Abstract
To fight COVID-19, much effort has been directed toward in vitro drug repurposing. Here, we investigate the impact of colloidal aggregation, a common screening artifact, in these repurposing campaigns. We tested 56 drugs reported as active in biochemical assays for aggregation by dynamic light scattering and by detergent-based enzyme counter screening; 19 formed colloids at concentrations similar to their literature IC50’s, and another 14 were problematic. From a common repurposing library, we further selected another 15 drugs that had physical properties resembling known aggregators, finding that six aggregated at micromolar concentrations. This study suggests not only that many of the drugs repurposed for SARS-CoV-2 in biochemical assays are artifacts but that, more generally, at screening-relevant concentrations, even drugs can act artifactually via colloidal aggregation. Rapid detection of these artifacts will allow the community to focus on those molecules that genuinely have potential for treating COVID-19.
Graphical Abstract
INTRODUCTION
Drug repurposing is an attractive idea in the face of a global pandemic, when rapid antiviral drug development is crucial. Although the historical pragmatism of this approach has drawn scrutiny,1,2 drug repurposing has the potential to dramatically cut both the time and cost needed to develop a new therapeutic.3 Repurposing campaigns typically screen curated libraries of thousands of approved drugs and investigational new drugs (INDs), and several assays have been developed to test these libraries for activity against SARS-CoV-2.4–6 Most high throughput, biochemical screens were developed to detect activity against two proteins that are used in viral infection and maturation: the human ACE-2 (angiotensin converting enzyme 2) and 3CL-Pro,7 the major polypeptide processing protease of SARS-2-CoV-2.
When testing molecules for biochemical activity at micromolar concentrations, it is important to control for artifacts8–12 including colloidal aggregation, which is perhaps the single most common artifact in early drug discovery.13,14 Drugs, though in many ways de-risked, are not immune to aggregation and artifactual behavior when screened at micromolar concentrations15,16 (though they are not expected to aggregate at on-target relevant concentrations). Knowing this, we wondered if colloidal aggregation was causing false positives in some COVID-19 drug repurposing studies, especially since several known aggregators, such as manidipine and methylene blue, were reported as apparently potent hits for COVID-19 targets.17,18
Aggregation is a common source of false positives in early drug discovery,19 arising from spontaneous formation of colloidal particles when organic, drug-like molecules are introduced into aqueous media.15,16,19,20 The resulting liquid particles are densely packed spheres21 that promiscuously inhibit proteins by sequestering them on the colloid surface,22 where they suffer partial unfolding.23 The resulting inhibition is reversible by disruption of the colloid and is characterized by an incubation effect on an order of several minutes due to enzyme crowding on the surface of the particle.24 Colloids often can be disrupted by the addition of small amounts, often sub-critical micelle concentrations, of non-ionic detergent such as Triton X-100.25 Accordingly, addition of detergent is a common perturbation to rapidly detect aggregates in counter screens against model enzymes such as AmpC β-lactamase or malate dehydrogenase (MDH). Aggregation can be physically detected by biophysical techniques such as nuclear magnetic resonance (NMR)26 and by dynamic light scattering (DLS), as the colloids typically form particles in the 50 to 500 nm radius size range, which is well suited to measurement by the latter technique.
Here, we investigate the role of colloidal aggregation as a source of false positives in drug repurposing studies for SARS-CoV-2 targets. We focused on in vitro ACE2 and 3CL-Pro screens since these are relevant for aggregation. We searched the literature and compiled hits from 12 sources18,27–37 where drug activities were in the micromolar and sub-micromolar range typical of colloidal aggregation. Drugs with cLogP values over 3.0 (most of those selected) or with conjugated ring systems conducive to stacking, such as methylene blue, chiniofon, and theaflavin (most of the remaining), were prioritized for testing. How the results of this study may impact the design of future repurposing screens both for SARS-2 and for other indicators will be considered.
RESULTS
Colloidal Aggregators are Common Hits in Drug Repurposing Screens for SARS-CoV-2.
We tested 56 drugs for colloidal aggregation that had been reported to be active in biochemical repurposing screens against SARS-CoV-218,27–30,32,38 (Table S1 and Experimental Section for a description of the literature search). In short, the 2D structures of compounds with reported activities in the micromolar range typical of colloidal aggregation were visually inspected for molecular features in known aggregators (e.g., multiple conjugated ring systems or calculated LogP (cLogP) >3). Five criteria were used to investigate whether reported hits formed colloidal aggregates: (a) particle formation indicated by scattering intensity, (b) clear autocorrelation curves, (c) an MDH IC50 value in the micromolar–high nanomolar range, (d) restoration of MDH activity with the addition of detergent, and less stringently (e) high Hill slopes in the inhibition concentration response curves (Figure 1).
Figure 1.
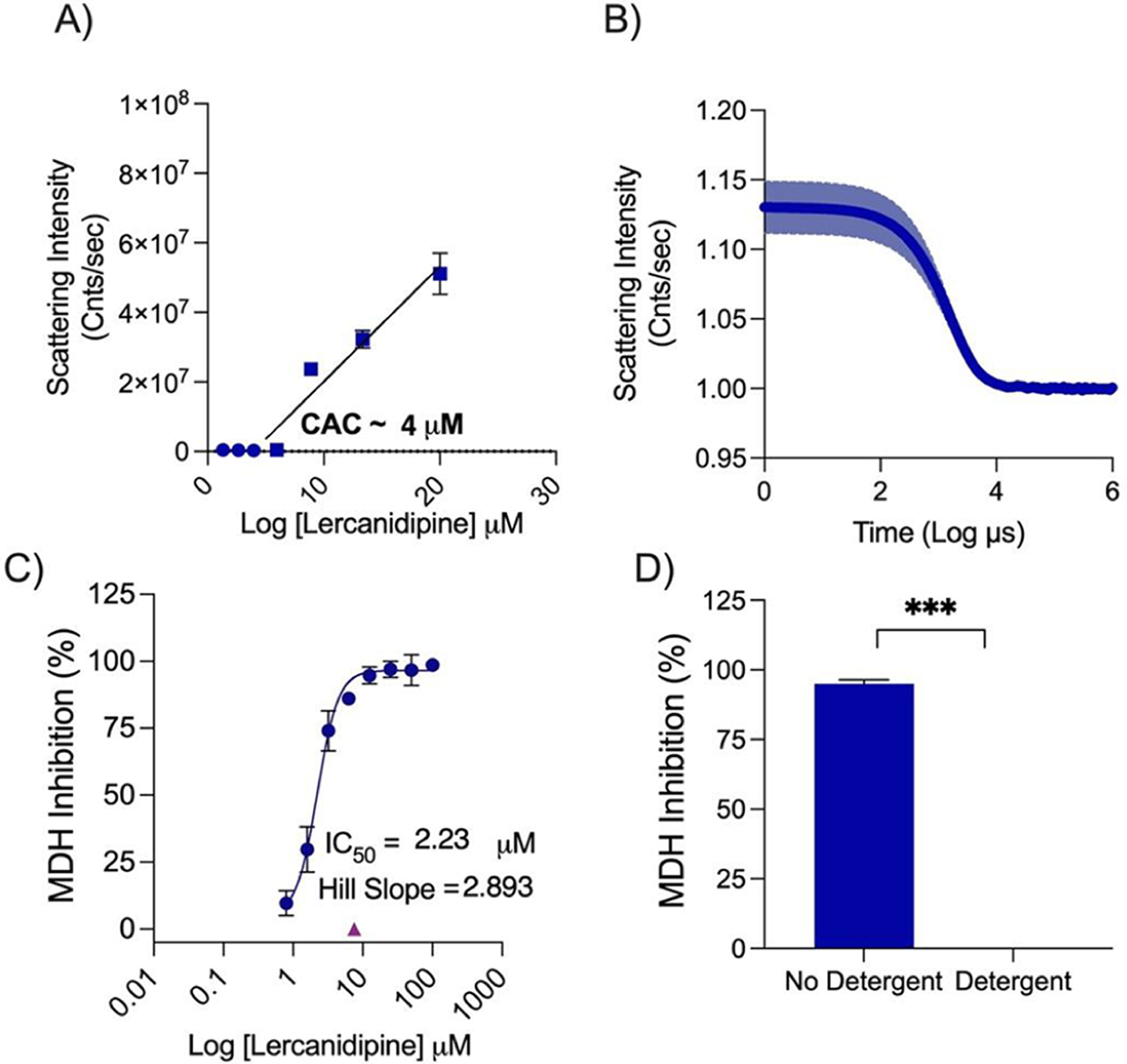
Lercanidipine’s behavior as an aggregator. (A) Critical aggregation concentration determined using scattering intensity measured on DLS. (B) Autocorrelation curve from DLS at 100 μM. (C) Dose response measured against MDH and showing the Hill slope. (D) MDH inhibition measured with or without 0.01% Triton X-100 at 7.5 μM.
Using the literature reported IC50 for the repurposed drugs as a starting point, we tested each drug for MDH inhibition and calculated the IC50 and Hill slope. We used IC50 values from the MDH concentration response curves and tested for detergent sensitivity at threefold the MDH IC50 (Figure 2). Next, we calculated the critical aggregation concentration (CAC) by measuring normalized scattering intensity on the DLS; any point above 1 × 106 was considered to be from the aggregated form. By plotting a best fit line for aggregating concentrations and non-aggregating concentrations, the CAC was given by the point of intersection (Figure 3). We also measured the DLS autocorrelation curve as a criterion: if this was well formed, it gave further confidence (Figure S1).
Figure 2.
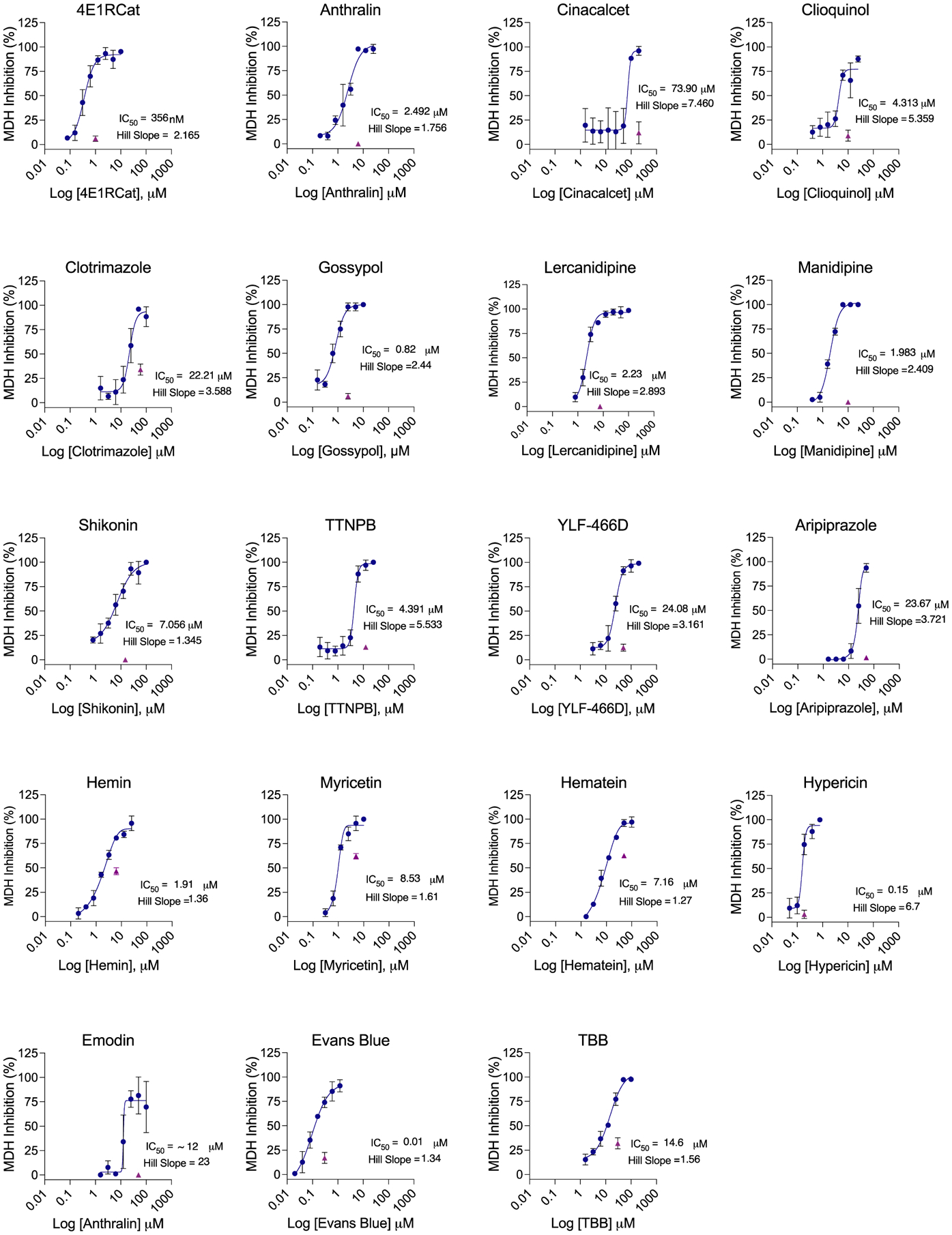
MDH inhibition concentration–response curves for literature active drugs. IC50 and Hill slopes are shown. Purple triangles indicate single-point MDH inhibition with the addition of 0.01% Triton X-100, tested at 3 times IC50. All measurements are in triplicate.
Figure 3.
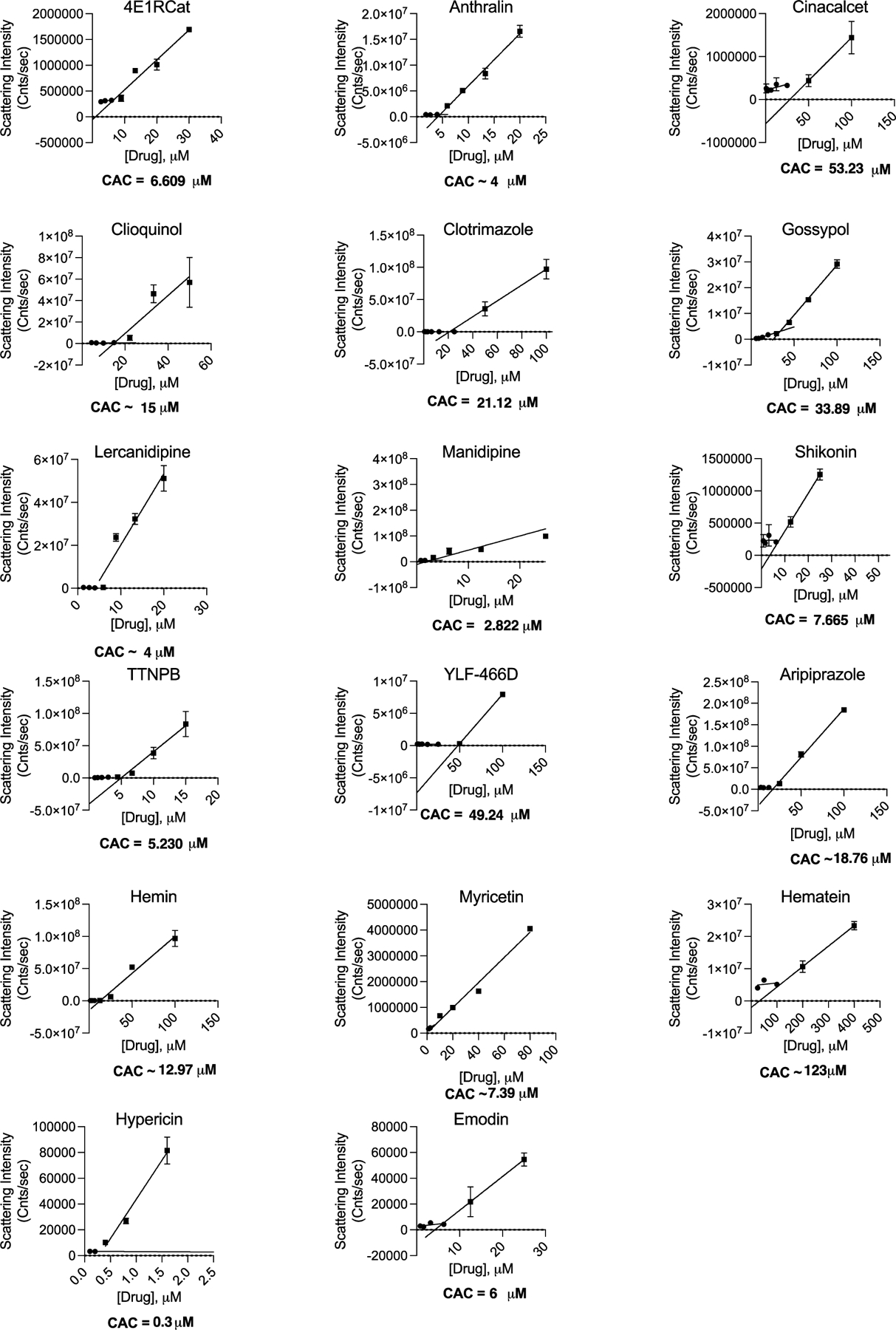
Critical aggregation concentrations for literature active drugs. The CAC is determined by finding the intersection of two best-fit lines for points with scattering intensity above or below 1 × 106. All measurements are in triplicate.
Nineteen molecules formed well-behaved particles by DLS with clean autocorrelation curves and inhibited MDH in the absence of, but not the presence of, 0.01% Triton X-100; these seem to be clear colloidal aggregators (Table 1 and Figure 2 and 3). Both DLS-based critical aggregation concentrations and MDH IC50 values were in the range of the IC50’s reported in the literature against the two SARS-CoV-2 enzymes; indeed, molecules like gossypol, manidipine, and TTNPB inhibited MDH even more potently than they did either ACE2 or 3CL-Pro. For most of the 19 drugs, the Hill slopes were high, though for several clear aggregators, such as Hemin and Shikonin, they were only in the 1.3–1.4 range. The Hill slope depends on the ratio of enzyme concentration to true KD and can vary from assay to assay39 and from aggregator to aggregator;13 while many consider it as a harbinger of aggregation, we take it as a soft criterion.13 Finally, two molecules, Evans blue and TBB, did not show particles by DLS, perhaps for spectral reasons, but did pass the other four criteria. To investigate them further, we asked whether they could be precipitated by gentle centrifugation. We tested these molecules for MDH inhibition before and after centrifugation (Figure S2) and found that enzyme activity was restored after centrifugation. This suggests that these molecules are forming colloidal aggregates, which can be spun down unlike small molecules that are genuinely in solution.22,23
Table 1.
Literature SARS-CoV-2 Repurposing Hits Shown to Cause Colloidal Aggregation
| Compound | Literature IC50a (μM) | MDH IC50 (μM) | % Reduction in MDH inhibition on detergent additionb | CACc (μM) | Colloid radius ±SD (nm) | Structure |
|---|---|---|---|---|---|---|
| 4E1RCat | 18.3 | 0.36 | 75% | 6.6 | 102 ± 8 |
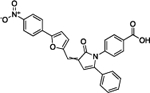
|
| Anthralin | Z ≤ −2 | 2.5 | 93% | 4.5 | 1014 ± 53 |
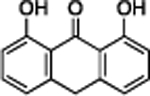
|
| Cinacalcet | 28.2 | 74 | 81% | 53 | 77 ± 4 |
|
| Clioquinol | 5.6, 2.8 | 4.3 | 57% | 15 | 1818 ± 177 |
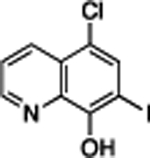
|
| Clotrimazole | 39.8 | 22 | 53% | 21 | 283 ± 15 |
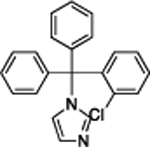
|
| Gossypol | 39.8 | 0.82 | 92% | 34 | 70 ± 6 |
|
| Lercanidipine | 16.2 | 2.2 | 95% | 4 | 853 ± 147 |
|
| Manidipine | 4.8 | 2.0 | 92% | 2.8 | 929 ± 75 |
|
| Shikonin | 15.8 | 7.0 | 92% | 7.7 | 169 ± 9 |
|
| TTNPB | 35.5 | 4.4 | 83% | 5.2 | 72 ± 3 |
|
| YLF-466D | 35.5 | 24 | 22% | 49 | 40 ± 0.4 |
|
| Hemin | 9.7 | 1.9 | 39% | 13 | 84 ± 0.9 |
|
| Aripiprazole | 26(min)d | 24 | 91% | 19 | 222 ± 5. |
|
| Hematein | 10d | 7.6 | 24% | 123 | 64 ± 1 |
|
| Myricetin | 10d | 8.5 | 32% | 7.4 | 100 ± 4 |
|
| Emodin | 51.23 | 12.7 | 59% | 6 | 58 ± 17 |
|
| Hypericin | 19.34 | 0.15 | 83% | 0.3 | 143 ± 6 |
|
| Evans Blue | 4.8 | 0.01 | 64% | NDe | NDe |
|
| TBB | 4.0 | 14.6 | 61% | NDe | NDe |
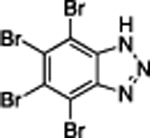
|
IC50’s measured against mPro or ACE2 in a variety of assays; see citations in Table S1.
Single-point Triton X 0.01% reversal assay performed at approximately 3× MDH IC50.
Critical aggregation concentration.
No IC50 available, single point or retention time.
Not Detectable by DLS, but precipitation by centrifugation suggests that inhibitory particles were present.
A characteristic example of a reported drug that is likely acting artifactually through colloidal aggregation is the calcium channel blocker lercanidipine, which has been reported to inhibit 3CL-Pro with an IC50 of 16.2 μM.18 Lercanidipine satisfies our five criteria for aggregation: in aqueous buffer, it forms particles that can be detected by a 10-fold increase in DLS scattering intensity (Cnts/sec), by a clearly defined autocorrelation curve in the DLS; it inhibits the counter-screening enzyme MDH with an IC50 of 2.2 μM, while MDH activity is restored upon addition of 0.01% Triton X-100 detergent (Figure 1). In the absence of detergent, lercanidipine inhibits MDH with a Hill slope of 2.9.
In addition to the 19 molecules that passed all five criteria for aggregation, another 14 molecules were more ambiguous, either forming particles by DLS but not inhibiting MDH or inhibiting MDH in a detergent-dependent manner but not forming particles detectable by DLS (Table S1). These 14 drugs may also be acting artifactually; however, further investigation is needed to determine their exact mechanisms. For this study, we focused only on clear colloidal aggregators.
Molecules Repurposed for 3CL-Pro Show Little Activity against That Enzyme in the Presence of Detergent.
In addition to testing the repurposed molecules against a counter-screening enzyme like MDH, we also tested the 12 that had been repurposed against 3CL-Pro against that enzyme itself. Because 3CL-Pro is unstable in buffer without either the presence of detergent or substantial amounts of serum albumin—both of which disrupt colloids40–42—we could not investigate the impact of detergent with 3CL-Pro as we could do with MDH. Still, we could ask whether the drugs repurposed for 3CL-Pro inhibited the enzyme in the presence of 0.05% Tween-20 used to keep the enzyme stable. Of the 12 drugs tested, only two had detectable potency below 200 μM in the presence of detergent, and for one of these two, 4E1RCat, the inhibition was reduced fivefold over its literature values (18.28 to 100 μM) (Table 2 and Figure S3). Only hemin continued to inhibit 3CL-Pro substantially, with an IC50 of 25 μM (but even this was 2.6-fold less potent than its literature value). As hemin’s inhibition of MDH was disrupted by detergent (Table 3) and it formed clear particles by DLS (Figure 3 and Figure S1), we further tested it against the model counter-screening enzyme AmpC β-lactamase. Hemin inhibited AmpC with an IC50 of 23 μM; at 25 μM hemin, addition of 0.01% (v/v) Triton X-100 fully restored enzyme activity—inhibition was abolished. Taken together, these observations further support the aggregation-based activity of these 12 repurposed drugs.
Table 2.
Literature Repurposing Hits Do Not Potently Inhibit 3CL-Pro in the Presence of Detergent
| compound | literature IC50a (μM) | 3CL-Pro IC50 with 0.05% Tween-20 IC50 (μM) |
|---|---|---|
| 4E1RCat | 18.3 | ~100 |
| anthralin | Z ≤ 2 | >200 |
| clotrimazole | 39.8 | >200 |
| gossypol | 39.8 | >200 |
| lercanidipine | 16.2 | >200 |
| manidipine | 4.8 | >100a |
| shikonin | 15.8 | ~200 |
| TTNPB | 35.5 | >200 |
| YLF-466D | 35.5 | >200 |
| hemin | 9.7 | 25 |
| hematein | 10a | >200 |
| emodin | 51.2 | >200 |
100 μM was the highest concentration used for manidipine, instead of 200 μM.
Table 3.
Six Drugs from a Repurposing Library Aggregate at Screening-Relevant Concentrationsb
| Compound | MDH IC50 (μM) | % Change in MDH Inhibition in presence of Triton-Xa | CACc (μM) | Colloid Radius ± SD (nm) | Structures |
|---|---|---|---|---|---|
| Adapalene | 14 | 77% | 9.2 | 422 ± 65 |
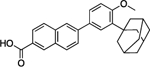
|
| Buparvaquone | 8.2 | 90% | 32 | 186 ± 15 |
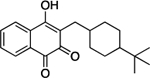
|
| Bifonazole | 17 | 87% | 25 | 184 ± 80 |
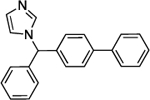
|
| Alpha-Tochopherol | 0.079 | 69% | 1.5 | 466 ± 5.6 |
|
| Bazedoxifene | 18 | 70% | 26 | 2822 ± 814 |
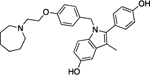
|
| Dracorhodin | 7.7 | 86% | 22 | 101 ± 22 |
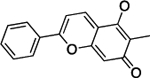
|
Single-point Triton X 0.01% reversal assay performed at approximately 3 times MDH IC50.
Indicates the drug concentration at which colloid radius measurements were made.
Critical aggregation concentration.
Colloidal Aggregators in Repurposing Libraries.
Target-based drug repurposing screens are common not only for SARS-CoV-2 but for many other viruses and indeed other indications. We thought it interesting to explore, if only preliminarily, the occurrence of colloidal aggregators in drug repurposing libraries. We prioritized drugs in the widely used SelleckChem FDA-approved library as potential aggregators, using a simple chemoinformatics approach.43 Library molecules were compared to a database of known aggregators using the Aggregator Advisor43 command line tool, which calculates molecular similarity (Tanimoto coefficients; Tc) between the two sets of molecules (Table S2). Molecules similar to a known aggregator (1 > Tc’s > 0.65) that were also hydrophobic (cLogP > 4) were drawn, inspected for diversity from one another and for the presence of features in known aggregators such as conjugated ring systems, and were prioritized for testing. Of the 2336 unique drugs in the library, 73 are already known aggregators and another 356 (16%) closely resemble known aggregators. We selected 15 of the latter for aggregation: six of these drugs satisfied our five criteria for aggregation; they inhibited MDH in the absence of, but not in the presence of, 0.01% Triton X-100 (Figure 4) and formed well-behaved particles detectable by DLS (Figure 5) with clean autocorrelation curves (Figure S4), often with steep Hill slopes. Taken together, these data suggest that these six drugs are prone to colloidal aggregation at screening-relevant concentrations (Table 3).
Figure 4.
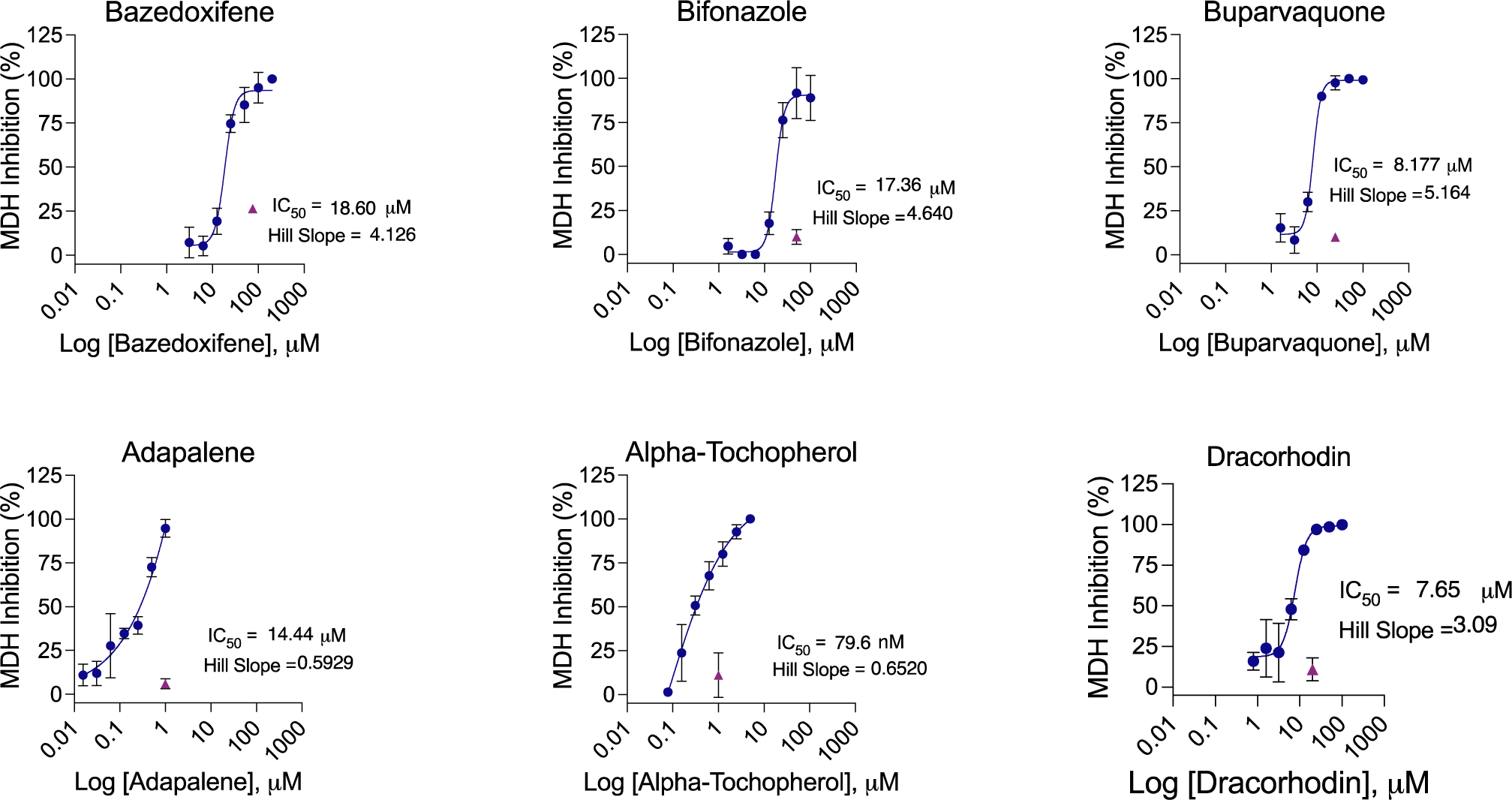
MDH inhibition dose–response curves for drugs drawn from a repurposing library. All measurements were in triplicate.
Figure 5.
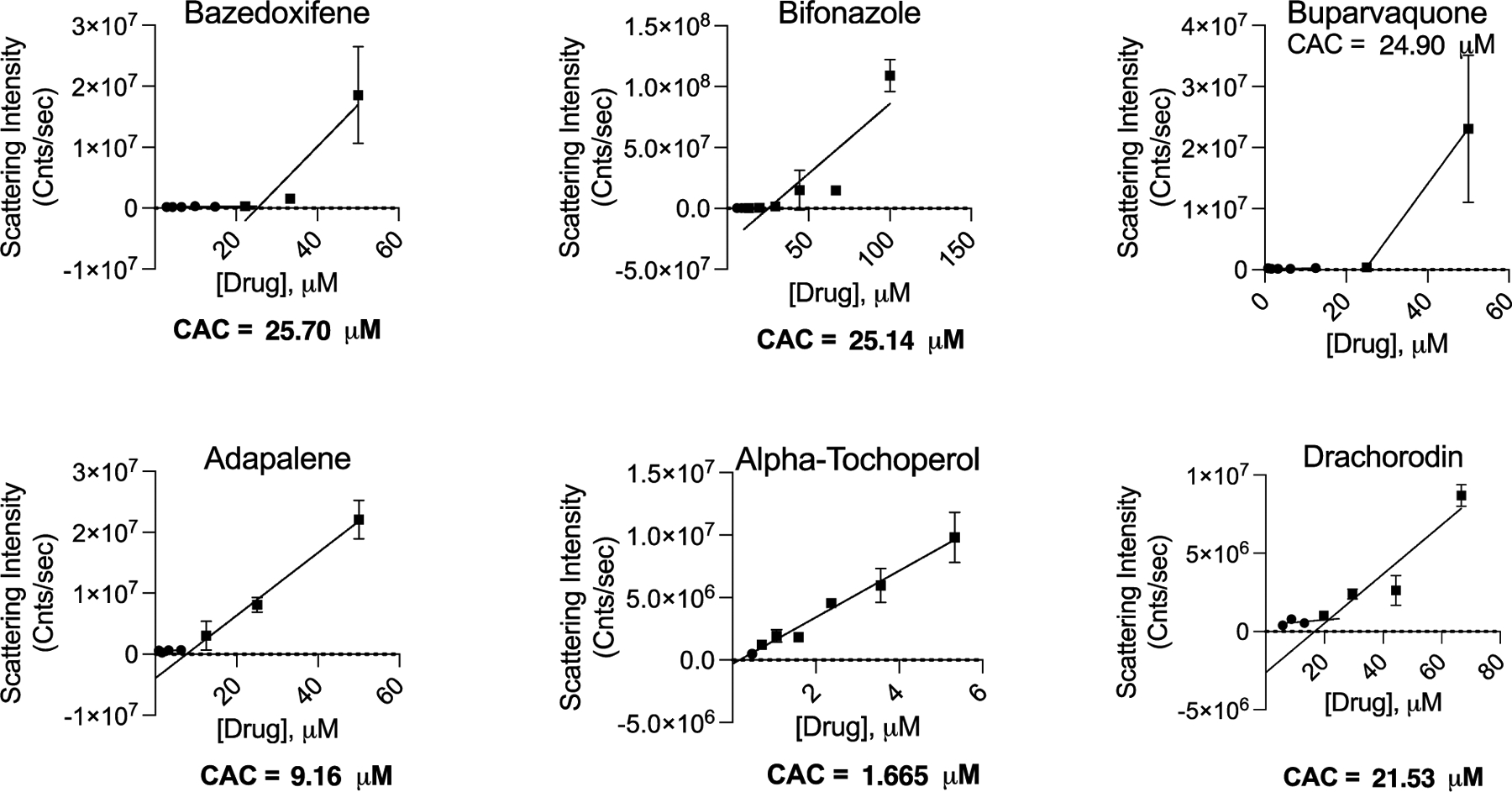
Critical aggregation concentrations for drugs drawn from a repurposing library. The CAC is determined by the intersection of two best-fit lines, for points with scattering intensity above or below 1 × 106. All measurements were in triplicate.
DISCUSSION AND CONCLUSIONS
Two broad observations from this study merit emphasis. First, many drugs repurposed for COVID-19 aggregate and inhibit counter-screening enzymes promiscuously at concentrations relevant to their reported IC50’s against the COVID-19 targets (ACE2 and 3CL-Pro). Of the 56 drugs tested, 19 fulfilled all five of our criteria for acting via colloidal aggregation: (i) they formed particles that were scattered strongly by DLS with (ii) well-behaved autocorrelation curves, (iii) they inhibited the counter-screening enzyme malate dehydrogenase—unrelated to either ACE2 or 3CL-Pro—at relevant concentrations in the absence, but (iv) not the presence, of detergent, and (v) they typically inhibited with steep Hill slopes. Each of these criteria individually is a harbinger of colloidal aggregation; when combined, they strongly support its occurrence. The other 14 of the 56 drugs fulfilled only some of these criteria, for instance, forming particles at relevant concentrations but not inhibiting MDH in a detergent-dependent manner. Some of these 14 drugs may also be aggregators, while others, like those that inhibit MDH but cannot be reversed by detergent, like tannic acid, may be acting as pan assay interference compounds (PAINS). A second observation from this study is that these artifacts are not so much a feature of SARS-CoV-2 repurposing but rather reflect the behavior of drugs at screening relevant concentrations. Thus, 6 of 15 drugs investigated from a general repurposing library were also aggregators at micromolar concentrations. An attraction of drug repurposing is that the molecules are thought to be de-risked from the pathologies of early discovery. However, at micromolar concentrations, drugs, which are often larger and more hydrophobic than the lead-like molecules found in most high-throughput screening and virtual libraries, are if anything more likely to aggregate, something that earlier studies also support.15,16
For 4 of the 19 aggregators found in this study, Triton X-100 detergent was already present in the reaction buffer used in the original publication (Table S1), reflecting the care of those studies. However, while it is commonly thought that detergent addition protects against aggregation from the outset, in fact, detergent often only right-shifts the onset of aggregation-based inhibition. Thus, even screens that control for aggregation by including detergent in the reaction buffer may consider +/− detergent controls during hit confirmation. On the other hand, several of the aggregators, including emodin, hemin, and hypericin (Table 1), notwithstanding their provenance from a drug repurposing library, have features that would ordinarily give medicinal chemists pause. Sometimes the “drugs” in drug repurposing libraries are not actually drugs, and despite their origins as phytochemical natural products, as with these molecules, they can have features, e.g., multiple phenolic groups in conjugated ring systems, that might prejudice them against further study.
Certain caveats should be mentioned. We do not pretend to have undertaken a comprehensive study of the increasingly large literature around drug repurposing for COVID-19. The molecules tested here represent only a subset of those investigated, drawn from an analysis of some of the literature then available. Also, we have not demonstrated that aggregation is actually occurring in the ACE-2 assay itself, though the lack of inhibition of 3CL-Pro in the presence of detergent fortifies our conclusions for the 12 molecules that inhibited this enzyme. Finally, it is important to note that just because some repurposed drugs aggregate at micromolar concentrations, the repurposing enterprise is not sunk. There are, after all, examples of drugs successfully repurposed, even for COVID-19, and some have even begun from screening hits (though typically they are subsequently modified chemically44).
These caveats should not obscure the main observations from this study. Many drugs repurposed for COVID-19 in biochemical assays are aggregators—still, others may be inhibiting through other artifactual mechanisms—and their promise as leads for treating the disease merits reconsideration. Indeed, while some repurposed drugs have advanced further into development,44 the aggregators described here do not seem have been further progressed. More broadly, drugs in repurposing libraries, though de-risked for whole body toxicity, pharmacokinetic exposure, and metabolism, are not de-risked for artifactual activity at screening relevant concentrations. More encouragingly, what this study illuminates is a series of facile assays that can rapidly distinguish drugs acting artifactually via colloidal aggregation from those drugs with true promise for treating SARS-CoV-2, and from pandemics yet to be faced.
EXPERIMENTAL SECTION
Literature Search and Chemoinformatic Selection of Potential Aggregators.
We used two approaches to identify drugs with the potential to form colloidal aggregates from repurposing screens: (1) literature searches of published SARS-CoV-2 biochemical drug screening papers including chemoinformatic analysis of the NCATS COVID-19 OpenData Portal37 3CL-Pro and ACE2 biochemical drug screens and (2) chemoinformatic predictions of potential aggregators found in the SelleckChem FDA-approved drug library using the Aggregation Advisor tool.43 Literature-based keyword searches were performed using variations of the keywords “SARS-CoV-2” and “drug repurposing” or “drug screen”. Inhibitors from biochemical drug-repurposing screens were visually inspected and prioritized for testing if they had cLogP values >3 or were highly conjugated. Next, data from the NCATS COVID-19 OpenData Portal37 drug-repurposing screens for modulators of 3CL-Pro and ACE2 activities were retrieved (accessed on September 28, 2020). In total, 12,262 and 3405 annotations were found for compounds screened against 3CL-Pro and ACE2, respectively. Molecules annotated with PubChem45 substance identifiers that had activities (AC50s) less than 50 μM but typically greater than 5 μM were selected. Simplified molecular input line entry system (SMILES) data for each compound were retrieved using the PubChemPy API (https://pubchempy.readthedocs.io) and used to calculate cLogP values using RDkit-2019.09.3.0 (http://www.rdkit.org). Molecules with cLogP > 3 were drawn, visually inspected for the presence of molecular features seen in known aggregators (e.g., multiple conjugated ring systems, overall hydrophobicity, and no covalent warheads or PAINs), and prioritized for testing. Finally, the SMILES of 2336 unique desalted molecules were selected from the SelleckChem library and were analyzed with Aggregation Advisor,43 a command line tool that calculates molecular similarity (Tanimoto coefficients; Tc) between a list of molecules and a database of known aggregators (Table S2). Molecules with 1 > Tc’s > 0.65 to a known aggregator and cLogP > 4 were drawn, inspected for structural diversity from one another and for the presence of molecular features seen in known aggregators (e.g., multiple conjugated ring systems), and prioritized for testing. Percentages were calculated relative to the 2336 unique molecules in the library with identified SMILES.
Compounds.
All compounds are >95% pure by HPLC, as reported by the vendors. Compounds were ordered from Sigma-Aldrich, SelleckChem, Cayman Chemical, or Medchem Express.
Dynamic Light Scattering.
To detect and quantify colloids, a DynaPro Plate Reader II (Wyatt Technologies) with a 60 mW laser at 830 nm wavelength and a detector angle of 158° was used; the beam size of the instrument was increased by the manufacturer to better enable detection of the colloids, which are larger than protein aggregates for which the instrument was designed. Samples were measured in 384-well plates with 30 μL loading and 10 acquisitions per sample. Compounds were dissolved in DMSO at 100 times their final concentration and were diluted into filtered 50 mM KPi, pH 7.0, to obtain a final 1% DMSO concentration. Compounds were first tested at 3 times the IC50 reported in the literature, and if active, they were further investigated in concentration–response tests. If no IC50 was available, compounds were tested at 100 μM. To calculate a CAC, each compound was serially diluted until substantial scattering disappeared; aggregating (>106 scattering intensity) and non-aggregating (<106 scattering intensity) portions of the data were fitted with separate nonlinear regression curves, and the point of intersection was determined using GraphPad Prism software version 9.1.1 (San Diego, CA).
Enzyme Inhibition.
MDH inhibition assays were performed at room temperature on a HP8453a spectrophotometer in kinetic mode using UV–vis Chemstation software (Agilent Technologies) in methacrylate cuvettes (Fisher Scientific, 14955128) with a final volume of 1 mL for both control and test reactions. MDH (from porcine heart, 901643, Sigma-Millipore) was added to a 50 mM KPi pH 7 buffer for a final concentration of 2 nM. Compounds were dissolved in DMSO at 100 times concentration; 10 μL of compound was used for a final DMSO concentration of 1%. After compound addition, the cuvette was mixed by pipetting up and down 5 times with a p1000, and the cuvette was then incubated for 5 min at room temperature. The reaction was initiated by the addition of 200 μM nicotinamide adenine dinucleotide (54839, Sigma-Aldrich) and 200 μM oxaloacetic acid (324427, Sigma-Aldrich), and the rate was monitored at 340 nm. A negative control was included in each run, in which 10 μL of DMSO without the compound was added. The reactions were monitored for 90 s, and the initial rates were divided by the initial rate of the negative control to obtain the % inhibition and % enzyme activity. For dose–response curves, three replicates were done for each concentration, the graphs were generated using GraphPad Prism version 9.1.1 (San Diego, CA).
3CL-Pro Kinetics Inhibition Assay.
A fluorescence-quenched substrate with the sequence rr-K(MCA)-ATLQAIAS-K(DNP)-COOH was synthesized via the Fmoc solid-phase peptide synthesis as described.46 Recombinant, active 3CL-Pro was expressed and purified as described.47 Kinetic measurements were carried out in Corning black 384-well flat-bottom plates and read on a BioTek H4 multimode plate reader. The quenched fluorogenic peptide had a final concentration of KM = 10 μM, and 3CL-Pro had a final concentration of 50 nM. The reaction buffer was 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.05% Tween-20 (v/v), and 1 mM DTT, pH 7.4. Drugs were incubated with protease prior to substrate addition at 37 °C for 1 h. After incubation, the substrate was added, and kinetic activity was monitored for 1 h at 37 °C. Initial velocities were calculated at 1 to 45 min in RFU/s. Velocities were corrected by subtracting the relative fluorescence of a substrate-only control, and fraction activity was calculated using a substrate-corrected no-inhibitor control where DMSO was added instead of a drug. Kinetics measurements were carried out in triplicate.
Colloid Centrifugation.
DMSO stocks of drugs were prepared and diluted to 100:1 into 1 mL of 50 mM KPi buffer, pH 7, in a 1.5 mL Eppendorf tube. This was mixed by pipetting and centrifuging at 14,000 rpm for 1 h at 4 °C in a benchtop microfuge. The supernatant (900 μL of 1 mL) was then tested for MDH inhibition as previously described.
Supplementary Material
ACKNOWLEDGMENTS
Supported by grants from the Defense Advanced Research Projects Agency HR0011-19-2-0020 and the NIH R35GM122481 (to B.K.S.) and from NIAID (P50AI150476 to C.S.C.). We thank Khanh Tang and John Irwin for help with Aggregation Advisor and Isabella Glenn for help with aggregation assays.
ABBREVIATIONS USED
- ACE2
angiotensin-converting enzyme 2
- CAC
critical aggregation concentration
- DLS
dynamic light scattering
- INDs
investigational new drugs
- NMR
nuclear magnetic resonance
- MDH
malate dehydrogenase
- PAINS
pan assay interference compounds
- Tc
Tanimoto coefficients
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.1c01547.
Table of all compounds tested, figure of autocorrelation curves for all compounds considered as aggregators, concentration response curves of the 17 literature compounds against 3CL-Pro in the presence of 0.05% Tween-20, centrifugation data for Evans blue and TBB, aggregate advisor analysis table (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.jmedchem.1c01547
The authors declare no competing financial interest.
Contributor Information
Henry R. O’Donnell, Department of Pharmaceutical Chemistry, University of California San Francisco (UCSF), San Francisco, California 94158-2550, United States
Tia A. Tummino, Department of Pharmaceutical Chemistry, University of California San Francisco (UCSF), San Francisco, California 94158-2550, United States; Graduate Program in Pharmaceutical Sciences and Pharmacogenomics, UCSF, San Francisco, California 94158-2550, United States; QBI COVID-19 Research Group (QCRG), San Francisco, California 94158-2550, United States
Conner Bardine, Graduate Program in Chemistry & Chemical Biology, UCSF, San Francisco, California 94158-2550, United States;.
Charles S. Craik, Department of Pharmaceutical Chemistry, University of California San Francisco (UCSF), San Francisco, California 94158-2550, United States;; QBI COVID-19 Research Group (QCRG), San Francisco, California 94158-2550, United States;
Brian K. Shoichet, Department of Pharmaceutical Chemistry, University of California San Francisco (UCSF), San Francisco, California 94158-2550, United States;; QBI COVID-19 Research Group (QCRG), San Francisco, California 94158-2550, United States;
REFERENCES
- (1).Edwards A What are the odds of finding a COVID-19 drug from a lab repurposing screen? J. Chem. Inf. Model 2020, 60, 5727–5729. [DOI] [PubMed] [Google Scholar]
- (2).Edwards A; Hartung IV No shortcuts to SARS-CoV-2 antivirals. Science 2021, 373, 488–489. [DOI] [PubMed] [Google Scholar]
- (3).Papapetropoulos A; Szado C Inventing new therapies without reinventing the wheel: the power of drug repurposing. Br. J. Pharmacol 2018, 175, 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Dotolo S; Marabotti A; Facchiano A; Tagliaferri R A review on drug repurposing applicable to COVID-19. Brief Bioinform 2021, 22, 726–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hossain MS; Hami I; Sawrav MSS; Rabbi MF; Saha O; Bahadur NM; Rahaman MM Drug repurposing for prevention and treatment of COVID-19: A clinical landscape. Discoveries 2020, 8, No. e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Singh TU; Parida S; Lingaraju MC; Kesavan M; Kumar D; Singh RK Drug repurposing approach to fight COVID-19. Pharmacol Rep 2020, 72, 1479–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Saxena A Drug targets for COVID-19 therapeutics: Ongoing global efforts. J. Biosci 2020, 45, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Baell JB; Holloway GA New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem 2010, 53, 2719–2740. [DOI] [PubMed] [Google Scholar]
- (9).Baell JB Feeling nature’s PAINS: Natural products, natural product drugs, and Pan Assay Interference Compounds (PAINS). J. Nat. Prod. 2016, 79, 616–628. [DOI] [PubMed] [Google Scholar]
- (10).Dahlin JL; Inglese J; Walters MA Mitigating risk in academic preclinical drug discovery. Nat Rev Drug Discov 2015, 14, 279–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Thorne N; Auld DS; Inglese J Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol 2010, 14, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lloyd MD High-throughput screening for the discovery of enzyme Inhibitors. J. Med. Chem 2020, 63, 10742–10772. [DOI] [PubMed] [Google Scholar]
- (13).Feng BY; Simeonov A; Jadhav A; Babaoglu K; Inglese J; Shoichet BK; Austin CP A high-throughput screen for aggregation-based inhibition in a large compound library. J. Med. Chem 2007, 50, 2385–2390. [DOI] [PubMed] [Google Scholar]
- (14).Jadhav A; Ferreira RS; Klumpp C; Mott BT; Austin CP; Inglese J; Thomas CJ; Maloney DJ; Shoichet BK; Simeonov A Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J. Med. Chem 2010, 53, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Doak AK; Wille H; Prusiner SB; Shoichet BK Colloid formation by drugs in simulated intestinal fluid. J. Med. Chem 2010, 53, 4259–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Seidler J; McGovern SL; Doman TN; Shoichet BK Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem 2003, 46, 4477–4486. [DOI] [PubMed] [Google Scholar]
- (17).Bojadzic D; Alcazar O; Buchwald P Methylene blue inhibits the SARS-CoV-2 spike-ACE2 protein-protein interaction-a mechanism that can contribute to its antiviral activity against COVID-19. Front Pharmacol 2021, 11, 600372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ghahremanpour MM; Tirado-Rives J; Deshmukh M; Ippolito JA; Zhang CH; Cabeza de Vaca I; Liosi ME; Anderson KS; Jorgensen WL Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett 2020, 11, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).McGovern SL; Shoichet BK Kinase inhibitors: not just for kinases anymore. J. Med. Chem 2003, 46, 1478–1483. [DOI] [PubMed] [Google Scholar]
- (20).Ganesh AN; Donders EN; Shoichet BK; Shoichet MS Colloidal aggregation: from screening nuisance to formulation nuance. Nano Today 2018, 19, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Blevitt JM; Hack MD; Herman KL; Jackson PF; Krawczuk PJ; Lebsack AD; Liu AX; Mirzadegan T; Nelen MI; Patrick AN; Steinbacher S; Milla ME; Lumb KJ Structural basis of small-molecule aggregate induced inhibition of a protein-protein interaction. J. Med. Chem 2017, 60, 3511–3517. [DOI] [PubMed] [Google Scholar]
- (22).McGovern SL; Helfand BT; Feng B; Shoichet BK A specific mechanism of nonspecific inhibition. J. Med. Chem 2003, 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- (23).Coan KED; Maltby DA; Burlingame AL; Shoichet BK Promiscuous aggregate-based inhibitors promote enzyme unfolding. J. Med. Chem 2009, 52, 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lak P; O’Donnell H; Du X; Jacobson MP; Shoichet BK A Crowding barrier to protein inhibition in colloidal aggregates. J. Med. Chem 2021, 64, 4109–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Feng BY; Toyama BH; Wille H; Colby DW; Collins SR; May BCH; Prusiner SB; Weissman J; Shoichet BK Small-molecule aggregates inhibit amyloid polymerization. Nat. Chem. Biol 2008, 4, 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).LaPlante SR; Aubry N; Bolger G; Bonneau P; Carson R; Coulombe R; Sturino C; Beaulieu PL Monitoring drug self-aggregation and potential for promiscuity in off-target in vitro pharmacology screens by a practical NMR strategy. J. Med. Chem 2013, 56, 7073–7083. [DOI] [PubMed] [Google Scholar]
- (27).Baker JD; Uhrich RL; Kraemer GC; Love JE; Kraemer BC A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PLoS One 2021, 16, No. e0245962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Jin Z; Du X; Xu Y; Deng Y; Liu M; Zhao Y; Zhang B; Li X; Zhang L; Peng C; Duan Y; Yu J; Wang L; Yang K; Liu F; Jiang R; Yang X; You T; Liu X; Yang X; Bai F; Liu H; Liu X; Guddat LW; Xu W; Xiao G; Qin C; Shi Z; Jiang H; Rao Z; Yang H Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [DOI] [PubMed] [Google Scholar]
- (29).Zhu W; Xu M; Chen CZ; Guo H; Shen M; Hu X; Shinn P; Klumpp-Thomas C; Michael SG; Zheng W Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol Transl Sci 2020, 3, 1008–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Olaleye OA; Kaur M; Onyenaka C; Adebusuyi T Discovery of Clioquinol and analogues as novel inhibitors of Severe Acute Respiratory Syndrome Coronavirus 2 infection, ACE2 and ACE2 - Spike protein interaction in vitro. Heliyon 2021, 7, No. e06426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Han Y; Duan X; Yang L; Nilsson-Payant BE; Wang P; Duan F; Tang X; Yaron TM; Zhang T; Uhl S; Bram Y; Richardson C; Zhu J; Zhao Z; Redmond D; Houghton S; Nguyen DT; Xu D; Wang X; Jessurun J; Borczuk A; Huang Y; Johnson JL; Liu Y; Xiang J; Wang H; Cantley LC; teneOver BR; Ho DD; Pan FC; Evans T; Chen HJ; Schwartz RE; Chen S Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tripathi PK; Upadhyay S; Singh M; Raghavendhar S; Bhardwaj M; Sharma P; Patel AK Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int. J. Biol. Macromol 2020, 164, 2622–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fu W; Chen Y; Wang K; Hettinghouse A; Hu W; Wang JQ; Lei ZN; Chen ZS; Stapleford KA; Liu CJ Repurposing FDA-approved drugs for SARS-CoV-2 through an ELISA-based screening for the inhibition of RBD/ACE2 interaction. Protein Cell 2021, 12, 586–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ge S; Wang X; Hou Y; Lv Y; Wang C; He H Repositioning of histamine H1 receptor antagonist: Doxepin inhibits viropexis of SARS-CoV-2 Spike pseudovirus by blocking ACE2. Eur. J. Pharmacol 2021, 896, 173897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lin C; Li Y; Zhang Y; Liu Z; Mu X; Gu C; Liu J; Li Y; Li G; Chen J Ceftazidime is a potential drug to inhibit SARS-CoV-2 infection in vitro by blocking spike protein-ACE2 interaction. Signal Transduction Targeted Ther. 2021, 6, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jang WD; Jeon S; Kim S; Lee SY Drugs repurposed for COVID-19 by virtual screening of 6,218 drugs and cell-based assay. Proc. Natl. Acad. Sci. U. S. A 2021, 118, No. e2024302118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Brimacombe KR; Zhao T; Eastman RT; Hu X; Wang K; Backus M; Baljinnyam B; Chen CZ; Chen L; Eicher T; Ferrer M; Fu Y; Gorshkov K; Guo H; Hanson QM; Itkin Z; Kales SC; Klumpp-Thomas C; Lee EM; Michael S; Mierzwa T; Patt A; Pradhan M; Renn A; Shinn P; Shrimp JH; Viraktamath A; Wilson KM; Xu M; Zakharov AV; Zhu W; Zheng W; Simeonov A; Mathe EA; Lo DC; Hall MD; Shen M An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv 2020, DOI: 10.1101/2020.06.04.135046. [DOI] [Google Scholar]
- (38).White MA; Lin W; Cheng X Discovery of COVID-19 inhibitors targeting the SARS-CoV2 Nsp13 Helicase. bioRxiv 2020, 9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Shoichet BK Interpreting steep dose-response curves in early inhibitor discovery. J. Med. Chem 2006, 49, 7274–7277. [DOI] [PubMed] [Google Scholar]
- (40).McGovern SL; Caselli E; Grigorieff N; Shoichet BK A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem 2002, 45, 1712–1722. [DOI] [PubMed] [Google Scholar]
- (41).McGovern SL; Helfand BT; Feng B; Shoichet BK A Specific mechanism for non-specific inhibition. J. Med. Chem 2003, 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- (42).Coan KED; Shoichet BK Stability and equilibria of promiscuous aggregates in high protein milieus. Mol. BioSyst 2007, 3, 208–213. [DOI] [PubMed] [Google Scholar]
- (43).Irwin JJ; Duan D; Torosyan H; Doak AK; Ziebart KT; Sterling T; Tumanian G; Shoichet BK An Aggregation advisor for ligand discovery. J. Med. Chem 2015, 58, 7076–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang CH; Spasov KA; Reilly RA; Hollander K; Stone EA; Ippolito JA; Liosi ME; Deshmukh MG; Tirado-Rives J; Zhang S; Liang Z; Miller SJ; Isaacs F; Lindenbach BD; Anderson KS; Jorgensen WL Optimization of triarylpyridinone inhibitors of the main protease of SARS-CoV-2 to low-nanomolar antiviral potency. ACS Med. Chem. Lett 2021, 12, 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kim S; Chen J; Cheng T; Gindulyte A; He J; He S; Li Q; Shoemaker BA; Thiessen PA; Yu B; Zaslavsky L; Zhang J; Bolton EE PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ravalin M; Theofilas P; Basu K; Opoku-Nsiah KA; Assimon VA; Medina-Cleghorn D; Chen YF; Bohn MF; Arkin M; Grinberg LT; Craik CS; Gestwicki JE Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases. Nat. Chem. Biol 2019, 15, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhang L; Lin D; Sun X; Curth U; Drosten C; Sauerhering L; Becker S; Rox K; Hilgenfeld R Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 2020, 368, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


