Abstract
As the COVID-19 pandemic spread globally, the consumption of antibiotics increased. However, no studies exist evaluating the effect of antibiotics use on the antibiotic resistance of intestinal flora in COVID-19 patients during the pandemic. To explore this issue, we collected 15 metagenomic data of fecal samples from healthy controls (HCs) with no use history of antibiotics, 23 metagenomic data of fecal samples from COVID-19 patients who received empirical antibiotics [COVID-19 (abx+)], 18 metagenomic data of fecal samples from antibiotics-naïve COVID-19 patients [COVID-19 (abx-)], and six metagenomic data of fecal samples from patients with community-acquired pneumonia [PC (abx+)] from the Sequence Read Archive database. A total of 513 antibiotic-resistant gene (ARG) subtypes of 18 ARG types were found. Antibiotic treatment resulted in a significant increase in the abundance of ARGs in intestinal flora of COVID-19 patients and markedly altered the composition of ARG profiles. Grouped comparisons of pairs of Bray-Curtis dissimilarity values demonstrated that the dissimilarity of the HC versus the COVID-19 (abx+) group was significantly higher than the dissimilarity of the HC versus the COVID-19 (abx-) group. The mexF, mexD, OXA_209, major facilitator superfamily transporter, and EmrB_QacA family major facilitator transporter genes were the discriminative ARG subtypes for the COVID-19 (abx+) group. IS621, qacEdelta, transposase, and ISCR were significantly increased in COVID-19 (abx+) group; they greatly contributed toward explaining variation in the relative abundance of ARG types. Overall, our data provide important insights into the effect of antibiotics use on the antibiotic resistance of COVID-19 patients during the COVID-19 epidemic.
Keywords: COVID-19 patients, Empirical antibiotic exposure, Fecal microbiota, Metagenomic analysis, Antibiotic resistance genes, Mobile genetic elements
1. Introduction
Coronavirus disease 2019 (COVID-19, or SARS-CoV-2) has rapidly spread worldwide (Auerswald et al., 2021), and it is exacting substantial medical and economic tolls upon humanity. This disease has been declared as a “Public Health Emergency of International Concern” by the World Health Organization (WHO) and is being treated with various antivirals, antibiotics, and antifungals (Miranda et al., 2020). For example, many COVID-19 patients presenting with mild disease without pneumonia or moderate disease with pneumonia are treated with antibiotics (WHO, 2020). Severely ill COVID-19 patients are treated with a variety of antibiotics to take control of the condition (Fehr and Perlman, 2015). A recent review showed that 72% (1450/2010) of hospitalized COVID-19 patients received antibiotics (Rawson et al., 2020). Current evidence indicates low rates of bacterial/fungal co-infection in patients with COVID-19, yet high prescription rates of broad-spectrum antibiotic agents have been reported (Rawson et al., 2020). In the short term, antibiotics exert a variety of roles during the pandemic, but the long-term use of antibiotics and their misuse will contribute to the development of antibiotic resistance (Miranda et al., 2020). This should be expected a marked increase in antibiotic resistance during this pandemic.
Human intestinal flora are considered an important reservoir of antibiotic-resistant genes (ARGs), which have posed immense threats to public health over the last two decades (Feng et al., 2018; Laxminarayan et al., 2013). The bacteria carrying out antibiotic resistance can persist in human intestines for several years, even after short periods of antibiotic intake (Clemente et al., 2015; Hu et al., 2013). Moreover, it is generally recognized that horizontal gene transfer (HGT) is an important evolutionary force for the spread of the ARGs among bacteria (Forsberg et al., 2014; Goossens et al., 2005). In the COVID-19 pandemic, it is unclear whether the use of empirical antibiotics will have a net-positive or net-negative impact on the emergence and development of antibiotic resistance of intestinal flora. Knowing the impact of the use of empirical antibiotics on the antibiotic resistance of intestinal flora is essential for designing antibiotic stewardship programs during the COVID-19 pandemic.
In the work described herein, we performed a metagenomic analysis on 62 fecal samples obtained from the U.S. National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database. To the best of our knowledge, this is the first study to characterize the alterations of fecal resistome in COVID-19 patients after empirical antibiotic exposure. This study aimed to report the impact of antibiotic use on antibiotic resistance during this pandemic to characterize the broad implications of the pandemic for health and social care systems. This issue must be approached with caution so that the long-term challenges of antibiotic resistance are addressed while preserving access to effective drugs.
2. Materials and methods
2.1. Data collection and information
To explore this issue, we selected and downloaded a total of 62 fecal metagenomic datasets from NCBI BioProject No. PRJNA624223 (Zuo et al., 2020), including 15 metagenomic data of fecal samples from healthy individuals with no history of antibiotic intake in the past 3 months, 23 metagenomic data of fecal samples from COVID-19 patients who received empirical antibiotics and without discontinued use of antibiotics during stool collection, 18 metagenomic data of fecal samples from antibiotics-naïve COVID-19 patients during stool collection, and six metagenomic data of fecal samples from community-acquired pneumonia patients who received empirical antibiotics. See Supplementary Table 1 for detailed accession number information relating to specimens. The basic information of subjects is provided in Supplementary Table 1. FastQ files for all SRA accessions were obtained from the NCBI SRA repository using prefetch and fastq-dump from the SRA toolkit (https://github.com/ncbi/sra-tools).
2.2. Bioinformatics analysis
High-quality microbial sequencing data were generated by removing low-quality and short reads and human host sequences using the “metaWRAP read_qc” module (Uritskiy et al., 2018). Metagenomic sequencing yielded 33.1 million microbial reads per sample on average, with data analysis performed using MetaPhlAn3 software, a bioinformatics pipeline developed for taxonomic classification from metagenomic sequence data (Beghini et al., 2021). ARG-like reads were determined in all samples using ARGs-OAP v2.0 (Yin et al., 2018). The default parameters of ARGs-OAP v2.0 were used, i.e., a sequence identity of 80%, an E-value cutoff of 10−7, and an alignment length of more than 25 amino acids. The Structured Antibiotic Resistance Genes (SARG) v2.0 database, provided by the ARGs-OAP v2.0 software developers, was used to search ARGs. The SARG database was created by concatenating the Antibiotic Resistance Genes Database (ARDB; http://ardb.cbcb.umd.edu/) (Liu and Pop, 2009) and the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca/) (Jia et al., 2017), which contained 24 ARG types (e.g., tetracycline-resistant gene) and 1244 ARG subtypes (e.g., tetA and tetB).
ARG-like and 16S rRNA gene sequences were prescreened using customized Perl scripts. We calculated and normalized ARG abundance (copies of ARG per 16S rRNA) using the following equation (Li et al., 2015):
where Ni(ARG-like sequence) is the number of the ARG-like sequences aligned to one specific ARG reference sequence; Li(ARGs reference sequence) is the length of the corresponding specific ARG reference sequence; N16S sequence is the number of the 16S rRNA gene sequence identified from the metagenomic reads; L16S sequence is the mean length of 16S rRNA genes sequence (1432 bp) in the Greengenes reference database (http://greengenes.secondgenome.com/downloads/database/13_5) (DeSantis et al., 2006); n is the number of the mapped ARG reference sequence belonging to the ARG type or subtype; Lreads is the sequence length of the metagenomic reads (150 nt).
MGE-like reads were identified and annotated in all samples by an aligned custom MGE database (https://github.com/KatariinaParnanen/MobileGeneticElementDatabase) (Pärnänen et al., 2018). Relatively strict criteria for MGE-like reads annotation with a sequence identity of 80%, an E-value cutoff of 10−7, and an alignment length of more than 25 amino acids were used to reduce the number of false positives. We calculated and normalized MGE abundance (copies of ARG per 16S rRNA) using the following equation:
where Ni(MGE-like sequence) is the number of the MGE-like sequence aligned to one specific MGE reference sequence; Li(MGEs reference sequence) is the length of the corresponding specific MGE reference sequence; N16S sequence is the number of the 16S rRNA gene sequence identified from the metagenomic reads; L16S sequence is the mean length of 16S rRNA genes sequence (1432 bp) in the Greengenes reference database (http://greengenes.secondgenome.com/downloads/database/13_5) (DeSantis et al., 2006); n is the number of the mapped MGE reference sequence belonging to the MGE type or subtype; Lreads is the sequence length of the metagenomic reads (150 nt).
2.3. Statistical analysis
Statistical analysis and graphical plotting were performed using R software. Alpha and beta diversity, principal coordinates analysis (PcoA), Procrustes analysis, permutational multivariate analysis of variance (PERMANOVA), and partial redundancy analysis (pRDA) were computed using the Vegan R package. We used the Kruskal-Wallis rank-sum test to compare the four groups and applied the Wilcoxon rank-sum test to assess the statistical significance between the two groups. LEfSe was performed to compare the characteristics of different resistance genes from the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+). Spearman's rank correlation between the microbiota and ARG was calculated with Hmisc in R. Network visualization was conducted using the Gephi platform (https://gephi.org/). The correlations of the relative abundance of ARGs and MGEs were evaluated by correlation and best random forest model.
3. Result
3.1. Antibiotic resistome differences among HC group, COVID-19 (abx-) group, COVID -19 (abx+) group, and PC (abx+) group
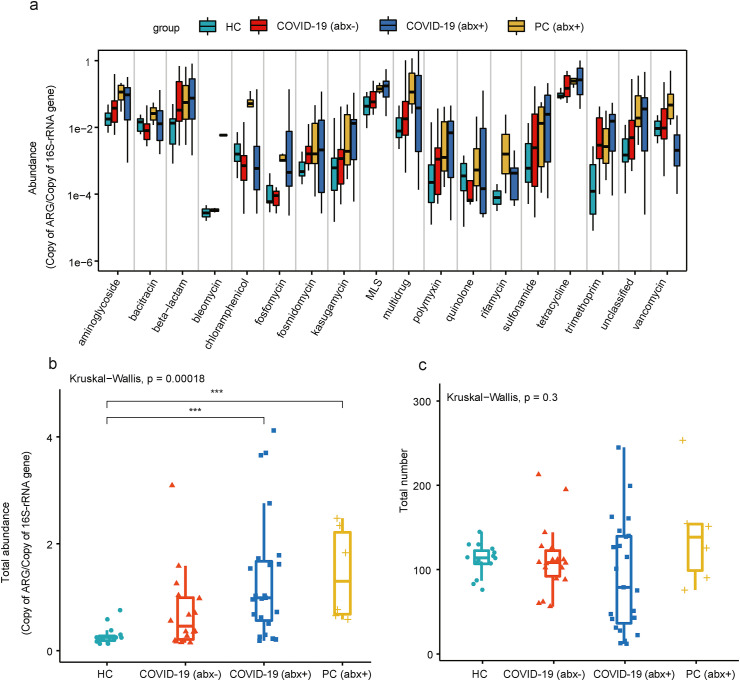
In total, we detected 18 ARG types and 513 ARG subtypes in the fecal microbiota of the study samples. Tetracycline, multidrug, beta-lactam, macrolide-lincosamide-streptogramin (MLS), aminoglycoside, and bacitracin resistance genes were the main classes of ARGs with high relative abundance [Fig. 1 (a)]. The total relative abundance of ARGs did not show any significant difference between the HC group and COVID-19 (abx-) group. Notably, the relative abundances of total ARGs in the COVID-19 (abx+) group and PC (abx+) group were significantly higher than those in the HC group [Fig. 1(b)]. The average relative abundances of ARGs in the COVID-19 (abx+) group and PC (abx+) group were 4.77 and 5.08 times higher than the HC group, respectively. No significant differences were identified in the number of ARGs between any two of the four groups [Fig. 1(c)].
Fig. 1.
(a) Comparison of antibiotic resistance gene (ARG) abundance at type level. Boxplots of (b) total relative abundance and (c) number of ARGs in four groups included in present study.
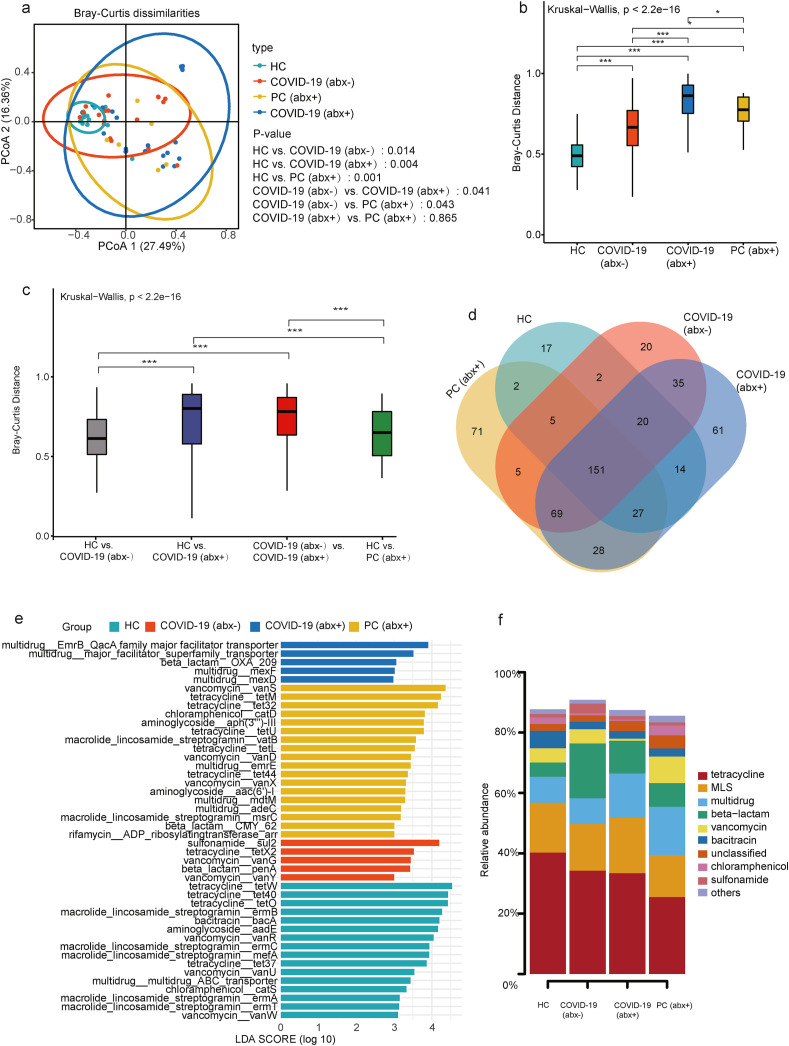
Beta diversity was analyzed using PCoA by Bray-Curtis, showing that no significant difference only between the COVID-19 (abx+) and PC (abx+) groups was observed [Fig. 2 (a)]. Note that empirical antibiotic exposure modifies not only the abundance of ARGs, but also their composition in the gut microbiome. We compared the within-group variation of the ARG composition based on Bray-Curtis distance and found that the dissimilarity among the subjects in the COVID-19 (abx+) group was statistically higher than those of other groups [Fig. 2(b)]. The magnitudes of the Bray-Curtis distance of four groups fall in the order COVID-19 (abx+)>PC (abx+)>COVID (abx-)>HC. Further grouped comparisons of pairs of Bray-Curtis dissimilarity values demonstrated that the similarity of the HC versus the COVID-19 (abx+) group was significantly lower than the similarity of the HC versus the COVID-19 (abx-) group [Fig. 2(c)]. In addition, the dissimilarity of the HC versus the COVID-19 (abx+) group was significantly higher than the dissimilarity of the HC versus the PC (abx+) group, which revealed that antibiotic treatment has a greater impact on COVID-19 patients compared to CAP patients.
Fig. 2.
(a) Beta diversity was analyzed using principal coordinates analysis (PCoA) by Bray Curtis, showing segregation of the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. Beta-diversity boxplots based on Bray-Curtis distances of metagenomics samples from the (b) same group (intra-beta diversity) and (c) between groups (inter-beta diversity). (d) Linear discriminant analysis effect size (LEfSe) histogram used to compare the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. (e) Venn diagram showing shared and unique ARG subtypes amongst four groups. (f) Shared ARGs and abundance comparisons in the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group. In the stacked bar plot, the percentage (%) of a specific ARG in one of the four groups is equal to the ratio of its corresponding abundance to the sum of the abundances of the ARG in the four groups.
Next, we compared differential enrichment of ARGs (linear discriminant analysis). The discriminative ARGs were differentially colored for the different groups. The OXA_209, major facilitator superfamily transporter, EmrB_QacA family major facilitator transporter genes, mexF, and mexD were the representative ARG subtypes for the COVID-19 (abx+) group. The mean relative abundance of major facilitator superfamily transporter, EmrB_QacA family major facilitator transporter genes, mexF, and mexD increased by more than 625.79-, 179.32-, 116.89-, and 341.18-fold, respectively, in the COVID-19 (abx+) group than in the HC group. The emergence and dissemination of multi-drug resistant (MDR) bacteria pose a grave public health problem (Nagarajan et al., 2018). In addition, sul2, vanG, tetX2, penA, and VanY genes displayed discriminativity in COVID-19 (abx-) subjects. We found that a greater proportion of vanS, tetM, tet32, catD, aph(3''')-III, tetU, vatB, tetL, vanD, emrE, tet44, vanX, aac(6′)-I, mdtM, adeC, msrC, CMY_62, and ADP_ribosylatingtransferase_arr were detected in the PC (abx+) group [Fig. 2(e)].
3.2. Shared ARGs among HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group
Fig. 2(d) is a Venn diagram showing shared and unique ARG subtypes amongst all groups. A total of 151 ARGs were common to the four groups despite the total abundance levels. The total number of ARGs in the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group and PC (abx+) was 238, 307, 405, and 358, respectively. The total number of ARGs in the COVID-19 (abx+) group was higher than in the HC group, COVID-19 (abx-) group, and PC (abx+). We found 17 unique ARG subtypes in the HC group and identified 20, 61, and 71 unique ARG subtypes in the COVID-19 (abx-), COVID-19 (abx+), and PC (abx+) groups, respectively. More unique ARGs were identified in the COVID-19 (abx+) and PC (abx+) groups [Fig. 2(d)]. Notably, compared with the HC group, the COVID-19 (abx+) group had 193 unique ARGs, including 156 beta-lactam resistance genes, 16 MDR genes, four aminoglycoside resistance genes, five MLS resistance genes, three trimethoprim resistance genes, two tetracycline resistance genes, two chloramphenicol resistance genes, one bleomycin resistance gene, one vancomycin resistance gene, one rifamycin resistance gene, one polymyxin resistance gene, and one unclassified resistance gene. A stacked column plot further showed detailed information on the types of shared ARGs and their abundance comparison among the HC, COVID-19 (abx-), COVID-19 (abx+), and PC (abx+) groups. Among these shared ARGs, MDR genes were more abundant in the COVID-19 (abx+) and PC (abx+) groups [Fig. 2(f)].
3.3. Correlation network of cooccurring ARG subtypes and microbial taxa
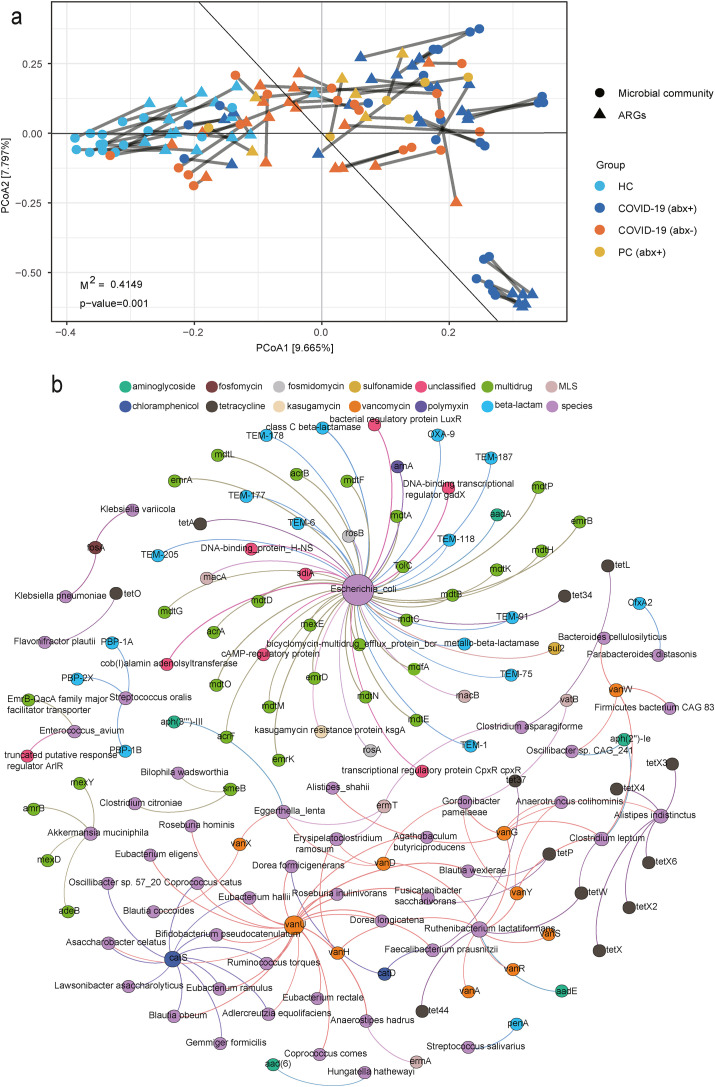
Procrustes tests depicted a correlation of 0.804 between ARGs and microbial communities (Procrustes sum of squares, 0.415; P = 0.001; number of permutations, 999) [Fig. 3 (a)]. This result indicates that the ARG profile overall was significantly correlated with microbial communities. The microbial community composition shapes ARG distribution in the gut microbiota. Network analysis showed the detailed co-occurrence patterns between the specific ARG subtypes and microbial taxa. It was indicated that the potential hosts of ARGs could be tracked using non-random co-occurrence patterns in network analysis (Li et al., 2015; Pehrsson et al., 2016; Su et al., 2015). The correlation network consisting of 150 edges and 145 nodes was constructed based on the significant positive correlations (q-value <0.01, r > 0.6) between ARG subtypes and microbial taxa that occurred in at least 20% of samples [Fig. 3(b)]. The detailed cooccurrence patterns between microbial taxa and ARG subtypes are presented in Supplementary Table 2.
Fig. 3.
(a) Procrustes analysis of correlations between ARGs and the microbial community. Statistical significance was verified by comparing the similarity of squared distances between matched samples in Procrustes analyses. P-value <0.05 indicated a significant correlation between antibiotic resistance and microbial community composition. (b) Network analysis showing co-occurrence pattern between microbial taxa and ARG subtypes based on Spearman's correlation analysis. An edge represents a strong (Spearman's r > 0.6) and significant (P < 0.01) correlation. The size of each node is proportional to the number of connections. The purple nodes represent microbial species, while the other nodes are colored according to ARG types. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We speculated that 48 bacterial species were the possible hosts of 97 ARG subtypes conferring resistance to 12 kinds of antibiotics based on the co-occurrence results. Among them, E. coli was predicted to harbor 54 ARGs, 25 of which were MDR genes (TolC, acrA, acrB, acrF, emrA, emrB, emrD, emrK, mdfA, and mdtA et al.), with the balance being one kasugamycin resistance gene (kasugamycin resistance protein ksgA), one aminoglycosides resistance gene (aadA), one sulfonamide resistance gene (sul2), two fosmidomycin resistance genes (rosB and rosA), two tetracycline resistance genes (tet34 and tetA), one polymyxin resistance gene (arnA), two MLS resistance genes (macA and macB), 12 beta-lactam resistance genes (OXA-9, TEM-1, TEM-118, TEM-177, TEM-178, TEM-187, TEM-205, TEM-6, TEM-75, TEM-91, class C beta-lactamase, and metallo-beta-lactamase), and several unclassified resistance genes. Some species (Akkermansia muciniphila, Alistipes indistinctus, Clostridium leptum, Eggerthella lenta, Gordonibacter pamelaeae, Ruthenibacterium lactatiformans, and Streptococcus oralis) were also predicted to harbor four or more ARG subtypes. Other bacterial species were speculated to carry three or one ARG subtypes. For instance, K. pneumoniae carried one ARG subtypes to fosfomycin (fosA), Prevotella copri carried one ARG subtype to tetracycline (tet37), and Erysipelatoclostridium ramosum carried three ARG subtypes to vancomycin (vanH and vanD) and MLS (ermT).
3.4. MGEs in fecal microbiome among HC group, COVID-19 (abx-) group, COVID -19 (abx+) group, and PC (abx+) group
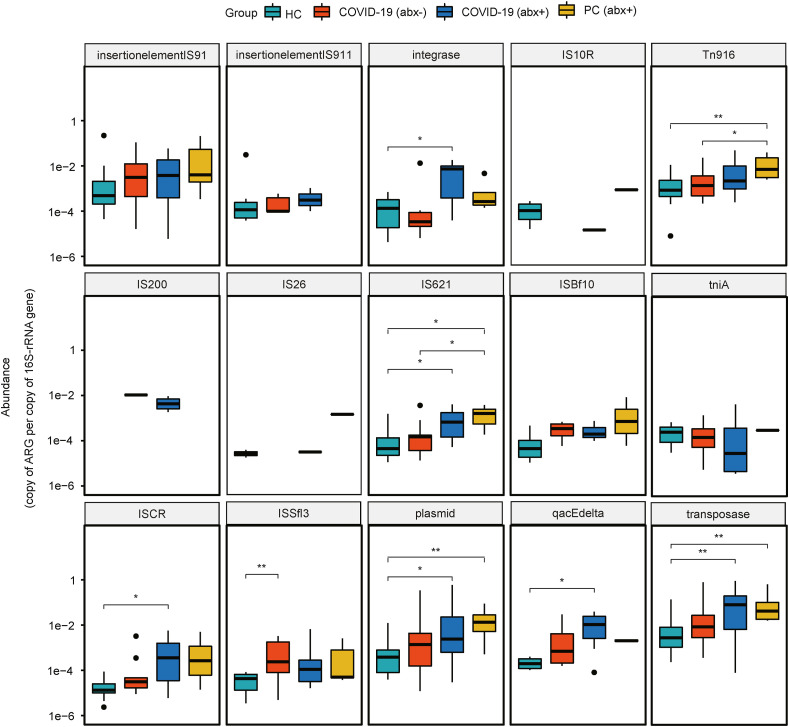
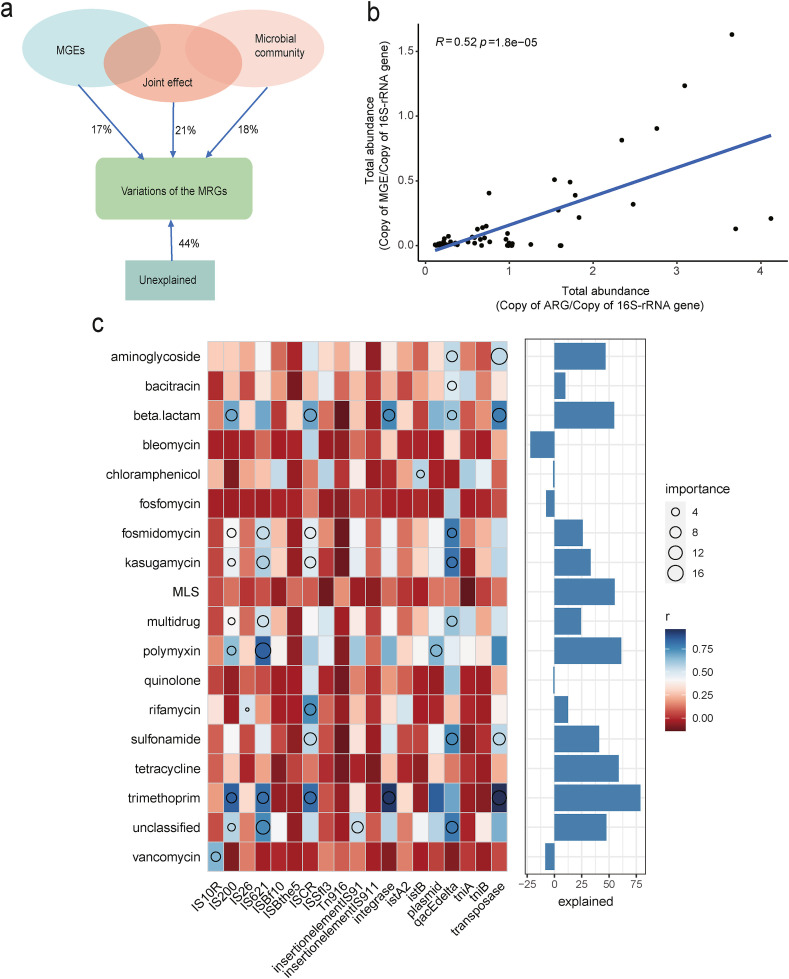
According to metagenomic analysis, we detected a total of 19 MGE types among all samples. Transposase had the highest relative abundance, followed by the plasmid. Here, we show the top 15 abundant MGEs and their differences among the four groups. Notably, the COVID-19 (abx+) group had a significantly higher abundance of integrase, IS621, ISCR, plasmid, qacEdelta, and transposase than the HC group. The PC (abx+) group had a significantly higher abundance of Tn916, IS621, plasmid, and transposase compared to the HC group (Fig. 4 ). We evaluated the relative role of MGEs and phylum-level microbial composition to explain the relative abundance of ARGs based on partial redundancy analysis (pRDA). The MGEs and microbial community showed contribution to the ARGs of 17% and 18%, respectively, and their joint effect was 21%. Our pRDA analyses showed that the explanatory proportion of MGEs and microbial community for the total ARG composition was close to equal [Fig. 5 (a)].
Fig. 4.
Boxplots showing relative abundances and differences of MGEs among the HC group, COVID-19 (abx-) group, COVID-19 (abx+) group, and PC (abx+) group.
Fig. 5.
(a) Total abundance of MGEs significantly correlated with the total abundance of detected ARGs based on Pearson's correlation. (b) pRDA differentiating the effect of microbial communities (at the phylum level) and MGEs on the profile of ARGs. (c) Associations of the relative abundance of ARGs and MGEs evaluated by correlation and best random-forest model. Circle size represents the variable importance (that is, proportion of explained variability estimated with out-of-bag cross-validation). Colors represent Spearman correlations. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The normalized abundance of MGEs was significantly positively correlated with the normalized abundance of ARGs detected [Fig. 5(b)]. Subsequently, we revealed the correlation between ARG types and MGEs in the fecal microbiome. Transposase, qacEdelta, IS621, IS200, and ISCR contributed more significantly toward explaining variation in the relative abundance of ARG types. Transposase was significantly related to aminoglycoside, beta-lactam, sulfonamide, and trimethoprim resistance genes. QacEdelta was significantly associated with aminoglycoside, bacitracin, beta-lactam, fosmidomycin, kasugamycin, multidrug, sulfonamide, and unclassified resistance genes. IS621 was significantly correlated with fosmidomycin, kasugamycin, multidrug, polymyxin, trimethoprim, and unclassified resistance genes. IS200 was significantly related to beta-lactam, fosmidomycin, kasugamycin, multidrug, polymyxin, trimethoprim, and unclassified resistance genes. ISCR was significantly correlated with beta-lactam, fosmidomycin, kasugamycin, rifamycin, sulfonamide, and trimethoprim resistance genes [Fig. 5(c)].
4. Discussion
Up to 70% of the patients with COVID-19 receive antibiotic treatment either in the outpatient or inpatient setting (Langford et al., 2021). There are two main reasons that patients with COVID-19 potentially receive antibiotic therapy. First, COVID-19 symptoms may be similar to bacterial pneumonia. Second, patients with COVID-19 may acquire a secondary bacterial infection that requires antibiotic treatment (Knight et al., 2021). There are potential threats during the COVID-19 pandemic that could drive antibiotic resistance. Since the beginning of the COVID-19 pandemic, the dynamics of antibiotic resistance have remained uncertain. Meanwhile, the impact of empirical antibiotic exposure on fecal resistome in COVID-19 patients remains largely unknown.
Consistent with past research, tetracycline resistance genes were found to be dominant in gut flora (Hu et al., 2021; Pal et al., 2016; Feng et al., 2018). Interestingly, the total ARG abundances in the COVID (abx+) group were the highest when compared with the HC and COVID (abx-) groups. This finding confirmed the severe emergence of ARGs in the gut microbiota of COVID-19 patients after empirical antibiotic exposure. In addition to gut microbiota, a similar result was recently reported that ARGs dramatically increased in the oropharyngeal microbiome of COVID-19 patients (Ma et al., 2021). In addition, we found that the total ARG abundances in the PC (abx+) group were also significantly higher than those in the HC group. Furthermore, the results of a recent study indicated that the total relative abundance of detected ARGs was significantly higher in individuals with diseases (non-COVID-19) who received antibiotic therapy for 1 month than that in healthy individuals (Duan et al., 2020). Collectively, these results highlight the need for the rational use of antibiotics in clinical settings.
Although antibiotic resistance is a common phenomenon, antibiotic consumption is a leading cause of the emergence of antibiotic resistance as well as of the acquisition, development, and spread of the resistome (Forslund et al., 2013; Wright, 2007). As with most infections, the initial antibiotic choice in COVID-19 patients is often empirical (Huang et al., 2020). For patients, broad-spectrum empirical antibiotic therapy increases not only antibiotic drug resistance but also the risk of adverse side effects (Llor and Bjerrum, 2014). It is worth stressing that antibiotic-resistant infections pose a major and growing threat to global health, and these infections kill an estimated 700,000 people worldwide annually (O'Neill, 2016). If appropriate measures are not taken, this number is expected to rise to 10 million per year by 2050 (Roope et al., 2019). Our results emphasize the necessity for decreasing unnecessary antibiotic use in the clinical management of COVID-19.
Antibiotic use favors the selection of resistant microbes. Increasing antibiotic resistance in intestinal flora of COVID-19 (abx+) patients can be explained by bystander selection (Tedijanto et al., 2018). Although antibiotic treatment is focused on controlling the pathogenic bacteria that cause co-infection in the lungs of COVID-19 patients, currently available antibiotics usually have antibacterial activity against many bacterial species and disseminate extensively throughout the body (Sullivan et al., 2001). Thus, the bacteria that constitute the human microbiome are subject to the selection pressure exerted by antibiotic consumption (Gustafsson et al., 2003; Lindgren et al., 2009; Nyberg et al., 2007). Hence, we speculate that antibiotic resistance in the microbiota of other body sites may also increase in addition to the gut. This is worthy of further research in the future.
COVID-19 has resulted in an exponential increase in global biocide use, possibly inducing further antibiotic selection pressure and contributing to the selection and development of highly resistant micro-organisms (Getahun et al., 2020; Ruiz, 2021). Compared with the HC group, a vast majority of unique ARGs in the COVID-19 (abx+) group are against beta-lactam antibiotics. A recent study by Peng et al. also found that beta-lactam resistance genes have increased in abundance during the COVID-19 pandemic (Peng et al., 2021). Four MDR genes (mexD, mexF, major facilitator superfamily transporter, and EmrB_QacA family major facilitator transporter) were significantly more abundant among COVID-19 (abx+) patients. MDR transporters, or efflux pumps, play a prominent role in acquired clinical antibiotic resistance (Higgins, 2007; Webber and Piddock, 2003). The emergence and dissemination of MDR bacteria can impair the clinical utility of major antibiotic agents, which could pose serious consequences (Kumar et al., 2013). The worldwide advent of MDR bacteria represents a serious public health issue. Previous research based on metagenomic analysis revealed ARG exchange events between environmental bacteria and clinical pathogens (Forsberg et al., 2012). This implies that receiving antibiotic administration not only increases the COVID-19 patient's own MDR genes of intestinal flora, but may also pollute the surrounding environment. In the future, with the wide spread of MDR bacteria, more clinically available antibiotics may prove to be ineffective.
Within-individual heterogeneity of ARG composition in the COVID-19 (abx+) group was found to be significantly higher compared with the HC, COVID-19 (abx-) groups, and PC (abx+). Meanwhile, the heterogeneity of ARG composition between the HC and COVID-19 (abx+) groups was significantly higher than between the HC and COVID-19 (abx-) groups. Procrustes analysis demonstrated that the microbial community composition shapes ARG distribution in the gut microbiota. We surmise that this heterogeneity found in ARG composition may also exist in intestinal flora. This serious disorder may have a negative impact on subsequent treatment, which is not conducive to the recovery of COVID-19 patients. Compared with antibiotics-naïve COVID-19 patients, the ARG composition of COVID-19 patients receiving antibiotic administration could require more time to return to the level of a healthy population. Whether this effect persists in the long term must be confirmed in investigations with a long-term follow-up.
Infections due to antibiotic-resistant bacteria are a major public health threat. The co-occurrence pattern network implied that 48 species might be potential hosts of 97 ARG subtypes. Among them, E. coli harbored maximum numbers of ARGs, 60% of which were MDR genes. A survey of antimicrobial resistances of fecal Enterobacteriacea in healthy people, short-term hospital patients, and long-term hospital patients also found that E. coli were the most highly representative carriers of resistance in all three groups (Osterblad et al., 2000). These results are similar to those of Feng et al. (2018), but E. coli increased the resistance to aminoglycoside, sulfonamide, and kasugamycin in our results. We posit that this could imply the emergence of E. coli with stronger antibiotics resistance. Considering that E. coli can co-infect COVID-19 patients (Zhu et al., 2020), the prevalence of ARGs is of special concern.
According to previous studies, Streptococcus oralis (Byers et al., 2000), Klebsiella pneumoniae (Martin and Bachman, 2018), Klebsiella quasipneumoniae (Brisse et al., 2014), Klebsiella variicola (Maatallah et al., 2014), Streptococcus salivarius (Olson et al., 2019), Enterococcus avium (Lee et al., 2004), Prevotella copri (Posteraro et al., 2019), Alistipes indistinctus (Parker et al., 2020), Erysipelatoclostridium ramosum (Zakham et al., 2019), Eggerthella lenta (Bo et al., 2020), and Anaerotruncus colihominis (Lau et al., 2006) can be pathogens or potential pathogens that can cause diseases in humans. The fact that these ARG-carrying pathogens are able to resist antibiotics taken by humans represents a high risk to human health.
HGT is a common event in the microbial ecosystem of the human intestinal tract (McInnes et al., 2020). A concerning result is that, compared to the HC group, the abundances of IS621, qacEdelta, transposase, and ISCR were significantly increased in the COVID-19 (abx+) group. These four MGEs also contributed significantly toward explaining variations in the relative abundance of ARG types. For COVID-19 patients, the elevated abundance of these MGEs may increase the opportunity for horizontal gene transfer of ARGs in gut microbiota (Abe et al., 2020; Lu et al., 2018; Larsson et al., 2018). What is more worrying is that HGT in this niche has the potential to mediate ARG transfer from commensal organisms into potential pathogens (Broaders et al., 2013). Consequently, it is possible for E. coli, K. pneumoniae, enterococci, and other opportunistic pathogens inhabiting the human gut to acquire resistance determinants from other members of gut flora (McInnes et al., 2020). ARGs that are carried on mobile genetic elements in pathogens could give rise to an immediate threat to the successful treatment of clinical bacterial infections.
5. Conclusions
The results of this study indicate that the use of empirical antibiotics has a significantly negative impact on the antibiotic resistance of intestinal flora. To reduce the potential long-lasting effects on antibiotic resistance and promote access to effective antibiotics, the development of treatment guidelines to limit unnecessary antibiotic exposure and of measures to maintain conventional surveillance of antibiotic resistance must be at the forefront of research in the post-COVID-19 era.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2019YFC1200601-6 and 2019YFC1200700) and the National Natural Science Foundation of China (82073624).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijheh.2021.113882.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Auerswald H., Yann S., Dul S., In S., Dussart P., Martin N.J., Karlsson E.A., Garcia-Rivera J.A. Assessment of inactivation procedures for SARS-CoV-2. J. Gen. Virol. 2021;102:1539. doi: 10.1099/jgv.0.001539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K., Nomura N., Suzuki S. Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020;96(5) doi: 10.1093/femsec/fiaa031. fiaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S., Passet V., Grimont P. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 2014;64(Pt 9):3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M., Valles-Colomer M., Weingart G., Zhang Y., Zolfo M., Huttenhower C., Franzosa E.A., Segata N. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife. 2021;10 doi: 10.7554/ELIFE.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers H.L., Tarelli E., Homer K.A., Beighton D. Isolation and characterisation of sialidase from a strain of Streptococcus oralis. J. Med. Microbiol. 2000;49:235–244. doi: 10.1099/0022-1317-49-3-235. [DOI] [PubMed] [Google Scholar]
- Broaders E., Gahan C.G., Marchesi J.R. Mobile genetic elements of the human gastrointestinal tract: potential for spread of antibiotic resistance genes. Gut Microb. 2013;4(4):271–280. doi: 10.4161/gmic.24627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo J., Wang S., Bi Y., Ma S., Wang M., Du Z. Eggerthella lenta bloodstream infections: two cases and review of the literature. Future Microbiol. 2020;15:981–985. doi: 10.2217/fmb-2019-0338. [DOI] [PubMed] [Google Scholar]
- Clemente J.C., Pehrsson E.C., Blaser M.J., Sandhu K., Gao Z., Wang B., Magris M., Hidalgo G., Contreras M., Noya-Alarcón Ó., Lander O., McDonald J., Cox M., Walter J., Oh P.L., Ruiz J.F., Rodriguez S., Shen N., Song S.J., Metcalf J., Knight R., Dantas G., Dominguez-Bello M.G. The microbiome of uncontacted Amerindians. Sci. Adv. 2015;1(3) doi: 10.1126/SCIADV.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li B., Jiang X., Yang Y., Wells G.F., Zhang T., Li X. Antibiotic resistome in a large-scale healthy human gut microbiota deciphered by metagenomic and network analyses. Environ. Microbiol. 2018;20(1):355–368. doi: 10.1111/1462-2920.14009l. [DOI] [PubMed] [Google Scholar]
- Forsberg K.J., Patel S., Gibson M.K., Lauber C.L., Knight R., Fierer N., Dantas G. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509(7502):612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Sunagawa S., Kultima J.R., Mende D.R., Arumugam M., Typas A., Bork P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013;23:1163–1169. doi: 10.1101/GR.155465.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K.J., Reyes A., Wang B., Selleck E.M., Sommer M.O., Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/SCIENCE.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens H., Ferech M., Vander Stichele R., Elseviers M., ESAC Project Group Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet (London, England) 2005;365:579–587. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- Gustafsson I., Sjölund M., Torell E., Johannesson M., Engstrand L., Cars O., Andersson D.I. Bacteria with increased mutation frequency and antibiotic resistance are enriched in the commensal flora of patients with high antibiotic usage. J. Antimicrob. Chemother. 2003;52:645–650. doi: 10.1093/JAC/DKG427. [DOI] [PubMed] [Google Scholar]
- Getahun H., Smith I., Trivedi K., Paulin S., Balkhy H.H. Tackling antimicrobial resistance in the COVID-19 pandemic. Bull. World Health Organ. 2020;98(7) doi: 10.2471/BLT.20.268573. 442-442A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C.F. Multiple molecular mechanisms for multidrug resistance transporters. Nature. 2007;446:749–757. doi: 10.1038/NATURE05630. [DOI] [PubMed] [Google Scholar]
- Hu Y., Yang X., Qin J., Lu N., Cheng G., Wu N., Pan Y., Li J., Zhu L., Wang X., Meng Z., Zhao F., Liu D., Ma J., Qin N., Xiang C., Xiao Y., Li L., Yang H., Wang J., Yang R., Gao G.F., Wang J., Zhu B. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013;4:2151. doi: 10.1038/NCOMMS3151. [DOI] [PubMed] [Google Scholar]
- Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N., Doshi S., Courtot M., Lo R., Williams L.E., Frye J.G., Elsayegh T., Sardar D., Westman E.L., Pawlowski A.C., Johnson T.A., Brinkman F.S., Wright G.D., McArthur A.G. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/NAR/GKW1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight G.M., Glover R.E., McQuaid C.F., Olaru I.D., Gallandat K., Leclerc Q.J., Fuller N.M., Willcocks S.J., Hasan R., van Kleef E., Chandler C.I. Antimicrobial resistance and COVID-19: intersections and implications. Elife. 2021;10:1–27. doi: 10.7554/ELIFE.64139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Mukherjee M.M., Varela M.F. Modulation of bacterial multidrug resistance efflux pumps of the major facilitator superfamily. Int. J. Bacteriol. 2013:1–15. doi: 10.1155/2013/204141. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford B.J., So M., Raybardhan S., Leung V., Soucy J.R., Westwood D., Daneman N., MacFadden D.R. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27:520–531. doi: 10.1016/J.CMI.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llor C., Bjerrum L. Antimicrobial resistance: risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. drug Saf. 2014;5:229–241. doi: 10.1177/2042098614554919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Ferguson D.A., Jr., Laffan J.J. Vancomycin-resistant Enterococcus avium infections: report of 2 cases and a review of Enterococcus avium infections. Infect. Dis. Clin. Pract. 2004;12:239–244. [Google Scholar]
- Li B., Yang Y., Ma L., Ju F., Guo F., Tiedje J.M., Zhang T. Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 2015;9(11):2490–2502. doi: 10.1038/ismej.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Pop M. ARDB—antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443. doi: 10.1093/NAR/GKN656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren M., Lofmark S., Edlund C., Huovinen P., Jalava J. Prolonged impact of a one-week course of clindamycin on Enterococcus spp. in human normal microbiota. Scand. J. Infect. Dis. 2009;41:215–219. doi: 10.1080/00365540802651897. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R., Duse A., Wattal C., Zaidi A.K., Wertheim H.F., Sumpradit N., Vlieghe E., Hara G.L., Gould I.M., Goossens H., Greko C., So A.D., Bigdeli M., Tomson G., Woodhouse W., Ombaka E., Peralta A.Q., Qamar F.N., Mir F., Kariuki S., Bhutta Z.A., Coates A., Bergstrom R., Wright G.D., Brown E.D., Cars O. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- Lu J., Wang Y., Li J., Mao L., Nguyen S.H., Duarte T., Coin L., Bond P., Yuan Z., Guo J. Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ. Int. 2018;121(Pt 2):1217–1226. doi: 10.1016/j.envint.2018.10.040. [DOI] [PubMed] [Google Scholar]
- Larsson D., Andremont A., Bengtsson-Palme J., Brandt K.K., de Roda Husman A.M., Fagerstedt P., Fick J., Flach C.F., Gaze W.H., Kuroda M., Kvint K., Laxminarayan R., Manaia C.M., Nielsen K.M., Plant L., Ploy M.C., Segovia C., Simonet P., Smalla K., Snape J., Topp E., van Hengel A.J., Verner-Jeffreys D.W., Virta M.P.J., Wellington E.M., Wernersson A.S. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Woo G.K., Fung A.M., Ngan A.H., Song Y., Liu C., Summanen P., Finegold S.M., Yuen K. Bacteraemia caused by Anaerotruncus colihominis and emended description of the species. J. Clin. Pathol. 2006;59(7):748–752. doi: 10.1136/jcp.2005.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes R.S., McCallum G.E., Lamberte L.E., van Schaik W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 2020;53:35–43. doi: 10.1016/j.mib.2020.02.002. [DOI] [PubMed] [Google Scholar]
- Miranda C., Silva V., Capita R., Alonso-Calleja C., Igrejas G., Poeta P. Implications of antibiotics use during the COVID-19 pandemic: present and future. J. Antimicrob. Chemother. 2020;75:3413–3416. doi: 10.1093/JAC/DKAA350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatallah M., Vading M., Kabir M.H., Bakhrouf A., Kalin M., Nauclér P., Brisse S., Giske C.G. Klebsiella variicola is a frequent cause of bloodstream infection in the stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS One. 2014;9(11) doi: 10.1371/JOURNAL.PONE.0113539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R.M., Bachman M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018;8:4. doi: 10.3389/FCIMB.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Zhang F., Zhou F., Li H., Ge W., Gan R., Nie H., Li B., Wang Y., Wu M., Li D., Wang D., Wang Z., You Y., Huang Z. Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target Ther. 2021;6(1):191. doi: 10.1038/s41392-021-00614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan D., Nagarajan T., Roy N., Kulkarni O., Ravichandran S., Mishra M., Chakravortty D., Chandra N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018;293:3492–3509. doi: 10.1074/JBC.M117.805499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg S.D., Osterblad M., Hakanen A.J., Löfmark S., Edlund C., Huovinen P., Jalava J. Long-term antimicrobial resistance in Escherichia coli from human intestinal microbiota after administration of clindamycin. Scand. J. Infect. Dis. 2007;39:514–520. doi: 10.1080/00365540701199790. [DOI] [PubMed] [Google Scholar]
- O'Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. [Google Scholar]
- Olson L.B., Turner D.J., Cox G.M., Hostler C.J. Streptococcus salivarius prosthetic joint infection following dental cleaning despite antibiotic prophylaxis. Case Rep. Infect. Dis. 2019;2019:1–4. doi: 10.1155/2019/8109280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterblad M., Hakanen A., Manninen R., Leistevuo T., Peltonen R., Meurman O., Huovinen P., Kotilainen P. A between-species comparison of antimicrobial resistance in enterobacteria in fecal flora. Antimicrob. Agents Chemother. 2000;44(6):1479–1484. doi: 10.1128/AAC.44.6.1479-1484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B.J., Wearsch P.A., Veloo A., Rodriguez-Palacios A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front. Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pärnänen K., Karkman A., Hultman J., Lyra C., Bengtsson-Palme J., Larsson D., Rautava S., Isolauri E., Salminen S., Kumar H., Satokari R., Virta M. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018;9(1):3891. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrsson E.C., Tsukayama P., Patel S., Mejía-Bautista M., Sosa-Soto G., Navarrete K.M., Calderon M., Cabrera L., Hoyos-Arango W., Bertoli M.T., Berg D.E., Gilman R.H., Dantas G. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533(7602):212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro P., De Maio F., Menchinelli G., Palucci I., Errico F.M., Carbone M., Sanguinetti M., Gasbarrini A., Posteraro B. First bloodstream infection caused by Prevotella copri in a heart failure elderly patient with Prevotella-dominated gut microbiota: a case report. Gut Pathog. 2019;11:1–6. doi: 10.1186/s13099-019-0325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C., Bengtsson-Palme J., Kristiansson E., Larsson D.G. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4(1):54. doi: 10.1186/s40168-016-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Zhang D., Chen T., Xia Y., Wu P., Seto W.K., Kozyrskyj A.L., Cowling B.J., Zhao J., Tun H.M. Gut microbiome and resistome changes during the first wave of the COVID-19 pandemic in comparison with pre-pandemic travel-related changes. J. Trav. Med. 2021;28(7) doi: 10.1093/jtm/taab067. taab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roope L., Smith R.D., Pouwels K.B., Buchanan J., Abel L., Eibich P., Butler C.C., Tan P.S., Walker A.S., Robotham J.V., Wordsworth S. The challenge of antimicrobial resistance: what economics can contribute. Science (New York, N.Y.) 2019;364(6435) doi: 10.1126/science.aau4679. [DOI] [PubMed] [Google Scholar]
- Ruiz J. Enhanced antibiotic resistance as a collateral COVID-19 pandemic effect? J. Hosp. Infect. 2021;107:114–115. doi: 10.1016/j.jhin.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson T.M., Moore L., Zhu N., Ranganathan N., Skolimowska K., Gilchrist M., Satta G., Cooke G., Holmes A. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin. Infect. Dis. 2020;71:2459–2468. doi: 10.1093/CID/CIAA530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J.Q., Wei B., Ou-Yang W.Y., Huang F.Y., Zhao Y., Xu H.J., Zhu Y.G. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 2015 doi: 10.1021/acs.est.5b01012. [DOI] [PubMed] [Google Scholar]
- Sullivan A., Edlund C., Nord C.E. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001;1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- Tedijanto C., Olesen S.W., Grad Y.H., Lipsitch M. Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11988. doi: 10.1073/PNAS.1810840115. –E11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP - a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6(1):158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber M.A., Piddock L.J. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003;51:9–11. doi: 10.1093/JAC/DKG050. [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2020. Clinical Management of COVID-19 Interim Guidance-May 2020. 2020. [Google Scholar]
- Wright G.D. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 2007;5(3):175–186. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- Yin X., Jiang X.T., Chai B., Li L., Yang Y., Cole J.R., Tiedje J.M., Zhang T. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics. 2018;34(13):2263–2270. doi: 10.1093/bioinformatics/bty053. [DOI] [PubMed] [Google Scholar]
- Zuo T., Zhang F., Lui G., Yeoh Y.K., Li A., Zhan H., Wan Y., Chung A., Cheung C.P., Chen N., Lai C., Chen Z., Tso E., Fung K., Chan V., Ling L., Joynt G., Hui D., Chan F., Chan P., Ng S.C. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955. doi: 10.1053/J.GASTRO.2020.05.048. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., Zhu F., Zhu B., Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakham F., Pillonel T., Brunel A.S., Zambelli P.Y., Greub G., Croxatto A., Bertelli C. Molecular diagnosis and enrichment culture identified a septic pseudoarthrosis due to an infection with Erysipelatoclostridium ramosum. Int. J. Infect. Dis. 2019;81:167–169. doi: 10.1016/J.IJID.2019.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.