Abstract
Ursolic acid (UA) is a triterpenoid phytochemical with strong anti-cancer effect. The metabolic rewiring, epigenetic reprogramming and chemopreventive effect of UA in prostate cancer (PCa) remain unknown. Herein, we investigated the efficacy of UA in PCa xenograft, and its biological effects on cellular metabolism, DNA methylation, and transcriptomic using multi-omics approaches. The metabolomics was quantified by liquid-chromatography mass-spectrometry (LC-MS) while epigenomic CpG methylation in parallel with transcriptomic gene expression was studied by next-generation sequencing technologies. UA administration attenuated the growth of transplanted human VCaP-Luc cells in immunodeficient mice. UA regulated several cellular metabolites and metabolism-related signaling pathways including S-adenosylmethionine (SAM), methionine, glucose 6-phosphate, CDP-choline, phosphatidylcholine biosynthesis, glycolysis, and nucleotide sugars metabolism. RNA-seq analyses revealed UA regulated several signaling pathways, including CXCR4 signaling, cancer metastasis signaling, and NRF2-mediated oxidative stress response. Epigenetic reprogramming study with DNA Methyl-seq uncovered a list of differentially methylated regions (DMRs) associated with UA treatment. Transcriptome-DNA methylome correlative analysis uncovered a list of genes, of which changes in gene expression correlated with the promoter CpG methylation status. Altogether, our results suggest that UA regulates metabolic rewiring of metabolism including SAM potentially driving epigenetic CpG methylation reprogramming, and transcriptomic signaling resulting in the overall anti-cancer chemopreventive effect.
Keywords: ursolic acid, metabolome, epigenome, transcriptome, cancer
1. INTRODUCTION
Prostate cancer (PCa) remains the most common cancer and the fifth leading cause of cancer death among men, despite extensive research that has been performed over the past few decades 1. Drug resistance, toxicity, and side effects are common in the treatment of prostate cancer. Hence clinicians treating PCa patients would need to think outside the box in terms of prevention and treatment of PCa. This has reawakened interest in dietary phytochemicals because they have been shown to be effective against certain oncogenic signaling pathways with relatively few side effects 2.
Ursolic acid (UA) is a triterpenoid phytochemical with strong antioxidant, anti-inflammatory and anti-cancer effects 3–5. UA’s anti-cancer activity includes inhibiting cellular proliferation, dysregulating cell-cycle progression, inducing apoptosis, and inhibiting metastasis 3,6,7. An integrated study of CpG methylome and RNA transcriptome also showed that UA regulated the antioxidant, anti-inflammatory, and anti-cancer pathways during UVB-induced carcinogenesis 8. VCaP prostate cancer cell line was derived from a vertebral bone metastasis from a patient with hormone refractory prostate cancer. It was passaged as xenografts in mice then cultured in vitro 9. We have shown that dietary administration of a naturally occurring mixture of tocotrienols inhibited the growth of xenografted human VCaP prostate tumors 10. We have reported that UA can regulate the antioxidant, anti-inflammatory, and epigenetic genes including DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) in rat leukocytes 11. However, the specific role of UA in PCa remains unclear.
Alterations in specific metabolic activities support cellular transformation, tumor initiation, and progression 12. Cancer cells have unique metabolic adaptations, which meet the anabolic needs of cell proliferation and adapt to the stress related to tumor growth and metastasis 13. Metabolic reprogramming is considered to be a hallmark of malignancy and the differential metabolic dependencies of tumors are emerging as a potential cancer therapeutic target 14,15. Metabolomics can observe the changes of metabolism before and after the intervention of bioactive compounds and discover effective metabolites 16. Triterpenoids have the potential to target cellular metabolic pathways. Studies have shown that cellular metabolic reprogramming occurs during the development of prostate cancer and that triterpenoids can intervene multiple metabolism pathways of prostate cancer cells 17. It has been reported that UA alters the glycolysis of breast cancer cells with different phenotypes 18.
However, there are few studies on the metabolic, epigenome and transcriptome mechanisms of UA in PCa tumor models. Herein, we performed liquid-chromatography mass-spectrometry (LC-MS) to quantify metabolites and next-generation sequencing (NGS) to quantify the impact of UA on transcriptomic and DNA CpG methylation in VCaP cells implanted xenograft tumor in immunodeficient nude mice. Our results suggest that UA alters metabolome, DNA CpG methylome and transcriptome contributing to the anti-cancer efficacy in human PCa VCaP implanted xenograft tumor.
2. MATERIALS AND METHODS
2.1. Cell line and culture
VCaP human prostate cancer cells were purchased from the American Type Tissue Collection (ATCC, Rockville, MD, USA) and cultured in RPMI 1640 medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. The VCaP cells were transfected with a luciferase construct and a single VCaP-Luc clone was selected for subcutaneous implantation as previously described 10.
2.2. Cell viability and proliferation assay
VCaP-Luc cells (3 × 103 cells per well) were maintained in 96-well cell culture plates for 24 h. Then, the cells were exposed to either DMSO (0.1%) or UA at concentrations ranging from 5 to 50 μM in DMEM supplemented with 1% FBS for 1 day. To determine cell viability and proliferation, 20 μL of MTS reagent (Promega, Madison, WI, USA) was added to each well of the 96-well plates. The absorbance at 490 nm was recorded after 1 hour of incubation.
2.3. Tumor xenograft in immunodeficient mice
Male NCr immunodeficient mice (7–8 weeks old) were purchased from Taconic Farms. Mice were kept at the Rutgers Animal Facility with a 12/12 h light/dark cycle and offered food and water ad libitum. The animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC). Mice were randomly divided into two groups: the control group and the UA treatment group. There were 10 mice in each group. VCaP cells (5 × 106 cells per 0.1 ml per mouse) were suspended in a mixture of Matrigel and RPMI 1640 medium (1:1) and injected subcutaneously into the back of the mouse. 0.1% UA diet (w/w) (Research Diets, New Brunswick, NJ) was administered orally in the AIN-93M mature rodent diet for 8 weeks based on published data 11,19. During the experiment, the tumor size was measured weekly using an IVIS imaging system and caliper. Before using the IVIS system for bioluminescence imaging, mice were anesthetized with isoflurane and injected with D-luciferin at a concentration of 3 mg per mouse. Images were captured with the mice in a prone position for 1 min, beginning at 8 min after the D-luciferin injection, and the luminescence signal was reported as the photon flux (ph/s). Tumor volume (V) was calculated as V = L × W2 × 0.52, where L is the length and W is the width of a xenograft. Bodyweight was measured once a week. The tumors were collected and weighed immediately at the end of the study.
2.4. Metabolite extraction and LC-MS
To extract metabolites from cultured cells, VCaP-Luc cells were seeded in 60 mm dishes with equal cell numbers (1 × 106 cells). After cultured overnight, the cells were exposed to various concentrations of UA without serum for 3 hours. Cell metabolites were be extracted by lysing the cells on the plate with 1 mL ice-cold lysis buffer consisting of 40:40:20 methanol:acetonitrile: water with 0.5% formic acid. The plates were incubated on ice for 5 min, then 50 uL of 15% NH4HCO3 was added to neutralize the acetic acid. Cells were scraped off from plates and collected and centrifuged at 15,000 × g for 10 min at 4 °C. Then, the supernatant containing the metabolites was transferred to new tubes and stored at −80 °C for LC-MS. Each condition was repeated in triplicate. The LC-MS method is the same as previously published 20. Conditions were optimized on an HPLC-ESI-MS system fitted with a Vanquish Horizon UHPLC and a Thermo Q Exactive Plus MS. The MS scans were obtained in both positive and negative ionization modes with a nominal mass resolution of 70,000 (defined at m/z 200), in addition to an automatic gain control target of 3 × 106 and m/z scan range of 72 to 1000. Metabolite data were obtained using the MAVEN software package 21. Statistical and pathway analysis (SMPDB) were performed on the MetaboAnalyst (V5.0) (www.metaboanalyst.ca) 22.
2.5. RNA-seq and computational analysis
RNA-seq was conducted by Genewiz Inc (South Plainfield, NJ, USA), as we described earlie8,23,24. Briefly, RNA was extracted with the AllPrep DNA/RNA Mini Kit. Illumina TruSeq RNA library preparation kit was used to construct the library following the manufacturer’s instructions. The samples were sequenced with 75 bp paired-end reads on a NextSeq 500 instrument (Illumina), with a minimum depth of 25–30 million reads per sample. The RNA-seq reads were compared with the human reference genome by HISAT2 software and analyzed by DEGSeq in R version 3.6.
2.6. Ingenuity pathway analysis (IPA)
To elucidate the signaling pathways associated with the differentially expressed genes (DEGs), the gene expression levels with |log2 fold changes| ≥ 0.3 and with a p-value less than 0.05 were analyzed by IPA software.
2.7. Methyl-Seq and computational analysis
The Methyl-Seq analysis was performed as described earlier 8,23,24. Briefly, the Methyl-Seq library was carried out with the Agilent SureSelect human Methyl-Seq kit. The captured Methyl-Seq library was bisulfite converted using the EZ DNA Methylation-Gold kit. The Methyl-Seq library was sequenced on an Illumina HiSeq 2000 platform with 75 bp single-end reads to a minimum sequencing depth of 40 to 50 million reads per sample. This Methyl-Seq was performed by Genewiz Inc. The reads were aligned to the in silico bisulfite-converted human genome with the Bismark (version 0.15.0) alignment algorithm 25. After alignment, DMRfinder 26 was used to extract methylation counts and identify differentially methylated regions (DMRs) with each DMR containing at least three CpG sites. ChIPseeker was used for genomic annotation. DNA methylation level was measured by methyl cytosine/(methyl cytosine + unmethyl cytosine). We then measured the DNA methylation difference by subtracting the individual DNA methylation value in the control group from the UA group.
2.8. Correlation between methylation and gene expression
The gene expression levels from RNA-seq were selected with a cutoff of |log2 (Fold Change)| ≥ 1 and p-value < 0.05. Then, the sorted genes from the RNA-seq were further sorted by Cutoff threshold DNA methylation differences ≥ 0.1. The genes of promoter DNA hypermethylation/RNA down-regulation, or promoter DNA hypomethylation/RNA up-regulation were used as genes of interest for further analysis. The results were plotted in R.
2.9. Quantitative real-time PCR (qPCR)
0.5 μg of RNA was used for reverse transcription by Taqman Reverse Transcription Reagents (Thermo Fisher Scientific, USA). The real-time PCR reactions were performed in triplicate with PowerUp SYBR Green Master Mix Kit (Thermo Fisher Scientific, USA) on the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific, USA). The levels of mRNA expressions were quantified by the ΔΔCt method and normalized against β-Actin levels. Primer sequences were listed in Supplementary Table S1.
2.10. Statistical analysis
Data were expressed as mean ± standard deviation. The student t-test was used for statistical analysis between the two groups. P-value < 0.05 was considered statistically significant unless otherwise noted.
3. RESULTS
3.1. UA attenuated tumor growth in xenograft animal model
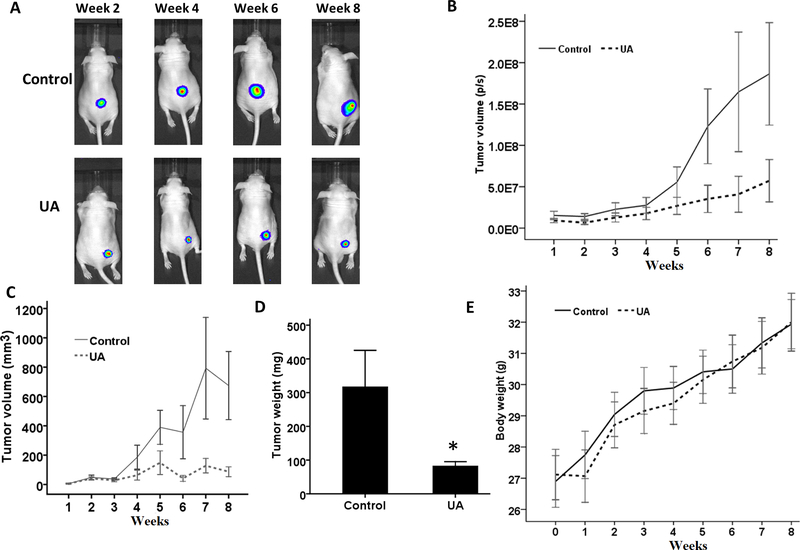
To evaluate the anti-cancer effects of UA, we applied a prostate tumor xenograft model to measure the activity of UA in immunodeficient nude mice in vivo. Male Ncr immunodeficient mice were injected subcutaneously with VCaP-Luc cells and then feed with a UA diet. As shown in Figure 1A, the luminescence images showed that the tumor volume of both groups increased with time. However, the tumor volume in the UA group increased at a much slower pace. According to the quantitative results, the xenograft volume of VCap-Luc cells in the UA group was relatively small (Figure 1B). Measurements by the caliper, as shown in Figure 1C, showed similar results with the luminescent image results. Besides, the average tumor weight of the UA treatment was lower than that in the control group (Figure 1D). There was no statistical significance in the average body weight of the two groups during the experiment (Figure 1E). Our results indicate that dietary UA administration can attenuate the growth of transplanted VCaP-Luc cells in immunodeficient mice.
FIGURE 1. Dietary UA administration attenuates the growth of human prostate tumors in immunodeficient mice.
(A) Representative bioluminescence IVIS images of subcutaneously implanted tumors in mice. (B) Strength of the bioluminescence signal (photons/s) from 1 to 8 weeks. (C) Tumor volumes were measured by the caliper during the treatment. (D) Weight of tumor mass. (E) Bodyweight during the experiment. The data are represented as the mean ± standard deviation.
3.2. Metabolic alterations after UA treatment
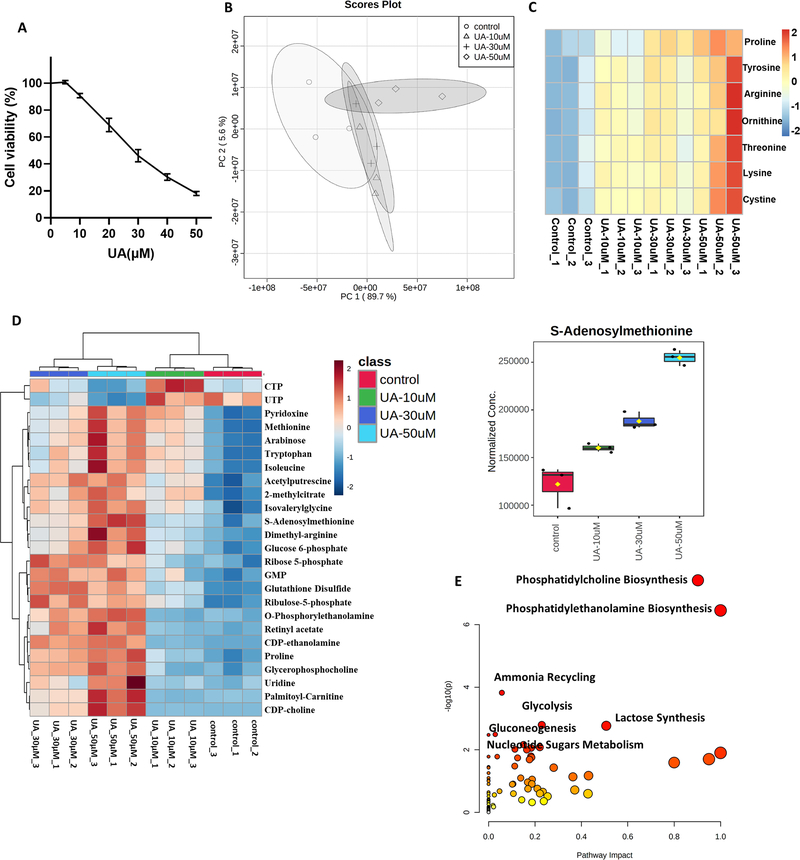
To study the cytotoxicity of UA on VCap-Luc cells, we analyzed the role of UA on cell viability using MTS. As shown in Figure 2A, cell viability was significantly reduced after UA exposure in a dose-dependent manner. Next, we measured the changes of metabolites in VCAP-Luc cells after UA treatment. Principal Component analysis (PCA) showed that the high concentration UA group could be clearly distinguished (Figure 2B). Interestingly, we found that a group of metabolites, mainly amino acids, changed significantly after UA treatment. The levels of amino acids (AA) such as lysine, arginine, proline, threonine, ornithine, tyrosine, cystine were increased in VCap-Luc cells exposed to UA (Figure 2C). In addition, Figure 2D showed that the most significant top 25 metabolites changed after UA treatment, including S-adenosylmethionine (SAM), methionine, glucose 6-phosphate, ribose 5-phosphate, CDP-choline, and ribulose-5-phosphate. Pathway analysis showed that the changes of metabolites after UA exposure were mainly related to phosphatidylcholine biosynthesis, phosphatidylethanolamine biosynthesis, ammonia recycling, glycolysis, lactose synthesis, gluconeogenesis, and nucleotide sugars metabolism (Figure 2E). These data suggest that these altered metabolites and metabolism-related signaling pathways may play a role in cell viability and epigenetic reprogramming coupled with gene expression alterations.
FIGURE 2. Metabolite profile of VCaP-Luc cells treated with UA.
(A) Cell viability assay of VCaP-Luc cells treated with UA for 1 day. (B) PCA plot for UA exposed cells. (C) Heatmap of amino acid levels regulated by UA at 3 hours (n=3 per group). (D) Top 25 metabolites in VCaP-Luc cells treated with UA for 3 hours (n=3 per group). (E) Identification of significantly enriched pathways based on statistically significant changes in metabolite concentrations with UA treatment.
3.3. Gene expression changes after UA treatment
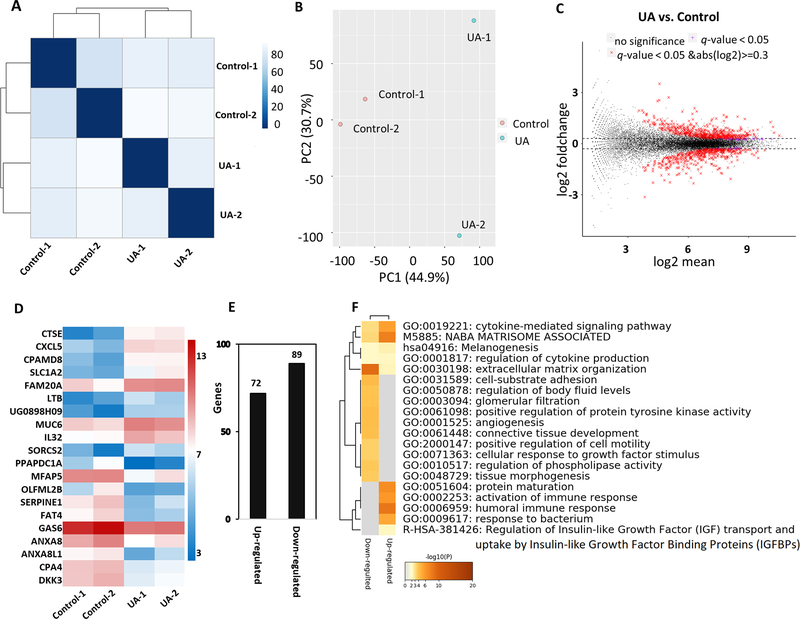
To understand the effects of UA in vivo, we used RNA-seq to analyze the tumors in the two groups. Euclidean distance clustering and principal component analysis (PCA) were performed to measure the variation in RNA-seq data in the control and UA-treated tumor samples. As shown in Figure 3A and 3B, the features of the two groups were separated. Next, we compared the global gene expression in the two groups using a cutoff threshold |log2 fold change| ≥ 0.3 and FDR adjusted p-value ˂ 0.05. As shown in Figure 3C, compared with the control group, there were 950 genes considered differentially expressed with 519 up- and 431 down-regulated in the UA group. Next, we generated a heatmap containing the top 20 genes based on FDR adjusted p-value less than 0.05 that were most up-regulated and down-regulated to account for the transcriptional profiles of UA-treated VCaP cells (Figure 3D). Of the top 10 most up-regulated genes, 6 belonged to the matrisome, including cathepsin E (CTSE), lymphotoxin beta (LTB), mucin 6, oligomeric mucus/gel-forming (MUC6), C-X-C motif chemokine ligand 5 (CXCL5), C3 and PZP like alpha-2-macroglobulin domain containing 8 (CPAMD8), and FAM20A golgi associated secretory pathway pseudokinase (FAM20A). Of the top 10 most downregulated genes, 4 belonged to the negative regulation of endopeptidase activity, including growth arrest specific 6 (GAS6), serpin family E member 1 (SERPINE1), annexin A8 (ANXA8), and annexin A8 like 1 (ANXA8L1)). Besides, after UA treatment, 161 genes were considered differentially expressed with 72 up- and 89 down-regulated, using an FDR adjusted p-value less than 0.05, and the |log2 fold change| ≥ 1 cutoff (Figure 3E). Next, the 161 genes were submitted to Metascape 27 (http://metascape.org) to understand the functional significance of dysregulated genes. Metascape analysis showed that multiple pathways associated with UA were significantly enriched, such as the cytokine-mediated signaling pathway, extracellular matrix organization, angiogenesis, and activation of immune response (Figure 3F). To further explore the biological function of DEGs, we did the IPA analysis to provide a broad overview of the regulation related to UA. A total of 1817 genes identified with a cutoff of |log2 fold change| ≥ 0.3 with p-value < 0.05 were mapped to the IPA. Based on the -log (p-value) and activation z scores, we selected the top canonical signaling pathways from the IPA. As shown in Table 1, 12 pathways were activated and 12 pathways were inhibited by UA treatment. Among those pathways, pathways such as oxidative ethanol degradation III, acute phase response signaling, Wnt/β-catenin signaling, and NRF2-mediated oxidative stress response were activated by UA, but pathways such as CXCR4 signaling, TGF-β signaling, colorectal cancer metastasis signaling, and IL-8 signaling were inhibited by UA.
FIGURE 3. Gene expression pattern of tumor in immunodeficient mice after dietary UA administration.
(A) Euclidean distance clustering between two groups. (B) PCA plot showed a comparison of transcriptome data between the two groups. (C) MA plot for differential expression analysis of UA vs. control. (D) Heatmap showing top 10 up and down-regulated genes based on both q-value and p-value < 0.05. Color scale represents log2FPKM of these for genes. (E) The number of differentially up and down-regulated genes based on p-value and q-value less than 0.05, and the |log2 fold change| ≥ 1. (F) Metascape analysis showing the top 20 enriched clusters.
Table 1.
Canonical pathways were identified using the IPA software. A positive Z score indicates activation of the pathway and a negative Z score indicates inhibition of the pathway.
| Ingenuity Canonical Pathways | −log(p−value) | z-score |
|---|---|---|
|
| ||
| PPARα/RXRα Activation | 5.98 | 2.41 |
| Acute Phase Response Signaling | 5.68 | 0.63 |
| Osteoarthritis Pathway | 5.64 | 0.71 |
| Wnt/β-catenin Signaling | 5.12 | 0.56 |
| Aryl Hydrocarbon Receptor Signaling | 4.97 | 1.07 |
| Dopamine Receptor Signaling | 4.80 | 0.71 |
| Synaptic Long-Term Potentiation | 4.38 | 0.21 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 3.99 | 0.38 |
| NRF2-mediated Oxidative Stress Response | 3.91 | 0.24 |
| Dopamine-DARPP32 Feedback in cAMP Signaling | 3.54 | 1.63 |
| LXR/RXR Activation | 3.47 | 2.14 |
| Oxidative Ethanol Degradation III | 3.47 | 1.13 |
| Colorectal Cancer Metastasis Signaling | 7.91 | −1.41 |
| PI3K Signaling in B Lymphocytes | 6.81 | −1.23 |
| Glioblastoma Multiforme Signaling | 6.03 | −1.57 |
| ILK Signaling | 5.92 | −2.19 |
| Cardiac Hypertrophy Signaling (Enhanced) | 5.69 | −1.52 |
| Protein Kinase A Signaling | 5.41 | −0.15 |
| Cardiac Hypertrophy Signaling | 4.81 | −1.72 |
| IL-8 Signaling | 4.67 | −1.83 |
| CXCR4 Signaling | 4.56 | −2.29 |
| FAT10 Cancer Signaling Pathway | 4.55 | −0.28 |
| TGF-β Signaling | 4.47 | −1.21 |
| LPS/IL-1 Mediated Inhibition of RXR Function | 4.36 | −0.58 |
3.4. DNA methylation changes after UA treatment
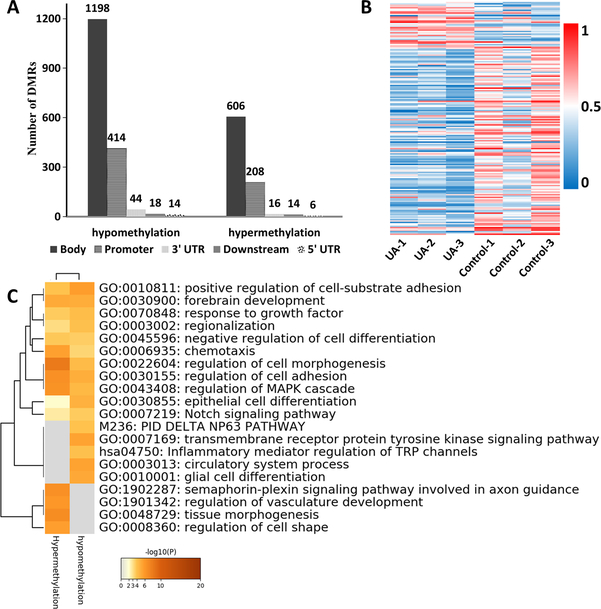
Next, we performed SureSelect Methyl-Seq on tumor samples to observe whether UA treatment affected the profiling of DNA methylome. Cutoff threshold DNA methylation differences ≥ 0.1 was used to select a subgroup of DMRs. In total, there were 2538 genes related to DMRs, divided into 850 hypermethylated genes and 1688 hypomethylated genes (Figure 4A). According to the annotation region analysis, the 850 hypermethylated regions are mainly distributed in the gene body (606), promoter (208), 3’UTR (16), downstream (14), and 5’UTR (6). For the 1688 hypomethylated regions, DMRs were identified in the gene body (1198), promoter (414), 3’UTR (44), downstream (18), and 5’UTR (14) (Figure 4A and Supplementary Table S2). Next, a heat map with a methylation cutoff level of 0.2 was generated to illustrate changes in UA-treated tumors. As shown in a heatmap (Figure 4B), a total of 177 DMRs (143 hypomethylated DMRs and 34 hypermethylated DMRs) were detected between the UA treatment and the control. Those genes associated with UA treatment include PI3 (peptidase inhibitor 3), CHI3L2 (chitinase 3 like 2), and GFAP (glial fibrillary acidic protein). Next, the 2538 genes filtered by Cutoff threshold DNA methylation differences ≥ 0.1 were submitted to the Metascape to explore the functional significance of dysregulated genes. Metascape analysis showed that pathways associated with UA were significantly enriched, such as positive regulation of cell-substrate adhesion, Notch signaling pathway, regulation of cell morphogenesis, inflammatory mediator regulation of TRP channels, and regulation of vasculature development (Figure 4C).
FIGURE 4. DNA methylation changes after the dietary UA administration.
(A) The number of differentially hypermethylated and hypomethylated DMRs. (B) Heat map showing significant changes in DMRs between UA group and control group. A total of 177 DMRs were identified at a methylation difference was 0.2. (C) Top 20 enriched clusters that were regulated by UA. Enriched clusters were identified by Metascape analysis using the list of 177 regulated genes associated with DMRs.
3.5. Correlation between methylation and gene expression
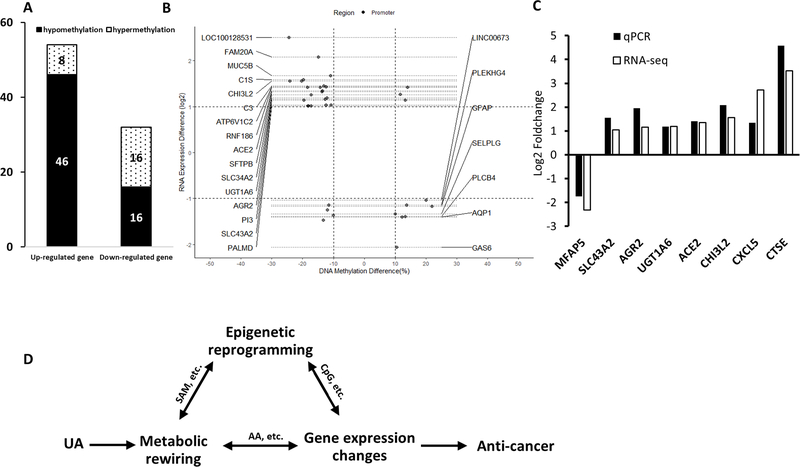
Next, the relevance between gene expression profiles and methylation profiles was studied to investigate the link between DNA methylation and transcriptome. With a cutoff of methylation ratio difference ≥ 0.1 for DNA methylation changes and |log2 (fold change)| ≥ 1 with p-value ˂ 0.05 for RNA expression, a total of 86 DEGs/DMRs were found. As shown in Figure 5A, 46 genes were identified between the up-regulated DEGs and hypomethylated genes, and 16 genes were identified between the down-regulated DEGs and hypermethylated genes. Compared with the control group, 23 DMRs in the promoter regions of the UA group were negatively correlated with the expression of corresponding genes (Figure 5B and Supplementary Table S3). These DMRs-related genes include ACE2 (angiotensin I converting enzyme 2), UGT1A6 (UDP glucuronosyltransferase family 1 member A6), SLC43A2 (solute carrier family 43 member 2), and AGR2 (anterior gradient 2, protein disulphide isomerase family member).
FIGURE 5. Correlation between methylation and gene expression.
(A) The number of differentially hypermethylated and hypomethylated DMRs with a cutoff of methylation ratio difference ≥ 0.1 for CpG methylation changes and |log2 (fold change)| ≥ 1 with p-value ˂ 0.05 for RNA expression. (B) Correlation between gene expression and CpG methylation in the promoter. Annotation genes had at least one CpG cluster in the promoter region, and the change of CpG methylation level was negatively correlated with the change of RNA expression. (C) Validation of RNA-seq results by qPCR. The expression of 8 selected genes was analyzed. The bar chart shows the expression of each candidate gene in the UA group compared to the control group. (D) The schematic diagram summarizing the potential correlations between the metabolic rewiring, epigenetic reprogramming, and gene expression profiles.
3.6. Validation of RNA-seq data with qPCR
To validate the results of the RNA-seq, qPCR was used to analyze the significantly expressed gene after UA treatment. We examined the expression of 8 DEGs in the two groups. The relative mRNA expression levels of these genes were presented in Figure 5C. For these 8 genes, the changes in gene expression in qPCR were consistent with the RNA-seq results, indicating that the DEGs from the RNA-seq analysis were highly reliable.
4. DISCUSSION
Studies have shown that UA inhibits the proliferation of cancer cells in both in vitro and in vivo cancer models 6,28–32. Herein, we conducted a comprehensive study with LC-MS and NGS technologies on the impact of UA on the metabolic rewiring and epigenetic reprogramming in the human PCa VCaP xenograft model. We found that dietary administration of 0.1% UA attenuated tumor growth of transplanted VCaP xenograft in immunodeficient mice. UA regulates metabolic remodeling and drives epigenetic CpG methylation reprogramming and transcriptome signaling pathways to achieve its anti-cancer effects. The schematic diagram illustrating the results is shown in the Figure 5D.
During cell proliferation, extensive metabolic rewiring takes place for cells to obtain sufficient nutrients, such as glucose, amino acids, lipids, and nucleotides, which are necessary to support cell growth and to cope with the redox challenges 33. Since we ran out of tumor samples, especially those treated with UA, we could not perform LC-MS metabolomics analysis on tumor samples, so we used cell line VCaP-Luc cells. In our metabonomic study, we identified several metabolites and metabolism-related signaling pathways with the treatment of UA that are critical to the growth and survival of cancer cells. One of the rewired mitochondrial metabolites SAM is the main biological methyl donor synthesized in all mammalian cells and is the methyl donor of all basic epigenetic methylation reactions, including CpG and histone methylation, that drive epigenetic reprogramming 34,35. In our current study, we also found that SAM increased significantly with increasing UA dose, which would lead to changes in the epigenetic methylation process as illustrated with alterations of DNA methylome. Moreover, it has been found that amino acids are not only cellular signaling molecules, but also regulatory molecules of gene expression and protein phosphorylation cascade 36. In this study, we found that a group of amino acids, including arginine, proline and tryptophan, changed significantly after UA treatment. It is increasingly recognized that some amino acids regulate key metabolic pathways necessary for maintenance, growth, reproduction, and immunity 36,37. Further analysis of these amino acids and the regulatory role of amino acids in metabolic rewiring, gene expression and epigenetic reprogramming will contribute to our understanding of UA. Moreover, pathway analysis of metabolome data showed that the top signaling pathways such as phosphatidylcholine biosynthesis, phosphatidylethanolamine biosynthesis, ammonia recycling, glycolysis, lactose synthesis, gluconeogenesis, and nucleotide sugars metabolism were identified. The biological functions of UA regulating metabolic pathways need to be further studied.
The previous study showed that the tumor masses of the UA treatment group were smaller than that of the control group 32. Another study showed that UA supplemented diet can decrease tumor cell proliferation and tumor size by decreasing the activation of the PI3K/AKT pathway and MAPK signaling pathway 19. The inhibitory effect of UA on the xenograft mouse model is consistent with previous reports. We also showed that a subset of differential expression genes was identified between the UA and control groups through RNA-seq. Pathway analysis of RNA-seq data showed that the top signaling pathways such as NRF2-mediated oxidative stress response, colorectal cancer metastasis signaling, CXCR4 signaling, and TGF-β signaling were identified (Table 1). These pathways may reflect one of the anti-cancer mechanisms of UA. For instance, UA has antioxidant effects including scavenging free radicals and regulating antioxidant enzymes 3, which is also consistent with our current findings.
Our previous study has shown that UA reduces the expression of epigenetic modifying enzymes including DNMTs, HDACs, and HDACs activity in vitro cell model 38. However, it is unclear whether UA administration changes the DNA methylation level in tumor cells in vivo. From our metabolomic results, mitochondrial metabolites including SAM are increased with increasing doses of UA (Figure 2D), thereby would affect epigenetic CpG methylation reprogramming. Here, we obtained a list of DMRs associated with UA treatment using SureSelect CpG Methyl-Seq. Our study further showed that dysregulated DMRs-related genes are closely related to the positive regulation of cell-substrate adhesion, the regulation of Notch signaling pathway, the regulation of cell morphogenesis, the inflammatory mediator regulation of TRP channels, and the regulation of vasculature development (Figure 4). Previously published studies have shown that UA suppressed metastasis via regulating MMP-2, MMP-9, VEGF, and integrin αVβ5 39–43. The present findings are consistent with previous studies showing that UA is a strong candidate as an anti-metastatic agent. From our Metascape analysis of RNA-seq and Methyl-Seq data, we found the top clusters were mostly involved in changes in matrisome associated, extracellular matrix organization, positive regulation of cell motility, angiogenesis, and positive regulation of cell-substrate adhesion. Next, by examining the relationship between DNA methylation and transcriptome, we identified an important gene subgroup associated with UA as shown in Figure 5.
In conclusion, the results of this study provide experimental basis for the anti-cancer effect of UA in human PCa, and reveal its molecular mechanism through multi-omics analysis of cell lines and xenograft tumor models. Our results suggest that UA regulates metabolic rewiring, reprograms CpG methylation sites, drives gene expression changes and activates signaling pathways, which contribute to anti-cancer efficacy in human PCa VCaP xenografts.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by R01AT009152 from the National Center for Complementary and Integrative Health (NCCIH) and R01CA200129 from the National Cancer Institute (NCI) (AK) and NIH grants P30CA072720-5923 (XS). We thank all members of Dr. Ah-Ng Kong’s lab for helpful discussions and preparation of the manuscript.
Funding information
National Center for Complementary and Integrative Health, Grant/Award Number: R01 AT009152; National Cancer Institute, Grant/Award Numbers: R01 CA200129; NIH grants P30CA072720-5923 (XS).
Abbreviations:
- AA
amino acids
- DEGs
differentially expressed genes
- DMRs
differentially methylated regions
- DNMTs
DNA methyltransferases
- HDACs
histone deacetylases
- IPA
ingenuity pathway analysis
- LC-MS
liquid-chromatography mass-spectrometry
- NGS
next-generation sequencing
- PCa
prostate cancer
- SAM
S-adenosylmethionine
- UA
ursolic acid
Footnotes
CONFLICT OF INTERESTS
The authors declare that they have no conflict of interests.
Supplementary material
Supplementary data are available.
DATA AVAILABILITY
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
REFERENCES
- 1.Rawla P Epidemiology of prostate cancer. World journal of oncology. 2019;10(2):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mokbel K, Wazir U, Mokbel K. Chemoprevention of prostate cancer by natural agents: Evidence from molecular and epidemiological studies. Anticancer research. 2019;39(10):5231–5259. [DOI] [PubMed] [Google Scholar]
- 3.Yin R, Li T, Tian JX, Xi P, Liu RH. Ursolic acid, a potential anticancer compound for breast cancer therapy. Critical reviews in food science and nutrition. 2018;58(4):568–574. [DOI] [PubMed] [Google Scholar]
- 4.Seo DY, Lee SR, Heo J-W, et al. Ursolic acid in health and disease. The Korean Journal of Physiology & Pharmacology. 2018;22(3):235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikeda Y, Murakami A, Ohigashi H. Ursolic acid: An anti-and pro-inflammatory triterpenoid. Molecular nutrition & food research. 2008;52(1):26–42. [DOI] [PubMed] [Google Scholar]
- 6.Hsu Y-L, Kuo P-L, Lin C-C. Proliferative inhibition, cell-cycle dysregulation, and induction of apoptosis by ursolic acid in human non-small cell lung cancer A549 cells. Life sciences. 2004;75(19):2303–2316. [DOI] [PubMed] [Google Scholar]
- 7.Prasad S, Yadav VR, Sung B, et al. Ursolic acid inhibits growth and metastasis of human colorectal cancer in an orthotopic nude mouse model by targeting multiple cell signaling pathways: chemosensitization with capecitabine. Clinical cancer research. 2012;18(18):4942–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Yin R, Wu R, et al. DNA methylome and transcriptome alterations and cancer prevention by triterpenoid ursolic acid in UVB-induced skin tumor in mice. Molecular carcinogenesis. 2019;58(10):1738–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korenchuk S, Lehr J, MClean L, et al. VCaP, a cell-based model system of human prostate cancer. In vivo (Athens, Greece). 2001;15(2):163–168. [PubMed] [Google Scholar]
- 10.Huang Y, Wu R, Su Z-Y, et al. A naturally occurring mixture of tocotrienols inhibits the growth of human prostate tumor, associated with epigenetic modifications of cyclin-dependent kinase inhibitors p21 and p27. The Journal of nutritional biochemistry. 2017;40:155–163. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Wang C, Li W, et al. Pharmacokinetics and pharmacodynamics of the triterpenoid ursolic acid in regulating the antioxidant, anti-inflammatory, and epigenetic gene responses in rat leukocytes. Molecular pharmaceutics. 2017;14(11):3709–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks OD, Berkers CR, Mason SM, et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Z, Liu X, Cheng C, Yu W, Yi P. Metabolism of Amino Acids in Cancer. Frontiers in Cell and Developmental Biology. 2020;8:1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luengo A, Gui DY, Vander Heiden MG. Targeting metabolism for cancer therapy. Cell chemical biology. 2017;24(9):1161–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J, Zhang LL, Li W, et al. Application of metabolomics for revealing the interventional effects of functional foods on metabolic diseases. Food chemistry. 2021;367:130697. [DOI] [PubMed] [Google Scholar]
- 17.Mamouni K, Kallifatidis G, Lokeshwar BL. Targeting Mitochondrial Metabolism in Prostate Cancer with Triterpenoids. International journal of molecular sciences. 2021;22(5):2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewinska A, Adamczyk-Grochala J, Kwasniewicz E, Deregowska A, Wnuk M. Ursolic acid-mediated changes in glycolytic pathway promote cytotoxic autophagy and apoptosis in phenotypically different breast cancer cells. Apoptosis. 2017;22(6):800–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Angel RE, Smith SM, Glickman RD, Perkins SN, Hursting SD. Antitumor effects of ursolic acid in a mouse model of postmenopausal breast cancer. Nutrition and cancer. 2010;62(8):1074–1086. [DOI] [PubMed] [Google Scholar]
- 20.Su X, Chiles E, Maimouni S, Wondisford FE, Zong W-X, Song C. In-source CID ramping and covariant ion analysis of hydrophilic interaction chromatography metabolomics. Analytical chemistry. 2020;92(7):4829–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Current protocols in bioinformatics. 2012;37(1):14.11. 11–14.11. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang Z, Chong J, Zhou G, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49(W1):W388–w396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Wu R, Sargsyan D, et al. UVB drives different stages of epigenome alterations during progression of skin cancer. Cancer letters. 2019;449:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Wu R, Sargsyan D, et al. CpG methyl-seq and RNA-seq epigenomic and transcriptomic studies on the preventive effects of Moringa isothiocyanate in mouse epidermal JB6 cells induced by the tumor promoter TPA. J Nutr Biochem. 2019;68:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaspar JM, Hart RP. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC bioinformatics. 2017;18(1):528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature communications. 2019;10(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonaccorsi I, Altieri F, Sciamanna I, et al. Endogenous reverse transcriptase as a mediator of ursolic acid’s anti-proliferative and differentiating effects in human cancer cell lines. Cancer letters. 2008;263(1):130–139. [DOI] [PubMed] [Google Scholar]
- 29.Andersson D, Liu J-J, Nilsson A, Duan R-D. Ursolic acid inhibits proliferation and stimulates apoptosis in HT29 cells following activation of alkaline sphingomyelinase. Anticancer research. 2003;23(4):3317–3322. [PubMed] [Google Scholar]
- 30.Zhang D-M, Tang PM-K, Chan JY-W, et al. Anti-proliferative effect of ursolic acid on multidrug resistant hepatoma cells R-HepG2 by apoptosis induction. Cancer Biology & Therapy. 2007;6(9):1377–1385. [Google Scholar]
- 31.Es-Saady D, Simon A, Ollier M, Maurizis J, Chulia A, Delage C. Inhibitory effect of ursolic acid on B16 proliferation through cell cycle arrest. Cancer letters. 1996;106(2):193–197. [DOI] [PubMed] [Google Scholar]
- 32.Zhang RX, Li Y, Tian DD, et al. Ursolic acid inhibits proliferation and induces apoptosis by inactivating Wnt/β-catenin signaling in human osteosarcoma cells. International journal of oncology. 2016;49(5):1973–1982. [DOI] [PubMed] [Google Scholar]
- 33.Zhu J, Thompson CB. Metabolic regulation of cell growth and proliferation. Nature reviews Molecular cell biology. 2019;20(7):436–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shyh-Chang N, Locasale JW, Lyssiotis CA, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R, Li S, Hudlikar R, et al. Redox signaling, mitochondrial metabolism, epigenetics and redox active phytochemicals. Free radical biology & medicine. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu G Amino acids: metabolism, functions, and nutrition. Amino acids. 2009;37(1):1–17. [DOI] [PubMed] [Google Scholar]
- 37.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nature reviews Drug discovery. 2019;18(5):379–401. [DOI] [PubMed] [Google Scholar]
- 38.Kim H, Ramirez CN, Su Z-Y, Kong A-NT. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. The Journal of nutritional biochemistry. 2016;33:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang HC, Huang CY, Lin-Shiau SY, Lin JK. Ursolic acid inhibits IL-1β or TNF‐α‐induced C6 glioma invasion through suppressing the association ZIP/p62 with PKC-ζ and downregulating the MMP-9 expression. Molecular Carcinogenesis: Published in cooperation with the University of Texas MD Anderson Cancer Center. 2009;48(6):517–531. [DOI] [PubMed] [Google Scholar]
- 40.Tang Q, Liu Y, Li T, et al. A novel co-drug of aspirin and ursolic acid interrupts adhesion, invasion and migration of cancer cells to vascular endothelium via regulating EMT and EGFR-mediated signaling pathways: multiple targets for cancer metastasis prevention and treatment. Oncotarget. 2016;7(45):73114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C-Y, Lin C-Y, Tsai C-W, Yin M-C. Inhibition of cell proliferation, invasion and migration by ursolic acid in human lung cancer cell lines. Toxicology in vitro. 2011;25(7):1274–1280. [DOI] [PubMed] [Google Scholar]
- 42.Ruan JS, Zhou H, Yang L, et al. Ursolic Acid Attenuates TGF-β1-Induced Epithelial-Mesenchymal Transition in NSCLC by Targeting Integrin αVβ5/MMPs Signaling. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2019;27(5):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Son J, Lee SY. Ursonic acid exerts inhibitory effects on matrix metalloproteinases via ERK signaling pathway. Chemico-Biological Interactions. 2020;315:108910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.