Abstract
Mutations in enhancers have been shown to often underlie natural variation but the evolved differences in enhancer activity can be difficult to identify in vivo. Threespine sticklebacks (Gasterosteus aculeatus) are a robust system for studying enhancer evolution due to abundant natural genetic variation, a diversity of evolved phenotypes between ancestral marine and derived freshwater forms, and the tractability of transgenic techniques. Previous work identified a series of polymorphisms within an intronic enhancer of the Bone morphogenetic protein 6 (Bmp6) gene that are associated with evolved tooth gain, a derived increase in freshwater tooth number that arises late in development. Here, we use a bicistronic reporter construct containing a genetic insulator and a pair of reciprocal two-color transgenic reporter lines to compare enhancer activity of marine and freshwater alleles of this enhancer. In older fish, the two alleles drive partially overlapping expression in both mesenchyme and epithelium of developing teeth, but the freshwater enhancer drives a reduced mesenchymal domain and a larger epithelial domain relative to the marine enhancer. In younger fish, these spatial shifts in enhancer activity are less pronounced. Comparing Bmp6 expression by in situ hybridization in developing teeth of marine and freshwater fish reveals similar evolved spatial shifts in gene expression. Together, these data support a model in which the polymorphisms within this enhancer underlie evolved tooth gain by shifting the spatial expression of Bmp6 during tooth development, and provide a general strategy to identify spatial differences in enhancer activity in vivo.
Keywords: enhancer, cis-regulation, evolution, transgene, transgenesis, insulator, fish, stickleback, development, tooth
Introduction
The process of development is largely orchestrated by developmental regulatory genes whose spatial and temporal patterns of transcription are controlled by enhancers, cis-regulatory elements that bind transcription factors and promote transcription of target genes (Furlong and Levine 2018; Gasperini et al. 2020). Most developmental regulatory genes are pleiotropic, and function repeatedly at different times and in different tissues during development (Sabarís et al. 2019). Thus, mutations in enhancers of developmental regulatory genes are often more tolerated than coding sequence mutations due to having fewer pleiotropic effects, as the impacts of enhancer mutations are more likely to be restricted in time and/or space, compared to the anatomically more widespread impacts of coding mutations (Carroll 2008). The importance of enhancers in regulating morphological evolution, natural variation, and disease phenotypes in humans is well established (Rebeiz and Tsiantis 2017; Rickels and Shilatifard 2018). However, a growing need has emerged for methods and approaches to compare the activity of molecularly divergent enhancer alleles.
Cis-regulatory changes have been shown to underlie the evolution of multiple morphological traits in threespine stickleback fish (Gasterosteus aculeatus). Threespine sticklebacks live in both marine and freshwater environments in the Northern Hemisphere, repeatedly forming populations in rivers, streams, ponds, and lakes from ancestral marine populations (Bell and Foster 1994; McKinnon and Rundle 2002). Following a freshwater colonization event, a suite of traits has been observed to typically evolve such as reduction in armor (Bell and Foster 1994; Cresko et al. 2004; Colosimo et al. 2005) and changes in body shape (Walker 1997; Walker and Bell 2000; Albert et al. 2008; Reid and Peichel 2010). Other traits that typically evolve major differences are those associated with feeding morphology, likely an adaptation to different diets of larger prey in freshwater environments relative to marine ancestral environments (Hagen 1967; Gross and Anderson 1984; Lavin and McPhail 1986; Schluter and McPhail 1992; Bell and Foster 1994). High-resolution genetic mapping studies have implicated cis-regulatory changes as underlying several phenotypes that have evolved in freshwater, including the reduction of armor plates (Colosimo et al. 2005; O’Brown et al. 2015; Indjeian et al. 2016; Archambeault et al. 2020), pelvic spines (Chan et al. 2010), and pigmentation (Miller et al. 2007), and increases in branchial bone length (Erickson et al. 2018), and pharyngeal tooth number (Cleves et al. 2014, 2018).
Tooth development is orchestrated by reciprocal signaling between dental epithelium and dental mesenchyme (Balic and Thesleff 2015). Tooth competence initially resides in the dental epithelium and is subsequently transferred to dental mesenchyme (Lumsden 1988). These two tissues coordinate tooth morphogenesis, with inner dental epithelial cells generating ameloblasts that secrete enamel and/or enameloid that covers the outside of the tooth, and mesenchymal cells generating odontoblasts that secrete the dentine that comprises the inner mineralized part of a tooth. Within the inner dental epithelium, future tooth cusp regions locally express different patterns of growth factors and are called enamel knots (Thesleff et al. 2001). Overall, the roles of dental epithelia and mesenchyme during tooth development are poorly understood. For example, in some experiments, dental mesenchyme positively regulates tooth number (Cai et al. 2007) while in other experiments dental mesenchyme negatively regulates tooth number (Munne et al. 2009).
Most work on tooth development has been done in monophyodont rodents that do not replace teeth. In polyphyodont vertebrates such as fish, teeth are regenerated throughout adult life. Most fish have two sets of jaws, both of which constantly regenerate teeth: an oral jaw in their first or mandibular segment and pharyngeal jaws usually in the seventh pharyngeal segment (Fraser et al. 2009; Ellis et al. 2016). Oral and pharyngeal teeth develop and replace similarly, and in sticklebacks, are morphologically indistinguishable and display similar patterns of gene expression (Ellis et al. 2016). Little is known about the molecular genetic circuitry regulating tooth replacement, but genetic analysis in sticklebacks has provided a powerful system to address this question.
Increases in pharyngeal tooth number have evolved independently in multiple freshwater stickleback populations (Ellis et al. 2015). Comparing lab-reared marine fish and freshwater fish from the benthic (bottom-dwelling) population of Paxton Lake revealed that a divergence in tooth number occurs late in development (around ∼20 mm total length, when fish are juveniles and about half of their adult length). This difference in tooth number continues to increase and becomes more significantly different at adult stages (Cleves et al. 2014). Quantitative trait loci (QTL) mapping identified a large effect QTL that underlies this evolved tooth gain. An F2 cross between a low-toothed Japanese marine fish and a high-toothed benthic Paxton Lake freshwater fish identified a QTL peak on chromosome 21 that explained approximately 30% of the variance in tooth number within the cross (Miller et al. 2014). The peak contained the candidate gene Bone morphogenetic protein 6 (Bmp6) which is dynamically expressed in developing teeth. In situ hybridization revealed Bmp6 expression early in inner dental epithelium, as well as in underlying dental mesenchyme, followed by a decrease in expression in the epithelium before the tooth finally erupts into a functional tooth (Cleves et al. 2014; Ellis et al. 2016). Allele-specific expression (ASE) experiments identified cis-regulatory changes in Bmp6. In tooth tissue from F1 hybrids of high-toothed Paxton benthic fish and low-toothed marine fish, a 1.4-fold decrease in Bmp6 expression from the high-tooth freshwater Paxton benthic allele compared to the marine allele was reported (Cleves et al. 2014). Work in mice and fish has demonstrated an essential role for BMPs in developing teeth (Vainio et al. 1993; Bei et al. 2000; Wang et al. 2012; Jia et al. 2013; Cleves et al. 2018), suggesting a possible causative role of Bmp6 in evolved tooth gain.
Further refinement of the QTL interval identified a haplotype containing 10 single nucleotide polymorphisms (SNPs) within intron 4 of Bmp6 that vary concordantly with the presence or absence of the tooth QTL (Cleves et al. 2018). These variable positions define a high-tooth associated haplotype and low-tooth associated haplotype from the Paxton benthic freshwater and marine alleles, respectively. Six core SNPs lie within 468 bases upstream of the previously described minimally sufficient Bmp6 intron 4 tooth enhancer (Supplementary Figure S1) (Cleves et al. 2018). We hypothesized that these core QTL-associated SNPs are modifying the spatial and/or temporal activity of the adjacent tooth enhancer.
Comparing expression patterns of two different alleles of an enhancer through reporter constructs in an organismal context presents three major problems: (1) comparisons of enhancer variants integrated in two different organisms are difficult to fully control for developmental time and genetic background differences, (2) aspects of reporter expression may in part reflect genomic integration site rather than actual enhancer activity, and (3) different fluorophores are known to have different physical properties (Cranfill et al. 2016) and thus reporter gene differences with different fluorophores might reflect differences in fluorophore brightness, stability, etc. instead of differences in enhancer activity. A single bicistronic transgenic construct that contains both enhancer/reporter pairings could address the first problem by providing a comparison within the same animal (and thus both enhancers being compared are at the same stage and in the same genotype). Furthermore, a single bicistronic construct simultaneously reduces the number of genomic integration sites to one and thus reduces position effects, partially addressing the second problem. The placement of a genetic insulator between the enhancer-reporter pairings can reduce cross talk of an enhancer with the opposite paired reporter, creating a more accurate expression profile. Genetic insulators have been shown to be effective in zebrafish (Bessa et al. 2009; Shimizu and Shimizu 2013). A second alternative approach to a single bicistronic transgene is the use of doubly transgenic two-color lines that include both marine and freshwater enhancers paired with different reporters as parts of separate transgenes. This approach addresses the first problem by having both enhancers in the same animal. With this doubly transgenic two-color line approach, enhancers can be tested with reciprocal pairings (i.e., multiple transgenic reporter lines with different enhancers driving different fluorophores), to control for possible position effects and possible fluorophore differences. Here we use transgenic reporter assay experiments to test the hypothesis that the marine and freshwater Bmp6 intron 4 enhancers have different spatial and/or temporal activity in developing fish embryos, larvae, and adults. We tested this hypothesis in two ways: first, by using a bicistronic enhancer transgene to compare activities of two enhancers in the same fish, and second, by comparing doubly transgenic two-color fish in which the marine and freshwater enhancers drive different fluorophores from different genomic integrations. Lastly, we tested whether the spatial shifts in enhancer activity between marine and freshwater enhancers are also observed for endogenous patterns of Bmp6 expression during tooth development in marine and freshwater fish.
Materials and methods
Animal statement
All animal work was approved by UCB animal protocol #AUP-2015-01-7117-2. Fish were reared as previously described (Erickson et al. 2014).
Insulator containing bicistronic construct
Gibson assembly was used to create bicistronic constructs to determine insulator efficiency in sticklebacks. Two enhancers with distinct expression domains were used: a 1.3-kb fragment from intron 4 of Bmp6 (Cleves et al. 2018) and the stickleback ortholog of the R2 enhancer for Col2a1a, first identified in zebrafish and previously shown to drive similar embryonic expression in sticklebacks (Dale and Topczewski 2011; Erickson et al. 2016). These two enhancers were placed on opposite sides of a genetic insulator, each with a different reporter gene, either mCherry (mCh) or enhanced GFP (eGFP). The mouse tyrosinase GAB (Guanine-rich sequence with A and B boxes) insulator was PCR amplified from the 2pC_GS plasmid (Bessa et al. 2009), while the R2 Col2a1a enhancer was PCR amplified from a previously used reporter plasmid (Erickson et al. 2016). The intron 4 enhancer of Bmp6 was PCR amplified from a reporter plasmid containing either the freshwater allele from the benthic Paxton Lake population or the allele from the Little Campbell marine population (Cleves et al. 2018). All enhancers were PCR amplified simultaneously with the Hsp70l promoter as a single amplicon. eGFP and mCh were amplified from previously used reporter plasmids (O’Brown et al. 2015). Primers used and assembly steps are listed in the Supplemental Methods. All components were combined using a Gibson assembly reaction (New England Biolabs ref # E2611L) following the manufacturer’s protocol and transformed into XL1 blue competent cells (Agilent). Transformed cells were grown on ampicillin-containing LB plates and colony inserts were sequence verified by colony PCR. Positive colonies were used to start 50 ml cultures, which were grown overnight. Plasmids were then isolated by Qiagen midi-prep (#12145) and inserts fully verified by Sanger sequencing.
Tol2 transposase mRNA was transcribed using the plasmid pCS2-TP (Kawakami 2004) that had been linearized with NotI. The linear plasmid was used as template for in vitro transcription using the mMessage SP6 kit (#AM1340). The resulting mRNA was purified using Qiagen RNeasy columns (#74104). Transgene plasmids were co-injected with Tol2 mRNA into newly in vitro fertilized one-cell embryos as described (Erickson et al. 2016). Approximately 200 ng of plasmid in 1 µl was combined with 1 µl of 2M KCl, 0.5 µl of 0.5% phenol red, and approximately 1 µl of 350 ng/µl of Tol2 transposase mRNA, with water added to a final volume of 5 µl, yielding a total concentration of ∼40 ng/µl of plasmid and 70 ng/µl of mRNA. Embryos were generated from Rabbit Slough (RABS; Alaska) marine fish, and lines established and maintained by crossing to lab-reared fish from this same population.
Generation of single color and doubly transgenic two-color reporter lines
The previously described ∼1.3 kb Bmp6 intron 4 tooth enhancer (Cleves et al. 2018) was amplified from a Paxton Lake benthic fish and Little Campbell marine fish (Supplementary Figure S1) using the primer pairs MDS35/36 (GCCGGCTAGCGAGAGCATCCGTCTTGTGGG/GCCGGGATCCAGAGTCCTGATGGCCTCTCC) to create reporter plasmids containing the positive orientation (i.e., same 5′ to 3′ orientation as in endogenous locus) of the enhancer relative to the reporter gene or MDS27/28 (GCCGGCTAGCAGAGTCCTGATGGCCTCTCC/GCCGGGATCCGAGAGCATCCGTCTTGTGGG) to create reporter plasmids containing the negative orientation [i.e., the opposite 5′ to 3′ orientation as in the endogenous locus, and possibly more similar to the orientation that an enhancer 3′ to the promoter (e.g., an enhancer in intron 4) would be after looping to contact the promoter] of the enhancer. The fragments were then cloned in both possible 5′ to 3′ orientations into a Tol2 reporter construct upstream of the zebrafish Hsp70l promoter and either eGFP or mCh using BamHI and NheI in the previously generated reporter constructs. Fish that were transgenic for both the marine and the freshwater reporter alleles were generated in one of two ways: (1) crossing of stable lines each containing a single transgene and (2) injection of one reporter construct into a stable transgenic line of the opposite (i.e., different population and fluorophore) allele.
Detecting enhancer activity by fluorescence microscopy
Enhancer activity of the transgenic constructs was imaged by fluorescence microscopy. Previous work demonstrated a cis-regulatory difference in Bmp6 expression between marine and freshwater alleles, with the difference arising late in development (Cleves et al. 2014). As both a divergence in tooth number attributed to the QTL and allele ASE differences arise late in development, post-20 mm total length (Cleves et al. 2014; 2018), reporter positive fish were dissected at total lengths pre- and post-tooth number divergence (20 mm total length) as previously described (Ellis and Miller 2016). Tooth plates were then fixed in 4% PFA (paraformaldehyde) in 1× phosphate-buffered saline (PBS) for 60 min, washed through a graded series of 3:1, 1:1, 1:3 PBS and glycerol solutions into 100% glycerol, flat-mounted, and imaged. Comparisons were made across the different alleles and orientations on a Leica M165FC dissecting microscope with filters GFP1 (#10447447) and RhodB (#10447360), and a Leica DM2500 compound microscope with filters GFP (#11532366) and TX2 (#11513885). To compare enhancer activity in fish before and after tooth divergence, ventral tooth plates and dorsal tooth plates were imaged and enhancer activity was assessed in the dental epithelium and mesenchyme of each tooth, in each of three pre-divergence sized fish (between 16 and 18.5 mm total length) and three post-divergence sized fish (between 30 and 48 mm total length) in two different sets of integrations and enhancer/reporter pairings. If the QTL-associated SNPs are responsible for the QTL peak and therefore tooth number differences observed late in development, as well as the ASE differences, we would expect the enhancers to have different activity in >20 mm fish compared to <20 mm fish. We would also expect the enhancers to have similar activity earlier in development, when ASE was not significantly different between the freshwater and marine alleles (Cleves et al. 2014).
Quantification of enhancer activity differences across tooth development
As we hypothesized that the QTL-associated intronic polymorphisms result in differential enhancer activity in the dental mesenchyme and/or epithelium, we characterized enhancer activity in both tissues across multiple tooth plates. The stage of each tooth was scored as either early (late cap to early bell stages in which mesenchyme has condensed under the epithelium but no mineralization has occurred), middle (mineralization of the forming tooth has started to occur, also called late bell stage), or late [a fully formed tooth has erupted, also called functional stage (Ellis et al. 2015)]. The activity for each enhancer allele was recorded as either present or absent in the epithelium (early and middle stages) and mesenchyme (all three stages). Additionally, we also recorded if either allele (marine or freshwater) drove more robust or extensive expression in each domain, indicating an allelic bias.
To quantify the expression domain sizes of marine and freshwater tooth enhancer alleles, we used the “measure” function in ImageJ (Schneider et al. 2012) on scaled fluorescence images of developing tooth germs. We measured the 2D (X/Y) mesenchymal domain areas for both the freshwater and marine alleles in three tooth germs each from 12 fish: one tooth germ for each of the three tooth stages (“early,” “middle,” and “late”), analyzed for both reporter construct orientations/fluorophore combinations, and assayed on fish of both <20 and >20 mm total length (for a total of 36 tooth germs, from 12 different fish). The epithelial measurements were taken using the same set of tooth germ images but excluding the “late” stage of tooth germ development because the epithelium becomes ruptured and degrades at this stage (for a total of 24 tooth germs, from 12 different fish).
To test if transgene construct orientation and fluorophore combination significantly affected expression domain size, Wilcoxon rank-sum two-tailed tests were used in R (R Core Team 2020) on mesenchymal and epithelial expression domain areas for marine and freshwater enhancer alleles at each tooth stage (n = 6 vs 6 teeth in all tests, combining fish from early and late fish stages, with each tooth from a different fish). To test if the freshwater allele drove a reduced mesenchymal area and an expanded epithelial domain relative to the marine allele, Wilcoxon signed-rank one-tailed tests were used in R, paired for each tooth germ assessed (n = 12 for each tooth germ stage, with each tooth from a different fish). To test if reduction in the mesenchymal area or expansion in epithelial area for the freshwater allele relative to the marine allele were more significant at >20 mm fish stages than <20 mm fish stages, we used Wilcoxon rank-sum one-tailed tests in R (n = 6 teeth for each tooth germ stage at each fish stage, with each tooth from a different fish).
In situ hybridization on sections
Stickleback adult (∼40 mm standard length) pharyngeal tissues were prepared, sectioned, and assayed by in situ hybridization (ISH) in parallel to compare the spatial distribution of Bmp6 mRNA. Adults derived from marine (RABS) and freshwater [Paxton Benthic (PAXB)] populations were euthanized, and their pharyngeal tissues were fixed overnight in 4% formaldehyde (Sigma P6148) in 1× PBS at 4°C with heavy agitation, washed 3× for 20 min with PBST (1x PBS, 0.1% Tween) on a nutator, then decalcified for 5 days in 20% ethylenediaminetetraacetic acid (EDTA, pH 8.0) at room temperature on a nutator. Marine and freshwater fish were always collected and prepared in parallel such that all storage and preparation intervals were equivalent. The ISH for Bmp6 was carried out as described previously (Square et al. 2021), with some modifications to ensure maximally comparable assays were performed on marine and freshwater samples in parallel. A previously published Bmp6 riboprobe was used in this study (Cleves et al. 2014; Square et al. 2021). The Bmp6 riboprobe was synthesized with digoxygenin-labeled UTP and added at a concentration of ∼300 ng/ml in 20 ml of hybridization buffer, split between two different LockMailer slide containers (Sigma-Aldrich), and agitated overnight in a rotating hybridization oven at 67°C. Slides from marine and freshwater fish were cohoused in the hybridization buffers to ensure equal exposure to the riboprobe between marine and freshwater samples. Hybridization buffer washes, blocking, and antibody incubation steps were as previously described (Square et al. 2021). Signal development was carried out for 2, 3, or 7 days to visualize mRNA localization. Marine and freshwater slides were developed in parallel (in the same solutions, in the same LockMailer containers), and only those sections that experienced the same coloration reaction were compared (i.e., we only directly compared sections that were prepared in parallel). To prepare slides for imaging, they were counterstained with DAPI, rinsed then washed 3× for 5+ min with deionized H2O, coverslipped with deionized H2O, and imaged on a Leica DM2500 microscope. The procedure outlined in this section was replicated three times, each replication used two marine and two freshwater adults, for a total of n = 6 fish from each background.
Results
Two ways to compare enhancers in transgenic fish
We used two strategies to compare enhancer alleles in the same transgenic fish. First, we used a single bicistronic construct with a genetic insulator separating two enhancer/reporter pairs. Second, we used two separate transgenic constructs, independently integrated in the same fish line and each containing a single enhancer allele (marine or freshwater, Supplementary Figure S1) with a distinct fluorescent reporter (eGFP or mCh), to generate doubly transgenic two-color fish.
Insulator efficiency in F0 fish
To test the first strategy of a bicistronic construct separated by an insulator, a bicistronic construct was generated using two enhancers that drive expression in non-overlapping domains. In sticklebacks, the Col2a1a R2 enhancer drives expression in the developing notochord with expression seen by the third day post fertilization (dpf) (Erickson et al. 2016). By 8 dpf, we observed R2 reporter expression in the developing craniofacial skeleton, including Meckel’s cartilage, the hyosympletic, and the ceratohyal (Supplementary Figure S2), similar to the reported enhancer activity in zebrafish (Dale and Topczewski 2011). The Bmp6 intron 4 tooth enhancer has not been reported to drive expression in the domains seen in the R2 Col2a1a enhancer. In addition, the previously described tooth and early fin domains (Cleves et al. 2018), as well as the presently described late fin domains, are not domains in which the Col2a1a enhancer has been observed to drive expression. Thus, to our knowledge, these two enhancers drive distinct and non-overlapping expression domains within these embryonic and larval tissues, providing multiple locations that can test for insulation within the construct.
Three clutches were injected with a Col2a1a enhancer/Bmp6 tooth enhancer bicistronic construct (Figure 1A) for a total of 228 injected embryos, of which 92 were scoreable at 7 dpf. Four domains (left and right pectoral fins, median fin fold, and notochord) were scored for insulation efficiency (0–2 for no to complete insulation, see Supplementary Figure S3 and Supplemental Methods). Across all domains, the average insulator score was 0.94 (Supplementary Table S1). Overall, the bicistronic construct using the mouse tyrosinase insulator element (GAB) moderately prevented reporter genes from being activated by nearby enhancers when placed between the elements. Within the same F0 fish, we observed both insulated and uninsulated domains, with insulation even varying within a domain (Figure 1B). For example, insulation was observed in the median fin and most of the left pectoral fin, but not within some regions of the right pectoral fin of a 7-dpf embryo in which both mCh and eGFP were observed. To control for enhancer/reporter pairing, the inverse construct was created, with the Col2a1a enhancer driving eGFP and the Bmp6 tooth enhancer driving mCh. A total of 154 fish were injected across two clutches, with 30 surviving to 7 dpf that were scoreable, with an average score of 0.64 (Supplementary Table S2). Overall, both insulator constructs demonstrate the ability to drive some degree of separate expression domains of two enhancers concurrently, consistent with results reported in zebrafish that showed insulators can block enhancer-promoter crosstalk (Bessa et al. 2009).
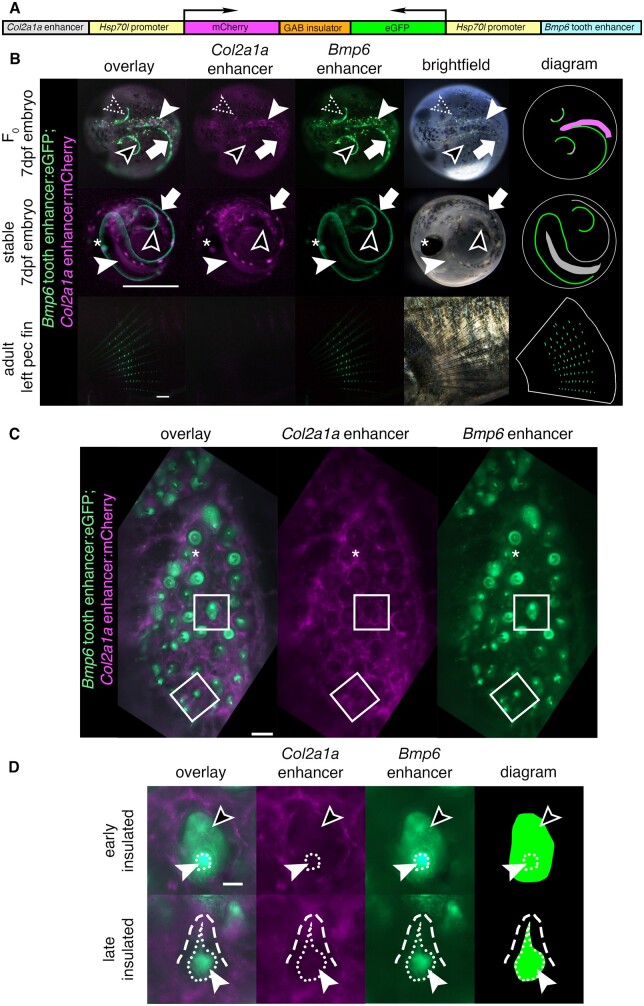
Figure 1.
An insulated bicistronic construct reports separate expression patterns from two different enhancers. (A) Bicistronic construct with a Col2a1a enhancer and Hsp70l promoter driving mCh and the freshwater Bmp6 intronic tooth enhancer and Hsp70l promoter driving eGFP, separated by the mouse tyrosinase insulator (GAB). (B) Transgenic fish show a separation of domains in red and green overlay, red channel only, green channel only, brightfield, and diagram (left to right). Top: In 7 days post fertilization (dpf) F0 embryos (dorsal view), insulation was observed in some but not all domains. Both mCh and eGFP were observed in the same area in the right pectoral fin (dotted arrowhead), indicating incomplete or failed separation of domains, while in other areas of the pectoral fin only eGFP was observed (black arrowhead). Within the notochord (solid white arrowhead), only mCh was observed, while in the median fin (white arrow) only eGFP was observed, indicating insulation in both domains. Middle: In 7 dpf stable F1 embryos (lateral view), only eGFP was observed in the pectoral fins (black arrowhead) indicating successful insulation in those domains, while both fluorophores were detected in the median fin (white arrow) and in the notochord (solid white arrowhead) indicating a lack of insulation. Both fluorophores were detected in the lens of the eye (asterisk), a domain driven by the Hsp70l promoter. Bottom: in adult pectoral fins (lateral view), eGFP but not mCh expression was detected. Diagram: schematic of eGFP and mCh expression in fins and notochord, with overlap shown in gray. Spheres trace the outline of the chorion (top, middle), and white lines trace the pectoral fin (bottom). (C and D) Dorsal pharyngeal tooth plate (C) and representative teeth of early and late stages (D) from adult stable transgenic fish. (C) Insulator effectiveness was observed with eGFP restricted to predicted tooth domains and mCh primarily present in the surrounding tissue. In some teeth, faint mCh appeared to be expressed in the dental mesenchyme (asterisk). (D) eGFP expression was detected in the dental mesenchyme (solid arrowhead and extent of mesenchyme as white dotted line) and dental epithelium (black arrowhead) of developing teeth, while mCh was expressed in the surrounding tissue (white dashed line outlines a mineralized tooth). Scale bars=1 mm (B), 100 µm (C), 25 µm (D). n=92 F0 embryos, >50 F1 embryos, >3 adult fish per time point and 3 teeth per fish for adult stage.
Insulator effectiveness in stable fish
Variation in insulator effectiveness across an individual F0 fish may be due to different genomic integrations of the bicistronic constructs. To determine the effectiveness of a single bicistronic transgene, F0 fish were outcrossed to create stable F1 individuals for the Col2a1a R2: mCh; Bmp6 tooth enhancer: eGFP bicistronic construct. In 7 dpf F1 embryos, complete fin domains of the Bmp6 enhancer were observed, with insulation apparent in some but not all domains (Figure 1B). In adults, Bmp6 enhancer activity was observed in the intersegmental joints of fins (described below), however, no mCh was observed, suggesting effective insulation in that domain (Figure 1B). Insulator activity was also observed in pharyngeal teeth (Figure 1C). The Bmp6 enhancer was observed to drive expression in the mesenchyme and inner dental epithelium of pharyngeal teeth (Figure 1D), consistent with previous reports. mCh was not observed in nearly all tooth domains, suggesting effective insulation in adult teeth. Thus, in stable transgenic adults, the insulator can separate the activity of the two enhancers, including within the dental epithelial and mesenchymal domains of the Bmp6 enhancer.
Bicistronic construct reveals spatial shifts in mesenchymal and epithelial activity of Bmp6 enhancer alleles
Since the GAB genetic insulator can block enhancer-promoter crosstalk in bicistronic constructs, a bicistronic construct with both the marine and freshwater alleles (Figure 2A) was used to create a stable line as a first test for enhancer activity differences. The marine allele, paired with mCh, appeared to drive a more robust mesenchymal domain compared to the freshwater allele (Figure 2, B and C). In contrast, within the inner dental epithelium, more GFP than mCh signal was detected, suggesting an expanded epithelial domain driven by the freshwater enhancer compared to the marine allele. Thus, in developing teeth from fish with this bicistronic transgene, the marine allele drove more expression in the mesenchyme while the freshwater allele drove more expression in the epithelium.
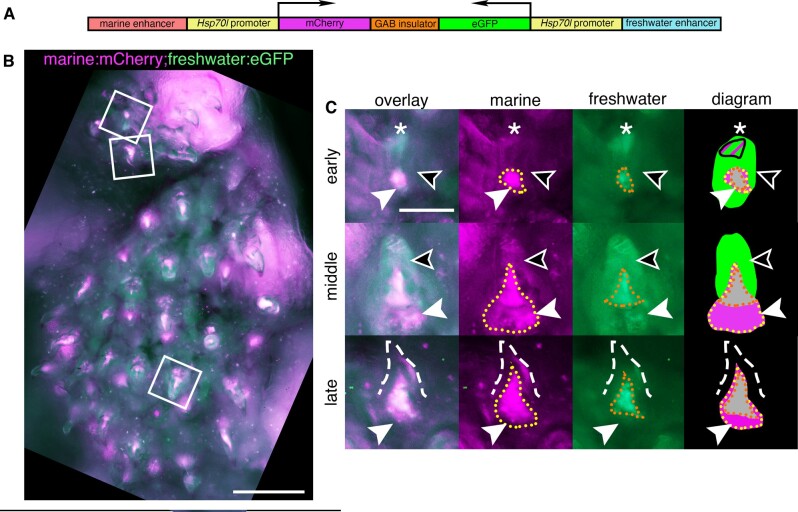
Figure 2.
A bicistronic construct using a genetic insulator separates the expression domains of the marine and freshwater alleles of the Bmp6 tooth enhancer. (A) Bicistronic construct with the marine allele of the intron 4 Bmp6 enhancer/Hsp70l promoter driving mCh and the freshwater allele/Hsp70l promoter driving eGFP separated by the mouse tyrosinase GAB insulator. (B) Dorsal pharyngeal tooth plate from a fish transgenic with construct (A), and representative teeth (white boxes) from early, middle, and late stages (early bell, late bell, and functional, respectively) (C). Early: epithelium expressed eGFP throughout (black arrowhead) while a concentrated tip (asterisk) was observed to contain both marine and freshwater activity. In the mesenchyme (white arrowhead) the marine allele had a more robust and larger expression domain (yellow dotted line) compared to the freshwater allele (orange dotted line). Middle: epithelium had freshwater expression while the marine allele continued to drive more robust expression in the mesenchyme compared to the freshwater allele. Late: As in the other stages, the freshwater allele had a more restricted expression domain in mesenchyme of erupted mineralized teeth (dashed line). Diagram: summary of tooth epithelial and mesenchymal domains. Overlapping mesenchyme domain is gray, and expanded marine mesenchyme is marked with white arrowhead. Scale bars=200 µm (B), 50 µm (C). n=3 fish, 3 teeth per fish.
Doubly transgenic fish confirm expanded freshwater epithelial Bmp6 enhancer activity in post-divergence fish
As a second method to compare the spatial and temporal activity of marine and freshwater enhancer alleles, we generated stable two-color transgenic lines with the two different alleles of the Bmp6 intron 4 tooth enhancer on separate constructs: marine: mCh; freshwater: eGFP, in the opposite 5′ to 3′ direction as the endogenous locus, and marine: eGFP; freshwater: mCh, in the same 5′ to 3′ direction as the endogenous locus. First, we present qualitative assessments of enhancer activity in these lines, and then below present quantitative analyses. In adult fish, both marine and freshwater enhancers were observed to drive dynamic expression in the inner dental epithelium, more intensely at earlier stages, and diminishing as development of the tooth approaches eruption (Figure 3, A–C and Supplementary Figure S4, A–C), consistent with Bmp6 expression detected by whole-mount ISH (Cleves et al. 2014; Ellis et al. 2016). In multiple tooth germs, a brighter focus was observed at the distal tip of the epithelium with both enhancers (Figure 3, A–C and Supplementary Figure S4, A–C), a domain resembling the localized distal epithelial expression of Fgf10 and putative enamel knot in developing shark teeth (Rasch et al. 2016). This distal epithelial domain was the last epithelial region to drive reporter expression prior to cessation in the epithelium. While both enhancers were observed to drive expression in the epithelium, the freshwater allele drove seemingly more robust expression of the reporter, both in terms of intensity as well as spatial extent of the domain (Figure 3, B and C and Supplementary Figure S4, B and C).
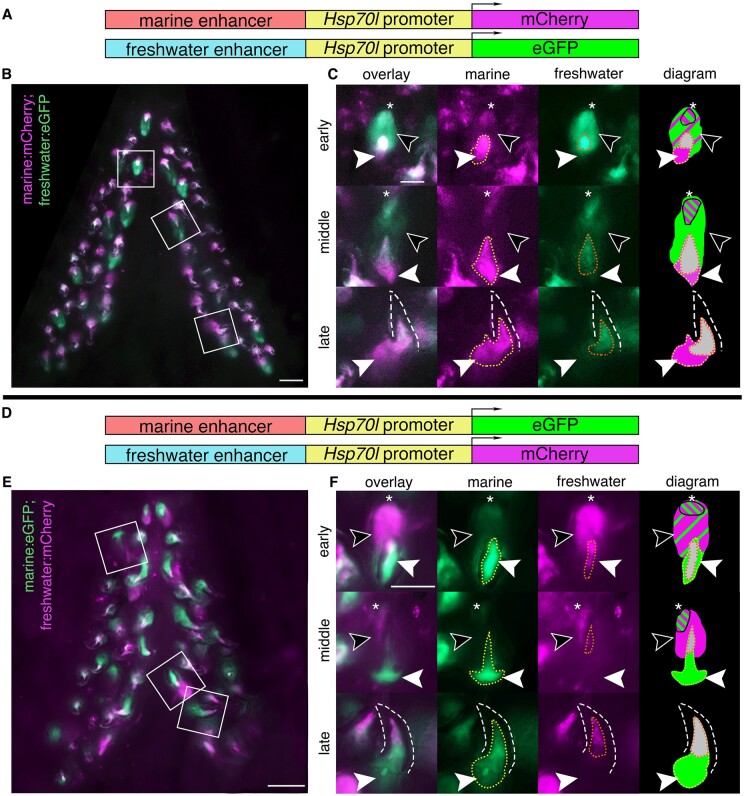
Figure 3.
Reduced mesenchymal and expanded epithelial expression of freshwater enhancer relative to marine enhancer in developing ventral pharyngeal teeth. Ventral pharyngeal tooth plates from fish doubly transgenic for two alleles of the Bmp6 intron 4 enhancer driving two different reporter genes (A, D): the marine enhancer driving mCh with the freshwater enhancer driving eGFP (B, C) and the marine enhancer driving eGFP with the freshwater enhancing driving mCh (E, F). Bilateral ventral pharyngeal tooth plates (B, E) are shown, next to representative teeth from three stages (C, F): early (early bell), middle (late bell), and late (functional) highlighted by white boxes in B, E. (C, F) Early: freshwater and marine enhancer drove expression in the epithelium (black arrowheads), with concentrated expression in the tip (asterisk), and more overall epithelial expression from the freshwater enhancer. Both enhancers also drove expression in the mesenchyme (solid white arrowhead) with a larger expression domain of the marine allele (yellow dotted line) compared to the freshwater allele (orange dotted line) seen in both genotypes. Middle: freshwater allele still drove expression in the epithelium while marine allele had reduced or undetectable expression outside concentrated tip. The marine allele drove more robust mesenchymal expression compared to the freshwater allele. Late: marine allele drove robust expression in the mesenchyme compared to freshwater allele in mineralized tooth (dashed line). Diagram: summary of tooth epithelial and mesenchymal domains. The relative sizes of green and magenta hatched lines correspond to the approximate relative strength of expression in the epithelium. Overlapping mesenchyme domain is gray, and expanded marine mesenchyme is marked with white arrowhead. Scale bars=100 µm (B, E), 50 µm (C, F). n=3 fish per genotype (6 total fish), >25 teeth per fish (304 total teeth).
Doubly transgenic fish confirm reduced freshwater mesenchymal Bmp6 enhancer activity in post-divergence fish
Reporter expression from the two alleles appeared in the mesenchyme of teeth across all stages. In pre-eruption (early and middle stage) tooth germs, condensed mesenchyme was observed to show activity of both enhancers (Figure 3, B and C and Supplementary Figure S4, B and C). In fully formed, erupted, late-stage teeth, reporter expression was observed in the mesenchymal core, extending from the tip of the core down to the base of the tooth where expression widened. Deeper mesenchyme was observed to consistently display marine but not freshwater enhancer activity. The deeper, broader, and more robust mesenchymal expression domain driven by the marine allele compared to the freshwater allele was also observed in stages of tooth development prior to eruption (Figure 3, B and C and Supplementary Figure S4, B and C).
Reciprocal reporter/enhancer pairing in second doubly transgenic two-color line support epithelial and mesenchymal shifts in enhancer activity
To determine if the previous observations were artifacts due to factors such as transgene position effects, fluorophore used, or enhancer orientation, next we made constructs where each enhancer had an opposite enhancer orientation and drove the other fluorophore (Figure 3D). These constructs were then randomly integrated by Tol2-mediated transgenesis, representing independent genomic integrations of oppositely oriented enhancers with alternate fluorophores, simultaneously controlling for genomic position effect, enhancer orientation, and fluorophore. Using these reciprocal constructs, we again observed the epithelial and mesenchymal differences seen in the bicistronic construct and the first double transgenic line, suggesting that the QTL-associated freshwater SNPs reduce mesenchymal and expand epithelial enhancer activity (Figure 3, E and F and Supplementary Figure S4, E and F).
Less pronounced enhancer activity differences in early fish
Allele specific differences in the expression levels of the freshwater and marine alleles of Bmp6, as well as tooth number, have been shown to arise later in development (>20 mm fish length). We hypothesized that if the SNPs found within the freshwater and marine haplotypes contribute to the ASE differences, and subsequent tooth number differences, the differences in enhancer expression should be more pronounced in larger fish compared to smaller fish. Fish smaller than the tooth divergence point (∼16–18.5 mm juveniles, see Materials and methods) were dissected from each genotype and tooth plates were fixed and imaged (Figure 4). While the epithelial and mesenchymal expression differences observed in the older post-divergence stages were still present in both the dental epithelium and mesenchyme (Figure 4, C and F), the enhancer differences were less pronounced. In multiple early and middle stage teeth, the epithelium showed similar activity from both alleles (Figure 4, C and F), unlike the expanded freshwater epithelial domain that was observed in larger fish. Overall, the expression patterns of the two enhancers appeared more similar in pre-divergence fish, consistent with previous ASE and tooth number results (Cleves et al. 2014).
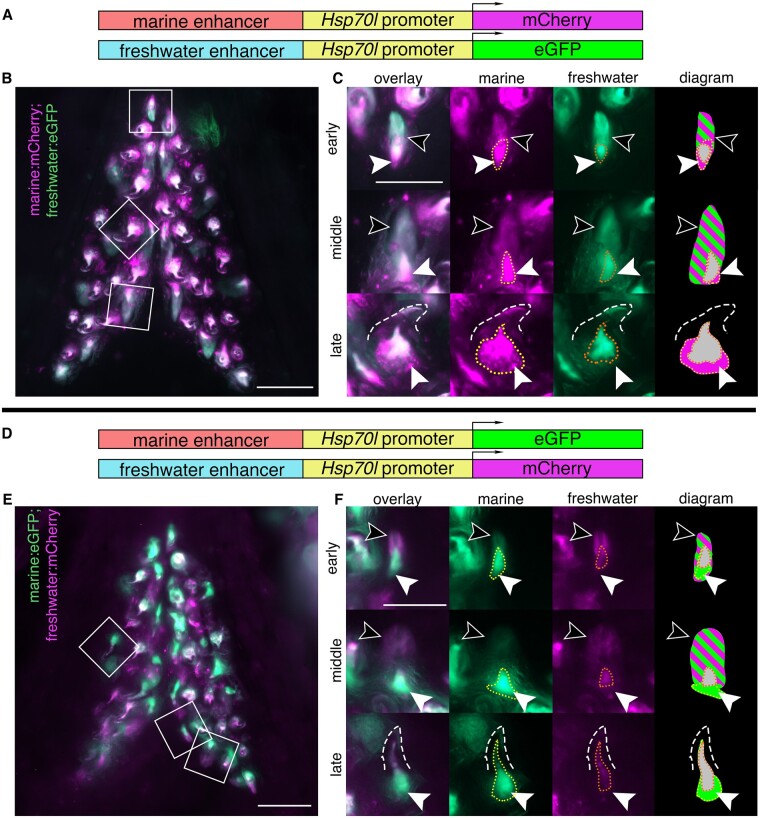
Figure 4.
Marine and freshwater Bmp6 enhancers drive more similar spatial patterns in younger fish. Ventral pharyngeal tooth plates from <20 mm (pre-tooth number divergence) fish doubly transgenic for two alleles of the Bmp6 intron 4 enhancer driving two different reporter genes (A, D): the marine enhancer driving mCh with the freshwater enhancer driving eGFP (B, C) and the marine enhancer driving eGFP with the freshwater enhancer driving mCh (E, F). Bilateral ventral tooth plates (B, E) are shown next to representative teeth from the three stages (C, F): early, middle, and late highlighted by white boxes in B, E. Early: both freshwater and marine enhancer drove expression robustly in the epithelium (black arrowheads), while both enhancers drove expression in the mesenchyme (white arrowheads), the marine enhancer drove a broader domain (yellow dotted line) compared to the freshwater enhancer (orange dotted line). Middle: both enhancers continued to drive robust, apparently similar levels of expression in the epithelium (black arrows). In the mesenchyme (white arrowheads) the domain of the freshwater enhancer was reduced compared to the marine allele. Late: marine allele continued to drive a broader domain within the mesenchyme of mineralized teeth (dashed line). The relative sizes of green and magenta hatched lines correspond to the approximate relative strength of expression in the epithelium. Overlapping mesenchyme domain is gray, and expanded marine mesenchyme is marked with white arrowhead. Scale bars=100 µm (B, E), 50 µm (C, F). n=3 fish per genotype (6 total fish), >25 teeth per fish (249 total teeth).
Quantification of epithelial and mesenchymal expression patterns
To quantify the spatial extent of enhancer activity, we used ImageJ to quantify the reporter gene expression area of tooth mesenchyme and tooth epithelium driven by the marine and freshwater enhancers in both reciprocal two-color lines. We measured the 2D area of each transgene expression domain and then expressed the ratio of the freshwater domain area divided by the marine domain area. We first asked whether the area of enhancer expression was significantly different between the two reciprocal two-color lines (Supplementary Figure S5). In both mesenchyme (Supplementary Figure S5A) and epithelium (Supplementary Figure S5B), the expression domain areas were not significantly different between marine: mCh; freshwater: eGFP and marine: eGFP; freshwater: mCh fish.
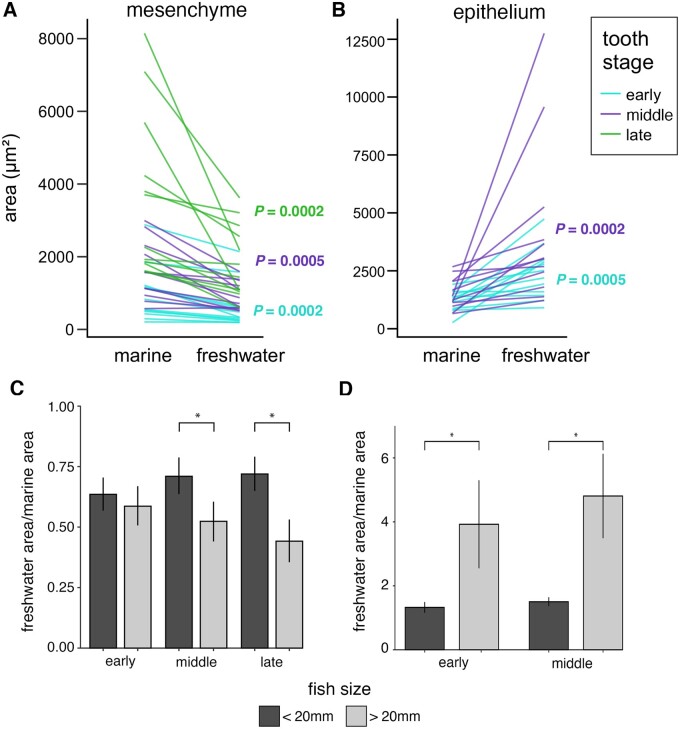
We next tested the hypotheses, based upon our imaging data (Figures 2–4), that the freshwater enhancer drives a reduced mesenchymal but expanded epithelial domain relative to the marine enhancer (Figure 5). In mesenchyme, the freshwater enhancer drove a significantly smaller area than the marine enhancer (Figure 5A) at all three (early, middle, late) tooth stages. In epithelium, the freshwater enhancer drove a significantly larger area than the marine enhancer (Figure 5B) at both early and middle tooth stages.
Figure 5.
Quantification of enhancer domains supports evolved spatial and temporal shifts in enhancer activity. Area of expression domains of both the marine and freshwater enhancers in the mesenchyme (A, C) and epithelium (B, D) for one tooth in each tooth stage (early, middle, late) in three fish per genotype (marine: mCh; freshwater: eGFP, marine: eGFP; freshwater: mCh) at early and late fish stages (<20 mm, >20 mm fish total length) (total n = 12 fish, 36 teeth). (A) In tooth mesenchyme, the freshwater enhancer drove significantly smaller area of reporter gene expression relative to the marine allele at all three tooth stages (early, middle, late). (B) In tooth epithelium, the freshwater enhancer drove significantly larger expression domains relative to the marine allele at both early and middle tooth stages. (A–B) P values show Wilcoxon signed-rank one-tailed tests testing the hypotheses that the freshwater allele is reduced in the mesenchyme and expanded in the epithelium (Figures 2–4). (C) Combining genotypes but maintaining separate categories for tooth stage, the same trend was observed with the reduced mesenchymal expression domain of the freshwater allele becoming more significant in late (>20 mm) stage fish relative to early stage fish (<20 mm). Differences were significant at both middle tooth stage (P = 0.046) and late tooth stage (P = 0.02). (D) Combined genotypes for both early and middle stage teeth showed a significant difference when comparing <20 mm and >20 mm fish (early stage: P = 0.007, middle stage: P = 0.01). (C and D) P-values show Wilcoxon rank sum one-tailed tests testing the hypotheses that the evolved reduction in mesenchymal area (C) and expansion in epithelium (D) are greater at >20 mm fish stages than at <20 mm fish stages. Error bars show standard error of the mean and asterisks denote P < 0.05. n = 3 fish per genotype per fish stage (12 total fish), 3 teeth per fish (36 total teeth).
Pooling the data of both reciprocal genotypes revealed that the relative mesenchymal area of the freshwater enhancer became significantly reduced at both middle and late tooth stages at late (>20 mm) fish stages compared to early (<20 mm) fish stages (Figure 5C). The expansion of the freshwater epithelial domain relative to the marine domain was significantly greater at late fish stages than early fish stages for both early and middle stage teeth (Figure 5D). Comparing enhancer activity between dorsal and ventral pharyngeal teeth revealed a trend toward more evolved shifts in mesenchymal expression in ventral teeth than dorsal teeth (Supplementary Figure S6A), consistent with the more pronounced phenotypic effects of the Bmp6 QTL on ventral than dorsal tooth number (Miller et al. 2014).
Overall, these quantitative data support greater spatial differences of marine and freshwater enhancers at late fish stages than early fish stages, and also strongly support the conclusions that the freshwater enhancer drives a smaller mesenchymal domain and a larger epithelial domain relative to the marine enhancer.
Pectoral and caudal fin expression differences
The Bmp6 intron 4 enhancer was previously known to drive expression in the developing fin margins of the pectoral and median fins early in development, starting approximately 4 dpf (Cleves et al. 2018). In pre-hatching fish, 6 dpf, the domains of the two enhancers appear to be identical (Supplementary Figure S7A). We found that enhancer activity persists at later stages in both the pectoral and caudal fins, specifically in the intersegmental joints. The fin rays of all fins in sticklebacks consist of a series of repeated segments, made up of hemi-segments encasing a mesenchymal core like other teleosts (Haas 1962; Santamaría et al. 1992). In the caudal fin of both genotypes (marine: mCh; freshwater: eGFP; and marine: eGFP; freshwater: mCh), the freshwater enhancer was observed to have activity in multiple intersegmental joints, while the activity of the marine enhancer was detected in few or no joints (Supplementary Figure S7B). A similar pattern is observed in the pectoral fins (Supplementary Figure S7C). With both enhancers, more basal joints were observed to have expression, while fluorophore intensity diminished as the joints became more distal. Overall, across both fin types, the freshwater allele appeared to be active in a larger number of intersegmental joints. While more proximal intersegmental joints were more likely to have activity from both enhancers, the most proximal joint was observed to be lacking detectable reporter expression in some fin rays (Supplementary Figure S8, A and B), suggesting a dynamic cycle of initial inactivity in newly formed, distal, intersegmental joints, followed by a period of activity in most joints as they adopt a more proximal identity, and a final transition to inactivity in the proximal most joints just prior to the ultimate fusion of the basalmost segment to the next segment.
Bmp6 expression differences between marine and freshwater fish
Given the consistent differences in reporter gene activity observed for the marine and freshwater enhancers, we next asked if endogenous Bmp6 expression differed in tooth germs between marine and freshwater animals in a similar fashion. To answer this, we performed ISH on thin sections of pharyngeal tissues from marine (RABS) and freshwater (PAXB) adults (∼40 mm standard length). Marine and freshwater samples were collected, prepared, and assayed in parallel to ensure maximal comparability of the resulting data (see Materials and methods). While early bud and cap stage tooth germs did not show any consistent differences in gene expression, we did observe more widespread mesenchymal expression in marine tooth germs at early and late bell stages, and consistently widespread inner dental epithelial expression in freshwater epithelium relative to marine epithelium at late bell stages (Figure 6 and Supplementary Figure S9). These ISH results corroborate the reporter construct activity, suggesting that the regulation of Bmp6 mRNA in tooth germs varies in the same direction as the variation in activity seen between the marine and freshwater Bmp6 intron 4 enhancers.
Figure 6.
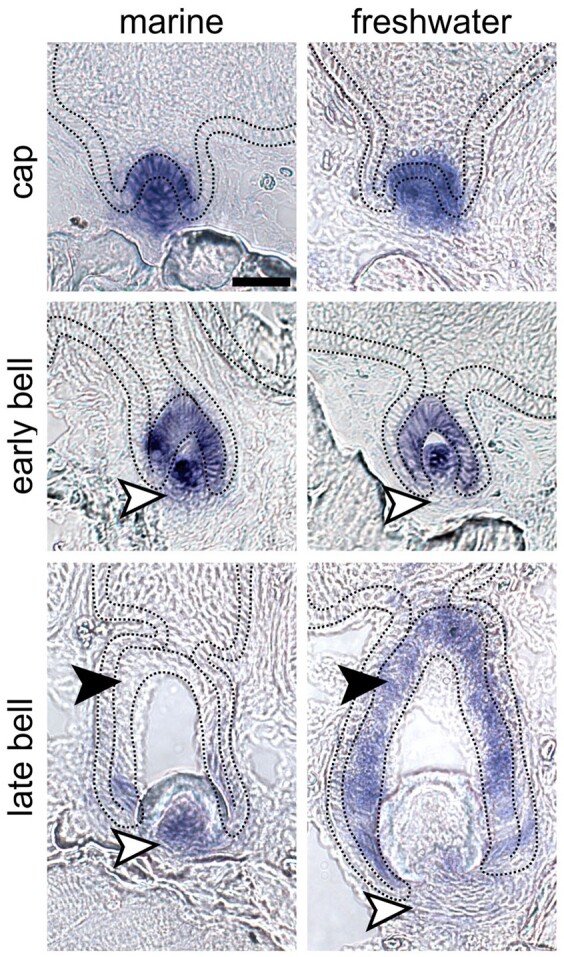
In situ hybridization (ISH) illustrates that Bmp6 expression shifts mirror enhancer activity differences in marine and freshwater backgrounds. ISH of Bmp6 expression on thin sections of marine (left column) and freshwater (right column) fish suggest that marine fish exhibit expanded mesenchymal expression at early and late bell stages (white arrowheads in middle and bottom rows, respectively), while freshwater fish exhibit relatively broader expression in the inner dental epithelium of late bell stage teeth (black arrowheads in bottom row). No expression domain differences were observed in cap stage tooth germs (top row). Marine and freshwater strains are derived from population in Rabbit Slough, AK, USA (RABS), and Paxton Lake, BC, Canada (PAXB), respectively. Black dotted lines demarcate the basalmost layer of epithelium, adjacent to the basement membrane, which includes the inner and outer dental epithelium. See Supplementary Figure S6 for DAPI counterstains and ISH images without markup. Scale bar=20 µm and applies to all panels. n=6 fish per population, >10 teeth per fish.
Discussion
Freshwater and marine alleles of Bmp6 tooth enhancer drive expression differences in developing teeth
Throughout the development of a tooth, multiple pathways and signals, including Bone Morphogenetic Proteins (BMPs), are involved in organ initiation and growth. Knocking out the receptor Bmpr1a in the dental epithelium of mice leads to arrested development of the tooth at the bud stage, demonstrating a key activating role for BMP signaling during tooth development (Andl et al. 2004). Overexpressing Noggin, a BMP antagonist, in the epithelium also results in arrest at the placode stage (Wang et al. 2012). In addition, in Msx1 mutant mice, exogenous Bmp4 can rescue tooth development (Bei et al. 2000). Together, these results suggest a dynamic role of Bmp signaling in tooth development in promoting tooth development at different stages. Bmp6 is dynamically expressed during stickleback tooth development. Expression is detected early in the overlying inner dental epithelium as well as in the condensing underlying odontogenic mesenchyme, with a subsequent cessation of expression in the epithelium, and continuous expression in the mesenchyme of the mineralizing tooth (Cleves et al. 2014; Ellis et al. 2016). Freshwater sticklebacks homozygous for mutations in Bmp6 have reductions in tooth number, showing Bmp6 is required for aspects of tooth development in fish (Cleves et al. 2018).
A previously identified freshwater high-toothed associated haplotype within intron 4 of Bmp6 underlies an evolved increase in tooth number. The core haplotype is defined by six polymorphic sites in the 468 bp region upstream of a minimally sufficient Bmp6 tooth enhancer, potentially modifying enhancer activity. Three lines of evidence (the bicistronic line, and two lines of reciprocal two-color lines) support the hypothesis that the associated polymorphisms upstream of the Bmp6 tooth enhancer result in evolved spatial shifts in enhancer activity between the marine and freshwater alleles (Figures 2–4). Both alleles drove expression in the epithelium of early developing teeth, and in dental mesenchyme throughout development, similar to the expression pattern of the adjacent minimally sufficient 511 bp tooth enhancer previously reported (Cleves et al. 2018) as well as the reported expression of the endogenous Bmp6 gene during tooth development (Cleves et al. 2014). In all three different transgenic comparisons, we observed that the freshwater, high-toothed associated enhancer allele maintained a larger and more robust expression domain in the overlying epithelium for a longer portion of a tooth’s development compared to the marine, low-toothed associated allele. Conversely, the freshwater allele appeared to drive reporter expression in a smaller domain in the underlying mesenchyme in a large proportion of teeth. As this reduced mesenchymal and expanded epithelial activity of the freshwater enhancer relative to the marine enhancer was observed in all three transgenic lines, the enhancer differences are unlikely to be due to differences in genomic integration, enhancer orientation, or different fluorophore properties. We additionally found that marine and freshwater endogenous Bmp6 gene expression domains differed in a manner that was consistent with the reporter gene results. Specifically, we observed larger mesenchymal domains in marine relative to freshwater fish and expanded epithelial domains in freshwater relative to marine fish, especially in late bell stage tooth germs. Together, these data support the hypothesis that the intron 4 enhancer variants associated with tooth number differences drive Bmp6 expression differences in tooth germs of >20 mm fish, which in turn leads to evolved tooth gain in freshwater fish (Figure 7). Outstanding questions include what these deep mesenchymal cells are and whether the expanded marine mesenchymal domain might include quiescent mesenchymal cells involved in tooth replacement.
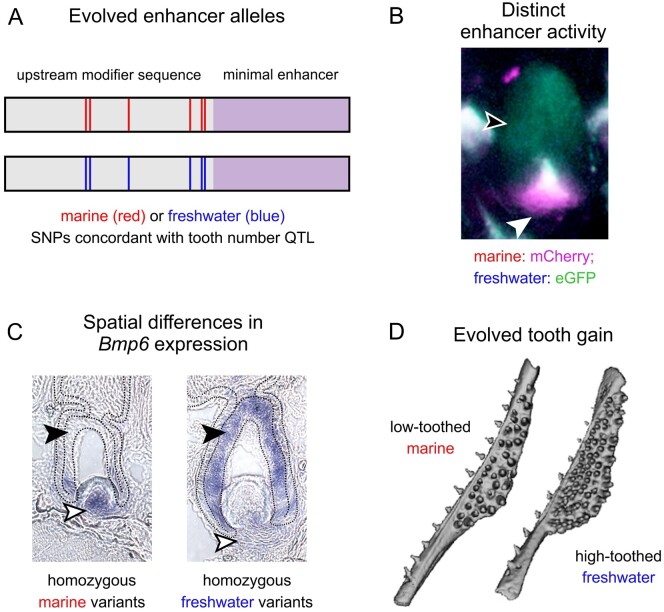
Figure 7.
A model for the role of Bmp6 cis-regulatory changes in underlying evolved tooth gain in sticklebacks. (A) Quantitative trait loci (QTL) and fine mapping previously revealed variants in intron 4 of Bmp6 that were associated with evolved tooth gain in freshwater fish (Cleves et al. 2014, 2018; Miller et al. 2014). These variants are adjacent to a previously characterized minimal enhancer (lavender) that was shown to drive expression in tooth epithelium and mesenchyme (Cleves et al. 2018). Six core single nucleotide polymorphisms (SNPs, depicted as red and blue lines within the modifier sequence) showed complete concordance with a large effect tooth number QTL (Cleves et al. 2018). (B) Marine and freshwater enhancers have different spatial activity, with the derived freshwater allele driving less mesenchymal expression, but more epithelial expression relative to the marine allele. (C) Consistent with the different enhancer activity, Bmp6 expression by in situ hybridization is reduced in the mesenchyme but expanded in the epithelium in freshwater teeth relative to marine teeth. (D) We hypothesize that the enhancer alleles (A) have spatially shifted enhancer activity (B), resulting in shifts in Bmp6 expression overall (C), and evolved tooth gain in freshwater fish (D).
One major unanswered question remains how the spatial and temporal differences in Bmp6 enhancer activity reported here could regulate the increases in tooth number and accelerated tooth replacement rates previously reported for freshwater fish relative to ancestral marine fish. We previously hypothesized that tooth regeneration might be regulated similarly to mammalian hair regeneration (Cleves et al. 2018), where BMP signaling promotes epithelial stem cell quiescence, and reducing BMP signaling in mouse skin accelerates hair regeneration (Kandyba et al. 2013). Multiple lines of gene expression data have supported this hypothesis (Cleves et al. 2018; Hart et al. 2018; Square et al. 2021). However, several other compelling, and not necessarily mutually exclusive, alternative hypotheses include (1) tooth regeneration might be regulated by coordinated cyclic waves involving BMP signaling, similar to the cyclic nature of hair regeneration previously reported in mice (Plikus et al. 2008), and (2) tooth regeneration might be regulated by a reaction-diffusion system in which BMPs act as inhibitors (Kondo and Miura 2010), similar to a proposal previously made for shark denticle formation (Cooper et al. 2018). This first alternative hypothesis of cyclic waves coordinating tooth replacement is reminiscent of the decades old Zahnreihen theory that posits that tooth replacement in polyphyodonts is coordinated across the dentition, usually occurring at alternating tooth positions (Edmund 1960). For all these hypotheses, the increased epithelial and/or the decreased mesenchymal enhancer activity of Bmp6 could be the causative change leading to evolved tooth gain. Future experiments will continue to test these hypotheses, test whether the epithelial and mesenchymal Bmp6 enhancer shifts are regulated by the same or different mutations, and ultimately determine how the intronic Bmp6 enhancer haplotype identified by our previous genetic mapping studies regulates increases in tooth number.
Previous ASE experiments demonstrated a 1.4-fold reduction in the freshwater Bmp6 allele compared to the marine in F1 hybrid adult tooth tissue that included the entire ventral pharyngeal jaw, and thus both tooth epithelial and mesenchymal cells (Cleves et al. 2014). The mesenchymal biases in reporter expression are consistent with the ASE result, with more robust mesenchymal expression driven by the marine allele compared to the freshwater allele potentially responsible for the higher expression of the marine allele in the ASE experiments. In contrast, the expanded freshwater epithelial enhancer domain is not consistent with the overall ASE result in which freshwater alleles had cis-regulatory downregulation relative to marine alleles. Since the reduced mesenchymal domain in the freshwater enhancer relative to the marine enhancer was the most striking qualitative difference, it is possible that the epithelial bias, with a stronger signal driven by the freshwater enhancer, is quantitatively canceled out by the bias in the mesenchyme, explaining the overall reduction of freshwater Bmp6 expression compared to marine Bmp6 expression in F1 hybrids.
The enhancer expression differences were significantly greater in larger, post-tooth number divergence fish compared to smaller, pre-tooth number divergence fish. While the mesenchyme appeared to have a somewhat reduced difference of expression between the two alleles, the epithelium demonstrated less pronounced differences in activity between the alleles in pre-divergence fish. These observations are consistent with ASE results and the divergence in tooth number in marine and freshwater fish. While the mesenchymal difference was still observable early, it is possible that there are other regulatory regions which act as repressors for the marine Bmp6 allele or enhancers for the freshwater Bmp6 allele early in development and so mask the mesenchymal bias of the intron 4 enhancer. For example, we previously reported a 5′ Bmp6 tooth enhancer that also contributes to the overall pattern of Bmp6 expression in developing teeth (Erickson et al. 2015).
Future experiments to measure ASE in isolated tissues, with epithelium and mesenchyme separated, could test whether opposing quantitative differences are present in dental epithelium vs mesenchyme, as the new data presented here suggest. A quantitative method could be used to further test the hypothesis that the two enhancers drive differing levels of expression, such as pyrosequencing (Wittkopp 2012) with the two enhancers both driving identical fluorophores, with a single synonymous mutation distinguishing the two. Alternatively, single-cell RNA-seq in the dental epithelium and mesenchyme, tracking the respective reporters of each enhancer, could determine if there are quantifiable expression differences between the two enhancers.
QTL-associated sequence difference in alleles may underlie expression domain differences and evolved tooth gain
There are 14 point mutations and three indels distinguishing a low-toothed marine (Little Campbell) allele from the high-toothed Paxton Lake allele of the intron 4 enhancer in our reporter constructs. Previous experiments identified ten SNPs that co-occur consistently with the presence or absence of a tooth number QTL and of these ten, the core six are present in the enhancer reporter constructs tested here (Cleves et al. 2018). From our results, we are unable to distinguish whether these six polymorphisms contribute to the expression differences we observed. While it is possible that the three indels or the eight non-QTL-associated SNPs may contribute, it is an attractive and parsimonious hypothesis that the same SNPs that co-occur with the tooth QTL are also responsible for the reporter expression differences, and the previously described ASE results. Of the six QTL-associated SNPs tested here, of special interest is the second QTL-associated SNP, which in the freshwater allele, creates a predicted NFATc1 binding site (Cleves et al. 2018). NFATc1 was shown to be required for balancing of quiescent and actively dividing stem cells in hair follicles (Horsley et al. 2008) which share homology with teeth (Pispa and Thesleff 2003; Biggs and Mikkola 2014; Ahn 2015), and so a difference in NFATc1 binding may potentially play a role in the Bmp6 ASE and enhancer activity differences observed previously and here. Supporting this hypothesis, Nfatc1b expression was recently shown to be present in stickleback tooth germs and functional tooth mesenchyme (Square et al. 2021).
To better determine which polymorphisms may underlie the expression differences we observed, hybrid enhancers can be made. For example, if the creation of an NFATc1 binding site is at least partially responsible for the observed differences, a marine allele with the SNP converted to the freshwater identity, from a “C” to a “T,” may recapitulate the freshwater enhancer expression patterns. By creating and testing hybrid enhancers, future experiments could test which enhancer polymorphisms alone and in combination contribute to the expression differences reported here.
Fin expression differences
In addition to the reporter expression differences driven by the two enhancers during tooth development, we observed distinct expression patterns in the pectoral and caudal fins. It was previously known that the minimal 511 base pair enhancer drove expression in the margins of early pectoral and median fins, but expression in adult fins had not been described. BMP signaling plays a role in fin regeneration, with BMP inhibition reducing osteoblast differentiation in new cells arising at the leading edge of the regenerating fin (Stewart et al. 2014). During zebrafish fin regeneration, bmp2b, bmp4, and bmp6 are expressed, and are thought to be important (Laforest et al. 1998; Murciano et al. 2002; Quint et al. 2002; Smith et al. 2006). While both alleles of the Bmp6 enhancer drive expression in the pectoral and caudal fins of sticklebacks, the differing enhancer activities may result in developmental differences, through osteoblast function in the developing lepidotrichia and intersegmental joints, possibly leading to different fin morphologies and/or regenerative abilities. Differences in expression of bmp2 have been observed in the regeneration of different rays of the caudal fin in cichlids (Ahi et al. 2017), as well as the expression of the gene msxb, which is downstream of bmp signaling in the regenerating zebrafish fin (Smith et al. 2006).
Multiple studies have identified habitat specific differences in fin morphology (Taylor and McPhail 1986; Kristjánsson et al. 2005; Hendry et al. 2011). As the two enhancers are derived from populations with two distinct ecotypes, a benthic freshwater population, and a highly mobile anadromous population, it is possible that this enhancer may influence pectoral and caudal fin size and shape in an adaptive manner. Characterization of fin morphology using fish from either a population in which the high-toothed and low-toothed associated haplotypes are segregating, or those from a control cross in which both alleles were present in the founding, could test whether there is a fin morphology difference associated with the different alleles.
Bicistronic constructs and the use of genetic insulators
Simultaneous comparison of two enhancer alleles in a single organism via a bicistronic construct is an attractive means to compare molecularly divergent enhancers (e.g., pairs of enhancers that contain sequence variation across populations to determine if there are population-specific differences in enhancer activity). Previous work in zebrafish utilized genetic insulators as part of an enhancer trap as well as with two different tissue-specific promoters and demonstrated the effectiveness of the technique (Bessa et al. 2009; Shimizu and Shimizu 2013).
Here, we used a bicistronic construct with a Bmp6 enhancer and a Col2a1a enhancer driving different fluorophores in mosaically transgenic F0 fish to test whether the activities of two enhancers could be insulated from each other. Within the same F0 individual, some domains demonstrated a high degree of insulator effectiveness while others did not. There are at least two possible explanations: (1) the insulated vs non-insulated regions represent distinct and mosaic integration events, with the insulator effectiveness determined by the integration site in a particular subpopulation of cells, or (2) the same integration event can differ in insulator behavior stochastically or based on some context that differs from an insulated expression domain to an un-insulated domain. Regardless, examining enhancer activity in stable lines will still provide a more complete picture of the role of the regulatory element and has advantages over mosaic F0 analyses.
Genetic insulators have been reported to limit enhancer activity across the insulator boundary (Bessa et al. 2009; Shimizu and Shimizu 2013) as well as protect against position effects (Chung et al. 1993), while other experiments show a lack of protection (Grajevskaja et al. 2013). The insulator used here, from the 5′ end of the mouse tyrosinase locus, was reported to bind CTCF (CCCTC-binding factor), like the β-globin 5′-HS4 insulator from chicken, and is reported to prevent influences from nearby chromatin state and gene activity, the hallmarks of genetic insulators (Montoliu et al. 1996; Giraldo et al. 2003; Molto et al. 2009). As there are conflicting reports of the use of insulators to fully shield from nearby chromatin states and position effects, the combined use of a landing pad locus could help to further reduce these effects (Roberts et al. 2014). We recommend a multipronged approach utilizing multiple transgenic lines (e.g., either bicistronic constructs or multiple independent reciprocal two-color lines where each enhancer drives a different fluorophore in the same animal). Similar methods in doubly transgenic animals should allow future dissection of spatial differences in enhancer alleles, with the two methods acting as means of independent verification.
Changes in cis-regulation of developmental genes can be an important driver of morphological evolution, as well as human disease. The impact of mutations in cis-regulatory regions can be difficult to predict, and if the effect is subtle or slight, also to detect. The use of two enhancers in the same individual, either as parts of two independent transgenes or within a single bicistronic construct, can both control for the trans-environment and make even slight differences in expression activity apparent due to simultaneous imaging of reporter genes driven by both enhancers. Such an approach allows for directly comparing molecularly divergent regulatory elements, potentially identifying causal polymorphisms with important developmental and evolutionary implications.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We thank Phillip Cleves, James Hart, Priscilla Erickson, and Ana Shaughnessy for helpful discussions, and Sophie Archambeault and Alyssa Bormann for comments on the manuscript.
Funding
M.D.S. was supported by a National Science Foundation Graduate Research Fellowship; T.A.S. was supported by National Institutes of Health (NIH) Fellowship F32-DE027871 to T.A.S. and C.T.M.; M.D.S., T.A.S., and C.T.M. were supported by NIH Grant R01-DE021475 to C.T.M.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Ahi EP, Richter F, Sefc KM.. 2017. A gene expression study of ornamental fin shape in Neolamprologus brichardi, an African cichlid species. Sci Rep. 7:17398.doi:10.1038/s41598-017-17778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn Y. 2015. Signaling in tooth, hair, and mammary placodes. Curr Top Dev Biol. 111:421–459. doi: 10.1016/bs.ctdb.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, et al. 2008. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 62:76–85. doi:10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, et al. 2004. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 131:2257–2268. doi:10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- Archambeault SL, Bärtschi LR, Merminod AD, Peichel CL.. 2020. Adaptation via pleiotropy and linkage: association mapping reveals a complex genetic architecture within the stickleback Eda locus. Evol Lett. 4:282–301. doi:10.1002/evl3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balic A, Thesleff I.. 2015. Tissue interactions regulating tooth development and renewal. Curr Top Dev Biol. 115:157–186. doi:10.1016/bs.ctdb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL.. 2000. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 127:4711–4718. [DOI] [PubMed] [Google Scholar]
- Bell MA, Foster SA, editors. 1994. The Evolutionary Biology of the Threespine Stickleback. Oxford, NY: Oxford University Press. [Google Scholar]
- Bessa J, Tena JJ, de la Calle-Mustienes E, Fernández-Miñán A, Naranjo S, et al. 2009. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis -regulatory regions in zebrafish. Dev Dyn. 238:2409–2417. doi:10.1002/dvdy.22051. [DOI] [PubMed] [Google Scholar]
- Biggs LC, Mikkola ML.. 2014. Early inductive events in ectodermal appendage morphogenesis. Semin Cell Dev Biol. 25–26:11–21. doi:10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Cai J, Cho S-W, Kim J-Y, Lee M-J, Cha Y-G, et al. 2007. Patterning the size and number of tooth and its cusps. Dev Biol. 304:499–507. doi:10.1016/j.ydbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 134:25–36. doi:10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Shapiro MD, et al. 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 327:302–305. doi:10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G.. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 74:505–514. doi:10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, et al. 2014. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc Natl Acad Sci U S A. 111:13912–13917. doi:10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Hart JC, Agoglia RM, Jimenez MT, Erickson PA, et al. 2018. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet. 14:e1007449. doi:10.1371/journal.pgen.1007449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G Jr., Dickson M, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 307:1928–1933. doi:10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Cooper RL, Thiery AP, Fletcher AG, Delbarre DJ, Rasch LJ, et al. 2018. An ancient Turing-like patterning mechanism regulates skin denticle development in sharks. Sci Adv. 4:eaau5484. doi:10.1126/sciadv.aau5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranfill PJ, Sell BR, Baird MA, Allen JR, Lavagnino Z, et al. 2016. Quantitative assessment of fluorescent proteins. Nat Methods. 13:557–562. doi:10.1038/nmeth.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, et al. 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci U S A. 101:6050–6055. doi:10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RM, Topczewski J.. 2011. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol. 357:518–531. doi:10.1016/j.ydbio.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmund AG. 1960. Tooth Replacement Phenomena in the Lower Vertebrates. Toronto: Royal Ontario Museum.
- Ellis NA, Donde NN, Miller CT.. 2016. Early development and replacement of the stickleback dentition. J Morphol. 277:1072–1083. doi:10.1002/jmor.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Glazer AM, Donde NN, Cleves PA, Agoglia RM, et al. 2015. Distinct developmental genetic mechanisms underlie convergently evolved tooth gain in sticklebacks. Development. 142:2442–2451. doi:10.1242/dev.124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Miller CT.. 2016. Dissection and flat-mounting of the threespine stickleback branchial skeleton. J. Vis. Exp. 54056. doi:10.3791/54056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Baek J, Hart JC, Cleves PA, Miller CT.. 2018. Genetic dissection of a supergene implicates Tfap2a in craniofacial evolution of threespine sticklebacks. Genetics. 209:591–605. doi:10.1534/genetics.118.300760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Cleves PA, Ellis NA, Schwalbach KT, Hart JC, et al. 2015. A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev Biol. 401:310–323. doi:10.1016/j.ydbio.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Glazer AM, Cleves PA, Smith AS, Miller CT.. 2014. Two developmentally temporal quantitative trait loci underlie convergent evolution of increased branchial bone length in sticklebacks. Proc Biol Sci. 281:20140822.doi:10.1098/rspb.2014.0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson PA, Ellis NA, Miller CT.. 2016. Microinjection for transgenesis and genome editing in threespine sticklebacks. J. Vis. Exp. 54055. doi:10.3791/54055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, et al. 2009. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 7:e31. doi:10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, Levine M.. 2018. Developmental enhancers and chromosome topology. Science. 361:1341–1345. doi:10.1126/science.aau0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Tome JM, Shendure J.. 2020. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nat Rev Genet. 21:292–310. doi:10.1038/s41576-019-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo P, Martinez A, Regales L, Lavado A, Garcia-Diaz A, et al. 2003. Functional dissection of the mouse tyrosinase locus control region identifies a new putative boundary activity. Nucleic Acids Res. 31:6290–6305. doi:10.1093/nar/gkg793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajevskaja V, Balciuniene J, Balciunas D.. 2013. Chicken β-globin insulators fail to shield the nkx2.5 promoter from integration site effects in zebrafish. Mol Genet Genomics. 288:717–725. doi:10.1007/s00438-013-0778-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross HP, Anderson JM.. 1984. Geographic variation in the gillrakers and diet of European threespine sticklebacks, Gasterosteus aculeatus. Copeia. 1984:87.doi:10.2307/1445038. [Google Scholar]
- Haas HJ. 1962. Studies on mechanisms of joint and bone formation in the skeleton rays of fish fins. Dev Biol. 5:1–34. doi:10.1016/0012-1606(62)90002-7. [DOI] [PubMed] [Google Scholar]
- Hagen DW. 1967. Isolating mechanisms in threespine sticklebacks (Gasterosteus). J Fish Res Bd Can. 24:1637–1692. doi:10.1139/f67-138. [Google Scholar]
- Hart JC, Ellis NA, Eisen MB, Miller CT.. 2018. Convergent evolution of gene expression in two high-toothed stickleback populations. PLoS Genet. 14:e1007443.doi:10.1371/journal.pgen.1007443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry AP, Hudson K, Walker JA, Räsänen K, Chapman LJ.. 2011. Genetic divergence in morphology-performance mapping between Misty Lake and inlet stickleback: Genetic divergence in morphology-performance mapping. J Evol Biol. 24:23–35. doi:10.1111/j.1420-9101.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E.. 2008. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 132:299–310. doi:10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Kingman GA, Jones FC, Guenther CA, Grimwood J, et al. 2016. Evolving new skeletal traits by cis -regulatory changes in Bone Morphogenetic Proteins. Cell. 164:45–56. doi:10.1016/j.cell.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Zhou J, Gao Y, Baek J-A, Martin JF, et al. 2013. Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development. 140:423–432. doi:10.1242/dev.081927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandyba E, Leung Y, Chen Y-B, Widelitz R, Chuong C-M, et al. 2013. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Proc Natl Acad Sci U S A. 110:1351–1356. doi:10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. 2004. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 77:201–222. doi:10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kondo S, Miura T.. 2010. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 329:1616–1620. doi:10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- Kristjánsson BK, Skúlason S, Noakes DLG.. 2005. Unusual number of pectoral fin rays in an Icelandic population of threespine stickleback (Gasterosteus aculeatus) recently isolated in freshwater. Evol Ecol. 18:379–384. doi:10.1007/s10682-004-2679-5. [Google Scholar]
- Laforest L, Brown CW, Poleo G, Géraudie J, Tada M, et al. 1998. Involvement of the sonic hedgehog, patched1 and bmp2 genes in patterning of the zebrafish dermal fin rays. Development. 125:4175–4184. [DOI] [PubMed] [Google Scholar]
- Lavin PA, McPhail JD.. 1986. Adaptive divergence of trophic phenotype among freshwater populations of the threespine stickleback (Gasterosteus aculeatus). Can J Fish Aquat Sci. 43:2455–2463. doi:10.1139/f86-305. [Google Scholar]
- Lumsden AG. 1988. Spatial organization of the epithelium and the role of neural crest cells in the initiation of the mammalian tooth germ. Development. 103 Suppl:155–169. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Rundle HD.. 2002. Speciation in nature: the threespine stickleback model systems. Trends Ecol Evol. 17:480–488. doi:10.1016/S0169-5347(02)02579-X. [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, et al. 2007. cis-regulatory changes in Kit Ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 131:1179–1189. doi:10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Glazer AM, Summers BR, Blackman BK, Norman AR, et al. 2014. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics. 197:405–420. doi:10.1534/genetics.114.162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molto E, Fernandez A, Montoliu L.. 2009. Boundaries in vertebrate genomes: different solutions to adequately insulate gene expression domains. Brief Funct Genomic Proteomic. 8:283–296. doi:10.1093/bfgp/elp031. [DOI] [PubMed] [Google Scholar]
- Montoliu L, Umland T, Schütz G.. 1996. A locus control region at −12 kb of the tyrosinase gene. Embo J. 15:6026–6034. doi:10.1002/j.1460-2075.1996.tb00991.x. [PMC free article] [PubMed] [Google Scholar]
- Munne PM, Tummers M, Järvinen E, Thesleff I, Jernvall J.. 2009. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 136:393–402. doi:10.1242/dev.025064. [DOI] [PubMed] [Google Scholar]
- Murciano C, Fernández TD, Durán I, Maseda D, Ruiz-Sánchez J, et al. 2002. Ray–interray interactions during fin regeneration of Danio rerio. Dev Biol. 252:214–224. doi:10.1006/dbio.2002.0848. [DOI] [PubMed] [Google Scholar]
- O'Brown NM, Summers BR, Jones FC, Brady SD, Kingsley DM.. 2015. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. Elife. 4:e05290. doi:10.7554/eLife.05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pispa J, Thesleff I.. 2003. Mechanisms of ectodermal organogenesis. Dev Biol. 262:195–205. doi:10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, et al. 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 451:340–344. doi:10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint E, Smith A, Avaron F, Laforest L, Miles J, et al. 2002. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 99:8713–8718. doi:10.1073/pnas.122571799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R Foundation for Statistical Computing. Vienna, Austria: R: A language and environment for statistical computing. https://www.R-project.org/. [Google Scholar]
- Rasch LJ, Martin KJ, Cooper RL, Metscher BD, Underwood CJ, et al. 2016. An ancient dental gene set governs development and continuous regeneration of teeth in sharks. Dev Biol. 415:347–370. doi:10.1016/j.ydbio.2016.01.038. [DOI] [PubMed] [Google Scholar]
- Rebeiz M, Tsiantis M.. 2017. Enhancer evolution and the origins of morphological novelty. Curr Opin Genet Dev. 45:115–123. doi:10.1016/j.gde.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DT, Peichel CL.. 2010. Perspectives on the genetic architecture of divergence in body shape in sticklebacks. Integr Comp Biol. 50:1057–1066. doi:10.1093/icb/icq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, Shilatifard A.. 2018. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 28:608–630. doi:10.1016/j.tcb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Miguel-Escalada I, Slovik KJ, Walsh KT, Hadzhiev Y, et al. 2014. Targeted transgene integration overcomes variability of position effects in zebrafish. Development. 141:715–724. doi:10.1242/dev.100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabarís G, Laiker I, Preger-Ben Noon E, Frankel N.. 2019. Actors with multiple roles: pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 35:423–433. doi:10.1016/j.tig.2019.03.006. [DOI] [PubMed] [Google Scholar]
- Santamaría JA, Marí-Beffa M, Becerra J.. 1992. Interactions of the lepidotrichial matrix components during tail fin regeneration in teleosts. Differentiation. 49:143–150. doi:10.1111/j.1432-0436.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- Schluter D, McPhail JD.. 1992. Ecological character displacement and speciation in sticklebacks. Am Nat. 140:85–108. doi:10.1086/285404. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9:671–675. doi:10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu A, Shimizu N.. 2013. Dual promoter expression system with insulator ensures a stringent tissue-specific regulation of two reporter genes in the transgenic fish. Transgenic Res. 22:435–444. doi:10.1007/s11248-012-9653-8. [DOI] [PubMed] [Google Scholar]
- Smith A, Avaron F, Guay D, Padhi BK, Akimenko MA.. 2006. Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev Biol. 299:438–454. doi:10.1016/j.ydbio.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Square TA, Sundaram S, Mackey EJ, Miller CT.. 2021. Distinct tooth regeneration systems deploy a conserved battery of genes. EvoDevo. 12:4.doi:10.1186/s13227-021-00172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Gomez AW, Armstrong BE, Henner A, Stankunas K.. 2014. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 6:482–498. doi:10.1016/j.celrep.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, McPhail JD.. 1986. Prolonged and burst swimming in anadromous and freshwater threespine stickleback, Gasterosteus aculeatus. Can J Zool. 64:416–420. doi:10.1139/z86-064. [Google Scholar]
- Thesleff I, Keränen S, Jernvall J.. 2001. Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 15:14–18. doi:10.1177/08959374010150010401. [DOI] [PubMed] [Google Scholar]
- Vainio S, Karavanova I, Jowett A, Thesleff I.. 1993. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 75:45–58. doi:10.1016/S0092-8674(05)80083-2. [PubMed] [Google Scholar]
- Walker JA. 1997. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L. (Gasterosteidae) body shape. Biol J Linn Soc. 61:3–50. doi:10.1111/j.1095-8312.1997.tb01777.x. [Google Scholar]
- Walker JA, Bell MA.. 2000. Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus). J Zool. 252:293–302. doi:10.1111/j.1469-7998.2000.tb00624.x. [Google Scholar]
- Wang Y, Li L, Zheng Y, Yuan G, Yang G, et al. 2012. BMP activity is required for tooth development from the lamina to bud stage. J Dent Res. 91:690–695. doi:10.1177/0022034512448660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ. 2012. Using pyrosequencing to measure allele-specific mRNA abundance and infer the effects of cis- and trans-regulatory differences. In: Orgogozo V, Rockman MV, editors. Molecular Methods for Evolutionary Genetics, Methods in Molecular Biology. Totowa, NJ: Humana Press. p. 297–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.