Abstract
Background:
Diabetic patients have a greater incidence of adhesive capsulitis (AC) and a more protracted disease course than patients with idiopathic AC. The purpose of this study was to compare gene expression differences between AC with diabetes mellitus and AC without diabetes mellitus.
Methods:
Shoulder capsule samples were prospectively obtained from diabetic or nondiabetic patients who presented with shoulder dysfunction and underwent arthroscopy (N = 16). Shoulder samples of AC with and without diabetes (n = 8) were compared with normal shoulder samples with and without diabetes as the control group (n = 8). Shoulder capsule samples were subjected to whole-transcriptome RNA sequencing, and differential expression was analyzed with EdgeR. Only genes with a false discovery rate < 5% were included for further functional enrichment analysis.
Results:
The sample population had a mean age of 47 years (range, 24–62 years), and the mean hemoglobin A1c level for nondiabetic and diabetic patients was 5.18% and 8.71%, respectively. RNA-sequencing analysis revealed that 66 genes were differentially expressed between diabetic patients and nondiabetic patients with AC whereas only 3 genes were differentially expressed when control patients with and without diabetes were compared. Furthermore, 286 genes were differentially expressed in idiopathic AC patients, and 61 genes were differentially expressed in diabetic AC patients. On gene clustering analysis, idiopathic AC was enriched with multiple structural and muscle-related pathways, such as muscle filament sliding, whereas diabetic AC included a greater number of hormonal and inflammatory signaling pathways, such as cellular response to corticotropin-releasing factor.
Conclusions:
Whole-transcriptome expression profiles demonstrate a fundamentally different underlying pathophysiology when comparing diabetic AC with idiopathic AC, suggesting that these conditions are distinct clinical entities. The new genes expressed explain the differences in the disease course and suggest new therapeutic targets that may lead to different treatment paradigms in these 2 subsets.
Level of evidence:
Basic Science Study; Molecular Biology
Keywords: Frozen shoulder, diabetes, RNA sequencing, adhesive capsulitis, whole genome sequencing, inflammation, corticosteriods
Adhesive capsulitis (AC), otherwise known as “frozen shoulder,” is a common affliction, affecting 2%−5% of the general population.21 The disease process of AC is known to include fibroproliferative changes in the shoulder associated with capsular inflammation causing pain and limitation in motion, particularly external rotation. Magnetic resonance imaging and arthroscopic visualization have consistently characterized a thickening and irritation of the rotator cuff interval among patients with this disorder.47 Some studies have also suggested the possibility of a chondrogenic or neuro-dystrophic etiology for AC.15
The underlying pathophysiology of AC is not yet completely understood, and treatments for the condition have remained relatively unchanged and continue to be chiefly marginally effective. Patients generally continue to be treated with physical therapy and often endure years of limited motion and pain.31 Frequently, this conservative approach is accompanied by permanently reduced range of motion. Previous studies of human capsular samples of AC have led to inconsistent histologic results,9–11,19,27,29,35,50,52 making consensus characterizing the disease difficult to achieve.3,12,33 Cell types, such as the myofibroblast, and specific signaling pathways have been identified in numerous studies, with heterogeneous findings1,9,13,21,29,30,32,37,40,47,52; notably, the inciting events of the underlying condition remain incompletely understood.3,12,33 Furthermore, previous studies have not pointed to a unified homogeneous disease process as results have varied with treatment. It is clear that thickening and irritation of the joint capsule occur and become typically unpredictably responsive to anti-inflammatory treatments such as nonsteroidal anti-inflammatory drugs or corticosteroid injections.10,41 In the most protracted cases, symptoms continue for months or years and may result in permanent disability.49
Although AC most commonly presents with no precisely attributable etiology (idiopathic AC), several risk factors have been described in the literature. AC more commonly occurs in patients with thyroid disorders,14 autoimmune dysfunction,8 serum lipid alterations,43 and Parkinson disease,45 as well as other fibroproliferative diseases such as Dupuytren contracture or Lederhosen disease.33,45,51 Notably, patients with diabetes and associated conditions such as hyperglycemia and metabolic syndrome have a much higher prevalence of AC compared with the general population.5,36 The degree of hyperglycemia correlates with both the prevalence and the prognosis of AC, with patients with insulin-dependent diabetes presenting with AC at appreciably greater rates than patients receiving oral hypoglycemic agents.53 Similarly, diabetic patients, especially those with poorly controlled diabetes, fare much less well with nonoperative management than idiopathic AC patients.46 Diabetic patients in whom AC develops also typically experience a more protracted disease course, often with failure of conservative management after months or years of physical therapy and the eventual need for arthroscopic capsular release.31
The purpose of this study was to compare genome-wide expression differences between AC with diabetes mellitus and AC without diabetes mellitus. Given that diabetes-associated AC generally presents with a more severe degree of stiffness and is less responsiveness to conservative treatment, we hypothesized that idiopathic AC and diabetic AC are distinct clinical entities with unique underlying pathophysiology, even though the histologic and gross appearance of tissue may be similar for both types of AC.2 Understanding the pathophysiology of AC is essential to direct more targeted treatments of this widespread condition, which affects many patients. Our hypothesis is supported by previous clinical findings that have shown differential outcomes of AC based on patient risk factors. We used RNA sequencing to compare genome-wide expression profiles for patient samples taken from AC patients and control patients both with and without carrying a confirmed diagnosis of diabetes mellitus. Our study focuses on a profoundly important clinical problem for many patients and reveals new transcriptional differences in these disorders that notably could translate readily into clinical practice paradigms.
Materials and methods
Study design
Thirty-five patients presenting with shoulder pain or dysfunction to a large academic medical center and undergoing subsequent arthroscopy were prospectively enrolled in this study from 2015 to 2018. The inclusion criteria consisted of patients with AC undergoing arthroscopy or patients without AC undergoing arthroscopy owing to shoulder dysfunction attributable to other glenohumeral pathology, such as a rotator cuff tear. AC was defined as dysfunctional shoulder pain with restricted active and passive range of motion without other contributing glenohumeral pathology seen during preoperative imaging or arthroscopy. The exclusion criteria were patients with a history of thyroid disease, those with a history of surgery for AC, and those with other pathology affecting the shoulder capsule.
In total, 16 patients were included for analysis in this study and were stratified by the presence or absence of diabetes, determined through review of the electronic medical record or preoperative A1c screening if patients had relevant risk factors, such as a family history or obesity. All diabetic patients in this study had type 2 diabetes, and no patients had a history of type 1 (insulin-dependent) diabetes. Diabetic patients with AC (diabetic AC, n = 4) were matched with control diabetic patients without AC (n = 4) for analysis. Similarly, nondiabetic patients with AC (idiopathic AC, n = 4) were matched with nondiabetic control patients without AC (n = 4) for analysis.
Patient demographic characteristics
The patients had a mean age of 47 years (range, 24–62 years). The mean duration of AC for diabetic and nondiabetic patients was 9.5 months and 9.7 months, respectively. The right shoulder was involved in 50% of diabetic AC patients and 75% of idiopathic AC patients. Diagnoses in control patients without AC included rotator cuff tear or sprain (n = 5), shoulder instability (n = 2), and arthritis (n = 1). When patients with AC were compared with control patients without AC, 7 of 8 patients with AC had received at least a single intra-articular steroid injection vs. 2 of 8 patients without AC (Supplementary Table SI).
The mean hemoglobin A1c level for nondiabetic and diabetic patients was 5.18% and 8.71%, respectively. Diabetic patients with and without AC had a mean duration of diabetes of 9.1 years and 11.1 years, respectively. The mean A1c level of diabetic patients with and without AC was 7.8% and 9.6%, respectively.
Capsular tissue procurement
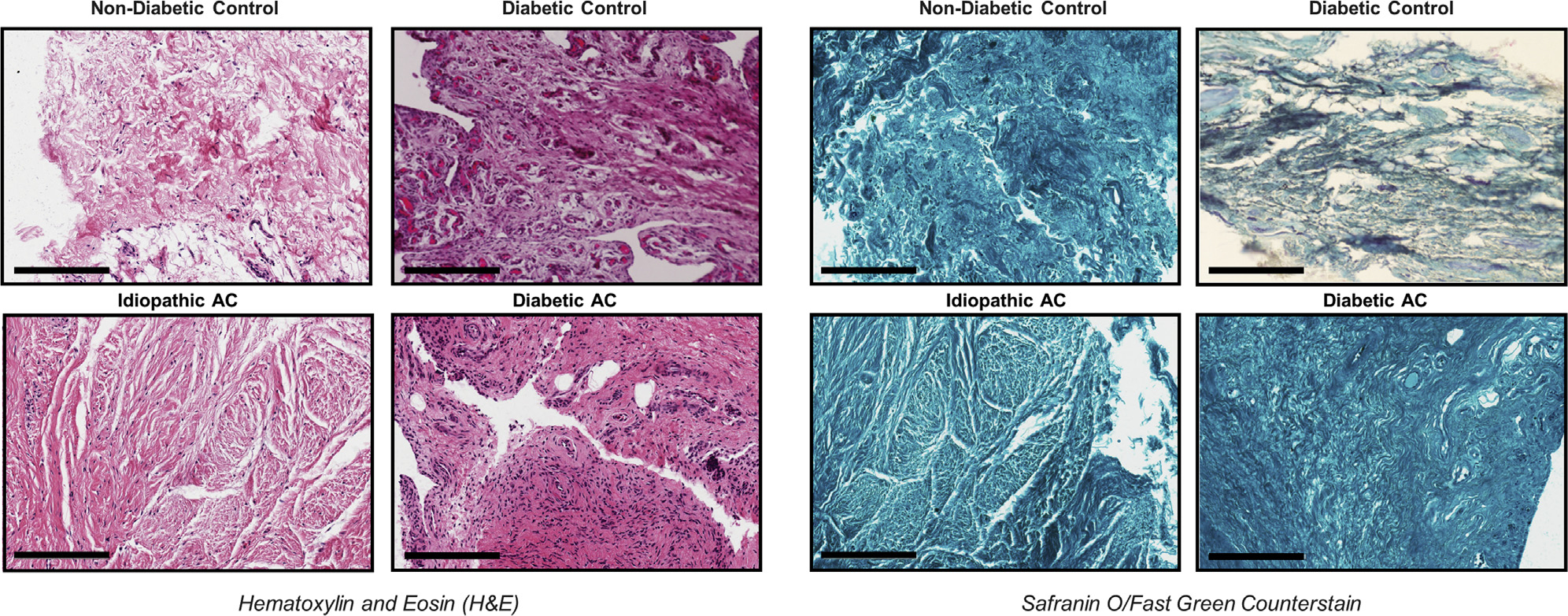
Shoulder capsular tissue was obtained from the rotator interval intraoperatively, with some portions of capsular tissue placed in ribonuclease-free microcentrifuge tubes, snap frozen in liquid nitrogen, and stored at −80°C. Remaining portions of capsular tissue were rinsed in sterile phosphate-buffered saline solution and fixed in 4% paraformaldehyde at 4°C for 4–5 days, followed by automatic tissue processing. Seven 1-μm-thick sections were stained with hematoxylin-eosin staining as well as safranin O with fast green counterstaining using routine procedures.
Tissue dissociation and RNA isolation and sequencing
Dissociation of capsular tissue for RNA extraction was performed using a customized protocol and the Miltenyi Biotec gentleMACS Dissociator (Gaithersburg, MD, USA). The small biopsy specimen from previously snap-frozen capsular tissue (0.5–1.0 mg) was transferred to specialized M Tubes (Miltenyi Biotec) with grinding units, along with 0.5 mL of Qiazol (Qiagen, Germantown, MD, USA). Following the manufacturer’s instructions, the M Tube cap was closed tightly to the second click and then attached to the instrument, cap down. The preset program “RNA02” for 30 seconds was used for all frozen tissue. Samples were then transferred to microtubes for immediate RNA extraction by the standard phenol-chloroform method using Qiazol. The resulting RNA fraction was further purified using a Qiagen RNeasy mini spin column per the manufacturer’s instructions. The eluate volume was 20 μL, and the resulting RNA concentration was determined using a spectrophotometer (NanoDrop 1000; Thermo Fisher Scientific, Waltham, MA, USA). The RNA quality was characterized by electrophoresis of 1.0 ng on a Bioanalyzer RNA chip (Agilent Instruments, Santa Clara, CA, USA). Assessment of the purity of all RNA preparations used in RNA sequencing showed RNA integrity numbers > 6.0.
Sequencing libraries were generated using 200 mg of mRNA via the TruSeq Stranded Total RNA high-throughput library preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer’s recommendations. The libraries were sequenced on an Illumina HiSeq 4000 system to 100 base pairs, single ended, and the sequencing depth was between 8.3 and 11.7 million reads per sample. Reads were aligned to the human genome (hg19) using the RNA-seq Unified Mapper (RUM) pipeline,18 which also produced read-count quantifications of transcript expression. Differential expression was performed using EdgeR.38 The resulting P values were converted to false discovery rates (FDRs) using the R function “p.adjust” (R Foundation for Statistical Computing, Vienna, Austria). The RNA-sequencing analysis was carried out at the Generation Sequencing Core (Perelman School of Medicine, Philadelphia, PA, USA; ngsc.med.upenn.edu). The data are available at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO).
Data analysis and statistics
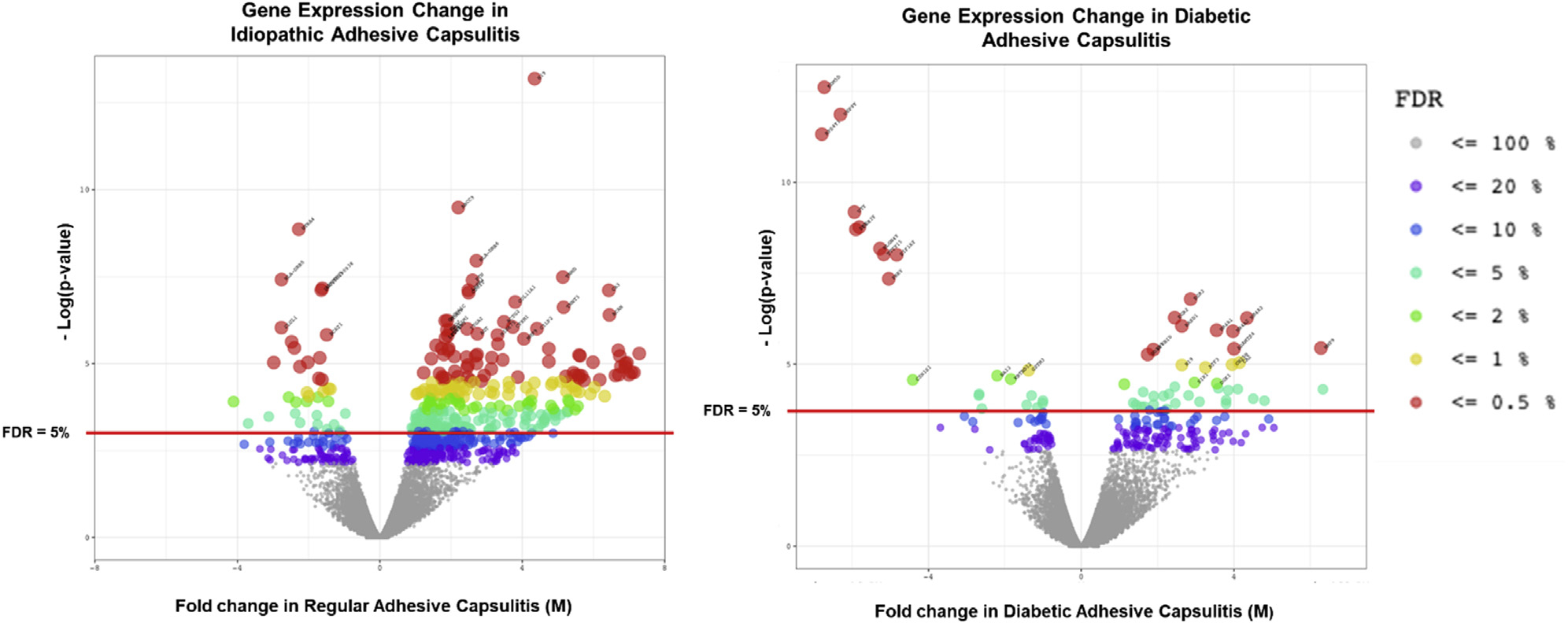
The functional and enrichment analysis of genes in idiopathic and diabetic AC patients was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID)24,25 with Gene Ontology (GO) annotations,4,17 using Benjamini-adjusted P values to determine the significance of enriched biological pathways. RNA expression levels in patients with idiopathic AC were compared with those in nondiabetic patients without AC. Similarly, genetic expression levels in diabetic patients with AC were compared with those in diabetic patients without AC. Only genes with significantly different expression in idiopathic or diabetic AC patients, defined as having an FDR < 5%, were included for further analysis (Fig. 1).
Figure 1.
Genetic expression differences in idiopathic and diabetic adhesive capsulitis patients compared with controls without AC. The RNA-sequencing results of gene expression differences in patients with adhesive capsulitis as compared with diabetic patients with adhesive capsulitis are shown. Patients with regular adhesive capsulitis were compared with normal control patients without adhesive capsulitis, and diabetic patients with adhesive capsulitis were compared with diabetic patients without adhesive capsulitis. FDR, false discovery rate; M, fold change.
Significant, differentially expressed genes in idiopathic AC were compared with the RNA expression profile in the diabetic AC cohort, and genes that differed by a fold change (FC) of at least ±2 were classified as specific to idiopathic AC. Significant genes in idiopathic AC patients that were within ±2 FC in the diabetic AC cohort, with an FDR < 50%, were classified as having similar expression in both subtypes of AC. The same process was repeated for differentially expressed genes in diabetic AC patients to classify genes specific to diabetic AC and genes with similar expression in both subtypes of AC.
Results
Histology
To confirm the location and examine differences in histopathologic findings, small portions of the rotator interval joint capsule taken for RNA preparation were taken for routine histologic analysis. Visualized with hematoxylin-eosin staining as well as safranin O with fast green counterstaining, the tissues were imaged and the tissue and cellular characteristics were evaluated (Fig. 2). All specimens showed characteristic dense layers of fibrous connective tissue with no marked differences in tissue architecture or cellularity except for diabetic AC patients’ samples, in which there were larger infiltrations of inflammatory cells.
Figure 2.
Representative histologic sections of capsular tissue in idiopathic and diabetic adhesive capsulitis (AC) demonstrating tissue and cellular characteristics. Staining was performed with hematoxylin-eosin (left panels) and safranin O with fast green counterstaining (right panels). Scale bars represent 200 μm.
RNA sequencing
RNA-sequencing results of shoulder capsular tissue demonstrated that 286 genes were significantly (FDR < 5%) differentially expressed in idiopathic AC as compared with 61 genes in diabetic AC when compared with nondiabetic controls and diabetic controls (Fig. 1). After removal of sex chromosome–linked genes from the data set, the expression levels of genes significant to idiopathic AC and diabetic AC were compared (Fig. 3). Most differentially expressed genes were upregulated in idiopathic AC (n = 251) and diabetic AC (n = 39). Of these upregulated genes, 92 of 251 were specific to idiopathic AC and 15 of 39 were specific to diabetic AC. Moreover, 54 of the upregulated genes had a similar FC in both cohorts, and 8 of these genes were significantly upregulated in both subtypes of AC: MMP9, MMP13, DGKI, KCNJ6, ADAM12, H19, POSTN, and ACAN. In contrast, there were smaller numbers of significantly downregulated genes in idiopathic AC (n = 35) and diabetic AC (n = 12). Of these downregulated genes, 7 of 35 were specific to idiopathic AC and 2 of 12 were specific to diabetic AC. Furthermore, only 2 downregulated genes had a similar FC in both cohorts (CSN1S1 and TRIM14), but neither was significantly downregulated in both subtypes of AC.
Figure 3.
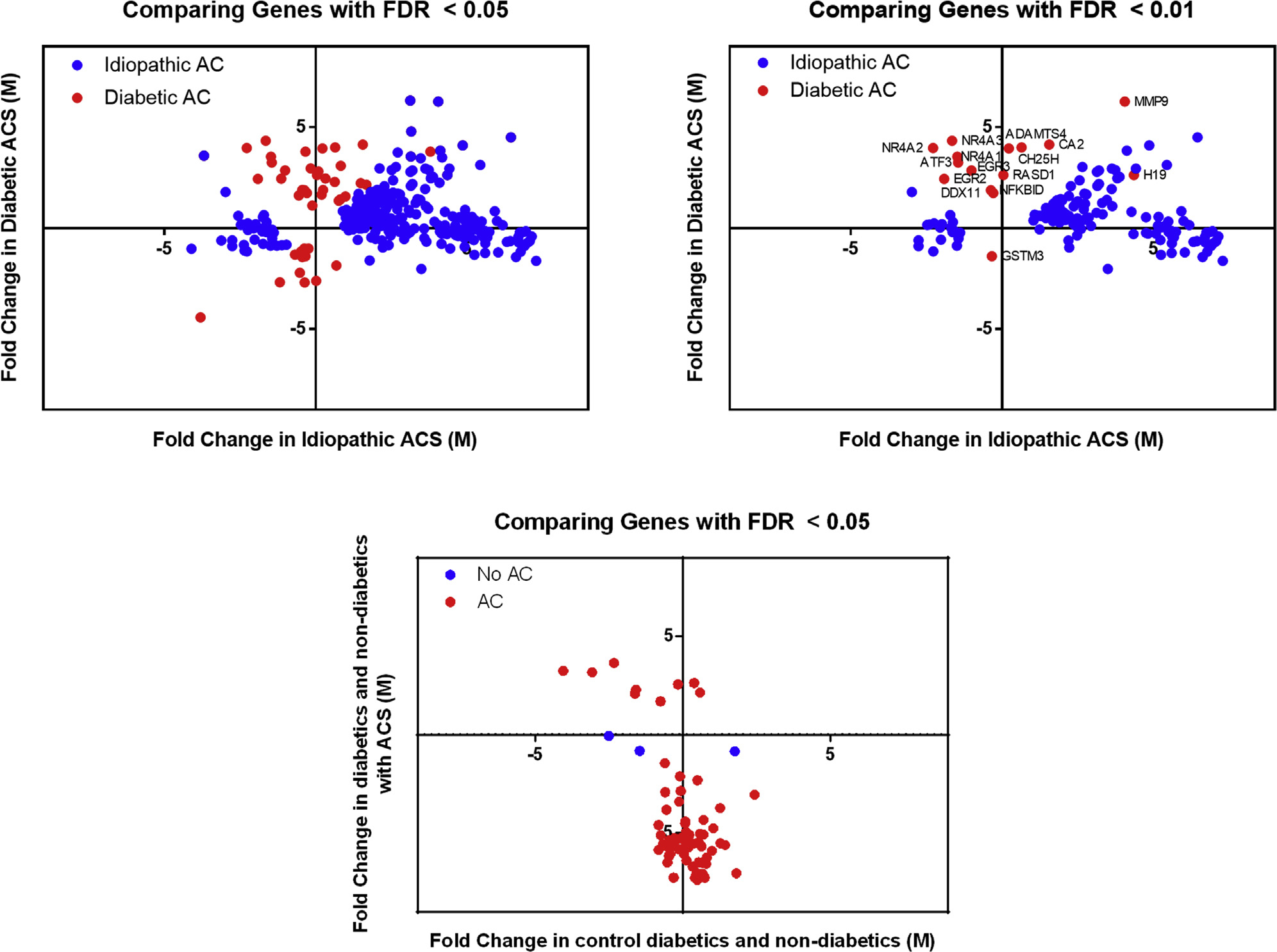
Comparison of genetic expression levels across various cohorts. The fold change of genes that were significantly (false discovery rate [FDR] < 0.05) differentially expressed in idiopathic or diabetic adhesive capsulitis (ACS/AC) were compared across patient cohorts. Additionally, the fold change of significantly differentially expressed genes in patients with AC or without AC were compared across cohorts. M, fold change.
There was minimal change in RNA expression when comparing diabetic and nondiabetic patients without AC. Specifically, comparison of nonpathologic shoulder tissue demonstrated that only 3 genes were significantly differentially expressed. In contrast, diabetic and nondiabetic patients with AC had a greater change in RNA expression. On comparison of pathologic shoulder capsular tissue, 66 genes were differentially expressed between diabetic and nondiabetic patients (Fig. 3).
Gene enrichment
Enriched biological pathways identified in diabetic AC were related to hormonal and transcriptional changes, including cellular response to corticotropin-releasing factor (CRF) stimulus, steroid hormone–mediated signaling pathway, response to glucocorticoid, response to insulin, intracellular receptor signaling pathway, and positive regulation of transcription from RNA polymerase (Pol) II promoter (Fig. 4). Other pathways in diabetic AC were relevant to cell differentiation and regulation, including skeletal muscle cell differentiation, fat cell differentiation, cellular response to fibroblast growth factor stimulus, and inflammatory response. The enriched biological pathways in idiopathic AC involved muscle cells and extracellular matrix and included pathways such as muscle filament sliding, muscle contraction, and extracellular matrix organization. Three pathways were common in both subtypes of AC: extracellular matrix disassembly, skeletal system development, and cell adhesion.
Figure 4.
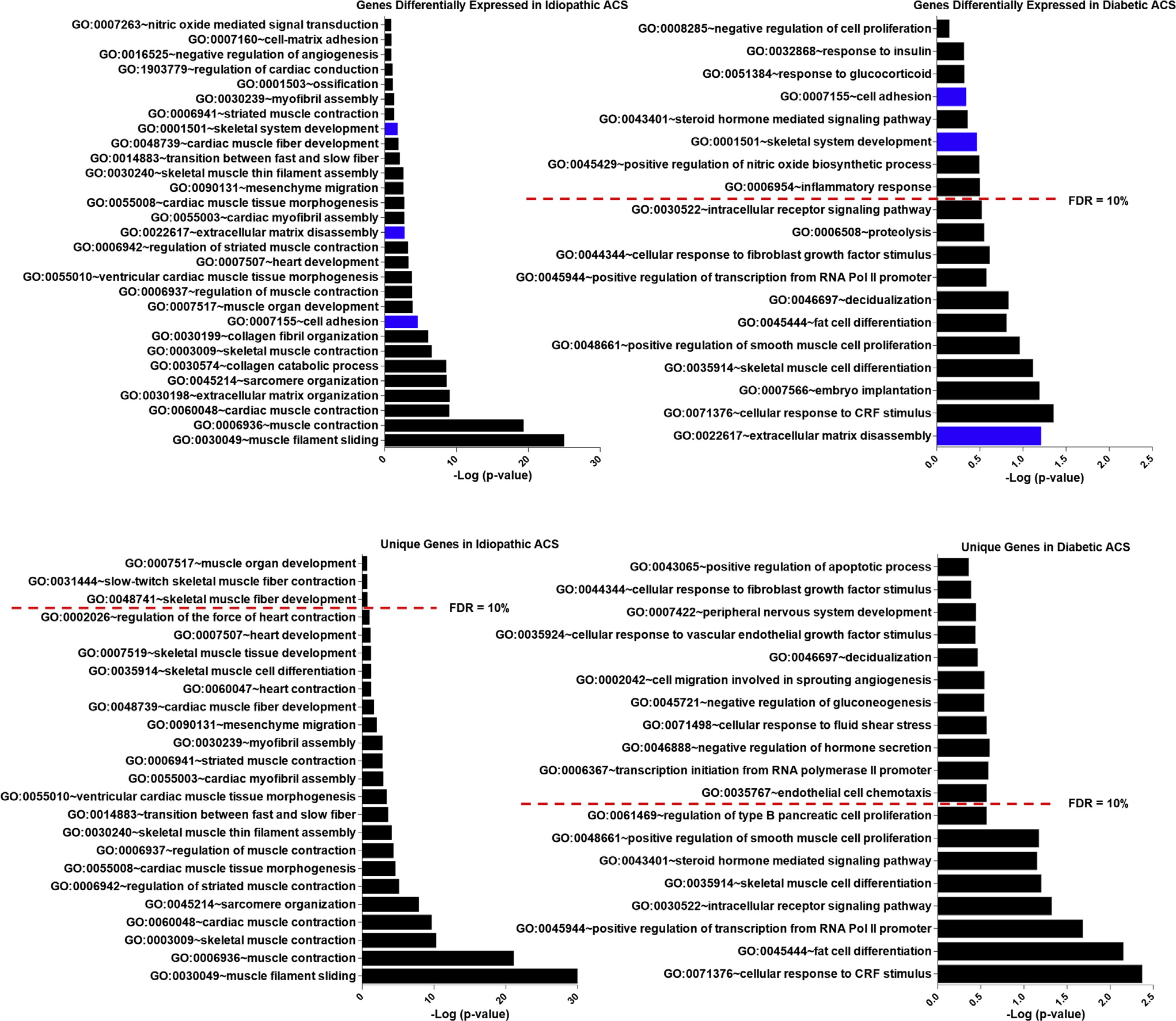
Enriched biological pathways in adhesive capsulitis (ACS): gene enrichment analysis of genes significantly associated with idiopathic and diabetic adhesive capsulitis. Biological pathways in blue were implicated in both idiopathic adhesive capsulitis and diabetic adhesive capsulitis. GO, Gene Ontology; FDR, false discovery rate; Pol, polymerase; CRF, corticotropin-releasing factor.
The most significantly enriched biological pathways were compared in AC (Figs. 5 and 6) and found to be distinct. The pathways with the higher-fold enrichment in idiopathic AC were associated predominantly with structural element pathways, whereas the pathways with higher-fold enrichment in diabetic AC included fewer structural pathways and demonstrated greater association with hormonal and cell signaling pathways. In idiopathic AC, the highest number of significantly upregulated genes expressed belonged to the muscle contraction pathway (n = 26). In diabetic AC, the highest number of significantly upregulated genes expressed belonged to the transcription from the Pol II promoter pathway (n = 10). When only the unique genes were analyzed, the pathway with the highest-fold enrichment in idiopathic AC was skeletal muscle thin filament assembly, and in diabetic AC, the pathway with the highest-fold enrichment was cellular response to CRF stimulus. The related genes and biological pathways for idiopathic AC and diabetic AC are shown in Tables I and II and Supplementary Tables SII and S3.
Figure 5.
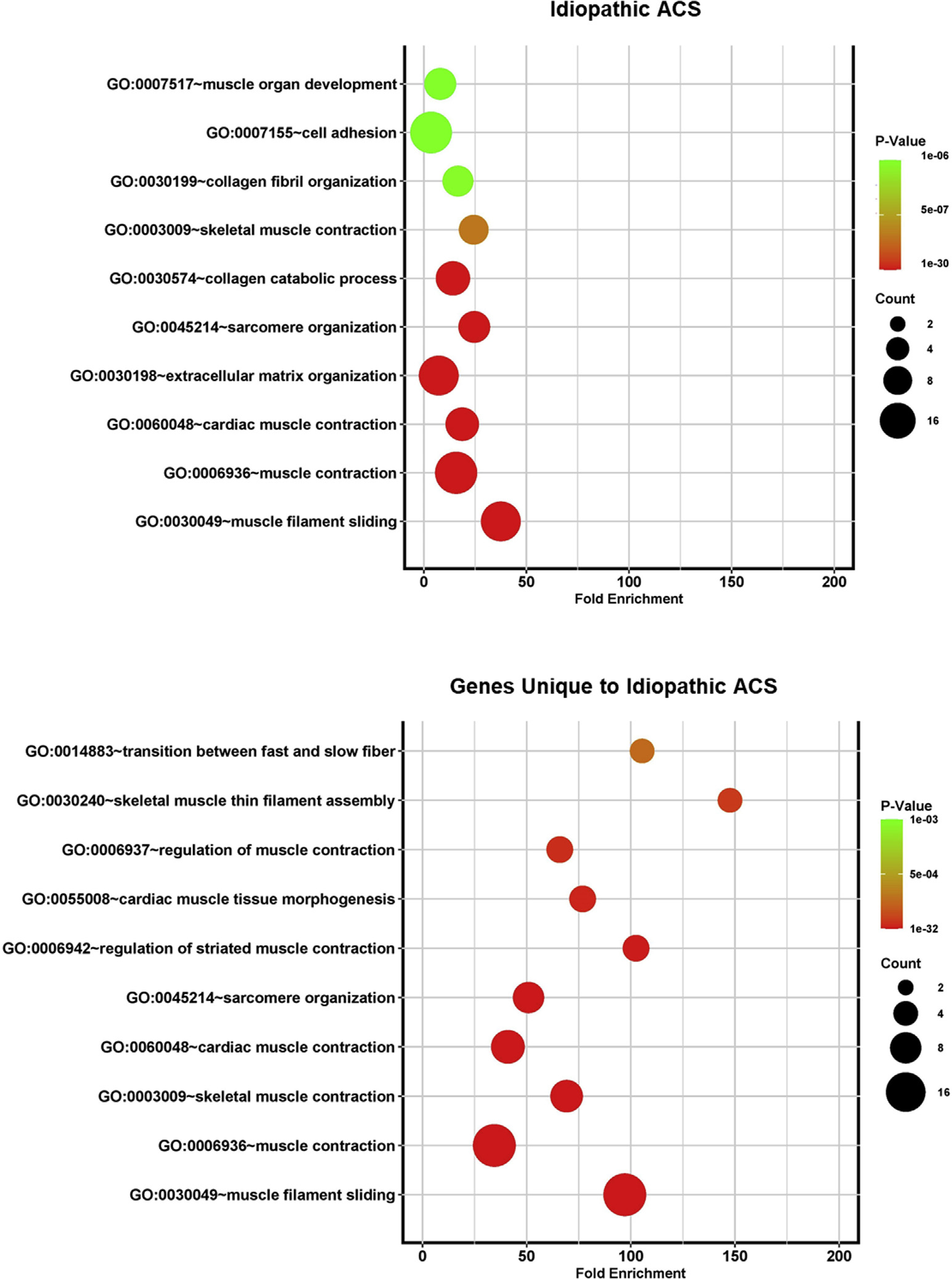
Bubble plots demonstrating fold enrichment for the most statistically significant biological pathways identified by Gene Ontology (GO) analysis for idiopathic adhesive capsulitis (ACS).
Figure 6.
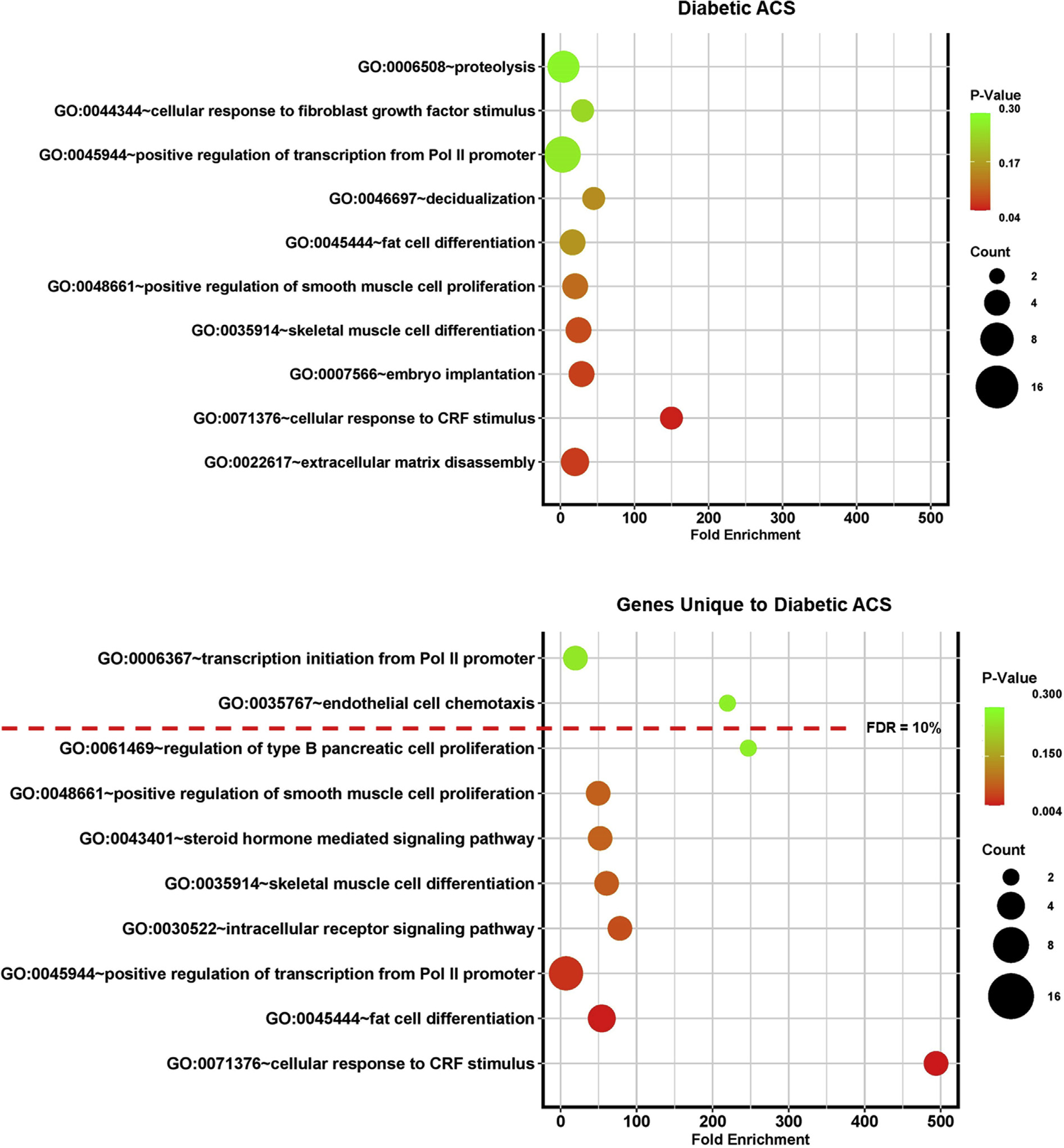
Bubble plots demonstrating fold enrichment for the most statistically significant biological pathways identified by Gene Ontology (GO) analysis for diabetic adhesive capsulitis (ACS). Pol, polymerase; CRF, corticotropin-releasing factor.
Table I.
Differentially expressed genes related to 10 most statistically significant biological pathways identified by GO analysis for idiopathic AC
| Biological pathway | Related genes |
|---|---|
| GO:0030049—muscle filament sliding | ACTA1, ACTC1, ACTN2, DES, MYBPC2, MYBPC1, MYH2, MYH7, MYL1, MYL2, MYL3, NEB, TTN, TCAP, TPM1, TPM2, TNNC1, TNNC2, TNNI1, TNNI2, TNNT1, TNNT3 |
| GO:0006936—muscle contraction | ACTA1, ACTA2, ACTG2, CACNA1S, CKMT2, DES, DYSF, HRC, LMOD2, LMOD3, MYOM1, MYOM2, MYBPC2, MYH1, MYH2, MYH7, MYL1, MYLK, MYLPF, MYOT, TTN, TRDN, TRIM63, TPM1, TPM2, VIPR1 |
| GO:0060048—cardiac muscle contraction | ACTC1, MYH7, MYL1, MYL2, MYL3, RYR2, SCN4B, TTN, TCAP, TPM1, TNNC1, TNNI1, TNNI2 |
| GO:0030198—extracellular matrix organization | ACAN, COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL5A3, COL11A1, COL16A1, EMILIN1, ELN, ITGA2, ITGA7, ITGA8, LOX, NDNF, NID1, NID2, OLFML2B, POSTN, SPARC |
| GO:0045214—sarcomere organization | LDB3, ACTN2, ANKRD1, CASQ1, FHOD3, KLHL41, LMOD2, OBSCN, TTN, TCAP, TPM1 |
| GO:0030574—collagen catabolic process | COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL5A3, COL6A5, COL11A1. COL18A1, MMP1, MMP13, MMP16, MMP3, MMP9 |
| GO:0003009—skeletal muscle contraction | STAC3, MYH7, TCAP, TNNC1, TNNC2, TNNI1, TNNI2, TNNT1, TNNT3 |
| GO:0030199—collagen fibril organization | ACAN, COL1A1, COL1A2, COL3A1, COL5A1, COL5A2, COL5A3, COL11A1, LOX, SERPINH1 |
| GO:0007155—cell adhesion | ADAM12, NUAK1, THY1, ACTN2, ACAN, CDH11, COL1A1, COL5A1, COL6A5, COL16A1, COL18A1, EMILIN1, ITGA2, ITGA7, ITGA8, LRRN2, MYBPC2, MYBPC1, NCAM1, NID2, POSTN, PCDH17, RELN, SPON1, STAB2 |
| GO:0007517—muscle organ development | CHODL, FHL3, HBEGF, ITGA7, MAPK12, MKX, MURC, NEB, SMTN, TAGLN, UNC45B |
GO, Gene Ontology; AC, adhesive capsulitis.
Table II.
Differentially expressed genes related to 10 most statistically significant biological pathways identified by GO analysis for diabetic AC
| Biological pathway | Related genes |
|---|---|
| GO:0022617—extracellular matrix disassembly | ADAMTS4, ACAN, MMP13, MMP9, SPP1 |
| GO:0071376—cellular response to CRF stimulus | NR4A1, NR4A2, NR4A3 |
| GO:0007566—embryo implantation | LIF, MMP9, PTGS2, SPP1 |
| GO:0035914—skeletal muscle cell differentiation | ATF3, EGR1, EGR2, NR4A1 |
| GO:0048661—positive regulation of smooth muscle cell proLiferation | AIF1, IL6, NR4A3, PTGS2 |
| GO:0045444—fat ceU differentiation | EGR2, NR4A1, NR4A2, NR4A3 |
| GO:0046697—decidualization | LIF, PTGS2, SPP1 |
| GO:0045944—positive regulation of transcription from RNA Pol II promoter | FOSL2, ATF3, CSRNP1, EGR1, EGR2, IL6, LIF, NR4A1, NR4A2, NR4A3 |
| GO:0044344—cellular response to fibroblast growth factor stimulus | EGR3, NR4A1, POSTN |
| GO:0006508—proteolysis | ADAM12, ADAMTS1, ADAMTS4, ACAN, C1QA, MMP13, MMP9 |
GO, Gene Ontology; AC, adhesive capsulitis; CRF, corticotropin-releasing factor; Pol, polymerase.
Discussion
The currently accepted paradigm is that there is a presumed inciting stimulus for the development of AC that leads to differential expression of genes in both diabetic and normoglycemic cohorts. On comparison of nondiabetic and diabetic shoulder capsule specimens from patients without AC, only 3 genes were differentially expressed. In contrast, 66 distinctly different genes were expressed when nondiabetic and diabetic samples from patients with AC were compared. Thus, the presence of diabetes alone does not fully explain the transcriptomic differences in shoulder capsular tissue seen in AC. Future studies may reveal what the possible inciting factors are for the development of AC and its subsequent unique gene expression. Triggering elements could be physical and epigenetic factors including subtle trauma, surges in blood glucose levels, autoimmune factors, stress, or even a mild infectious agent.
In idiopathic AC, our analysis found that the collagen family of genes was involved in numerous pathways, including extracellular matrix organization, collagen catabolic process, collagen fibril organization, and cell adhesion.44 Other relevant genes of interest found in idiopathic AC included the myosin, troponin, and actin family. In diabetic AC, the NR4A1, NR4A2, and NR4A3 genes were represented in a number of the enriched pathways, including cellular response to CRF stimulus, fat cell differentiation, and positive regulation of transcription from RNA Pol II promoter.42,54,55 The early growth response (EGR) family of transcription factors, including EGR1, EGR2, and EGR3, was also involved in a number of enriched biological pathways, including skeletal muscle cell differentiation and fat cell differentiation.7 It appears, then, that idiopathic AC generally manifests with increased expression of genes related to muscle or collagen metabolism whereas diabetic AC is characterized by a panoply of DNA transcriptional (POL2), cell division regulation (NR4A), and hormonal (CRF) alterations.
When we more closely examine similarities and differences in the idiopathic AC and diabetic AC cohorts, some generalizations may be made. Both groups manifested upregulation of extracellular matrix disassembly, cell adhesion, and skeletal system development, suggesting perhaps an attempt at increased tissue remodeling.23 For the idiopathic AC cohort, many genes were relevant to muscle tissue and included muscle filament sliding, muscle contraction, sarcomere organization, muscle organ development, skeletal muscle thin filament assembly, collagen fibril and extracellular matrix organization, and myofibril assembly, consistent with the anabolic and increased connective tissue production seen.22 However, in diabetic AC, many upregulated genes are related to hormonal and transcriptional changes, including cellular response to CRF stimulus, steroid hormone–mediated signaling pathway, response to glucocorticoid, response to insulin, intracellular receptor signaling pathway, and positive regulation of transcription from RNA Pol II promoter.48
Numerous pathways have been implicated in the pathogenesis of AC, but the underlying pathophysiology remains unclear. Upregulation of inflammatory and fibroproliferative pathways has long been held as fundamental to the process; however, angiogenesis, neurogenesis, and cartilage differentiation have also been implicated. The specific pathways responsible for AC have been investigated in previous studies, with heterogeneous results, likely owing to variation in timing and tissue location, as well as inclusion of patients with varying risk factors.3,12,33 Moreover, studies exploring the pathophysiology of AC have often failed to make a distinction between patients with diabetes and those without diabetes. Other studies that did made a distinction have included only either idiopathic AC patients or diabetic AC patients without comparing the 2 cohorts. It is important to note that in this study, we report findings that confirm the distinct pathophysiology and synergistic effect of diabetes on AC in patients as compared with those without known risk factors for the disease.
In a previous study, a comparison of gene expression and translation among 12 patients with AC, including 2 patients with diabetes, Hagiwara et al19 found various differentially regulated genes, 11 of which were consistent with those in our study. These genes included ACAN, COL1A1, and SPARC, which were upregulated in both idiopathic AC and diabetic AC; EGR1, EGR2, NR4A1, ATF3, LIPG, and C1QTNF6, which were upregulated only in diabetic AC; and GPX3 and SEMA3E, which were only downregulated in idiopathic AC.19 On further proteomic analysis of AC, which excluded diabetic AC patients, Hagiwara et al20 found several upregulated pathways that were consistent with our findings identified in idiopathic AC but not in diabetic AC. These pathways included sarcomere organization, collagen catabolic process, collagen fibril organization, extracellular matrix organization, cell adhesion, and cell-matrix adhesion.20 These data further validate our findings that tissue repair, collagen metabolism, cell-cell adhesion, and cell-matrix adhesion are important processes for idiopathic AC whereas diabetic AC has a separate underlying pathophysiology.
In a recent genome-wide expression study using RNA sequencing of 22 patients with AC, without the identification of diabetic status, 50 enriched pathways were identified.28 Regarding these identified pathways, we also found that 6 of 29 biological process pathways were upregulated in our idiopathic AC group whereas 3 of 11 biological process categories were similarly upregulated in our diabetic AC group. Concordant pathways for idiopathic AC included collagen catabolic process, collagen fibril organization, extracellular matrix disassembly, and ventricular cardiac muscle tissue development, whereas concordant pathways for diabetic AC included extracellular matrix disassembly, skeletal muscle cell differentiation, and response to glucocorticoid.
Previous studies have described the upregulation of IL-6 and various other interleukins in shoulder capsule samples and joint fluid from patients with idiopathic AC.1,35 Our results showed that IL-6 was similarly upregulated in the diabetic AC cohort, suggesting that there may be a common upstream inflammatory pathway in both subtypes of AC. Kim et al32 also reported on the upregulation of intercellular adhesion molecule 1 (ICAM-1) in capsular tissue and synovial fluid from patients with AC. It is interesting to note that ICAM-1 was not directly upregulated in either subtype of AC in our study, but EGR, a transcription factor known to regulate the expression of ICAM-1, was upregulated in idiopathic AC.16 In our study, EGR1, EGR2, EGR3, and NR4A1 were all upregulated in diabetic AC but not idiopathic AC, and ACAN was upregulated in both diabetic AC and idiopathic AC. This finding suggests that ACAN, a cartilage-related gene, may play a similar chondrogenic role underlying some of the pathophysiology in both subtypes of AC. In contrast, it is likely that the transcription factors comprising the EGR family, which have been implicated as key mediators for inflammation and fibrosis through transforming growth factor β regulation, play a more important role in diabetic AC.7 These results point to the possibility of developing anti-EGR therapies targeted toward diabetic AC. Furthermore, NR4A1—an orphan nuclear receptor that has been implicated in various metabolic processes such as carbohydrate and lipid metabolism—has also recently been implicated as a key mediator of transforming growth factor β signaling and fibrosis.42,54,55 As such, the nuclear orphan receptor (NR4A) superfamily presents a promising therapeutic opportunity in diabetes-induced AC and warrants further study.
Taken together, our findings identify distinct underlying pathophysiology between idiopathic AC and diabetic AC. Our findings in idiopathic AC patients are consistent with findings in the existing literature from studies of AC; however, our findings in diabetic patients are unique.20 Furthermore, our data suggesting a distinct pathophysiology between idiopathic AC and diabetic AC are supported by the disparate outcomes observed clinically in diabetic patients with AC, including more frequent failure of conservative management and need for arthroscopic capsule release.31,46 Despite the unique upstream pathophysiology, there are some common pathways regarding structural elements between both subtypes of AC, potentially explaining the similar initial clinical presentation of idiopathic AC and diabetic AC. In our study, we found that important extracellular matrix proteases MMP1, MMP13, MMP16, MMP3, and MMP9 were upregulated in idiopathic AC and MMP9 and MMP13 were also upregulated in diabetic AC.
Diabetes mellitus has been known to affect numerous organ systems34 and influence multiple hormonal pathways.39 We have presented yet another way in which diabetes may alter CRF, insulin, and glucocorticoid transcription. Other pathways in diabetic AC identified in our study were relevant to cell differentiation and regulation, including skeletal muscle cell differentiation, fat cell differentiation, cellular response to fibroblast growth factor stimulus, and inflammatory response. Of note, the upregulation of fat cell differentiation in diabetic AC samples is of special interest and merits further study. It has been clearly established that diabetes mellitus is linked to obesity and increased dietary free fatty acid levels.6 However, impaired fat cell differentiation has also been shown to be strongly associated with insulin resistance and diabetes mellitus.26
The more virulent clinical course of diabetic AC can be explained by the consequential protein and hormonal gene upregulation demonstrated in this work. Studies undertaking biological evaluation of AC should take great care in recognizing the different patient risk factors that may have an impact on underlying pathophysiology. The difference in underlying pathophysiology for idiopathic AC and diabetic AC suggests that medical treatment for frozen shoulder may be preferentially targeted to certain subtypes of AC. As such, there is biological justification for developing different treatment algorithms for diabetic patients and nondiabetic patients with frozen shoulder. With identification of the transcriptomic constitution of each disease entity, new and targeted treatment strategies can be developed.
Although sex differences were not the focus of this study, it is important to keep in mind that our study was performed in subject populations of both sexes, with the exception that diabetic AC shoulder capsule samples were only obtained from female patients. However, all significantly differentially expressed sex chromosome–specific genes were removed from the data set following RNA-sequencing analysis. Moreover, the enriched biological pathways identified by gene ontology (GO) analysis have not been found in previous studies comparing RNA-sequencing results between male and female patients. A previous study also found that sex did not have a major impact on gene expression levels from shoulder samples in patients with AC.28
Another limitation is that the mean hemoglobin A1c level in diabetic patients was 8.7%, suggesting that many patients had uncontrolled diabetes. As such, the level of diabetic control preceding the collection of shoulder capsule specimens could have significantly varied between patients, thus affecting the results of this study. There was also variation in treatment history between patients with AC and those without AC. The majority of all patients with AC had received at least a single intra-articular steroid injection whereas only 2 patients without AC had received at least a single intra-articular injection, potentially affecting shoulder capsule gene expression results. However, this is unlikely to have affected the comparison of gene expression between diabetic AC and idiopathic AC as both diabetic and nondiabetic patients with AC were more likely to have received steroid injections compared with their respective control groups. Furthermore, the presence of metabolic syndrome or dyslipidemia was not evaluated in every subject. It is possible that the presence of dyslipidemia or metabolic syndrome may have been a confounding variable in this study, and further studies should evaluate the impact of dyslipidemia and metabolic syndrome on AC pathophysiology. Similarly, 3 nondiabetic patients were not screened for A1c levels because they had no symptoms of diabetes or relevant risk factors, such as a family history or obesity. However, we cannot definitively exclude the possibility that these 3 patients did not have unrecognized diabetes, which also could have affected the results of this study.
Conclusion
This study has shown for the first time that transcriptome expression profiles demonstrate a fundamentally different underlying pathophysiology for diabetic AC and idiopathic AC. These findings suggest that these conditions are distinct clinical entities. In translational terms, the new genes expressed reveal potentially new therapeutic targets, and further characterization may lead to different treatment paradigms in these 2 clinical subsets.
Supplementary Material
Acknowledgments
The authors thank Ms. Kendra Miller and Dr. Jerahme Martinez for their assistance with shoulder tissue histologic analysis.
Disclaimer
The study was funded by the Arthroscopy Association of North America (AANA). Additional funding was received from the National Institutes of Health (NIAMS R01 AR071340) (G.R.D.) and the Penn Center for Musculoskeletal Disease Histology and Biomechanics Cores (P30 AR069619).
Joshua A. Gordon received grant funding from AANA.
Ali S. Farooqi has a pending invention patent related to the study.
G. Russell Huffman received grant funding from AANA.
John D. Kelly received grant funding from AANA.
George R. Dodge indicates that funding was received from the National Institutes of Health (NIAMS R01 AR071340) and the Penn Center for Musculoskeletal Disease Histology Core (P30 AR069619).
The other authors, their immediate families, and any research foundations with which they are affiliated have not received any financial payments or other benefits from any commercial entity related to the subject of this article.
Footnotes
Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jse.2021.06.016.
References
- 1.Akbar M, McLean M, Garcia-Melchor E, Crowe LA, McMillan P, Fazzi UG, et al. Fibroblast activation and inflammation in frozen shoulder. PLoS One 2019;14:e0215301. 10.1371/journal.pone.0215301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando A, Sugaya H, Hagiwara Y, Takahashi N, Watanabe T, Kanazawa K, et al. Identification of prognostic factors for the nonoperative treatment of stiff shoulder. Int Orthop 2013;37:859–64. 10.1007/s00264-013-1859-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andronic O, Ernstbrunner L, Jüngel A, Wieser K, Bouaicha S. Biomarkers associated with idiopathic frozen shoulder: a systematic review. Connect Tissue Res 2020;61:509–16. 10.1080/03008207.2019.1648445 [DOI] [PubMed] [Google Scholar]
- 4.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin DC, Gans I, Park MJ, Carey JL, Kelly JD IV. The association of metabolic syndrome markers with adhesive capsulitis. J Shoulder Elbow Surg 2014;23:1043–51. 10.1016/j.jse.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 6.Beck-Nielsen H, Groop LC. Metabolic and genetic characterization of prediabetic states. Sequence of events leading to non-insulin-dependent diabetes mellitus. J Clin Invest 1994;94:1714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol 2011; 30:235–42. 10.1016/j.matbio.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourji K, Gatto M, Cozzi F, Doria A, Punzi L. Rheumatic and autoimmune thyroid disorders: a causal or casual relationship? Autoimmun Rev 2015;14:57–63. 10.1016/j.autrev.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 9.Brown IDM, Kelly IG, McInnes PIB. OC15 detection of matrix metalloproteinases in primary frozen shoulders. Orthop Proc 2008;90-B:364. 10.1302/0301-620X.90BSUPP_II.0900364c [DOI] [Google Scholar]
- 10.Bunker TD, Anthony PP. The pathology of frozen shoulder. A Dupuytren-like disease. J Bone Joint Surg Br 1995;77:677–83. [PubMed] [Google Scholar]
- 11.Bunker TD, Reilly J, Baird KS, Hamblen DL. Expression of growth factors, cytokines and matrix metalloproteinases in frozen shoulder. J Bone Joint Surg Br 2000;82:768–73. [DOI] [PubMed] [Google Scholar]
- 12.Cho C-H, Song K-S, Kim B-S, Kim DH, Lho Y-M. Biological aspect of pathophysiology for frozen shoulder. Biomed Res Int 2018;2018: 7274517. 10.1155/2018/7274517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen C, Leal MF, Belangero PS, Figueiredo EA, Smith MC, Andreoli CV, et al. The roles of Tenascin C and Fibronectin 1 in adhesive capsulitis: a pilot gene expression study. Clinics (Sao Paulo) 2016;71:325–31. 10.6061/clinics/2016(06)07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen C, Tortato S, Silva OBS, Leal MF, Ejnisman B, Faloppa F. Association between frozen shoulder and thyroid diseases: strengthening the evidences. Rev Bras Ortop (Sao Paulo) 2020;55:483–9. 10.1055/s-0039-3402476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dias R, Cutts S, Massoud S. Frozen shoulder. BMJ 2005;331:1453. 10.1136/bmj.331.7530.1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dryden NH, Sperone A, Martin-Almedina S, Hannah RL, Birdsey GM, Khan ST, et al. The transcription factor Erg controls endothelial cell quiescence by repressing activity of nuclear factor (NF)-κB p65. J Biol Chem 2012;287:12331–42. 10.1074/jbc.M112.346791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gene Ontology Consortium. The Gene Ontology resource: 20 years and still going strong. Nucleic Acids Res 2019;47:D330–8. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 2011;27:2518–28. 10.1093/bioinformatics/btr427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagiwara Y, Ando A, Onoda Y, Takemura T, Minowa T, Hanagata N, et al. Coexistence of fibrotic and chondrogenic process in the capsule of idiopathic frozen shoulders. Osteoarthritis Cartilage 2012;20:241–9. 10.1016/j.joca.2011.12.008 [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara Y, Mori M, Kanazawa K, Ando A, Yabe Y, Koide M, et al. Comparative proteome analysis of the capsule from patients with frozen shoulder. J Shoulder Elbow Surg 2018;27:1770–8. 10.1016/j.jse.2018.03.010 [DOI] [PubMed] [Google Scholar]
- 21.Hand GCR, Athanasou NA, Matthews T, Carr AJ. The pathology of frozen shoulder. J Bone Joint Surg Br 2007;89-B:928–32. 10.1302/0301-620X.89B7.19097 [DOI] [PubMed] [Google Scholar]
- 22.Henderson CA, Gomez CG, Novak SM, Mi-Mi L, Gregorio CC. Overview of the muscle cytoskeleton. Compr Physiol 2017;7:891–944. 10.1002/cphy.c160033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honig B, Shapiro L. Adhesion protein structure, molecular affinities, and principles of cell-cell recognition. Cell 2020;181:520–35. 10.1016/j.cell.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 26.Jansson PA, Pellmé F, Hammarstedt A, Sandqvist M, Brekke H, Caidahl K, et al. A novel cellular marker of insulin resistance and early atherosclerosis in humans is related to impaired fat cell differentiation and low adiponectin. FASEB J 2003;17:1434–40. 10.1096/fj.02-1132com [DOI] [PubMed] [Google Scholar]
- 27.Kabbabe B, Ramkumar S, Richardson M. Cytogenetic analysis of the pathology of frozen shoulder. Int J Shoulder Surg 2010;4:75–8. 10.4103/0973-6042.76966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamal N, McGee SL, Eng K, Brown G, Beattie S, Collier F, et al. Transcriptomic analysis of adhesive capsulitis of the shoulder. J Orthop Res 2020;38:2280–9. 10.1002/jor.24686 [DOI] [PubMed] [Google Scholar]
- 29.Kanbe K, Inoue K, Inoue Y, Chen Q. Inducement of mitogen-activated protein kinases in frozen shoulders. J Orthop Sci 2009;14:56–61. 10.1007/s00776-008-1295-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilian O, Pfeil U, Wenisch S, Heiss C, Kraus R, Schnettler R. Enhanced alpha1 (I) mRNA expression in frozen shoulder and Dupuytren tissue. Eur J Med Res 2007;12:585. [PubMed] [Google Scholar]
- 31.Kim DH, Kim YS, Kim BS, Sung DH, Song KS, Cho CH. Is frozen shoulder completely resolved at 2 years after the onset of disease? J Orthop Sci 2020;25:224–8. 10.1016/j.jos.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 32.Kim YS, Kim JM, Lee YG, Hong OK, Kwon HS, Ji JH. Intercellular adhesion molecule-1 (ICAM-1, CD54) is increased in adhesive capsulitis. J Bone Joint Surg Am 2013;95:e181–8. 10.2106/jbjs.K.00525 [DOI] [PubMed] [Google Scholar]
- 33.Kraal T, Lübbers J, van den Bekerom MPJ, Alessie J, van Kooyk Y, Eygendaal D, et al. The puzzling pathophysiology of frozen shoulders—a scoping review. J Exp Orthop 2020;7:91. 10.1186/s40634-020-00307-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni S, Sharda S, Watve M. Bi-stability in type 2 diabetes mellitus multi-organ signalling network. PLoS One 2017;12:e0181536. 10.1371/journal.pone.0181536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lho YM, Ha E, Cho CH, Song KS, Min BW, Bae KC, et al. Inflammatory cytokines are overexpressed in the subacromial bursa of frozen shoulder. J Shoulder Elbow Surg 2013;22:666–72. 10.1016/j.jse.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 36.Lo SF, Chu SW, Muo CH, Meng NH, Chou LW, Huang WC, et al. Diabetes mellitus and accompanying hyperlipidemia are independent risk factors for adhesive capsulitis: a nationwide population-based cohort study (version 2). Rheumatol Int 2014;34:67–74. 10.1007/s00296-013-2847-4 [DOI] [PubMed] [Google Scholar]
- 37.Lubis AM, Lubis VK. Matrix metalloproteinase, tissue inhibitor of metalloproteinase and transforming growth factor-beta 1 in frozen shoulder, and their changes as response to intensive stretching and supervised neglect exercise. J Orthop Sci 2013;18:519–27. 10.1007/s00776-013-0387-0 [DOI] [PubMed] [Google Scholar]
- 38.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288–97. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mraz M, Bartlova M, Lacinova Z, Michalsky D, Kasalicky M, Haluzikova D, et al. Serum concentrations and tissue expression of a novel endocrine regulator fibroblast growth factor-21 in patients with type 2 diabetes and obesity. Clin Endocrinol 2009;71:369–75. 10.1111/j.1365-2265.2008.03502.x [DOI] [PubMed] [Google Scholar]
- 40.Nago M, Mitsui Y, Gotoh M, Nakama K, Shirachi I, Higuchi F, et al. Hyaluronan modulates cell proliferation and mRNA expression of adhesion-related procollagens and cytokines in glenohumeral synovial/capsular fibroblasts in adhesive capsulitis. J Orthop Res 2010; 28:726–31. 10.1002/jor.21075 [DOI] [PubMed] [Google Scholar]
- 41.Neviaser AS, Neviaser RJ. Adhesive capsulitis of the shoulder. J Am Acad Orthop Surg 2011;19:536–42. 10.5435/00124635-201109000-00004 [DOI] [PubMed] [Google Scholar]
- 42.Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med 2015;21:150–8. 10.1038/nm.3777 [DOI] [PubMed] [Google Scholar]
- 43.Park HB, Gwark J-Y, Jung J. What serum lipid abnormalities are associated with adhesive capsulitis accompanied by diabetes? Clin Orthop Relat Res 2018;476:2231–7. 10.1097/CORR.0000000000000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ricard-Blum S The collagen family. Cold Spring Harbor Perspect Biol 2011;3:a004978. 10.1101/cshperspect.a004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riley D, Lang AE, Blair RD, Birnbaum A, Reid B. Frozen shoulder and other shoulder disturbances in Parkinson’s disease. J Neurol Neurosurg Psychiatry 1989;52:63–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rill BK, Fleckenstein CM, Levy MS, Nagesh V, Hasan SS. Predictors of outcome after nonoperative and operative treatment of adhesive capsulitis. Am J Sports Med 2010;39:567–74. 10.1177/0363546510385403 [DOI] [PubMed] [Google Scholar]
- 47.Rodeo SA, Hannafin JA, Tom J, Warren RF, Wickiewicz TL. Immunolocalization of cytokines and their receptors in adhesive capsulitis of the shoulder. J Orthop Res 1997;15:427–36. [DOI] [PubMed] [Google Scholar]
- 48.Russell G, Lightman S. The human stress response. Nat Rev Endocrinol 2019;15:525–34. 10.1038/s41574-019-0228-0 [DOI] [PubMed] [Google Scholar]
- 49.Shaffer B, Tibone JE, Kerlan RK. Frozen shoulder. A long-term follow-up. J Bone Joint Surg Am 1992;74:738–46. [PubMed] [Google Scholar]
- 50.Uhthoff HK, Boileau P. Primary frozen shoulder: global capsular stiffness versus localized contracture. Clin Orthop Relat Res 2007; 456:79–84. 10.1097/BLO.0b013e318030846d [DOI] [PubMed] [Google Scholar]
- 51.Warner JJP. Frozen shoulder: diagnosis and management. J Am Acad Orthop Surg 1997;5:130–40. [DOI] [PubMed] [Google Scholar]
- 52.Xue M, Gong S, Dai J, Chen G, Hu J. The treatment of fibrosis of joint synovium and frozen shoulder by Smad4 gene silencing in rats. PLoS One 2016;11:e0158093. 10.1371/journal.pone.0158093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yian EH, Contreras R, Sodl JF. Effects of glycemic control on prevalence of diabetic frozen shoulder. J Bone Joint Surg Am 2012;94: 919–23. 10.2106/jbjs.J.01930 [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Wang Q, Liu W, Liu F, Ji A, Li Y. The orphan nuclear receptor 4A1: a potential new therapeutic target for metabolic diseases. J Diabetes Res 2018;2018:9363461. 10.1155/2018/9363461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat Commun 2014;5:3388. 10.1038/ncomms4388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


