Abstract
Atherosclerosis is the underlying cause of heart attack, ischemic stroke and peripheral arterial disease, and genetic factors involved remain mostly unidentified. We previously identified a significant locus on mouse chromosome 17 for atherosclerosis, Ath49, in an intercross between BALB/c and SM strains. Ath49 partially overlaps in the confidence interval with Ath22 mapped in an AKR × DBA/2 intercross. Bioinformatics analysis prioritized Mep1a, encoding meprin 1α metalloendopeptidase, as a likely candidate gene for Ath49. To prove causality, Mep1a−/−Apoe−/− mice were generated and compared with Mep1a+/+Apoe−/− mice for atherosclerosis development. Mep1a was found abundantly expressed in atherosclerotic lesions but not in healthy aorta and liver of mice. Mep1a−/− Apoe−/− mice exhibited significant reductions in both early and advanced lesion sizes. Loss of Mep1a led to decreased necrosis but increased macrophage and neutrophil contents in advanced lesions, reduced plasma levels of CXCL5 and an oxidative stress biomarker. In addition, Mep1a−/− mice had significantly reduced triglyceride levels on a chow diet. Thus, Mep1a is a susceptibility gene for atherosclerosis and aggravates atherosclerosis partially through action on oxidative stress and inflammation.
Keywords: Atherosclerosis, Mep1a, metalloendopeptidase, oxidative stress, mice
Introduction
Atherosclerosis is the primary cause of heart attack, ischemic stroke, and peripheral arterial disease. It is a complex disease resulting from interactions between environmental and genetic factors (Mozaffarian et al. 2015). Genome-wide association studies (GWAS) have identified over 300 variants associated with coronary heart disease in humans [Coronary Artery Disease (C4D) Genetics Consortium 2011; CARDIoGRAMplusC4D Consortium et al. 2013; Nikpay et al. 2015; Nelson et al. 2017], with most of the variants implicated in pathways related to blood vessel morphogenesis, lipid metabolism, nitric oxide signaling, and inflammation (Nelson et al. 2017). However, the loci identified account for only 21% of the genetic heritability of coronary heart disease (Nelson et al. 2017), suggesting that many more loci remain to be discovered.
A complementary approach to human studies in finding novel genes and pathways involved in atherosclerosis is to study animal models, which have the advantage of allowing strict control over environmental influence and accurate phenotypic characterization of atherosclerotic lesions. Apolipoprotein E-null (Apoe−/−) and LDL receptor-null (Ldlr−/−) mouse models reproduce all phases of atherosclerotic lesions seen in humans (Ishibashi et al. 1994; Nakashima et al. 1994). Over a dozen intercrosses or backcrosses have been generated from atherosclerosis-susceptible and -resistant inbred strains carrying the mutant Apoe or Ldlr gene, leading to the identification of over 50 unique atherosclerosis susceptibility loci (http://www.informatics.jax.org/allele).
In an intercross between BALB/c-Apoe−/− and SM-Apoe−/− mice, we discovered a significant locus influencing atherosclerotic lesion size, named Ath49, near the major histocompatibility complex (MHC) on chromosome 17 (Grainger et al. 2016). Ath49 partially overlaps in the confidence interval with Ath22, a locus mapped in an AKR-Apoe−/− × DBA/2- Apoe−/− intercross (Smith et al. 2006). Using bioinformatics resources, we have prioritized Mep1a, encoding meprin 1α, as a potential candidate gene for Ath49. The Mep1a gene contains multiple SNPs that are predicted to affect the expression and function of its protein product (Grainger et al. 2016). Mep1a is a zinc-dependent metalloprotease that plays a role in inflammation and fibrosis (Arnold et al. 2017), two key events of atherogenesis (Gisterå and Hansson 2017). In addition, a nonspecific meprin inhibitor suppresses atherosclerotic plaque formation in mice (Gao et al. 2009). Here, we report the prioritization and characterization of Mep1a as a causal gene for Ath49 through the use of genetic and genomic means.
Materials and methods
Prioritization of candidate genes for Ath49
The confidence interval of Ath49 overlaps with that of Ath26 mapped in an intercross between AKR-Apoe−/− and DBA/2-Apoe−/− mice (Smith et al. 2006). As the two QTL for atherosclerosis were mapped in two crosses derived from different inbred mouse strains, we used genomic sequence variant data from the Mouse Genomes Project to prioritize positional candidate genes. Probable candidate genes were those that contain one or more nonsynonymous SNPs or a SNP in upstream regulatory regions and these SNPs are shared by the founder strains carrying the high allele but are different from the ones shared by the founder strains carrying the low allele at the QTL, as reported (Rowlan et al. 2013; Grainger et al. 2016). SNPs were queried via the Sanger Mouse Genomes Project database (https://www.sanger.ac.uk/sanger/Mouse_SnpViewer/rel-1505) for the inbred strains that were used to the identification of Ath26 and Ath49. These mouse strains include AKR, DBA/2, BALB/c, and SM. SIFT (Sorting Intolerant From Tolerant) score, which predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids (Vaser et al. 2016), was obtained through Ensembl Genome Browser (https://useast.ensembl.org/index.html). SIFT scores vary between 0 and 1, and an amino acid substitution with a SIFT score of ≤0.05 is considered to alter protein function. We used available eQTL for mouse atherosclerotic lesions (doi:10.1371/journal.pgen.1005711) to prioritize positional candidate genes that contain one or more SNPs in upstream regulatory regions that might affect transcript levels (Bennett et al. 2015).
Animals, experimental design, and procedures
Mep1a −/− mice, which were generated using embryonic stem cells derived from the 129 strain and subsequently backcrossed onto the C57BL/6J (B6) background (Banerjee et al. 2009). The Mep1a gene was disrupted by inserting a neomycin cassette in exon 7, the region that codes for the zinc-catalytic center of the gene product (Banerjee et al. 2009). As Ath49 was mapped in a cross derived from Apoe−/− mouse strains, we crossed Mep1a−/− mice with B6-Apoe−/− mice (The Jackson Laboratory, stock #: 002052) to create Mep1α−/−Apoe−/− mice. The Mep1a gene is adjacent to the H2 locus on chromosome 17 so we performed genotypic analysis of short tandem repeats to confirm the presence of the H2b haplotype in Mep1α−/−Apoe−/− mice. Mice of both sexes were weaned at 3 weeks of age onto a rodent chow diet. At 6 weeks of age, one group was started on a Western diet containing 21% fat, 34.1% sucrose, 0.15% cholesterol, and 19.5% casein by weight (TD 88137, Envigo) and maintained on the diet for 12 weeks to examine advanced plaque formation (Nakashima et al. 1994; Shi et al. 2000a). The other group remained on the chow diet and was euthanized at 12 weeks of age to determine early lesion (fatty streak) formation as reported (Tian et al. 2005; Miyoshi et al. 2006a; Manichaikul et al. 2011). Mice were fasted overnight (∼15 h) before retro-orbital blood was collected into tubes containing 8 μl of 0.5 mol/L ethylenediaminetetraacetic acid (EDTA) with the animals under isoflurane anesthesia. Plasma was prepared and stored at −80°C before use. Animal care and experimentation were carried out by following a protocol (#3109) approved by the Institutional Animal Care and Use Committee.
Measurements of plasma glucose and lipid levels
Fasting plasma glucose and lipid levels were measured by enzymatic methods, as reported (Tian et al. 2005; Wang et al. 2015; Shi et al. 2021). Plasma glucose was measured with a Sigma glucose (HK) assay kit. Six micro litter of plasma samples (3x dilution for plasma obtained from mice fed the Western diet) were incubated with 150 μl of assay reagent in a 96-well plate for 30 min at 30°C. The absorbance at 340 nm was read on a Molecular Devices (Menlo Park, CA, USA) plate reader. Total cholesterol, HDL cholesterol, and triglyceride were measured with the use of Thermo DMA assay kits (Thermo Electron Corp.). NonHDL cholesterol was calculated as the difference between total and HDL cholesterol.
Morphometric analysis of aortic atherosclerosis
Atherosclerotic lesion sizes in the aortic root of mice were measured as reported (Qiao et al. 1994; Su et al. 2006). Briefly, the vasculature of mice was perfusion-fixed with 4% PFA (paraformaldehyde) through the left ventricle of the heart. The basal portion of the heart and ascending aorta were harvested, embedded in OCT (optimal cutting temperature compound, Tissue-Tek) and cross-sectioned in 10-μm thickness. In the region from the appearance to the disappearance of aortic valves, every other section was collected. In all other regions, every fifth section was collected. Sections were stained with oil red O and hematoxylin and counterstained with fast green. Atherosclerotic lesion areas were measured using Zeiss AxioVision 4.8 software. For the chow diet group, aortic lesion areas from all stained sections were measured due to small sizes and the summation of all values measured was divided by the number of sections counted. For mice fed the Western diet, aortic lesion areas on 10 to 15 sections were measured for each animal as reported (Venegas-Pino et al. 2013). The top 5 largest c lesion areas were averaged for each mouse and this value was used for statistical analysis.
Plaque stability assessment
Metrics pertaining to the stability of advanced atherosclerotic lesions, including necrotic area, fibrous cap thickness, collagen content, and cellular composition, were measured on selected aortic sinus sections stained using the standard hematoxylin and eosin (H&E) procedure, Masson’s trichrome stain kit (Azer Scientific), and immunohistochemistry for mice fed the Western diet. Morphometric analyses were done using the Versatile Wand Tool plugin for ImageJ. The empty areas within atherosclerotic lesions on H&E stained sections were measured. The degree of necrosis was expressed as a percentage of the necrotic areas relative to the total lesion area on the same section. Fibrous cap thickness was measured on H&E stained sections and expressed as an average of measurements at 3 or more even spaced points along with an overlying cap. Collagen content in atherosclerotic lesions was determined on trichrome stained sections and expressed as a percentage of blue stain areas relative to the total lesion area on the same section.
Immunohistochemical analysis
Immunostaining was performed on frozen aortic sections as reported (Zhao et al. 2019). Briefly, sections were fixed in acetone, blocked with 10% normal donkey serum in 10% bovine serum albumin in saline, and then incubated with one or more primary antibodies, including rat anti-mouse CD107b (Mac3)(1:100 dilution; BD Pharmigen 550292), rat anti-mouse Ly6g (Gr-1) (1:200 dilution; clone RB6-8C5; eBioscience 14-5931-82), goat anti-mouse α-smooth muscle cell actin (SMC-A) (1:100 dilution; Ango Biosciences ARG63621), rabbit anti-mouse Mep1a (histo1 and Western2, 1:1000 dilution; provided by Christoph Becker-Pauly, Kiel University), goat anti-mouse LIX (CXCL5) (undiluted polyclonal LIX Conjugate P129149 from R&D Systems LIX ELISA Kit). Immunoreactivity was visualized with the Vectastain Elite ABC Kit (Vector Laboratories) or with one or more fluorescentsecondary antibodies, including goat anti-rat IgG (H + L) (1:250 dilution; Alexa Fluor 488; Cell Signaling 4416S), donkey anti-goat IgG (1:250 dilution; NL557; R&D Systems NL001), donkey anti-goat IgG (H + L) (1:250 dilution; Alexa Fluor 633/Cy3; Invitrogen A21082). Slides stained with fluorescent antibody were mounted with DAPI Fluoromount-G (Southern Biotech) and images were taken using a Zeiss LSM 800 confocal microscope.
Confocal images were analyzed using the Fiji package for ImageJ (Schindelin et al. 2012). Cells positive for Mac3 or SMC-A were counted and normalized to the total number of DAPI positive cells in aortic lesions to get the percent positive macrophages and smooth muscle cells. For the staining with the Ly6g and CXCL5 antibodies that were not confined to cells, the area stained was measured and normalized to the aortic lesion area.
Western blot analysis
Expression of Mep1a in the aorta and liver of 6-week-old Mep1a+/+Apoe−/− mice before foam cells developed was determined by western blot analysis. Protein samples were prepared as reported (Shi et al. 2004). After the vasculature was flushed thoroughly with saline, thoracic aorta, small pieces of liver and kidney tissues were harvested, mechanically broken up in liquid nitrogen, and dispersed in a lysis buffer. After centrifugation, the supernatant was collected and used for detection of Mep1a and ß-actin proteins. Protein samples (10 µg) were separated by electrophoresis on 4%–12% Tris poly-acrylamide gels and electrophoretically transferred onto a nitrocellulose membrane, which was probed with primary antibodies for Mep1a and ß-actin (Santa Cruz).
Circulating MCP-1 and CXCL5 assays
Plasma levels of MCP-1 and CXCL5 were measured with enzyme-linked immunosorbent assay (ELISA) kits by following the instructions of the manufacturer (R&D Systems, Minneapolis, MN, USA).
Measurement of malondialdehyde (MDA)
Plasma levels of MDA were measured with a Cayman TBARS (Thiobarbituric Acid Reactive Substances) kit (Cat. # 10009055), as reported (Fuller et al. 2020).
Statistical analysis
Data are presented as means ± SEM, with “n” indicating the number of mice. Two-tailed unpaired Student’s t-tests were performed to determine statistical significance for differences between two groups in various measurements. P-values ≤ 0.05 were considered statistically significant.
Results
Prioritization of candidate genes for Ath49
Two significant QTL for atherosclerotic lesions were mapped to chromosome 17, with Ath26 mapped to 33∼53 Mb in an intercross between AKR and DBA/2 Apoe−/− mice (Smith et al. 2006) and Ath49 mapped to 43∼65 Mb in an intercross between BALB/c and SM Apoe−/− mice (Grainger et al. 2016). The physical location of these two QTL overlaps between 43 to 53 Mb (Figure 1). BALB/c and DBA/2 alleles were associated with increased atherosclerotic lesion sizes at the QTL, while AKR and SM alleles were associated with decreased lesion sizes. The Sanger SNP database was queried to find SNPs that were shared by the high allele strains BALB/c and DBA/2 but were different from SNPs of the low allele strain AKR within the 43∼53 Mb common interval (Table 1). The low allele SM strain was not included due to lack of its DNA sequence information in the dataset. Mep1a and Tdrd6 are only two genes that contain one or more nonsynonymous SNPs leading to amino acid substitution in the region. Mep1a contains 14 nonsynonymous SNPs with 2 (rs218712173 and rs215987489) yielding extremely low SIFT scores of 0.02 and 0.01 for amino acid substitutions at residues 232 (N/S) and 247 (F/Y), respectively, in the range of producing detrimental effects on protein function (Table 1). In addition, an A/G SNP at 43,491,721 bp introduces a premature stop codon, leading to protein truncation at residue 151. Tdrd6 contains 2 nonsynonymous SNPs (rs33049652 and rs51245534), but their high SIFT scores [0.28 at residue 36 (V/A) and 0.58 at residue 1508 (A/T)] preclude significant impacts on protein function. Thus, of the two genes, Mep1a is the only one that contains nonsynonymous SNPs of functional importance.
Figure 1.
Overlapping of the confidence interval of QTL for atherosclerotic lesions on mouse chromosome 17 (Chr17). Ath49, mapped in an intercross between BALB-Apoe−/− and SM-Apoe−/− mice, overlaps with Ath22 mapped in AKR-Apoe−/− and DBA/2-Apoe−/− mice in the confidence interval from 43 to 53 Mb.
Table 1.
Positional candidate genes in the 43 ∼ 53 Mb region on Chr17 determined by haplotype analysis
| Chr | Position | Gene | dbSNP | Ref | BALB_cJ | DBA_2J | AKR_J | Variants | Amino acid position | SIFT score | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 43474462 | Mep1a | rs13467271 | C | T | T | — | — | |||
| 17 | 43482389 | Mep1a | rs48095932 | A | — | — | G | splice_region_variant | |||
| 17 | 43486313 | Mep1a | rs50539426 | C | — | — | T | V/I | 288 | 1 | |
| 17 | 43486366 | Mep1a | rs263416339 | C | — | — | T | splice_region_variant | |||
| 17 | 43486368 | Mep1a | rs224258235 | G | — | — | A | splice_region_variant | |||
| 17 | 43487139 | Mep1a | rs214680904 | G | — | — | A | P/L | 255 | 1 | |
| 17 | 43487163 | Mep1a | rs215987489 | A | — | — | T | F/Y | 247 | 0.01 | |
| 17 | 43487200 | Mep1a | rs252705192 | C | — | — | T | E/K | 235 | 0.18 | |
| 17 | 43487208 | Mep1a | rs218712173 | T | — | — | C | N/S | 232 | 0.02 | |
| 17 | 43487220 | Mep1a | rs229991911 | G | — | — | T | P/Q | 228 | 0.13 | |
| 17 | 43487282 | Mep1a | rs580757039 | G | — | — | T | D/E | 207 | 0.15 | |
| 17 | 43487302 | Mep1a | rs250429860 | G | — | — | A | H/Y | 201 | 0.06 | |
| 17 | 43487308 | Mep1a | rs221971200 | A | — | — | Ga | Y/H | 199 | 0.44 | |
| 17 | 43487318 | Mep1a | rs246176896 | A | — | — | G | splice_region_variant | |||
| 17 | 43491606 | Mep1a | rs46011116 | T | — | — | G | N/T | 189 | 0.9 | |
| 17 | 43491690 | Mep1a | rs50252056 | A | — | — | G | F/S | 161 | 0.21 | |
| 17 | 43491700 | Mep1a | rs255741402 | C | — | — | T | G/R | 158 | 0.41 | |
| 17 | 43491721 | Mep1a | rs48412763 | G | — | — | A | stop_gained | 151 | Q/a | |
| 17 | 43491733 | Mep1a | rs107621975 | G | — | — | C | Q/E | 147 | 0.28 | |
| 17 | 43491736 | Mep1a | rs108717471 | C | — | — | T | D/N | 133 | 0.76 | |
| 17 | 43500657 | Mep1a | rs47627745 | C | — | — | A | upstream_gene_variant | |||
| 17 | 43501101 | Mep1a | rs108408343 | C | — | — | T | upstream_gene_variant | |||
| 17 | 43501441 | Mep1a | rs46400553 | G | — | — | A | upstream_gene_variant | |||
| 17 | 43501628 | Mep1a | rs51680090 | C | — | — | A | upstream_gene_variant | |||
| 17 | 43501640 | Mep1a | rs226486190 | A | — | — | G | upstream_gene_variant | |||
| 17 | 43502090 | Mep1a | rs107953806 | C | — | — | T | upstream_gene_variant | |||
| 17 | 43544432 | Ankrd66 | rs33886189 | G | — | — | A | upstream_gene_variant | |||
| 17 | 43545402 | Ankrd66 | rs33166719 | A | — | — | G | upstream_gene_variant | |||
| 17 | 43546425 | Ankrd66 | rs218010348 | A | — | — | G | upstream_gene_variant | |||
| 17 | 43546868 | Ankrd66 | rs108186043 | T | — | — | C | upstream_gene_variant | |||
| 17 | 43547227 | Ankrd66 | rs33339990 | G | — | — | A | upstream_gene_variant | |||
| 17 | 43547392 | Ankrd66 | rs33409247 | T | — | — | C | upstream_gene_variant | |||
| 17 | 43547909 | Ankrd66 | rs33242891 | A | — | — | C | upstream_gene_variant | |||
| 17 | 43548285 | Ankrd66 | rs108489143 | C | — | — | T | upstream_gene_variant | |||
| 17 | 43565671 | Pla2g7 | rs33107182 | G | A | A | — | upstream_gene_variant | |||
| 17 | 43565744 | Pla2g7 | rs33404062 | T | C | C | — | upstream_gene_variant | |||
| 17 | 43566951 | Pla2g7 | rs33694139 | G | A | A | — | upstream_gene_variant | |||
| 17 | 43567598 | Pla2g7 | rs46595298 | T | C | C | — | upstream_gene_variant | |||
| 17 | 43567840 | Pla2g7 | rs48712550 | T | C | C | — | upstream_gene_variant | |||
| 17 | 43568147 | Pla2g7 | rs51948026 | C | Ta | Ta | — | 5_prime_utr_variant upstream_gene_variant | |||
| 17 | 43568245 | Pla2g7 | rs50592172 | T | C | C | — | upstream_gene_variant | |||
| 17 | 43624921 | Tdrd6 | rs33325152 | G | A | A | — | upstream_gene_variant | |||
| 17 | 43625218 | Tdrd6 | rs33570046 | G | A | A | — | upstream_gene_variant | |||
| 17 | 43625634 | Tdrd6 | rs51245534 | C | T | T | — | A/T | 1508 | 0.58 | |
| 17 | 43630049 | Tdrd6 | rs33049652 | A | G | G | — | V/A | 36 | 0.28 | |
| 17 | 43631121 | Tdrd6 | rs33210666 | C | T | T | — | upstream_gene_variant | |||
| 17 | 43632092 | Tdrd6 | rs33444914 | G | A | A | — | upstream_gene_variant | |||
| 17 | 43634739 | Tdrd6 | rs29523294 | A | G | G | — | upstream_gene_variant | |||
| 17 | 46538571 | Cul9 | rs33405663 | A | G | G | — | upstream_gene_variant | |||
| 17 | 53467577 | Efhb | rs107955170 | T | G | G | — | upstream_gene_variant | |||
| 17 | 53562719 | Kat2b | rs51135397 | C | T | T | — | upstream_gene_variant | |||
| 17 | 53582910 | Kat2b | rs47441203 | A | G | G | — | upstream_gene_variant | |||
| 17 | 53659473 | Kat2b | rs45714062 | A | G | G | — | nmd_transcript_variant | |||
| 17 | 53661589 | Kat2b | rs29535114 | C | G | G | — | nmd_transcript_variant | |||
| 17 | 53661819 | Kat2b | rs33435892 | G | A | A | — | nmd_transcript_variant | |||
| 17 | 53662994 | Kat2b | rs33719275 | C | T | T | — | nmd_transcript_variant | |||
| 17 | 53667321 | Kat2b | rs33451147 | C | T | T | — | nmd_transcript_variant | |||
| 17 | 53667799 | Kat2b | rs33316568 | G | T | T | — | nmd_transcript_variant | |||
| 17 | 53668820 | Kat2b | rs52501654 | T | C | C | — | nmd_transcript_variant | |||
| 17 | 53669174 | Kat2b | rs52173659 | T | G | G | — | nmd_transcript_variant | |||
| 17 | 53669819 | Kat2b | rs33530707 | G | A | A | — | nmd_transcript_variant | |||
| 17 | 53689244 | Sgol1 | rs33700085 | C | G | G | — | 5_prime_utr_variant | |||
Chr, chromosome; Ref, reference SNP; SIFT, Sorting Intolerant From Tolerant (intolerant SNP is highlighted in bold).
Upstream_gene_variant: A sequence variant located 5' of a gene.
NMD transcript variant: Nonsense mediated decay transcript variant.
Mep1a has 95 SNPs in upstream regions but only a few are shown to save space.
aDenotes likely SNP.
“—” same as reference SNP.
Eight genes within the 43∼53 Mb region contain one or more SNPs that potentially affect transcript levels between the high allele strains (BALB/c and DBA/2) and the low allele strain (AKR) (Table 1). These genes include Mep1a, Ankrd66, Pla2g7, Tdrd6, Cul9, Efhb, Kat2b, and Sgol1. We evaluated their associations with atherosclerosis using the gene expression dataset for the aorta and liver of over 100 inbred mouse strains from the Hybrid Mouse Diversity Panel (HMDP) (Bennett et al. 2015). None of them are among the genes whose transcript expression in the aorta or liver are positively or negatively correlated with atherosclerosis (no eQTL detected).
Mep1a expression in atherosclerotic lesions, healthy aorta, and liver
Immunohistochemical analysis revealed the expression of Mep1a in atherosclerotic lesions (Figure 2). The lesion areas that were intensely stained with oil red O showed abundant Mep1a expression. No notable Mep1a expression was observed in surrounding aortic walls. Western blot analysis showed Mep1a expression in the kidney but not in the aorta and liver of Mep1a+/+Apoe−/− mice. Mep1a−/−Apoe−/− mice showed no Mep1a expression in the kidney. In contrast, ß-actin was detectable in all the tissues tested from both genotypes of mice.
Figure 2.
A through F, Corresponding staining of adjacent cross-sections of the aortic root of an Apoe−/− mouse shows co-location of Mep1a and Oil Red O positive areas pointed by arrows. The top row is microimages that include the entire aortic cross section (original magnification: 4x), and the bottom row is microimages that show a representative region at a high magnification (10x). Scale bars are shown in the figure. G, Western blot analysis of Mep1a and ß-actin proteins in the aorta, liver, and kidney of 6-week-old Mep1a+/+Apoe−/− mice. Kidney samples from Mep1a−/− Apoe−/− mice were used as a negative control for Mep1a. Each lane represents an individual mouse. L, ladder.
Genetic ablation of Mep1a inhibits atherosclerosis
To determine the role of Mep1a in both early and advanced atherosclerotic lesion formation, Mep1a−/−Apoe−/− mice were generated and fed either a chow or Western diet. We and others documented that Apoe−/− mice develop early atherosclerotic lesions (fatty streaks) at 12 weeks of age on a chow diet and advanced lesions after 12 weeks of Western diet (Nakashima et al. 1994; Tian et al. 2005; Miyoshi et al. 2006a; Manichaikul et al. 2011). At 12 weeks of age on the chow diet, female Mep1a−/−Apoe−/− mice had an average lesion area of 9455 ± 1681 μm2/section (n = 10), which was significantly smaller than the lesion area of 27,910 ± 20,178 μm2/section in Mep1a+/+Apoe−/− mice (n = 16; P = 0.039) (Figure 3). After 12 weeks on the Western diet, which started at 6 weeks of age, female Mep1a−/−Apoe−/− mice had a mean lesion area of 441,661 ± 34,643 μm2/section (n = 16), significantly smaller than the lesion area of 615,987 ± 21,352 μm2/section in Mep1a+/+Apoe−/− mice (n = 30; P = 0.0002).
Figure 3.
Atherosclerotic lesion areas in the aortic root of female Mep1a−/−Apoe−/− and Mep1a+/+Apoe−/− control mice fed a chow or Western diet. Each circle represents the lesion size of an individual mouse. The horizontal short line represents the mean lesion size of an entire group. * P < 0.05 compared with Mep1a+/+Apoe−/− mice on the same diet.
Mep1a deficiency affects atherosclerotic plaque stability
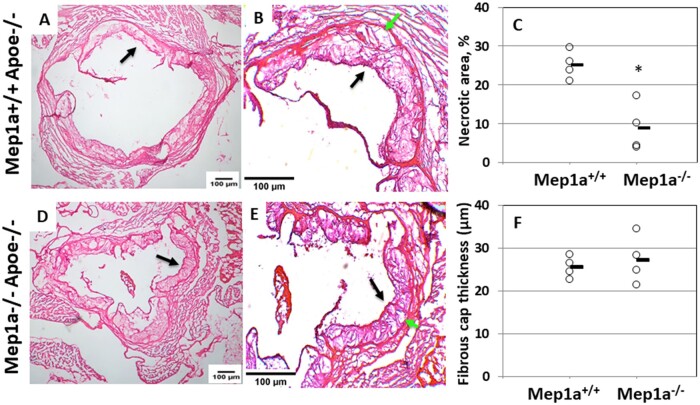
Advanced atherosclerotic lesions were analyzed to determine the influence of Mep1a on histological features reflecting plaque stability. On H&E stained sections, alterations in necrotic areas of atherosclerotic lesions were grossly noticeable (Figure 4). Mep1a−/−Apoe−/− mice showed a significant reduction in the percentage of necrotic lesion areas relative to Mep1a+/+Apoe−/− control mice (8.9 ± 3.1% vs 25.2 ± 1.8%; P = 0.0065) (Figure 4C). The thickness of the fibrous caps covering the luminal surface of atherosclerotic lesions at multiple locations (3∼9) on multiple sections was quantified for 4 mice per group. The average thickness of fibrous caps was not significantly altered in Mep1a−/−Apoe−/− mice (27.3 ± 2.8 vs 25.5 ± 1.2 µm; P = 0.59) (Figure 4F).
Figure 4.
Influence of Mep1a deficiency on stability of advanced atherosclerotic lesions in Apoe−/− mice. Sections were stained with H&E. A, B, D, E: Representative images of H&E stained sections of a Mep1a−/−Apoe−/− and a Mep1a+/+Apoe−/− mouse fed 12 weeks of Western diet. Black arrow points at the fibrous cap, and green arrow at necrotic area. A, D: 4x; B, E: 10x. C, Quantification of necrotic atherosclerotic lesion areas on H&E stained sections from Mep1a−/−Apoe−/− and Mep1a+/+Apoe−/− mice fed 12 weeks of Western diet. The magnitude of necrosis is expressed as a percentage of the unstained area relative to the entire lesion area on the same aortic cross section for 4 Mep1a+/+Apoe−/− and 4 Mep1a−/− Apoe−/− mice. * P < 0.05 vs Mep1a+/+Apoe−/− mice. F, Average fibrous cap thickness of advanced atherosclerotic lesions measured on H&E stained sections from Mep1a−/−Apoe−/− and Mep1a+/+Apoe−/− mice fed 12 weeks of Western diet. Each circle represents the measurement of an individual mouse, and the horizontal short line represents the mean value of an entire group.
Trichrome staining allows for quantification of collagen content (blue color) in atherosclerotic lesions. Blue stain areas in atherosclerotic lesions were measured for Mep1a+/+Apoe−/− and Mep1a−/−Apoe−/− mice fed 12 weeks of Western diet (Figure 5, A–E). The percentage of blue stain areas relative to the total lesion area was comparable between Mep1a+/+Apoe−/− mice and Mep1a−/−Apoe−/− mice (33.5 ± 5.0% vs 34.0 ± 2.6%; P > 0.05) (Figure 5E).
Figure 5.
The collagen content in atherosclerotic lesions determined by trichrome staining for Mep1a−/−Apoe−/− and Mep1a+/+Apoe−/− mice fed 12 weeks of Western diet. A, B, C, D: Representative images of trichrome stained atherosclerotic lesions of a Mep1a+/+Apoe−/− and a Mep1a−/− Apoe−/− mouse. Red and blue stain areas are highlighted by black and green arrows, respectively. A, C: 4x; B, D: 10x. E, Quantification of collagen content in atherosclerotic lesions on trichrome stained aortic root sections from Mep1a−/−Apoe−/− and Mep1a+/+Apoe−/− mice fed 12 weeks of Western diet. The amount of collagen content is expressed as a percentage of the blue stain area relative to the entire lesion area on the same section for 4 Mep1a+/+Apoe−/− or 4 Mep1a−/− Apoe−/− mice. Each circle represents the measurement of an individual mouse, and the horizontal short line represents the mean value of an entire group.
Mep1a deficiency increases macrophage content in atherosclerotic lesions
Immunofluorescent staining also showed focal expression of Mep1a antigen in atherosclerotic lesions, especially abundant in fatty streak lesions (Figure 6, A and C). Because the Mep1a gene is located proximal to the MHC locus, we investigated its influence on immune cell content of atherosclerotic lesions. Mep1a−/−Apoe−/− mice showed increased macrophage content (Mac3 positive cells) in advanced lesions relative to Mep1a+/+Apoe−/− mice (75 ± 4% vs 21 ± 4%; P < 0.0001) (Figure 6, C–E). Smooth muscle cell content (SMC-A positive cells) was also increased although the difference did not reach statistical significance (Mep1a−/−Apoe−/−: 12 ± 5% vs 5 ± 3% of control mice; P = 0.17) (Figure 6F).
Figure 6.
Immunofluorescence analysis of cellular components in atherosclerotic lesions of Mep1a−/− Apoe−/− and Mep1a+/+Apoe−/− mice. Representative immunofluorescence images showing staining for macrophages (green color), vascular smooth muscle cell (red), Mep1a (magenta), and nuclei (blue). Macrophages (E) and smooth muscle cells (F) in atherosclerotic lesions of Mep1a+/+Apoe−/− and Mep1a−/− Apoe−/− mice were quantified. The density of either cell type was expressed as a percentage of positive stained cells relative to all stained nuclei (DAPI) in an image. Results are expressed as individual values for 8 Mep1a+/+Apoe−/− mice and 8 Mep1α−/− Apoe−/− mice. The horizontal short line represents the mean value of an entire group. *P < 0.05.
Chromogenic immunostaining for Mac3 (macrophages) also showed an increased signal in atherosclerotic lesions of Mep1a−/−Apoe−/− mice compared to Mep1a+/+ Apoe−/− mice (Figure 7). As seen in fluorescent staining, atherosclerotic lesions of Mep1a+/+ Apoe−/− mice showed a weak Mac3 stain (Figure 7, A and E). In contrast, notable Mac3 staining was seen in the lesions of Mep1a−/−Apoe−/− mice (Figure 7, I and M).
Figure 7.
Chromogenic immunohistochemical analysis of Mac3 (macrophages), Ly6G (neutrophils), and CXCL5 antigens in advanced atherosclerotic lesions of Mep1a+/+ Apoe−/− (A through H) and Mep1a−/− Apoe−/− mice (I through P). Top row is microimages taken at a lower magnification (4x), bottom row is microimages taken at a high magnification (10x) for both genotypes of mice. Arrows point at stained lesion areas. Note the difference in the intensity of Mac3 staining between the two genotypes of mice.
Mep1a deficiency increases neutrophil content in atherosclerotic lesions
The presence of neutrophils and CXCL5 in atherosclerotic lesions was determined by immunofluorescent staining. We observed a significant increase in neutrophil content (Ly6g positive cells) in advanced lesions of Mep1a−/−Apoe−/− mice relative to Mep1a+/+ Apoe−/− mice (5.9 ± 1.4% vs 0.4 ± 0.1%; P = 0.017) (Figure 8). CXCL5, a major cytokine for neutrophil recruitment, was expressed in atherosclerotic lesions of Mep1a+/+ Apoe−/− mice, and the staining was continuously distributed along the luminal surface of the lesions. In Mep1a−/−Apoe−/− mice, CXCL5 staining exhibited a patchy distribution and colocalization with neutrophils within atherosclerotic lesions. Quantitative analysis showed no significant difference in CXCL5 expression levels between Mep1a−/−Apoe−/− and Mep1a+/+ Apoe−/− mice (14.4 ± 4.1% vs 16.3 ± 7.5%; P = 0.83).
Figure 8.
Immunofluorescence detection of CXCL5 and neutrophils (Ly6g) in atherosclerotic lesions of Mep1a−/− Apoe−/− and Mep1a+/+Apoe−/− mice. Representative immunofluorescence images showing staining for CXCL5 and neutrophils in atherosclerotic lesions of Mep1a+/+Apoe−/− (A–C) and Mep1a−/− Apoe−/− mice (D–F). CXCL5 was stained red, neutrophils stained green, and nuclei stained blue. G, Neutrophils in atherosclerotic lesions were quantified and expressed as a percentage of positive stained area relative to the lesion area in the same images for 6 Mep1a+/+Apoe−/− mice and 7 Mep1a−/− Apoe−/− mice. H, Expression of CXCL5 was expressed as a percentage of positive stained area over the lesion area on the same images for 4 Mep1a+/+Apoe−/− mice and 5 Mep1a−/− Apoe−/− mice. * P < 0.05. Each circle represents the measurement of a mouse, and the horizontal short line represents the mean value of a group. Arrows point at stained areas.
Chromogenic immunostaining also showed scare signals for Ly6G in atherosclerotic lesions of Mep1a−/−Apoe−/− mouse (Figure 7, J and N). The two groups showed extensive staining for CXCL5 in atherosclerotic lesions (Figure 7, C, G, K, and O), but no staining was seen outside atherosclerotic lesions.
Mep1a deficiency lowers systemic levels of CXCL5 and oxidative stress
Plasma levels of MCP-1 were comparable between Mep1a−/−Apoe−/− and Mep1a+/+ Apoe−/− mice on chow (20.4 ± 11.5 vs 14.1 ± 8.7 pg/dl; P = 0.22) or Western diet (72.0 ± 6.0 vs 75.7 ± 45.4 pg/dl; P = 0.82) (Figure 9A). Mice of both genotypes showed significantly elevations in plasma MCP-1 levels on the Western diet over the chow diet (P < 0.002). Surprisingly, Mep1a−/−Apoe−/− mice showed a 75% decrease in plasma CXCL5 level on the chow diet (451.0 ± 250.8 vs 1855.0 ± 1292.4 pg/dl; P = 0.008) and a 57% decrease on the Western diet (595.6 ± 582.4 vs 1385.6 ± 649.3 pg/dl; P = 0.01) compared to Mep1a+/+ Apoe−/− mice (Figure 9B). Mep1a−/−Apoe−/− mice exhibited a reduced plasma CXCL5 level and Mep1α+/+ Apoe−/− mice exhibited a slightly decreased CXCL5 level on the Western diet relative to the chow diet, although the difference was not statistically significant (P > 0.32).
Figure 9.
Plasma levels of MCP-1 (A), CXCL5 (B), and MDA (C) in Mep1a−/− Apoe−/− and Mep1a+/+Apoe−/− mice when fed a chow or Western diet. Results are means ± SE of 7 to 10 mice per group. * P < 0.05 vs Mep1a+/+Apoe−/− mice; + P < 0.05 vs chow diet.
Plasma levels of MDA, a marker of oxidative stress, was determined by TBARS (Thiobarbituric Acid Reactive Substances) assay. Mep1a−/−Apoe−/− mice showed a significant reduction in plasma MDA levels compared to Mep1a+/+ Apoe−/− mice on both chow (33.2 ± 0.7 vs 39.6 ± 2.6 μM; P = 0.04) and Western diets (53.4 ± 3.1 vs 76.7 ± 6.2 μM; P = 0.006) (Figure 9C). Both Mep1a−/−Apoe−/− and Mep1a+/+ Apoe−/− mice had significant elevations in plasma MDA levels on the Western diet compared to the chow diet (P < 0.0002).
Influence of Mep1a deficiency on plasma lipid profile and body weight
Mep1a −/− Apoe−/− mice had similar plasma levels of HDL, nonHDL cholesterol and glucose as Mep1a+/+ Apoe−/− mice on either chow or Western diet (Figure 10). Plasma triglyceride levels were significantly lower in Mep1a−/−Apoe−/− mice than Mep1a+/+ Apoe−/− mice on the chow diet (73.1 ± 4.4 vs 118.4 ± 11.8 mg/dl; P = 0.008) but not on the Western diet (146.5 ± 7.6 vs 153.7 ± 10.4 mg/dl; P = 0.6). In addition, Mep1a−/−Apoe−/− mice had smaller body weight than Mep1a+/+ Apoe−/− mice on either chow (18.5 ± 0.3 vs 19.9 ± 0.5 g; P = 0.034) or Western diet (19.5 ± 0.4 vs 22.9 ± 0.6 g; P = 0.00073).
Figure 10.
Plasma levels of non-HDL, HDL cholesterol, triglyceride, glucose, and body weight in Mep1a-/- Apoe-/- and Mep1a+/+ Apoe-/- mice when fed a chow or Western diet. Results are means ± SE of 8 to 23 mice per group. * p < 0.05 vs. Mep1a+/+ Apoe-/- mice.
Discussion
In this study, we used bioinformatics resources to prioritize Mep1a as the most likely candidate gene for Ath49, an atherosclerosis susceptibility locus on mouse chromosome 17, and then used knockout mice to confirm the causality. There were multiple SNPs in both coding and regulatory regions of Mep1a between the founder strains that were used to map Ath49. Genetic ablation of Mep1a significantly decreased both early and advanced atherosclerotic lesion sizes in Apoe−/− mice, and altered histological features associated with plaque stability such as reduced intraplaque necrosis and increased inflammatory cell content. In addition, Mep1a deficiency lowered circulating levels of CXCL5 and oxidative stress but had no influence on HDL and nonHDL cholesterol levels in Apoe−/− mice.
We took advantage of rich genetic and genomic resources of the mouse to prioritize positional candidate genes for Ath49. Because Ath49 overlaps with Ath26 in the confidence interval and the two QTL were mapped in intercrosses derived from different mouse strains (BALB/c, SM, AKR, and DBA/2) (Smith et al. 2006; Grainger et al. 2016), we were able to use genomic sequence variant data from the Mouse Genomes Project to reduce the number of candidates to 8 genes, including Mep1a, Ankrd66, Pla2g7, Tdrd6, Cul9, Efhb, Kat2b, and Sgol1. All these genes contain one or more SNPs that are shared by the high allele strains (BALB/c and DBA/2) but are different from ones of the low allele strain (AKR). This analysis is based on the finding that 97% of the genetic variants between common mouse strains are ancestral, and thus the causal variants for QTL are most likely among common polymorphisms shared by multiple strains (Wiltshire et al. 2003). As QTL for a quantitative trait are derived from changes in the function or quantity of a gene product, we focused on candidate genes that carry a nonsynonymous SNP or one in upstream regulatory region segregating between the high and low allele strains. Of the 8 candidate genes, only Mep1a contains nonsynonymous SNPs that impact protein function. As the 8 candidate genes all contain one or more SNPs in the upstream regulatory regions, we further used the gene expression data from the HMDP for the aorta and liver of over 100 inbred mouse strains to determine their potential association with atherosclerosis (Bennett et al. 2015). The finding that mRNA expression levels of these 7 genes in the liver and aorta showed no significant association with atherosclerosis do not support the possibility of changed gene expression underlying Ath49.
We demonstrated that Mep1a is a susceptibility gene contributing to the development of atherosclerosis in mice. Indeed, Mep1a was found abundantly expressed in atherosclerotic lesions and ablation of Mep1a resulted in significant reductions in both early and advanced atherosclerotic lesions of Apoe−/− mice. The latter finding complements a previous report that Apoe−/− mice treated with actinonin developed smaller advanced atherosclerosis (Gao et al. 2009), although this meprin inhibitor may exert its anti-atherosclerotic effect through action on Mep1β and other proteases as well as deformylases. Foam cell lesions start to develop in the aorta of Apoe−/− mice at 8 weeks of age on a chow diet (Nakashima et al. 1994). We determined Mep1a expression in the aorta, liver, and kidney of 6-week-old Apoe−/− mice before foam cell lesions had developed in the aorta. The absence of Mep1a protein expression observed in the aorta and liver of mice is consistent with the previous findings made at the transcript level, which also show the expression of Mep1a in the kidney, intestine, and certain leukocytes but not in the brain, heart, skeletal muscle, lung, spleen, and the testis (Jiang et al. 1993; Bankus and Bond 1996; Sun et al. 2009).
A likely mechanism by which Mep1a accelerates atherosclerosis is through its action on oxidative stress in inflammatory cells. We observed that Mep1a deficiency significantly lowered plasma levels of MDA, a reactive aldehyde yielded from peroxidation of unsaturated fatty acids, in Apoe−/− mice fed either chow or Western diet. Elevated levels of MDA suggest an occurring of oxidative stress, which can be caused by increased reactive oxygen species (ROS) production or a decreased antioxidant capacity. On the Western diet, Apoe−/− mice developed severe hyperglycemia and hyperlipidemia, both of which are major drivers of oxidative stress (Fuller et al. 2020), and showed significantly elevated plasma MDA levels. As Mep1a deficiency had no effect on plasma lipid and glucose levels of Apoe−/− mice on the Western diet, the reduced MDA level of Mep1a−/−Apoe−/− mice shouldn’t be attributable to these measures. Nevertheless, the reduced triglyceride level may partially be responsible for decreased MDA levels of Mep1a−/− mice on the chow diet. Hyperlipidemia also promotes LDL deposition in the arterial wall (Brown et al. 2004; Zhao et al. 2019). Accumulated LDL in the arterial wall can be readily oxidized by ROS to become oxidized LDL, which stimulates arterial wall cells to produce MCP-1, VCAM-1, and other proinflammatory molecules (Shi et al. 2000b;Miyoshi et al. 2006b) and subsequently promotes immune cell recruitment and foam cell formation.
Another mechanism underlying the proatherogenic effect of Mep1a is through its action on cytokines and inflammatory molecules. Intriguingly, we observed a 50%–80% reduction in plasma CXCL5 levels of Mep1a−/− mice fed a chow or Western diet. CXCL5, also called epithelial-derived neutrophil-activating peptide 78 (ENA-78), is an inflammatory chemokine produced upon stimulation with IL-1 or TNF-α (Chang et al. 1994). We found that both CXCL5 and Mep1a are expressed in atherosclerotic lesions, so are IL-1 and TNF-α (Fatkhullina et al. 2016). Mep1a proteolytically processes IL-1β from its inactive proform to biologically functional proinflammatory cytokine (Herzog et al. 2009). Thus, lack of Mep1a is expected to hinder the production of active IL-1β during hyperlipidemia-induced inflammation and the subsequent production of CXCL5. Indeed, both fluorescence and chromogenic immunohistochemical analyses showed a reduced signal for CXCL5 in atherosclerotic lesions of Mep1a−/− mice although the reduction was not statistically significant. IL-1β promotes the development of atherosclerosis in Apoe−/− mice (Vromman et al. 2019). IL-18 is another pro-inflammatory cytokine similar in structure to IL-1β and plays a role in inflammatory processes and atherosclerosis (Whitman et al. 2002; Dinarello 2018). Mep1a also cleaves IL-6, IL-6 receptor, nidogen 1, and thymosin-β4, which are involved in inflammation (Keiffer and Bond 2014; Arnold et al. 2017). MCP-1, a pro-inflammatory cytokine produced by macrophages, endothelial cells, and smooth muscle cells and involved in macrophage recruitment (Miyoshi et al. 2006b; Yuan et al. 2009), was not altered in Mep1a−/− mice. In addition, plasma triglyceride levels were significantly lower in Mep1a−/− mice on the chow diet and thus could be partially responsible for the decreased lesion formation. As no changes in plasma lipid levels were observed in Mep1a−/− mice on the Western diet, other factors should be responsible for the attenuated advanced plaque formation.
Besides inhibiting plaque growth, Mep1a deficiency affected plaque stability at multiple aspects, including reductions in intraplaque necrosis and increases in inflammatory cell content. Unstable plaques have the features of a thin fibrous cap, a large necrotic core, and a high inflammatory cell content (Falk et al. 2013). A physiological function of meprins is to regulate the maturation of fibrillar procollagens I and III through cleavage of the N- and C-terminal prodomains, a prerequisite for collagen fibril assembly (Kronenberg et al. 2010; Broder et al. 2013). Collagen types I and III are a critical component of atherosclerotic lesions (Rekhter 1999). Nevertheless, Mep1a deficiency showed little impact on collagen amount in advanced atherosclerotic lesions of Apoe−/− mice. One putative explanation is that Mep1b is also expressed in atherosclerotic lesions and is capable of compensating for the functional loss of Mep1a in collagen synthesis.
We observed a significant decrease in the necrotic area of advanced plaques in Mep1a−/− mice. This finding is consistent with a previous report that Apoe−/− mice receiving meprin inhibition had fewer apoptotic cells in atherosclerotic lesions (Gao et al. 2009). Necrosis is a prominent feature of advanced plaques and a major contributor to plaque destabilization (Fok 2012). The reduced necrotic area and the increased density of macrophages suggest a diminished macrophage death in the advanced plaques of Mep1a−/−Apoe−/− mice. Smooth muscle cells are also a major cellular component of advanced atherosclerosis, but their density was not altered in Mep1a−/−Apoe−/− mice. Necrosis can be induced by multiple factors such as oxidative stress and high concentrations of cytokines (Gao et al. 2009). Mep1a−/−Apoe−/− mice showed reduced oxidative stress and cytokine production (CXCL5), which may explain the reduced necrosis of advanced plaques. The smaller plaque sizes of Mep1a−/−Apoe−/− mice can also be a factor, as the volume of necrosis is greater in larger plaques (Xu et al. 2010).
We observed an increased number of neutrophils in advanced plaques of Mep1a−/−Apoe−/− mice. CXCL5 is a major cytokine involved in neutrophil trafficking during inflammation (Mei et al. 2010), and abundantly expressed in advanced lesions of Apoe−/− mice. Paradoxically, Mep1a−/−Apoe−/− mice had lower systemic and comparable local levels of CXCL5 but showed more neutrophil infiltration in atherosclerotic lesions. However, as CXCL5 has an inhibitory effect on macrophage foam cell formation (Rousselle et al. 2013), its lower level could partially be responsible for the increased macrophage number in atherosclerotic lesions of Mep1a−/−Apoe−/− mice. In this study, we found that the Western diet, which accelerates atherogenesis, had no influence on systemic levels of CXCL5 in Apoe−/− mice. This finding is consistent with a previous report that also showed increased CXCL5 mRNA expression in the aorta (Rousselle et al. 2013).
In summary, we have identified Mep1a as a novel susceptibility gene for atherosclerosis in mice. Removal of Mep1a suppressed both early and advanced atherosclerotic lesion formation and affected plaque stability in the aorta of Apoe−/− mice. We have also identified Mep1a as a major gene contributing to oxidative stress and CXCL5 production. Thus, Mep1a can be potential target for developing anti-atherosclerosis and anti-oxidative stress therapy. It is noteworthy that the observed effect of Mep1a on atherosclerosis is only restricted to the aortic root. We and others have observed vascular site-specific effects of genetic factors on atherosclerosis in both mice and humans (Franceschini et al. 2018; Jones et al. 2020). Thus, further studies are needed to determine whether Mep1a affects atherosclerosis in other arterial sites.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript and supplementary material available at GENETICS online.
Author contributions
A.T.G. and W.S. initiated, designed, performed the study, and wrote the manuscript. N.P., J.L., M.-H.C., A.M.A. performed experiments, C.B.-P. contributed Mep1a knockout mice, anti-Mep1a antibodies, and revised the manuscript.
Funding
The work was supported by NIH grants R01 DK116768 and HL112281 and Commonwealth Health Research Board (CHRB) Virginia.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Arnold P, Otte A, Becker-Pauly C.. 2017. Meprin metalloproteases: molecular regulation and function in inflammation and fibrosis. Biochim Biophys Acta Mol Cell Res. 1864:2096–2104. doi:10.1016/j.bbamcr.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Oneda B, Yap LM, Jewell DP, Matters GL, et al. 2009. MEP1A allele for meprin A metalloprotease is a susceptibility gene for inflammatory bowel disease. Mucosal Immunol. 2:220–231. doi:10.1038/mi.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankus JM, Bond JS.. 1996. Expression and distribution of meprin protease subunits in mouse intestine. Arch Biochem Biophys. 331:87–94. doi:10.1006/abbi.1996.0286. [DOI] [PubMed] [Google Scholar]
- Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, et al. 2015. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet. 11:e1005711.doi:10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broder C, Arnold P, Vadon-Le Goff S, Konerding MA, Bahr K, et al. 2013. Metalloproteases meprin α and meprin β are C- and N-procollagen proteinases important for collagen assembly and tensile strength. Proc Natl Acad Sci USA. 110:14219–14224. doi:10.1073/pnas.1305464110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Jin L, Jien ML, Matsumoto AH, Helm GA, et al. 2004. Lipid retention in the arterial wall of two mouse strains with different atherosclerosis susceptibility. J Lipid Res. 45:1155–1161. doi:10.1194/jlr.M400092-JLR200. [DOI] [PubMed] [Google Scholar]
- Chang MS, McNinch J, Basu R, Simonet S.. 1994. Cloning and characterization of the human neutrophil-activating peptide (ENA-78) gene. J Biol Chem. 269:25277–25282. [PubMed] [Google Scholar]
- Coronary Artery Disease (C4D) Genetics Consortium. 2011. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 43:339–344. doi:10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. ; CARDIoGRAMplusC4D Consortium. 2013. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 45:25–33. doi:10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello CA. 2018. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev. 281:8–27. doi:10.1111/imr.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk E, Nakano M, Bentzon JF, Finn AV, Virmani R.. 2013. Update on acute coronary syndromes: the pathologists’ view. Eur Heart J. 34:719–728. doi:10.1093/eurheartj/ehs411. [DOI] [PubMed] [Google Scholar]
- Fatkhullina AR, Peshkova IO, Koltsova EK.. 2016. The role of cytokines in the development of atherosclerosis. Biochemistry (Mosc). 81:1358–1370. doi:10.1134/S0006297916110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok P-W. 2012. Growth of necrotic cores in atherosclerotic plaque. Math Med Biol. 29:301–327. doi:10.1093/imammb/dqr012. [DOI] [PubMed] [Google Scholar]
- Franceschini N, Giambartolomei C, de Vries PS, Finan C, Bis JC, et al. ; MEGASTROKE Consortium. 2018. GWAS and colocalization analyses implicate carotid intima-media thickness and carotid plaque loci in cardiovascular outcomes. Nat Commun. 9:5141–5114. doi:10.1038/s41467-018-07340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DT, Grainger AT, Manichaikul A, Shi W.. 2020. Genetic linkage of oxidative stress with cardiometabolic traits in an intercross derived from hyperlipidemic mouse strains. Atherosclerosis. 293:1–10. doi:10.1016/j.atherosclerosis.2019.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Guo R, Chen J, Chen Y, Wang H, et al. 2009. A meprin inhibitor suppresses atherosclerotic plaque formation in ApoE-/- mice. Atherosclerosis. 207:84–92. doi:10.1016/j.atherosclerosis.2009.04.036. [DOI] [PubMed] [Google Scholar]
- Gisterå A, Hansson GK.. 2017. The immunology of atherosclerosis. Nat Rev Nephrol. 13:368–380. doi:10.1038/nrneph.2017.51. [DOI] [PubMed] [Google Scholar]
- Grainger AT, Jones MB, Li J, Chen M-H, Manichaikul A, et al. 2016. Genetic analysis of atherosclerosis identifies a major susceptibility locus in the major histocompatibility complex of mice. Atherosclerosis. 254:124–132. doi:10.1016/j.atherosclerosis.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog C, Haun RS, Kaushal V, Mayeux PR, Shah SV, et al. 2009. Meprin A and meprin α generate biologically functional IL-1β from pro-IL-1β. Biochem Biophys Res Commun. 379:904–908. doi:10.1016/j.bbrc.2008.12.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Goldstein JL, Brown MS, Herz J, Burns DK.. 1994. Massive xanthomatosis and atherosclerosis in cholesterol-fed low density lipoprotein receptor-negative mice. J Clin Invest. 93:1885–1893. doi:10.1172/JCI117179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Sadler PM, Jenkins NA, Gilbert DJ, Copeland NG, et al. 1993. Tissue-specific expression and chromosomal localization of the alpha subunit of mouse meprin A. J Biol Chem. 268:10380–10385. doi:10.1016/S0021-9258(18)82212-8. [PubMed] [Google Scholar]
- Jones MB, An A, Shi LJ, Shi W.. 2020. Regional variation in genetic control of atherosclerosis in hyperlipidemic mice. G3 (Bethesda). 10:4679–4689. doi:10.1534/g3.120.401856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiffer TR, Bond JS.. 2014. Meprin metalloproteases inactivate interleukin 6. J Biol Chem. 289:7580–7588. doi:10.1074/jbc.M113.546309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg D, Bruns BC, Moali C, Vadon-Le Goff S, Sterchi EE, et al. 2010. Processing of procollagen III by Meprins: new players in extracellular matrix assembly? J Invest Dermatol. 130:2727–2735. doi:10.1038/jid.2010.202. [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Wang Q, Shi YL, Zhang Z, Leitinger N, et al. 2011. Characterization of Ath29, a major mouse atherosclerosis susceptibility locus, and identification of Rcn2 as a novel regulator of cytokine expression. Am J Physiol Circ Physiol. 301:H1056–61. doi:10.1152/ajpheart.00366.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei J, Liu Y, Dai N, Favara M, Greene T, et al. 2010. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 33:106–117. doi:10.1016/j.immuni.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Li Y. D, Shih M, Wang X, Laubach VE, et al. 2006a. Deficiency of inducible NO synthase reduces advanced but not early atherosclerosis in apolipoprotein E-deficient mice. Life Sci. 79:525–531. doi:10.1016/j.lfs.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tian J, Matsumoto AH, Shi W.. 2006b. Differential response of vascular smooth muscle cells to oxidized LDL in mouse strains with different atherosclerosis susceptibility. Atherosclerosis. 189:99–105. doi:10.1016/j.atherosclerosis.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, et al. 2015. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 131:29–322. [DOI] [PubMed] [Google Scholar]
- Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R.. 1994. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb. 14:133–140. [DOI] [PubMed] [Google Scholar]
- Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, et al. ; EPIC-CVD Consortium. 2017. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 49:1385–1391. doi:10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, et al. 2015. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary arterydisease. Nat Genet. 47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao JH, Xie PZ, Fishbein MC, Kreuzer J, Drake TA, et al. 1994. Pathology of atheromatous lesions in inbred and genetically engineered mice. Genetic determination of arterial calcification. Arterioscler Thromb J Vasc Biol Am Heart Assoc. 14:1480–1497. [DOI] [PubMed] [Google Scholar]
- Rekhter MD. 1999. Collagen synthesis in atherosclerosis: too much and not enough. Cardiovasc Res. 41:376–384. doi:S0008-6363(98)00321-6. [pii] [DOI] [PubMed] [Google Scholar]
- Rousselle A, Qadri F, Leukel L, Yilmaz R, Fontaine J-F, et al. 2013. CXCL5 limits macrophage foam cell formation in atherosclerosis. J Clin Invest. 123:1343–1347. doi:10.1172/JCI66580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlan JS, Li Q, Manichaikul A, Wang Q, Matsumoto AH, et al. 2013. Atherosclerosis susceptibility Loci identified in an extremely atherosclerosis-resistant mouse strain. J Am Heart Assoc. 2:e000260.doi:10.1161/JAHA.113.000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Haberland ME, Jien ML, Shih DM, Lusis AJ.. 2000b. Endothelial responses to oxidized lipoproteins determine genetic susceptibility to atherosclerosis in mice. Circulation. 102:75–81. [DOI] [PubMed] [Google Scholar]
- Shi W, Wang NJ, Shih DM, Sun VZ, Wang X, et al. 2000a. Determinants of atherosclerosis susceptibility in the C3H and C57BL/6 mouse model: evidence for involvement of endothelial cells but not blood cells or cholesterol metabolism. Circ Res. 86:1078–1084. [DOI] [PubMed] [Google Scholar]
- Shi W, Wang X, Wong J, Hedrick CC, Wong H, et al. 2004. Effect of macrophage-derived apolipoprotein E on hyperlipidemia and atherosclerosis of LDLR-deficient mice. Biochem. Biophys. Res Commun. 317:223–229. doi:10.1016/j.bbrc.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Shi LJ, Tang X, He J, Shi W.. 2021. Hyperlipidemia influences the accuracy of glucometer-measured blood glucose concentrations in genetically diverse mice. Am J Med Sci. 362:297–302. doi:10.1016/j.amjms.2021.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Bhasin JM, Baglione J, Settle M, Xu Y, et al. 2006. Atherosclerosis susceptibility loci identified from a strain intercross of apolipoprotein E-deficient mice via a high-density genome scan. ATVB. 26:597–603. doi:10.1161/01.ATV.0000201044.33220.5c. [DOI] [PubMed] [Google Scholar]
- Su Z, Li Y, James JC, McDuffie M, Matsumoto AH, et al. 2006. Quantitative trait locus analysis of atherosclerosis in an intercross between C57BL/6 and C3H mice carrying the mutant apolipoprotein E gene. Genetics. 172:1799–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Jin H-J, Bond JS.. 2009. Disruption of the meprin alpha and beta genes in mice alters homeostasis of monocytes and natural killer cells. Exp Hematol. 37:346–356. doi:10.1016/j.exphem.200810.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Pei H, James JC, Li Y, Matsumoto AH, et al. 2005. Circulating adhesion molecules in apoE-deficient mouse strains with different atherosclerosis susceptibility. Biochem Biophys Res Commun. 329:1102–1107. [DOI] [PubMed] [Google Scholar]
- Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC.. 2016. SIFT missense predictions for genomes. Nat Protoc. 11:1–9. doi:10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- Venegas-Pino DE, Banko N, Khan MI, Shi Y, Werstuck GH.. 2013. Quantitative analysis and characterization of atherosclerotic lesions in the murine aortic sinus. J Vis Exp. 82:50933. doi:10.3791/50933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vromman A, Ruvkun V, Shvartz E, Wojtkiewicz G, Masson GS, et al. 2019. Stage-dependent differential effects of interleukin-1 isoforms on experimental atherosclerosis. Eur Heart J. 40:2482–2491. doi:10.1093/eurheartj/ehz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Grainger AT, Manichaikul A, Farber E, Onengut-Gumuscu S, et al. 2015. Genetic linkage of hyperglycemia and dyslipidemia in an intercross between BALB/cJ and SM/J Apoe-deficient mouse strains. BMC Genet. 16:133. doi:10.1186/s12863-015-0292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman SC, Ravisankar P, Daugherty A.. 2002. Interleukin-18 enhances atherosclerosis in apolipoprotein E(-/-) mice through release of interferon-gamma. Circ Res. 90:E34–E38. doi:10.1161/hh0202.105292. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, et al. 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci USA. 100:3380–3385. doi:10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z-S, Lee BK, Park D-W, Lee S-W, Kim Y-H, et al. 2010. Relation of plaque size to compositions as determined by an in vivo volumetric intravascular ultrasound radiofrequency analysis. Int J Cardiovasc Imaging. 26:165–171. doi:10.1007/s10554-009-9520-8. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Miyoshi T, Bao Y, Sheehan JP, Matsumoto AH, et al. 2009. Microarray analysis of gene expression in mouse aorta reveals role of the calcium signaling pathway in control of atherosclerosis susceptibility. Am J Physiol Circ Physiol. 296:H1336–43. doi:10.1152/ajpheart.01095.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huangfu C, Chang Z, Grainger AT, Liu Z, et al. 2019. Atherogenesis in the carotid artery with and without interrupted blood flow of two hyperlipidemic mouse strains. J Vasc Res. 56:241–254. doi:10.1159/000502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the manuscript are represented fully within the manuscript and supplementary material available at GENETICS online.