Abstract
The unstructured nature of Real-World (RW) data from onco-hematological patients and the scarce accessibility to integrated systems restrain the use of RW information for research purposes. Natural Language Processing (NLP) might help in transposing unstructured reports into standardized electronic health records. We exploited NLP to develop an automated tool, named ARGO (Automatic Record Generator for Onco-hematology) to recognize information from pathology reports and populate electronic case report forms (eCRFs) pre-implemented by REDCap. ARGO was applied to hemo-lymphopathology reports of diffuse large B-cell, follicular, and mantle cell lymphomas, and assessed for accuracy (A), precision (P), recall (R) and F1-score (F) on internal (n = 239) and external (n = 93) report series. 326 (98.2%) reports were converted into corresponding eCRFs. Overall, ARGO showed high performance in capturing (1) identification report number (all metrics > 90%), (2) biopsy date (all metrics > 90% in both series), (3) specimen type (86.6% and 91.4% of A, 98.5% and 100.0% of P, 92.5% and 95.5% of F, and 87.2% and 91.4% of R for internal and external series, respectively), (4) diagnosis (100% of P with A, R and F of 90% in both series). We developed and validated a generalizable tool that generates structured eCRFs from real-life pathology reports.
Subject terms: Translational research, B-cell lymphoma, Data acquisition, Data integration, Pathology, Diagnostic markers, B-cell lymphoma
Introduction
Over the last few years, the complexity of clinical and biological data for a proper diagnosis and prognostication of onco-hematological diseases has remarkably increased, especially in the field of lymphomas1,2. In parallel, novel therapeutics found continue approvals from large, controlled trials, but missed parallel validation in the Real-World (RW) settings3. This major controversy claims for an urgent improvement of the capability to collect and share RW data with the final goal to support clinical and translational research4. Frequently, RW data are derived from fragmented sources as medical registries, electronic records, computerized patient order entries, individual databases, paper notes, as well as monocentric bio-banking-related annotations. Moreover, the common dearth of specialized data-entry professionals and the uneasy accessibility to data-extraction systems for most physicians accentuate the need for tools that facilitate the process of health data recording 5.
Natural Language Processing (NLP) is a consolidated technique to extract essential unstructured data from text, e.g. from diagnostic and prognostic notes6–10, widely adopted also in onco-hematology11–16. REDCap (Research Electronic Data CAPture) is a recognized platform of electronic case report forms (eCRFs) enabling rapid, high-quality and standardized annotation of data17,18. A potential bridge between NLP and eCRFs population is interposed by Optical Character Recognition (OCR), namely a state-of-the-art technology able to convert paper-based reports into digital formats to be further structured—possibly through NLP techniques—in electronic health records (EHR). Thus, OCR overcomes the need of integration between textual reporting and digital storage systems19,20.
Here, we describe the development of a NLP-based tool, named ARGO (Automatic Record Generator for Onco-hematology), to automatically convert RW reports in standardized eCRFs for data collection. To test its generalizability, we applied ARGO to a multicentric set of RW pathology reports of Non-Hodgkin Lymphomas (NHL) and validated its functionality, performance, and suitability for future translation into the daily practice.
Results
Electronic data collection workflow
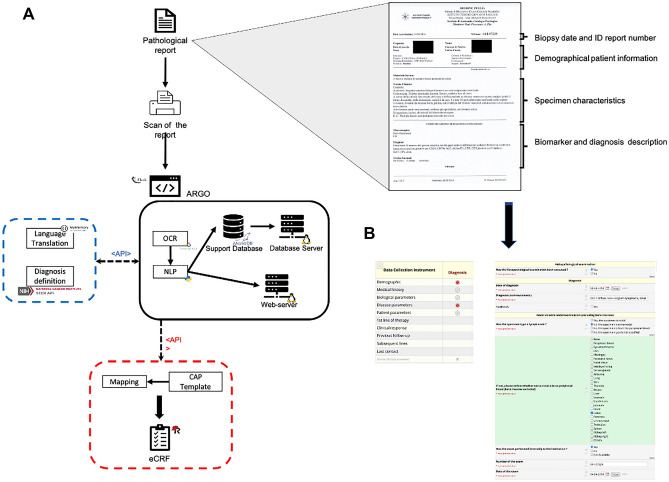
The capacity of ARGO to effectively automize eCRF generation was tested on both internal and external cohorts of NHL paper-based pathology reports, including Diffuse Large B-Cell Lymphoma (DLBCL), Follicular Lymphoma (FL), and Mantle Cell Lymphoma (MCL). ARGO read all the words in each template currently adopted at the Pathology Unit of the IRCCS Istituto Tumori “Giovanni Paolo II” as well as at Pathology Units of six additional Italian Hospitals. As illustrated in Fig. 1A, each histopathology report included several data organized in four main sections: (1) biopsy date and Identification (ID) report number; (2) patient demographical information; (3) specimen characteristics; and (4) biomarker and diagnosis description.
Figure 1.
Graphical description of the framework. (A) Each paper-based report is manually transformed into an image file by a common digital scanner (right upside, an example of paper-based report from the Pathology Unit of the IRCCS Istituto Tumori “Giovanni Paolo II” of Bari, Italy). Then, the image is uploaded into ARGO through a web interface (black block), transformed in structured text through OCR and saved (by an NLP approach) as structured data in a database via webserver. “Diagnosis” attribution is carried out via API connecting ARGO with SEER servers (blue block). Finally, ARGO automatically populates eCRFs via API (red block). (B) Representative picture of REDCap dashboard for a single case report including “Demography” and “Disease parameters” forms (red bullets). Abbreviations. ARGO: Automatic Record Generator for Onco-hematology, OCR: Optical Character Recognition, NLP: Natural Language Processing, SEER: Surveillance, Epidemiology, and End Results, eCRFs: electronic Case Report Forms, API: Application Programming Interface, REDCap: Research Electronic Data-Capture, ID: Identification.
The first implementation step of ARGO pipeline consisted in the advantageous transformation of each paper-based report into an image file (.jpg extension) by using a common digital scanner. Thus, each report was uploaded on the ARGO application, which saved structured text into a support database, retrieved all the relevant data from the text, and transferred them directly into dedicated eCRFs. 233 out of 239 reports of the internal series and all those belonging to the external series (n = 93) were successfully converted in eCRF records. ARGO failed in converting six reports due to either low optical quality or their length (> 1 paper page). Figure 1B shows the main sections of each eCRF, which included both “demographic” and “disease” modules (see also Figure S1 from Supplementary Appendix), in a way consistent with the content of the corresponding original paper report. A video demonstrates ARGO’s functionality in Multimedia Appendix.
Data retrieved from diagnostic reports
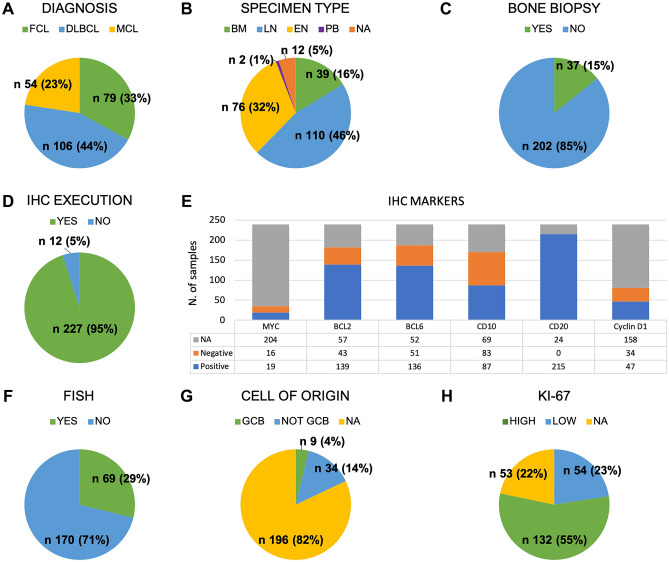
Among the 239 paper-based reports retrieved from the internal series, 106, 79, and 54 were conclusive for a diagnosis of DLBCL, FL, and MCL, respectively (Fig. 2A). Overall, 110 diagnostic specimens were obtained from a tissue extracted from a lymph-node (LN), 76 were extra-nodal (EN), and 39 from bone marrow (BM), 2 from peripheral blood (PB), while for 12 cases this information was not available (Fig. 2B). In 85% of cases, a matched bone marrow biopsy was not available (Fig. 2C). Results from immunohistochemistry (IHC) staining for MYC, BCL2, BCL6, cluster of differentiation (CD)10, CD20, and Cyclin-D1 were available in 227 out of 239 cases and included a qualitative (positive/negative) assessment for the most relevant markers (Fig. 2D-E). A FISH (Fluorescent in situ hybridization) analysis (for MYC, BCL2, BCL6 or Cyclin-D1) appeared in the 29% of reports (Fig. 2F), whereas Cell of Origin (COO) categorization was reported in nearly 18% of cases (Fig. 2G). Of note, 186 out of 239 reports included the quantitative value of the Ki-67. Among these, 54 reported a value lower than 30% (Fig. 2H). Table 1 shows the full reports characteristics from the internal series.
Figure 2.
Characteristics of data retrieved from diagnostic reports. Graphical representation of diagnostic features, subdivided into specific fields, captured by ARGO from a total of n. 239 paper-based pathology reports of the internal series. Abbreviations. ARGO: Automatic Record Generator for Onco-hematology, FCL: Follicular Lymphoma, DLBCL: Diffuse Large B-cell Lymphoma, MCL: Mantle Cell Lymphoma, BM: Bone Marrow, LN: Lymph-Node, EN: Extra-Nodal, PB: Peripheral Blood, NA: Not Available, IHC: Immunohistochemistry, N: number, CD: Cluster of Differentiation, FISH: Fluorescent in situ hybridization, GCB: Germinal Center B-like.
Table 1.
Characteristics of pathology reports.
| Internal series | External series | |||
|---|---|---|---|---|
| N | (%) | N | (%) | |
| 239 | (100.0) | 93 | (100.0) | |
| Diagnosis | ||||
| DLBCL | 106 | (44.4) | 49 | (52.7) |
| FCL | 79 | (33.1) | 24 | (25.8) |
| MCL | 54 | (22.6) | 20 | (21.5) |
| Specimen type | ||||
| LN | 110 | (46.0) | 53 | (57.0) |
| EN | 76 | (31.8) | 28 | (30.1) |
| BM | 39 | (16.3) | 12 | (12.9) |
| PB | 2 | (0.8) | 0 | (0.0) |
| NA | 12 | (5.0) | 0 | (0.0) |
| Bone biopsy | ||||
| Yes | 37 | (15.5) | 14 | (15.1) |
| No | 202 | (84.5) | 79 | (84.9) |
| IHC Execution | ||||
| Yes | 227 | (95.0) | 93 | (100.0) |
| No | 12 | (5.0) | 0 | (0.0) |
| IHC Markers | ||||
| MYC | ||||
| Positive | 19 | (7.9) | 30 | (32.3) |
| Negative | 16 | (6.7) | 8 | (8.6) |
| NA | 204 | (85.4) | 55 | (59.1) |
| BCL2 | ||||
| Positive | 139 | (58.2) | 72 | (77.4) |
| Negative | 43 | (18.0) | 6 | (6.5) |
| NA | 57 | (23.8) | 15 | (16.1) |
| BCL6 | ||||
| Positive | 136 | (56.9) | 59 | (63.4) |
| Negative | 51 | (21.3) | 11 | (11.8) |
| NA | 52 | (21.8) | 23 | (24.7) |
| CD10 | ||||
| Positive | 87 | (3.6) | 40 | (4.3) |
| Negative | 83 | (34.7) | 39 | (41.9) |
| NA | 69 | (28.9) | 14 | (15.1) |
| CD20 | ||||
| Positive | 215 | (90.0) | 84 | (90.3) |
| Negative | 0 | (0.0) | 2 | (2.2) |
| NA | 24 | (10.0) | 7 | (7.5) |
| Cyclin D1 | ||||
| Positive | 47 | (19.7) | 21 | (22.6) |
| Negative | 34 | (14.2) | 22 | (23.7) |
| NA | 158 | (66.1) | 50 | (53.8) |
| FISH | ||||
| Yes | 69 | (28.9) | 5 | (5.4) |
| No | 170 | (71.1) | 88 | (94.6) |
| Cell of origin | ||||
| GCB | 9 | (3.8) | 14 | (15.1) |
| Not GCB | 34 | (14.2) | 19 | (20.4) |
| NA | 196 | (82.0) | 60 | (64.5) |
| Ki-67 | ||||
| < 30 | 54 | (22.6) | 17 | (18.3) |
| ≥ 30 | 132 | (55.2) | 58 | (62.4) |
| NA | 53 | (22.2) | 18 | (19.4) |
ARGO: Automatic Record Generator for Onco-hematology, FCL: Follicular Lymphoma, DLBCL: Diffuse Large B-cell Lymphoma, MCL: Mantle Cell Lymphoma, BM: Bone Marrow, LN: Lymph-Node, EN: Extra-Nodal, PB: Peripheral Blood, NA: Not Available, IHC: Immunohistochemistry, N: number, CD: Cluster of Differentiation, FISH: Fluorescent in situ hybridization, GCB: Germinal Center B-like.
Among the 93 paper-based reports retrieved externally from other six Italian centers, 49, 24, and 20 were conclusive for a diagnosis of DLBCL, FL, and MCL, respectively (Table 1). Overall, 53 diagnostic specimens were obtained from LN, 28 were EN, and 12 from BM. In 85% of cases, a matched bone marrow biopsy was not available. Results from IHC staining for MYC, BCL2, BCL6, CD10, CD20, and Cyclin-D1 were available in 93 out of 93 cases and in external series included a qualitative (positive/negative) assessment for the most relevant biomarkers. A FISH analysis (for MYC, BCL2, BCL6 or Cyclin-D1) appeared in the 5.4% of reports, whereas COO categorization was reported in nearly 35.5% of cases. Of note, 93 out of 93 reports included the quantitative value for Ki-67. Among these, 17 reported a value lower than 30%. Table 1 shows the full reports characteristics from the external series.
Internal vs external validation
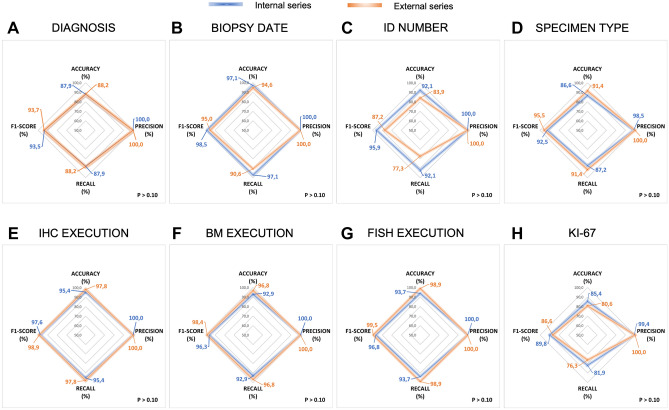
Overall, ARGO detected 127,578 terms of interest and successfully generated EHR from 326 out of 332 processed histopathology reports. Figure 3 shows the comparative (internal vs. external series) post-hoc validation of ARGO performance for all the study data fields. For the “DIAGNOSIS” field, ARGO reached 88.9% vs. 87.9% (p > 0.05) of accuracy and recall, 93.5% and 93.7% of F1-score, also achieving 100% of precision in both series. For the “BIOPSY DATE” and the “ID NUMBER” fields of the internal series, all the applied metrics were > 90%. In comparison, accuracy, F1-score, and recall of external series for the “BIOPSY DATE” field were 94.6%, 95.0%, and 90.6%, respectively (P > 0.10), whereas “ID NUMBER” field ranged between 77.3% (recall from the external series) and 100.0% (precision from both internal and external series) (P > 0.10). For the “SPECIMEN TYPE” field, ARGO reached 86.6% vs. 91.4% of accuracy, 98.5% vs. 100.0% of precision, 92.5% vs. 95.5% of F1-score and 87.2% vs. 91.4% of recall (P > 0.10 in all instances). Similar high performance was observed for the “IHC EXECUTION” field (95.4% vs. 97.8% of accuracy and recall, 97.6% vs. 98.9% F1-score, and 100.0% of precision [P > 0.10]), although accuracy, recall and F1-score, but not precision (100.0%), slightly decreased as for single biomarker analyses (Supplementary Table S1). Similar results were also recorded the “BM AND FISH EXECUTION” fields. Finally, ARGO allowed the detection of “Ki-67”-related information with 85.4% vs. 80.6% (P > 0.10), 99.4% vs. 100.0% (P > 0.10), 81.9% vs. 76.3% (P > 0.10), and 89.8% vs. 86.6% (P > 0.10) of accuracy, precision, recall and F1-score, respectively. Overall, no significant differences between internal vs. external series were found in 14 out of 15 tested data fields (Supplementary Table S1). Of note, there is significant improvement (P < 0.01) in detecting the CD10 biomarker from the internal (67.4% of accuracy, 96.9% of precision, 55.9% of recall, and 70.9% of F1-score) compared to the external series (82.8% of accuracy, 98.5% of precision, 81.0% of recall, and 88.9% of F1-score).
Figure 3.
ARGO performance. Radar graphs indicate the performance metrics as percentage of accuracy, precision, recall and F1-score in different data fields for both internal and external series of pathology reports. Abbreviations. ID: Identification, IHC: Immunohistochemistry, BM: Bone Marrow, FISH: Fluorescent in situ hybridization.
To assess potential weaknesses of OCR in detecting data regarding single biomarkers, we selected 50 reports from the internal series with higher image resolution and reassessed the validation metrics (Table 2). Overall, recall and F1-score metrics improved of 12.9% and 9.1%, respectively. Moreover, we assessed the NLP performance on the internal series, independently of OCR. Interestingly, we observed an incremental trend of the recall for 7 of the 8 variables analyzed (“DIAGNOSIS” 87.9% vs. 90.0%; “ID NUMBER” 92.1% vs. 96.2%, “SPECIMEN TYPE” 87.2% vs. 92.7, “IHC EXECUTION” 95.4% vs. 95.8%, “FISH EXECUTION” 93.7% vs. 97.5%, “BM EXECUTION” 92.9% vs. 97.1%, and “KI-67” 81.9% vs. 89.4%). Only the field “BIOPSY DATE” showed a slight decrease from 97.1 to 96.2%, which we considered not relevant (Table 3).
Table 2.
Comparison of ARGO performance in the whole vs. the top 50 reportsa.
| PRECISION (P) = TP/(TP + FP) | RECALL (R) = TP/(TP + FN) | F1-SCORE = 2*(P*R)/(P + R) | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All reports, N = 239 | Top reports, N = 50a | Diff | All reports, N = 239 | Top reports, N = 50a | Diff | All reports, N =239 | Top reports N = 50a | Diff | |||||||||||||
| DATA-FIELD | TP, N | FP, N | TP + FP | P, % | TP, N | FP, N | TP + FP, N | P, % | % | TP, N | FN, N | TP + FN, N | R, % | TP, N | FN, N | TP + FN, N | R, % | % | F1, % | F1, % | % |
| MYC | 20 | 0 | 20 | 100.0 | 13 | 0 | 13 | 100.0 | 0.0 | 20 | 15 | 35 | 57.1 | 13 | 4 | 17 | 69.2 | 12.1 | 72.7 | 81.8 | 9.1 |
| BCL2 | 130 | 2 | 132 | 98.5 | 28 | 0 | 28 | 100.0 | 1.5 | 130 | 55 | 185 | 71.4 | 28 | 5 | 33 | 84.8 | 13.4 | 82.2 | 91.8 | 9.6 |
| BCL6 | 115 | 1 | 116 | 99.1 | 27 | 0 | 27 | 100.0 | 0.9 | 115 | 51 | 166 | 61.5 | 27 | 5 | 32 | 84.4 | 22.9 | 75.9 | 91.5 | 15.6 |
| CD10 | 95 | 3 | 98 | 96.9 | 25 | 0 | 25 | 100.0 | 3.1 | 95 | 75 | 170 | 55.9 | 25 | 7 | 32 | 78.1 | 22.2 | 70.9 | 87.7 | 16.8 |
| CD20 | 164 | 1 | 165 | 99.4 | 36 | 0 | 36 | 100.0 | 0.6 | 164 | 51 | 215 | 76.3 | 36 | 3 | 39 | 92.3 | 16.0 | 86.3 | 96.9 | 10.6 |
| Cyclin D1 | 58 | 0 | 58 | 100.0 | 5 | 0 | 5 | 100.0 | 0.0 | 58 | 23 | 81 | 71.6 | 5 | 3 | 8 | 62.5 | -9.1 | 83.5 | 76.3 | -7.2 |
| – | Mean (std) | 1.0 (1.2) | Mean (std) | 12.9 (11.7) | Mean (std) | 9.1 (8.6) | |||||||||||||||
ARGO: Automatic Record Generator for Onco-hematology, TP: True Positive, FP: False Positive, FN: False Negative, CD: Cluster of Differentiation, Diff: difference, std: standard deviation.
aTop 50 reports (internal series) with the highest optical resolution.
Table 3.
Comparison of ARGO performance using OCR + NLP and NLP alone (internal series).
| DATA-FIELD | PRECISION (%) | RECALL (%) | F1-SCORE (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OCR + NLP | NLP | Diff | OCR + NLP | NLP | Diff | OCR + NLP | NLP | Diff | |
| DIAGNOSIS | 100.0 | 100.0 | 0.0 | 87.9 | 90.0 | 2.1 | 93.5 | 94.7 | 1.2 |
| BIOPSY DATE | 100.0 | 100.0 | 0.0 | 97.1 | 96.2 | – 0.8 | 98.5 | 98.1 | – 0.4 |
| EXAM NUMBER | 100.0 | 100.0 | 0.0 | 92.1 | 96.2 | 4.2 | 95.9 | 96.2 | 0.3 |
| SPECIMEN TYPE | 98.5 | 99.5 | 1.0 | 87.2 | 92.7 | 5.5 | 92.5 | 96.0 | 3.5 |
| IHC EXECUTION | 100.0 | 100.0 | 0.0 | 95.4 | 95.8 | 0.4 | 97.6 | 95.8 | – 1.8 |
| FISH EXECUTION | 100.0 | 100.0 | 0.0 | 93.7 | 97.5 | 3.8 | 96.8 | 97.9 | 1.1 |
| BM EXECUTION | 100.0 | 100.0 | 0.0 | 92.9 | 97.1 | 4.2 | 96.3 | 98.7 | 2.4 |
| KI-67 | 99.4 | 99.4 | 0.0 | 81.9 | 89.4 | 7.5 | 89.8 | 94.2 | 4.4 |
ARGO: Automatic Record Generator for Onco-hematology, OCR: Optical Character Recognition, NLP: Natural Language Processing, IHC: Immunohistochemistry, FISH: Fluorescent in situ Hybridization, BM: Bone Marrow, Diff: difference.
Discussion
In the study, we aimed at designing a pipeline to automate the collection of RW onco-hematological data, about lymphoma diagnoses. Leveraging well-recognized technologies as OCR and NLP we developed a new tool, called ARGO, and provided a “proof” for its reliability in generating eCRFs directly from unstructured histopathology reports. We successfully tested ARGO performance, in terms of accuracy, precision, recall and F1 score, on a multicentric cohort of 326 lymphoma cases including DLBCL, FL, and MCL from seven independent centers.
ARGO generalizability stands on three assumptions: (1) the implementation of a function fully dedicated to recognize each input template independently of the clinical features collected in the report (a new template form an additional center might be easily read by adding few NLP regular expressions to the header_function.py); (2) ARGO is able to detect the set of clinically relevant terms for the diagnosis definition by matching standard criteria, that might be tailored to every clinical field (for example, other subtypes of lympho-proliferative disorders); (3) the choice at developing eCRFs on the College of American Pathologist (CAP) templates conferring high level of standardization to the clinical content.
In comparison with other applications in oncology, ARGO confirmed super-imposable performances in data field detection6,11,14,16,21,22, while overcoming some limitations. For instance, in the work by Nguyen et al., each metric decreases as the number of classes describing a certain data field increases11. This trend is globally confirmed in our experience, and even for data fields with high number of classes, such as “SPECIMEN TYPE”, we achieved a very high precision level. Also, to potentiate the OCR performance, we created three separate thesauri for “BIOMARKERS”, “SPECIMEN TYPE” and “DIAGNOSIS”. As shown in Tanenblatt et al.22, we first included officially-recognized nomenclatures in the “biomarkers” and “diagnosis” dictionaries, referring to the “International Statistical Classification of disease and related health problems 10th revision” (ICD10, version 2019, World Health Organization) classification23. Then, we manually added synonyms, abbreviations and other uncommon expressions noticed in our set of reports. Nevertheless, ARGO failed in converting six reports as a direct consequence of OCR-based limitations in reading reports with both low-quality optical resolution and describing IHC analyses from multiple samples. At this regard, the improvement of ARGO performance observed excluding OCR from the pipeline indicates a potential pitfall, which can be easily overcome by a manual supervision by a dedicated data-entry/manager.
From a more applicative point of view, ARGO might maximize the use of clinical data in translational research by boosting the adoption of EHR. Especially in onco-hematology, the public healthcare system still lacks standardized models of RW data collection, and several gaps exist concerning how to electronically collect unstructured information. Application of a computerized approach to extract data from paper-based reports and directly populate eCRFs provides two main advantages, such as the standardization of data collection and the data integration between Institutions and research networks. Finally, our system takes advantage from two levels of personalization related to REDCap, i) the designing of graphic interfaces directly by the clinical investigators according to specific clinical endpoints; and ii) the easily population of eCRFs via Application Programming Interface (API). Therefore, ARGO appeared as a valid tool for a precise and time-saving recording of clinical data when compared to manual abstraction16. Our approach results feasible in the daily practice, facilitating consultation, filtering, and management of RW data. This step is crucial to study wide proportions of onco-hematological patients who have no access to clinical trials and support national research networking.
Main limitations of the study could be the language of histopathology reports. However, current pathology reporting systems allow the use of personalized data fields according to shared templates and translating software, e.i. as “MyMemory” software, enable the easy switch up across languages. Moreover, providing the set of regular NLP rules used into ARGO might easily address this issue simply translating from Italian to other languages all words researched in the text included in each report.
Given the accuracy and efficiency in generating correct electronic records for multicentric subsets of different lymphoma types, our approach could be tailored to additional disease models in oncology and could set the basis to validate novel biomarkers for translational research.
Methods
Data collection
Overall, 332 histopathology paper-based reports were collected between 2014 and 2020 at the Pathology Unit of the IRCCS Istituto Tumori 'Giovanni Paolo II' in Bari, Italy (239) and from six different Italian centers (93) from Unit of Hematology, Azienda Ospedaliero-Universitaria Policlinico Umberto I in Rome, Italy, Hematology, AUSL/IRCCS of Reggio Emilia in Reggio Emilia, Italy, Division of Hematology 1, AOU “Città della Salute e della Scienza di Torino” in Turin, Italy, Division of Hematology, Azienda Ospedaliero-Universitaria Maggiore della Carità di Novara in Novara, Italy, Department of Medicine, Section of Hematology, University of Verona in Verona, Italy, and Division of Diagnostic Haematopathology, IRCCS European Institute of Oncology in Milan, Italy. The internal series included 106 DLBCL, 79 FL, and 54 MCL, while the external one comprised 49 DLBCL, 24 FL, and 20 MCL.
A unique ID code was assigned to each report. According to the diagnostic criteria for each lymphoma subtype, reports included IHC results obtained from LN, EN, BM or PB specimens. Qualitative and quantitative information for IHC markers including MYC, BCL2, BCL6, CD10, CD20, Cyclin-D1 were reported. Some reports also included molecular data from FISH analysis, while some reports included either FISH results or the level tumor cell infiltration as addendum. For DLBCL, molecular classification according to the COO estimated by the Hans algorithm was also included24. Ki-67 proliferation index was also reported as quantitative value ranging from 5 to 100%.
The work was approved by the Institutional Review Board of the IRCCS Istituto Tumori “Giovanni Paolo II” hospital in Bari, Italy. All methods were carried out in accordance with relevant local regulations and after obtainment of dedicated informed consent.
Automated detection of relevant terms in paper-based reports
We aimed this step of the workflow at automating the detection of relevant terms to be extracted from the text fields of paper-based reports. ARGO exploits OCR25 and NLP26 techniques to convert images of reports into text and detect relevant words in the text based on an “ad-hoc” thesaurus.
The conversion from image to text has been implemented in Tesseract OCR© (version 4.1.1-rc2-20-g01fb). To improve conversion performance, each pathology report was firstly converted from pdf to image through Poppler library (version 0.26.5). Then, the image was translated in a grey scale of 8 bits (from 0 to 255 levels of grey).
Image transformation was developed in Python by OpenCV© software (version 4.2.0).
In ARGO, NLP techniques were adopted to automatically extract relevant terms for the disease diagnosis, to be transferred into the digitalized eCRFs. Thus, a set of NLP regular expressions were applied to extract information concerning the diagnosis, date of the report, report ID, type of the specimen, execution of BM biopsy, IHC, and FISH analyses, as well as quantitative and qualitative data of selected IHC markers (MYC, BCL2, BCL6, CD10, CD20, Cyclin-D1), COO subtypes and Ki-67 proliferation index (paragraph “ARGO function and NLP rules”).
The disease nomenclature was assigned based on the highest match between the pattern of detected biomarkers in each report and a reference pattern, as reported in the “Hematopoietic and Lymphoid Neoplasm Coding Manual guidelines” from the “Surveillance, Epidemiology and End Results (SEER) program” of the National Institute of Health27. The final diagnosis nomenclature was referred to the ICD10 classification23. Communication between ARGO and SEER official servers was flexibly dealt via API.
ARGO was developed in Flask©, version 1.1.2, the webserver was an Oracle© Linux Server 7.8 with kernel 4.14.35–1902.303.5.3.el7uek.x86_64. We used MariaDB© 5.5.68 as database. NLP algorithms were developed in Python 3.6.8. Translation from English to Italian language was dealt via API tool MyMemory© (version 3.5.0). To increase the detectability of biomarkers in the reports we also built three thesauri in Phyton with NLP regular expressions (Supplementary Appendix Source code S1 and Table S2). Despite the domain specificity of such thesauri, the technique of knowledge extraction by flexibly introducing a new thesaurus is a general feature of ARGO.
ARGO functions and NLP rules
ARGO was developed according to three functions: function_read.py, header_info.py, and params.py. Function_read.py was the main function and incorporated (1) the call to the header_info.py function to recognize the report template as input, (2) the set of NLP expressions to identify both biomarker and diagnosis description, and (3) the call to the params.py function which included two API tokens, the first to take data on biomarkers and diagnosis from the SEER database and the second provided from the REDCap project ID to allow automatic data entry. Supplementary Fig. S2A details the pseudocode to process a pathology report. ARGO embedded two main activities, namely i) the recognition of the template from the header section including the fields “BIOPSY DATE” and “ID NUMBER”, the demographical patient information (“NAME”, “SURNAME”, “DATE OF BIRTH”, “PLACE OF BIRTH”, “SEX”, and “SSN” [Social Security Number]), and the “SPECIMEN TYPE” (via header_info.py), and ii) the recognition of the “IHC MARKERS” (“POSITIVITY/NEGATIVITY” or “QUANTITY”) from the biological samples, the fields “FISH”, “DIAGNOSIS”, and “CELL OF ORIGIN” from the disease section (via function_read.py). Supplementary Fig. S2B shows an example of NLP input from the internal series. The regular expressions used to automatically recognize the header section for internal reports are reported in Table 4. Those for the external reports are detailed in Supplementary Table S3.
Table 4.
Set of NLP regular expressions embedded into the header_function.py for the internal reports.
| REDCap data label | BIOPSY DATE | ID NUMBER | SURNAME | NAME | DATE OF BIRTH | PLACE OF BIRTH | SEX | SSN | SPECIMEN TYPE | |
|---|---|---|---|---|---|---|---|---|---|---|
| REDCap data variable | nod_date_exam_req | nod_exam_num_req | pts_surname_demo | pts_name_demo | dob_demo | city_born_demo | sex_demo | ssn_demo | ln_specimen_dis | |
| REPORT TEMPLATE for internal reports | Internal | “Accettazione” or ”Pervenuto” or “Richiesta” del” or “Ricevimento” | "N. Esame" | "Cognome" | "Nome" | "Data di nascita" | "Comune di Nascita" | "Sesso" | "Codice Fiscale" | "Materiale Inviato" |
| NLP pattern | cettaz. +|ervenuto. +|ichiesta.*del. + [0–3][0–9]/[0–1][0–9]/2[0–9][0–9][0–9] | + same. *[0–3][0–9-.-d] | COGNOME.*|COGNOME.*DATA|COGNOME.*CITT | \\bNOME.*|\\bNOME.*DATA|\\bNOME.*CITT | . + asci. + [0–3][0–9]/[0–1][0–9]/[1, 2][0–9][0–9][0–9] | . + omu. + asci. + \w + | . + ess.{1,3}m | [A-Z]{6}[0–9][0–9][A-Z][0–9]{2}[A-Z][0–9]{3}[A-Z] | ate. + al. + via. + \n. + |
NLP: Natural Language Processing; ID: Identification; NA, Not Available, SSN, Social Security Number.
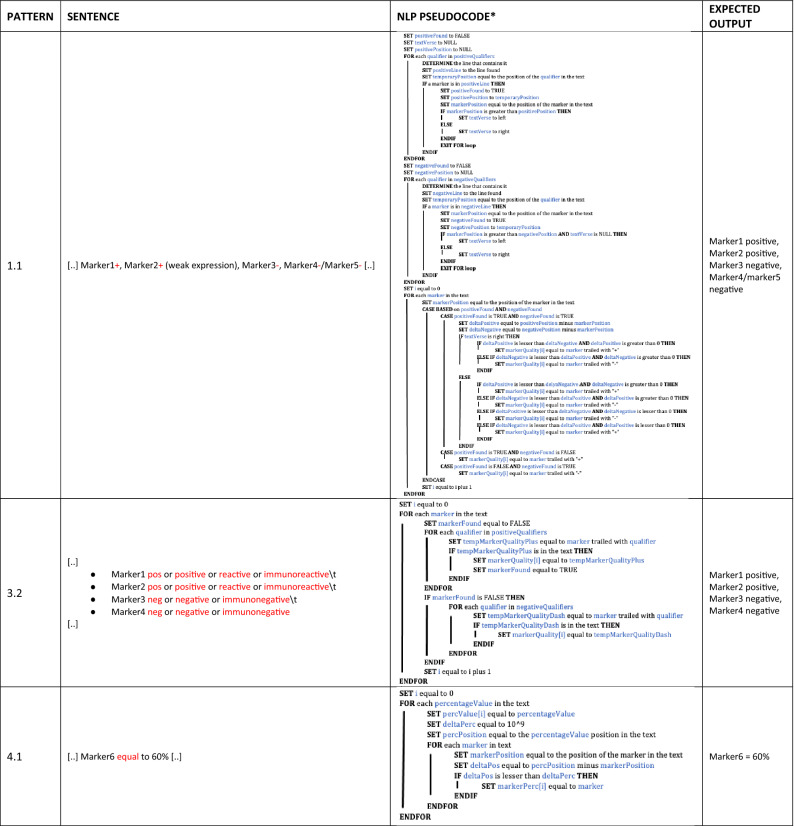
Concerning function_read.py, we identified the set of pathological description patterns according to the following four scenarios:
description of qualitative markers by symbolic qualifiers in a free text form (e.g. “ + ” for positivity and “-” for negativity);
description of qualitative markers by textual qualifiers in a free text form (e.g. “positive”, “reactive” or “immunoreactive” for positivity and “negative” or “immunonegative” for negativity);
description of both qualitative and quantitative markers by symbolic or textual qualifiers in a bullet form;
description of pure quantitative markers (as Ki-67).
Table 5 shows three representative patterns of description with their relative NLP pseudocodes and expected results. The whole set of patterns is detailed in Supplementary Table S4.
Table 5.
Representative sets of NLP rules embedded into the function_read.py for patterns 1.1, 3.2, and 4.1.
*Precondition. For each case we,
REMOVE SPACES;
REMOVE NEWLINES;
TRANSFORM "." in NEWLINES;
positiveQualifiers = [pos, positive, reactive, immunoreactive];
negativeQualifiers = [neg, negative, immunonegative].
Abbreviations. NLP: Natural Language Processing.
Data-mapping and automatic population of eCRFs
For a systematic collection of the diagnostic variables in this study, we designed dedicated eCRFs on REDcap17,18. eCRFs were suited to the synoptic templates provided and approved by the CAP. We referred to DLBCL, FL, and MCL templates28,29. The data-mapping between ARGO and the eCRFs was performed by providing the relevant data fields from the REDCap dictionary as a flexible input to the application (Supplementary Table S5). Finally, we used API technology for the automatic data entry and final upload of the information of interest into the eCRFs.
Validation metrics
ARGO performance, regarded as the level of consistency between data included in the original pathology reports and those automatically transferred into eCRFs, was assessed in terms of accuracy, precision, recall and F1 score30. To calculate each measure, we defined the cases in the following (1) true-positive: cases in which ARGO detected correctly the expected variables; (2) false-positive: cases in which ARGO detected variables even if not present in the original report; (3) true-negative: cases in which ARGO did not detect a variable not present in the original report; and (4) false-negative: cases in which ARGO failed in detecting a variable present in the original report.
Results for each data-field of internal and external series were statistically compared by a chi-square test.
Supplementary Information
Acknowledgements
Authors would thank the Scientific Board of IRCCS Istituto Tumori “Giovanni Paolo II” of Bari, Italy, Drs. G. Volpe, A. Negri, G. Gargano, A.S. Pappagallo, P. Mondelli, A. Bucci, Eng. G. Salomone and F. Pacoda (ICT of IRCCS Istituto Tumori “Giovanni Paolo II” of Bari, Italy) for their support. The study was funded by Italian Minister of Health (Grant RC2021) and the Apulia Region Grant “Tecnopolo per la medicina di precisione—CUP B84I18000540002”.
Author contributions
All authors had approved the submitted version and had agreed both to be personally accountable for the author's own contributions and to ensure that questions related to the accuracy or integrity of any part of the work. Substantial contributions to conception or design of the work: GMZ, VC, SC, AG, and SC. Drafting of the study: GMZ, VC, SC, AG, and SC. Creation of the tool: GMZ and VC. Acquisition, analysis, or interpretation of data for the work: all authors. Supervision: ALG, AG, and SC.
Funding
Tecnopolo per la medicina di precisione, B84I18000540002, Italian Minister of Health, RC2021.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-03204-z.
References
- 1.WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. (IARC Press, 2017).
- 2.Campo E, et al. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood. 2011;117:5019–5032. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khozin S, Blumenthal GM, Pazdur R. Real-world data for clinical evidence generation in oncology. J. Natl. Cancer Inst. 2017;109:1–5. doi: 10.1093/jnci/djx187. [DOI] [PubMed] [Google Scholar]
- 4.Zaccaria, G. M. et al. Applying data warehousing to a phase III clinical trial from the Fondazione Italiana Linfomi (FIL) ensures superior data quality and improved assessment of clinical outcomes. JCO Clin. Cancer Informatics 1–15 (2019). 10.1200/CCI.19.00049. [DOI] [PMC free article] [PubMed]
- 5.Zong, N. et al. Developing an FHIR-Based Computational Pipeline for Automatic Population of Case Report Forms for Colorectal Cancer Clinical Trials Using Electronic Health Records. JCO Clin. Cancer Inform. 201–209 (2020). 10.1200/cci.19.00116. [DOI] [PMC free article] [PubMed]
- 6.Xie F, et al. Characterization of patients with advanced chronic pancreatitis using natural language processing of radiology reports. PLoS ONE. 2020;15:1–13. doi: 10.1371/journal.pone.0236817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley JM, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J. Pathol. Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP) Nat. Protoc. 2019;14:3426–3444. doi: 10.1038/s41596-019-0227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkataraman GR, et al. FasTag: Automatic text classification of unstructured medical narratives. PLoS ONE. 2020;15:1–18. doi: 10.1371/journal.pone.0234647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryu, B. et al. Transformation of Pathology Reports into the Common Data Model with Oncology Module: Use Case for Colon Cancer (Preprint). J. Med. Internet Res.22, (2020). [DOI] [PMC free article] [PubMed]
- 11.Nguyen, A. N., Moore, J., O’Dwyer, J. & Philpot, S. Assessing the utility of automatic cancer registry notifications data extraction from free-text pathology reports. AMIAAnnu. Symp. proc. AMIA Symp.2015, 953–962 (2015). [PMC free article] [PubMed]
- 12.Spasić I, Livsey J, Keane JA, Nenadić G. Text mining of cancer-related information: Review of current status and future directions. Int. J. Med. Inform. 2014;83:605–623. doi: 10.1016/j.ijmedinf.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Lin FPY, Pokorny A, Teng C, Epstein RJ. TEPAPA: A novel in silico feature learning pipeline for mining prognostic and associative factors from text-based electronic medical records. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser AP, et al. Automated extraction of grade, stage, and quality information from transurethral resection of bladder tumor pathology reports using natural language processing. JCO Clin. Cancer Inf. 2018 doi: 10.1200/po.17.00128.2019.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odisho AY, et al. Natural language processing systems for pathology parsing in limited data environments with uncertainty estimation. JAMIA Open. 2020;3:431–438. doi: 10.1093/jamiaopen/ooaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odisho AY, et al. Automating the capture of structured pathology data for prostate cancer clinical care and research. JCO Clin. Cancer Inf. 2019 doi: 10.1200/cci.18.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris PA, et al. Research electronic data capture (REDCap)-A metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris PA, et al. The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moon, S. et al. Medical concept intersection between outside medical records and consultant notes: A case study in transferred cardiovascular patients. Proc. - 2017 IEEE Int. Conf. Bioinforma. Biomed. BIBM 20172017, 1495–1500 (2017).
- 20.Qader, W. A. & Ameen, M. M. Diagnosis of Diseases from Medical Check-up Test Reports Using OCR Technology with BoW and AdaBoost algorithms. Proc. 5th Int. Eng. Conf. IEC 2019 205–210 (2019). 10.1109/IEC47844.2019.8950605.
- 21.Hanauer DA, Miela G, Chinnaiyan AM, Chang AE, Blayney DW. The registry case finding engine: an automated tool to identify cancer cases from unstructured, free-text pathology reports and clinical notes. J. Am. Coll. Surg. 2007;205:690–697. doi: 10.1016/j.jamcollsurg.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Tanenblatt, M., Coden, A. & Sominsky, I. The ConceptMapper approach to named entity recognition. Proc. 7th Int. Conf. Lang. Resour. Eval. Lr. 2010 546–551 (2010).
- 23.World Health Organization. Classification of diseases (ICD). https://www.who.int/classifications/classification-of-diseases.
- 24.Hans CP, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 25.Schantz, H. F. The history of OCR, optical character recognition. (Recognition Technologies Users Association, 1982).
- 26.Eisenstein, J. Introduction to Natural Language Processing. Adaptive Computation and Machine Learning serie. (MIT Press, 2019).
- 27.Ruhl, J., Adamo, M. P., Dickie, L. & Negoita, S. Hematopoietic and Lymphoid Neoplasm Coding Manual. https://seer.cancer.gov/tools/heme/Hematopoietic_Instructions_and_Rules.pdf (2020).
- 28.Duncavage E, et al. Template for reporting results of biomarker testing of specimens from patients with diffuse large B-cell lymphoma, not otherwise specified. Arch. Pathol. Lab. Med. 2016;140:1225–1227. doi: 10.5858/arpa.2015-0418-CP. [DOI] [PubMed] [Google Scholar]
- 29.Ellis, D. W. Protocol for the examination of specimens from patients with hodgkin lymphoma. 1–15 (2013).
- 30.Resnik, P. & Lin, J. Evaluation of NLP Systems. in The Handbook of Computational Linguistics and Natural Language Processing (eds. Clark, A., Fox, C. & Lappin, S.) 271–295 (Wiley-Blackwell, 2010). doi:10.1002/9781444324044.ch11.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.