Abstract
Purpose
Multiple studies demonstrate MRI-targeted biopsy detects more clinically significant cancer than systematic biopsy, however some clinically significant cancers are detected by systematic biopsy only. While these events are rare, we sought to perform a retrospective analysis of these cases to ascertain the reasons that MRI-targeted biopsy missed clinically significant cancer, which was subsequently detected on systematic prostate biopsy.
Methods
Patients were enrolled in a prospective study comparing cancer detection rates by transrectal MRI-targeted fusion biopsy and systematic 12-core biopsy. Patients with an elevated PSA, abnormal digital rectal exam, or imaging findings concerning for prostate cancer underwent prostate MRI and subsequent MRI-targeted and systematic biopsy in the same setting. The subset of patients with grade group (GG) ≥3 cancer found on systematic biopsy and GG≤2 cancer (or no cancer) on MRI-targeted biopsy were classified as MRI-targeted biopsy misses. A retrospective analysis of the MRI and MRI-targeted biopsy real-time screen captures determined the cause of MRI-targeted biopsy miss. Multivariable logistic regression analysis compared baseline characteristics of patients with MRI-targeted biopsy misses to GG-matched patients whose clinically significant cancer was detected by MRI-targeted biopsy.
Results
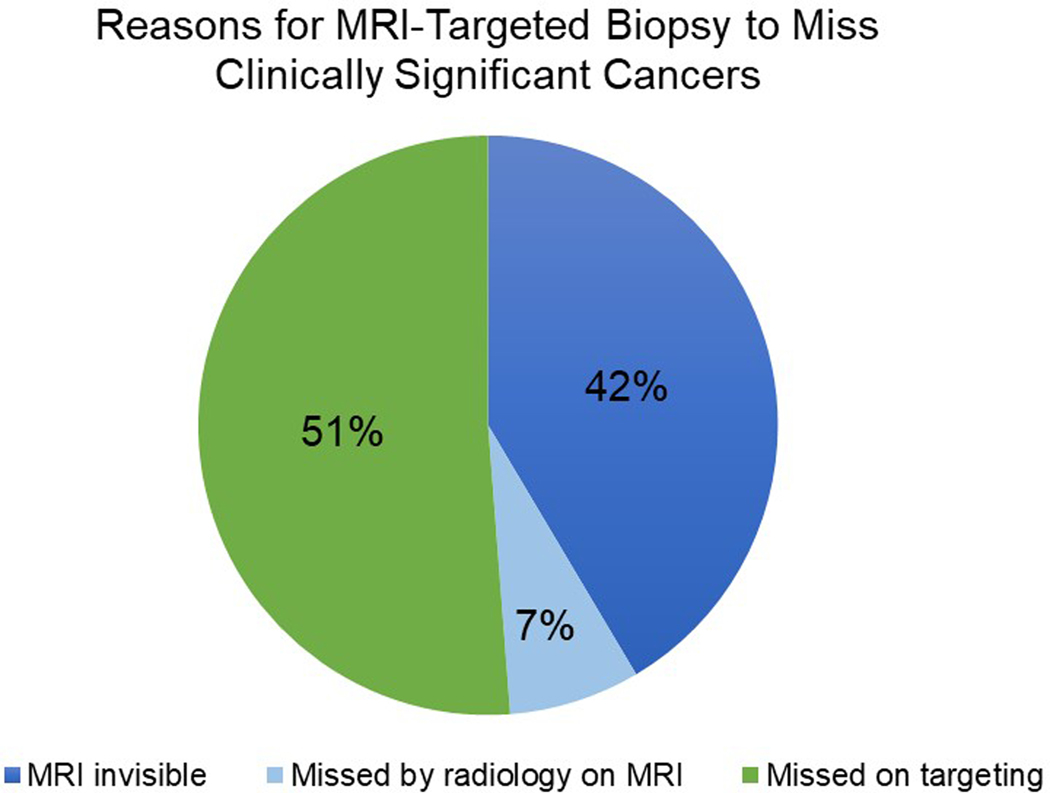
Over the study period of 2007 to 2019, 2103 patients met study inclusion criteria and underwent combined MRI-targeted and systematic prostate biopsies. 41 (1.9%) men were classified as MRI-targeted biopsy misses. Most MRI-targeted biopsy misses were due to errors in lesion targeting (n=21, 51.2%), followed by MRI-invisible lesions (n=17, 40.5%), and MRI lesions missed by the radiologist (n=3, 7.1%). On logistic regression analysis, lower PI-RADS score was associated with having clinically significant cancer missed on MRI-targeted biopsy.
Conclusion
While uncommon, most MRI-targeted biopsy misses are due to errors in lesions targeting, which highlights the importance of accurate co-registration and targeting when using software-based fusion platforms. Additionally, some patients will harbor MRI-invisible lesions which are un-targetable by MRI-targeted platforms. The presence of a low PI-RADS score despite a high PSA is suggestive of harboring an MRI-invisible lesion.
Background
Multiple recent high-quality studies have demonstrated that MRI-targeted biopsy results in improved detection of clinically significant prostate cancer relative to systematic biopsy. 1–4 However, despite these studies which demonstrate that most clinically significant cancers are detected by multiparametric MRI, evidence also demonstrates that MRI-targeted biopsy alone still misses clinically significant cancers. 1, 2, 5
Despite this finding, few investigators have attempted to define the cause of these “MRI misses.” Previous data has clearly demonstrated that 7% of grade group (GG)≥3 cancers can be missed by MRI alone, and approximately 2% of clinically significant cancers are detected by systematic biopsy alone when MRI-targeted and systematic biopsy are performed in the same setting. 2, 6, 7
While MRI-targeted biopsy can miss clinically significant cancers, the reasons for these misses such as radiologist misinterpretation, poor targeting during MRI-targeted biopsy, or lesion MRI-invisibility can all contribute to these MRI-targeted biopsy-missed diagnosis of clinically significant cancer. Additionally, patient factors such as Prostate Imaging–Reporting and Data System (PI-RADS) score, index-lesion size, or prostate volume, may be associated with having a MRI-targeted biopsy miss.8 Before MRI-targeted biopsy can replace systematic biopsy in reliably detecting and ruling-out clinically significant cancer, it is important to define the reasons for MRI-targeted biopsy and identify patients at risk for harboring an undetected, clinically significant lesion.
In this study we perform a post hoc analysis of the patients from the Trio study 2 in order to define the causes for MRI-targeted biopsy missing clinically significant cancer. Furthermore, we describe baseline patient characteristics that may predict having an MRI-targeted biopsy missed lesion.
Methods
Study Population
The detailed methods and results of the Trio study have been previously published.2 In brief, patients with an elevated prostate specific antigen (PSA) (>4.0 ng/mL) or abnormal digital rectal exam were enrolled in a nationally registered clinical trial (NCT00102544), and underwent a multiparametric 3T MRI with an endorectal coil (when possible) using technical standards from PI-RADS v 2.1.9 In MRI scans with motion artifact, a repeat sequences was always obtained. Men found to have any MRI prostate lesions concerning for prostate cancer, based on Likert suspicion scale or PI-RADS ≥2, underwent both MRI-targeted and systematic 12-core extended sextant biopsies in the same setting.10, 11 Men without MRI suspicion of prostate cancer were excluded from the analysis.
Parent Study Biopsy Protocol
MRI-targeted biopsies were performed using the UroNav fusion biopsy software (Philips, USA). Image targets were assigned to lesions reported by the reviewing radiologist on MRI preoperatively. At the time of biopsy, the radiologist or urologist performing the biopsy obtained 2 biopsy cores, axial and sagittal, of each assigned target. Systematic biopsies were then performed in the same setting by a second physician who was instructed to ignore any possible hemorrhage tracts. All biopsies were performed transrectally and within 1 year of the MRI.
Post Hoc Study Design
We performed a post hoc analysis on all patients in the parent study who had clinically significant cancer detected on systematic biopsy but not detected by MRI-targeted biopsy. Clinically significant cancer was defined as having GG ≥3 in this post-hoc analysis and the parent study.2 These patients, defined as “MRI-targeted biopsy misses,” then underwent an unblinded review of their prebiopsy MRI and UroNav biopsy tracking recording. Upon re-review, one expert genitourinary radiologist used the location of the tracked positive systematic biopsy core to identify the location of the prostate cancer within the MRI. This was one of the two radiologists who read MRIs in the original parent study. The radiologist then retrospectively identified if a lesion was missed due to one of three reasons: a lesion that was missed on targeted biopsy but seen on MRI (Targeting error, Figure 1), a lesion that was initially undetected on MRI, but MRI-visible on re-review (Radiology Miss, Figure 2), or if no lesion was seen even retrospectively (MRI-invisible, Figure 3). Each of these possibilities was recorded for each patient and reported here. The radiologist also assigned each patient a maximum PI-RADS v2 score (if one had not been previously assigned) and reported the size and location of the missed lesions that contained GG≥3 cancer. Lesion size could not be described among patients with MRI-invisible lesions.
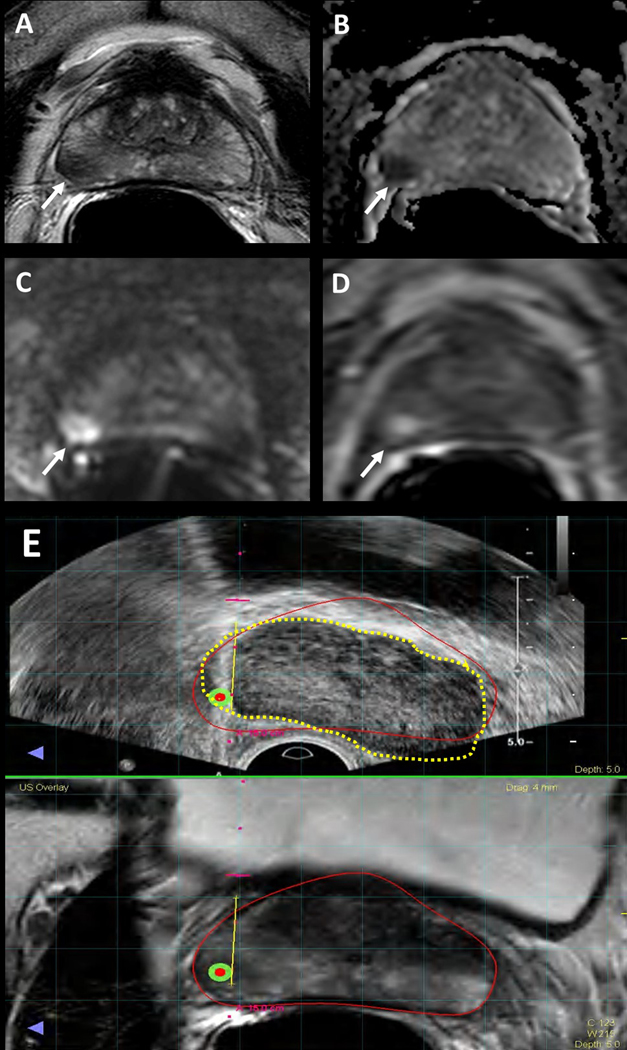
Figure 1.
Multiparametric MRI image example of clinically significant cancer that was missed due to MRI-targeted biopsy targeting error. 56-year-old male with a serum PSA of 5.9ng/ml. Axial T2-Weighted (T2W) MRI shows a hypointense lesion in the right mid-peripheral zone (arrow) (A), the lesion shows diffusion restriction on ADC map (B) and b2000 DW MRI (C) and early enhancement on DCE MRI (D) (arrows). Targeted biopsy revealed Gleason 3+4 prostate cancer in this lesion, whereas systematic biopsy yielded Gleason 4+4 prostate adenocarcinoma in the right peripheral zone. (E) Screen captures of the TRUS/MRI fusion guided biopsy procedure (E) demonstrates a registration error between MRI (inked in red) vs. TRUS (inked in dashed yellow), which is likely the reason for the under-sampling of the right mid peripheral zone lesion during TRUS/MRI fusion guided biopsy.
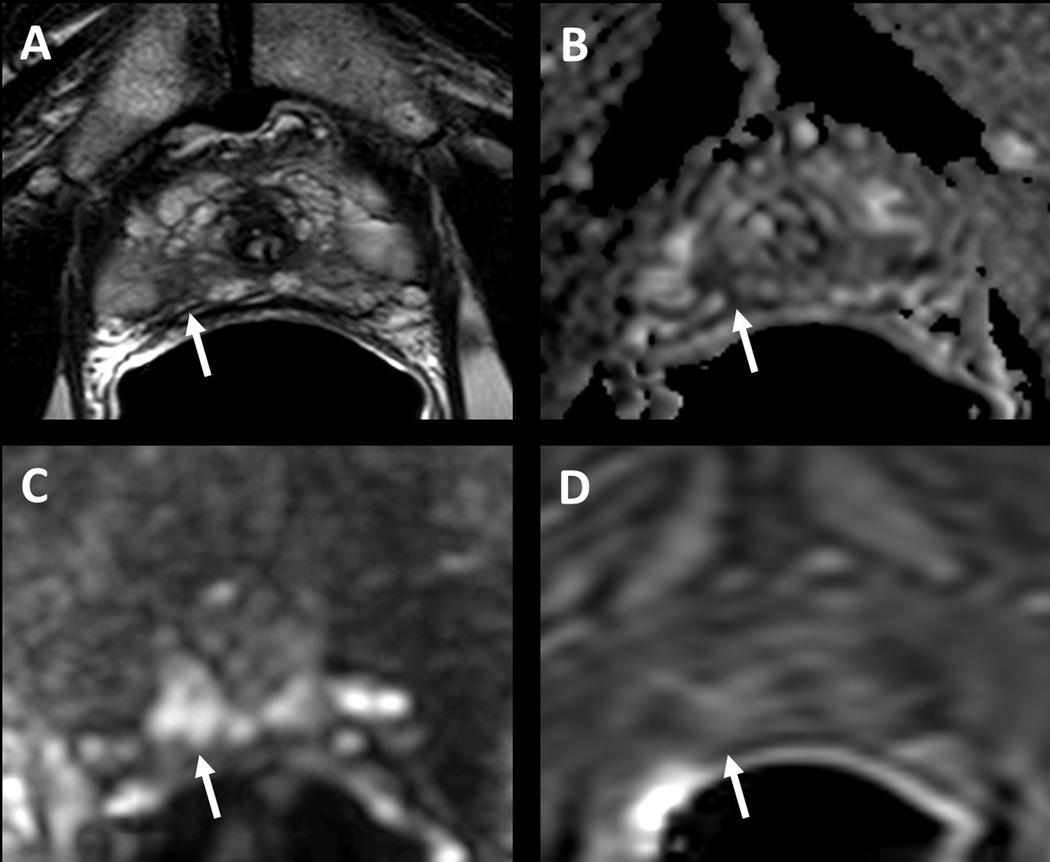
Figure 2.
Multiparametric MRI image example of clinically significant cancer missed by initial radiologist read. 71-year-old male with a serum PSA of 14.2ng/ml. Axial T2W MRI shows a hypointense lesion in the right apical peripheral zone (arrow) (A), the lesion shows diffusion restriction on ADC map (B) and b2000 DW MRI (C) and early enhancement on DCE MRI (D) (arrows). This lesion was not reported in the prospective radiology read-out, however systematic biopsy yielded Gleason 4+3 prostate adenocarcinoma in the right apical peripheral zone.
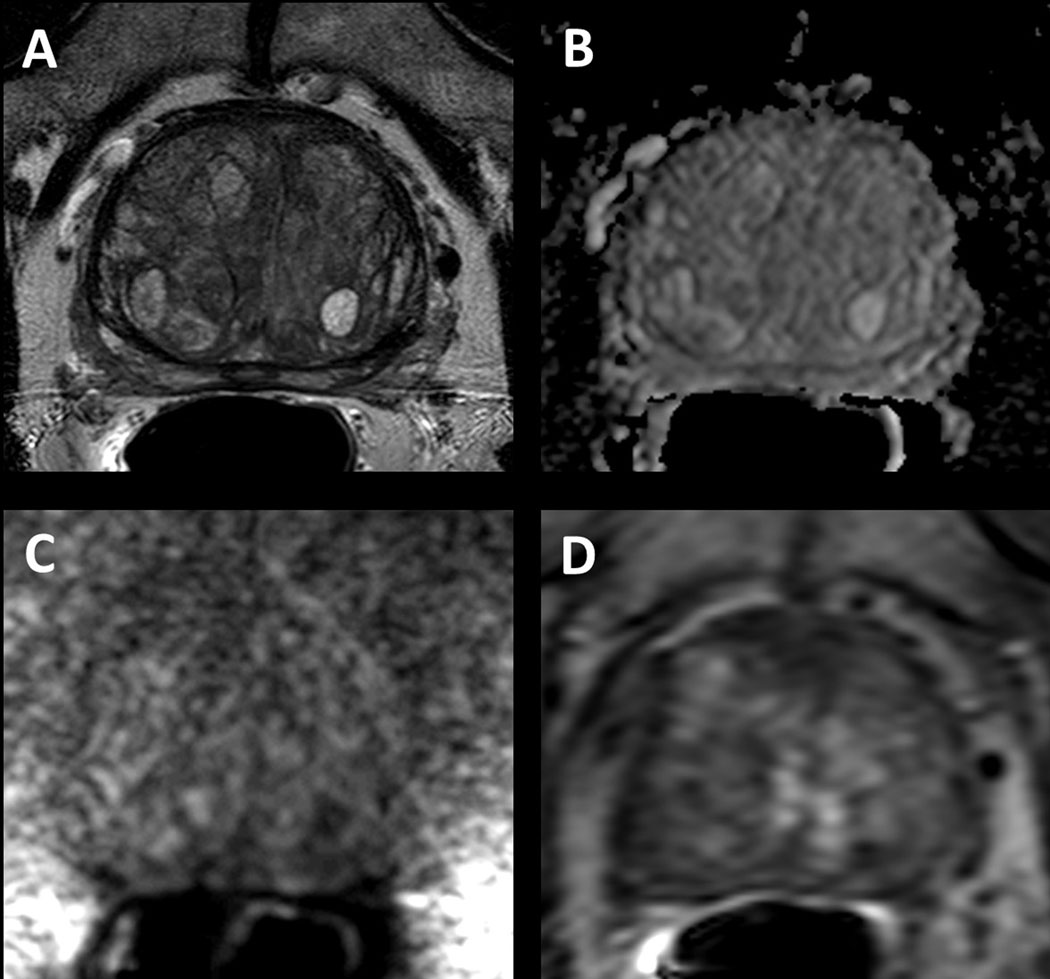
Figure 3.
Multiparametric MRI image example of clinically significant cancer that was MRI-invisible. 72-year-old male with a serum PSA of 6.55ng/ml. Systematic biopsy revealed Gleason 4+4 prostate adenocarcinoma in the right base lateral region of the prostate, however axial T2W MRI (A), ADC map (B), b2000 DW MRI (C) and DCE MRI (D) does not demonstrate an MRI visible disease focus in the right base of the prostate.
systematic biopsy yielded Gleason 4+3 prostate adenocarcinoma in the right apical peripheral zone.
Statistical Analysis
Baseline differences between missed and not missed patients were compared using Mann-Whitney U tests and chi-square tests for continuous and categorical variables, respectively. Univariate and multivariate logistic regression identified potential variables that predicted having an MRI-targeted biopsy miss. Two-sided p-values of <0.05 were considered statistically significant. All statistical analyses were performed using R version 4.0.2. Comparative analysis was not performed between characteristics of patients with different reasons for MRI-targeted biopsy misses due to limited cohort size.
Results
Study population
Over the study period of 2007 to 2019, 2,103 patients met study inclusion criteria and underwent combined MRI-targeted and systematic prostate biopsies. In the parent study, 466 (22.5%) patients were ultimately diagnosed with GG≥3,2 and 41 (1.9%) patients had GG≥3 cancer that was detected by only systematic biopsy and missed by MRI-targets biopsy. These 41 men represented the study population for the post-hoc analysis (Table 1). Wholemount correlation was not available for all patients, however, for patients whose information was available(56%, n=23), this is presented in supplemental Table 1.
Table 1:
Baseline characteristics among patients with MRI-detected and MRI-targeted biopsy missed clinically significant cancer
| All patients | GG ≥ 3 on combined biopsy | ||||
|---|---|---|---|---|---|
| N = 2,103 | N | MRI-detected n = 425 | MRI-targeted biopsy missed n = 41 | p-value1 | |
| Age | 64.0 (58.0, 69.0) | 466 | 66.0 (61.0– 71.0) | 65.0 (58.0– 71.0) | 0.3 |
| Race | 466 | 0.7 | |||
| White | 1,653 (79%) | 313 (74%) | 28 (68%) | ||
| Black | 289 (14%) | 72 (17%) | 11 (27%) | ||
| Asian | 75 (3.6%) | 22 (5.2%) | 1 (2.4%) | ||
| Hispanic | 23 (1.1%) | 4 (0.9%) | 0 (0%) | ||
| Other | 39 (1.9%) | 6 (1.4%) | 1 (2.4%) | ||
| Unknown | 24 (1.1%) | 8 (1.9%) | 0 (0%) | ||
| PSA | 466 | <0.001 | |||
| Median (25%–75%) | 6.7 (4.6–10.2) | 10.5 (6.3–17.6) | 7.8 (4.9–11.8) | ||
| Minimum-Maximum | 0.3–231.6 | 1.1–231.6 | 2.6–26.2 | ||
| Clinical Stage (0=no cancer detected) | 466 | 0.1 | |||
| 0 | 791 (38%) | — | — | ||
| T1c | 1,127 (54%) | 319 (75%) | 39 (95%) | ||
| T2a | 155 (7.4%) | 83 (20%) | 2 (4.9%) | ||
| T2b | 13 (0.6%) | 9 (2.1%) | 0 (0%) | ||
| T2c | 16 (0.8%) | 13 (3.1%) | 0 (0%) | ||
| T4 | 1 (<0.1%) | 1 (0.2%) | 0 (0%) | ||
| Prior Biopsy | 466 | 0.2 | |||
| Naive | 436 (21%) | 117 (28%) | 7 (17%) | ||
| Negative | 873 (42%) | 142 (33%) | 13 (32%) | ||
| Positive | 794 (38%) | 166 (39%) | 21 (51%) | ||
| Volume of Prostate on MRI | 451 | <0.001 | |||
| Median (25%–75%) | 51.0 (37.0–71.0) | 42.0 (33.0–58.0) | 50.0 (40.0–61.0) | ||
| Minimum-Maximum | 5.7–420.0 | 13.0–178.0 | 21.7–260.0 | ||
| Number of targets on MRI | 2.0 (1.0– 3.0) | 466 | 3.0 (2.0– 4.0) | 2.0 (1.0– 3.0) | 0.006 |
| GG of MRI-targeted Biopsy | 466 | ||||
| No cancer | 1019 (49%) | — | 9 (22%) | ||
| GG1 | 289 (15%) | — | 8 (20%) | ||
| GG2 | 370 (18%) | — | 24 (59%) | ||
| Highest PI-RADS | 215 | <0.001 | |||
| 2 | 52 (7.0%) | 3 (1.7%) | 2 (4.9%) | ||
| 3 | 89 (12%) | 3 (1.7%) | 5 (12%) | ||
| 4 | 356 (48%) | 48 (28%) | 22 (54%) | ||
| 5 | 249 (33%) | 120 (69%) | 12 (29%) | ||
| Number of cores on FBx | 4.0 (2.0– 6.0) | 466 | 6.0 (4.0– 8.0) | 4.0 (2.0– 6.0) | 0.006 |
| Number of Positive Cores on FBx | 1.0 (0.0– 3.0) | 466 | 4.0 (2.0– 6.0) | 2.0 (1.0– 2.0) | <0.001 |
| Number of Cores on SBx | 12.0 (12.0– 12.0) | 466 | 12.0 (12.0– 12.0) | 12.0 (12.0– 12.0) | 0.032 |
| Number of Positive Cores on SBx | 1.0 (0.0– 3.0) | 466 | 4.0 (1.0– 6.0) | 3.0 (2.0– 5.0) | 0.3 |
| Total # of Cores | 16.0 (15.0– 18.0) | 466 | 18.0 (16.0– 20.0) | 16.0 (14.0– 18.0) | 0.005 |
| Total % Positive Cores | 11.1 (0.0– 31.8) | 466 | 41.4 (26.7– 61.1) | 31.2 (20.0– 43.8) | 0.001 |
Statistical tests performed: Mann Whitney U test; chi-square test of independence
Differences between patients with MRI-targeted biopsy misses and accurate diagnoses by MRI-targeted biopsy
Men with lesions missed by MRI-targeted biopsy tended to have lower PSA (7.8 ng/mL (IQR: 4.9–11.8) vs 10.5 ng/mL, (IQR: 6.3–17.6); p <0.001), larger prostate volumes (median 50.0 ml (IQR: 40.0–61.0) vs 42.0 (IQR: 33.0–58.0); p <0.001), a lower maximum PI-RADS score (p<0.001), and fewer median biopsy targets (3.0 vs 2.0; p<0.001 ).
Univariate and Multivariate logistic regression of predictors for having an MRI-targeted biopsy miss
On univariate logistic regression, lower baseline PSA(ng/mL) (OR: 0.092, CI 0.87–0.97; p = 0.006), lower maximum PI-RADS score (1-point) (OR: 0.35, CI 0.21–0.55; p <0.001), fewer number of MRI targets (OR: 0.82, CI 0.71–0.94; p=0.3), and higher prostate volume on MRI (mL)(OR: 1.05, CI 1.0–1.11; p=0.006) predicted having any kind of MRI-targeted biopsy missed lesion (Table 2). On multivariate regression, lower maximum PI-RADS score was independently predictive for MRI-targeted biopsy missing a clinically significant lesion (OR: 0.42, CI 0.24–0.70; p<0.001).
Table 2:
Logistic regression predicting having a clinically significant cancer missed by MRI-targeted biopsy
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| OR1 | 95% CI1 | p-value | OR1 | 95% CI1 | p-value | |
| Age | 0.98 | 0.94, 1.02 | 0.27 | — | — | — |
| PSA | 0.92 | 0.87, 0.97 | 0.006 | 0.95 | 0.88, 1.00 | 0.089 |
| Race | ||||||
| White | — | — | — | — | — | — |
| Black | 1.49 | 0.67, 3.11 | 0.3 | — | — | — |
| Asian | 0.49 | 0.03, 2.47 | 0.49 | — | — | — |
| Other | 1.79 | 0.09, 11.0 | 0.59 | — | — | — |
| PI-RADS score (2–5) | 0.35 | 0.21, 0.55 | <0.001 | 0.42 | 0.24, 0.70 | 0.001 |
| Volume of Prostate on MRI | 1.06 | 1.01, 1.11 | 0.023 | 1.06 | 0.99, 1.15 | 0.092 |
| Number of Targets on targeted biopsy | 0.82 | 0.71, 0.94 | 0.007 | 0.99 | 0.84, 1.15 | 0.9 |
| Prior Biopsy | 1.85 | 0.84, 4.66 | 0.15 | — | — | — |
Reasons for MRI-targeted biopsy miss on Retrospective Review
The majority of MRI targeted biopsy misses were due to errors in lesion targeting (21/41, 51.2%), followed by MRI-invisible lesions (17/41, 41.4%) and MRI lesions missed by the radiologist (3/41, 7.3%) (Figure 4). Among the 41 patients with MRI-targeted biopsy miss, we described patient characteristics among each of the reasons for MRI-targeted biopsy miss (Table 3).
Figure 4.
Proportion of each reason for MRI-targeted biopsy missing clinically significant cancer
Table 3:
Characteristics among patients with lesions undetected by MRI-targeted
| N | Missed by Radiologist n = 3 | Missed on Targeting n = 21 | MRI-Invisible n = 17 | ||
|---|---|---|---|---|---|
| Age | 41 | 71.0 (68.0–73.5) | 65.0 (58.0–71.0) | 65.0 (59.0–68.0) | |
| Race | 41 | ||||
| White | 3 (100%) | 16 (76%) | 9 (53%) | ||
| Black | 0 (0%) | 4 (19%) | 7 (41%) | ||
| Asian | 0 (0%) | 0 (0%) | 1 (5.9%) | ||
| Other | 0 (0%) | 1 (4.8%) | 0 (0%) | ||
| Prior Bx Hx | 41 | ||||
| Naive | 1 (33%) | 3 (14%) | 3 (18%) | ||
| Negative | 1 (33%) | 7 (33%) | 5 (29%) | ||
| Positive | 1 (33%) | 11 (52%) | 9 (53%) | ||
| PSA | 41 | 7.2 (5.7–10.2) | 7.8 (5.3–11.6) | 8.0 (4.9–11.9) | |
| DRE Clinical Stage | 41 | ||||
| NA | 0 (0%) | 1 (4.8%) | 1 (5.9%) | ||
| T1c | 3 (100%) | 20 (95%) | 15 (88%) | ||
| T2a | 0 (0%) | 0 (0%) | 1 (5.9%) | ||
| MRI volume (ml) | 41 | 49.0 (43.5–56.9) | 50.0 (40.0–59.0) | 55.0 (46.0–66.0) | |
| # of targets | 41 | 2.0 (1.5–3.5) | 2.0 (1.0–3.0) | 3.0 (2.0–4.0) | |
| # of cores on targeted Bx | 41 | 4.0 (3.0–7.0) | 4.0 (2.0–6.0) | 6.0 (4.0–8.0) | |
| # positive cores on targeted Bx | 41 | 1.0 (0.5–3.0) | 2.0 (1.0–3.0) | 2.0 (0.0–2.0) | |
| # of Random cores | 41 | 12.0 (12.0–12.0) | 12.0 (12.0–12.0) | 12.0 (12.0–12.0) | |
| # positive Random cores | 41 | 4.0 (3.0–6.0) | 3.0 (2.0–5.0) | 2.0 (1.0–5.0) | |
| PI-RADS of visible MRI index lesion | 41 | ||||
| 2 | 0 (0%) | 0 (0%) | 2 (12%) | ||
| 3 | 1 (33%) | 1 (4.8%) | 3 (18%) | ||
| 4 | 1 (33%) | 12 (57%) | 9 (53%) | ||
| 5 | 1 (33%) | 8 (38%) | 3 (18%) | ||
| Size of MRI index lesion (cm) | 41 | 1.2 (1.1–1.4) | 1.1 (0.9–1.8) | 1.0 (0.8–1.2) | |
| Size of missed lesion (cm) | 24 | 0.6 (0.6–0.9) | 1.1 (0.9–1.3) | — | |
| Location of missed lesion | 24 | ||||
| Zone | Peripheral | 2 (67%) | 18 (86%) | — | |
| Transition | 1 (33) | 3 (14%) | — | ||
| Apex-Base | Apical | 2 (67%) | 6 (29%) | — | |
| Apical-mid | 0 | 5 (24%) | — | ||
| Base | 0 | 1 (4.8%) | — | ||
| Mid | 1 (33%) | 5 (24%) | — | ||
| Mid-base | 0 | 4 (19%) | — | ||
| Laterality | Left | 2 (67%) | 7 (33%) | — | |
| Midline | 0 | 3 (14%) | — | ||
| Right | 1 (33%) | 11 (52%) | — | ||
| Anterior-Posterior | Anterior | 1 (33%) | 6 (29%) | — | |
| Posterior | 2 (67%) | 15 (71%) | — | ||
Statistics presented: Median (25%–75%); n (%)
Characteristics of men with MRI-Invisible lesions
Prostate volume was higher in patients with MRI-invisible lesions than patients with other reasons for miss ((52.5mL (IQR: 44.0mL-67.0mL) vs 50.0mL (IQR: 35.0–59.0) for patients missed by radiologist and 50.0mL (IQR: 35.0–59.0) for patients missed on targeting). Additionally, the rate of clinically significant MRI-Invisible lesions was significantly higher among Black patients than patients of other racial backgrounds (8.5% for Black patients vs. 2.6% for non-Black patients; p = 0.009). There was one patient with hemorrhage artifact leading to the lesion being missed.
Characteristics of men with Targeting Errors
Patients with targeting errors leading to MRI-targeted biopsy miss had lesions predominantly located in the peripheral zone, apex, and posterior gland. Patients with targeting errors additionally had larger missed lesion sizes (1.1 cm (IQR: 0.9–1.3) vs 0.6 cm (IQR: 0.6–0.9)) and higher PI-RADS scores than patients with lesions missed by radiologists.
Characteristics of men with lesions missed by radiologist
In addition to having smaller missed-lesion sizes, men with lesions missed by radiologists were older (71.0 (IQR: 68–73.5) vs. 65 (IQR: 58.0–71.0) and 65 (IQR: 59.0–68.0)), and had lower PSAs (7.2 ng/mL (IQR: 5.7–10.2) vs 7.8 ng/mL (IQR: 5.3–11.6) and 8.0 ng/mL (IQR: 4.9–11.9)), when compared to patients with targeting miss or MRI-invisible lesions.
Discussion
High-quality studies in recent years demonstrate the superiority of MRI-targeted biopsy in detecting clinically significant prostate cancers compared to systematic biopsy alone 2. However, MRI-targeted biopsy does not capture all clinically significant lesions which leads to the potential for missed detection of aggressive disease. Notably, there is heterogeneity in the definition of clinically significant cancer across the literature. The definition in this study was based on identifying the most clinically important targeted biopsy failures (i.e. GG≥3). The most recently pooled negative predictive value (NPV) of MRI-targeted fusions biopsy for detecting clinically significant disease is 86–96% 12. While the NPV of MRI-targeted biopsy is high, prior publications have shown up to 8.8% of clinically significant cancers can be missed with the use of MRI targeted biopsy alone.13 Little research has been dedicated to defining the reasons for these MRI-targeted biopsy failures.
Here we define the possible causes of MRI-targeted biopsy failure: MRI-invisible lesions, radiology missed detection of lesions, and targeting error. We found targeting errors are a primary limitation of MRI-targeted biopsy for detecting clinically significant cancer. 14 In addition, MRI-invisible lesions can still contain clinically significant cancer, representing 42% of the MRI-targeted biopsy misses. Furthermore, men with Black race, larger prostate volume, or a low PI-RADS score despite an elevated PSA were more likely to have MRI-targeted biopsy misses suggesting that increased suspicion of an MRI-targeted biopsy miss should be considered in these patient groups. These patients may have increased benefit from combined biopsy or increasing the number of biopsy cores obtained at each MRI target. 15
We found that targeting error was the most common reason for MRI-targeted biopsy misses. This finding is consistent with a previous study by Coker et al who reported that targeting error accounted for 42% (15/35) of patients who had any grade prostate cancer detected by systematic biopsy but missed by MRI fusion-guided biopsy. Notably, only 1/35 of the MRI-targeted biopsy missed cancers in their cohort ultimately had GG≥3. 16 The current study expanded on these results by examining a larger cohort of patients who all had clinically significant MRI-targeted biopsy missed cancers, and this study showed that these previously identified reasons for missed lesions can be responsible for the misdiagnosis of aggressive disease.
Targeting errors are predominantly due to misregistration of the fusion biopsy platforms which currently report a margin for registration error of 2 −3mm.17 This error, which causes a misalignment of the superimposed MRI image over the TRUS image, may be due to differences in prostate shape between MRI and TRUS which may arise from prostate deformation by the endorectal coil or TRUS probe, glandular changes between the time of MRI and biopsy, or variability in bladder fullness between images. 18, 19 To address this common limitation, biopsy protocols combining visual registration and image-fusion platforms now seek to improve cancer detection rate by minimizing the effect of registration errors. 20
In this study, patients with MRI-targeted biopsy misses also had significantly larger volume prostates than patients who had accurately detected cancers. These results corroborate a prior study that identified larger prostate volume predicted greater biopsy needle tip deflection thus affecting biopsy accuracy.21
The second most common reason for MRI-targeted biopsy miss was MRI-invisible lesions. MRI-invisible lesions may generally represent more low-grade lesions 2; however, our results indicated that a relatively high frequency of clinically significant MRI-targeted biopsy missed lesions are due to MRI-invisibility. Identifying these patients is critical because, among clinically significant cancers, MRI-invisible lesions are associated with a higher risk of biological aggressiveness. 22–24 Histopathological and molecular features associated with MRI-invisibility include Gleason pattern 4 cribriform architecture, more aggressive tumor microenvironments, and tumors with increased mutation density. 22–24 Consequently, concerns about MRI-invisible lesions are a contributor to the continued use of systematic biopsy. This finding reinforces the Trio study finding of the added value of random biopsy cores in addition to targeted cores. Finally, the least common reason for MRI-targeted biopsy miss was that lesions were initially missed by the radiologist, and this was rare (3/41). This may be correlated with radiologist experience, as prior studies have demonstrated a learning curve in prostate MRI interpretation. 25 Prostate MRI has a well-characterized, steep learning curve, and radiologist experience drastically influences cancer detection rates. 26
Among all patients with MRI-targeted biopsy misses, patients harboring high-risk MRI-invisible lesions had lower median PI-RADS scores and lower index lesion sizes, but similar PSA to men with missed lesions for other reasons. In these patients, the highest-grade lesion was an MRI-invisible tumor, so PI-RADS and maximum MRI-index lesion size were not valuable predictors of their risk of harboring disease. Therefore, this pattern of low PI-RADS, despite an elevated PSA may characterize patients with suspicion for MRI-invisible lesions who warrant a systematic biopsy.
Black men were the only racial group in this study whose representation was higher among the MRI-targeted biopsy miss patients than among the MRI-targeted biopsy detected patients. Black patients also had significantly more MRI-invisible lesions than other races. These findings contrast with Shin et al who found no significant difference in the MRI-targeted biopsy cancer detection rates between White and Black men. However, this study differed in its definition of clinically significant cancer being GG ≥2. 27 It is well-characterized that Black men with prostate cancer have worse clinical outcomes and higher cancer stages at diagnosis. 28 This may be attributed to disparate access to care; however, the role of MRI-visibility and targeted biopsy accuracy has not yet been widely analyzed in this population. Our study’s findings warrant further investigation.
In addition to its retrospective design, limitations of our study include the lack of information on ultimate lesion volume and location on final wholemount pathology for the entire cohort. 29In the present study, lesion size was entirely based on MRI. Therefore, missed lesion size and location could not be described among patients with MRI-invisible lesions, and wholemount localization using patient-specific sectioning molds will be a focus of subsequent studies.30 There are varied definitions of clinically significant cancer across studies, so our use of GG≥3 may limit the generalizability of our results. Since GG3 and above was considered clinically significant for our study, our data is likely to underreport the number of MRI misses that would be seen if GG2 or above was used as the clinically significant threshold. This occurs because MRI has a poorer detection rate of GG2 disease, than GG3 disease.2 Also, although physicians performing the systematic biopsy were instructed to ignore any hemorrhage tracts from the prior MRI-targeted biopsy, this may have still influenced free-hand selection of sextant regions. PI-RADS criteria used in this analysis was limited by not accounting for intralesional heterogeneity, potentially contributing to missed radiology reads and targeting errors. Another weakness is that MRIs were re-read by a single reader involved in the parent study, and MRIs prior to 2015 were retroactively assigned PI-RADS scores unblinded. This could have introduced a conformation bias to some re-reads and PI-RADS scores. Finally, in this study, there were very low frequency of lesions that were missed due to radiologist oversight, which may not be generalized to all institutions.
Conclusion
This study describes the largest reported cohort of biopsy-mapped clinically significant lesions missed by targeted biopsy. Though these are rare events, the reasons MRI-targeted biopsy missed a small proportion of clinically significant lesions were targeting errors, MRI-invisible lesions, and being initially missed by the radiologist. These results further elucidate the biopsy and patient factors that could inform providers’ suspicion of a patient having an MRI-targeted biopsy missed lesion.
Supplementary Material
Acknowledgments
Funding This research supported by the National Institutes of Health (NIH) Intramural Research Program, and the NIH Medical Research Scholars Program, a public-private partnership supported jointly by the NIH and generous contributions to the Foundation for the NIH from the Doris Duke Charitable Foundation, the American Association for Dental Research, the Colgate-Palmolive Company, Genentech, alumni of student research pro-grams, and other individual supporters via contributions to the Foundation for the National Institutes of Health
Conflicts of interest/Competing interests NIH and Philips have a Cooperative Research and Development Agreement. NIH has intellectual property in the field, including among other patents and patent applications, Patent: "System, methods, and instrumentation for image guided prostate treatment" US Patent number: 8948845, with inventors/authors including PLC, BW and PP. NIH and Philips (InVivo Inc) have a licensing agreement. NIH and authors PLC, BW and PP receive royalties from the US government for a licensing agreement with Philips/InVivo Inc. NIH does not endorse or recommend any commercial products, processes, or services. The views and personal opinions of authors expressed herein do not necessarily reflect those of the US Government, nor reflect any official recommendation nor opinion of the NIH nor NCI.
Ethics approval This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Institutional Review Board of The National Cancer Institute
Key of Abbreviations:
- mpMRI
Multiparametric Magnetic Resonance Imaging
- PI-RADS
Prostate Imaging–Reporting and Data System
- TRUS
Transrectal ultrasound
- PSA
Prostate specific antigen
- GG
Grade Group
- T2W
T2-Weighted
References
- 1.Rouviere O, Puech P, Renard-Penna R. et al. : Use of prostate systematic and targeted biopsy on the basis of multiparametric MRI in biopsy-naive patients (MRI-FIRST): a prospective, multicentre, paired diagnostic study. Lancet Oncol, 20: 100, 2019 [DOI] [PubMed] [Google Scholar]
- 2.Ahdoot M, Wilbur AR, Reese SE et al. : MRI-Targeted, Systematic, and Combined Biopsy for Prostate Cancer Diagnosis. The New England Journal of Medicine, 382: 917, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siddiqui MM, Rais-Bahrami S, Turkbey B. et al. : Comparison of MR/ultrasound fusion-guided biopsy with ultrasound-guided biopsy for the diagnosis of prostate cancer. JAMA, 313: 390, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasivisvanathan V, Rannikko AS, Borghi M. et al. : MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med, 378: 1767, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed HU, El-Shater Bosaily A, Brown LC et al. : Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet (London, England), 389: 815, 2017 [DOI] [PubMed] [Google Scholar]
- 6.Norris JM, Carmona Echeverria LM, Bott SRJ et al. : What Type of Prostate Cancer Is Systematically Overlooked by Multiparametric Magnetic Resonance Imaging? An Analysis from the PROMIS Cohort. Eur Urol, 78: 163, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salami SS, Ben-Levi E, Yaskiv O. et al. : In patients with a previous negative prostate biopsy and a suspicious lesion on magnetic resonance imaging, is a 12-core biopsy still necessary in addition to a targeted biopsy? BJU Int, 115: 562, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Muthigi A, George AK, Sidana A. et al. : Missing the Mark: Prostate Cancer Upgrading by Systematic Biopsy over Magnetic Resonance Imaging/Transrectal Ultrasound Fusion Biopsy. J Urol, 197: 327, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkbey B, Rosenkrantz AB, Haider MA et al. : Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur Urol, 76: 340, 2019 [DOI] [PubMed] [Google Scholar]
- 10.Rais-Bahrami S, Siddiqui MM, Turkbey B. et al. : Utility of multiparametric magnetic resonance imaging suspicion levels for detecting prostate cancer. J Urol, 190: 1721, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaur S, Harmon S, Mehralivand S. et al. : Prospective comparison of PI-RADS version 2 and qualitative in-house categorization system in detection of prostate cancer. J Magn Reson Imaging, 48: 1326, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathianathen NJ, Omer A, Harriss E. et al. : Negative Predictive Value of Multiparametric Magnetic Resonance Imaging in the Detection of Clinically Significant Prostate Cancer in the Prostate Imaging Reporting and Data System Era: A Systematic Review and Meta-analysis. Eur Urol, 78: 402, 2020 [DOI] [PubMed] [Google Scholar]
- 13.Gold SA, Hale GR, Bloom JB et al. : Follow-up of negative MRI-targeted prostate biopsies: when are we missing cancer? World J Urol, 37: 235, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Klotz L, Zhang L, Lam A. et al. : Clinical results of long-term follow-up of a large, active surveillance cohort with localized prostate cancer. J Clin Oncol, 28: 126, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Lahoud J, Doan P, Kim L. et al. : Perilesional Biopsies Increase Detection of Significant Prostate Cancer in Men with PI-RADS 4/5 Lesions: Validation of the PI-RADS Steering Committee Recommendation. Eur Urol, 2021 [DOI] [PubMed] [Google Scholar]
- 16.Coker MA, Glaser ZA, Gordetsky JB et al. : Targets missed: predictors of MRI-targeted biopsy failing to accurately localize prostate cancer found on systematic biopsy. Prostate Cancer Prostatic Dis, 21: 549, 2018 [DOI] [PubMed] [Google Scholar]
- 17.Xu S, Kruecker J, Turkbey B. et al. : Real-time MRI-TRUS fusion for guidance of targeted prostate biopsies. Comput Aided Surg, 13: 255, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tewes S, Hueper K, Hartung D. et al. : Targeted MRI/TRUS fusion-guided biopsy in men with previous prostate biopsies using a novel registration software and multiparametric MRI PI-RADS scores: first results. World J Urol, 33: 1707, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Costa DN, Pedrosa I, Donato F Jr. et al. : MR Imaging-Transrectal US Fusion for Targeted Prostate Biopsies: Implications for Diagnosis and Clinical Management. Radiographics, 35: 696, 2015 [DOI] [PubMed] [Google Scholar]
- 20.Hamid S, Donaldson IA, Hu Y. et al. : The SmartTarget Biopsy Trial: A Prospective, Within-person Randomised, Blinded Trial Comparing the Accuracy of Visual-registration and Magnetic Resonance Imaging/Ultrasound Image-fusion Targeted Biopsies for Prostate Cancer Risk Stratification. Eur Urol, 75: 733, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halstuch D, Baniel J, Lifshitz D. et al. : Assessment of Needle Tip Deflection During Transrectal Guided Prostate Biopsy: Implications for Targeted Biopsies. J Endourol, 32: 252, 2018 [DOI] [PubMed] [Google Scholar]
- 22.Truong M, Hollenberg G, Weinberg E. et al. : Impact of Gleason Subtype on Prostate Cancer Detection Using Multiparametric Magnetic Resonance Imaging: Correlation with Final Histopathology. J Urol, 198: 316, 2017 [DOI] [PubMed] [Google Scholar]
- 23.Houlahan KE, Salmasi A, Sadun TY et al. : Molecular Hallmarks of Multiparametric Magnetic Resonance Imaging Visibility in Prostate Cancer. Eur Urol, 76: 18, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Houdt PJ, Ghobadi G, Schoots IG et al. : Histopathological Features of MRI-Invisible Regions of Prostate Cancer Lesions. J Magn Reson Imaging, 51: 1235, 2020 [DOI] [PubMed] [Google Scholar]
- 25.Gaziev G, Wadhwa K, Barrett T. et al. : Defining the learning curve for multiparametric magnetic resonance imaging (MRI) of the prostate using MRI-transrectal ultrasonography (TRUS) fusion-guided transperineal prostate biopsies as a validation tool. BJU Int, 117: 80, 2016 [DOI] [PubMed] [Google Scholar]
- 26.Ruprecht O, Weisser P, Bodelle B. et al. : MRI of the prostate: interobserver agreement compared with histopathologic outcome after radical prostatectomy. Eur J Radiol, 81: 456, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Shin T, Smyth TB, Ukimura O. et al. : Detection of prostate cancer using magnetic resonance imaging/ultrasonography image-fusion targeted biopsy in African-American men. BJU Int, 120: 233, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGinley KF, Tay KJ, Moul JW: Prostate cancer in men of African origin. Nat Rev Urol, 13: 99, 2016 [DOI] [PubMed] [Google Scholar]
- 29.Tan N, Margolis DJ, Lu DY et al. : Characteristics of Detected and Missed Prostate Cancer Foci on 3-T Multiparametric MRI Using an Endorectal Coil Correlated With Whole-Mount Thin-Section Histopathology. AJR Am J Roentgenol, 205: W87, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trivedi H, Turkbey B, Rastinehad AR et al. : Use of patient-specific MRI-based prostate mold for validation of multiparametric MRI in localization of prostate cancer. Urology, 79: 233, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.