months) | Females in cross | Total pups | Total litters | Avg. pups/litter | OFM | LFM | Pups lost |
|---|---|---|---|---|---|---|---|---|
| (A) Fertility tests: Prdm9EP/EP females with a B6 genetic background | ||||||||
| B6 | 6 | 1 | 5 | 1 | 5 | 0.83 | 0.17 | 0 |
| B6 | 6 | 1 | 28 | 5 | 5.6 | 4.67 | 0.83 | 5 |
| B6 | 6 | 1 | 10 | 2 | 5 | 1.67 | 0.33 | 0 |
| B6 | 6 | 1 | 10 | 2 | 5 | 1.67 | 0.33 | 2 |
| B6 | 6 | 1 | 24 | 5 | 4.8 | 4.00 | 0.83 | 9 |
| B6 | 6 | 1 | 20 | 5 | 4 | 3.33 | 0.83 | 2 |
| AVG | 2.69 | 0.56 | 3 | |||||
| (B) Fertility tests: Prdm9EP/EP females with a B6xWSB-F2 genetic background | ||||||||
| B6xWSB-F2 | 6 | 1 | 24 | 6 | 4 | 4.00 | 1.00 | 0 |
| B6xWSB-F2 | 6 | 2 | 41 | 10 | 4.1 | 3.42 | 0.83 | 0 |
| B6xWSB-F2 | 6 | 2 | 74 | 13 | 5.7 | 6.17 | 1.08 | 3 |
| B6xWSB-F2 | 6 | 2 | 41 | 8 | 5.1 | 3.42 | 0.67 | 1 |
| B6xWSB-F2 | 6 | 1 | 28 | 6 | 4.7 | 4.67 | 1.00 | 1 |
| B6xWSB-F2 | 7+ | 2 | 79 | 9 | 8.8 | 5.64 | 0.64 | 5 |
| AVG | 4.55 | 0.87 | 1.67 | |||||
| (C) Fertility tests: Prdm9EK/EK females with a B6 genetic background | ||||||||
| B6 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| B6 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| B6 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| B6 | 6 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| B6 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| B6 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | N/A |
| AVG | 0 | 0 | N/A | |||||
(A, B) We previously reported fertility data for Prdm9EP/EP females elsewhere; we reproduce it here for comparison with fertility data for Prdm9EK/EK females (C). Females were crossed at 5 weeks of age with a sexually mature (>8 weeks), wild-type B6 male, in pairs or trios as noted. Productive crosses were allowed to continue for at least 6 months, and longer if necessary until the females stopped producing offspring. Unproductive crosses were allowed to continue at least 3 months. The number of pups in each cross that died before wean are noted in the last column; all other offspring appeared grossly normal and healthy at wean. All offspring in the B6 background, and the offspring from at least 3 l per cross in the B6xWSB-F2 background, were genotyped for Prdm9EP to confirm maternal homozygosity. As expected, all genotyped offspring were heterozygous for Prdm9EP.
OFM, offspring per female per month; LFM, litters per female per month.
Discussion
Prdm9 was identified as the mammalian recombination activator in 2010 (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). Since then, it has amassed a degree of fame as its many curious properties have come to light. It is unique among mammalian histone modifying enzymes in several respects. First, it is the only known gene that specifically combines the KRAB, PR/SET, and ZnF functional domains. Each of these domains has unique properties. The KRAB domain binds proteins that help to open local chromatin (Parvanov et al. 2017; Tian et al. 2021). The PR/SET domain has the intrinsic capacity to de novo trimethylate two different lysine residues, thereby creating a unique epigenetic signature at recombination hotspots (Hayashi et al. 2005; Baker et al. 2014; Eram et al. 2014; Powers et al. 2016). The ZnF DNA-binding domain is among the most rapidly evolving protein-coding regions known in vertebrates, in contrast to other ZnF proteins where these domains are mostly conserved (Najafabadi et al. 2015; Wolf et al. 2020). Second, it is the only mammalian hybrid sterility gene identified to date (Mihola et al. 2009; Bhattacharyya et al. 2013; Lustyk et al. 2019). Third, its loss was thought to lead to unconditional infertility, but the growing list of exceptions to this rule suggests modifiers and different degrees of dependence on PRDM9 for successful meiosis among mammalian species, subspecies, and populations (Narasimhan et al. 2016; Mihola et al. 2019; Powers et al. 2020; Mihola et al. 2021). This study now adds to understanding of the unique functions of PRDM9 in two ways: first, by pointing out the interdependence of PRDM9 domains in vivo and in vitro, and second, by elaborating the molecular and phenotypic effects of these two unique mutations, we provide new information of theoretical and potentially practical significance.
The purpose of this study was twofold. Fundamentally, we sought to fully characterize and directly compare the catalytic activities, in vitro and in vivo, of two catalytic mutations of PRDM9 at the same residue. In terms of reproduction and evolutionary biology, the main question is why are Prdm9EP/EP B6 females fertile, but Prdm9EK/EK B6 females infertile? The fact that Prdm9EK is a slightly more severe hypomorph based on H3K4me3 ChIP-seq in males and the fertility phenotype in females in vivo, together with the fact that catalytically inactive PRDM9 causes infertility in B6 females (Diagouraga et al. 2018), suggests that Prdm9EP/EP and Prdm9EK/EK females are, respectively, above and below a catalytic activity threshold required for fertility. Technically, we sought to contribute resources of potentially practical value as tools for genetic mapping and targeting specific sites for H3K4 trimethylation de novo.
The structure of Prdm9 suggests a modular protein: three well-separated domains with theoretically distinct functions. The catalytic PR/SET domain and the DNA-binding ZnF domain, which retain their functions in vitro when isolated from the rest of the protein, had indeed seemed particularly autonomous (Billings et al. 2013; Eram et al. 2014; Walker et al. 2015; Powers et al. 2016). The disparity of the effects of these mutations on the isolated PR/SET domain vs full-length PRDM9, however, shows this not to be the case. Clearly, the tertiary structure of full-length PRDM9 imposes an enzymatic context on the PR/SET domain that dramatically alters its response to catalytic mutations. In retrospect, this pseudomodularity is perhaps not surprising in a protein encoded by such an evolutionary labile gene. Prdm9 belongs to the KRAB-ZnF family of transcription factors; these genes are generally characterized by rapid evolution via gene duplications and repurposing for new functions through subsequent ZnF array diversification (Lupo et al. 2013; Imbeault et al. 2017). In contrast, Prdm9 evolves a multitude of alleles of the same gene while preserving its unique function as a recombination regulator (Parvanov et al. 2010; Hinch et al. 2011; Buard et al. 2014; Kono et al. 2014). Evolutionary analysis indicates that PRDM9-directed recombination may be present in lower taxa, and that it may have been lost in several vertebrate lineages, as it was lost in the canid lineage (Baker et al. 2017). A structurally complete version of Prdm9 is present in most placental mammals and such organisms as turtles, snakes, lizards, and some fish, while partial versions missing one or more functional domains are present in many taxa. A complete version of Prdm9 can be found in such early vertebrates as jawless fish and coelacanth, suggesting that its absence from other taxa resulted from repeated loss or repurposing of Prdm9 (Baker et al. 2017). In a protein whose functions change frequently over evolutionary time, functional domains are expected to merge or separate frequently, and sometimes incompletely. Such may be the case with PRDM9; despite the modularity suggested by its structure, functional autonomy of its catalytic domain evidently has not happened.
Apart from using these two mutations as cautionary examples against assuming domain modularity when engineering enzymes, it is instructive to contemplate the results of these enzymatic manipulations. First, the in vivo effects of these mutations differ from those in both the isolated PR/SET domains and the full-length proteins in vitro. This is not entirely unexpected, because PRDM9 does not operate in a vacuum during meiosis. Unlike the situation in vitro, PRDM9 is bound to DNA when it trimethylates H3K4 and H3K36 in vivo. Additionally, multiple other proteins have been found to interact with PRDM9 during meiosis, including cohesin complexes containing the meiosis-specific proteins REC8 and STAG3 (Parvanov et al. 2017; Bhattacharyya et al. 2019). The RNA-binding protein EWSR1 (Tian et al. 2021) and the chromatin remodeling factor HELLS (Spruce et al. 2020) also interact with PRDM9, and both are required for efficient PRDM9-dependent catalytic activity in vivo. These complex interactions have not yet been placed in a temporal time-course, and, moreover, could reorient either the PR/SET domain or the substrate, allowing for the residual activity we observe with full-length PRDM9EP and PRDM9EKin vivo, but not in vitro. Second, these two mutations alter the same residue: glutamate 360 to proline and lysine, respectively. The mutations were deliberately designed based on a published crystal structure of the PR/SET domain with docked histone tail peptides (Eram et al. 2014). We chose proline and lysine on the principles of changing the local shape of the binding pocket and reversing the charge of the residue, respectively. Interestingly, changing the shape (Glu > Pro) was more effective than charge reversal (Glu > Lys) in separating the two methyltransferase functions. In fact, we did create a PR/SET domain that, when cloned separately, can trimethylate H3K4, but not H3K36 de novo. If this domain retains the same capacity when expressed alone in vivo, it might be possible to artificially trimethylate H3K4 at selected genomic sites by tethering it to a DNA-binding domain or to inactivated Cas9.
The unexpected female fertility phenotype of Prdm9EP added another dimension to the hypomorphic nature of the two mutations. The phenotypic contrast between Prdm9EP and Prdm9EK in females makes these two mutations especially fascinating, as their catalytic effects differ only slightly in severity in males (Table 1; Figure 3, C–F). Of course, a caveat to this study is that we assessed in vivo catalytic activity in spermatocytes, while this phenotype involves oocytes. It would be ideal to directly assess the in vivo effects of these mutations on oocytes, but the low numbers of oocytes per ovary and the timing of meiosis (during fetal development) make ChIP-seq in oocytes a heroic endeavor to say the least (Brick et al. 2018). Such an experiment would be worthwhile for future work; in the meantime, our assumption that the catalytic effects of these mutations are similar in spermatocytes and oocytes is supported by the in vitro data.
As we demonstrated previously, in females homozygous for PRDM9EP, meiotic DSBs occur at both PRDM9-dependent and PRDM9-independent sites. In some of the offspring of these females, PRDM9-independent DSBs gave rise to crossovers, indicating the presence of a PRDM9-independent crossover pathway in female mice (Powers et al. 2020). However, this pathway is not sufficient for successful meiosis in B6 females in the absence of Prdm9. The fact that Prdm9EP, but not Prdm9EK, is compatible with some fertility in B6 females suggests the possibility of a minimum threshold of PRDM9-dependent meiotic DSBs, or some other complex phenotype modification. There is evidence that inefficient repair of PRDM9-independent meiotic DSBs contributes to meiotic arrest in Prdm9-null animals, and this has been speculated to be its primary cause. In females, DNA damage surveillance unequivocally plays a role, as removing the checkpoint kinase Chk2 permits marginal rescue of female fertility in Prdm9-null B6 mice—albeit with a high incidence of pregnancy loss and a high perinatal death rate (Powers et al. 2020). If Prdm9-null meiotic arrest is indeed due to inefficient DSB repair, our results suggest a model in which oocytes with “methyltransferase-comatose” PRDM9 can complete meiosis if enough meiotic DSBs are PRDM9-dependent, allowing the oocyte to evade DNA damage checkpoint activation. We propose that in the case of Prdm9EP homozygotes, a minority of oocytes complete meiosis, but that in the case of Prdm9EK homozygotes, the slightly lower catalytic activity of PRDM9EK shifts the balance too far in favor of inefficiently repaired PRDM9-independent DSBs, and few or no oocytes evade the checkpoint. A key difference between PRDM9-dependent and PRDM9-independent DSBs is the presence of H3K36me3 at the former, but not the latter. Since H3K36me3 is known to promote homologous recombination repair in somatic cells (Aymard et al. 2014; Pfister et al. 2014), one might speculate that inefficient repair at ectopic DSBs is due to its absence, and that H3K36 trimethylation activity could be a determinant of whether an oocyte passes the checkpoint.
Prdm9 EP and Prdm9EK straddle the female fertility threshold for loss of PRDM9 functionality in B6 mice. Notably, as we previously observed, female fertility in Prdm9EP homozygotes is higher in F2 crosses between B6 and WSB/EiJ (WSB), another domesticus strain, indicating an effect of genetic modifiers and/or hybrid vigor (Powers et al. 2020). Examination of the effects of these mutations in other genetic backgrounds and in outbred mice would be an interesting approach to determine the nature of threshold fertility. It is possible, for instance, that the activity of PRDM9EP is not sufficient for female fertility in some strains, and that the lower activity PRDM9EK is sufficient for female fertility in others. A particularly interesting case would be that of PWD/Ph, a musculus strain in which Prdm9-null males are able to produce small amounts of sperm, while the Prdm9-null females are infertile (Mihola et al. 2019). This strain appears not to follow the trend of female robustness to loss of PRDM9 function seen in domesticus and castaneus strains, and thus the effect of Prdm9EP and Prdm9EK on both spermatogenesis and oogenesis in these mice would be informative to assess. Catalytic Prdm9 hypomorphs such as these, and Prdm9EP in particular, may also have some practical utility for genetic mapping. Because it allows for healthy offspring with PRDM9-independent crossovers in domesticus strains, Prdm9EP might permit recombination events within “cold spots” refractory to PRDM9-dependent recombination.
In summary, the Prdm9EP and Prdm9EK mutations have revealed informative insights into the catalytic properties of PRDM9. Their dramatically different effects on the isolated PR/SET domain vs the full-length protein show that PRDM9 does not exhibit autonomous modular function despite a structure that suggests it might. The moderately different effects of these two mutations on the full-length protein in vitro vs In vivo suggest a regulatory and/or determinative role for proteins that interact with PRDM9 at the sites of hotspot activation. The different phenotypic effects of these two mutations reveal the minimum threshold of PRDM9-dependent catalytic activity for female fertility in the B6 genetic background. As intriguing as these discoveries are, PRDM9 likely has more peculiarities yet to be uncovered, and these mutations will continue to be useful for further study of this exceptional and fascinating gene.
Data availability
Strains and plasmids are available upon request. All sequence data newly reported in this study are available at the Gene Expression Omnibus (GEO) under accession number GSE181479.
Acknowledgments
We would like to thank Mary Ann Handel for critical reading, the Genome Technologies and Genetic Engineering Technology cores at The Jackson Laboratory for their invaluable expertise and assistance with this project.
Funding
This work was supported by National Institutes of Health Grants R01 GM078452 to P.M.P., R01 GM125736 to P.M.P., P01 GM99640 to K.P., and Cancer Core Grant CA34196 to the Jackson Laboratory. N.R.P. was supported in part by NICHD T32 Training Program in Developmental Genetics (T32 HD007065 to the Jackson Laboratory).
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Auton A, Rui Li Y, Kidd J, Oliveira K, Nadel J, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9:e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F, Bugler B, Schmidt CK, Guillou E, Caron P, et al. 2014. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat Struct Mol Biol. 21:366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K.. 2014. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 24:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Z, Schumer M, Haba Y, Bashkirova L, Holland C, et al. 2017. Repeated losses of PRDM9-directed recombination despite the conservation of PRDM9 across vertebrates. Elife. 6:e24133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 327:836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, et al. 2010. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 42:859–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, et al. 2013. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci U S A. 110:e468–e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Walker M, Powers NR, Brunton C, Fine AD, et al. 2019. Prdm9 and meiotic cohesin proteins cooperatively promote DNA double-strand break formation in mammalian spermatocytes. Curr Biol. 29:1002–1018.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, et al. 2013. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol. 14:r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV.. 2012. Genetic recombination is directed away from functional genomic elements in mice. Nature. 485:642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Thibault-Sennett S, Smagulova F, Lam KG, Pu Y, et al. 2018. Extensive sex differences at the initiation of genetic recombination. Nature. 561:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, et al. 2014. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 9:e85021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagouraga B, Clement JAJ, Duret L, Kadlec J, de Massy B, et al. 2018. PRDM9 Methyltransferase activity is essential for meiotic DNA double-strand break formation at its binding sites. Mol Cell. 69:853–865.e6. [DOI] [PubMed] [Google Scholar]
- Eram MS, Bustos SP, Lima-Fernandes E, Siarheyeva A, Senisterra G, et al. 2014. Trimethylation of histone H3 lysine 36 by human methyltransferase PRDM9 protein. J Biol Chem. 289:12177–12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y.. 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 438:374–378. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, et al. 2011. The landscape of recombination in African Americans. Nature. 476:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Baudat F, Taillepierre M, Stanzione M, Toth A, et al. 2017. The PRDM9 KRAB domain is required for meiosis and involved in protein interactions. Chromosoma. 126:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid PY, Trono D.. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 543:550–554. [DOI] [PubMed] [Google Scholar]
- Irie S, Tsujimura A, Miyagawa Y, Ueda T, Matsuoka Y, et al. 2009. Single-nucleotide polymorphisms of the PRDM9 (MEISETZ) gene in patients with nonobstructive azoospermia. J Androl. 30:426–431. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jasin M, Keeney S.. 2013. How much is enough? Control of DNA double-strand break numbers in mouse meiosis. Cell Cycle. 12:2719–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh-Stenta X, Joy J, Poulsen A, Li R, Tan Y, et al. 2014. Characterization of the histone methyltransferase PRDM9 using biochemical, biophysical and chemical biology techniques. Biochem J. 461:323–334. [DOI] [PubMed] [Google Scholar]
- Kono H, Tamura M, Osada N, Suzuki H, Abe K, et al. 2014. Prdm9 polymorphism unveils mouse evolutionary tracks. DNA Res. 21:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, et al. 2013. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics. 14:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustyk D, Kinsky S, Ullrich KK, Yancoskie M, Kasikova L, et al. 2019. Genomic structure of Hstx2 modifier of Prdm9-dependent hybrid male sterility in mice. Genetics. 213:1047–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Landa V, Pratto F, Brick K, Kobets T, et al. 2021. Rat PRDM9 shapes recombination landscapes, duration of meiosis, gametogenesis, and age of fertility. BMC Biol. 19:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Pratto F, Brick K, Linhartova E, Kobets T, et al. 2019. Histone methyltransferase PRDM9 is not essential for meiosis in male mice. Genome Res. 29:1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J.. 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 323:373–375. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Koh E, Sakugawa N, Sato H, Hayashi H, et al. 2008. Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 25:553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fuentes V, Di Rienzo A, Vila C.. 2011. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. 6:e25498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, et al. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 327:876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, et al. 2015. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 33:555–562. [DOI] [PubMed] [Google Scholar]
- Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, et al. 2016. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 352:474–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov PM.. 2018. PRDM9 and its role in genetic recombination. Trends Genet. 34:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K.. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science. 327:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Tian H, Billings T, Saxl RL, Spruce C, et al. 2017. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol Biol Cell. 28:488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, et al. 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 7:2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers NR, Dumont BL, Emori C, Lawal RA, Brunton C, et al. 2020. Sexual dimorphism in the meiotic requirement for PRDM9: a mammalian evolutionary safeguard. Sci Adv. 6:eabb6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, et al. 2016. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet. 12:e1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Brick K, Pu Y, Camerini-Otero RD, Petukhova GV.. 2016. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 30:266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruce C, Dlamini S, Ananda G, Bronkema N, Tian H, et al. 2020. HELLS and PRDM9 form a pioneer complex to open chromatin at meiotic recombination hot spots. Genes Dev. 34:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Billings T, Petkov PM.. 2021. EWSR1 affects PRDM9-dependent histone 3 methylation and provides a link between recombination hotspots and the chromosome axis protein REC8. Mol Biol Cell. 32:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Billings T, Baker CL, Powers N, Tian H, et al. 2015. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin. 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, de Iaco A, Sun MA, Bruno M, Tinkham M, et al. 2020. KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. Elife. 9:e56337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mathioudakis N, Diagouraga B, Dong A, Dombrovski L, et al. 2013. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 5:13–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. All sequence data newly reported in this study are available at the Gene Expression Omnibus (GEO) under accession number GSE181479.