Abstract
In the treatment of acute myelogenous leukemia (AML) with allogeneic hematopoietic cell transplantation (HCT), we previously demonstrated that there is a greater protection from relapse of leukemia when the HCT donor has either the Cen B/B KIR genotype or a genotype having two or more KIR B gene segments. In those earlier analyses, KIR genotyping could only be assessed at the low resolution of gene presence or absence. To give the analysis greater depth, we developed high resolution KIR sequence-based typing, which defines all the KIR alleles and distinguishes the expressed alleles from those that are not expressed. We now describe and analyze high resolution KIR genotypes for 890 donors of this human transplant cohort. Cen B01 and Cen B02 are the common CenB haplotypes, with Cen B02 having evolved from Cen B01 by deletion of the KIR2DL5, 2DS3/5, 2DP1 and 2DL1 genes. We observed a consistent trend for Cen B02 providing stronger protection against relapse than Cen B01. This correlation indicates that protection depends on the donor having inhibitory KIR2DL2 and/or activating KIR2DS2, and is enhanced by the donor lacking inhibitory KIR2DL1, 2DL3 and 3DL1. High resolution KIR typing has allowed us to compare the strength of the interactions between the recipient’s HLA class I and the KIR expressed by the donor derived NK cells and T cells, but no clinically significant interactions were observed. The trend observed between donor Cen B02 and reduced relapse of leukemia points to the value of studying ever larger transplant cohorts.
Introduction
Natural killer (NK) cell function is controlled by the interaction of many types of NK receptor with their ligands expressed on tissue cells (1). The most variable of these interactions are those between Killer-cell immunoglobulin-like receptors (KIR) and HLA class I ligands (2). Both the receptors and the ligands are encoded by gene families, some members of which are highly polymorphic (2). These gene families are located on different chromosomes and segregate independently, providing an additional level of variation distinguishing individuals (2). The impact of NK cell responses on clinical outcomes following hematopoietic cell transplantation (HCT) became a focus for investigation following the observation of improved survival in haploidentical transplants having a KIR ligand mismatch (3).
The initial study of Ruggeri et al (3) showed a reduction in relapse of leukemia and improved survival for AML patients receiving a haploidentical, T-cell depleted transplant from a family member, when the donor has a KIR ligand not present in the recipient. The proposed mechanism is that a subset of donor-derived NK cells kills the recipient’s leukemia cells because the recipient cells lack an inhibitory ligand for KIR present in the donor. In that study, reduced graft-versus-host disease (GVHD) was also observed and proposed to be caused by donor NK cells killing recipient dendritic cells (4, 5). These results led to several investigations of the effect of NK cells and KIR ligand mismatch in other transplant settings (6–10). Emerging from such studies was the finding that NK cell effects were predominantly associated with transplantation treatment for AML and that they were influenced by various clinical factors including donor type (either related or unrelated), preparative regimen and graft characteristics including source and T-cell content (6–12).
Our previous investigations demonstrated a significant association of protection from relapse with a donor having Cen B/B and/or two or more KIR B segments in their KIR genotype (13–15) that was enhanced by the recipient having of a C1-bearing HLA-C allotype (14). At that time, further refinement of the association was not possible because high resolution KIR genotyping had yet to be developed. Greater understanding of the mechanism that provides protection from relapse could help facilitate the development of therapies that can harness the beneficial effects of NK cells in preventing relapse of leukemia. Developing NK cell therapies in place of, or in addition to, transplantation could reduce the reliance on donor selection, which restricts the pool of available donors based on KIR genotype (16).
In the study presented here, we performed high resolution KIR genotyping on a subset of 890 donors from our original retrospective cohort of 1532 (14). We successfully discriminated the two major forms of Cen B, as well as distinguishing the KIR alleles that specify functional proteins from those alleles that are not expressed. The KIR allele data were also used to develop interaction scores that provide measures of the strength and diversity of the KIR:HLA interactions. These variables were used to test immunogenetic associations with clinical outcomes.
Materials and methods
Samples.
We studied 890 patients with AML, who comprise a subset of those analyzed previously (14). Between 1988 and 2009, these patients received myeloblative preparation for a URD HCT facilitated by the National Marrow Donor Program. DNA samples were obtained from the National Marrow Donor Program Research Sample Repository. Outcome data were obtained from the Center for International Blood and Marrow Transplant Research. The demographics of the cohort are shown in Figure 1. DNA samples and clinical data were obtained with informed consent and approval from the National Marrow Donor Program and University of Minnesota Institutional Review Boards.
Fig. 1. Demographic characteristics of the transplant cohort.
Shown are the characteristics of the transplant cohort. Subgroups correspond to KIR Cen genotypes with B/B the sum of B01/B01, B01/B02 and B02/B02. Values shown are N (%). P-value calculation type is indicated by the superscript, A ANOVA F-test, C Chi-square.
KIR genotyping.
To prepare libraries for high throughput sequencing, genomic DNA was fragmented and enriched for those fragments originating from the KIR genomic region and the HLA class I genes, using a pool of oligonucleotide probes (17). Improvements to the library preparation were subsequently made (18). The captured fragments were subjected to paired-end sequencing using a MiSeq instrument and V3 sequencing chemistry (Illumina, San Diego, CA). KIR gene content and KIR allele identities were determined using the PING bioinformatics pipeline (17). HLA-A, -B and -C alleles were determined using the NGSengine 1.7.0 software (GenDx, Utrecht, the Netherlands) with the IPD-IMGT/HLA Database (19). Results were compared to the previous KIR and HLA genotyping of these samples (14).
Statistical analysis.
We tested the same clinical outcomes as in the previous analysis: overall survival, disease free survival, transplant related mortality, relapse, acute GVHD grades II-IV, acute GVHD grades III-IV and chronic GVHD. In the multivariable models we used, all clinical variables were tested first for the affirmation of the proportional hazard assumption. Factors violating this assumption were adjusted through stratification. A stepwise forward-backward selection procedure was then performed to determine the adjusted clinical variables (with a threshold of 0.05 for both entry and retention in the model). KIR variables were tested individually. There were 27 variables tested. Those grouped into four broad categories, refinements of previous analyses(13–15), specific KIR:HLA interactions (20), number of potential HLA:KIR interactions (21–23) and strength of the HLA:KIR interactions. The interaction score variables were tested as both continuous variables and as categorical variables discretized into tertiles. All other variables were tested as categorical variables. To adjust for multiple testing of 27 variables, a threshold of the overall P<0.05/27=0.0018 was used for determining statistical significance.
KIR:HLA interaction scores.
There were two different HLA:KIR interaction scoring models used to assess functional diversity. The first counted the number of interactions and the second used previously published HLA:KIR binding and expression data to calculate a score reflecting the strength and diversity of the interactions. In the case of homozygosity, the interaction was counted twice. As there were a number of mismatched transplants included in this cohort, analyses were performed for the combination of donor KIR with either donor or recipient HLA to determine if there were differential effects.
KIR and HLA genotypes were used for counting the number of possible interactions between the donor KIR and either donor or recipient HLA class I, as described previously (21–23). Binding partners have been described for the inhibitory KIR, 3DL1 (24, 25), 3DL2 (26, 27), and 2DL1, 2DL2, 2DL3 (28–30), and the activating KIR, 2DS1 (30), 2DS2 (31), 2DS4 (32) and 2DS5 (33). The interactions counted are shown in Supplemental Figure 1. Activating and inhibitory interactions were counted and tested separately. Due to the small numbers of individuals with potential activating KIR:HLA interactions, the test was also performed as the presence or absence of activating KIR:HLA interaction.
The allele level data were also used to calculate an interaction score based on the observed binding of KIR and HLA allotype pairs (30, 33–35). The complete scoring matrix is shown in Supplemental Figure 1. Only inhibitory interactions were scored. The centromeric score values come from published binding studies (30, 34) where they were reported as absolute binding values. Certain 2DL1 allotypes have been reported to have low cell surface expression (12, 36–38). The effect of altered expression was included by multiplying the binding values for those allotypes by 0.5 to account for the expression level difference. Telomeric interaction scores are based on the binding of 3DL1 and were developed using the binding data from Saunders et al (35). KIR3DL1 allotypes were assigned to one of four groups (K001, K004, K005, K015) based on similarity. These groups correspond to the non-expressed KIR3DL1 exemplified by 3DL1*004 (39, 40)(K004), the two deeply diverged lineages exemplified by 3DL1*005 and 3DL1*015 (41)(K005 and K015) and the interlineage recombinants exemplified by 3DL1*001 (41)(K001). These values were originally reported as percent of maximum and not as absolute binding values. We normalized them to the centromeric values so that the 100% score of Saunders was equivalent to the maximum centromeric value. KIR and/or HLA of the donor and/or recipient were given the value of the closest allele if they did not appear in the matrix. In homozygous individuals the interaction was counted twice, once for each allele. A total interaction score was computed by summing the centromeric and telomeric scores.
The interaction score reflected the aggregate strength of potential interactions and provides the possibility for a few strong interactions to score similarly to a larger number of weaker interactions. We also calculated average scores for the centromeric, telomeric and total interactions by dividing the interaction score by the number of interactions. This gives a value representing the overall average strength of the potential KIR:HLA interactions.
Results
High resolution KIR genotyping distinguishes CenB01 and CenB02.
The human KIR gene family maps to human chromosome 19q13.4 (42) and consists of distinctive centromeric (Cen) and telomeric (Tel) regions which are separated by a 13kb segment that lacks KIR genes. This region is rich in repetitive elements and is a hotspot for recombination (17, 42, 43) (Fig. 2A). Based upon KIR gene and allele content, the centromeric and telomeric regions are further distinguished by being part of a KIR A or B haplotype (17, 43, 44). The Cen A and Tel A regions are of fixed gene content, whereas the B regions vary in gene content (17, 43, 44). This feature is exemplified by the different KIR gene content of CenB01 and Cen B02, the two most frequent forms of Cen B. Common to Cen B01 and Cen B02 are the KIR3DL3, 2DS2, 2DL2/3 and 3DP1 genes. Where they differ is in the genomic segment containing the KIR2DL5, 2DS3/5, 2DP1 and 2DL1 KIR genes. This segment is present in Cen B01, but absent from Cen B02 (Fig. 2A). In addition to the two frequent Cen B haplotypes, there is a variety of low frequency haplotypes that either lack one or more KIR genes or have one or more duplicated KIR genes (45–48).
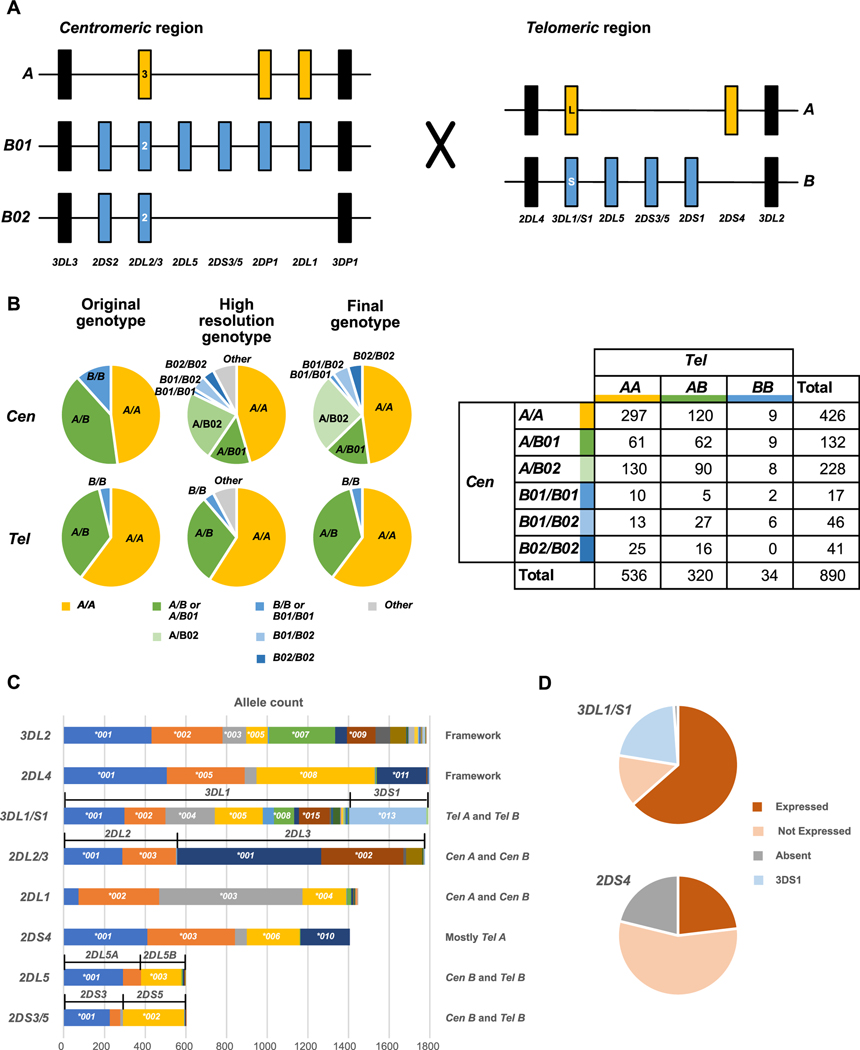
Fig. 2. Allele level KIR genotyping resolves haplotypes and alleles encoding functional differences.
Panel A) Schematic representation of different centromeric (Cen) and telomeric (Tel) KIR haplotype structures. KIR A haplotype specific alleles/genes are in gold, KIR B haplotype specific alleles/genes are in blue, framework genes are in black. The large X indicates the central repetitive region which facilitates recombination between the Cen and Tel segments. Panel B) Genotypes were assigned as combinations of one of the major structural groups shown in Panel A. (Left) Pie charts showing the proportion of each genotype identified in the original genotyping (left), full genotyping (center) and final genotyping (right). In the full genotyping ~7% of genotypes differed from those formed by the major structural groups shown in Panel A by either deletion or duplication of genes. These were assigned to one of the major structural groups (Supplemental Figure 2) for the final genotyping. (Right) Table of Cen/Tel combinations present in the dataset. Panel C) Number of alleles present in the dataset. The greater number of 2DL4 alleles compared to 3DL2 is due to the duplication genotypes that contain additional 2DL4 genes. To the right of the graph, the location of genes in the major structural types (Panel A) are indicated. Panel D) For 3DL1/S1 and 2DS4, the percent of expressed, non-expressed and absent alleles are shown.
Previous low resolution KIR genotyping analysis of this transplant cohort could not distinguish Cen B01 from Cen B02 (14) as in the absence of gene content or allelic information, presence of Cen B02 could be masked by Cen B01 and unambiguous assignment of Cen B01 in the presence of Tel B was not possible. These two Cen haplotypes differ significantly in their KIR gene content, which is likely to result in different functional phenotypes. Shown on the left half of Figure 2B are pie charts showing the proportions of the different types of Cen and Tel KIR segments. The ‘Low resolution’ genotypes are those described in our original study (14), which did not distinguish Cen B01 from Cen B02. The ‘High resolution’ KIR genotypes are those defined in this study, in which Cen B01 and Cen B02 are distinguished. The ‘Final’ KIR genotypes are the genotypes used for the statistical analysis. In this group, genotypes with duplications or deletions were assigned to one of the major genotype groups based on similarity (Supplemental Figure 2).
Of the 890 donors, 821 of them could be assigned genotypes that are combinations of the most frequent KIR haplotype segments shown in Figure 2A (Supplemental Figure 2). This gives rise to six centromeric genotypes and three telomeric genotypes. A minority subset of 69 donors (7.7% of the cohort) have KIR genotypes that are not combinations of the most frequent KIR haplotypes. These donors have at least one KIR haplotype that differs from a frequent haplotype by duplication or deletion of one or more KIR genes. For the analysis, each of these donors was included in one of the frequent genotype groups, based on their gene and allele content (Supplemental Figure 2). For the centromeric genotypes Cen A/A is defined as having only 3DL3–2DL3–2DP1–2DL1–3DP1 in the centromeric interval, regardless of gene copy number. Cen A/B02 is defined by the addition of 2DS2 and/or 2DL2 to the Cen A/A genotype. Cen A/B01 is defined by having 2DL5 and 2DS3/5, as well as 2DS2 and/or 2DL2. Distinguishing Cen B01/B01, Cen B01/B02 and Cen B02/B02 is the presence of 2 copies of 2DL1 in Cen B01/B01, one copy of 2DL1 in Cen B01/B02, and absence of 2DL1 in Cen B02/B02. In addition, Cen B02/B02 is characterized by the lack of 2DL5–2DS3/5. For the telomeric genotypes, Tel A/A is defined as having only 2DL4–3DL1–2DS4–3DL2 in the telomeric interval, regardless of gene copy number. Tel B/B is defined by presence of 3DS1, 2DL5, 2DS3/5 and/or 2DS1 in the telomeric interval, combined with absence of 3DL1 and 2DS4. All other combinations of KIR are considered to be Tel A/B. A complete list of all of the duplication and deletion haplotypes and their assignments is given in Supplemental Figure 2.
The number of occurrences for individual KIR alleles is shown in Fig. 2C (allele and phenotype frequencies are shown in Supplemental Figure 3) and highlights the allelic variability in the KIR genes. The donor cohort is comprised primarily of individuals of European ancestry (Fig. 1) and the common alleles and allele frequencies are consistent with those observed in other European populations (http://allelefrequencies.net/) (49). The number of common alleles (>5% frequency) varies between the genes with some having a single dominant allele (e.g. 2DL5, 2DS3 or 2DS5) and others having several, with the largest number observed for 3DL1/S1 (eight alleles present at frequencies greater than 5%). It is expected that the allele frequencies and common alleles will vary depending on the population of origin. For example, one of the common 2DL3 alleles in this cohort, 2DL3*002, is present at a frequency of 22%. In comparison it is rare in an African population with a reported frequency of less than 1% (23).
This extensive variability also produces high levels of heterozygosity for some of the KIR genes. Together these features result in numerous subgroups, each comprising a small number of individuals, when individual KIR alleles are considered separately. Even with our cohort of 890 transplants the numbers were too small for a robust analysis, when individual alleles were assessed. We therefore used the allelic information to develop interaction scores, which were used to test hypotheses that different strengths of interaction, or diversity of interactions, correlated with differences in transplant outcome. We also performed an analysis of functional presence/absence of 3DL1/S1 and of 2DS4. In Europeans, these two KIR genes have a high frequency of non-expressed alleles (Fig. 2D). In our previous analysis (13) of association of presence/absence of individual genes with outcome these non-expressed alleles were considered to be ‘present’. In this revised analysis, the non-expressed alleles are considered to be absent. We found no association for the presence or absence of 3DL1/S1 or 2DS4 with any of the transplant outcomes tested (Fig. 3A).
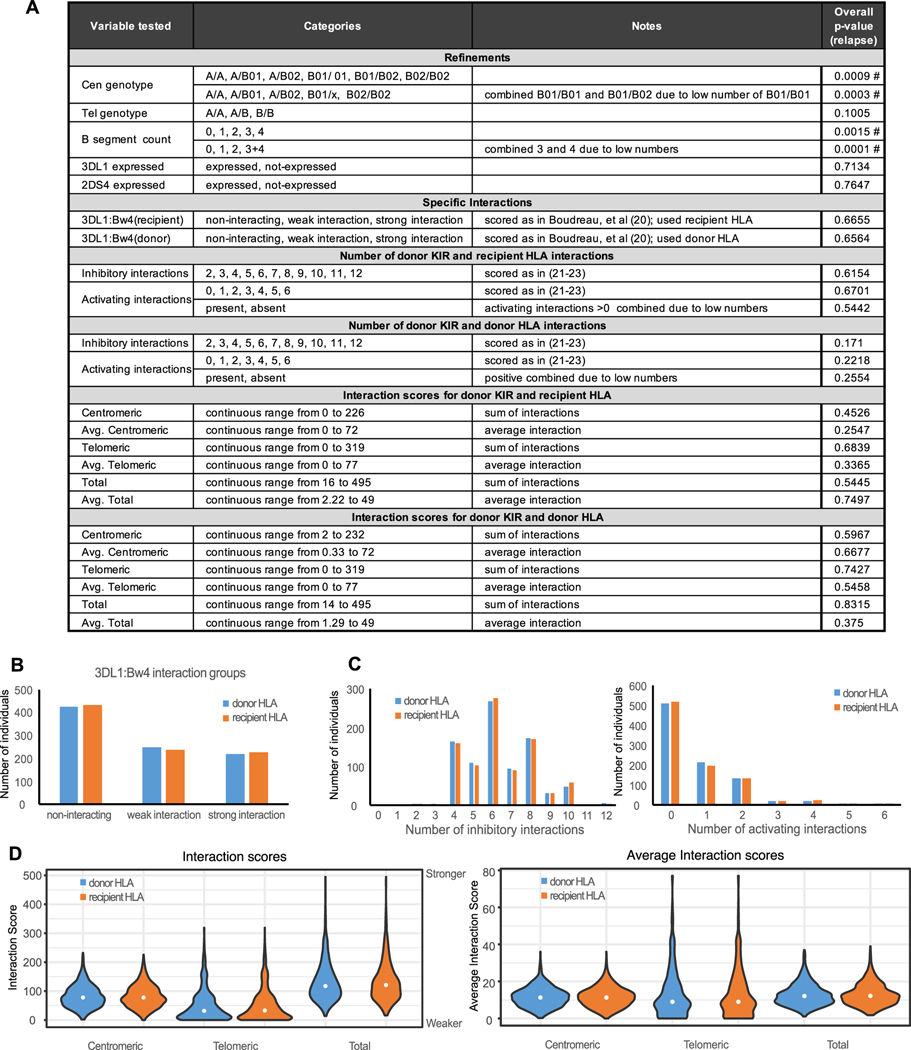
Fig. 3. Cen B/B and a B segment count of two or more correlate with protection from relapse.
Panel A) Individual variables tested in the statistical model are shown for each category investigated. The overall p-values for the analysis of association with relapse are shown in the rightmost column; those with statistical significance are indicated by a #. Complete results are in Supplemental Figure 4. Panel B) Distribution of the classes of 3DL1:Bw4 interaction (20) considering donor 3DL1 and either donor (blue) or recipient (orange) HLA-B. Panel C) Distribution of the number of inhibitory and/or activating interactions (21–23) considering donor KIR with either donor (blue) or recipient (orange) HLA class I. Panel D) Distribution of interaction scores (centromeric, telomeric and total) and average interaction scores (centromeric, telomeric and total) considering donor KIR with either donor (blue) or recipient (orange) HLA class I.
Confirmation that Cen genotype and B-segment count both correlate with relapse protection.
We tested 27 variables in our statistical model (Fig. 3A and Supplemental Figure 4). They are of four broad categories; refinement of previous tests, tests of specific interactions, tests of the diversity of interactions and tests of overall interaction strength. As the transplant cohort contained samples from both HLA matched and HLA mismatched samples, we tested the combination of donor KIR and donor HLA as well as the combination of donor KIR and recipient HLA for all the interaction variables.
Boudreau et al found that combinations of KIR3DL1 and HLA-B that either interact weakly, or do not interact at all, correlate with reduced incidence of relapse following allogeneic hematopoietic cell transplant for AML (20). In that study, most transplant donors and recipients were HLA matched, whereas in our cohort, more transplants were mismatched and the extent of their mismatch was generally greater. For this reason, we tested the combination of the donor KIR with both the donor and recipient HLA (Fig. 3B). In neither test did we find a correlation with any of the endpoints tested. These results are consistent with those of Schetelig et al (50), who saw no correlation of 3DL1:HLA-B combinations with relapse or overall survival. The discordant findings of these three investigations could reflect differences in the transplants studied. Our study cohort comprises transplants primarily derived from bone marrow and having a higher overall degree of HLA mismatch (Fig. 1). The cohort of Boudreau et al was split between bone marrow (55%) and PBSC (45%) transplants and were either 9/10 (44%) or 10/10 (56%) HLA matched (20). The cohort of Schetelig et al comprised transplants that were 96% PBSC and having 9/10 (21%) or 10/10 (78 %) HLA match (50). These differences could result in distinct immune environments following transplant in which specific NK receptor:ligand pairs have a dominant effect. Further analysis of larger and contemporary cohorts will be needed to examine these effects.
In our previous study we correlated protection from relapse with Cen B genotype and B-segment count (14). Those results were replicated in the current analysis (Fig. 3A and Supplemental Fig. 4). We hypothesized that this correlation could be due either to the strength and/or the diversity of KIR:HLA interactions. In testing this hypothesis we used two systems for scoring the interactions. The first measured the diversity of KIR:HLA interactions by determining the number of potential KIR:HLA interactions between the allotypes encoded by the donor KIR and donor, or recipient, HLA class I allotypes. Each transplant was scored by counting potential interactions between donor KIR and both donor and recipient HLA class I (Supplemental Fig. 1). Interactions involving inhibitory KIR and activating KIR were scored separately. The distribution of the scores is shown in Fig. 3C. Because of the low number of activating KIR interactions, mainly due to the high frequency of non-functional 2DS4 alleles, the activating interaction score was also tested as presence/absence of activating KIR:HLA interaction without further stratification. None of these variables showed statistically significant association with relapse (Fig. 3A and Supplemental Fig. 4), nor any other endpoint (not shown).
The second type of interaction score we used was based on experimentally determined strengths of KIR-HLA class I interactions (30, 33–35). This analysis examined only the inhibitory KIR: KIR2DL1, 2DL2, 2DL3 and 3DL1. This score was assessed for the allotypes encoded by centromeric KIR2DL1, 2DL2, and 2DL3, for allotypes encoded by telomeric KIR3DL1 and for the combination of centromeric and telomeric allotypes (Fig. 3D). The scores were also adjusted to obtain an average interaction score (Fig. 3D). The combination of donor KIR with either donor or recipient HLA was tested. Neither score showed a correlation with relapse that is statistically significant (Fig. 3A).
Comparing the effect of CenB01 versus CenB02.
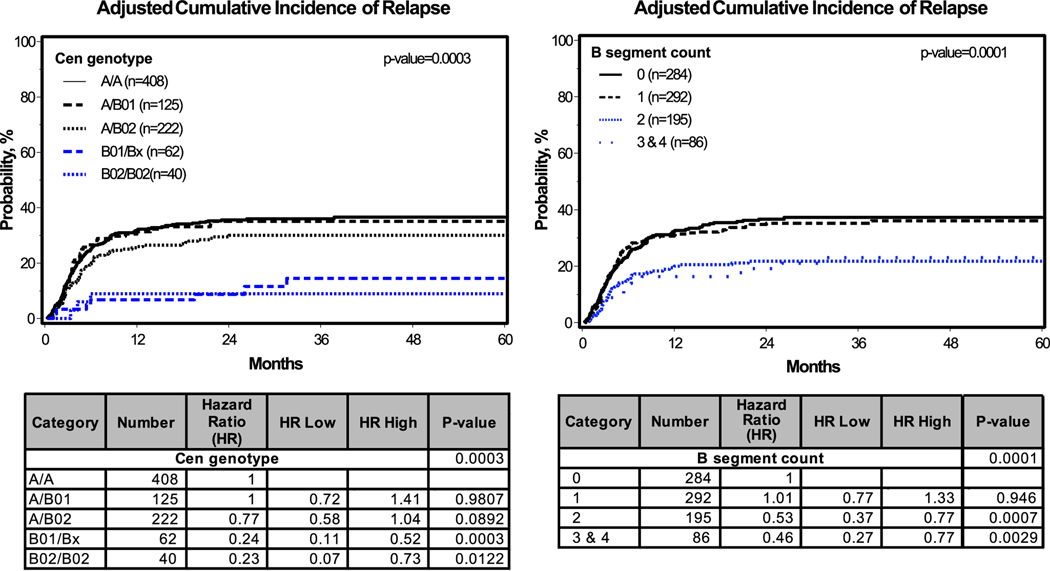
Protection from relapse was first observed for transplant donors who were either Cen B/B or had two or more KIR B gene segments. We further stratified the Cen B genotypes into Cen B01 and Cen B02 (Fig. 2A and B) and improved the assessment of B segment counts by determining the deletion and duplication events that alter the B segment count. Greater protection against relapse was associated with Cen B01/Bx and Cen B02/B02 compared to all other Cen genotypes (Fig. 4, left panels). This additional refinement produced no statistically significant difference between Cen B01 and Cen B02 (Fig. 4, left panels). However, in both the Cen B02/B02 versus Cen B01/Bx and the Cen A/B02 vs Cen A/B01 groups there is a clear trend for Cen B02 to confer greater protection against relapse (HR 0.77, CI 0.58–1.04, P=0.0892) (Fig. 4, left panels).
Fig. 4. Cumulative incidence of leukemia relapse for Cen genotype and B segment count.
Shown is the graph of adjusted cumulative incidence (top) and stratified hazard ratios and p-values (bottom). Tests for interaction showed no interaction between the variables. In the Cen genotype model, B01/B01(n=17) and B01/B02 (n=45) were combined into B01/Bx because of the low frequency of B01. Similarly, the B-segment counts of 3 (n=71) and 4 (n=15) were combined, because of the low number of donors having four B segments. For the centromeric genotype there is a trend toward significance for the A/B02 genotype compared to either the A/A or A/B01 genotypes.
Protection from relapse was also seen for donors having two or more B segments (Fig. 4, right panels). Testing for interaction between B segment count, Cen genotype and adjusted covariates showed no significant interaction. Because of the low number of Tel B/B genotypes, the majority of two B-segment genotypes have a Cen B segment. This points to a mechanism requiring the presence of 2DL2 and/or 2DS2 in a genomic background that encodes fewer or weaker inhibitory KIR that interact with HLA.
Discussion
Our previous studies of HCT treatment for AML (13–15) demonstrated a significant protection from relapse that is associated with a donor having the Cen B/B KIR genotype and/or two or more KIR B gene segments. Providing the best protection were donors having the Cen B/B genotype and recipients having C1+ HLA-C. This observation is consistent with a mechanism that requires a donor with one or two Cen B gene segments as there is no significant association with donors that are Tel B/B and have no Cen B. Further refinement of this model to consider allelic differences was not possible without the higher resolution of the genotyping data provided in this study.
Two overlapping hypotheses arise from this study. First, protection from relapse is associated with one or more of the specific genes or alleles found in the Cen B segment. Supporting this hypothesis, is relapse protection was strongly associated with the presence of two Cen B segments and the association is retained in the presence of two or more B segments. While this latter group includes individuals who are Tel B/B, this group is very small. It is therefore possible that this association is driven by individuals who have at least one Cen B segment and is combined with one or two Tel B segments. The second hypothesis is that the association with protection from relapse reflects an association with strength or diversity of KIR:HLA interactions, and that it is interactions of the products encoded by the Cen B segment that more strongly influence the interaction.
Allotypic diversity of the KIR and their HLA ligands produces a range of interaction strengths. We developed an interaction score that accounts for binding strength and the expression levels of 2DL1, 2DL2, 2DL3 and 3DL1/S1 with their known ligands. We calculated an interaction score that reflects the sum of all interactions (range 0–495) and an average interaction score (range 0–39) (Fig. 3D). The statistical analysis incorporated the scores as continuous variables, as well as when the values are divided into tertiles. Although no statistically significant correlations with transplant outcome were observed, there is a clear trend toward significance when the data are analyzed as tertiles. It is possible that the cohort size of 890 was too small to detect a significant association and that a larger, more contemporary cohort with opportunity to adjust for critical clinical variables such as preparatory regimen and graft source could better inform this question.
The overall diversity of interactions was examined by counting the number of distinct KIR:HLA interactions that are predicted by the genotype data. We analyzed activating and inhibitory interactions separately. A large part of the cohort had no activating KIR:HLA interactions due to the high percentage (56%, Fig. 2D) of 2DS4 alleles that are not-expressed. No statistically significant associations were identified. This may reflect the small number of individuals in some of the numeric categories (Fig. 3C).
Boudreau et al (20) reported that protection from relapse is associated with combinations of KIR and HLA class I that predict weak or non-inhibitory interactions. We similarly divided our cohort using the criteria in their study and examined the donor KIR genotype paired with either the donor HLA genotype or the recipient HLA genotype separately. Despite having a similarly sized transplant cohort as Boudreau et al, we found no significant association with relapse for either combination. Although differences in preparatory regimen were included in our statistical model, it is possible that other clinical correlates are responsible for the differences observed in our study and that of Boudreau et al.
Two main Cen B structures (Fig. 2A) differ in KIR gene content. With only presence/absence data available for our previous analyses, we (13, 14) could not discriminate Cen B01 from Cen B02 for all individuals. With high resolution KIR genotyping we have now resolved the Cen B content for all individuals tested (Fig. 2B). Although not statistically significant, there is a trend to significance indicating that Cen B02 gives more effective protection from relapse than Cen B01. This is most apparent in the comparison of Cen A/B01 individuals with Cen A/B02 individuals (Fig. 4). Cen A/B01 and Cen A/A individuals are not distinguished whereas Cen A/B02 individuals show an increase in protection from relapse.
The confirmed association of donors with 2 or more B-segments, particularly Cen B homozygous donors, with relapse protection, supports the interpretation that KIR2DL2 and/or KIR2DS2 are necessary for protection. Consistent with this interpretation is the absence of any association with the Tel genotype. In the 3 and 4 B-segment groups all individuals have at least one Cen B and most have two (Fig. 2B). Even the 2 B-segment group is biased toward individuals having at least one Cen B (96% total, 23% B/B, 73% A/B). The effect of 2DL2 and/or 2DS2 is enhanced when the contribution of 2DL1, 2DL3 and 3DL1 to NK cell inhibitory potential is decreased or absent. We could not subset our cohort further to determine if there was a predictable hierarchy of contribution for each of these covariates. Testing the degree of contribution from each of these will require analysis of a much larger transplant cohort.
Supplementary Material
Key Points.
KIR Cen B is associated with protection from relapse following HSCT
KIR Cen B02 provides stronger protection against relapse
Protection from relapse associates with presence of less inhibitory KIR
Acknowledgments
1 This work was supported by NIH grant P01 CA111412 to J.M.
References
- 1.Kumar S. 2018. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 154: 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augusto DG, and Petzl-Erler ML. 2015. KIR and HLA under pressure: evidences of coevolution across worldwide populations. Hum Genet 134: 929–940. [DOI] [PubMed] [Google Scholar]
- 3.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, and Velardi A. 2002. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295: 2097–2100. [DOI] [PubMed] [Google Scholar]
- 4.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, Stern M, Pende D, Perruccio K, Burchielli E, Topini F, Bianchi E, Aversa F, Martelli MF, and Velardi A. 2007. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood 110: 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, and Velardi A. 1999. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood 94: 333–339. [PubMed] [Google Scholar]
- 6.Hsu KC, Gooley T, Malkki M, Pinto-Agnello C, Dupont B, Bignon J-D, Bornhäuser M, Christiansen F, Gratwohl A, Morishima Y, Oudshoorn M, Ringden O, van Rood JJ, Petersdorf E, and International Histocompatibility Working Group. 2006. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant 12: 828–836. [DOI] [PubMed] [Google Scholar]
- 7.Malmberg K-J, Schaffer M, Ringdén O, Remberger M, and Ljunggren H-G. 2005. KIR-ligand mismatch in allogeneic hematopoietic stem cell transplantation. Mol Immunol 42: 531–534. [DOI] [PubMed] [Google Scholar]
- 8.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, Maccario R, Bonetti F, Wojnar J, Martinetti M, Frassoni F, Giorgiani G, Bacigalupo A, and Holowiecki J. 2003. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood 102: 814–819. [DOI] [PubMed] [Google Scholar]
- 9.Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, Boudreau C, Nelson G, Oudshoorn M, van Rood J, Velardi A, Maiers M, Setterholm M, Confer D, Posch PE, Anasetti C, Kamani N, Miller JS, Weisdorf D, Davies SM, and KIR Study Group, Center for International Blood and Marrow Transplantation Research. 2006. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant 12: 876–884. [DOI] [PubMed] [Google Scholar]
- 10.Leung W. 2011. Use of NK cell activity in cure by transplant. Br J Haematol 155: 14–29. [DOI] [PubMed] [Google Scholar]
- 11.Bultitude WP, Schellekens J, Szydlo RM, Anthias C, Cooley SA, Miller JS, Weisdorf DJ, Shaw BE, Roberts CH, Garcia-Sepulveda CA, Lee J, Pearce RM, Wilson MC, Potter MN, Byrne JL, Russell NH, MacKinnon S, Bloor AJ, Patel A, McQuaker IG, Malladi R, Tholouli E, Orchard K, Potter VT, Madrigal JA, Mayor NP, and Marsh SGE. 2020. Presence of donor-encoded centromeric KIR B content increases the risk of infectious mortality in recipients of myeloablative, T-cell deplete, HLA-matched HCT to treat AML. Bone Marrow Transplant 55: 1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubreuil L, Maniangou B, Chevallier P, Quéméner A, Legrand N, Béné MC, Willem C, David G, Alizadeh M, Makanga DR, Cesbron A, Gendzekhadze K, Gagne K, and Retière C. 2020. Centromeric KIR AA Individuals Harbor Particular KIR Alleles Conferring Beneficial NK Cell Features with Implications in Haplo-Identical Hematopoietic Stem Cell Transplantation. Cancers 12: 3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh SGE, Guethlein LA, Parham P, Miller JS, and Weisdorf DJ. 2009. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 113: 726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SGE, Spellman S, Haagenson MD, Saeturn K, Ladner M, Trachtenberg E, Parham P, and Miller JS. 2014. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 192: 4592–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh SGE, Geraghty D, Spellman S, Haagenson MD, Ladner M, Trachtenberg E, Parham P, and Miller JS. 2010. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116: 2411–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisdorf D, Cooley S, Wang T, Trachtenberg E, Haagenson MD, Vierra-Green C, Spellman S, Spahn A, Vogel J, Kobusingye H, Fehninger T, Woolfrey A, Devine S, Ross M, Waller EK, Sobecks R, Parham P, Guethlein LA, Marsh SGE, Miller J, and participating center writing committee. 2019. KIR Donor Selection: Feasibility in Identifying better Donors. Biol Blood Marrow Transplant 25: e28–e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Marin WM, Norberg SJ, Ashouri E, Jayaraman J, Wroblewski EE, Trowsdale J, Rajalingam R, Oksenberg JR, Chiaroni J, Guethlein LA, Traherne JA, Ronaghi M, and Parham P. 2016. Defining KIR and HLA Class I Genotypes at Highest Resolution via High-Throughput Sequencing. Am J Hum Genet 99: 375–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemat-Gorgani N, Hilton HG, Henn BM, Lin M, Gignoux CR, Myrick JW, Werely CJ, Granka JM, Möller M, Hoal EG, Yawata M, Yawata N, Boelen L, Asquith B, Parham P, and Norman PJ. 2018. Different Selected Mechanisms Attenuated the Inhibitory Interaction of KIR2DL1 with C2+ HLA-C in Two Indigenous Human Populations in Southern Africa. J Immunol 200: 2640–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, and Marsh SGE. 2020. IPD-IMGT/HLA Database. Nucleic Acids Res 48: D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudreau JE, Giglio F, Gooley TA, Stevenson PA, Le Luduec J-B, Shaffer BC, Rajalingam R, Hou L, Hurley CK, Noreen H, Reed EF, Yu N, Vierra-Green C, Haagenson M, Malkki M, Petersdorf EW, Spellman S, and Hsu KC. 2017. KIR3DL1/HLA-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation. J Clin Oncol 35: 2268–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemat-Gorgani N, Guethlein LA, Henn BM, Norberg SJ, Chiaroni J, Sikora M, Quintana-Murci L, Mountain JL, Norman PJ, and Parham P. 2019. Diversity of KIR, HLA Class I, and Their Interactions in Seven Populations of Sub-Saharan Africans. J Immunol 202: 2636–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, Gignoux CR, Guethlein LA, and Parham P. 2015. Loss and Gain of Natural Killer Cell Receptor Function in an African Hunter-Gatherer Population. PLoS Genet 11: e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, Koram KA, Riley EM, Abi-Rached L, and Parham P. 2013. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet 9: e1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foley BA, Santis DD, Van Beelen E, Lathbury LJ, Christiansen FT, and Witt CS. 2008. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: implications for patient and donor suitability for haploidentical stem cell transplantations. Blood 112: 435–443. [DOI] [PubMed] [Google Scholar]
- 25.Gumperz JE, Litwin V, Phillips JH, Lanier LL, and Parham P. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. Journal of Experimental Medicine 181: 1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Döhring C, Scheidegger D, Samaridis J, Cella M, and Colonna M. 1996. A human killer inhibitory receptor specific for HLA-A1,2. The Journal of Immunology 156: 3098–3101. [PubMed] [Google Scholar]
- 27.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland‐Jones S, and Braud VM. 2004. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. European Journal of Immunology 34: 1673–1679. [DOI] [PubMed] [Google Scholar]
- 28.Wagtmann N, Rajagopalan S, Winter CC, Peruui M, and Long EO. 1995. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity 3: 801–809. [DOI] [PubMed] [Google Scholar]
- 29.Winter CC, and Long EO. 1997. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. The Journal of Immunology 158: 4026–4028. [PubMed] [Google Scholar]
- 30.Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, and Parham P. 2015. Polymorphic HLA-C Receptors Balance the Functional Characteristics of KIR Haplotypes. J Immunol 195: 3160–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moesta AK, Graef T, Abi-Rached L, Aguilar AMO, Guethlein LA, and Parham P. 2010. Humans Differ from Other Hominids in Lacking an Activating NK Cell Receptor That Recognizes the C1 Epitope of MHC Class I. The Journal of Immunology 185: 4233–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, and Parham P. 2009. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. Journal of Experimental Medicine 206: 2557–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, Chazara O, Moffett A, and Parham P. 2017. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun Inflamm Dis 5: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi-Rached L, Norman PJ, Guethlein LA, Fleischhauer K, and Parham P. 2012. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol 189: 1418–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O’Connor GM, Loiacono F, Widjaja J, Price DA, Falco M, Mingari MC, Moretta L, McVicar DW, Rossjohn J, Brooks AG, and Vivian JP. 2016. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med 213: 791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bari R, Bell T, Leung W-H, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, and Leung W. 2009. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine245. Blood 114: 5182–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huhn O, Chazara O, Ivarsson MA, Retière C, Venkatesan TC, Norman PJ, Hilton HG, Jayaraman J, Traherne JA, Trowsdale J, Ito M, Kling C, Parham P, Ghadially H, Moffett A, Sharkey AM, and Colucci F. 2018. High-Resolution Genetic and Phenotypic Analysis of KIR2DL1 Alleles and Their Association with Pre-Eclampsia. The Journal of Immunology 201: 2593–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Luduec J-B, Boudreau JE, Freiberg JC, and Hsu KC. 2019. Novel Approach to Cell Surface Discrimination Between KIR2DL1 Subtypes and KIR2DS1 Identifies Hierarchies in NK Repertoire, Education, and Tolerance. Front. Immunol. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, and Parham P. 2001. Different NK Cell Surface Phenotypes Defined by the DX9 Antibody Are Due to KIR3DL1 Gene Polymorphism. The Journal of Immunology 166: 2992–3001. [DOI] [PubMed] [Google Scholar]
- 40.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, and Parham P. 2003. The Protein Made from a Common Allele of KIR3DL1 (3DL1*004) Is Poorly Expressed at Cell Surfaces due to Substitution at Positions 86 in Ig Domain 0 and 182 in Ig Domain 1. The Journal of Immunology 171: 6640–6649. [DOI] [PubMed] [Google Scholar]
- 41.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CVF, Chandanayingyong D, Chang Y-H, Crespí C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Milà J, Park MH, Pitchappan RM, Ramdath DD, Shiau M-Y, Stephens HAF, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, and Parham P. 2009. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 19: 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, and Wilson MJ. 2001. The genomic context of natural killer receptor extended gene families. Immunol Rev 181: 20–38. [DOI] [PubMed] [Google Scholar]
- 43.Pyo C-W, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, Marsh SGE, Miller JS, Parham P, and Geraghty DE. 2010. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One 5: e15115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, and Parham P. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity 7: 753–763. [DOI] [PubMed] [Google Scholar]
- 45.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, Middleton D, Carrington M, and Trowsdale J. 2010. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet 19: 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF, Cookson WO, Trowsdale J, and Traherne JA. 2012. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res 22: 1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roe D, Vierra-Green C, Pyo C-W, Eng K, Hall R, Kuang R, Spellman S, Ranade S, Geraghty DE, and Maiers M. 2017. Revealing complete complex KIR haplotypes phased by long-read sequencing technology. Genes Immun 18: 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roe D, Vierra-Green C, Pyo C-W, Geraghty DE, Spellman SR, Maiers M, and Kuang R. 2020. A Detailed View of KIR Haplotype Structures and Gene Families as Provided by a New Motif-Based Multiple Sequence Alignment. Front Immunol 11: 585731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez-Galarza FF, McCabe A, dos Santos EJM, Jones J, Takeshita L, Ortega-Rivera ND, Cid-Pavon GMD, Ramsbottom K, Ghattaoraya G, Alfirevic A, Middleton D, and Jones AR. 2020. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Research 48: D783–D788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schetelig J, Baldauf H, Heidenreich F, Massalski C, Frank S, Sauter J, Stelljes M, Ayuk FA, Bethge WA, Bug G, Klein S, Wendler S, Lange V, de Wreede LC, Fürst D, Kobbe G, Ottinger HD, Beelen DW, Mytilineos J, Fleischhauer K, Schmidt AH, and Bornhäuser M. 2020. External validation of models for KIR2DS1/KIR3DL1-informed selection of hematopoietic cell donors fails. Blood 135: 1386–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.