Abstract
Resistance to platinum-based chemotherapy is a major clinical challenge in ovarian cancer, contributing to the high mortality-to-incidence ratio. Management of the platinum-resistant disease has been difficult due to diverse underlying molecular mechanisms. Over the past several years, research has revealed several novel molecular targets that are being explored as biomarkers for treatment planning and monitoring of response. The therapeutic landscape of ovarian cancer is also rapidly evolving, and alternative therapies are becoming available for the recurrent platinum-resistant disease. This review provides a snapshot of platinum resistance mechanisms and discusses liquid-based biomarkers and their potential utility in effective management of platinum-resistant ovarian cancer.
Keywords: Platinum resistance, Ovarian cancer, Biomarkers, Paclitaxel, DNA repair, Cisplatin, Epigenetic
1. Introduction
Ovarian cancer (OC) is the seventh most common cancer worldwide and ranks eighth in women’s cancer-related mortalities. Moreover, its incidence is much higher in developed countries than in developing countries [1]. In the United States, OC is the second most common gynecological malignancy, causing more deaths than any other cancer of the female reproductive system [2]. Most OCs are of epithelial origin (epithelial ovarian cancer, EOC) and classified into five major histological subtypes: high-grade serous carcinoma (HGSC), low-grade serous carcinoma (LGSC), endometrioid carcinoma (EC), clear cell carcinoma (CCC), and mucinous carcinoma (MC). These distinct EOC histological subtypes differ in their prevalence, clinicopathological characteristics, and molecular features [3–5].
Clinical management of EOC has been difficult due to the lack of effective screening and prognostic biomarkers and lack of effective treatment strategies for the advanced disease. In the prostate, lung, colorectal, and ovarian cancer screening (PLCO) trial, CA-125 and transvaginal ultrasound failed to provide the mortality benefit of screening to the EOC patients [6]. Similarly, platinum-based drugs used in combination with taxanes as a primary line of therapy are not very effective against the advanced EOC [7]. The response to chemotherapy varies among different EOC subtypes due to inherent or acquired chemoresistance. Non-serous subtypes such as CCC and MC generally exhibit a more insufficient response to platinum therapy due to their genetic landscape. HGSC, on the other hand, initially respond well to platinum-based treatments, but the disease eventually relapses in a significant majority of patients due to therapy failure.
Developing a precise understanding of inherent or acquired chemoresistance mechanisms is highly critical to achieving optimal clinical management of OC. Over the years, we have uncovered several pathways involved in the platinum resistance of OC. This information can be useful in developing alternative therapies or enhancing existing platinum-based treatments by targeting resistance pathways. Further, advances in molecular biology techniques, such as next-generation sequencing (NGS), are revolutionizing the therapeutic management of cancer by providing information on the genetic contexture of the disease. NGS can provide information on multiple genomic and transcriptomic alterations at once with high precision and therefore being increasingly used in oncology for improved therapeutic planning [8].
The use of liquid biopsies to determine the molecular features of cancer is gaining momentum due to its several advantages. In contrast to tissue biopsy, liquid biopsy is minimally invasive and can be performed repeatedly without complications as required for disease monitoring. It is also less cumbersome and less expensive and has a shorter turnaround time if we consider the longer scheduling time needed for the tissue biopsy [9]. More importantly, liquid biopsy can theoretically capture tumor heterogeneity and the overall tumor burden more efficiently than the tumor biopsy [10, 11]. In the following sections, we provide a background of platinum-based therapy and underlying drug resistance mechanisms and focus on potential clinical applications of liquid biopsies and treatment paradigms of platinum-resistant ovarian cancer.
2. Platinum-based therapy and associated chemoresistance mechanisms
Platinum drugs function by forming DNA adducts that halt DNA replication and induce cell death in rapidly dividing tumor cells [12]. They are the mainstay of treatment for patients with advanced EOC in combination with other drugs; however, despite an early response, tumor recurrence is fairly common due to inherent or acquired resistance mechanisms (Figure 1). Below we discuss the existing knowledge on these resistance mechanisms.
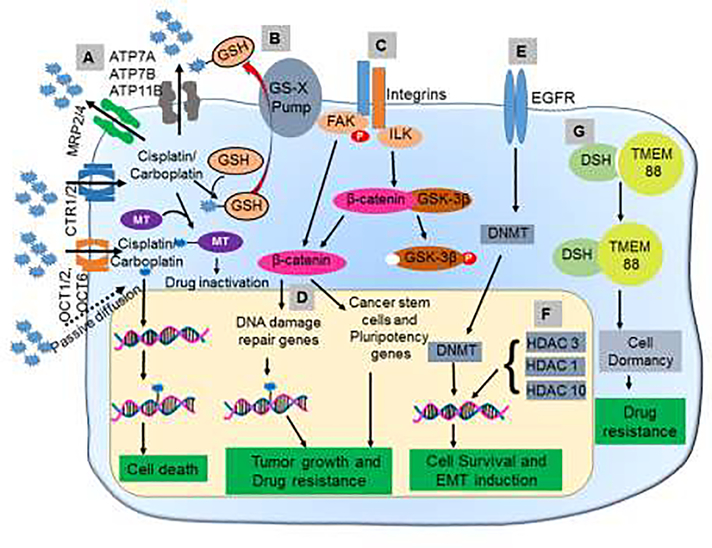
Figure 1. Mechanisms underlying platinum resistance in ovarian cancer.
(A) Dysregulation of transporters associated with cisplatin/carboplatin leads to the reduced influx and increased efflux. (B) GSH and metallothionein mediate the inactivation of platinum compounds via the formation of drug conjugates. (C) Interaction between integrin and ECM promotes platinum resistance by inducing DNA damage repair proteins and CSC genes. (D) Upregulation of DNA damage repair proteins such as BRCA1 enables cancer cells to repair the DNA damage induced by platinum-based drugs and become resistant. (E) The upregulation of EGFR enhances the activity of DNMTs and leads to global methylation and downregulation of proteins regulating proliferation, migration, and invasion. (F) HDACs downregulate genes responsible for apoptosis and proliferation. (G) Inhibition of Wnt signaling mediated through the binding of Dishevelled to TMEM88 leads to cell dormancy and thus resists the action of the cytotoxic drugs. MT=Metallothionein; DSH=Dishevelled; FAK=focal adhesion kinase; ILK=Integrin-linked kinase; OCT=organic cation transporter; GSH=Glutathione; GS-X=Glutathione conjugate export pump; GSK= Glycogen synthase kinase 3; DNMT= DNA Methyltransferase; HDAC=Histone deacetylases; TMEM=Transmembrane Protein 88; EGFR=Epidermal Growth Factor Receptor
2.1. Drug influx and efflux pathways
Platinum-based drugs enter the cancer cells either by passive diffusion or through facilitated transport [13]. Cancer cells can effectively expel out these drugs to protect themselves. Varieties of transporter proteins regulate the influx and efflux of platinum compounds, and thus their altered expression may induce chemoresistance. Transporters associated with the influx of platinum drugs are members of the solute carrier superfamily, such as organic cation transporter (OCT) 1, 2, and 6 and copper transporter 1/2 (CTR1/2) [14, 15]. Since both OCT and CTR proteins facilitate the entry of cisplatin inside the cells, their downregulation reduces the treatment efficacy of platinum-based drugs. Although there has been no significant study reporting the downregulation of OCT1/2 in platinum resistance, OCT6 downregulation directly correlates with cisplatin resistance in lung cancer [16]. Similarly, decreased expression of CTR1 is associated with poor disease-free survival after platinum-based therapy [17, 18].
Copper exporters (ATP7A and ATP7B) also play a critical role in the induction of cisplatin resistance as they facilitate the expulsion of cisplatin from OC cells, thus reducing the effective cisplatin concentration [19]. Overexpression of ATP7A and ATP7B is also linked to the resistance to the cytotoxicity caused by cisplatin and its analogs [20, 21]. Platinum resistance can also be manifested due to the altered expression and action of the multi-drug resistance proteins (MRP). These proteins belong to the ATP binding cassette superfamily and are significant in non-specific drug resistance [22]. High expression of MRP2 and MRP4 in OC has been associated with cisplatin resistance and poor clinical outcome [23]. A distinct mechanism involving an overexpression of cell trafficking protein, annexin A4, has been reported in CCC in another study, which enhances platinum efflux causing its insufficient accumulation [24].
2.2. Intracellular redox balance and drug modification
Reactive oxygen species (ROS) are byproducts of aerobic metabolism and can also be generated under stress conditions [25]. The physiological balance of ROS is maintained by ROS-induced adaptive upregulation of antioxidant systems [26]. Compared to normal cells, cancer cells have higher intracellular ROS levels, which play a significant role in pathogenesis [27, 28]. However, excess generation of ROS caused by ionizing radiation and chemotherapeutic agents is associated with tumor cell death [29]. To counter the detrimental effect of ROS, cancer cells overexpress antioxidant enzymes that provide them a survival benefit. Elevated glutathione (GSH) levels are reported in several cancers including OC and linked with chemoresistance [30–32]. A high level of GSH in tumor cells is associated with increased expression and activity of GSH-related enzymes and exporter proteins, such as γ-glutamyl-cysteine ligase (GCL), γ-glutamyl-transpeptidase (GGT), and GSH-transporting export pumps [33, 34]. GSH also has a high affinity toward platinum and forms cisplatin-thiol conjugates reducing cisplatin cytotoxicity [35]. It was recently shown that a plant-derived agent, sanguinarine, reduced the GSH levels in PROC cells and sensitized them to cisplatin toxicity [36]. Also, cellular chelating agents such as metallothioneins caused sequestration of platinum compounds, reducing their availability and contributing towards platinum resistance [37].
2.3. Tumor microenvironment, cell adhesion molecules, and stem cell-associated mechanism
The tumor microenvironment (TME) consists of various host cells, including fibroblasts, adipocytes, mesothelial cells, and immune cells (neutrophils, natural killer cells, macrophages, polymorphonuclear neutrophils, myeloid-derived suppressor cells, etc). It also has biomolecules secreted by tumor and TME-associated host cells referred to as the extracellular matrix (ECM) [38]. Clinical studies have reported a correlation between platinum resistance and interaction between ECM and cell adhesion molecules like CD44, CD117, CD133, and β-integrin with poor prognosis in OC patients [39]. Binding with ECM components like the integrin family of proteins seems to provide a survival advantage for tumor cells when exposed to cytostatic drugs. Activation of integrin-linked kinase (ILK) is significantly higher in PROC as compared to platinum-sensitive OC [40]. Enhanced activity of focal adhesion kinase (FAK), a downstream effector of integrin signaling, is reported in OC exhibiting resistance to carboplatin and paclitaxel [41]. Phosphorylation of FAK increases after the cisplatin treatment and activates β-catenin signaling to support pluripotency and DNA repair [41]. Tumor-infiltrating lymphocytes and the presence of high Treg (regulatory T cells) count are associated with platinum resistance and disease recurrence [42]. Cytokines secreted by tumor-associated macrophages (TAMs) also promote platinum resistance by inducing epithelial to mesenchymal transition (EMT) and ECM remodeling [43, 44]. An increased presence of cancer stem cells (CSCs) has also been implicated in therapy resistance and cancer relapse [45]. Overexpression of CSC markers is associated with enhanced EMT features and multi-drug resistance in OC [46]. Cisplatin induces the expression of EMT and CSC markers and promotes aggressiveness and chemoresistance of OC cells [39]. Several stem cell markers, including aldehyde dehydrogenase (ALDH), CD44, CD24, CD133, CD117, and CXCR4, are overexpressed in PROC as compared to the platinum-sensitive ones [46–48]. ALDH1A1 expression is further correlated with lower progression-free survival of OC patients [49, 50]. An enhanced expression of stemness-maintaining proteins such as Nanog, Oct4, and CD73 is also linked to aggressive and chemoresistant OC [51–53]. Thus, inherent or TME-induced stemness potential is also an effective mechanism of platinum resistance in OC.
2.4. DNA damage repair machinery
As previously described, platinum-based drugs confer cytotoxicity by forming DNA adducts [12]. Thus, resistance to platinum therapy could be induced by the upregulation of DNA repair proteins that catalyze the removal of platinum adducts and repair the damage [7]. Indeed, most platinum-resistant cancers show an upregulation of DNA damage repair proteins, including but are not limited to BRCA1/2, mismatch repair proteins (MSH1, MSH2), excision repair cross-complementing (ERCC) member proteins, RAD51 and Fanconi Anemia Complementation Group D2 [7, 54]. Even the presence of mutant BRCA1 is sufficient to initiate DNA damage repair by loading of RAD51 protein on the DNA [55]. Loss of proteins essential for replication fork stabilization, such as pax transactivation-domain interacting protein (PTIP), also renders the cells resistant to platinum-based drugs. OC patients diagnosed with the BRCA1 mutation, but having high PTIP expression, show a more prolonged progression-free survival than those with low PTIP [56]. Replication protein A (RPA) is crucial for stabilizing DNA during replication and cells deficient in RPA show impaired nucleotide excision repair (NER), and thus less protection of replication fork making OC cells sensitive to platinum therapy [57]. Ataxia-telangiectasia and Rad3-related protein (ATR) is an important DNA damage sensor, which activates the homologous recombination (HR) pathway in response to DNA damaging agents [58]. ATR thus initiates the repair of cisplatin treatment-induced stalled replication forks, thereby rendering the cells therapy-resistant [59]. Translesion DNA synthesis (TLS) is a process by which cells allow themselves to replicate despite harboring errors in the underlying DNA sequence. Thus, the upregulation of proteins involved in TLS confers platinum resistance by sustaining the replication of damaged DNA [60]. DNA polymerase eta (Pol η) is crucial for the TLS after the formation of 8-Oxo-Guanine, making it a potential therapeutic target [61].
2.5. Epigenetic mechanisms
Epigenetic silencing of BRCA1 is suggested to be an additional mechanism for its loss of function in cancer. However, the data associating BRCA1 methylation in OC with clinical hallmarks and outcomes after platinum chemotherapy is inconsistent. While some studies suggest statistically significant association of BRACA1 promoter methylation with patient survival following platinum therapy [62, 63], others report an inverse trend or no significant association [64–66]. OC cells show a high expression profile of enhancer of zeste homolog 2 (EZH2), associated with methylation of H3K27 and gene silencing [67]. EZH2 is also predicted to be responsible for BRCA1 downregulation and shift towards an advanced OC stage, platinum resistance, and poor patient survival [68, 69]. ARID1A, a subunit of SWI/SNF chromatin remodeling complex, is frequently mutated or repressed in CCC and associated with platinum resistance [70, 71]. Cisplatin treatment also induces epidermal growth factor receptor (EGFR)-mediated upregulation in DNA methyltransferase (DNMT) [72]. Enhanced DNMT1 activity affects the expression of genes associated with the critical cellular processes, such as proliferation, migration and invasion, and chemoresistance [73]. Abrogation of EGFR expression augmented cisplatin sensitivity of the OC cells accompanied by halted DNMT activity and DNA methylation. The methylation status of p27, a CDK inhibitor, is also linked to cisplatin sensitivity. PROC cells show significant downregulation of p27 due to promoter hypermethylation, overcoming cell cycle arrest induced by cisplatin treatment [74]. Administration of demethylating agents, 5-aza-2’-deoxycytidine, restores p27 expression and increases cisplatin sensitivity [74]. Promoter hypermethylation of ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) has been linked to the upregulation of anti-apoptotic proteins BCl-2 and XIAP and thereby promotes cisplatin resistance in OC cells [75]. In another report, ten eleven translocation (TET) proteins belonging to the family of cytosine demethylases were also associated with platinum-resistance, as evident from their ability to regulate the expression of vimentin, one of the critical players in EMT [76].
Promoter hypomethylation of TMEM88 is associated with platinum resistance in all OC histological subtypes [77]. TMEM88 causes downregulation of canonical Wnt signaling by interacting with its effector protein, Dishevelled, to induce dormancy and resist the cytotoxic effects of platinum therapy [78]. Cisplatin-resistant OC cells show a higher expression of HDAC1, and its knockdown leads to growth arrest and apoptosis in cisplatin-resistant OC cells [79]. HDAC1 inhibition decreases c-MYC expression and upregulates tumor suppressor miR-34a linking the HDAC1/c-MYC/miR34a axis to cisplatin sensitivity [79]. Another histone deacetylase, HDAC10, also supports platinum resistance by facilitating DNA damage repair even in BRCA1 negative background [80]. SIRT5, a member of the sirtuin class of histone deacetylases, is upregulated in various cancers, and its overexpression is reported in cisplatin-resistant OC cells where it abrogates cisplatin-induced ROS to sustain cell survival [81].
MicroRNAs (miRNAs) represent another important class of epigenetic regulators that has been associated with platinum resistance. miR-214 promotes platinum resistance in OC by downregulating PTEN expression [82]. PTEN is targeted by other deregulated miRNAs in OC, such as miR-130a and miR-186 and thus appears to be a significant determinant of sensitivity to platinum drugs [83, 84]. Decreased expression of miR-137 is also associated with platinum resistance that causes upregulation of MYC and EZH2 [85]. miR-21–3p promotes platinum resistance via downregulation of neuron navigator 3 [86]. In other reports, PROC cells are shown to have a downregulated expression of miR-1294 and miR-139–5p, whose restoration increased platinum sensitivity by inhibiting insulin-like growth factor 1 and c-jun expression, respectively [87, 88]. miR-199a, which targets mTOR and hypoxia-inducible factor-alpha, is also associated with cisplatin resistance [89, 90]. Thus, miRNAs could serve as clinically useful therapeutic targets to overcome platinum resistance in OC.
3. Molecular biomarkers of platinum resistance in ovarian cancer
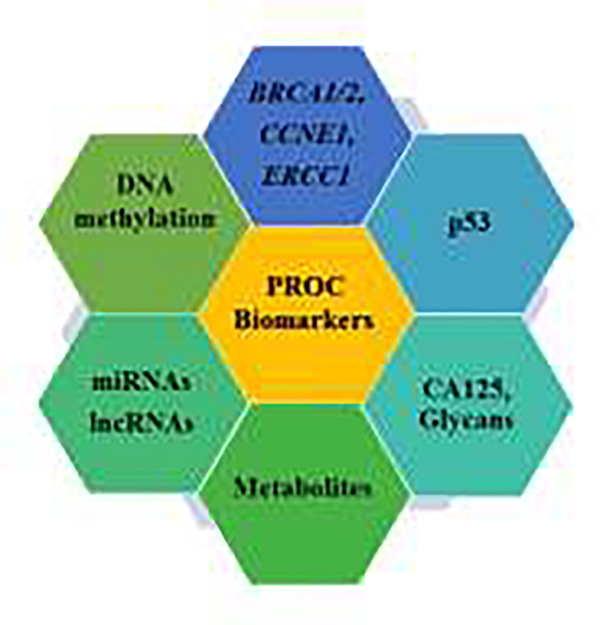
Identification of molecular biomarkers associated with platinum resistance could significantly improve the clinical management of PROC by aiding in the early prediction of inherent resistance to guide therapeutic selection. These markers can also be very useful in monitoring the response of OC to platinum therapy and signal acquisition of chemoresistance. Below we discuss several potential biomarkers that have shown a strong association with platinum resistance in mechanistic and clinical studies. These candidate biomarkers can be explored further for PROC diagnosis, monitoring, and treatment guidance (Figure 2).
Figure 2. Biomarkers of platinum-resistant ovarian cancer.
Molecular analysis of clinical specimens and laboratory studies have identified several molecular alterations associated with platinum-resistant ovarian cancer. These signature alterations can be exploited as molecular biomarkers to predict the nature of platinum resistance and monitor the response of ovarian cancer to therapy.
3.1. BRCA1/2, CCNE1, and ERCC1
BRCA1/2 mutations are frequently reported in HSGC and associated with sensitivity to platinum therapy [91, 92]. In several cases, reversion mutations in BRCA1/2 that restore the gene functionality have been reported and associated with acquired resistance to platinum-based drugs and PARP inhibitors [92, 93]. In another study, amplification of CCNE1, a gene encoding for cyclin E1, was reported in OC independent of BRCA1/2 mutations and associated with resistance to platinum-based drugs [94]. Increased expression of excision repair cross complementation group-1 (ERCC1) along with BRCA1 is also suggested to serve as a biomarker to identify patients at high risk for developing platinum resistance. Increased expression of a larger novel transcript of ERCC1, initiated upstream of the normal transcription initiation site, was observed in cisplatin-treated OC cells [95]. Higher expression of this novel transcript was also found in the PROC patients compared to the platinum-sensitive ones, suggesting that it could be used to predict the outcome of platinum therapy in OC patients [95]. In a comparative analysis, CCC, which are more likely to exhibit de novo resistance to platinum-based drugs, were found to express higher levels of ERCC compared to other subtypes supporting its utility as a potentially useful biomarker [96].
3.2. ARID1A, TERT and HNF-1β
Loss of expression or inactivating mutation in tumor suppressor gene, AT-rich interaction domain 1A (ARID1A), are reported in CCC and correlated with poor overall survival of the patients treated with platinum therapy [71]. Mutations in telomerase reverse transcriptase (TERT) promoter leading to increased TERT mRNA levels are also reported in non-serous subtypes of OC [97]. These mutations correlate with the loss of ARID1A expression in CCC, promote cancer stem cell-like properties and induce platinum resistance [98, 99]. Hepatocyte nuclear factor-1 beta (HNF-1β) appears to be more frequently expressed in CCC tissue than other OC histological subtypes [100]. Functional studies have shown that HNF1β confers platinum resistance by upregulating glutathione synthesis and supporting cell survival [101]. Thus, HNF-1β could potentially be exploited as a biomarker of platinum resistance in OC.
3.3. TP53
TP53 is more frequently mutated in HGSC than other OC subtypes, and its loss of function due to inactivating mutations is shown to confer platinum resistance in ovarian tumor cells [102, 103]. However, there appears to be ambiguity about the role of p53 since another study reports greater efficacy of cisplatin in OC cells having no or mutant TP53 compared to those with wild-type TP53 [104]. In another study, it was found that platinum treatment led to an additional mutation, S185G, in the mutant TP53 (R273H) and was associated with acquired chemoresistance in OC cells [105]. Therefore, the TP53 mutational frequency could serve as a biomarker of inherent or acquired platinum resistance and assist in therapeutic planning. TP53 clonal somatic variants have also been identified in patients with HGSOC. Notably, these variants were the same identified six years prior to OC diagnosis in the Papanicolaou tests of the afflicted individuals [106]. Thus, these clonal variants should be explored as the risk predictor for HGSOC development. In a more recent study, TP53 mutations were identified in a significant majority of OC with a greater frequency in HGSOC. Moreover, they often co-occurred with BRCA1 or BRCA2 mutations and associated with platinum sensitivity, but not the overall survival [107]. Thus, it appears that BRCA mutations may serve as major confounding factors in determining the clinicopathologic association of TP53 mutations with disease status and outcomes. All of these observations indicate that TP53 could be a useful biomarker of platinum resistance, but a more precise understanding of its function in different molecular contexts is required before clinical exploitation.
3.4. DNA methylation
Aberrant DNA methylation can control both ovarian tumorigenesis and platinum resistance [108]. Thus, measuring the nature and extent of methylation could serve as plausible means for biomarker development for early OC diagnosis and prediction of platinum resistance. A comparative genome-wide study of CCC and non-CCC subtypes suggested that the CCC had a distinct methylation profile. It showed hypomethylation of the stress-associated genes while the genes involved in tumor development, such as estrogen receptor alpha, were hypermethylated [109]. Hypomethylation of human endogenous retrovirus K is also observed in CCC and correlated with platinum resistance [110]. In other reports, hypermethylation in the promoter region of a mismatch repair gene hMSH2 (muts homolog 2) or hypomethylation of CpG sites within the Msh homeobox 1 gene (MSX1) have been associated with platinum resistance in OC [111, 112]. A recent CpG methylome and transcriptome study also identified a panel of differentially expressed genes (IL6, IL6ST, SMAD3, KLF4, TGFBR1, EGF, JUN, PPARG, PPARGC1A, and AR) in cisplatin-resistant OC cells. Among these genes, KLF4 and IL6 were the strongest candidates for predicting platinum resistance [113]. Another study suggested a role of hypermethylation of MLH1 and ZIC1 gene promoters in cisplatin resistance of OC cells [114]. A phase II clinical trial conducted to test the therapeutic efficacy of low-dose decitabine (demethylation agent) before carboplatin-treatment revealed that the number of demethylated genes in ovarian tumor samples was directly proportional to progression-free survival [115]. Furthermore, the study suggested that the demethylation in a set of genes (RASSF1A, AKT1S1, MLH1, HOXA10, and HOXA11) [115] could be used as a biomarker to stratify PROC patients as candidates for the treatment with demethylation agents. Hypermethylation is also reported in BRCA1 promoter in OC patients and associated with sensitivity to platinum drugs [116].
3.5. MicroRNAs and long noncoding RNAs
A substantial number of studies have demonstrated the functional roles of microRNAs and other noncoding RNAs in OC cancer pathogenesis and platinum resistance [117, 118]. Marked loss of let-7g in OC tissues and sera is associated with platinum resistance, suggesting its utility as a biomarker for treatment guidance and therapeutic failure [119]. Another member of the let-7 family, miR-98–5p, is highly expressed in cisplatin-resistant ovarian tumor cells and associated with inadequate therapy response [117]. miR-622 is shown to induce resistance to platinum drugs and PARP inhibitors in BRCA1/2 mutant OC by activating homologous recombination dependent DNA double-strand breaks repair in the treated cells [120]. miR-622 is highly expressed in HGSC and is associated with worse outcomes after platinum drug treatment [120] and thus could be used as a predictive biomarker for treatment planning and monitoring the therapy-response. In other studies, overexpression of miR-454–3p, miR-98–5p, miR-183–5p, and miR-22–3p and downregulation of miR-484, miR-642, and miR-217 has also been associated with platinum resistance [121, 122]. Mining of the cancer genome atlas (TCGA) identified 32 dysregulated long noncoding RNAs (lncRNAs) in PROC, suggesting their clinical utility as biomarkers [123]. Another study found an eight lncRNAs signature as a predictor of treatment response in OC patients [124]. The expression of noncoding RNA, HOTAIR, was also associated with poor survival in carboplatin-treated OC patients [125].
3.6. CA-125 and glycan biomarkers
CA-125, a serum-based biomarker, is used to monitor therapy response and disease relapse in OC patients [126]. CA-125 is also known as mucin 16 (MUC16), a member of the mucin glycoprotein family. Differential expression of CA-125/MUC16 and MUC1 is reported in PROC cases [127]. An increase in CA-125 level following oral glucose intake is also suggested as a multi-drug resistance predictive biomarker [128]. To predict a decline in CA-125 level with the duration of treatments in OC patients, a unique model referred to as CA-125 elimination rate constant K (KELIM) was developed, and a high KELIM value was suggested to be an indicator of better therapeutic response [129]. CA-125, IL6, and leptin or serum CA-125 and ascites leptin can also be used as potential novel biomarkers to predict baseline clinical resistance to first-line treatment and poor outcome in HGSC patients [130, 131]. A recent study conducted to determine the predictive and prognostic potential of CA-125 in PROC patients, who received second-line therapy (platinum-based drug combined with bevacizumab), suggested its utility as an independent predictor of therapeutic response and OC recurrence [132].
Aberrant glycosylation of proteins, including mucins, is reported in several malignancies and used as a biomarker [133, 134]. Increased levels of poly-LacNAc and low levels of α2–6-linked sialic structures are reported in cisplatin-resistance OC cells, potentially suggesting their utility as PROC biomarker [135]. An altered expression in five glyco-subclasses and five single glycans is reported in recurrent and PROC patients. Further, a panel of three glycans, Lewis type N-glycans, α2,3 sialic acid, and multi-branch glycans (tri- and tetra-antennary glycans), was significantly altered in OC patients with the response of platinum-based drug [136]. Recently, N-glycomic profiling was carried out in tissue and serum samples obtained from advanced-stage high-grade OC patients. Data suggested a differential expression of N-glycans in OC samples and further found a positive association between tissue N-glycans and platinum resistance compared to serum N-glycans [137]. Thus, these glycans could be used as a predictor of chemotherapeutic response in PROC patients.
3.7. Metabolomic biomarkers
Metabolomics is the large-scale study of small metabolites in biological samples that can provide comprehensive information on cellular metabolism and be exploited for biomarker development [138]. Human Metabolome Database (https://hmdb.ca/) contains information on >25,000 small molecule metabolites present in the human body and serves as a valuable resource for biochemical studies and biomarker discovery. A clinical trial is ongoing to generate multi-omics (exome sequencing, transcriptomics, and metabolomics) findings on EOC patients of different clinical stages and pathological subtypes (NCT03742856). Another trial is aimed at determining the presence of prostaglandin metabolite in urine samples of OC patients (NCT00900523). Several urine metabolites have been identified to exhibit differential levels in OC patients and healthy subjects suggesting their potential utility as disease biomarkers [139]. Studies also suggest that metabolic alterations are associated with cancer progression and chemotherapy resistance [140, 141]. Metabolic profiling of platinum-sensitive and -resistant OC cells revealed an association of altered cysteine and methionine metabolism with platinum resistance [142]. In another study, metabolic comparison between serous (OVCAR3) and non-serous (ES2) cancer cells suggested the presence of a distinct amino acid pool such as L-threonine, L-tyrosine and L-phenylalanine, and L-asparagine and ornithine (non-proteinogenic amino acid). Furthermore, carboplatin treatment facilitated increased GSH accumulation in non-serous OC compared to serous and imparted platinum resistance by forming platinum-thiol conjugates [101]. A study uncovered differences in lipid composition between platinum-resistant and -sensitive OC cells. PROC cells had lower lipids, for example, PC38:4, PC38:6, dehydrosphinganine, and lactosyl-ceramide, compared to the platinum-sensitive OC cells, suggesting an increased lipid utilization or decreased biosynthesis [143]. Levels of amino acid and associated metabolites such as histidine, tryptophan, phenylalanine, and kynurenine, among others, are reported to be significantly high in the plasma of relapsed OC patients [144]. Serum metabolome profiling also identified differential levels of six metabolites, calycanthidine, 1-monopalmitin, ricinoleic, methlester, polyoxyethylene-(600) mono-ricinoleate/glycidyl stearate and dodemorph, in PROC patients, which could be further examined for their potential use as biomarkers [145]. Thus, metabolites have good potential to be developed as biomarkers of disease progression and therapeutic response.
4. Liquid biopsy for ovarian cancer management
“Liquid biopsy” is a new concept that has gained substantial attention due to its several potential benefits over classical “tumor biopsy.” A liquid biopsy collects a sample of body fluid that is later analyzed to gather molecular information on the tumor type, its severity, and its response to therapeutic interventions [11, 146]. It overcomes several challenges associated with solid biopsy procedures, such as the tumor’s location and size, risk of potential spread, etc. Further, a liquid biopsy can be cost-effective, performed with ease, and potentially capture tumor heterogeneity more effectively than a solid biopsy [146, 147]. Liquid biopsy contains circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), extracellular microRNA (ex-miRNA), proteins, and metabolites that can be used for biomarker development and applications, as discussed below.
4.1. Circulating tumor cells (CTCs)
Disseminated tumor cells or CTCs can be originated from the primary or metastatic tumor sites and travel to other parts of the body through the bloodstream or lymphatic system [148]. Various methods have been studied to detect CTCs in liquid biopsy, but the CELLSEARCH® CTC Test is the only test that has received FDA clearance for detecting CTCs in the patients’ blood samples. Another method, CellCollector, has also been developed, which showed improved efficacy over CellSearch® in a comparative analysis in patients with neuroendocrine tumors [149]. However, both of these methods solely depend on the presence of EpCAM and certain cytokeratins on CTCs. A recent study tested an electrically conducive chip, incorporating a nano-roughened microfluidic platform to capture CTCs expressing different antigens (EpCAM, TROP-2, EGFR, vimentin, and N-cadherin) in OC patients. The CTC-cluster with a cutpoint of ≥3 CTCs showed a reduction in progression-free survival and positivity correlated with platinum resistance [150]. In another study, Wimberger and colleagues reported the prevalence of EpCAM-positive disseminated cancer cells in the bone marrow of OC patients treated with platinum-based drugs, which suggested a high chance of disease relapse [151]. Molecular characterization of circulating OC tumor cells has indicated that patients treated with platinum-based therapy show a more predominant expression of peptidylprolyl isomerase C (PPIC) than the EpCAM expression. Furthermore, OC patients with PPIC-positive CTCs were shown to have a poorer overall- and disease-free survival compared to PPIC negative patients [152]. Another study reported that platinum-sensitive OC patients with PPIC+ CTCs had high levels of neopterin and Kyn/Trp, associated with advanced disease, peritoneal carcinomatosis, ascites, sub-optimal debulking, inadequate response to therapy, and worse outcome. Moreover, neopterin and Kyn/Trp levels were heightened in CTC-positive patients, both at diagnosis and at follow-up in platinum-sensitive disease [153]. Interestingly, while ERCC1 expression in tumor tissue did not show any clinical significance [154], the presence of ERCC1- positive CTCs at the time of primary diagnosis served as an independent predictor of patients’ survival and platinum resistance [155]. Therefore, characterization of CTCs for different antigens or a combination of antigens can be an effective tool for OC diagnosis and predicting the chances of therapeutic success or failure and disease recurrence.
4.2. Cell-free circulating tumor DNA (ctDNA)
Cell-free circulating tumor DNA (ctDNA) has also been studied for monitoring drug response and disease recurrence in OC patients. In a recent study, genome-wide copy number alterations in ctDNA were examined as novel biomarkers in high-grade serous OC patients [156]. Shallow whole-genome sequencing of liquid biopsy demonstrated a gain in clonal regions, 3q26.2 and 8q24.3, in a significant majority of plasma samples, mirroring solid biopsies from the same cases. A heterogeneous pattern of genomic amplification was also observed in OC patients before and after platinum-based therapy in the 19p31.11 and 19q13.42 regions. Further, ctDNA analysis outperformed CA-125 in anticipating clinical and radiological progression, suggesting its utility to monitor disease evolution following treatment and disease relapse prediction [156]. BRCA1/2 reversion mutations have also been detected in the ctDNA in the plasma of PROC patients [157, 158]. In a cohort of gynecologic cancer patients, ctDNA analysis of PROC cases (78%) showed mutations in many genes, including EGFR, KRAS, TP53, PIK3CA, MYC, BRAF, MET, CCNE1, and CDK6. Moreover, a higher ctDNA mutation frequency was associated with worse overall survival [159]. Similarly, TP53 mutations have been detected in circulating tumor DNA (ctDNA) samples of OC patients, and a marked reduction in mutant TP53 in ctDNA is reported following treatment with platinum and non-platinum drugs in HGSC patients [160, 161]. CpG methylation has also been reported in the peripheral blood DNA of OC patients treated with platinum-based drugs by performing bisulfite sequencing during the first relapse. Therefore, blood-based DNA methylation can be used as a novel biomarker to predict the overall survival of PROC patients [162]. A clinical trial is currently ongoing to evaluate ctDNA as an early biomarker of disease recurrence and platinum-based treatment efficacy in OC patients (NCT03302884).
4.3. Extracellular Vesicles (EVs) and microRNAs (ex-miRNA)
Tumor cells release tiny membrane vesicles in the extracellular spaces to exert biological functions locally and at distant sites [28, 163]. These extracellular vesicles (EVs) harbor tumor-specific biomolecules (DNA, RNA, protein, and metabolites) and can be easily isolated from body fluids such as blood, urine, etc. In recent years, several novel methods have been developed for the characterization of EVs for clinical application. A label-free, high-throughput approach for quantitative analysis of exosomes or smaller-sized EVs was developed and used to analyze ascites samples from OC patients. This nano-plasmonic exosome (nPLEX) assay was based on transmission surface plasmon resonance through periodic nanohole arrays functionalized with antibodies to enable profiling of exosome surface proteins. CD24- and EpCAM- positive exosomes were identified using this method, suggesting its utility for biomarker development and future clinical applications [164]. Another technique, ExoCounter, has also been used to quantify the number of exosomes in the serum of OC patients using CD9, CD63, CD147, and HER2 as surface markers [165]. Since EVs derived from PROC cells have been shown to impart chemoresistance in platinum-sensitive cells [166], they could potentially be exploited to predict chemoresistance and monitor therapeutic responses. Characterization of embedded biomaterial in EVs can also be a valuable source for developing precise tumor-specific biomarkers.
Circulating extracellular miRNAs (ex-miRNAs) have been examined as biomarkers for platinum resistance in OC patients. Cf-miRNAs are quite stable as they remain protected from nuclease activity by high-density lipoprotein, miRNA-binding proteins, argonaute2, or EV encapsulation [167–169]. cf-miRNAs can be isolated from various body fluids and profiled using high-throughput approaches, such as miRNA microarrays or next-generation sequencing (NGS). Previous studies, including ours, have shown altered release of EVs and encapsulated miRNAs in response to tumor hypoxia and exposure to chemotherapeutic drug supporting their clinical utility as liquid biopsy-based biomarkers [28, 170]. A major limitation; however, is the lack of defined endogenous controls for serum miRNAs. Nonetheless, miR-16, RNU-6B, and RNU-48 or let-7d, let-7 g, and let-7i have often been used as referenced genes and yielded reliable normalization [169]. NGS-based detection of EV-derived miRNAs in plasma samples revealed that miR-1908, miR-181a, miR-486, miR223, and miR-21 were overexpressed in PROC patients [171]. In another study, PROC patients showed higher serum miR-130a, which correlated with tumor progression and lymph node metastasis [83]. Increased serum level of U2–1 snRNA fragment (RNU2–1f) was also reported in OC patients but did not show any association with histological subtype or platinum resistance [172]. The enhanced level of serum miR-622 associated significantly with lower progression-free and overall survival of OC patients and was an independent predictive biomarker of response to platinum-based drugs both in newly diagnosed and relapsed OC cases [173]. Another study showed a lower level of serum miR-125b in patients exhibiting chemoresistance than those who responded favorably to chemotherapy. Moreover, serum miR-125b combined with serum CA125 improved sensitivity and specificity of EOC diagnosis [174]. A reduction in the level of circulating miR-148–5p has been reported in OC patients treated with chemotherapeutic drugs, carboplatin and decitabine, and associated with poor progression-free survival [175]. Clearly, cf-miRNAs hold immense potential to be exploited as biomarkers for inherent and acquired chemoresistance as well as disease diagnosis.
4.4. Serum proteins
Teng and et al. identified 16 differentially expressed proteins in the secretome of the platinum-sensitive and -resistant OC cells. Among these proteins, COL11A1 was highly expressed in PROC cells [176]. Similarly, another study demonstrated increased levels of soluble PD-L1 (sPD-L1) and decreased levels of sPD-L2 in serum samples of OC patients. Enhanced sPD-L1 was associated significantly with residual tumor burden, while reduced sPD-L2 levels predicted platinum resistance. Furthermore, sPD-L1 levels were negatively associated with overall and progression-free survival, although only in platinum-sensitive patients [177]. Therefore, both sPD-L1 and sPD-L2 should be investigated further in larger cohorts to explore their utility as serum biomarkers of unfavorable disease outcomes despite drug sensitivity. Serum lactate dehydrogenase (LDH) has been used to monitor radiotherapy and chemotherapy responses in many cancer types. Recently, Ikeda et al. reported high serum LDH in PROC patients and suggested its use as a marker for platinum-resistant disease [178]. Robust reduction in serum VEGF-A is also associated with inadequate therapeutic response and worse prognosis in PROC patients treated with gemcitabine combined with anti-angiogenic drug, bevacizumab [179]. Thus, serum VEGF-A could be exploited as a liquid biopsy biomarker for therapeutic response prediction in PROC patients. Calcium-binding protein calretinin was present in the serum of OC patients at significantly higher levels than the healthy subjects and was an independent biomarker of platinum resistance [180]. Annexin A3, a member of the calcium and phospholipid-binding protein family, is highly overexpressed in PROC cell lines and tissues but does not show any significant clinical association in PROC patients [181]. It could be due to tumor heterogeneity or interaction of Annexin A3 with other serum factors interfering with its capture and measurement. Therefore, not all secreted proteins, overexpressed in tumor tissues and exhibiting clinicopathological association, could be used as liquid biopsy biomarkers. Moreover, the detection procedure may also need to be optimized for each protein biomarker.
5. Conclusion and future perspective
Platinum-based therapy remains the first line of treatment for OC patients, but disease relapse is inevitable in most cases due to inherent or acquired chemoresistance [7, 182]. Years of research have identified several molecular mechanisms underlying platinum resistance, as discussed earlier, and this information has helped develop alternative therapeutic strategies. While most genes implicated in platinum resistance belong to DNA damage repair machinery, some other proteins also make a substantial contribution. In many instances, the development of platinum resistance could be multifactorial due to disease heterogeneity. Thus, understanding the molecular nature and then formulating the best combination therapy regimen would be crucial in achieving the best possible clinical outcome.
Molecular markers suggestive of different underlying platinum-resistance mechanism(s) could be of immense help in the treatment planning. Since platinum resistance could also be dynamic or some mechanisms could dominate over others at initial presentation, constant monitoring of therapeutic responses using biomarker sets will be required throughout the treatment and afterward. This greatly emphasizes the need for reliable, mechanism-specific biomarkers and approaches to assist in repeated monitoring of the disease. Although we have several good leads for biomarker development, we do not yet have the bonafide ones for accurate tracking, surveillance, or treatment guidance of PROC. The use of approaches that can capture the tumor heterogeneity is also crucial to better plan the therapeutic course of action. In that regard, liquid biopsy-based biomarkers could be ideal, requiring less invasive procedures and theoretically providing comprehensive information on the molecular landscape of the disease. However, discrepancies across studies could arise due to the use of different sampling and analytical methods. Lack of established endogenous controls is also a significant bottleneck in cases where quantification of the biomarker is required to diagnose or base the therapeutic decision. We have also had limited success in having a superior treatment choice to improve patients’ survival. Treatment response varies considerably among different subtypes of OC or even within a subtype due to inherent genetic, epigenetic, and microenvironmental differences supporting distinct chemoresistance pathways. Multifocal and integrative genomic/epigenomic profiling in a subtype-specific manner could help identify the compendium of altered genetic and epigenetic landscape therein. This, in turn, will provide a rationale to move forward with functional validation to ultimately define the target(s) for therapeutics.
It is high time that more efforts are directed towards biomarker testing in larger cohorts in randomized trials. A multicenter comprehensive analysis of carefully chosen sample cohorts will be required to achieve this goal. The approaches described above for genome and epigenome profiling could serve as a unique platform for biomarker discovery followed by stringent validation. However, it could be a difficult task to achieve since platinum drugs are delivered in combination with other agents, and their joint action could complicate mechanistic interpretations. Therefore, molecular and/or immune profiling before and after such treatments may aid in refining biomarkers and developing alternate treatment strategies. Immunotherapy is currently under the reincarnation phase to successfully treat human malignancies, and its thoughtful use could help overcome platinum resistance. To achieve an optimal outcome, it would be necessary to first stratify the patients that would benefit from specific immunotherapeutic modalities based on existing and novel biomarkers under development. At this time, clinical and basic research endeavors ought to be combined while wisely utilizing the technological advancements to achieve paradigm-shifting progress in the management of platinum-resistant ovarian cancer.
Acknowledgments
We would like to acknowledge the funding from NIH/NCI [R01CA224306, R01CA175772, U01CA185490 (to APS) and R01CA204801, R01CA231925 (to SS)] and USAMCI (to APS and SS).
Footnotes
Conflict of Interest
The authors have no conflict of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin (2021). [DOI] [PubMed] [Google Scholar]
- [2].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2020, CA Cancer J Clin 70(1) (2020) 7–30. [DOI] [PubMed] [Google Scholar]
- [3].Devouassoux-Shisheboran M, Genestie C, Pathobiology of ovarian carcinomas, Chin J Cancer 34(1) (2015) 50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Prat J, New insights into ovarian cancer pathology, Ann Oncol 23 Suppl 10 (2012) x111–7. [DOI] [PubMed] [Google Scholar]
- [5].Gadducci A, Cosio S, Therapeutic Approach to Low-Grade Serous Ovarian Carcinoma: State of Art and Perspectives of Clinical Research, Cancers (Basel) 12(5) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pinsky PF, Yu K, Kramer BS, Black A, Buys SS, Partridge E, Gohagan J, Berg CD, Prorok PC, Extended mortality results for ovarian cancer screening in the PLCO trial with median 15years follow-up, Gynecol Oncol 143(2) (2016) 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Damia G, Broggini M, Platinum Resistance in Ovarian Cancer: Role of DNA Repair, Cancers (Basel) 11(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ross JS, Cronin M, Whole cancer genome sequencing by next-generation methods, Am J Clin Pathol 136(4) (2011) 527–39. [DOI] [PubMed] [Google Scholar]
- [9].Smania MA, Liquid biopsy for cancer screening, diagnosis, and treatment, J Am Assoc Nurse Pract 32(1) (2020) 5–7. [DOI] [PubMed] [Google Scholar]
- [10].Lo YMD, Lam WKJ, Towards multi-cancer screening using liquid biopsies, Nat Rev Clin Oncol 17(9) (2020) 525–526. [DOI] [PubMed] [Google Scholar]
- [11].Corcoran RB, Liquid biopsy versus tumor biopsy for clinical-trial recruitment, Nat Med 26(12) (2020) 1815–1816. [DOI] [PubMed] [Google Scholar]
- [12].Stornetta A, Zimmermann M, Cimino GD, Henderson PT, Sturla SJ, DNA Adducts from Anticancer Drugs as Candidate Predictive Markers for Precision Medicine, Chem Res Toxicol 30(1) (2017) 388–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gately DP, Howell SB, Cellular accumulation of the anticancer agent cisplatin: a review, Br J Cancer 67(6) (1993) 1171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Buss I, Hamacher A, Sarin N, Kassack MU, Kalayda GV, Relevance of copper transporter 1 and organic cation transporters 1–3 for oxaliplatin uptake and drug resistance in colorectal cancer cells, Metallomics 10(3) (2018) 414–425. [DOI] [PubMed] [Google Scholar]
- [15].Gao H, Zhang S, Hu T, Qu X, Zhai J, Zhang Y, Tao L, Yin J, Song Y, Omeprazole protects against cisplatin-induced nephrotoxicity by alleviating oxidative stress, inflammation, and transporter-mediated cisplatin accumulation in rats and HK-2cells, Chem Biol Interact 297 (2019) 130–140. [DOI] [PubMed] [Google Scholar]
- [16].Kunii E, Oguri T, Kasai D, Ozasa H, Uemura T, Takakuwa O, Ohkubo H, Takemura M, Maeno K, Niimi A, Organic cation transporter OCT6 mediates cisplatin uptake and resistance to cisplatin in lung cancer, Cancer Chemother Pharmacol 75(5) (2015) 985–91. [DOI] [PubMed] [Google Scholar]
- [17].Ishida S, McCormick F, Smith-McCune K, Hanahan D, Enhancing tumor-specific uptake of the anticancer drug cisplatin with a copper chelator, Cancer Cell 17(6) (2010) 574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kilari D, Guancial E, Kim ES, Role of copper transporters in platinum resistance, World J Clin Oncol 7(1) (2016) 106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakayama K, Kanzaki A, Ogawa K, Miyazaki K, Neamati N, Takebayashi Y, Copper-transporting P-type adenosine triphosphatase (ATP7B) as a cisplatin based chemoresistance marker in ovarian carcinoma: comparative analysis with expression of MDR1, MRP1, MRP2, LRP and BCRP, Int J Cancer 101(5) (2002) 488–95. [DOI] [PubMed] [Google Scholar]
- [20].Chisholm CL, Wang H, Wong AH, Vazquez-Ortiz G, Chen W, Xu X, Deng CX, Ammonium tetrathiomolybdate treatment targets the copper transporter ATP7A and enhances sensitivity of breast cancer to cisplatin, Oncotarget 7(51) (2016) 84439–84452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mangala LS, Zuzel V, Schmandt R, Leshane ES, Halder JB, Armaiz-Pena GN, Spannuth WA, Tanaka T, Shahzad MM, Lin YG, Nick AM, Danes CG, Lee JW, Jennings NB, Vivas-Mejia PE, Wolf JK, Coleman RL, Siddik ZH, Lopez-Berestein G, Lutsenko S, Sood AK, Therapeutic Targeting of ATP7B in Ovarian Carcinoma, Clin Cancer Res 15(11) (2009) 3770–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Januchowski R, Sterzynska K, Zaorska K, Sosinska P, Klejewski A, Brazert M, Nowicki M, Zabel M, Analysis of MDR genes expression and cross-resistance in eight drug resistant ovarian cancer cell lines, J Ovarian Res 9(1) (2016) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Arts HJ, Katsaros D, de Vries EG, Massobrio M, Genta F, Danese S, Arisio R, Scheper RJ, Kool M, Scheffer GL, Willemse PH, van der Zee AG, Suurmeijer AJ, Drug resistance-associated markers P-glycoprotein, multidrug resistance-associated protein 1, multidrug resistance-associated protein 2, and lung resistance protein as prognostic factors in ovarian carcinoma, Clin Cancer Res 5(10) (1999) 2798–805. [PubMed] [Google Scholar]
- [24].Kim A, Enomoto T, Serada S, Ueda Y, Takahashi T, Ripley B, Miyatake T, Fujita M, Lee CM, Morimoto K, Fujimoto M, Kimura T, Naka T, Enhanced expression of Annexin A4 in clear cell carcinoma of the ovary and its association with chemoresistance to carboplatin, Int J Cancer 125(10) (2009) 2316–22. [DOI] [PubMed] [Google Scholar]
- [25].Schieber M, Chandel NS, ROS function in redox signaling and oxidative stress, Curr Biol 24(10) (2014) R453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O, Oxidative stress and antioxidant defense, World Allergy Organ J 5(1) (2012) 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Khan MA, Gahlot S, Majumdar S, Oxidative stress induced by curcumin promotes the death of cutaneous T-cell lymphoma (HuT-78) by disrupting the function of several molecular targets, Mol Cancer Ther 11(9) (2012) 1873–83. [DOI] [PubMed] [Google Scholar]
- [28].Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC, Singh S, Khushman M, Singh AP, Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK, Br J Cancer 116(5) (2017) 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Redza-Dutordoir M, Averill-Bates DA, Activation of apoptosis signalling pathways by reactive oxygen species, Biochim Biophys Acta 1863(12) (2016) 2977–2992. [DOI] [PubMed] [Google Scholar]
- [30].Gamcsik MP, Kasibhatla MS, Teeter SD, Colvin OM, Glutathione levels in human tumors, Biomarkers 17(8) (2012) 671–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Oien DB, Chien J, Cheng N, Regulation of chemo-sensitivity in ovarian cancer via a stroma dependent glutathione pathway, Transl Cancer Res 5(Suppl 3) (2016) S514–S519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen HH, Kuo MT, Role of glutathione in the regulation of Cisplatin resistance in cancer chemotherapy, Met Based Drugs 2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Estrela JM, Ortega A, Obrador E, Glutathione in cancer biology and therapy, Crit Rev Clin Lab Sci 43(2) (2006) 143–81. [DOI] [PubMed] [Google Scholar]
- [34].O’Brien ML, Tew KD, Glutathione and related enzymes in multidrug resistance, Eur J Cancer 32A(6) (1996) 967–78. [DOI] [PubMed] [Google Scholar]
- [35].Mistry P, Kelland LR, Abel G, Sidhar S, Harrap KR, The relationships between glutathione, glutathione-S-transferase and cytotoxicity of platinum drugs and melphalan in eight human ovarian carcinoma cell lines, Br J Cancer 64(2) (1991) 215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sarkhosh-Inanlou R, Molaparast M, Mohammadzadeh A, Shafiei-Irannejad V, Sanguinarine enhances cisplatin sensitivity via glutathione depletion in cisplatin-resistant ovarian cancer (A2780) cells, Chem Biol Drug Des 95(2) (2020) 215–223. [DOI] [PubMed] [Google Scholar]
- [37].Andrews PA, Murphy MP, Howell SB, Metallothionein-mediated cisplatin resistance in human ovarian carcinoma cells, Cancer Chemother Pharmacol 19(2) (1987) 149–54. [DOI] [PubMed] [Google Scholar]
- [38].Khan MA, Zubair H, Anand S, Srivastava SK, Singh S, Singh AP, Dysregulation of metabolic enzymes in tumor and stromal cells: Role in oncogenesis and therapeutic opportunities, Cancer Lett 473 (2020) 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Latifi A, Abubaker K, Castrechini N, Ward AC, Liongue C, Dobill F, Kumar J, Thompson EW, Quinn MA, Findlay JK, Ahmed N, Cisplatin treatment of primary and metastatic epithelial ovarian carcinomas generates residual cells with mesenchymal stem cell-like profile, J Cell Biochem 112(10) (2011) 2850–64. [DOI] [PubMed] [Google Scholar]
- [40].Reyes-Gonzalez JM, Quinones-Diaz BI, Santana Y, Baez-Vega PM, Soto D, Valiyeva F, Marcos-Martinez MJ, Fernandez-de Thomas RJ, Vivas-Mejia PE, Downstream Effectors of ILK in Cisplatin-Resistant Ovarian Cancer, Cancers (Basel) 12(4) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Diaz Osterman CJ, Ozmadenci D, Kleinschmidt EG, Taylor KN, Barrie AM, Jiang S, Bean LM, Sulzmaier FJ, Jean C, Tancioni I, Anderson K, Uryu S, Cordasco EA, Li J, Chen XL, Fu G, Ojalill M, Rappu P, Heino J, Mark AM, Xu G, Fisch KM, Kolev VN, Weaver DT, Pachter JA, Gyorffy B, McHale MT, Connolly DC, Molinolo A, Stupack DG, Schlaepfer DD, FAK activity sustains intrinsic and acquired ovarian cancer resistance to platinum chemotherapy, Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ojalvo LS, Thompson ED, Wang TL, Meeker AK, Shih IM, Fader AN, Cimino-Mathews A, Emens LA, Tumor-associated macrophages and the tumor immune microenvironment of primary and recurrent epithelial ovarian cancer, Hum Pathol 74 (2018) 135–147. [DOI] [PubMed] [Google Scholar]
- [43].Nowak M, Klink M, The Role of Tumor-Associated Macrophages in the Progression and Chemoresistance of Ovarian Cancer, Cells 9(5) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ghoneum A, Almousa S, Warren B, Abdulfattah AY, Shu J, Abouelfadl H, Gonzalez D, Livingston C, Said N, Exploring the clinical value of tumor microenvironment in platinum-resistant ovarian cancer, Semin Cancer Biol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T, Drug Resistance Driven by Cancer Stem Cells and Their Niche, Int J Mol Sci 18(12) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Terraneo N, Jacob F, Dubrovska A, Grunberg J, Novel Therapeutic Strategies for Ovarian Cancer Stem Cells, Front Oncol 10 (2020) 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen X, Zhang J, Zhang Z, Li H, Cheng W, Liu J, Cancer stem cells, epithelial-mesenchymal transition, and drug resistance in high-grade ovarian serous carcinoma, Hum Pathol 44(11) (2013) 2373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Meng E, Long B, Sullivan P, McClellan S, Finan MA, Reed E, Shevde L, Rocconi RP, CD44+/CD24- ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival, Clin Exp Metastasis 29(8) (2012) 939–48. [DOI] [PubMed] [Google Scholar]
- [49].Landen CN Jr., Goodman B, Katre AA, Steg AD, Nick AM, Stone RL, Miller LD, Mejia PV, Jennings NB, Gershenson DM, Bast RC Jr., Coleman RL, Lopez-Berestein G, Sood AK, Targeting aldehyde dehydrogenase cancer stem cells in ovarian cancer, Mol Cancer Ther 9(12) (2010) 3186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA, Palle K, Rocconi RP, ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling, PLoS One 9(9) (2014) e107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lupia M, Angiolini F, Bertalot G, Freddi S, Sachsenmeier KF, Chisci E, Kutryb-Zajac B, Confalonieri S, Smolenski RT, Giovannoni R, Colombo N, Bianchi F, Cavallaro U, CD73 Regulates Stemness and Epithelial-Mesenchymal Transition in Ovarian Cancer-Initiating Cells, Stem Cell Reports 10(4) (2018) 1412–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liu S, Sun J, Cai B, Xi X, Yang L, Zhang Z, Feng Y, Sun Y, NANOG regulates epithelial-mesenchymal transition and chemoresistance through activation of the STAT3 pathway in epithelial ovarian cancer, Tumour Biol 37(7) (2016) 9671–80. [DOI] [PubMed] [Google Scholar]
- [53].Samardzija C, Quinn M, Findlay JK, Ahmed N, Attributes of Oct4 in stem cell biology: perspectives on cancer stem cells of the ovary, J Ovarian Res 5(1) (2012) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G, Molecular mechanisms of cisplatin resistance, Oncogene 31(15) (2012) 1869–83. [DOI] [PubMed] [Google Scholar]
- [55].Chen JJ, Silver D, Cantor S, Livingston DM, Scully R, BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway, Cancer Res 59(7 Suppl) (1999) 1752s–1756s. [PubMed] [Google Scholar]
- [56].Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A, Replication fork stability confers chemoresistance in BRCA-deficient cells, Nature 535(7612) (2016) 382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Belanger F, Fortier E, Dube M, Lemay JF, Buisson R, Masson JY, Elsherbiny A, Costantino S, Carmona E, Mes-Masson AM, Wurtele H, Drobetsky E, Replication Protein A Availability during DNA Replication Stress Is a Major Determinant of Cisplatin Resistance in Ovarian Cancer Cells, Cancer Res 78(19) (2018) 5561–5573. [DOI] [PubMed] [Google Scholar]
- [58].Mei L, Zhang J, He K, Zhang J, Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: where we stand, J Hematol Oncol 12(1) (2019) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Huntoon CJ, Flatten KS, Wahner Hendrickson AE, Huehls AM, Sutor SL, Kaufmann SH, Karnitz LM, ATR inhibition broadly sensitizes ovarian cancer cells to chemotherapy independent of BRCA status, Cancer Res 73(12) (2013) 3683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vaisman A, Masutani C, Hanaoka F, Chaney SG, Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta, Biochemistry 39(16) (2000) 4575–80. [DOI] [PubMed] [Google Scholar]
- [61].McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA, The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases delta and eta, Nucleic Acids Res 37(9) (2009) 2830–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ignatov T, Eggemann H, Costa SD, Roessner A, Kalinski T, Ignatov A, BRCA1 promoter methylation is a marker of better response to platinum-taxane-based therapy in sporadic epithelial ovarian cancer, J Cancer Res Clin Oncol 140(9) (2014) 1457–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bai X, Fu Y, Xue H, Guo K, Song Z, Yu Z, Jia T, Yan Y, Zhao L, Mi X, Wang E, Zheng Z, Zhao H, Yao W, Wei M, BRCA1 promoter hypermethylation in sporadic epithelial ovarian carcinoma: Association with low expression of BRCA1, improved survival and co-expression of DNA methyltransferases, Oncol Lett 7(4) (2014) 1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chiang JW, Karlan BY, Cass L, Baldwin RL, BRCA1 promoter methylation predicts adverse ovarian cancer prognosis, Gynecol Oncol 101(3) (2006) 403–10. [DOI] [PubMed] [Google Scholar]
- [65].Kalachand RD, Stordal B, Madden S, Chandler B, Cunningham J, Goode EL, Ruscito I, Braicu EI, Sehouli J, Ignatov A, Yu H, Katsaros D, Mills GB, Lu KH, Carey MS, Timms KM, Kupryjanczyk J, Rzepecka IK, Podgorska A, McAlpine JN, Swisher EM, Bernards SS, O’Riain C, O’Toole S, O’Leary JJ, Bowtell DD, Thomas DM, Prieske K, Joosse SA, Woelber L, Chaudhry P, Hafner N, Runnebaum IB, Hennessy BT, BRCA1 Promoter Methylation and Clinical Outcomes in Ovarian Cancer: An Individual Patient Data Meta-Analysis, J Natl Cancer Inst 112(12) (2020) 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kalachand RD, O’Riain C, Toomey S, Carr A, Timms KM, O’Toole S, Madden S, Bates M, O’Leary JJ, Gleeson N, O’Donnell D, Grogan L, Breathnach O, Farrelly A, Stordal B, Hennessy BT, Prevalence of tumor BRCA1 and BRCA2 dysfunction in unselected patients with ovarian cancer, Obstet Gynecol Sci 63(5) (2020) 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jones BA, Varambally S, Arend RC, Histone Methyltransferase EZH2: A Therapeutic Target for Ovarian Cancer, Mol Cancer Ther 17(3) (2018) 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Rizzo S, Hersey JM, Mellor P, Dai W, Santos-Silva A, Liber D, Luk L, Titley I, Carden CP, Box G, Hudson DL, Kaye SB, Brown R, Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2, Mol Cancer Ther 10(2) (2011) 325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hu S, Yu L, Li Z, Shen Y, Wang J, Cai J, Xiao L, Wang Z, Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo, Cancer Biol Ther 10(8) (2010) 788–95. [DOI] [PubMed] [Google Scholar]
- [70].Caumanns JJ, Wisman GBA, Berns K, van der Zee AGJ, de Jong S, ARID1A mutant ovarian clear cell carcinoma: A clear target for synthetic lethal strategies, Biochim Biophys Acta Rev Cancer 1870(2) (2018) 176–184. [DOI] [PubMed] [Google Scholar]
- [71].Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Nakayama N, Ishikawa M, Ishibashi T, Iida K, Kobayashi H, Otsuki Y, Nakayama S, Miyazaki K, Loss of ARID1A expression is related to shorter progression-free survival and chemoresistance in ovarian clear cell carcinoma, Mod Pathol 25(2) (2012) 282–8. [DOI] [PubMed] [Google Scholar]
- [72].Granados ML, Hudson LG, Samudio-Ruiz SL, Contributions of the Epidermal Growth Factor Receptor to Acquisition of Platinum Resistance in Ovarian Cancer Cells, PLoS One 10(9) (2015) e0136893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cacan E, Ali MW, Boyd NH, Hooks SB, Greer SF, Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance, PLoS One 9(1) (2014) e87455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhao Y, Li Q, Wu X, Chen P, Upregulation of p27Kip1 by demethylation sensitizes cisplatin-resistant human ovarian cancer SKOV3 cells, Mol Med Rep 14(2) (2016) 1659–66. [DOI] [PubMed] [Google Scholar]
- [75].Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N, Lin B, UCHL1 Is a Putative Tumor Suppressor in Ovarian Cancer Cells and Contributes to Cisplatin Resistance, J Cancer 4(8) (2013) 662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Han X, Zhou Y, You Y, Lu J, Wang L, Hou H, Li J, Chen W, Zhao L, Li X, TET1 promotes cisplatin-resistance via demethylating the vimentin promoter in ovarian cancer, Cell Biol Int 41(4) (2017) 405–414. [DOI] [PubMed] [Google Scholar]
- [77].de Leon M, Cardenas H, Vieth E, Emerson R, Segar M, Liu Y, Nephew K, Matei D, Transmembrane protein 88 (TMEM88) promoter hypomethylation is associated with platinum resistance in ovarian cancer, Gynecol Oncol 142(3) (2016) 539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang X, Yu X, Jiang G, Miao Y, Wang L, Zhang Y, Liu Y, Fan C, Lin X, Dong Q, Han Q, Zhao H, Han Y, Han X, Rong X, Ding S, Wang E, Wang E, Cytosolic TMEM88 promotes invasion and metastasis in lung cancer cells by binding DVLS, Cancer Res 75(21) (2015) 4527–37. [DOI] [PubMed] [Google Scholar]
- [79].Liu X, Yu Y, Zhang J, Lu C, Wang L, Liu P, Song H, HDAC1 Silencing in Ovarian Cancer Enhances the Chemotherapy Response, Cell Physiol Biochem 48(4) (2018) 1505–1518. [DOI] [PubMed] [Google Scholar]
- [80].Islam MM, Banerjee T, Packard CZ, Kotian S, Selvendiran K, Cohn DE, Parvin JD, HDAC10 as a potential therapeutic target in ovarian cancer, Gynecol Oncol 144(3) (2017) 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Sun X, Wang S, Gai J, Guan J, Li J, Li Y, Zhao J, Zhao C, Fu L, Li Q, SIRT5 Promotes Cisplatin Resistance in Ovarian Cancer by Suppressing DNA Damage in a ROS-Dependent Manner via Regulation of the Nrf2/HO-1 Pathway, Front Oncol 9 (2019) 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ, MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN, Cancer Res 68(2) (2008) 425–33. [DOI] [PubMed] [Google Scholar]
- [83].Chen C, Wang HJ, Yang LY, Jia XB, Xu P, Chen J, Liu Y, [Expression of MiR-130a in Serum Samples of Patients with Epithelial Ovarian Cancer and Its Association with Platinum Resistance], Sichuan Da Xue Xue Bao Yi Xue Ban 47(1) (2016) 60–3. [PubMed] [Google Scholar]
- [84].Xiang Y, Chen YJ, Yan YB, Liu Y, Qiu J, Tan RQ, Tian Q, Guan L, Niu SS, Xin HW, MiR-186 bidirectionally regulates cisplatin sensitivity of ovarian cancer cells via suppressing targets PIK3R3 and PTEN and upregulating APAF1 expression, J Cancer 11(12) (2020) 3446–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Sun J, Cai X, Yung MM, Zhou W, Li J, Zhang Y, Li Z, Liu SS, Cheung ANY, Ngan HYS, Li Y, Dai Z, Kai Y, Tzatsos A, Peng W, Chan DW, Zhu W, miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer, Oncogene 38(4) (2019) 564–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pink RC, Samuel P, Massa D, Caley DP, Brooks SA, Carter DR, The passenger strand, miR-21–3p, plays a role in mediating cisplatin resistance in ovarian cancer cells, Gynecol Oncol 137(1) (2015) 143–51. [DOI] [PubMed] [Google Scholar]
- [87].Zhang Y, Huang S, Guo Y, Li L, MiR-1294 confers cisplatin resistance in ovarian Cancer cells by targeting IGF1R, Biomed Pharmacother 106 (2018) 1357–1363. [DOI] [PubMed] [Google Scholar]
- [88].Jiang Y, Jiang J, Jia H, Qiao Z, Zhang J, Recovery of miR-139–5p in Ovarian Cancer Reverses Cisplatin Resistance by Targeting C-Jun, Cell Physiol Biochem 51(1) (2018) 129–141. [DOI] [PubMed] [Google Scholar]
- [89].Wang Z, Ting Z, Li Y, Chen G, Lu Y, Hao X, microRNA-199a is able to reverse cisplatin resistance in human ovarian cancer cells through the inhibition of mammalian target of rapamycin, Oncol Lett 6(3) (2013) 789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Feng X, Liu N, Deng S, Zhang D, Wang K, Lu M, miR-199a modulates cisplatin resistance in ovarian cancer by targeting Hif1alpha, Onco Targets Ther 10 (2017) 5899–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Madariaga A, Lheureux S, Oza AM, Tailoring Ovarian Cancer Treatment: Implications of BRCA1/2 Mutations, Cancers (Basel) 11(3) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Eoh KJ, Kim HM, Lee JY, Kim S, Kim SW, Kim YT, Nam EJ, Mutation landscape of germline and somatic BRCA1/2 in patients with high-grade serous ovarian cancer, BMC Cancer 20(1) (2020) 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Barber LJ, Sandhu S, Chen L, Campbell J, Kozarewa I, Fenwick K, Assiotis I, Rodrigues DN, Reis Filho JS, Moreno V, Mateo J, Molife LR, De Bono J, Kaye S, Lord CJ, Ashworth A, Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor, J Pathol 229(3) (2013) 422–9. [DOI] [PubMed] [Google Scholar]
- [94].Etemadmoghadam D, Weir BA, Au-Yeung G, Alsop K, Mitchell G, George J, Australian G Ovarian Cancer Study, S. Davis, A.D. D’Andrea, K. Simpson, W.C. Hahn, D.D. Bowtell, Synthetic lethality between CCNE1 amplification and loss of BRCA1, Proc Natl Acad Sci U S A 110(48) (2013) 19489–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liu J, Zhang L, Mao P, Jiang G, Liu L, Wang J, Yang W, Owusu L, Li W, Functional characterization of a novel transcript of ERCC1 in chemotherapy resistance of ovarian cancer, Oncotarget 8(49) (2017) 85759–85771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Reed E, Yu JJ, Davies A, Gannon J, Armentrout SL, Clear cell tumors have higher mRNA levels of ERCC1 and XPB than other histological types of epithelial ovarian cancer, Clin Cancer Res 9(14) (2003) 5299–305. [PubMed] [Google Scholar]
- [97].Wu RC, Ayhan A, Maeda D, Kim KR, Clarke BA, Shaw P, Chui MH, Rosen B, Shih Ie M, Wang TL, Frequent somatic mutations of the telomerase reverse transcriptase promoter in ovarian clear cell carcinoma but not in other major types of gynaecological malignancy, J Pathol 232(4) (2014) 473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Huang HN, Chiang YC, Cheng WF, Chen CA, Lin MC, Kuo KT, Molecular alterations in endometrial and ovarian clear cell carcinomas: clinical impacts of telomerase reverse transcriptase promoter mutation, Mod Pathol 28(2) (2015) 303–11. [DOI] [PubMed] [Google Scholar]
- [99].Yamaguchi S, Maida Y, Yasukawa M, Kato T, Yoshida M, Masutomi K, Eribulin mesylate targets human telomerase reverse transcriptase in ovarian cancer cells, PLoS One 9(11) (2014) e112438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Li Q, Zeng X, Cheng X, Zhang J, Ji J, Wang J, Xiong K, Qi Q, Huang W, Diagnostic value of dual detection of hepatocyte nuclear factor 1 beta (HNF-1beta) and napsin A for diagnosing ovarian clear cell carcinoma, Int J Clin Exp Pathol 8(7) (2015) 8305–10. [PMC free article] [PubMed] [Google Scholar]
- [101].Lopes-Coelho F, Gouveia-Fernandes S, Goncalves LG, Nunes C, Faustino I, Silva F, Felix A, Pereira SA, Serpa J, HNF1beta drives glutathione (GSH) synthesis underlying intrinsic carboplatin resistance of ovarian clear cell carcinoma (OCCC), Tumour Biol 37(4) (2016) 4813–29. [DOI] [PubMed] [Google Scholar]
- [102].N. Cancer Genome Atlas Research, Integrated genomic analyses of ovarian carcinoma, Nature 474(7353) (2011) 609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, Kilian U, Jones LA, El-Naggar A, Minguillon C, Schonborn I, Reich O, Kreienberg R, Lichtenegger W, Press MF, Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer, Clin Cancer Res 7(10) (2001) 2984–97. [PubMed] [Google Scholar]
- [104].Hagopian GS, Mills GB, Khokhar AR, Bast RC Jr., Siddik ZH, Expression of p53 in cisplatin-resistant ovarian cancer cell lines: modulation with the novel platinum analogue (1R, 2R-diaminocyclohexane)(trans-diacetato)(dichloro)-platinum(IV), Clin Cancer Res 5(3) (1999) 655–63. [PubMed] [Google Scholar]
- [105].Lorenzon I, Pellarin I, Pellizzari I, D’Andrea S, Belletti B, Sonego M, Baldassarre G, Schiappacassi M, Identification and Characterization of a New Platinum-Induced TP53 Mutation in MDAH Ovarian Cancer Cells, Cells 9(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Paracchini L, Pesenti C, Delle Marchette M, Beltrame L, Bianchi T, Grassi T, Buda A, Landoni F, Ceppi L, Bosetti C, Paderno M, Adorni M, Vicini D, Perego P, Leone BE, D’Incalci M, Marchini S, Fruscio R, Detection of TP53 Clonal Variants in Papanicolaou Test Samples Collected up to 6 Years Prior to High-Grade Serous Epithelial Ovarian Cancer Diagnosis, JAMA Netw Open 3(7) (2020) e207566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ghezelayagh TS, Pennington KP, Norquist BM, Khasnavis N, Radke MR, Kilgore MR, Garcia RL, Lee M, Katz R, Leslie KK, Risques RA, Swisher EM, Characterizing TP53 mutations in ovarian carcinomas with and without concurrent BRCA1 or BRCA2 mutations, Gynecol Oncol 160(3) (2021) 786–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Hentze JL, Hogdall CK, Hogdall EV, Methylation and ovarian cancer: Can DNA methylation be of diagnostic use?, Mol Clin Oncol 10(3) (2019) 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Yamaguchi K, Huang Z, Matsumura N, Mandai M, Okamoto T, Baba T, Konishi I, Berchuck A, Murphy SK, Epigenetic determinants of ovarian clear cell carcinoma biology, Int J Cancer 135(3) (2014) 585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Iramaneerat K, Rattanatunyong P, Khemapech N, Triratanachat S, Mutirangura A, HERV-K hypomethylation in ovarian clear cell carcinoma is associated with a poor prognosis and platinum resistance, Int J Gynecol Cancer 21(1) (2011) 51–7. [DOI] [PubMed] [Google Scholar]
- [111].Tian H, Yan L, Xiao-Fei L, Hai-Yan S, Juan C, Shan K, Hypermethylation of mismatch repair gene hMSH2 associates with platinum-resistant disease in epithelial ovarian cancer, Clin Epigenetics 11(1) (2019) 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bonito NA, Borley J, Wilhelm-Benartzi CS, Ghaem-Maghami S, Brown R, Epigenetic Regulation of the Homeobox Gene MSX1 Associates with Platinum-Resistant Disease in High-Grade Serous Epithelial Ovarian Cancer, Clin Cancer Res 22(12) (2016) 3097–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lund RJ, Huhtinen K, Salmi J, Rantala J, Nguyen EV, Moulder R, Goodlett DR, Lahesmaa R, Carpen O, DNA methylation and Transcriptome Changes Associated with Cisplatin Resistance in Ovarian Cancer, Sci Rep 7(1) (2017) 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Fang F, Munck J, Tang J, Taverna P, Wang Y, Miller DF, Pilrose J, Choy G, Azab M, Pawelczak KS, VanderVere-Carozza P, Wagner M, Lyons J, Matei D, Turchi JJ, Nephew KP, The novel, small-molecule DNA methylation inhibitor SGI-110 as an ovarian cancer chemosensitizer, Clin Cancer Res 20(24) (2014) 6504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP, Epigenetic resensitization to platinum in ovarian cancer, Cancer Res 72(9) (2012) 2197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Stefansson OA, Villanueva A, Vidal A, Marti L, Esteller M, BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer, Epigenetics 7(11) (2012) 1225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X, Li Y, Wu S, miR-98–5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1, Cell Death Dis 9(5) (2018) 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Samuel P, Pink RC, Brooks SA, Carter DR, miRNAs and ovarian cancer: a miRiad of mechanisms to induce cisplatin drug resistance, Expert Rev Anticancer Ther 16(1) (2016) 57–70. [DOI] [PubMed] [Google Scholar]
- [119].Biamonte F, Santamaria G, Sacco A, Perrone FM, Di Cello A, Battaglia AM, Salatino A, Di Vito A, Aversa I, Venturella R, Zullo F, Costanzo F, MicroRNA let-7g acts as tumor suppressor and predictive biomarker for chemoresistance in human epithelial ovarian cancer, Sci Rep 9(1) (2019) 5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Choi YE, Meghani K, Brault ME, Leclerc L, He YJ, Day TA, Elias KM, Drapkin R, Weinstock DM, Dao F, Shih KK, Matulonis U, Levine DA, Konstantinopoulos PA, Chowdhury D, Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer, Cell Rep 14(3) (2016) 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vecchione A, Belletti B, Lovat F, Volinia S, Chiappetta G, Giglio S, Sonego M, Cirombella R, Onesti EC, Pellegrini P, Califano D, Pignata S, Losito S, Canzonieri V, Sorio R, Alder H, Wernicke D, Stoppacciaro A, Baldassarre G, Croce CM, A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis, Proc Natl Acad Sci U S A 110(24) (2013) 9845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Qi X, Yu C, Wang Y, Lin Y, Shen B, Network vulnerability-based and knowledge-guided identification of microRNA biomarkers indicating platinum resistance in high-grade serous ovarian cancer, Clin Transl Med 8(1) (2019) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zhu Y, Zhao Y, Dong S, Liu L, Tai L, Xu Y, Systematic identification of dysregulated lncRNAs associated with platinum-based chemotherapy response across 11 cancer types, Genomics 112(2) (2020) 1214–1222. [DOI] [PubMed] [Google Scholar]
- [124].Liu R, Zeng Y, Zhou CF, Wang Y, Li X, Liu ZQ, Chen XP, Zhang W, Zhou HH, Long noncoding RNA expression signature to predict platinum-based chemotherapeutic sensitivity of ovarian cancer patients, Sci Rep 7(1) (2017) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Teschendorff AE, Lee SH, Jones A, Fiegl H, Kalwa M, Wagner W, Chindera K, Evans I, Dubeau L, Orjalo A, Horlings HM, Niederreiter L, Kaser A, Yang W, Goode EL, Fridley BL, Jenner RG, Berns EM, Wik E, Salvesen HB, Wisman GB, van der Zee AG, Davidson B, Trope CG, Lambrechts S, Vergote I, Calvert H, Jacobs IJ, Widschwendter M, HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer, Genome Med 7 (2015) 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Soletormos G, Duffy MJ, Othman Abu Hassan S, Verheijen RH, Tholander B, Bast RC Jr., Gaarenstroom KN, Sturgeon CM, Bonfrer JM, Petersen PH, Troonen H, CarloTorre G, Kanty Kulpa J, Tuxen MK, Molina R, Clinical Use of Cancer Biomarkers in Epithelial Ovarian Cancer: Updated Guidelines From the European Group on Tumor Markers, Int J Gynecol Cancer 26(1) (2016) 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Budiu RA, Mantia-Smaldone G, Elishaev E, Chu T, Thaller J, McCabe K, Lenzner D, Edwards RP, Vlad AM, Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer, Cancer Immunol Immunother 60(7) (2011) 975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].He Y, Gu Z, Zhu Q, Chen M, He C, Huang Y, Li Q, Di W, CA125 over-release behavior following a 75-g oral glucose test as a predictive biomarker of multidrug resistance in patients with ovarian cancer, Int J Cancer 145(6) (2019) 1690–1700. [DOI] [PubMed] [Google Scholar]
- [129].You B, Robelin P, Tod M, Louvet C, Lotz JP, Abadie-Lacourtoisie S, Fabbro M, Desauw C, Bonichon-Lamichhane N, Kurtz JE, Follana P, Leheurteur M, Piano FD, Ferron G, De Rauglaudre G, Ray-Coquard I, Combe P, Chevalier-Place A, Joly F, Leary A, Pujade-Lauraine E, Freyer G, Colomban O, CA-125 ELIMination Rate Constant K (KELIM) Is a Marker of Chemosensitivity in Patients with Ovarian Cancer: Results from the Phase II CHIVA Trial, Clin Cancer Res (2020). [DOI] [PubMed] [Google Scholar]
- [130].Matte I, Garde-Granger P, Bessette P, Piche A, Serum CA125 and ascites leptin level ratio predicts baseline clinical resistance to first-line platinum-based treatment and poor prognosis in patients with high grade serous ovarian cancer, Am J Cancer Res 9(1) (2019) 160–170. [PMC free article] [PubMed] [Google Scholar]
- [131].Lane D, Matte I, Garde-Granger P, Laplante C, Carignan A, Rancourt C, Piche A, Inflammation-regulating factors in ascites as predictive biomarkers of drug resistance and progression-free survival in serous epithelial ovarian cancers, BMC Cancer 15 (2015) 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Bartl T, Karacs J, Kreuzinger C, Pfaffinger S, Kendler J, Ciocsirescu C, Wolf A, Reinthaller A, Meyer E, Brandstetter M, Postl M, Langthaler E, Braicu E, Vergote I, Cunnea P, Gourley C, Schmitt WD, Cacsire Castillo-Tong D, Christoph G, Tumor Growth Rate Estimates Are Independently Predictive of Therapy Response and Survival in Recurrent High-Grade Serous Ovarian Cancer Patients, Cancers (Basel) 13(5) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kailemia MJ, Park D, Lebrilla CB, Glycans and glycoproteins as specific biomarkers for cancer, Anal Bioanal Chem 409(2) (2017) 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Peixoto A, Relvas-Santos M, Azevedo R, Santos LL, Ferreira JA, Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks, Front Oncol 9 (2019) 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lin G, Zhao R, Wang Y, Han J, Gu Y, Pan Y, Ren C, Ren S, Xu C, Dynamic analysis of N-glycomic and transcriptomic changes in the development of ovarian cancer cell line A2780 to its three cisplatin-resistant variants, Ann Transl Med 8(6) (2020) 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Zhao R, Lin G, Wang Y, Qin W, Gao T, Han J, Qin R, Pan Y, Sun J, Ren C, Ren S, Xu C, Use of the serum glycan state to predict ovarian cancer patients’ clinical response to chemotherapy treatment, J Proteomics 223 (2020) 103752. [DOI] [PubMed] [Google Scholar]
- [137].Zahradnikova M, Ihnatova I, Lattova E, Uhrik L, Stuchlikova E, Nenutil R, Valik D, Nalezinska M, Chovanec J, Zdrahal Z, Vojtesek B, Hernychova L, Novotny MV, N-Glycome changes reflecting resistance to platinum-based chemotherapy in ovarian cancer, J Proteomics 230 (2021) 103964. [DOI] [PubMed] [Google Scholar]
- [138].Vermeersch KA, Styczynski MP, Applications of metabolomics in cancer research, J Carcinog 12 (2013) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zhang T, Wu X, Ke C, Yin M, Li Z, Fan L, Zhang W, Zhang H, Zhao F, Zhou X, Lou G, Li K, Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling, J Proteome Res 12(1) (2013) 505–12. [DOI] [PubMed] [Google Scholar]
- [140].Ke C, Li A, Hou Y, Sun M, Yang K, Cheng J, Wang J, Ge T, Zhang F, Li Q, Li J, Wu Y, Lou G, Li K, Metabolic phenotyping for monitoring ovarian cancer patients, Sci Rep 6 (2016) 23334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Allott EH, Howard LE, Cooperberg MR, Kane CJ, Aronson WJ, Terris MK, Amling CL, Freedland SJ, Serum lipid profile and risk of prostate cancer recurrence: Results from the SEARCH database, Cancer Epidemiol Biomarkers Prev 23(11) (2014) 2349–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Poisson LM, Munkarah A, Madi H, Datta I, Hensley-Alford S, Tebbe C, Buekers T, Giri S, Rattan R, A metabolomic approach to identifying platinum resistance in ovarian cancer, J Ovarian Res 8 (2015) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Alonezi S, Tusiimire J, Wallace J, Dufton MJ, Parkinson JA, Young LC, Clements CJ, Park JK, Jeon JW, Ferro VA, Watson DG, Metabolomic Profiling of the Effects of Melittin on Cisplatin Resistant and Cisplatin Sensitive Ovarian Cancer Cells Using Mass Spectrometry and Biolog Microarray Technology, Metabolites 6(4) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Ke C, Hou Y, Zhang H, Fan L, Ge T, Guo B, Zhang F, Yang K, Wang J, Lou G, Li K, Large-scale profiling of metabolic dysregulation in ovarian cancer, Int J Cancer 136(3) (2015) 516–26. [DOI] [PubMed] [Google Scholar]
- [145].Wu W, Wang Q, Yin F, Yang Z, Zhang W, Gabra H, Li L, Identification of proteomic and metabolic signatures associated with chemoresistance of human epithelial ovarian cancer, Int J Oncol 49(4) (2016) 1651–65. [DOI] [PubMed] [Google Scholar]
- [146].Bradley SH, Barclay ME, “Liquid biopsy” for cancer screening, BMJ 372 (2021) m4933. [DOI] [PubMed] [Google Scholar]
- [147].Bai Y, Zhao H, Liquid biopsy in tumors: opportunities and challenges, Ann Transl Med 6(Suppl 1) (2018) S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Dasgupta A, Lim AR, Ghajar CM, Circulating and disseminated tumor cells: harbingers or initiators of metastasis?, Mol Oncol 11(1) (2017) 40–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Mandair D, Vesely C, Ensell L, Lowe H, Spanswick V, Hartley JA, Caplin ME, Meyer T, A comparison of CellCollector with CellSearch in patients with neuroendocrine tumours, Endocr Relat Cancer 23(10) (2016) L29–32. [DOI] [PubMed] [Google Scholar]
- [150].Lee M, Kim EJ, Cho Y, Kim S, Chung HH, Park NH, Song YS, Predictive value of circulating tumor cells (CTCs) captured by microfluidic device in patients with epithelial ovarian cancer, Gynecol Oncol 145(2) (2017) 361–365. [DOI] [PubMed] [Google Scholar]
- [151].Wimberger P, Heubner M, Otterbach F, Fehm T, Kimmig R, Kasimir-Bauer S, Influence of platinum-based chemotherapy on disseminated tumor cells in blood and bone marrow of patients with ovarian cancer, Gynecol Oncol 107(2) (2007) 331–8. [DOI] [PubMed] [Google Scholar]
- [152].Obermayr E, Castillo-Tong DC, Pils D, Speiser P, Braicu I, Van Gorp T, Mahner S, Sehouli J, Vergote I, Zeillinger R, Molecular characterization of circulating tumor cells in patients with ovarian cancer improves their prognostic significance -- a study of the OVCAD consortium, Gynecol Oncol 128(1) (2013) 15–21. [DOI] [PubMed] [Google Scholar]
- [153].Gostner JM, Obermayr E, Braicu IE, Concin N, Mahner S, Vanderstichele A, Sehouli J, Vergote I, Fuchs D, Zeillinger R, Immunobiochemical pathways of neopterin formation and tryptophan breakdown via indoleamine 2,3-dioxygenase correlate with circulating tumor cells in ovarian cancer patients- A study of the OVCAD consortium, Gynecol Oncol 149(2) (2018) 371–380. [DOI] [PubMed] [Google Scholar]
- [154].Rubatt JM, Darcy KM, Tian C, Muggia F, Dhir R, Armstrong DK, Bookman MA, Niedernhofer LJ, Deloia J, Birrer M, Krivak TC, Pre-treatment tumor expression of ERCC1 in women with advanced stage epithelial ovarian cancer is not predictive of clinical outcomes: a Gynecologic Oncology Group study, Gynecol Oncol 125(2) (2012) 421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kuhlmann JD, Wimberger P, Bankfalvi A, Keller T, Scholer S, Aktas B, Buderath P, Hauch S, Otterbach F, Kimmig R, Kasimir-Bauer S, ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance, Clin Chem 60(10) (2014) 1282–9. [DOI] [PubMed] [Google Scholar]
- [156].Paracchini L, Beltrame L, Grassi T, Inglesi A, Fruscio R, Landoni F, Ippolito D, Delle Marchette M, Paderno M, Adorni M, Jaconi M, Romualdi C, D’Incalci M, Siravegna G, Marchini S, Genome-wide Copy-number Alterations in Circulating Tumor DNA as a Novel Biomarker for Patients with High-grade Serous Ovarian Cancer, Clin Cancer Res (2020). [DOI] [PubMed] [Google Scholar]
- [157].Weigelt B, Comino-Mendez I, de Bruijn I, Tian L, Meisel JL, Garcia-Murillas I, Fribbens C, Cutts R, Martelotto LG, Ng CKY, Lim RS, Selenica P, Piscuoglio S, Aghajanian C, Norton L, Murali R, Hyman DM, Borsu L, Arcila ME, Konner J, Reis-Filho JS, Greenberg RA, Robson ME, Turner NC, Diverse BRCA1 and BRCA2 Reversion Mutations in Circulating Cell-Free DNA of Therapy-Resistant Breast or Ovarian Cancer, Clin Cancer Res 23(21) (2017) 6708–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, Helman E, Radke MR, Say C, Vo LT, Mann E, Isaacson JD, Maloney L, O’Malley DM, Chambers SK, Kaufmann SH, Scott CL, Konecny GE, Coleman RL, Sun JX, Giordano H, Brenton JD, Harding TC, McNeish IA, Swisher EM, BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma, Cancer Discov 9(2) (2019) 210–219. [DOI] [PubMed] [Google Scholar]
- [159].Charo LM, Eskander RN, Okamura R, Patel SP, Nikanjam M, Lanman RB, Piccioni DE, Kato S, McHale MT, Kurzrock R, Clinical implications of plasma circulating tumor DNA in gynecologic cancer patients, Mol Oncol 15(1) (2021) 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, Freeman S, Moyle P, Sala E, Sayal K, Hosking K, Gounaris I, Jimenez-Linan M, Earl HM, Qian W, Rosenfeld N, Brenton JD, Exploratory Analysis of TP53 Mutations in Circulating Tumour DNA as Biomarkers of Treatment Response for Patients with Relapsed High-Grade Serous Ovarian Carcinoma: A Retrospective Study, PLoS Med 13(12) (2016) e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Otsuka J, Okuda T, Sekizawa A, Amemiya S, Saito H, Okai T, Kushima M, Detection of p53 mutations in the plasma DNA of patients with ovarian cancer, Int J Gynecol Cancer 14(3) (2004) 459–64. [DOI] [PubMed] [Google Scholar]
- [162].Flanagan JM, Wilson A, Koo C, Masrour N, Gallon J, Loomis E, Flower K, Wilhelm-Benartzi C, Hergovich A, Cunnea P, Gabra H, Braicu EI, Sehouli J, Darb-Esfahani S, Vanderstichele A, Vergote I, Kreuzinger C, Castillo-Tong DC, Wisman GBA, Berns EM, Siddiqui N, Paul J, Brown R, Platinum-Based Chemotherapy Induces Methylation Changes in Blood DNA Associated with Overall Survival in Patients with Ovarian Cancer, Clin Cancer Res 23(9) (2017) 2213–2222. [DOI] [PubMed] [Google Scholar]
- [163].Patel GK, Patton MC, Singh S, Khushman M, Singh AP, Pancreatic Cancer Exosomes: Shedding Off for a Meaningful Journey, Pancreat Disord Ther 6(2) (2016) e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H, Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor, Nat Biotechnol 32(5) (2014) 490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kabe Y, Suematsu M, Sakamoto S, Hirai M, Koike I, Hishiki T, Matsuda A, Hasegawa Y, Tsujita K, Ono M, Minegishi N, Hozawa A, Murakami Y, Kubo M, Itonaga M, Handa H, Development of a Highly Sensitive Device for Counting the Number of Disease-Specific Exosomes in Human Sera, Clin Chem 64(10) (2018) 1463–1473. [DOI] [PubMed] [Google Scholar]
- [166].Crow J, Atay S, Banskota S, Artale B, Schmitt S, Godwin AK, Exosomes as mediators of platinum resistance in ovarian cancer, Oncotarget 8(7) (2017) 11917–11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Vickers KC, Remaley AT, Lipid-based carriers of microRNAs and intercellular communication, Curr Opin Lipidol 23(2) (2012) 91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M, Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma, Proc Natl Acad Sci U S A 108(12) (2011) 5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T, Clinical relevance of circulating cell-free microRNAs in ovarian cancer, Mol Cancer 15(1) (2016) 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Patton MC, Zubair H, Khan MA, Singh S, Singh AP, Hypoxia alters the release and size distribution of extracellular vesicles in pancreatic cancer cells to support their adaptive survival, J Cell Biochem 121(1) (2020) 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Kuhlmann JD, Chebouti I, Kimmig R, Buderath P, Reuter M, Puppel SH, Wimberger P, Kasimir-Bauer S, Extracellular vesicle-associated miRNAs in ovarian cancer - design of an integrated NGS-based workflow for the identification of blood-based biomarkers for platinum-resistance, Clin Chem Lab Med 57(7) (2019) 1053–1062. [DOI] [PubMed] [Google Scholar]
- [172].Kuhlmann JD, Baraniskin A, Hahn SA, Mosel F, Bredemeier M, Wimberger P, Kimmig R, Kasimir-Bauer S, Circulating U2 small nuclear RNA fragments as a novel diagnostic tool for patients with epithelial ovarian cancer, Clin Chem 60(1) (2014) 206–13. [DOI] [PubMed] [Google Scholar]
- [173].Vigneron N, Vernon M, Meryet-Figuiere M, Lambert B, Briand M, Louis MH, Krieger S, Joly F, Lheureux S, Blanc-Fournier C, Gauduchon P, Poulain L, Denoyelle C, Predictive Relevance of Circulating miR-622 in Patients with Newly Diagnosed and Recurrent High-Grade Serous Ovarian Carcinoma, Clin Chem 66(2) (2020) 352–362. [DOI] [PubMed] [Google Scholar]
- [174].Chen Z, Guo X, Sun S, Lu C, Wang L, Serum miR-125b levels associated with epithelial ovarian cancer (EOC) development and treatment responses, Bioengineered 11(1) (2020) 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Benson EA, Skaar TC, Liu Y, Nephew KP, Matei D, Carboplatin with Decitabine Therapy, in Recurrent Platinum Resistant Ovarian Cancer, Alters Circulating miRNAs Concentrations: A Pilot Study, PLoS One 10(10) (2015) e0141279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Teng PN, Wang G, Hood BL, Conrads KA, Hamilton CA, Maxwell GL, Darcy KM, Conrads TP, Identification of candidate circulating cisplatin-resistant biomarkers from epithelial ovarian carcinoma cell secretomes, Br J Cancer 110(1) (2014) 123–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Buderath P, Schwich E, Jensen C, Horn PA, Kimmig R, Kasimir-Bauer S, Rebmann V, Soluble Programmed Death Receptor Ligands sPD-L1 and sPD-L2 as Liquid Biopsy Markers for Prognosis and Platinum Response in Epithelial Ovarian Cancer, Front Oncol 9 (2019) 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Ikeda A, Yamaguchi K, Yamakage H, Abiko K, Satoh-Asahara N, Takakura K, Konishi I, Serum lactate dehydrogenase is a possible predictor of platinum resistance in ovarian cancer, Obstet Gynecol Sci 63(6) (2020) 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Soyama H, Miyamoto M, Matsuura H, Iwahashi H, Kakimoto S, Ishibashi H, Sakamoto T, Hada T, Suminokura J, Takano M, Rapid decrease in serum VEGF-A levels may be a worse prognostic biomarker for patients with platinum-resistant recurrent ovarian cancer treated with bevacizumab and gemcitabine, Cancer Chemother Pharmacol 85(5) (2020) 941–947. [DOI] [PubMed] [Google Scholar]
- [180].Link T, Passek S, Wimberger P, Frank K, Vassileva YD, Kramer M, Kuhlmann JD, Serum calretinin as an independent predictor for platinum resistance and prognosis in ovarian cancer, Int J Cancer 146(9) (2020) 2608–2618. [DOI] [PubMed] [Google Scholar]
- [181].Jin Y, Feng LP, Jiang X, Wang YX, Yin J, Yang ZP, Li Y, Pan LY, Annexin A3 Is a Potential Predictor of Platinum Resistance in Epithelial Ovarian Cancer Patients in a Prospective Cohort, J Cancer 6(7) (2015) 678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Luvero D, Milani A, Ledermann JA, Treatment options in recurrent ovarian cancer: latest evidence and clinical potential, Ther Adv Med Oncol 6(5) (2014) 229–39. [DOI] [PMC free article] [PubMed] [Google Scholar]