Summary
Neural activity sculpts circuit wiring in many animals. In vertebrates, patterned spontaneous network activity (PaSNA) generates sensory maps and establishes local circuits 1–3. However, it remains unclear how PaSNA might shape neuronal circuits and behavior in invertebrates. Previous work in the developing Drosophila embryo discovered intrinsic muscle activity that did not require synaptic transmission, and hence was myogenic, preceding PaSNA 4–6. These studies, however, monitored muscle movement, not neural activity, and were therefore unable to observe how myogenic activity might relate to subsequent neural network engagement. Here we use calcium imaging to directly record neural activity and characterize the emergence of PaSNA. We demonstrate that the spatiotemporal properties of PaSNA are highly stereotyped across embryos, arguing for genetic programming. Neural activity begins well before it becomes patterned, emerging during the myogenic stage. Remarkably, inhibition of mechanosensory input, as well as inhibition of muscle contractions, results in premature and excessive PaSNA, demonstrating that muscle movement serves as a brake on this process. Finally, transient mechanosensory inhibition during PaSNA, followed by quantitative modeling of larval behavior, shows that mechanosensory modulation during development is required for proper larval foraging. This work provides a foundation for using the Drosophila embryo to study the role of PaSNA in circuit formation, provides mechanistic insight into how PaSNA is entrained by motor activity, and demonstrates that spontaneous network activity is essential for locomotor behavior. These studies argue that sensory feedback during the earliest stages of circuit formation can sculpt locomotor behaviors through innate motor learning.
Keywords: nervous system development, development of locomotor behavior, spontaneous network activity, neural circuit wiring, behavioral development, Drosophila embryo
Graphical Abstract
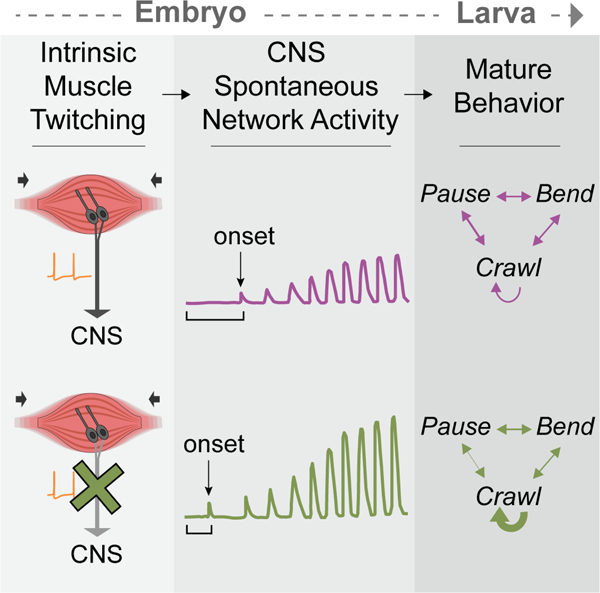
eTOC Blurb
Carreira-Rosario et al. show that stereotyped, spontaneous network activity in the Drosophila embryo shapes innate larval behavior. This activity is negatively modulated by preceding intrinsic muscle twitching and sensory input. This work highlights self-generated sensory feedback as a mechanism to shape circuits that control innate behaviors.
Results
PaSNA in the Drosophila embryo
Motor movements begin in the embryo as uncoordinated twitching at stage 16, followed by larger scale movements that progressively become stronger and more organized prior to hatching approximately 7 hours later (Figure 1A). To characterize the emergence of neural activity across these stages, we developed a wide-field imaging preparation in which we could monitor neural activity in 20–30 embryos simultaneously (Figure 1B; Methods). We expressed the genetically encoded calcium indicator GCaMP6s in all neurons, while co-expressing nuclear TdTomato to allow for ratiometric imaging, and acquired images every 7 seconds from the myogenic stage through hatching (Video S1). Under these imaging conditions, 95% of control animals hatched (n = 60), demonstrating that this preparation does not disrupt normal development. Finally, to correct for small variations in the developmental timing of individual embryos, we monitored ventral nerve cord (VNC) condensation and normalized developmental stage by computing the ratio of the length of the embryo to the length of the central nervous system (CNS) (Figure S1A), following standard methods 7,8.
Figure 1. Characterization of patterned spontaneous network activity in Drosophila embryo.
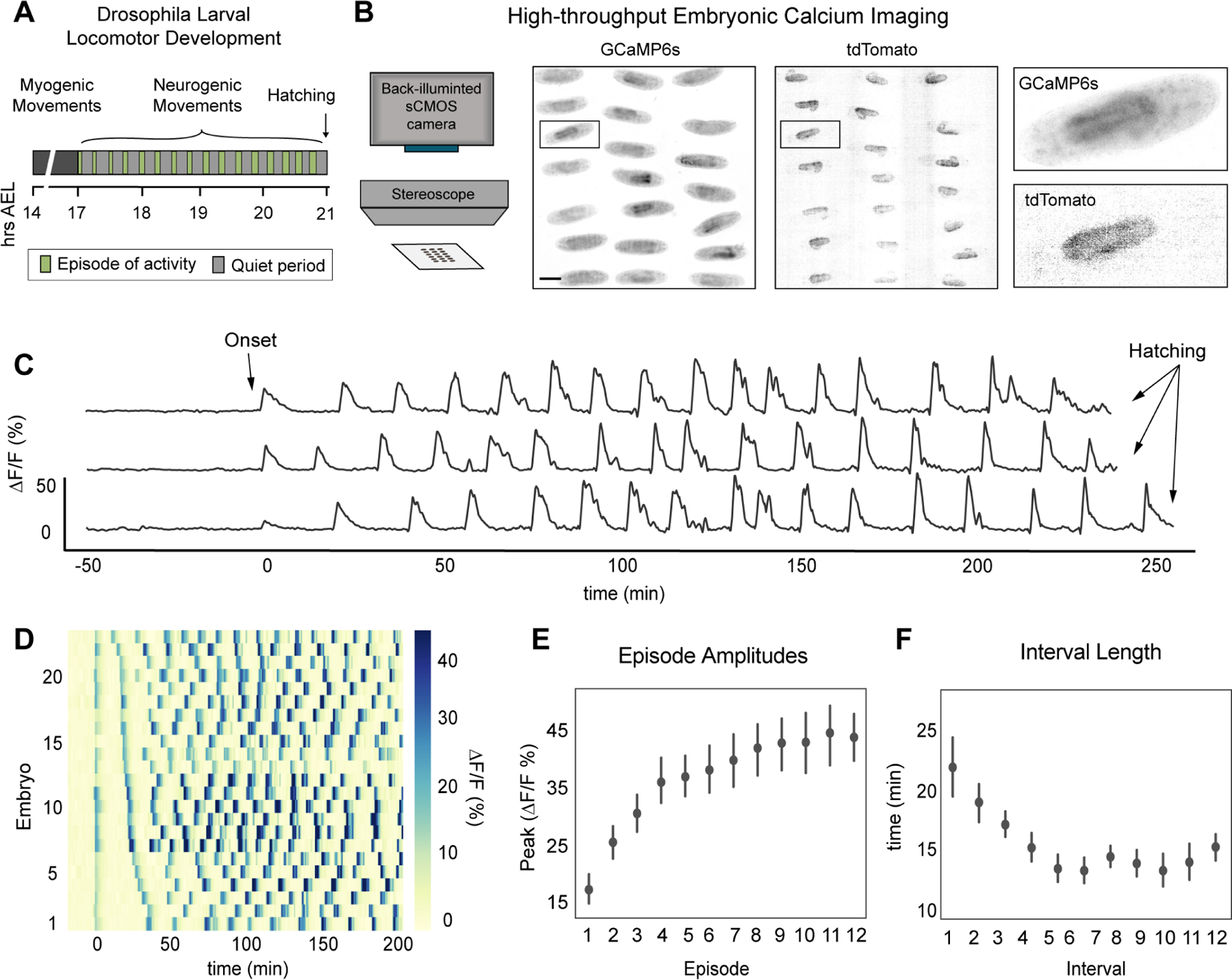
(A) Schematic of Drosophila larval locomotor development. Time in hours after egg laying (hrs AEL). (B) Schematic of high-throughput imaging system (left). Images of GCaMP6s and tdTomato signal across the imaging field, color inverted for visualization (right). Scale bar: 200 μm. (C) GCaMP6s:TdTomato ΔF/F traces from three individual embryos. (D) Raster plot for PaSNA trimmed at 200 minutes post-onset, sorted by distance between first and second peak, with each trace corresponding to an individual embryo. Increasingly strong movements prevent accurate measurements at later stages. ΔF/F heat map scale to the right. (E) ΔF/F peaks for episodes 1 through 12 (n = 23). (F) Quantification of the first eleven interbout interval lengths (n = 23). For (E) and (F), points represent mean and lines depict the 95% confidence interval (CI). See also Figure S1 and Video S1. For genotype information see Table S1.
Consistent with the pattern of muscle movements 5, we observed episodes in which intracellular calcium concentrations increased in many neurons and their neurites (Figure 1C, D; Video S1). Strikingly, the timing of the first large wave of neural activity was highly consistent from animal to animal, appearing at a length ratio of 2.2 (95%CI [0.06, 0.06]) (Figure S1B), corresponding to early stage 17. Aligning calcium traces by the timing of the first episode revealed that the overall PaSNA pattern was qualitatively and quantitatively similar across all embryos (Figure 1D-F). In particular, a total of 17 PaSNA episodes (95%CI [0.99, 0.99]) that occurred over 275 minutes (95%CI [18.3, 18.3]) preceded hatching (Figure S1B, C). Moreover, the size and duration of each wave of activity consistently increased over the first eight waves (Figure 1E). Finally, the interbout interval dramatically decreased over the first five episodes of PaSNA from 21.8 minutes (95% CI [2.3,2.5]) to 13.3 minutes (95% CI [1.1,1.2]) (Figure 1 F). Stereotypy analysis revealed that the observed intervals were significantly more stereotyped across embryos than expected by chance (Figure S1D). Taken together, these data show that PaSNA is highly stereotyped from embryo to embryo, suggesting that PaSNA is genetically encoded.
Spatiotemporal properties of the initial PaSNA episode
We focused next on the first episode of PaSNA, a period of particular interest given that it represents the transition from myogenic to neurogenic movement. To investigate the spatiotemporal patterns of neural activity during single episodes of PaSNA, we developed a two-photon (2P) microscopy preparation to image embryos expressing pan-neuronal GCaMP6s and TdTomato. This system allows for imaging of the VNC for two hours at cellular resolution, acquiring imaging volumes at 2.6 Hz. Embryos survive imaging, hatch and become adult flies (n = 8). Preceding the first episode of PaSNA, we observed sporadic neuronal firing throughout the VNC, an activity pattern we refer to as flickering. This activity was observed during the 30 minutes before the first PaSNA episode, thus appearing during the myogenic phase of movement 5. After this, the first PaSNA episode began, and comprised three phases (Figure 2A, B; Video S2). During the first phase, we observed increased levels of neural activity within a stereotyped region, marking the localized initiation of PaSNA and defining the start of neurogenic activity. During the second phase, we observed a single wave of neural activity that traversed the VNC and defined propagation. During the third phase, we observed a period of peak activity along the VNC that persisted for approximately 80 seconds. Activity then returned to basal levels where an interbout interval containing flickering activity persisted until the next episode.
Figure 2. Spatiotemporal and network properties of a single PaSNA episode.
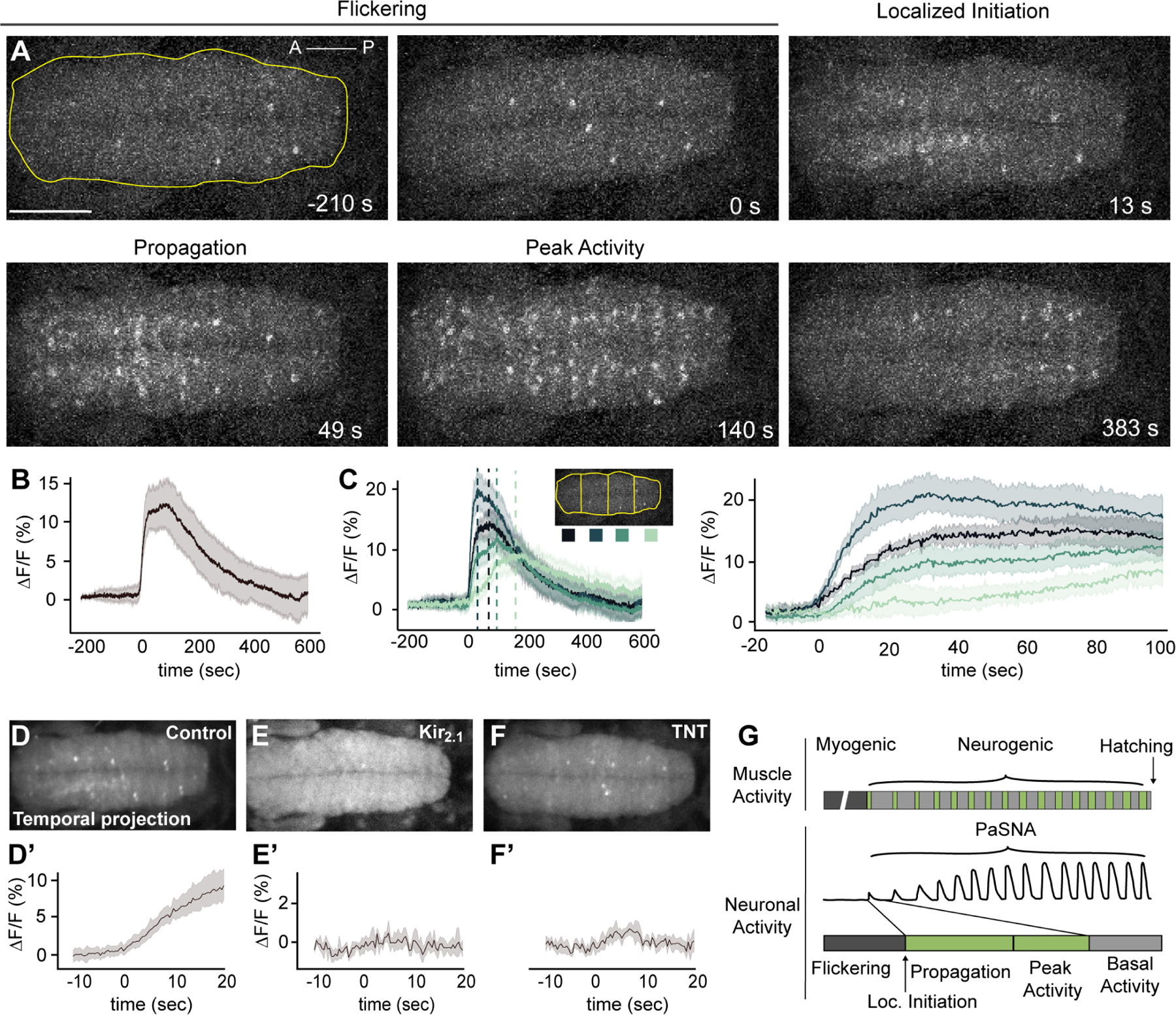
(A) Image frames during the first episode of PaSNA in an embryo. Phases labeled on top. Images are maximum intensity projections from an embryonic VNC expressing pan-neuronal GCaMP6s. Time stamps are relative to the positive inflection point caused by the activity burst. Yellow line delineates the VNC, with the ROI used for Panel B. Scale bar: 50 μm. (B) ΔF/F trace of the entire VNC during the first episode of PaSNA (n = 8). (C) ΔF/F of the color-coded four ROIs. Left displays −200 seconds to 600 seconds; right displays from −20 to 100 seconds relative to the initiation of PaSNA. (D-F’) Temporal projections (top) and ΔF/F VNC traces (bottom) for 30 seconds near the localized initiation time of the episode for control embryos (n = 8) (D), embryos expressing Kir2.1 pan-neuronally (E) (n = 5) and embryos expressing TNT pan-neuronally (F) (n = 6). (G) Schematic of Drosophila larval locomotor development showing activity at the muscle (top) and neuronal level (bottom). For all time series, dark lines represent the mean, while shading depicts the 95%CI. See also Video S2, S3, S4. For genotype information see Table S1.
Next, we asked whether the location of the focal activity observed during localized initiation was invariant across embryos. Analysis of neural activity within ROIs along the anterior-posterior (A-P) axis of the VNC showed that PaSNA always initiated in the anterior region of the VNC (Figure 2C) (n =8). Furthermore, in 100% of embryos, activity initiated in one of the two most anterior ROIs, a region spanning the thoracic segments. After initiation, activity always propagated along the A-P axis. Strikingly, the wave of neural activity propagated slowly, reaching the most posterior region of the embryo approximately 75 seconds after localized initiation, corresponding to a propagation speed of less than 2 μm per second. Lastly, in all embryos, the more posterior regions were the last to return to basal, flickering activity. Together, these observations demonstrate that the initial episode of PaSNA is spatiotemporally patterned.
The role of neural activity in initiating PaSNA
Next, we examined the role of neural activity in the initiation of PaSNA. To test whether neuronal depolarization caused the observed calcium transients, we inhibited depolarization through pan-neuronal expression of the inward-rectifier potassium channel Kir2.1 9. As expected, this abolished flickering during the myogenic phase, as well as all three phases of PaSNA, indicating that PaSNA is a voltage-dependent process (Figure 2E; Video S3). Next, we tested whether PaSNA is driven by chemical synapses by inhibiting synaptic transmission using tetanus toxin (TNT) 9. Pan-neuronal expression of TNT had no effect on flickering during the myogenic phase, but prevented all three phases of PaSNA including the propagating waves of neural activity (Figure 2F; Video S4). Together, these results show that while neuronal depolarization and chemical synaptic transmission are both crucial for PaSNA, only depolarization is required for flickering.
Mechanosensory input negatively modulates PaSNA
We next sought to determine whether the initial, myogenic phase of spontaneous muscle movement might be functionally coupled to the initiation of PaSNA. We reasoned that this coupling could occur through sensory feedback via proprioceptors. Therefore, we first asked which proprioceptive neurons are active during the myogenic phase. We used the calcium integrator system CaLexA, a method for transcriptionally labeling active neurons (Figure 3A) 10. We restricted this system to only those neurons that are active during the myogenic phase by pan-neuronal expression of TNT, which prevents embryos from entering PaSNA (Figure 2F; Figure 3A). Strikingly, every embryo displayed high levels of CaLexA expression in mechanosensory chordotonal (mechano-ch) neurons in most segments (n = 30 embryos) (Figure 3B-E). Specifically, observed expression in lch5 in every hemisegment as well as expression in lch1 and vchA/B in some segments. Notably, none of these embryos showed CaLexA signal in any other proprioceptive neurons.
Figure 3. Mechanosensory neurons modulate the amplitude of PaSNA episodes.
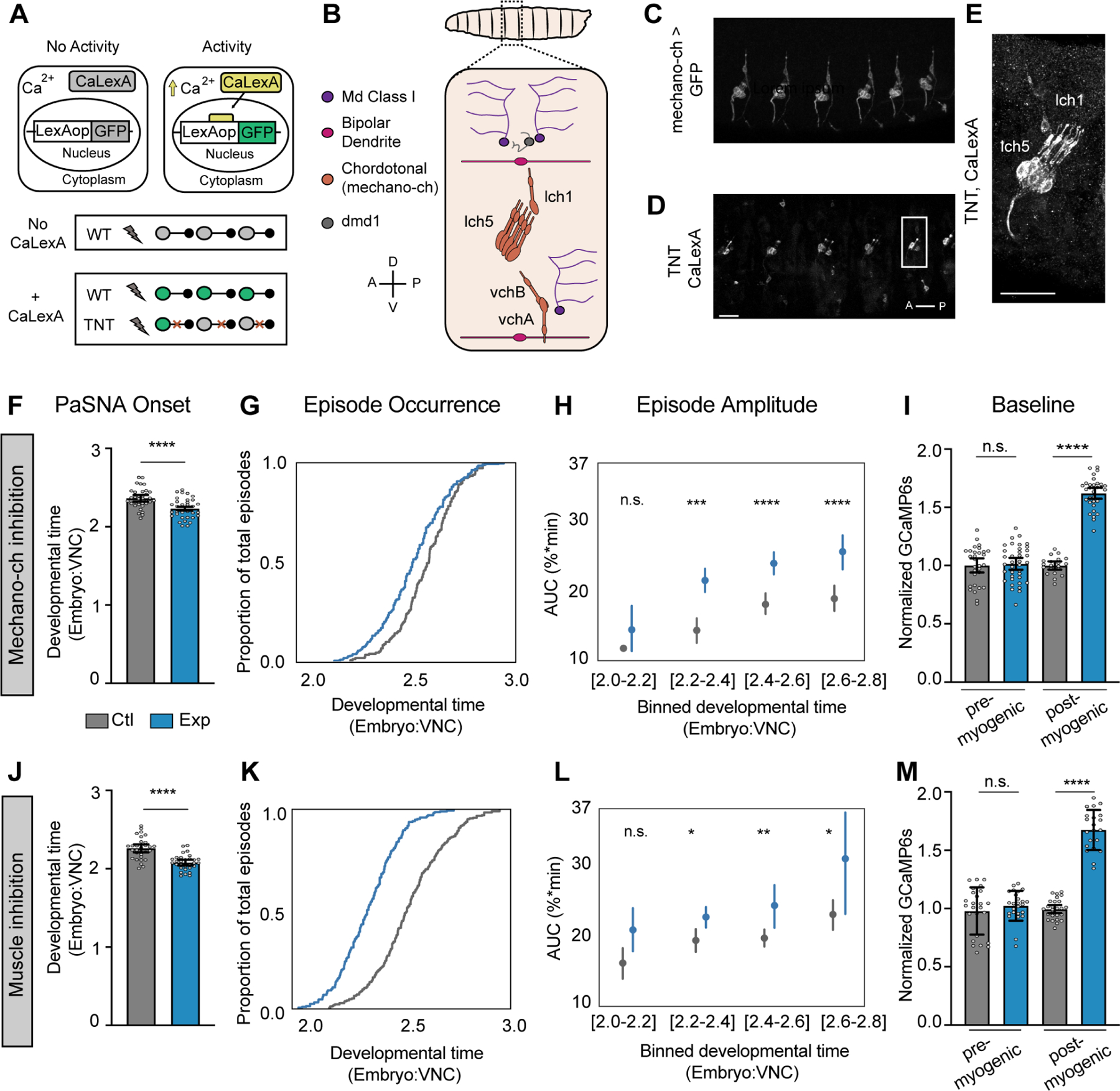
(A) Schematic illustration of the experiment using CaLexA to reveal neural activity during the myogenic phase. (B) Schematic of an embryonic anterior body wall hemisegment showing all proprioceptive neurons. There are eight mechanosensory chordotonal neurons (mechano-ch [orange]). Five of these form a laterally located cluster (lch5). A solitary mechano-ch is located dorsal to lch5 (lch1), and a pair of mechano-ch neurons is located ventrally (vchB and vchA). Anatomical coordinates: anterior (A), posterior (P), dorsal (D) and ventral (V). (C) Expression of the mechano-ch driver inactive (iav) along several body wall segments. (D-E) CaLexA driving GFP expression in a 19 hrs AEL embryo expressing pan-neuronal TNT (n= 30 embryos). Note expression in lch5 in every hemisegment as well as expression in lch1 and vchA/B in some segments. Scale bars: 20μm. (F-I) Measurements of the timing and intensity of PaSNA in control embryos (gray) and experimental embryos expressing TNT in mechano-ch neurons (blue). (F) Quantification of PaSNA onset (n = 36 control; n = 33 experimental). (G) Cumulative occurrence of the first twelve episodes plotted as the proportion of total episodes across developmental time (n = 17 control; n = 32 experimental). (H) Area under the peak curve (AUC) quantification for the first twelve episodes plotted against developmental time. Values were binned based on developmental time (n = 17 control; n = 32 experimental). (I) Quantification of GCaMP6s baseline levels normalized against control mean before (14hrs AEL; n = 30 control; n = 37 experimental) and after (10 minutes before PaSNA onset; n = 20 control; n= 32 experimental) the myogenic phase. (J-M) Measurements of the timing and intensity of PaSNA in control embryos (gray) and experimental embryos expressing Kir2.1 in muscles (blue). (J) Quantification of PaSNA onset (n = 36 control; n = 30 experimental). (K) Cumulative occurrence of the first twelve episodes plotted as the proportion of total episodes across developmental time (n = 28 control; n = 28 experimental). (L) AUC quantification for the first twelve episodes plotted against binned developmental time (n = 28, control; n = 28 experimental). (M) Quantification of GCaMP6s baseline levels normalized against control mean before (n = 25 control; n = 25 experimental) and after (n = 26 control; n = 21 experimental) the myogenic phase. For (H) and (L), points represent mean and lines depict the 95% confidence interval. For all bar graphs the mean and 95% CI are displayed. ****p<0.0001, ***p<0.001, **p<0.005, *p<0.05. For (F) and (J) we used two-sample t-tests. For (H) and (L) we used two-sample t-tests with Holm-Bonferroni correction. For (I) and (M) we used two-sample Welch’s t-tests to account for differences in variance. See also Figure S2. For genotype information see Table S1.
Mechano-ch neurons detect muscle stretch, relaying proprioceptive signals used to regulate larval crawling speed 11–14. Additionally, these neurons have been previously linked to embryonic neural circuit formation 12, making them ideal candidates for coupling muscle movements to PaSNA. To test this idea, we silenced mechano-ch neurons and examined whether this perturbation altered the timing or amplitude of PaSNA episodes. To do this, we used the inactive (iav) enhancer to express TNT in mechano-ch neurons, and monitored neural activity throughout PaSNA using wide-field imaging of pan-neuronally expressed GCaMP6s. Consistent with previous work 5, PaSNA was not abolished after mechano-ch silencing. Strikingly, however, PaSNA started prematurely in these embryos (Figure 3F). This led to embryos experiencing more episodes of PaSNA earlier in development (Figure 3G), as well as increasing PaSNA duration, and the total number of episodes (Figure S2A,B). The amplitude of most PaSNA episodes was increased in these embryos as compared to controls (Figure 3H). Lastly, interbout intervals remained largely unchanged (Figure S2C).
We reasoned that if mechano-ch neurons were coupling muscle contraction to PaSNA, there might be an effect of blocking the synaptic transmission in these cells during the myogenic phase. To test this, we examined the baseline fluorescence of GCaMP6s (relative to nuclear TdTomato expressed on the same RNA transcript), as a proxy for the intracellular calcium concentration and membrane excitability, before and after the myogenic phase. As expected, inhibiting mechano-ch neurons had no effect on baseline fluorescence when muscles had not yet contracted, before the myogenic phase (Figure 3I). Strikingly, inhibiting mechano-ch neurons dramatically increased the baseline fluorescence signal of GCaMP6s after muscle contraction during the myogenic phase (but before PaSNA began; Figure 3I). Such an increase in baseline GCaMP6s signal is consistent with higher intracellular calcium concentrations and increased membrane excitability, providing a potential explanation for the premature onset and increased amplitude of PaSNA seen when mechano-ch neurons are inhibited. We note that this change in baseline GCaMP6s fluorescence cannot be accounted for by changes in protein expression, as these measures are normalized relative to TdTomato in every cell. Thus, mechano-ch neurons act during the myogenic phase. Finally, to complement these results, we repeated these experiments in embryos expressing Kir2.1 in mechano-ch neurons. However, while we observed significant increases in the amplitudes of initial PaSNA episodes and GCaMP baseline (Figure S2G), overall effects were modest, suggesting that this functional inhibition was incomplete, and further obscured by genetic background effects that increased baseline GCaMP6s fluorescence (Figure S2L).
If mechano-ch neurons are coupling muscle movements to PaSNA, inhibiting muscle contractions should have similar effects to inhibiting mechano-ch neurons. To test this, we inhibited muscle contraction by expressing Kir2.1 using a muscle specific driver 15, while expressing GCaMP6s pan-neuronally. Strikingly, preventing muscle contraction caused premature PaSNA onset, increased the amplitude of PaSNA episodes, and led to higher baseline fluorescence in post-myogenic but not pre-myogenic embryos (Figure 3J-M). Thus silencing muscle contraction, or proprioceptive neuron function lead to premature onset of PaSNA and increased amplitude of PaSNA episodes, strongly suggesting that muscle contractions induce mechano-ch activity during the myogenic phase to negatively modulate PaSNA.
Developmental inhibition of mechanosensory input leads to abnormal larval behavior
Our observation that silencing mechano-ch neurons increased PaSNA raised the question of whether this change in PaSNA had behavioral consequences, and more specifically, whether these transient changes in PaSNA resulted in larval behavioral deficits. We inhibited mechano-ch activity transiently (from the late myogenic phase through to the end of PaSNA) by expressing the G. theta anion channelrhodopsin 1 (GtACR1) 16 in mechano-ch neurons, and examined larval behavior 24hrs after hatching using the Frustrated Total Internal Reflection-based imaging method (FIM; Figure 4A) 17. We employed Time REsolved BehavioraL Embedding (TREBLE) 18 to characterize potential behavioral differences between the optogenetically silenced condition and the control. TREBLE is a quantitative framework for identifying structure in behavior by collecting features (such as larval posture or velocity) into temporal windows and embedding these into a low-dimensional space (Figure 4B). As previously shown 18, we found that the major components of the larval foraging ethogram 19,20 can be captured in a 2-dimensional space using TREBLE (Figure 4B, C). In this 2-dimensional space, crawling is represented by an oscillator with directional movement (Figure S3) and is connected to regions corresponding to pausing and turning (Figure 4C).
Figure 4. Temporal embryonic inhibition of mechanosensory input leads to abnormal larval behavior.
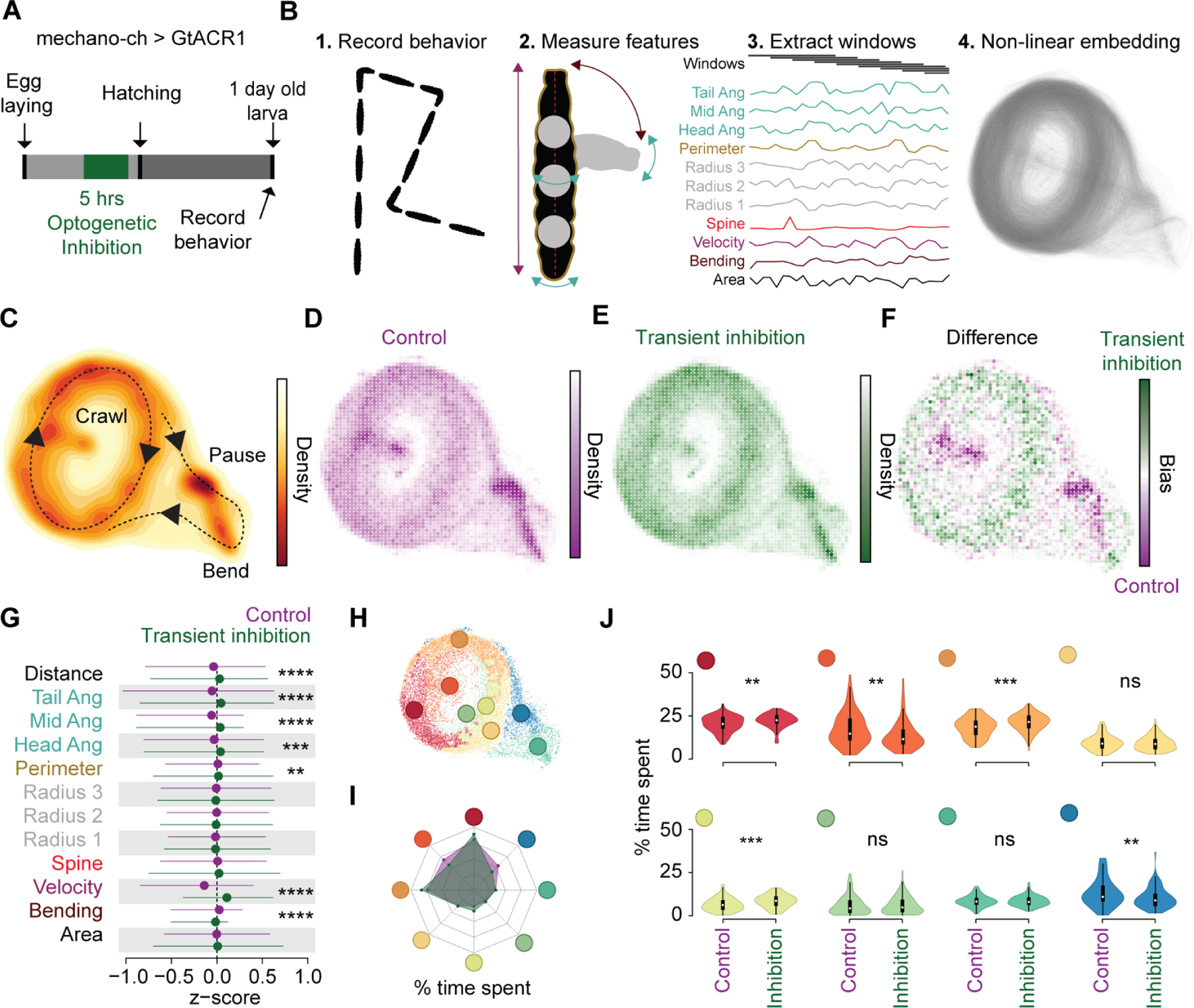
(A) Schematic of experimental design. (B) Workflow for Time Resolved BehavioraL Embedding (TREBLE) (Methods). (C) Probability density function of larval locomotor space plotted as a heatmap. Behaviors annotated qualitatively. Density scale to the right. (D-E) Bin-wise occurrence distributions for control (n= 84) (D) and transient inhibition (n = 97) (E) groups. (F) Difference map between control (purple) and transiently inhibited (green) animals. Bias scale to the right. (G) Comparison of primary behavioral features between control (purple) and transiently inhibited (green) larvae. (H) Behavioral space colored via Louvain clusters (Methods). (I) Radar chart comparing the percentage of time spent in each of the Louvain clusters for control (purple shade) and transient inhibition (green shade) groups. (J) Differences in occurrence in each behavioral cluster between control and transiently inhibited animals. **** p < 0.0001, *** p< 0.001, ** p < 0.01. For (G) we used trial-wise Kruskal-Wallis test with Bonferroni correction. For (J) we used trial-wise Kruskal-Wallis test. See also Figure S3. For genotype information see Table S1.
In the TREBLE approach, both control and transiently inhibited larvae were used to generate a single, common behavior space (n = 181 total; 84 control larvae, 97 transiently inhibited larvae); 179,409 windows; see Methods) where we could directly infer behavioral variation via differences in the likelihood that either control or experimental larvae occupied specific regions of the space (Figures 4D-E). Control and inhibited conditions displayed notably different occurrence distributions (Figures 4D-F), the biggest deviations of which were restricted to specific regions of behavior space (Figure 4F). Control larvae were more likely to visit parts of the behavior space that correspond to pauses and bends (Figure 4F; Figure S3) while inhibited larvae spent more time, proportionally, in the crawling oscillator (Figure 4F). To confirm these differences using a TREBLE independent approach, we compared the primary behavioral features themselves, as measured using the FIM system, and observed that inhibited larvae bend less, crawl further, and have an increased velocity distribution relative to control animals (Figure 4G).
Finally, to quantitatively compare control and transiently inhibited larvae in the TREBLE space, we clustered the behavior space based on similarity to identify discrete elements of behavior (see Methods). We then compare the frequency of occurrence of discrete behavioral motifs (Figure 4H). Reflecting our previous findings, the mechano-ch silenced and control animals displayed different overall distributions (Figure 4I) and significantly varied across a number of behavioral motifs (Figure 4J; trial-wise Kruskal-Wallis test). Specifically, controls were more likely to pause (Figure 4J; dark blue; p < 0.01) and head cast during crawling (Figure 4J; dark orange; p < 0.002) while the mechano-ch silenced larvae were more likely to be crawling (Figure 4J; red, orange, light green). These findings demonstrate that developmental inhibition of mechano-ch neurons leads to an apparent simplification of larval foraging behavior, biasing animals toward ongoing crawling as opposed to the typical sequence of crawling, pausing, and head casting.
Discussion
These studies demonstrate that PaSNA in the Drosophila embryo follows a stereotyped sequence of wave-like, large-scale network activation events interspersed by low-activity periods (Figure 1, 2). Strikingly, our data also demonstrate that muscle contractions shape the magnitude of PaSNA via mechanosensory input, beginning during the myogenic phase, but perhaps also continuing throughout each subsequent wave (Figure 3). Transiently disrupting this mechanosensory feedback during embryonic development results in deficits in larval locomotor behavior, arguing that neural activity plays a critical role in the functional organization of locomotor circuits (Figure 4). These results suggest that sensory inputs generated by spontaneous muscle contraction play a role in subsequent circuit establishment, thereby providing one of the earliest examples of sensory regulation of locomotor development in any context.
Our work has measured the trajectory of neural activity across embryonic development. Prior to PaSNA, individual CNS neurons display transient elevations in intracellular calcium levels (flickering) that depend on depolarization of the plasma membrane, but which are independent of synaptic input. In addition, as previous work has demonstrated, muscle twitching independent of neural activity also occurs, and based on our work, appears to lead to the selective activation of mechano-ch sensory neurons. The output of mechano-ch neurons then acts to negatively modulate the basal levels of intracellular calcium in the CNS, the onset of PaSNA, and the amplitude of PaSNA waves. While it is possible that the activation of mechano-ch neurons occurs independently of muscle contraction, given the phenotypic similarities revealed by muscle inhibition and mechano-ch inhibition (Figure 3), we favor a model where mechano-ch neuronal activity is driven by muscle contraction.
Our data demonstrate that the first episode of PaSNA invariably begins in the thoracic region (Figure 2). After this initial event, PaSNA proceeds through a stereotyped sequence of accelerating, intensifying waves through to hatching. Given the striking similarity in both the spatial and temporal properties of PaSNA in the embryo, as well as analogous observations in the Drosophila visual system 21, we hypothesize that this process is under tight genetic control. Similarly, observations across systems, including in the mammalian cortex, have led to speculation that genetic information underlies the spontaneous neuronal activity present in these developing circuits 22. Our characterization of the initiation and progression of PaSNA in the Drosophila embryo sets the stage for the dissection of these mechanisms at the level of specific circuits, cells, and molecules.
Most previous studies examining PaSNA function in shaping developing circuits rely on perturbations that abolish PaSNA. Interestingly, our study shows that an increase in PaSNA also leads to changes in behavior. This result suggests that organisms must quantitatively tune the level of PaSNA during circuit establishment. Supporting this, previous studies have demonstrated that excessive neuronal activity during larval circuit formation leads to hyperexcitable motor circuits that are prone to seizures 23,24. It is possible that the excessive PaSNA experienced after mechano-ch inhibition leads to hyperexcitability of specific neurons within the circuits that control foraging behavior. Identifying the neurons affected upon mechano-ch transient inhibition and probing their electrophysiological properties will test this idea.
Relatively little attention has been paid to examining the role of spontaneous neural activity in shaping innate behaviors. In that light, our finding that the activity of mechano-ch neurons during development shapes locomotor behavior is remarkable. Given that this behavioral effect is developmentally programmed, we hypothesize that mechano-ch input is needed to pattern connectivity or determine the physiological properties of specific cells in developing motor circuits. Indeed, blocking synaptic transmission in mechano-ch neurons throughout development changes the connectivity of these cells with their post-synaptic partners, cells that mediate behavioral responses to vibration 25. We hypothesize that the changes that mechano-ch inputs exert on developing circuits are, in fact, widespread, modifying circuits across the CNS through PaSNA. In vertebrates, motor feedback is crucial to shaping learned motor behaviors through activity-dependent mechanisms 26. It is tempting to speculate that the sculpting of innate foraging behavior by mechano-ch neuron activity in Drosophila reveals an analogous, evolutionarily ancient mechanism that may have been co-opted in other contexts to enable motor learning.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Requests for information and resources not made available here should be directed to and will be fulfilled by the lead contact, Arnaldo Carreira-Rosario (arnaldo@stanford.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All original code for Figures 1–3 has been deposited at https://github.com/ClandininLab/pasna-acr-currbio2021 and is publicly available as of the date of publication.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Stocks
All stocks were kept at 25°C on molasses-based food. The following stocks were used: UAS-IVS-Syn21-GCaMP6s-P2A-nls-tdTomato-p10 on JK66B was a gift from Marta Zlatic (MRC Laboratory of Molecular Biology). LexAop-Kir2.1 at VIE-260B, UAS-Kir2.1 at VIE-260B and LexAop-TNT on VIE−260B were gifts from Barry Dickson (The University of Queensland). pBDP-LexA:p65 on attp40 was a gift from T. Shirangi (Villanova University). UAS-GtACR1 at attP2 was a gift from A. Claridge-Chang (Duke-NUS Med School). The following stocks were obtained from the Bloomington Drosophila Stock Center: elav-GAL4.L on 3rd (BDSC# 8760), elav-GAL4.L on 2nd (BDSC# 8765), GMR44H10-lexa::p65 on attP40 (BDSC# 61543), elavc155-GAL4 (BDSC# 458), UAS-TeTxLC.tnt G2 (BDSC# 28838), UAS-mLexA-VP16-NFAT, LexAop-rCD2-GFP (CaLexA) (BDSC# 66542), LexAop-CD8-GFP-2A-CD8-GFP on 2nd ( BDSC# 66545), iav-lexA::p65 27 on VK00013 (BDSC# 52246), attP-9A VK000013 (BDSC# 9732) and iav-GAL4.K on 3rd ( BDSC# 52273).
METHOD DETAILS
Embryo collection for calcium imaging
For all imaging experiments, embryos were collected in 15–30 minute time windows the day before imaging, and grown at 25°C or 23°C on standard 3.0% agar molasses collection caps covered with a thin layer of wet yeast. Before imaging, embryos were dechorionated with double-sided tape and staged using elongation of the anterior midgut as a guide 4,7. To prevent dehydration, embryos were transferred into Halocarbon oil or saline no more than 5 minutes after dechorionation.
Wide-field imaging
Staged, dechorionated embryos were mounted ventral side up on double-sided tape, covered with Halocarbon oil (180 cSt) and imaged using a Leica M205 FA system with a Plan Apo Corr. 2X objective. Stereoscopic magnification was used to achieve a final magnification of 64X. Fluorescent signals were acquired using LED illumination (CooLED pE-300 white). GCaMP6s was excited and collected using an ET470/40x ET525/50m band-pass filter set, while tdTomato was excited and collected using an ET545/25x ET605/70m band-pass filter set, acquiring each signal sequentially. Each cycle of imaging acquisition was 7 seconds long. We used a back-thinned sCMOS camera (Orca-Fusion BT - Hamamatsu) to capture images at a 1024 × 1024 resolution (after 2×2 binning), corresponding to a pixel size of 2.0 μm × 2.0 μm. Imaging sessions were up to 9 hrs in duration, depending on the experiment, and were conducted at 24±1°C.
Two-photon imaging
Staged, dechorionated embryos were mounted ventral side up on Sylgard pads and imaged using a Bruker Ultima system. We used a Leica 20X HCX APO 1.0 NA water immersion objective lens, a piezo objective mount, resonant scanning and GaAsP PMTs. GCaMP6s and tdTomato signals were excited with a Chameleon Vision II laser (Coherent) at 920nm, and collected through a 525/50nm or a 595/50nm filter, respectively. Both signals were simultaneously collected using resonant scanning mode. Imaging volumes were acquired at an XY resolution of 358 × 148 (corresponding to a pixel size of 1.05 μm × 1.05 μm), with 41 z-sections separated by 1.5μm steps, at a volume rate of 2.6Hz. During the entire imaging session embryos were submerged in an oxygenated saline solution (as above), and kept at 25C°.
Immunostaining and confocal imaging
Immunostaining was performed as previously described 28. The 1° antibody used was chicken anti-GFP (1:2,000, Abcam). The 2° antibody used was anti-chicken Alexa 488 (1:500, Life Technologies/Thermo Fisher). Confocal image stacks were acquired on a Leica SP8, using 40X HC PL APO 40X 1.3NA oil objective and a HyD detector. Images were processed in Fiji (https://imagej.net/Fiji). Adjustments to brightness and contrast were applied uniformly to the entire image.
Behavior data collection
Parents were crossed and fed with wet yeast containing 0.5 mM all trans-Retinal (ATR) at least three days before embryo collection. ATR and yeast were replaced every day. Embryos were collected for 30 minutes on standard 3.0% agar molasses collection caps covered with a thin layer of wet yeast without ATR and incubated at 25°C in darkness. 15.5 hours later, embryos were placed under a 3.8uW/mm2 550nm LED for 5 hours. Light pulses 600ms long were delivered at one second intervals, as previously shown to induce inhibition in Drosophila embryos using halorhodopsin 23. Halorhodopsin, like GtACR1, is a silencing optogenetic tool that relies on chloride ions entering the cell. Control animals were kept in the same incubator, in darkness. One day after light exposure was terminated, at the L1 stage, animals were collected and transferred to a Petri dish with1.0% agar and relocated to a room kept at 23°C and 60% humidity. After 10 minutes of acclimation to the room, groups of 8 to 12 larvae were transferred to a 7.5 × 7.5 cm 1.0% agar arena. After 15 to 30 seconds, locomotion was recorded using a FIM imaging system 17 (https://www.uni-muenster.de/Informatik.AGRisse/media/fim-media.html) at 10 fps for 5 minutes. The FIM system was equipped with an azA2040–25gm camera (Basler) and a TEC-V7X macro zoom lens (Computar). Individual larvae were then tracked using FIMtrack software 17,29. Primary measurements from FIMtrack were used for behavioral analyses (see below).
QUANTIFICATION AND STATISTICAL ANALYSIS
Processing of calcium imaging data
After image acquisition, regions of interest (ROIs) were manually drawn on the ventral nerve cords and mean intensities were extracted using LAS X software (Leica Microsystems). For Figure 2 ROIs were drawn on Fiji (https://imagej.net/Fiji). To account for movement of the embryo and changes in gene expression over time, we encoded and recorded a structural fluorescent marker (tdTomato) in conjunction with the calcium sensor (GCaMP6s) and considered the ratio of the latter to the former as our measurement of calcium levels in the embryo. This ratiometric calcium signal was then converted into ΔF/F signal, dependent on a baseline signal computed separately for each embryo. For figure 2, the initial baseline for ΔF/F prior to peak detection was determined by calculating the mean of the 100 values lowest ratiometric values. For figures 1 and 3, the baseline for each time point in the ratiometric calcium signal was computed as a function of 16 minutes of the signal flanking the time point of interest (8 minutes prior to and 8 minutes after the time point). The 16-minute signal was divided into 20 bins of signal amplitude ranges. The bin with the largest number of samples was taken to primarily reflect the baseline, while other bins were taken to reflect deviations from the baseline. The choice of 20 bins was made empirically based on the sparsity of neuronal activity. The mean of the samples in the largest bin was considered the baseline value for the time point in the middle. At the two edges of the signal, where the full 8 minutes prior to or after the considered time point do not exist, linear fits were used as the baseline. A 150-second, quartic Savitzky–Golay filter was applied to the resulting ΔF/F signal.
Episode and peak detection
For figure 2, a 150-second, cubic Savitzky–Golay filter was first applied to the initial ΔF/F trace. Standard deviation for the filtered data was then calculated. Candidate first episodes were detected by finding the first instance where the filtered signal is equal to or greater than 1.2 times the standard deviation. Given that the intervals between episodes are at least 25 minutes, the large increase in signal must appear after a minimum of 30 minutes in order to be considered as a bona fide first episode. These candidate episodes were then manually curated for miss-called episodes due to small fluctuations that resulted in rapid increase in signal but were not sustained over longer than 20 seconds. Traces were then trimmed from −245 to 800 seconds (time series plots) relative to the initiation of the episode. These traces were used to calculate a new ΔF/F with a new baseline that was calculated as the mean of the 25 timepoints with the lowest signal. The Seaborn library was then used to plot traces.
For Figures 1 and 3, where episodes throughout PaSNA were monitored, peaks in the ΔF/F signal were detected using thresholds in the zeroth, first, and second derivatives of the signal. Each derivative signal was filtered with a 150-second, quartic Savitzky-Golay filter. The first derivative threshold was used to detect a rapid rise, while the second derivative was used to detect concavity. First, values crossing the zeroth derivative threshold were identified as peak candidates. A minimum peak distance of 500 seconds was enforced, following a greedy heuristic that kept candidates with the largest values first. Manually analyzed data showed that distinct episodes are invariably separated by more than 600 seconds. Then, of the remaining candidates, only those preceded by threshold crossings in both the first derivative and the second derivative within 210 seconds were kept and detected as episodes. Minimum thresholds of 0.06 for the zeroth derivative and 0.006 for the first derivative were derived empirically, while the maximum threshold of 0 for the second derivative was chosen to select for concavity. For Figure 3 and FigureS2 we calculated the area under the curve 20 seconds surrounding the peak using the trapezoidal rule.
Stereotypy of episode timing
We assessed the extent to which PaSNA episodes occurred with stereotyped interpeak intervals by comparing the episode interval distributions of embryos to those generated under a null model. Under the null model, episodes corresponding to each real embryo occurred randomly following a Poisson process with a rate parameter equal to the mean rate of the first twelve episodes in the real embryo. In our Monte Carlo sampling of interpeak intervals, we rejected those under 500 seconds, consistent with the minimum peak distance imposed in our peak detection algorithm. We sampled 1,000,000 model embryos for each of 23 real embryos such that more than 500,000 remained after the rejections. We then computed the root mean squared error (RMSE) from the mean for each model dataset of 23 embryos, and the resulting distribution was compared against the RMSE computed for each peak in the real dataset. We assessed significance by examining how many model datasets had RMSE lower than that of each peak in the real dataset, and corrected for multiple comparisons using the Holm-Bonferroni method.
Statistical analysis
Statistical tests for Figure 3F, I, J, M; Figure S2A, B, E, H, I, J were done with Graphpad Prism. Statistical tests for Figure 3H, L; FIgureS2C, D, G, K were done with scipy and statsmodels libraries. For two group comparisons with equal variance, we conducted unpaired-student’s t-test. For two group comparisons with unequal variance, we conducted Welch’s t-test. For three groups comparisons with unequal variance, we conducted Brown-Forsythe and Welch ANOVA followed by a Dunnett’s T3 multiple comparison test. For multiple comparison of two groups we used an unpaired student’s t-test with Holm-Bonferroni method.
Behavioral analysis
Primary measurements from FIMtrack 30 reflecting larval size, shape, and velocity were used for input, in addition to the angular velocity of the head, midpoint, and tail. Size measurements (i.e. area, perimeter, radii, spine length) were detrended using the ma function in the R package forecast (window size = 10) and converted to z-scores. Principal component analysis was used to control for potentially redundant information in the input features, yielding 8 principal components that explained >90% of the variance in the feature set. To find the appropriate timescale with which to analyze the behavioral features an empirical window search procedure was used (described in 18). We constructed behavior spaces using the top 8 PCs sweeping window sizes ranging between 100 ms and 5 seconds. For a given window size (denoted w), the windows were compiled as follows: given frame i, the 8 PCs corresponding to frames i:i+w were linearized and concatenated, resulting in a vector with length 8w. This was repeated for all windows in the data set and the resulting vectors were appended to produce a window matrix with 8w rows. A behavior space was then constructed by embedding this matrix into low-dimensional space via the UMAP algorithm 31. The appropriate window size was then determined by comparing the structural (Procrustes and Euclidean distance) and temporal features (recurrence) of behavior spaces produced from 20 random trials per window size. As was found before 18, a window size of 800ms was chosen.
We then created a behavior space encompassing the full control and transient inhibition datasets using this window size. Trials were first filtered to include those that were longer than 2.5 seconds and that traveled at least 50mm, resulting in 84 control and 97 transient inhibition trials and a total of 179,409 frames. The resulting behavior space captured the major components of the larval foraging ethogram (Figures 4B-C). Differences in behavior patterns between the conditions were inferred using 2-dimensional kernel density estimation (as in Figures 4B-C) computed over all trials for each condition. The difference map in Figure 4F was produced by first normalizing via division by the greatest value (to produce a range of values between 0 and 1) and then subtracting the transient inhibition map from the control map. Differences in individual feature distributions (as in Figure 4G) were assessed using a Kruskal-Wallis test comparing the mean value for each trial across conditions (for each test n = 84 control and n = 97 for inhibited). To control for the autocorrelation in behavior we sampled each measurement every 10th frame for a total of ~ 8,000 measurements per condition.
Louvain clustering was used to identify discrete components of behavior space. First, a graph was created with 2 sets of edges: the first representing the xy-coordinates in behavior space for each frame and the second corresponding to the xy-coordinates of the immediately following frame. This graph provides both information about the local neighborhood densities of the points in behavior space and the temporal sequencing between points over time. Louvain clustering was then run on this graph using the function cluster_louvain in the R package igraph 32. Differences in occurrence in each cluster between the conditions were assessed using a Kruskal-Wallis test, again comparing the occurrence density of all individual trials between the two conditions (n = 84 control and n = 97 for inhibited).
Supplementary Material
Video S1. High-throughput wide-field calcium imaging. Related to Figure 1. Pan-neuronal GCaMP6s (top) and TdTomato (bottom) signals of an individual representative embryo throughout PaSNA at 750 times real time speed. Video is an XY cropped region from the time-lapse used for the snapshot shown on Figure 1. Scale bar is 100μm and the timestamp is in minutes:seconds.
Video S2. Two-photon calcium imaging of the first PaSNA episode. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative control embryo imaged using two photon microscopy.
Video S3. PaSNA depends on depolarizations. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative embryo expressing Kir2.1 pan-neuronally, imaged using two photon microscopy.
Video S4. PaSNA depends on synaptic transmission. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative embryo expressing TNT pan-neuronally, imaged using two photon microscopy.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP | Abcam | ab13970 |
| Anti-chicken Alexa 488 | ThermoFisher | A-11039 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| All-trans-retinal | Sigma-Aldrich | R2500–100MG |
| Deposited Data | ||
| PaSNA data | This paper | https://github.com/ClandininLab/pasna-acr-currbio2021 |
| Experimental Models: Organisms/Strains | ||
| UAS-IVS-Syn21-GCaMP6s-P2A-nls-tdTomato-p10 (JK66B) | Gift from Marta Zlatic | N/A |
| LexAop- Kir2.1 (VIE-260B) | Gift from Barry Dickson | Feng et al. 33 |
| UAS- Kir2.1(VIE-260B) | Gift from Barry Dickson | Feng et al. 33 |
| LexAop-TNT (VIE-260B) | Gift from Barry Dickson | Sen et al. 34 |
| pBDP-LexA:p65 (attp40) | Gift from Troy Shirangi | Shirangi et al.35 |
| UAS-GtACR1(attP2) | Gift from Adam Claridge-Chang | Mohammad et al.16 |
| elav-GAL4.L on 3rd | Bloomington Drosophila Stock Center | BDSC 8760 |
| elav-GAL4.L on 2nd | Bloomington Drosophila Stock Center | BDSC 8765 |
| GMR44H10-lexa::p65 (attP40) | Bloomington Drosophila Stock Center | BDSC 61543 |
| elavc155-GAL4 | Bloomington Drosophila Stock Center | BDSC 458 |
| UAS-TeTxLC.tnt G2 | Bloomington Drosophila Stock Center | BDSC 28838 |
| UAS-mLexA-VP16-NFAT, LexAop-rCD2-GFP (CaLexA) | Bloomington Drosophila Stock Center | BDSC 66542 |
| LexAop-CD8-GFP-2A-CD8-GFP | Bloomington Drosophila Stock Center | BDSC 66545 |
| iav-lexA::p65 (VK00013) | Bloomington Drosophila Stock Center | BDSC 52246 |
| attP-9A (VK000013) | Bloomington Drosophila Stock Center | BDSC 9732 |
| iav-GAL4.K | Bloomington Drosophila Stock Center | BDSC 52273 |
| Software and Algorithms | ||
| FIMtrack | WWU Munster | http://www.uni-muenster.de/Informatik.AGRisse/media/fim-media.html |
| Fiji | ImageJ | http://imagej.net/software/fiji/ |
| Python3.6 | Python Software Foundation | https://www.python.org/ |
| Prism_9 | GraphPad | https://www.graphpad.com/ |
| R | The R Project for Statistical Computing | https://www.r-project.org/ |
| TREBLE | York et al. 18 | https://www.biorxiv.org/content/10.1101/2020.09.30.321406v2 |
| PaSNA analysis | This paper | https://github.com/ClandininLab/pasna-acr-currbio2021 |
Highlights.
PaSNA in the Drosophila embryonic CNS is spatiotemporally stereotyped
Mechanosensory neurons negatively modulate PaSNA
Embryonic PaSNA is required for innate locomotor behavior
Acknowledgements
We thank Estela Stephenson for technical support and members of the Clandinin lab and the Doe lab for support and helpful discussions on this work. We thank D. Berfin Azizoglu for comments on the manuscript. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Funding was provided by HHMI (CQD, AC-R), NIH HD27056 (CQD), F32NS105350–01A1, The Walter V. and Idun Berry Postdoctoral Fellowship, K99NS119295–01 (AC-R), the Stanford School of Medicine Dean’s Fellowship (RAY), The Simons Foundation (TRC), and the National Defense Science & Engineering Graduate Fellowship, Stanford Graduate Fellowship, and Stanford Mind, Brain, Computation and Technology Training Program (MC). Lastly, we want to acknowledge that our laboratory at Stanford University stands on the ancestral lands of the Muwekma Ohlone Tribe (http://muwekma.org/home.html).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare no competing interests.
References
- 1.Goodman CS, and Shatz CJ (1993). Developmental mechanisms that generate precise patterns of neuronal connectivity. Cell 72, 77–98. [DOI] [PubMed] [Google Scholar]
- 2.Kirkby LA, Sack GS, Firl A, and Feller MB (2013). A role for correlated spontaneous activity in the assembly of neural circuits. Neuron 80, 1129–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson A, Gribizis A, Chen C, and Crair MC (2017). Activity-dependent development of visual receptive fields. Current Opinion in Neurobiology 42, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereanu W, Spindler S, Im E, Buu N, and Hartenstein V. (2007). The Emergence of Patterned Movement During Late Embryogenesis of Drosophila. Dev Neurobiology, 1669–1685. [DOI] [PubMed]
- 5.Crisp S, Evers JF, Fiala A, and Bate M. (2008). The development of motor coordination in Drosophila embryos. Development 3717, 3707–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crisp SJ, Evers JF, and Bate M. (2011). Endogenous Patterns of Activity Are Required for the Maturation of a Motor Network. J. Neurosci 31, 10445–10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos-Ortega JA, and Hartenstein V. (2013). The Embryonic Development of Drosophila melanogaster (Springer Science & Business Media; ). [Google Scholar]
- 8.Olofsson B, and Page DT (2005). Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol 279, 233–243. [DOI] [PubMed] [Google Scholar]
- 9.Baines RA, Uhler JP, Thompson A, Sweeney ST, and Bate M. (2001). Altered Electrical Properties in DrosophilaNeurons Developing without Synaptic Transmission. J. Neurosci 21, 1523 LP–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masuyama K, Zhang Y, Rao Y, and Wang JW (2012). Mapping Neural Circuits with Activity-Dependent Nuclear Import of a Transcription Factor. J. Neurogenet 26, 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hehlert P, Zhang W, and Göpfert MC (2020). Drosophila Mechanosensory Transduction. Trends Neurosci. [DOI] [PubMed]
- 12.Fushiki A, Kohsaka H, and Nose A. (2013). Role of Sensory Experience in Functional Development of Drosophila Motor Circuits. PLoS One 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldwell JC, Miller MM, Wing S, Soll DR, and Eberl DF (2003). Dynamic analysis of larval locomotion in Drosophila chordotonal organ mutants. Proc. Natl. Acad. Sci. U. S. A 100, 16053–16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng LE, Song W, Looger LL, Jan LY, and Jan YN (2010). The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron 67, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zarin AA, Mark B, Cardona A, Litwin-Kumar A, and Doe CQ (2019). A multilayer circuit architecture for the generation of distinct locomotor behaviors in Drosophila. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammad F, Stewart J, Ott S, Chlebikova K, Chua JY, Koh T-W, Ho J, and Claridge-Chang A. (2017). Optogenetic inhibition of behavior with anion channelrhodopsins. Nat. Methods 14, 271–274. [DOI] [PubMed] [Google Scholar]
- 17.Risse B, Thomas S, Otto N, Löpmeier T, Valkov D, Jiang X, and Klämbt C. (2013). FIM, a Novel FTIR-Based Imaging Method for High Throughput Locomotion Analysis. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.York RA, Carreira-Rosario A, Giocomo LM, and Clandinin TR (2021). Flexible analysis of animal behavior via time-resolved manifold embedding. Cold Spring Harbor Laboratory, 2020.09.30.321406. [Google Scholar]
- 19.Green CH, Burnet B, and Connolly KJ (1983). Organization and patterns of inter- and intraspecific variation in the behaviour of Drosophila larvae. Anim. Behav 31, 282–291. [Google Scholar]
- 20.Clark MQ, McCumsey SJ, Lopez-Darwin S, Heckscher ES, and Doe CQ (2016). Functional Genetic Screen to Identify Interneurons Governing Behaviorally Distinct Aspects of Drosophila Larval Motor Programs. G3 6, 2023–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akin O, Bajar BT, Keles MF, Frye MA, and Zipursky SL (2019). Cell-type-Specific Patterned Stimulus-Independent Neuronal Activity in the Drosophila Visual System during Synapse Formation. Neuron 101, 894–904.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khazipov R, and Luhmann HJ (2006). Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 29, 414–418. [DOI] [PubMed] [Google Scholar]
- 23.Giachello CNG, and Baines RA (2015). Inappropriate Neural Activity during a Sensitive Period in Embryogenesis Results in Persistent Seizure-like Behavior. Curr. Biol 25, 2964–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giachello CNG, and Baines RA (2017). Regulation of motoneuron excitability and the setting of homeostatic limits. Curr. Opin. Neurobiol 43, 1–6. [DOI] [PubMed] [Google Scholar]
- 25.Valdes-Aleman J, Fetter RD, Sales EC, Heckman EL, Venkatasubramanian L, Doe CQ, Landgraf M, Cardona A, and Zlatic M. (2021). Comparative Connectomics Reveals How Partner Identity, Location, and Activity Specify Synaptic Connectivity in Drosophila. Neuron 109, 105–122.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino H, Hwang EJ, Hedrick NG, and Komiyama T. (2016). Circuit Mechanisms of Sensorimotor Learning. Neuron 92, 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shearin HK, Dvarishkis AR, Kozeluh CD, and Stowers RS (2013). Expansion of the gateway multisite recombination cloning toolkit. PLoS One 8, e77724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manning L, and Doe CQ (2017). Immunofluorescent antibody staining of intact Drosophila larvae. Nat. Protoc 12, 1–14. [DOI] [PubMed] [Google Scholar]
- 29.Risse B, Berh D, Otto N, Klämbt C, and Jiang X. (2017). FIMTrack: An open source tracking and locomotion analysis software for small animals. PLoS Comput. Biol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Risse B, Otto N, Berh D, Jiang X, and Klämbt C. (2014). FIM imaging and FIMtrack: Two new tools allowing high-throughput and cost effective locomotion analysis. J. Vis. Exp [DOI] [PMC free article] [PubMed]
- 31.McInnes L, Healy J, and Melville J. (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML].
- 32.Csardi G, Nepusz T, and Others (2006). The igraph software package for complex network research. InterJournal, complex systems 1695, 1–9. [Google Scholar]
- 33.Feng K, Palfreyman MT, Häsemeyer M, Talsma A, and Dickson BJ (2014). Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron 83, 135–148. [DOI] [PubMed] [Google Scholar]
- 34.Sen R, Wang K, and Dickson BJ (2019). TwoLumps Ascending Neurons Mediate Touch-Evoked Reversal of Walking Direction in Drosophila. Curr. Biol 29, 4337–4344.e5. [DOI] [PubMed] [Google Scholar]
- 35.Shirangi TR, Stern DL, and Truman JW (2013). Motor Control of Drosophila Courtship Song. Cell Rep. 5, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. High-throughput wide-field calcium imaging. Related to Figure 1. Pan-neuronal GCaMP6s (top) and TdTomato (bottom) signals of an individual representative embryo throughout PaSNA at 750 times real time speed. Video is an XY cropped region from the time-lapse used for the snapshot shown on Figure 1. Scale bar is 100μm and the timestamp is in minutes:seconds.
Video S2. Two-photon calcium imaging of the first PaSNA episode. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative control embryo imaged using two photon microscopy.
Video S3. PaSNA depends on depolarizations. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative embryo expressing Kir2.1 pan-neuronally, imaged using two photon microscopy.
Video S4. PaSNA depends on synaptic transmission. Related to Figure 2. Maximum intensity projections of pan-neuronal GCaMP6s signals in a representative embryo expressing TNT pan-neuronally, imaged using two photon microscopy.
Data Availability Statement
All original code for Figures 1–3 has been deposited at https://github.com/ClandininLab/pasna-acr-currbio2021 and is publicly available as of the date of publication.


