Abstract
Ninety-three Bacteroides fragilis isolates from different geographic locations were analyzed for the presence of an enterotoxin-encoding gene. It was shown that blood culture isolates were more likely to carry this gene than strains from other sources. All enterotoxin-positive strains belonged to the PCR fingerprint group I.
Bacteroides fragilis is an obligately anaerobic bacterium that can be isolated from a variety of human infections (2). In the mid-1980s it was recognized that some strains produce an enterotoxin (ET) that can cause acute diarrhea in humans, young lambs, calves, pigs, and foals (6, 7). Enterotoxigenic B. fragilis (ETBF) strains have also been isolated from the feces of children with diarrhea (9). Kato et al. (4) showed that B. fragilis blood culture isolates were more likely to be ETBF and suggested that ETBF strains are more virulent than enterotoxin-negative strains. Recently, the enterotoxin gene of B. fragilis has been cloned, sequenced, and identified as producing a zinc metalloprotease of 44.4 kDa (5).
In order to determine the relative frequency of ETBF in different geographic locations, 93 B. fragilis clinical isolates from Germany and Southern California were analyzed. Two PCR assays were used to detect two independent genetic sequences of the B. fragilis enterotoxin gene. In addition, the isolates were further analyzed for their PCR fingerprint profiles using a tRNA gene primer (T3B).
Twenty-nine strains (13 blood isolates and 16 wound isolates) from the R. M. Alden Research Laboratory (RMA, Santa Monica, Calif.), 32 strains (wound isolates) from the Institute of Medical Microbiology, Free University, Berlin (IMB), and one strain (wound isolate) from the Institute of Medical Microbiology, University of Munich were included in the study. An additional 31 strains were consecutive isolates (wound isolates) from the Institute of Medical Microbiology, University of Leipzig (IML). For reference purposes, B. fragilis ATCC 25285, an ET-negative strain, and ATCC 43858 and ATCC 43859, ET-positive strains, were used. Strains representing other Bacteroides species, B. distasonis (n = 5), B. ovatus (n = 5), B. thetaiotaomicron (n = 5), B. vulgatus (n = 5) and B. uniformis (n = 5), were included for comparison and came from the RMA collection. Isolates were identified and grown in culture as described previously (1).
Ten colonies of each isolate were used for preparation of the chromosomal DNA with a tissue kit (Qiagen, Hilden, Germany); 2.5 μl of the lysates was subjected to the PCR assay. The PCR fingerprint assay was performed as described previously (1), using the T3B primer, a tRNA gene sequence (5′-AGGTCGCGGTTCGAATCC-3′), as a single oligonucleotide (12).
For restriction fragment length polymorphism (RFLP) analysis of selected T3B-primed fragments, the restriction endonuclease CFO 1 was used for B. fragilis PCR group I, and HaeIII (Boehringer, Mannheim, Germany) was used for group II. Two units of enzyme was added to 1 μg of DNA (samples contained about 6 to 8 μg of DNA) and incubated at 37°C for 2 h.
The PCR assay for ETBF detection was accomplished using two different primer pairs, RS-3–RS-4 (RS-3, 5′-TGAAGTTAGTGCCCAGATGCAGG-3′; RS-4, 5′-GCTCAGCGCCCAGTATATGACC-3′; amplification of a 367-bp fragment) (10) and GBF 101-GBF 110 (GBF 101, 5′-GAGCCGAAGACGGTGTATGT-3′; GBF 110, 5′-TCCCACTGGCTTCAAAATCCGAAGC-3′; amplification of a 358-bp fragment) (4). Conditions were optimized as follows: 2 min at 95°C, 1 cycle; 2 min at 95°C, 1 min at 65°C, and 1 min at 72°C, 45 cycles; and 2.5 min at 72°C, 1 cycle. Product samples were held at 4°C prior to analysis and then concentrated to a volume of 20 μl prior to separation in 1.2% agarose gels (1). The amplified products were detected by staining with ethidium bromide (2 μg/ml).
Data were statistically analyzed using the Statistical Package for Social Science (SPSS) software for Microsoft Windows. Specifically, the chi-square test was used, and the confidence interval was determined.
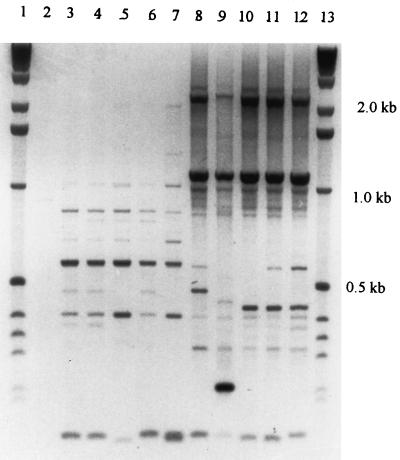
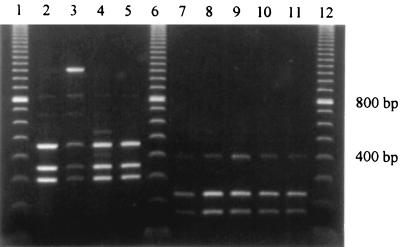
The results showed that 80 of the strains under study belonged to PCR fingerprint group I, which is similar to the type strain B. fragilis ATCC 25285, and 13 belonged to group II, which is similar to the strain VPI 2393 (reference strain for the DNA homology group II) (3) (Fig. 1). These results were underscored by RFLP analysis of selected PCR products (Fig. 2). Digestion of the group I 1,050-bp fragment (n = 80) with CFO 1 resulted in all cases in 3 fragments (470, 350, and 220 bp). For group II (n = 13), a typical 370-bp fragment was cut by HaeIII into 2 fragments (220 and 180 bp).
FIG. 1.
Clinical isolates of the species B. fragilis form two PCR fingerprint groups, amplified with the primer T3B. Lanes 1 and 13, 1-kb DNA ladder; lane 2, negative control without DNA; lanes 3 to 6, RMA 5912, 5935, 6600, and 0309 (multiresistant clinical isolates of PCR fingerprint group II); lane 7, VPI 2393 (reference strain of the DNA homology group II); lanes 8 to 11, clinical isolates of group I, RMA 5735, 5691, 6791, and 5081; lane 12, ATCC 25285 (type strain).
FIG. 2.
RFLP profiles of B. fragilis reference strains and clinical isolates of PCR fingerprint groups I and II. Lanes 1, 6, and 12, 100-bp DNA ladder. Group I (digested with the enzyme CFO 1): lanes 2 to 5, B. fragilis ATCC 25285 (type strain; enterotoxin negative) and RMA 5081, 5735, and 6791. Group II (digested with the enzyme HaeIII): lanes 7 to 11, B. fragilis VPI 2393 (DNA homology group II reference strain; enterotoxin negative) and RMA 0309, 6600, 5935, and 5912.
Using the two different sets of primers, the expected 367- and 358-bp DNA fragments of the ET gene were amplified in 10 of 93 clinical B. fragilis isolates (11%) and the two ET control strains. No discrepancy between the two PCR assays was demonstrated. Although the number of strains studied here is small, differences in the frequencies of ETBF strains among various clinical sources and the different geographic regions were noted. Nine percent (7 of 80) of non-blood culture isolates versus 23% (3 of 13) of blood culture isolates were ET positive, and 14% (4 of 29) of U.S. strains versus 9% (6 of 64) of German isolates were ET positive. However, with the chi-square test, these differences were not statistically significant (P ≥ 0.05).
All of the ET-positive isolates belonged to PCR fingerprint group I. However, in the T3B-amplified PCR fingerprint profiles, no additional major bands could distinguish ETBF from non-ETBF strains. Interestingly, all isolates of the PCR group II demonstrated increased resistance to carbapenems, as indicated by evaluated imipenem MICs. These strains represent the second taxon in the species B. fragilis as defined by Podglajen et al. (8).
ETBF is an emerging etiologic agent of diarrhea in humans and animals. ETBF may also be found in nondiarrheal infections. The results demonstrated here are somewhat different from those of studies from Hungary and Japan, which have analyzed strains from stool specimens and strains isolated from other sources and found 19 and 25% (4, 11), respectively, to be positive for enterotoxin. In this study, only 9% of German strains and 14% of U.S. strains were found to be ETBF. This suggests that there might be geographical differences in the prevalence of ETBF worldwide. However, the data did not reach statistical significance.
This study supports the findings of Kato et al. (4) that B. fragilis blood culture isolates are more likely to be ETBF than strains isolated from other sources. This is supportive of their theory that enterotoxin production may contribute to the virulence properties and pathogenicity of B. fragilis strains even in nondiarrheal infections.
Acknowledgments
We thank Sharon Hunt Gerardo for critical review and editorial assistance with the manuscript.
REFERENCES
- 1.Claros M C, Schönian G, Gräser Y, Montag T, Rodloff A C, Citron D M, Goldstein E J C. Identification and strain differentiation of ‘Bacteroides fragilis group’ species and Prevotella bivia by PCR fingerprinting. Anaerobe. 1995;1:209–217. doi: 10.1006/anae.1995.1020. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein E J C, Citron D M. Annual incidence, epidemiology, and comparative in vitro susceptibilities to cefoxitin, cefotetan, cefmetazole, and ceftizoxime of recent community-acquired isolates of the Bacteroides fragilis group. J Clin Microbiol. 1988;26:2361–2366. doi: 10.1128/jcm.26.11.2361-2366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson J L. Taxonomy of the Bacteroides. I. Deoxyribonucleic acid homologies among Bacteroides fragilis and other saccharolytic Bacteroides species. Int J Syst Bacteriol. 1978;28:245–256. [Google Scholar]
- 4.Kato N, Kato H, Watanabe K, Ueno K. Association of enterotoxigenic Bacteroides fragilis with bacteremia. Clin Infect Dis. 1996;23(Suppl. 1):83–86. doi: 10.1093/clinids/23.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 5.Kling J J, Wright R L, Van Tassell R L, Wilkins T D. Cloning and characterization of the gene for the metalloprotease enterotoxin of Bacteroides fragilis. FEMS Microbiol Lett. 1997;146:279–284. doi: 10.1016/s0378-1097(96)00488-0. [DOI] [PubMed] [Google Scholar]
- 6.Myers L L, Shoop D S. Association of enterotoxigenic Bacteroides fragilis with diarrheal disease in young pigs. Am J Vet Res. 1987;48:774–775. [PubMed] [Google Scholar]
- 7.Myers L L, Shoop D S, Stackhouse L L, Newman F S, Flaherty R J, Letson G W, Sack R B. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J Clin Microbiol. 1987;25:2330–2333. doi: 10.1128/jcm.25.12.2330-2333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podglajen I, Breuil J, Casin I, Collatz E. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J Bacteriol. 1995;177:5270–5275. doi: 10.1128/jb.177.18.5270-5275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sack R B, Albert M J, Allam K, Neogi P K B, Akbar M S. Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. J Clin Microbiol. 1994;32:960–963. doi: 10.1128/jcm.32.4.960-963.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shetab R, Cohen S H, Prindiville T, Tang Y J, Cantrell M, Rahmani D, Silva J., Jr Detection of Bacteroides fragilis enterotoxin gene by PCR. J Clin Microbiol. 1998;36:1729–1732. doi: 10.1128/jcm.36.6.1729-1732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szöke I, Dosa E, Nagy E. Enterotoxigenic Bacteroides fragilis in Hungary. Anaerobe. 1997;3:87–89. doi: 10.1006/anae.1997.0078. [DOI] [PubMed] [Google Scholar]
- 12.Welsh J, McClelland M. Genomic fingerprints produced by PCR with consensus tRNA gene primers. Nucleic Acids Res. 1991;19:861–866. doi: 10.1093/nar/19.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]