Abstract
Pathogenic variants in PINK1 cause early-onset Parkinson’s disease. Although many PINK1 variants have been reported, the clinical significance is uncertain for the majority of them. To gain insights into the consequences of PINK1 missense variants in a systematic manner, we selected 50 PINK1 missense variants from patient- and population-wide databases and systematically classified them using Sherloc, a comprehensive framework for variant interpretation based on ACMG-AMP guidelines. We then performed functional experiments, including mitophagy and Parkin recruitment assays, to assess the downstream consequences of PINK1 variants. Analysis of PINK1 missense variants based on Sherloc showed that the patient databases over-annotate variants as likely pathogenic. Furthermore, our study shows that pathogenic PINK1 variants are most often linked to a loss-of-function for mitophagy and Parkin recruitment, while this is not observed for variants of unknown significance. In addition to the Sherloc framework, the added layer of evidence of our functional tests suggests a reclassification of 9/50 missense variants. In conclusion, we suggest the assessment of multiple layers of evidence, including functional data on top of available clinical and population-based data, to support the clinical classification of a variant and show that the presence of a missense variant in PINK1 in a Parkinson’s disease case does not automatically imply pathogenicity.
Subject terms: Parkinson's disease, Mutation, Genetic testing
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder worldwide, characterized by neuronal loss, with a focus on dopaminergic neurons in the substantia nigra1. Most PD patients have an idiopathic form of the disease that has an age at onset above 60 years. However, a small minority of patients (±4%) develop early-onset PD (EOPD) that begins before 45 years of age. For the majority of EOPD patients, the disease is inherited in an autosomal recessive manner via variants in the PRKN, PINK1, or PARK7 genes that lead to a loss-of-function2,3. These genes are therefore often the first inspected using gene panels in genetic diagnostics of EOPD cases, with variants occasionally identified. With the advent of next-generation sequencing methods, an increasing number of sequence variants in these genes have been collected4–7. For instance, more than 300 variants of the PINK1 gene have been identified. However, the functional consequences and clinical significance of the majority of these variants are unknown, which poses major challenges in genetic counseling practices.
To overcome hurdles in variant interpretation, the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG-AMP)8,9 provide guidelines to systematically categorize pathogenicity using standard terminology. In addition, common frameworks such as Sherloc have been established on top of the guidelines set by the ACMG-AMP to objectively assess pathogenicity using specific scoring systems that minimize bias10. Variants that have been proven to be (likely) disease-causing based on clinical, population-wide and functional evidence are annotated as (likely) pathogenic, while variants that are (likely) not disease-causing based on these sources of evidence are annotated as (likely) benign. However, most variants cannot be clearly assigned to these groups because the evidence for (likely) pathogenic and (likely) benign are both insufficient and are thus designated as having uncertain significance. As there are still only few functional studies on rare variants, current annotations often rely heavily on data such as the minor allele frequency (MAF) of a variant in population cohorts and the segregation of the variant with disease (when available). In silico computational predictions and modeling using the corresponding protein structure may provide added guidance for the interpretation of variants. To more accurately categorize pathogenicity, we need multiple layers of evidence, including functional testing in the form of appropriate experimental assays that can determine the biochemical or cellular consequence of the variant. Although these steps are all necessary to improve genetic diagnostics, pathogenicity of variants may remain uncertain even when there is a multitude of different data. Recently, a study showed that only a minority of the numerous reported PRKN variants could be classified as pathogenic using Sherloc in combination with cellular functional studies11.
PINK1 is another example of a recessive PD gene with many reported genetic variants. PINK1 encodes PTEN-induced putative kinase 1 (PINK1), a serine-threonine kinase that works in concert with the E3 ubiquitin ligase Parkin (encoded by PRKN) to maintain mitochondrial quality12–15. Through a membrane potential-dependent process, PINK1 is imported from the outer mitochondrial membrane (OMM) into the inner mitochondrial membrane, where it is constitutively degraded by mitochondrial proteases16,17. However, PINK1 import and cleavage is blocked upon mitochondrial depolarization caused by damage (e.g. oxidative stress), resulting in the accumulation of PINK1 on the OMM. At the OMM, PINK1 activates Parkin through phosphorylation of Parkin and ubiquitin, leading to stable recruitment and activation of Parkin onto the mitochondrial surface17–19. By ubiquitinating different OMM substrates, Parkin then orchestrates different mitochondrial quality control pathways, from the degradation of individual proteins to the complete removal of damaged mitochondria via mitophagy (Fig. 1a)13–15.
Fig. 1. Most PINK1 missense variants are classified as uncertain significance using Sherloc.
a Schematic overview of the PINK1/Parkin pathway in healthy and damaged state. b Venn diagram showing the number of unique and overlapping PINK1 missense variants reported in disease-related databases (ClinVar and MDSgene) and the population-wide database gnomAD. c ACMG-AMP annotations of the 50 missense variants investigated in this study, as determined by the disease-related databases (left) or using Sherloc (right). Horizontal bar graphs show all the studied variants. Vertical bar graphs are stratified by the origin of the reported missense variant in (b). d Schematic overview of the functional domains of PINK1 with the location of the 50 missense variants. Variants are color-coded following the legend of (c). B = benign; LB = likely benign; VUS = variant of uncertain significance; LP = likely pathogenic; P = pathogenic; MTS = mitochondrial targeting sequence; TM = transmembrane domain; CTR = C-terminal region.
In the present study, we utilized the Sherloc framework to systematically annotate 50 missense variants in PINK1 and followed this up with functional in vitro assays to gain insights into the consequence of these variants, with the overall aim being to provide better guidance for the interpretation of rare and/or novel missense variants.
Results
Compilation of disease-associated and population-based missense variants in PINK1
To compile an overview of the PINK1 missense variants reported in disease-associated databases and population-based databases, we searched the Movement Disorder Society Genetic mutation database (MDSgene)5 and ClinVar6 databases for missense variants in PINK1 and identified 78 variants reported in EOPD or PD patients, of which 54 were also reported in the population-based Genome Aggregation Database (gnomAD)7 (Fig. 1b). For practical reasons, we selected 46 disease-associated variants (this included all pathogenic variants), of which 19 were not reported in gnomAD, and four rare population-based missense variants (<8 reported alleles) to obtain a list of missense variants that covers the whole coding region of PINK1 (Fig. 1b). Although the clinical significance of each of these 46 missense variants was assigned by the patient databases, the pathogenic evaluation and the presence of bias in the interpretation remained unclear. The reported clinical significance of these 46 missense variants showed enrichment for likely pathogenic (15/46) and pathogenic (15/46) and an underrepresentation of likely benign (1/46) and benign (4/46) variants, while 11/46 variants were of uncertain significance. The rare population-based variants were classified with uncertain significance (Fig. 1c).
Many disease-associated missense variants in PINK1 are of uncertain significance
In order to better classify these 50 missense variants, we used the most recent best practice guidelines for variant classification recommended by the ACMG-AMP9. Pathogenicity was determined based on several criteria with varying strengths of evidence from which the annotation is derived9. However, since several criteria could not be checked with certainty or were not applicable for the missense variants in this study (Supplementary Fig. 1), most variants remained of uncertain significance (Supplementary Table 1). To overcome this, we utilized the semiquantitative Sherloc framework, a comprehensive refinement of the ACMG-AMP criteria10, to re-analyze the clinical significance of the missense variants. Benign and pathogenic points were allocated to each variant based on four layers of evidence (Supplementary Fig. 2): (1) the rarity of the variant based on MAF and homozygosity status derived from population-wide databases gnomAD/dbSNP, (2) whether the variant was found in patients, derived from disease-associated databases ClinVar/MDSgene, (3) segregation of the variant based on detailed family-reports from literature, and (4) evidence from experimental studies collected from the literature (Supplementary Table 2). Supplementary Table 1 provides a detailed overview of the points assigned per missense variant, including the summed benign and pathogenic points and the clinical significance.
Using the Sherloc framework, ten likely pathogenic variants and only two pathogenic variants were reclassified to uncertain significance, but no benign variants were. Furthermore, three likely pathogenic variants became pathogenic, and the one likely benign variant became benign. No variants of uncertain significance were reclassified (Fig. 1c). Not unexpectedly, missense variants reported to be unique in patients were mostly classified as pathogenic (11/19; Fig. 1c), and variants present in both patient and population databases were mostly classified as uncertain significance (17/28) or benign (5/28), with fewer being classified as likely pathogenic (1/28) or pathogenic (5/28). Thus, based on our Sherloc reclassification, most missense variants in PINK1 that are reported in disease-associated databases are of uncertain significance.
Generation of a mitophagy reporter to study the consequences of PINK1 carrying missense variants
We then investigated if variants classified as pathogenic are more often located or specifically located in one of the several domains of PINK1, including the N-terminal mitochondrial targeting signal, transmembrane domain, and the kinase domain with three insertions20. Interestingly, we observed that most of the pathogenic variants (14/16) are located in the kinase domain (Fig. 1d), suggesting a critical role for this domain. Indeed, previous studies have shown that many pathogenic variants (as classified in these earlier studies) abrogate the kinase activity of PINK1, leading to a failure in Parkin recruitment to mitochondria upon mitochondrial damage (Supplementary Table 2)18,21,22. Consequently, PINK1 and Parkin are unable to activate mitophagy-mediating proteins at the OMM, leading to failure in mitophagy induction13.
We then hypothesized that we could use mitophagy induction as a read-out of PINK1 function/activity and that the results of this experimental study could add to the fourth layer of evidence in the Sherloc framework, thereby supplementing existing evidence for the 50 missense variants. This would allow us to further improve the classification of these variants. To do so, we generated a mitophagy reporter in Human cervix epithelioid carcinoma (HeLa) cells in which we genetically depleted PINK1 using Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9, after which we stably expressed a reporter construct with the fluorophore mt-mKeima and FLAG-Parkin (Supplementary Fig. 3). mt-mKeima allows for sensitive quantification of mitochondria in the cytosol and lysosome23, whereas FLAG-Parkin is necessary to study PINK-Parkin-induced mitophagy as HeLa cells do not express endogenous Parkin (Supplementary Fig. 3C).
Most (likely) pathogenic PINK1 variants affect mitophagy induction
To study PINK1-Parkin mitophagy, we treated the reporter cells transiently expressing wild type (WT) PINK1-HA or variant-PINK1-HA with protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which induces mitochondrial stress by dissipating the mitochondrial membrane potential that leads to the stabilization of PINK1 and Parkin recruitment to mitochondria17. After 24 h of 10 µM CCCP treatment, mitophagy induction was determined using FACS. CCCP treatment in WT-PINK1-HA-expressing reporter cells led to an increase in cells with mitochondria in acidic compartments (i.e., lysosomes), indicative of mitophagy induction (Fig. 2a; Supplementary Fig. 4A.i/ii). When we transiently expressed the different variant-PINK1-HA proteins, we observed that the majority of missense variants in PINK1 resulted in a wide range of mitophagy induction (Fig. 2a) that was not caused by differences in their protein levels, as assessed by western blotting (Supplementary Fig. 5A, B), except for variant p.Val317Ile that showed significantly increased PINK1 protein levels, which might possibly mask a reduced mitophagy induction due to loss of PINK1 kinase activity.
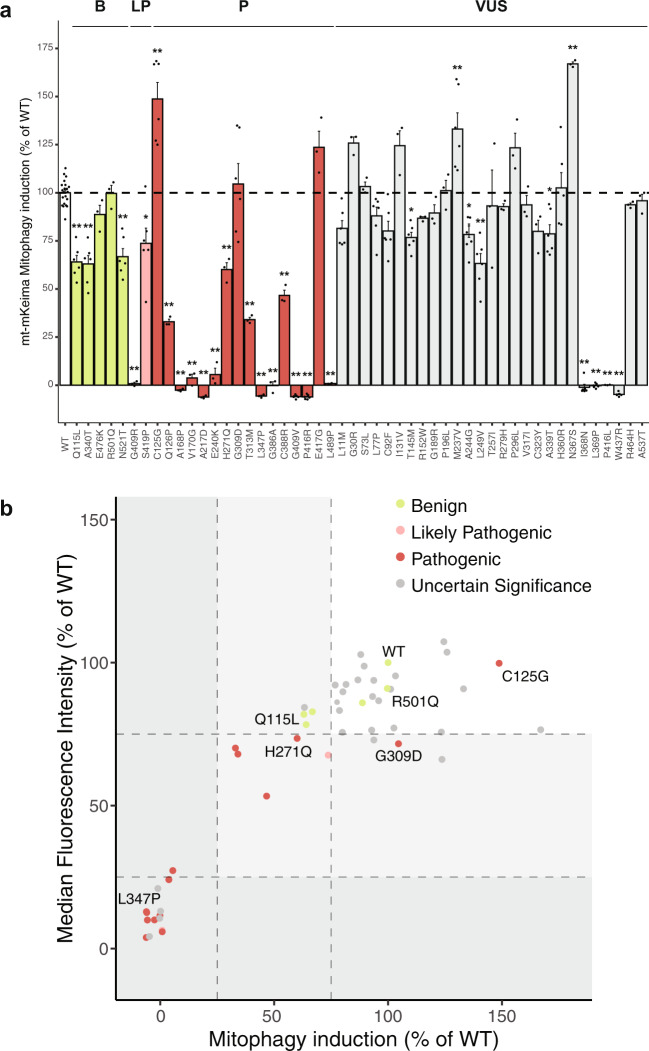
Fig. 2. Mitophagy induction is affected by most (likely) pathogenic PINK1 variants.
a Assessment of mitophagy induction upon CCCP treatment in HeLa PINK1 KO cells stably expressing mt-mKeima-FLAG-Parkin transfected with PINK1 WT or missense variants. All data shown are normalized against PINK1 WT control. Each dot represents a separate experiment. Bars are color-coded by the annotation as determined by Sherloc. Data analyzed using one-way ANOVA. *p < 0.05, ** p < 0.005. b Relation between mitophagy induction and median fluorescence intensity of acidic mt-mKeima for PINK1 WT or missense variants. The X-axis is mean data from (a). Y-axis is mean data from Supplementary Fig. 4B. Three groups of variants based on the amount of mitophagy function are highlighted in the areas in white (group 1), light gray (group 2), and dark gray (group 3). Each dot represents a variant and is color-coded by the Sherloc annotation.
We found that the PINK1 proteins carrying missense variants classified as benign all induced mitophagy upon CCCP treatment, although some did show a slightly decreased function (p.Gln115Leu, p.Ala340Thr, p.Asn521Thr) compared to WT PINK1 (Fig. 2a). In contrast, most missense variants in PINK1 annotated as (likely) pathogenic either caused a significant decrease in mitophagy induction (p.Ser419Pro, p.Gln126Pro, p.His271Gln, p.Thr313Met, and p.Cys388Arg) compared to WT PINK1 or completely abolished mitophagy induction (p.Gly409Arg, p.Ala168Pro, p.Val170Gly, p.Ala217Asp, p.Glu240Lys, p.Leu347Pro, p.Gly386Ala, p.Gly409Val, and p.Leu489Pro) upon CCCP treatment (Fig. 2a). Interestingly, PINK1 carrying the pathogenic missense variants p.Gly309Asp and p.Glu417Gly was able to induce mitophagy in a manner similar to WT PINK1, while PINK1 carrying the pathogenic missense variant p.Cys125Gly increased mitophagy induction compared to WT PINK1 upon CCCP treatment. This suggests that these missense variants are either not pathogenic or exert a different pathogenic mechanism that does not involve mitophagy. Four missense variants of uncertain significance in PINK1 (p.Thr145Met, p.Ala244Gly, p.Leu249Val, and p.Ala339Thr) exhibited a significant decrease compared to WT PINK1 upon CCCP treatment. Notably, four missense variants (p.Ile368Asn, p.Leu369Pro, p.Pro416Leu, and p.Trp437Arg) that were classified as uncertain significance completely abolished mitophagy induction. Based on this data, we conclude that these latter four variants affect PINK1 function that may lead to their reclassification as likely pathogenic. Overall, most missense variants of uncertain significance caused no significant alteration in mitophagy induction, and thus, using the mitophagy reporter, we found no additional evidence to reclassify these variants.
In addition, for some PINK1 variants, we observed that the mt-mKeima fluorescence signal was reduced even though the induction of mitophagy was unaltered compared to WT PINK1 treated with CCCP (e.g., Gly309Asp, Supplementary Fig. 4A.vii). Therefore, we determined the median fluorescence intensity (MFI) of mt-mKeima of cells expressing the variants of PINK1 after CCCP treatment. For most cells expressing variant PINK1, the MFI increased upon CCCP treatment in accordance with an increase in mitophagy induction (Fig. 2b, Supplementary Fig. 4B). However, a marked decrease in MFI was observed in cells expressing PINK1 carrying several variants (including p.Gly309Asp, p.Glu417Gly, p.Arg279His, p.Pro296Leu, p.Cys323Tyr, p.His360Arg, p.Asn367Ser, and p.Arg464His) that showed normal or increased mitophagy induction. This likely suggests that these missense variants do affect PINK1 function and lead to a decrease in mitophagy function. When using both mitophagy induction and MFI, three groups of missense variants were distinguished: (1) missense variants with normal or increased mitophagy function (Fig. 2b, white area), (2) missense variants that cause decreased (25–75%) mitophagy function (Fig. 2b, light gray area) and (3) missense variants that completely abolish mitophagy function (Fig. 2b, dark gray area). Importantly, (likely) pathogenic missense variants fell mostly in groups 2 and 3, further confirming that mitophagy deficits underlie PINK1 dysfunction.
Many PINK1 variants affect Parkin translocation
To further confirm that variant PINK1 mitophagy function was indeed affected via PINK1-Parkin‒dependent mechanisms15,17,18, we also investigated the mitochondrial translocation of Parkin. To this end, we transiently transfected WT-HA-tagged PINK1 (PINK1-HA) or variant-PINK1-HA in stably expressed EGFP-Parkin/PINK1 knock-out (KO) HeLa cells treated with 10 µM CCCP for 6 hours following immunofluorescence. CCCP treatment caused translocation of cytosolic EGFP-Parkin into “clustered perinuclear aggregates” (representing mitochondria17) that colocalized with WT PINK1 (Fig. 3a/b), as was also observed for all PINK1 proteins carrying benign variants. In contrast, cells expressing PINK1 variants designated as (likely) pathogenic had aberrant EGFP-Parkin localization that was mainly in the form of “punctate diffuse aggregates”, such as p.Gly309Asp (Fig. 3a), or remained “diffuse”, such as p.Leu347Pro (Fig. 3a). The latter correlated completely with abolished mitophagy induction (Fig. 2a). “Diffuse” EGFP-Parkin was also observed for four of the missense variants of uncertain significance that also showed abolished mitophagy induction (Fig. 2a). Interestingly, cells expressing PINK1 proteins carrying missense variants (mainly of uncertain significance) that showed “punctate diffuse aggregates” of EGFP-Parkin (p.Gln126Pro, p.Ile131Val, p.Met237Val, p.Leu249Val, p.His271Gln, p.Pro296Leu, p.Gly309Asp, p.His360Arg, p.Asn367Ser, p.Cys388Arg, p.Glu417Gly, p.Ser419Pro, and p.Arg464His, Supplementary Fig. 6) did induce mitophagy upon CCCP treatment, albeit some at decreased levels (Fig. 2a). Since mitochondrial clustering occurs as a result of the formation of poly-ubiquitin chains by Parkin15, we hypothesize that these missense variants (despite mitophagy induction) do impair PINK1 function, thereby affecting Parkin recruitment to mitochondria upon CCCP treatment, and thus may exert likely pathogenic effects. Overall, missense variants that caused a decrease in mitophagy function also led to impairments in EGFP-Parkin translocation, while variants with normal mitophagy function caused complete translocation of cytosolic EGFP-Parkin (Fig. 3b, c).
Fig. 3. PINK1 variants with disrupted mitophagy affect Parkin translocation.
a Representative fluorescence images of HeLa PINK1 KO cells stably expressing EGFP-Parkin transfected with PINK1-HA WT and PINK1-HA variants p.Gly309Asp and p.Leu347Pro and before or after treatment with CCCP showing examples of three types of Parkin localization: clustered perinuclear aggregates, punctate diffuse aggregates, and diffuse cytosolic localization. Upper panels show EGFP-Parkin, middle panels show PINK1, and lower panels show an overlay including DAPI to stain nuclei. Scale bar, 10 μm. b Quantification of Parkin translocation by PINK1 WT or PINK1 missense variants upon CCCP treatment. Mean values are shown from three independent experiments. Bars are color-coded by the annotation as determined by Sherloc. Data analyzed using one-way ANOVA. *p < 0.05, **p < 0.005. c Quantification of variants for each of the types of Parkin localization, color-coded by the Sherloc annotation.
Functional assays assist in the determination of pathogenicity but do not guarantee classification
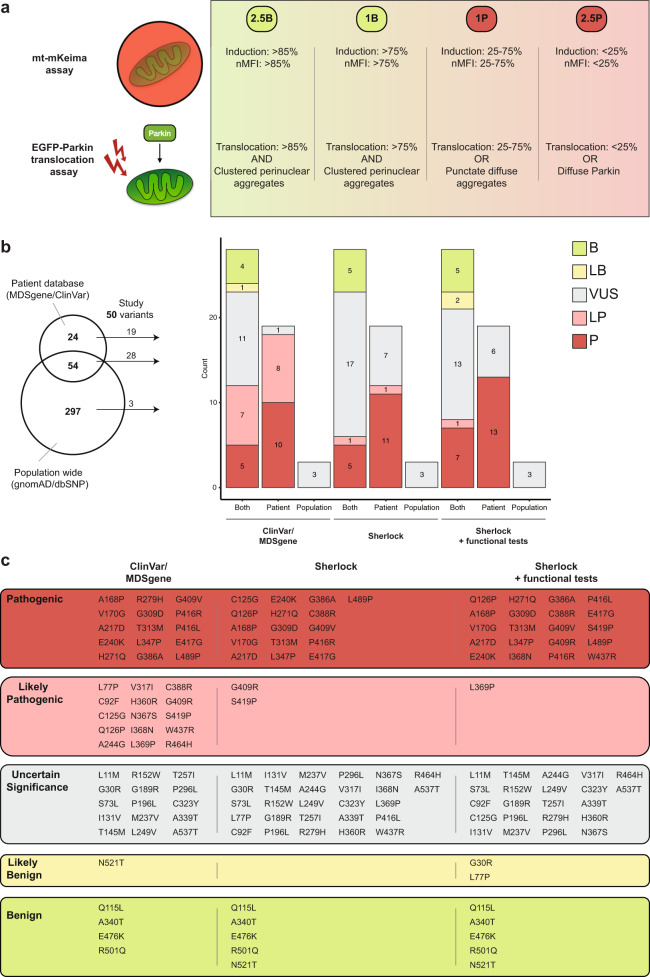
Since the PINK1 variants had varying functional consequences, we constructed a scoring system based on the observed PINK1 deficits (Fig. 4a). Specifically, points were assigned in a similar manner to the Sherloc framework (Supplementary Fig. 1D): 2.5 benign points = PINK1-variant similar to WT PINK1, 2.5 pathogenic points = PINK1-variant disables mitophagy function and Parkin translocation, one pathogenic point = PINK1-variant affects mitophagy function or Parkin translocation, or one benign point = PINK1-variant mildly reduced mitophagy function or Parkin localization compared to WT PINK1. Next, we complemented the original Sherloc scores with our functional assay scores. This led to a revision of the final classification for nine variants (9/50 = 18%) (Fig. 4b): six variants of uncertain significance were reclassified to pathogenic (p.Ile368Asn, p.Gly409Arg, p.Pro416Leu, p.Ser419Pro, and p.Trp437Arg) or likely pathogenic (p.Leu369Pro), two variants of uncertain significance (p.Gly30Arg and p.Leu77Pro) were reclassified to likely benign, and one pathogenic variant (p.Cys125Gly) was reclassified to uncertain significance. The classification of the benign variants and the four rare population-based variants did not change. In total, based on our data, 20 of the 46 clinically relevant variants are classified as pathogenic, one as likely pathogenic, two as likely benign, and five as benign (Table 1, Fig. 4c, Supplementary Fig. 7).
Fig. 4. Updating Sherloc-based classification with functional assay results in the reclassification of nine PINK1 variants.
a Scoring system based on the functional assays in this study. Points were assigned as shown, based on both the mitophagy assay (%induction and MFI) and the Parkin translocation assay (% translocation and type of Parkin translocation). b Updated ACMG-AMP annotations of the 50 missense variants as shown in Fig. 1c, including the updated Sherloc classification (right). Horizontal bar graphs show all the studied variants. Vertical bar graphs are stratified by the origin of the reported missense variant in the Venn diagram shown on the left. c Overview of all PINK1 missense variants assessed in this study and their annotation based on the patient databases, Sherloc and Sherloc after functional testing.
Table 1.
Final PINK1 variant reclassification using Sherloc and functional assays.
| PINK1 -Variant | Database | Clinical assignment | Final Sherloc score | |||
|---|---|---|---|---|---|---|
| ClinVar/MDSgene | Sherloc | Sherloc + assays | P | B | ||
| p.Leu11Met | ClinVar | Uncertain significance | Uncertain significance | Uncertain significance | 1 | 1 |
| p.Gly30Arg | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Likely Benign | – | 4 |
| p.Ser73Leu | ClinVar, MDSgene, gnomAD | Uncertain significance | Uncertain significance | Likely Benign | – | 3.5 |
| p.Leu77Pro | MDSgene | Likely Pathogenic | Uncertain significance | Uncertain significance | 3 | 1 |
| p.Cys92Phe | MDSgene | Likely Pathogenic | Uncertain significance | Uncertain significance | 3.5 | 1 |
| p.Gln115Leu | ClinVar, gnomAD | Benign | Benign | Benign | 7 | |
| p.Cys125Gly | MDSgene | Likely Pathogenic | Pathogenic | Uncertain significance | 2.5 | – |
| p.Gln126Pro | MDSgene, gnomAD | Likely Pathogenic | Pathogenic | Pathogenic | 9.5 | – |
| p.Ile131Val | gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 0.5 | – |
| p.Thr145Met | ClinVar, gnomAD, | Uncertain significance | Uncertain significance | Uncertain significance | 0.5 | 2.5 |
| p.Arg152Trp | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 2 | 2.5 |
| p.Ala168Pro | ClinVar, MDSgene, gnomAD | Pathogenic | Pathogenic | Pathogenic | 6.5 | – |
| p.Val170Gly | MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Gly189Arg | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | – | 1 |
| p.Pro196Leu | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | – | 1 |
| p.Ala217Asp | ClinVar, MDSgene | Pathogenic | Pathogenic | Pathogenic | 11.5 | – |
| p.Met237Val | gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 1.5 | – |
| p.Glu240Lys | MDSgene, gnomAD | Pathogenic | Pathogenic | Pathogenic | 6.5 | – |
| p.Ala244Gly | MDSgene | Likely Pathogenic | Uncertain significance | Uncertain significance | 2.5 | – |
| p.Leu249Val | ClinVar, MDSgene, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 1 | 1 |
| p.Thr257Ile | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | – | 2 |
| p.His271Gln | ClinVar, MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Arg279His | ClinVar, MDSgene, gnomAD | Pathogenic | Uncertain significance | Uncertain significance | – | 2 |
| p.Pro296Leu | ClinVar, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 1.5 | – |
| p.Gly309Asp | ClinVar, MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Thr313Met | ClinVar, MDSgene, gnomAD | Pathogenic | Pathogenic | Pathogenic | 14.5 | – |
| p.Val317Ile | MDSgene, gnomAD | Likely Pathogenic | Uncertain significance | Uncertain significance | 1.5 | 1 |
| p.Cys323Tyr | gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 0.5 | 1 |
| p.Ala339Thr | ClinVar, MDSgene, gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 1.5 | 1 |
| p.Ala340Thr | ClinVar, gnomAD | Benign | Benign | Benign | 7 | |
| p.Leu347Pro | ClinVar, gnomAD | Pathogenic | Pathogenic | Pathogenic | 15.5 | – |
| p.His360Arg | MDSgene | Likely Pathogenic | Uncertain significance | Uncertain significance | 2.5 | – |
| p.Asn367Ser | MDSgene, gnomAD | Likely Pathogenic | Uncertain significance | Uncertain significance | 2 | – |
| p.Ile368Asn | MDSgene, gnomAD | Likely Pathogenic | Uncertain significance | Pathogenic | 5 | – |
| p.Leu369Pro | MDSgene, gnomAD | Likely Pathogenic | Uncertain significance | Likely Pathogenic | 4 | – |
| p.Gly386Ala | MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Cys388Arg | ClinVar, MDSgene | Likely Pathogenic | Pathogenic | Pathogenic | 11.5 | – |
| p.Gly409Arg | MDSgene, gnomAD | Likely Pathogenic | Likely pathogenic | Pathogenic | 5.5 | – |
| p.Gly409Val | MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Pro416Leu | MDSgene | Pathogenic | Uncertain significance | Pathogenic | 5.5 | – |
| p.Pro416Arg | MDSgene | Pathogenic | Pathogenic | Pathogenic | 7 | – |
| p.Glu417Gly | MDSgene | Pathogenic | Pathogenic | Pathogenic | 5.5 | – |
| p.Ser419Pro | MDSgene | Likely Pathogenic | Likely pathogenic | Pathogenic | 5 | – |
| p.Trp437Arg | MDSgene | Likely Pathogenic | Uncertain significance | Pathogenic | 5.5 | – |
| p.Arg464His | MDSgene, gnomAD | Likely Pathogenic | Uncertain significance | Uncertain significance | 3 | – |
| p.Glu476Lys | ClinVar | Benign | Benign | Benign | – | 9.5 |
| p.Leu489Pro | MDSgene | Pathogenic | Pathogenic | Pathogenic | 5 | – |
| p.Arg501Gln | ClinVar, gnomAD | Benign | Benign | Benign | – | 9.5 |
| p.Asn521Thr | ClinVar, gnomAD | Likely benign | Benign | Benign | – | 7 |
| p.Ala537Thr | gnomAD | Uncertain significance | Uncertain significance | Uncertain significance | 0.5 | 2.5 |
Discussion
In this study, we systematically investigated EOPD-associated or population-based PINK1 missense variants. Using the Sherloc framework, supplemented with cellular assays, we investigated 50 missense variants in PINK1 that were mostly of uncertain significance. We found that 20 of the 46 patient-associated variants had sufficient evidence for a pathogenic annotation and were seldom found in the general population. Most of these pathogenic variants completely abolished mitophagy function and Parkin translocation. We also identified multiple missense variants that displayed intact mitophagy function but aberrant Parkin accumulation. Although our study provides insights into the functional consequences of the missense variants, interpretation of the pathological consequences remains challenging without also assessing clinical and population-based data with a comprehensive scoring system such as Sherloc. Therefore, we advise genetic diagnostic laboratories to acquire multiple layers of evidence including available functional data, to determine whether a variant truly is disease-causing or not and plea for the implementation of functional tests in the routine workflow of genetic diagnostic laboratories.
Even with the variant classification guidelines formulated by ACMG-AMP8, variant classification seems subject to personal interpretation10, especially for infrequently encountered variants. Our work shows that the clinical significance of many of the variants reported in the ClinVar and MDSgene patient databases were assumed to be more damaging than the evidence implied, as we have reclassified 11/46 variants reported as (likely) pathogenic to uncertain significance. However, using Sherloc, the classification of variants that were (likely) pathogenic was supported by multiple lines of evidence, such as a very low MAF, absence of homozygosity in population databases, segregation with disease and previously reported functional aberrations in different cellular assays.
With our systematic analysis of 50 PINK1 missense variants for mitophagy and Parkin translocation defects, we have shown that most variants classified as (likely) pathogenic by Sherloc caused loss of PINK’s downstream functionality, as reflected by our observations of no or significantly reduced mitophagy induction and/or Parkin translocation. This is consistent with the idea that disease-causing PINK1 variants confer pathogenicity through a loss-of-function mechanism leading to defects in PINK1 kinase activity2,3.
Not surprisingly, most (likely) pathogenic missense variants are localized in domains that are crucial for the kinase activity of PINK1. For instance, the PINK1 variants p.Ala168Pro, p.Val170Gly, p.Ala217Asp, and p.Leu369Pro, which were previously predicted to disrupt ATP-binding and kinase activity20, showed no mitophagy induction and/or Parkin recruitment in our functional studies. Likewise, the pathogenicPINK1 variant p.Thr313Met, which affects a phosphorylation site of PINK1 that is required for proper PINK1 activity24, showed significantly reduced mitophagy induction and loss of Parkin recruitment. Moreover, multiple pathogenic PINK1 missense variants that were previously predicted to change the protein fold of the kinase core (including p.Leu347Pro, p.Ile368Asn, p.Trp437Arg, and p.Leu489Pro) or the activation loop (including p.Gly386Ala, p.Gly388Arg, p.Gly409Arg, p.Gly409Val, p.Pro416Arg, and p.Pro416Leu)20 also showed abolished mitophagy function and Parkin recruitment. This data reinforces the idea that pathogenic PINK1 variants cause loss of PINK1 kinase activity.
However, not all (likely) pathogenic variants exhibit loss or reduced PINK1-induced mitophagy and Parkin recruitment, e.g., variants p.His271Gln, p.Gly309Asp, p.Glu417Gly, and p.Ser419Pro. Intriguingly, these and other variants of uncertain significance had aberrant Parkin localization in the form of diffuse punctate aggregates instead of the perinuclear clustering observed for WT PINK1. Perinuclear clustering of mitochondria occurs as a result of Parkin-mediated K63-linked polyubiquitination of mitochondrial substrates, including p6225–27. This can only ensue with the precise interplay between ubiquitin, PINK1, and Parkin. In agreement with our study, others have shown that several patient-associated variants in Parkin25 and PINK126 are unable to cluster mitochondria upon CCCP treatment. Therefore, we hypothesize that the missense variants that display diffuse punctate Parkin localization disturb the polyubiquitination mechanism. Of note are PINK1 variants p.Pro296Leu and p.Gly309Asp that reside in Insertion 3, a crucial element of the kinase domain that is directly involved in ubiquitin binding and impair ubiquitin phosphorylation20. As phosphorylated ubiquitin facilitates the confirmational change of Parkin from inactive to active state28, these missense variants likely somewhat hinder the activation of Parkin. It is possible that similar obstructions occur with the other variants that aberrantly localize Parkin. In contrast, all benign PINK1 variants were able to accumulate Parkin in the perinuclear region. Notably, different studies have shown that mitochondrial clearance occurs despite p62-mediated translocation of mitochondria25,27, confirming our observations that the missense variants that affect Parkin translocation are able to induce mitophagy.
In addition, most PINK1 variants of uncertain significance, including those predicted to affect kinase function (p.Met237Val, p.Ala244Gly, p.Val317Ile, and p.Ala339Thr)20, did not cause complete loss of mitophagy and were able to activate Parkin. This illustrates that a change in the structure that is predicted to be damaging does not abolish kinase function per se. Furthermore, although all benign PINK1 variants were able to induce mitophagy, three benign PINK1 variants with relatively high MAF, p.Gln115Leu, p.Ala340Thr, and p.Asn521Thr, were less able to induce mitophagy compared to WT PINK1. This may suggest that some degree of defective mitophagy is tolerated in cells before pathogenic effects occur. Likewise, similarly reduced mitophagy induction was observed for some PINK1 variants of uncertain significance. While one could argue that these variants with seemingly similar functional effects, as benign variants, should be regarded as not disease-causing, we cannot exclude that a mitophagy-unrelated disease-causing mechanism may be conferred via other downstream PINK1 pathways, especially for those variants with aberrant Parkin translocation. Indeed, additional PINK1-Parkin‒mediated pathomechanisms for EOPD have been proposed14,29. Next to mitophagy, PINK1 and Parkin regulate a plethora of different mitochondrial functions14,30, such as mitochondrial dynamics31–33, transport of mitochondria34,35, biogenesis of mitochondria36,37 and formation of mitochondrial-derived vesicles38. Furthermore, recent studies highlight a key role for PINK1 as a repressor of the immune system through the repression of mitochondrial antigen presentation39,40. By studying the two most robust pathways—mitophagy and Parkin recruitment—reported to be linked to PINK1’s actions, we may have missed the effects of PINK1 variants that impact other functions. For example, the PINK1 variants p.Gly309Asp and p.Leu347Pro are associated with perturbed mitochondrial dynamics41 (see Supplementary Table 2 for more examples). Yet, they also showed impaired Parkin recruitment in our study, suggesting that Parkin may play a role in many of PINK1’s functions not related to mitophagy14. Additionally, it remains to be determined whether PINK1 variants with normal mitophagy and Parkin recruitment may confer pathogenicity through impairments in one of those functions not related to mitophagy and thus cannot yet be classified as benign. The variants that showed a clear loss-of-function in our functional assays, however, are expected to be disease-causing, which is also supported by other studies (Supplementary Table 2). An additional limitation in this study is the use of the immortalized HeLa cell line in conjunction with overexpression of PINK1 and Parkin to study the effects of the PINK1 variants. As such, this artificial system differs considerably from the biological situation that causes disease and should be considered as a proxy for the real situation until it can be proven in models that more closely mimic the patient situation. Nonetheless, our approach allows for a practical investigation of the biochemical consequences of the missense variants that can be carried out in most laboratory settings. Finally, while it is possible to interrogate functional deficits for genes such as PINK1 and PRKN that act in a mechanistic pathway that can be studied in a relatively straightforward manner, it remains difficult to do so for other genes linked to EOPD (e.g., PARK7) where the molecular function is less known.
Functional data, however, is only one line of evidence to evaluate pathogenicity. In order to make final decisions on the clinical significance of variants, functional studies should be supplemented by clinical data and evidence from population genetics. The pathogenic variants in our study could be assigned as such because of the extensive data availability, and this data was absent for most variants of uncertain significance. The presence of a missense variant in PINK1 in a Parkinson’s disease case does not automatically imply pathogenicity. The annotations from patient databases ClinVar and MDSgene should thus be interpreted with care, and we recommend using a consistent framework such as Sherloc for future annotations of variants in disease-linked genes such as PINK1 found in patients.
Methods
Collecting PINK1 variants
All the variants in the PINK1 gene used in this study were collected from three publicly available databases (access date: 15 September 2020): MDSgene5, ClinVar6, and gnomAD version 2.1.17. At the time of collection, the patient-specific Parkinson disease Mutation Database was unavailable4. Annotations about clinical significance were taken from ClinVar and MDSgene. Since each database used different classifications, we unified them into the standard annotation recommended by the ACMG-AMP (Supplementary Table 1).
Annotation of PINK1 variants using Sherloc
All variants were classified using the Sherloc framework, as previously described10. Briefly, pathogenic or benign points were assigned to each variant based on four layers of evidence: (1) population evidence, with pathogenic or benign points scored based on the MAF of variants in gnomAD7 or the dbSNP database42, (2) evidence from case reports (if available), where pathogenic points were given to rare (MAF < 8 counts with no homozygous variants) variants found in patients, (3) family-segregation information (if available), where pathogenic points were given for segregating variants, and (4) experimental evidence reported in the literature, where pathogenic, benign or conflicting points were scored for functional proof that a variant affected downstream function (e.g., Parkin recruitment or substrate phosphorylation). With respect to this last layer, we curated 54 studies that investigated at least one of the 50 variants in our study (see Supplementary Table 2 for overview of studies and their variants).
Cell culture, transient transfections, and treatments
HeLa cells were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Waltham, MA) supplemented with 10% fetal bovine serum (Invitrogen) and 1% Penicillin–Streptomycin (Gibco, Waltham, MA) in a 37 °C incubator with 5% CO2. Cells were transiently transfected using polyethylenimine (Sigma-Aldrich, Saint Louis, MO) in a 1:5 ratio (DNA:polyethylenimine) for 48 h prior to performing the experiments. To induce mitochondrial depolarization, HeLa cells were treated with 10 µM CCCP (Sigma-Aldrich) for the indicated times, prior to cell harvesting or fixation. DMSO was used as a control treatment.
Generation of stable reporter PINK1 knock-out cell lines
PINK1 KO cells were obtained by CRISPR-Cas9‒mediated genome editing in HeLa cells, as previously described43. Briefly, a 20-nt sgRNA sequence that targets exon 2 of the PINK1 gene was cloned into the pSpCas9(BB)-2A-GFP (PX458) plasmid (a gift from Prof. Feng Zhang (Broad Institute, Cambridge, MA), Addgene plasmid #48138) using the BbsI restriction enzyme to form the targeting plasmid expressing Cas9-GFP. Following validation, the PX458-sgRNA plasmid was transfected into HeLa cells. GFP-positive cells were single-cell sorted 48 h post-transfection using an SH800S cell sorter (Sony Biotechnology, San Jose, CA) and grown in separate cultures that were subsequently screened for the presence of frameshift mutations leading to nonsense-mediated decay on both alleles. PINK1 KO was validated and confirmed using western blotting.
First, the cDNA of mt-mKeima and WT Parkin were obtained from the pCHAC-mt-mKeima13 (a gift from Prof. Richard Youle (National Institutes of Health, Bethesda, MD), Addgene plasmid #72342) and pEGFP-Parkin vectors44 (a gift from Prof. Edward Fon (McGill University, Montreal, Quebec, Canada), Addgene plasmid #45875), respectively. Second, the cDNA of mitochondria-targeting mKeima (mt-mKeima) and WT FLAG-Parkin were cloned into the pIRES vector (Clontech Laboratories, Mountain View, CA). mt-mKeima cDNA was cloned between restriction sites NheI and EcoRI and FLAG-tagged Parkin cDNA was cloned at restriction site NotI in multiple cloning sites 1 and 2, respectively.
To obtain stable PINK1 KO reporter cell lines for mKeima-FLAG-Parkin and EGFP-Parkin, PINK1 KO HeLa cells were transfected with either the mt-mKeima-FLAG-Parkin or pEGFP-Parkin plasmids. After 48 h, the growth medium was replaced with selection medium containing 800 ng/µl G-418 (Sigma-Aldrich), which was refreshed every 3 days for 14 days. Single, stably expressing cells were sorted using an SH800S cell sorter (Sony Biotechnology). These single cells were grown into separate cell lines that were used in the following experiments.
Cloning and site-directed mutagenesis of PINK1
To assess PINK1 function based on downstream functional consequences, we generated PINK1-HA expression vector by subcloning the pcDNA-DEST47 PINK1 C-GFP45 (a gift from Prof. Mark Cookson (National Institutes of Health, Bethesda, MD), Addgene plasmid #13316) into the pcDNA3.1(−) vector (Thermo Fischer Scientific, Waltham, MA) between the restriction sites EcoRI and HindIII. To introduce the 50 missense variants into the cDNA of PINK1, site-directed mutagenesis was performed using the QuickChange II XL kit following the manufacturer’s protocol (Agilent, Santa Clara, CA). The complete cDNA of PINK1 was subsequently analyzed using Sanger sequencing to validate the presence of the correct nucleotide change, as well as the absence of any other variants. The complete list of mutagenesis primers can be found in Supplementary Table 3.
FACS analysis of mitophagy
HeLa PINK1 KO cells stably expressing mt-mKeima-FLAG-Parkin were transiently transfected with WT-PINK1-HA or variant-PINK1-HA. Cells were treated with either DMSO or 10 µM CCCP (Sigma-Aldrich) for 24 h, after which the cells were rinsed, dissociated with 0.05% Trypsin-EDTA (Thermo Fischer Scientific), resuspended in 4′,6-diamidino-2-phenylindole (DAPI)-containing (100 ng/ml) medium and transferred to FACS tubes. FACS analysis was performed with a Novocyte Quanteon flow cytometer (Agilent). mt-mKeima was detected using 488 and 561 nm lasers with a 615 nm emission filter to quantify the mitochondria present in the cytosol or lysosome, respectively. At least 50,000 events were recorded for each experiment, and each event was gated for a cell that was DAPI-negative and mt-mKeima-positive. The population of cells with a high 561:488 nm ratio was determined by drawing a gate in the upper left quadrant of DMSO-treated untransfected cells, which was subsequently used for all samples in the same independent experiment. Mitophagy induction was quantified as the increase in this cell population upon CCCP treatment for each PINK1 variant minus the control condition (untreated cells) and normalized against cells expressing WT-PINK1-HA from the same experiment. In addition, MFI was determined for the gated cells, using a high 561:488 nm ratio as a second measure of the amount of mitophagy induction. Background MFI was determined by the MFI from the population of non-induced mKeima-positive cells (low 561:488 ratio) and was subtracted from the MFI of the induced mKeima-positive cells and then normalized against WT-PINK1-HA from the same independent experiment. Data analysis was performed using Kaluza Analysis software (Beckman Coulter, Brea, CA).
EGFP-Parkin localization using immunofluorescence
HeLa cells stably expressing EGFP-Parkin were seeded on glass coverslips in 24-well plates and transiently transfected with WT-PINK1-HA or variant-PINK1-HA. Subsequently, cells were treated with DMSO or 10 µM CCCP (Sigma-Aldrich) for 6 h following fixation in 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then permeabilized in 0.1% Triton X-100 in PBS for 10 min and blocked with 3% bovine serum albumin in PBS for 1 h. Coverslips were then incubated overnight at 4 °C with rat anti-HA IgG monoclonal antibody (3F10, Roche, Basel, Switzerland; 1:250), washed, and subsequently incubated for 1 h at room temperature with goat anti-Rat IgG (H+L) Alexa Fluor 594 secondary antibody (Invitrogen; 1:250). Coverslips were mounted onto glass slides in DAPI-containing mounting medium (Vector Laboratories, Burlingame, CA). The slides were analyzed using structured illumination microscopy (SIM). SIM images were acquired with an AxioObserver Z1 compound microscope (Carl Zeiss, Oberkochen, Germany) equipped with an Apotome, 63x oil objective, and an AxioCam MRm3 CCD camera (Carl Zeiss). Images were captured for each condition with identical exposure times. Translocation of EGFP-Parkin was assessed in cells that expressed both WT- or variant-PINK1-HA and EGFP-Parkin and scored by a blinded observer for either diffuse or aggregated EGFP-Parkin, as previously described44. At least 75 cells were scored per variant in three independent experiments.
Protein extraction and immunoblotting
Cells were harvested in 2% sodium dodecyl sulfate (SDS)/phosphate-buffered saline (PBS) buffer containing a proteinase inhibitor cocktail (Roche) and sonicated. Protein concentrations were quantified using the PierceTM BCA protein assay kit (Thermo Fischer Scientific), and samples were mixed with loading sample buffer containing 10% β-mercaptoethanol before being boiled at 95 °C for 5 min. Subsequently, equal amounts of total protein extracts were subjected to SDS-PAGE, transferred to nitrocellulose membranes, blocked for 1 h in skimmed milk, and incubated overnight with primary antibody at 4 °C and then with for the corresponding secondary antibody for 1 h at room temperature. Blots were imaged on a ChemidocTM MP Imaging System (Bio-Rad, Hercules, CA). All blots were derived from the same experiment and were processed in parallel. Original uncut blots are shown in Supplementary Fig. 8. The primary antibodies used were mouse anti-β-Actin (MP Biomedicals 8691001, 1:5000), mouse anti-Parkin (Santa-Cruz Biotechnology, Dallas, TX, sc-32282, 1:500), and rabbit anti-PINK1 (D8G3, Cell signaling, Danvers, MA,1:1000). The secondary antibodies were HRP-conjugated goat anti-mouse IgG (H + L) (Bio-Rad, 1:10,000) and HRP-conjugated goat anti-rabbit IgG (H + L) (Bio-Rad, 1:10,000).
Statistical analyses
The data obtained from the FACS and Parkin localization experiments were analyzed using a linear model by one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test. Data are means ± standard error of the mean from at least three independent experiments. A p-value < 0.05 was considered statistically significant. Statistical analyses were computed in R (version 1.3.959).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
D.S.V. is supported by a Rosalind Franklin Fellowship from the University of Groningen (UG). T.v.L. is supported by the Weston Brain Institute, Michael J. Fox Foundation, Hersenstichting, and the MD-PhD program of the UG. K.Y.M. is supported by the Jan Kornelis de Cock Stichting and the U4 Ph.D. program of the Behavioral and Cognitive Neuroscience Graduate School of the UG. None of the funding bodies were involved in the collection, analysis, and interpretation of data, nor in the writing of the manuscript. We would like to thank Johan Teunis for assisting with the flow cytometry experiments and Kate McIntyre for editing this manuscript.
Author contributions
K.Y.M. conceived and designed the project, created the cell lines, performed all in vitro experiments and fluorescence microscopy, analyzed the data, and wrote the manuscript. M.R.F. participated in creating the cell lines, performed cloning and mutagenesis of the plasmids. T.v.L. participated in the interpretation of results and helped revise the manuscript. D.S.V. designed and supervised the project, participated in the interpretation of results, and wrote and revised the manuscript. All authors read and approved the final manuscript.
Data availability
The raw data and fluorescence images are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41531-021-00258-8.
References
- 1.Olanow CW, Tatton WG. Etiology and pathogenesis of Parkinson’s disease. Annu. Rev. Neurosci. 1999;22:123–144. doi: 10.1146/annurev.neuro.22.1.123. [DOI] [PubMed] [Google Scholar]
- 2.Kilarski LL, et al. Systematic review and UK-based study of PARK2 (Parkin), PINK1, PARK7 (DJ-1) and LRRK2 in early-onset Parkinson’s disease. Mov. Disord. 2012;27:1522–1529. doi: 10.1002/mds.25132. [DOI] [PubMed] [Google Scholar]
- 3.Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat. Rev. Neurol. 2013;9:445–454. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- 4.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum. Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasten M, et al. Genotype-phenotype relations for the Parkinson’s disease genes Parkin, PINK1, DJ1: MDSGene systematic review. Mov. Disord. 2018;33:730–741. doi: 10.1002/mds.27352. [DOI] [PubMed] [Google Scholar]
- 6.Landrum MJ, et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellard, S. et al. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020 v4. https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (2020). [DOI] [PubMed]
- 10.Nykamp K, et al. Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2017;19:1105–1117. doi: 10.1038/gim.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi W, et al. The landscape of Parkin variants reveals pathogenic mechanisms and therapeutic targets in Parkinson’s disease. Hum. Mol. Genet. 2019;28:2811–2825. doi: 10.1093/hmg/ddz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: Much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 16.Greene AW, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 20.Schubert AF, et al. Structure of PINK1 in complex with its substrate ubiquitin. Nature. 2017;552:1–28. doi: 10.1038/nature24645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okatsu K, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun. 2012;3:1–10. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 2011;18:1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Matenia D, Hempp C, Timm T, Eikhof A, Mandelkow EM. Microtubule affinity-regulating kinase 2 (MARK2) turns on phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) at Thr-313, a mutation site in Parkinson disease: effects on mitochondrial transport. J. Biol. Chem. 2012;287:8174–8186. doi: 10.1074/jbc.M111.262287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okatsu K, et al. P62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harper JW, Ordureau A, Heo J-M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 2018;19:93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 29.Grenier K, McLelland GL, Fon EA. Parkin- and PINK1-dependent mitophagy in neurons: will the real pathway please stand up? Front. Neurol. 2013;4 JUL:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge, P., Dawson, V. L. & Dawson, T. M. PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 10.1186/s13024-020-00367-7 (2020). [DOI] [PMC free article] [PubMed]
- 31.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by Parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 32.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 33.Pryde KR, Smith HL, Chau KY, Schapira AHV. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. J. Cell Biol. 2016;213:163–171. doi: 10.1083/jcb.201509003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, et al. Parkinson’s disease–associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y, et al. PINK1 primes Parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 2017;18:918–932. doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens DA, et al. Parkin loss leads to Paris-dependent declines in mitochondrial mass and respiration. Proc. Natl Acad. Sci. USA. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matheoud D, et al. Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1 −/− mice. Nature. 2019;571:565–569. doi: 10.1038/s41586-019-1405-y. [DOI] [PubMed] [Google Scholar]
- 40.Matheoud D, et al. Parkinson’s disease-related proteins PINK1 and Parkin repress mitochondrial antigen presentation. Cell. 2016;166:314–327. doi: 10.1016/j.cell.2016.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Wang HL, et al. PARK6 PINK1 mutants are defective in maintaining mitochondrial membrane potential and inhibiting ROS formation of substantia nigra dopaminergic neurons. Biochim. Biophys. Acta. Mol. Basis Dis. 2011;1812:674–684. doi: 10.1016/j.bbadis.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Sherry ST, et al. DbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 45.Beilina A, et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc. Natl Acad. Sci. USA. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and fluorescence images are available from the corresponding authors upon reasonable request.