Summary
Laboratory mice comprise an expeditious model for preclinical vaccine testing; however, vaccine immunogenicity in these models often inadequately translates to humans. Reconstituting physiologic microbial experience to specific pathogen free (SPF) mice induces durable immunological changes that better recapitulate human immunity. We examined whether mice with diverse microbial experience better model human responses post-vaccination. We cohoused laboratory mice with pet store mice, which have varied microbial exposures, and then assessed immune responses to influenza vaccines. Human transcriptional responses to influenza vaccination are better recapitulated in cohoused mice. Although SPF and cohoused mice were comparably susceptible to acute influenza infection, vaccine-induced humoral responses were dampened in cohoused mice, resulting in poor control upon challenge. Additionally, protective heterosubtypic T cell immunity was compromised in cohoused mice. Because SPF mice exaggerated humoral and T cell protection upon influenza vaccination, reconstituting microbial experience in laboratory mice through cohousing may better inform preclinical vaccine testing.
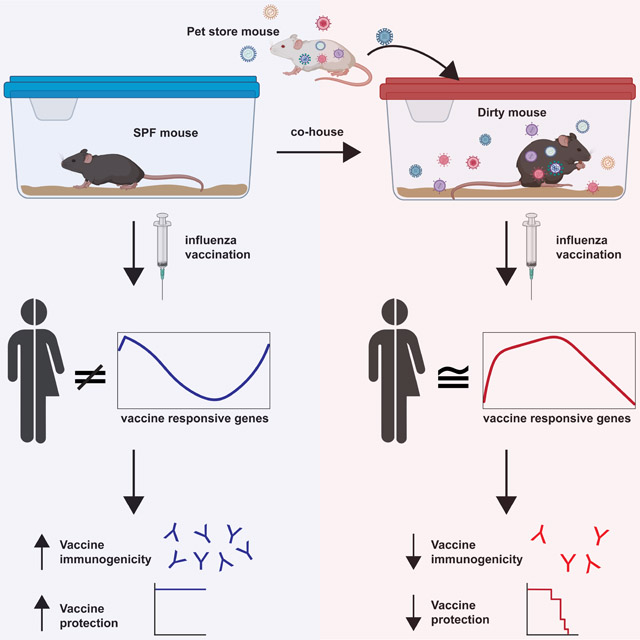
Graphical Abstract
eTOC blurb
Research animals are housed in clean facilities to reduce confounding variables from microbial exposure. Fiege et al demonstrate that natural microbial exposure of mice results in signatures post-vaccination that are more similar to humans. Additionally, they demonstrate that animals in clean facilities exaggerate the immunogenicity to influenza vaccination.
Introduction
Laboratory mice are often the first model organism for the assessment of new vaccine modalities. These animals have a number of advantages including cost, wealth of available tools for assessment, inbred genetics, and specific pathogen free (SPF) housing environments to increase reproducibility (Jameson and Masopust, 2018). However, immune stimuli that successfully prevent or treat disease in mice often fail to adequately translate to humans (Davis, 2008; Jameson and Masopust, 2018; Koff et al., 2013; Mestas and Hughes, 2004; Payne and Crooks, 2007; Rice, 2012; Rivera and Tessarollo, 2008; Seok et al., 2013; von Herrath and Nepom, 2005). The activation status and composition of the immune systems of SPF mice and humans are fundamentally different, potentially confounding comparisons of the responses to experimental vaccines. To improve the translatability of mouse studies we introduced pathogen experience into SPF animals by cohousing with pet store mice. These now ‘dirty mice’ have fundamentally altered immune systems that better recapitulate key aspects of human immunity (Beura et al., 2016; Hamilton et al., 2020; Huggins et al., 2019). Additionally, dirty, but not SPF mice, were able to recapitulate the clinical features of a human immunodeficiency disease (Takeda et al., 2019). In another model, wild mouse fecal transfer studies altered the immune system of laboratory mice impacting the outcome of influenza A virus (IAV) infections and predicting the failure of two immunological treatments in humans that were successful in SPF mice (Rosshart et al., 2019; Rosshart et al., 2017). Together these data provide examples that animals with diverse histories of microbial exposure can better mimic feature of the human immune system.
The value of preclinical vaccine model organisms depends on many variables, including species-specific pathogenesis, availability of reagents for evaluating immune responses and establishing mechanisms of protection, and the likelihood of recapitulating human vaccine reactogenicity and immunogenicity. This remains an empirical science, and there is no perfect, universal model organism. In practice, vaccine candidates are often tested first in mice and then vetted through species that incur higher material and logistical costs that constrain group sizes and the number of experimental conditions. Improvements in the predictive value of mouse models, including a reduction in the incidence of false positive results for immunogenicity, could enable more accurate down-selection of less desirable vaccine candidates.
New vaccine approaches and technologies must be tested for endemic pathogens that lack effective vaccines, emerging infectious agents, and for pathogens such as influenza viruses that constantly evolve. With respect to influenza virus, vaccine modalities that provide enhanced seasonal protection (current efficacy rates range from ~20-60% per year) as well as potential broadly acting ‘universal’ protection are in development. There is also the threat of the emergence of novel influenza strains from zoonotic reservoirs for which no vaccine may exist. Vaccine efficacy studies in humans are logistically challenging: the infection rate per year is ~10%, necessitating large cohorts; emerging IAV strains can be unpredictable; and human challenge studies present ethical challenges (Roestenberg et al., 2018; Zhou et al., 2018). Therefore, animal models have been essential in vaccine development to evaluate safety, immunogenicity, and to understand mechanisms/correlates of protection. Unfortunately, many vaccines and vaccine adjuvants that were successful in mice have failed to translate to humans (Bracci et al., 2006; Cooper et al., 2004; Couch et al., 2009; Manzoli et al., 2009; Moldoveanu et al., 1998; Proietti et al., 2002; Tregoning et al., 2018; Young et al., 2015).
We tested whether immune responses to influenza vaccination and infection differed between SPF mice and mice with more physiologic microbial experience. We observed that SPF mice exaggerated vaccine-elicited humoral and cellular responses and protective immunity compared to dirty mice and that the transcriptional signatures in response to vaccination in dirty mice more closely mimic responses in humans. These data provide a rationale for including dirty mice in preclinical vaccine candidate evaluation.
Results
Dirty mice recapitulate vaccine-induced transcriptional signatures observed in humans
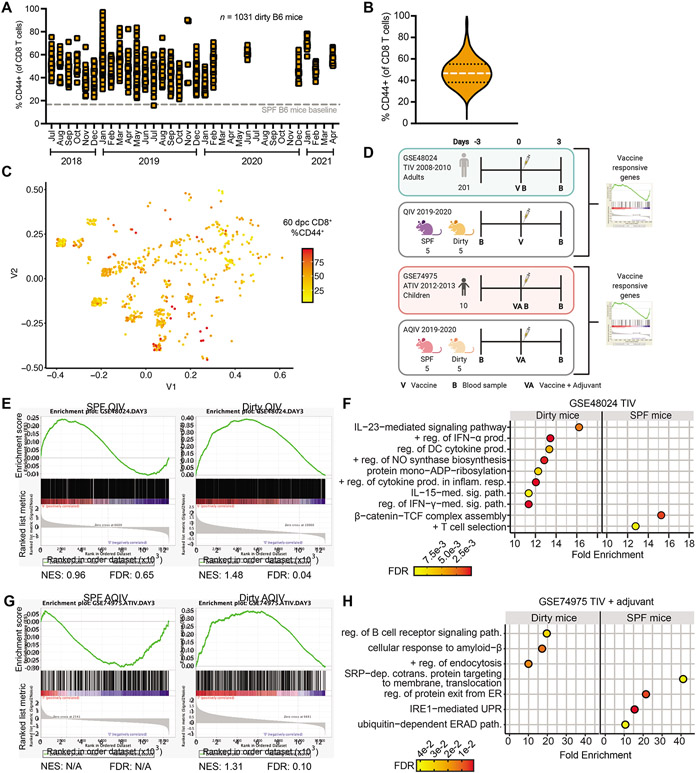
Cohousing adult pet store mice with inbred laboratory mice for 60 days results in the transmission of a diverse array of natural rodent microorganisms through physiological routes and doses, with profound alterations to the innate and adaptive immune systems (Beura et al., 2016; Hamilton et al., 2020; Huggins et al., 2019). All mice in our cohoused colony are screened for the activation state of CD8+ T cells, which we employ to provide a simple readout for microbial exposure. Given the potential heterogeneity in pathogens transmitted, and to ensure rigor and reproducibility of the immunological impacts of cohousing, we evaluated the impact of cohousing using mice from 3 different pet stores over a period of 34 months. There was a consistent influence on the immune system as measured by systemic CD8+ T cell activation levels (Figure 1A-B). Through serological testing, we uncovered 153 distinct combinations of pathogens transferred to the formerly SPF mice. However, no specific pathogen combination correlated with the increase in CD8+ T cell activation induced by cohousing (Figure 1C). One of the pathogens encountered, Mycoplasma pulmonis, is a chronic lung pathogen which could impact responses to influenza. We segregated mice out by Mycoplasma pulmonis status and found no impact on T cell activation or on any of our subsequent analyses (Figure S1A and data not shown). Males cannot be cohoused with pet store mice therefore we exposed males to contaminated bedding from pet store mice to determine the impact of transmission by fomites on basal immune status. Additionally, we cohoused another common laboratory strain, BALB/c. Both of these models demonstrated similar combinations of pathogens and impact on immune response (Figure S1B-C) suggesting that fomite transfer and other inbred strains can be used for these analyses. To further investigate any associations between pathogen exposure and T cell activation we used factor analysis of mixed data. This demonstrated that while some individual pathogens can contribute to differences between mice, the CD44 profile cannot be used to distinguish between animals (Figure S1B and D). These data support the consistency of the pet store cohousing approach and suggest that global exposure, rather than specific microbe combinations, drives a generalized immune experienced phenotype.
Figure 1: Cohousing better recapitulates vaccine-induced transcriptional signatures observed in humans.
(A and B) Mice from 3 different pet stores over a 34-month period were housed with SPF C57BL/6 mice. CD44 expression by bloodborne CD8+ T cells was determined 60 days after cohousing and (A) graphed by date of experiment or (B) volume plot of all animals combined, n=1031. 16.68% of CD8+ T cells expressed CD44 in age-matched SPF B6 mice (dotted line in A). (C) Multidimensional scaling plot of serology from cohoused mice at 60 days post cohousing, n=719. Distances represent similarities in past exposure to 18 pathogens. (D) Model demonstrating experimental design for comparing vaccinated SPF and dirty mice to vaccinated humans. (E and G) PBMCs were harvested on −3 and 3-days post vaccination with 2019-2020 QIV or AQIV. Vaccine responsive genes were generated comparing day 3 to −3 and these lists were queried humans vaccinated with TIV (GSE48024) or ATIV GSE74975 and compared by GSEA. Normalized enrichment score (NES) (F and H) Gene Ontology (GO) of mouse genes enriched in humans with and adjusted p value <0.01 using Panther. For (F) plotted GO terms had false discovery rate (FDR) <0.01 and fold enrichment (FE) >10. For (H) plotted GO terms had FDR <0.05 and FE >5.
Previous studies have demonstrated that exposure of SPF mice to microbes can result in baseline changes to the immune system that are more closely aligned to humans (Beura et al., 2016; Hamilton et al., 2020; Huggins et al., 2019; Reese et al., 2016; Rosshart et al., 2019; Rosshart et al., 2017; Takeda et al., 2019). Studies using SPF mouse models have often been derided for their ability to predict human responses to immune perturbations, including vaccines (Davis, 2008; Jameson and Masopust, 2018; Koff et al., 2013; Mestas and Hughes, 2004; Payne and Crooks, 2007; Rice, 2012; Rivera and Tessarollo, 2008; Seok et al., 2013; von Herrath and Nepom, 2005). Additionally, there are significant disparities in the response to vaccine adjuvants in SPF mice compared to humans and non-human primates (Bracci et al., 2006; Chioato et al., 2010; Cooper et al., 2004; Couch et al., 2009; Eisenbarth et al., 2008; Francica et al., 2017; Moldoveanu et al., 1998; Mosca et al., 2008; Proietti et al., 2002; Tregoning et al., 2018). Therefore, we hypothesized that dirty mice will better recapitulate the responses to vaccinations observed in people. To test this hypothesis, we transcriptionally profiled PBMCs from dirty and SPF mice prior to, and at 3-days post vaccination with the 2019/2020 influenza quadrivalent vaccine with and without adjuvant. To directly compare the response to humans we used gene set enrichment analysis (GSEA) (Mootha et al., 2003; Subramanian et al., 2005). We generated vaccine response genes from healthy adult humans vaccinated with trivalent vaccines from the 2007-2011 influenza seasons (Franco et al., 2013; Nakaya et al., 2011) and queried these gene sets against vaccine responsive genes from SPF and dirty mice from the same time point post vaccination (Figure 1D). This analysis demonstrated significant enrichment of human vaccine gene sets from dirty mice, but not SPF mice (Figure 1E). Genes driving this phenotype include many immune pathways, particularly those involved in cytokine signaling (Figure 1F). Similar results were found using data from two other independent human influenza vaccine studies (Nakaya et al., 2016; Nakaya et al., 2011) (Figure S1E-F). Additionally, we probed the response to AddaVax adjuvanted vaccine (as a surrogate for MF59) against human gene sets from pediatric patients vaccinated with MF59 adjuvanted trivalent vaccine from the 2012-2013 influenza season (Nakaya et al., 2016). GSEA also demonstrated that the human adjuvanted vaccine responsive genes are significantly in dirty mice and not in SPF mice, identifying B cell signaling pathway enrichment only in dirty mice (Figure 1G-H). Together these data suggest that dirty mice may provide a more reliable preclinical model in which to study the immune responses to influenza virus vaccines.
Cohousing does not alter disease following acute influenza infection.
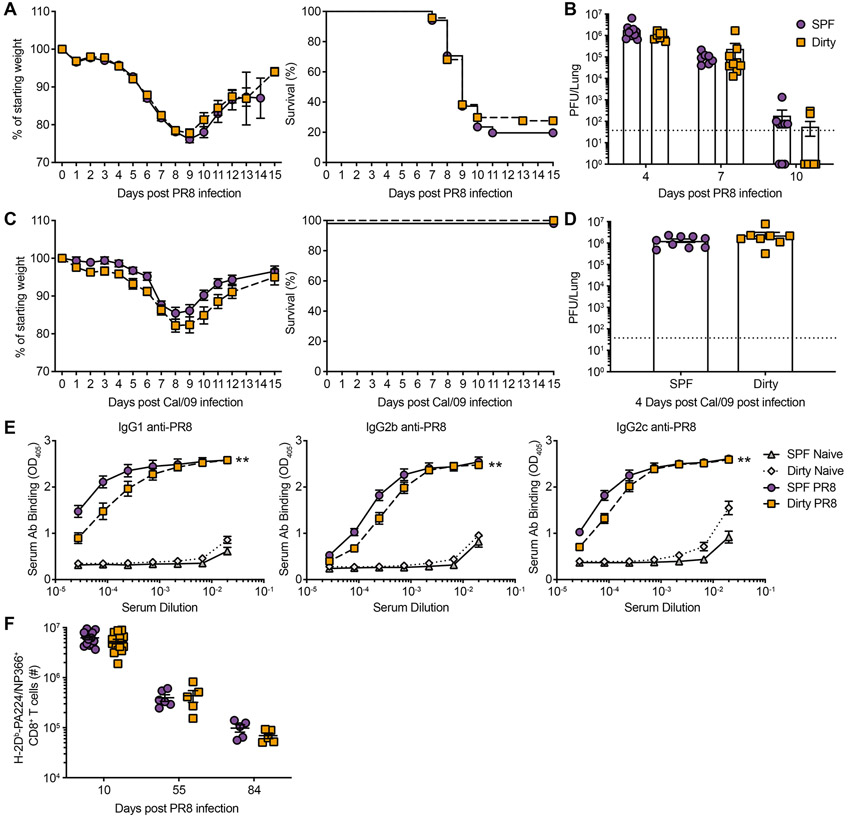
To determine how alterations in the basal immune state impact acute influenza infection, dirty and SPF mice were infected with mouse-adapted PR8 and pandemic H1N1 Cal/09 strains. Despite significant alterations in basal immunity between dirty and SPF mice, only minor differences in disease progression and viral loads were observed (Figure 2A-D). These data suggest that increased microbial experience and an altered immune system at baseline do not significantly impact outcome after acute IAV infection. We further evaluated the induction of IAV-specific adaptive immune responses to acute infection and observed only modest changes in IAV-specific antibodies (Figure 2E). Additionally, there were no significant alterations in IAV-specific CD8+ T cell numbers or surface molecule expression at acute or memory time points (Figure 2F and S2A-E). We also evaluated the CD4+ T cell response in the lung and draining lymph node at acute and memory time points. We did not observe any alterations in antigen-specific CD4+ T cell numbers or differentiation between dirty and SPF mice (Figure S2F-G). Together these data demonstrate that immune experience does not significantly impact the capacity to generate primary immune responses to pathogenic IAV infection, permitting evaluation of vaccine challenge experiments.
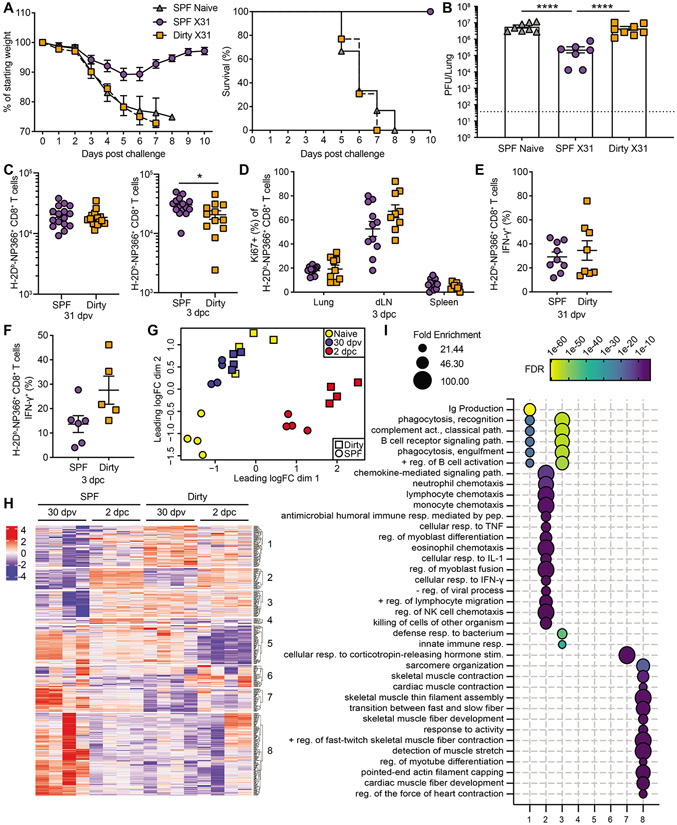
Figure 2: SPF and dirty mice exhibit similar primary responses to influenza virus.
Dirty and SPF mice were infected with either 40 PFU of PR8 (A to B and E to F) or 5000 PFU Cal/09 (C to D). Animals evaluated for weight loss (A and C) and pulmonary virus titers on indicated days post infection (dpi) (B and D). Dotted line, limit of detection (LOD) 37.5. At 50+ dpi, serum was assessed by ELISA to detect IgG1-, IgG2b- and IgG2c- PR8-specific antibodies (E). At 10, 55 and 84 dpi lungs were harvested and the number of H-2Db-PA224/NP366+ CD44+ CD8+ T cells from the lung was determined (F). Data (A to D) are representative of 4 and 2 independent experiments for PR8 and Cal/09, respectively with 4-9 mice per group. These data (E) are a combination of 5 independent experiments with at least 7 mice per group. These data (H) are a combination of 5 independent experiments with at least 5 mice per group. Significance (B, D, and F) was determined using student unpaired two-tailed t-test. Significance (E) was determined using AUC and one-way ANOVA. Error bars indicate mean ± SEM. *p < 0.05, ** p < 0.01, *** p < 0.001.
Reduced immunogenicity and efficacy of humoral vaccination in dirty mice
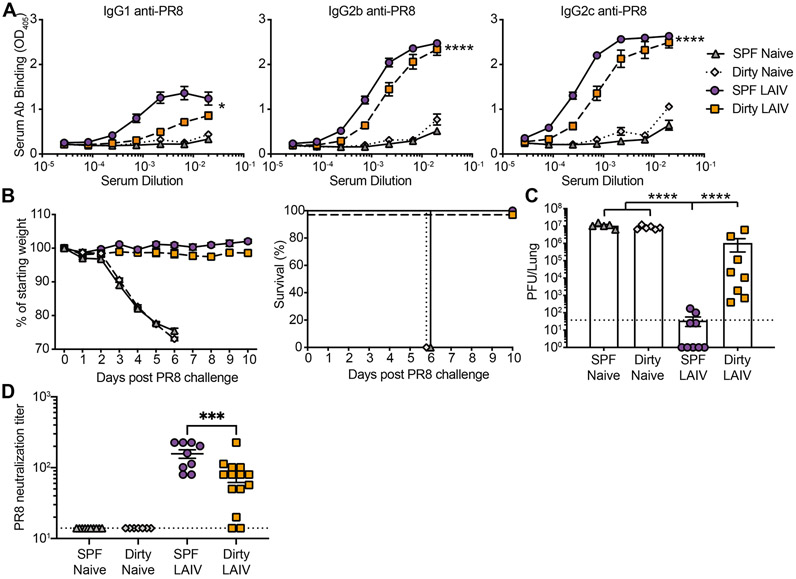
Humoral immunity is the primary target of seasonal IAV vaccination strategies. While sterilizing immunity is often used as a metric for vaccine efficacy in mice, this is rarely achieved in humans (Bouvier, 2018). To address vaccine-elicited humoral immunity, SPF and dirty mice were vaccinated intranasally with live attenuated influenza vaccine (LAIV) containing PR8-HA and -NA (Waring et al., 2018). IAV-specific circulating antibodies were evaluated 30 days post vaccination demonstrating a slight decrease in IAV-specific IgG2b and IgG2c in dirty animals (Figure 3A). We then challenged vaccinated dirty and SPF mice with a lethal dose of PR8. While we did not observe any differences in morbidity or mortality, there was a significant defect in generating sterilizing immunity in dirty mice (Figure 3B-C). Over three-quarters of vaccinated SPF mice had a viral load below the limit of detection three days after challenge, while all vaccinated dirty mice had detectable virus in the lungs. Consistent with these results, there was also a reduced capacity for serum antibodies to neutralize virus in dirty compared with SPF mice (Figure 3D). These results demonstrate a failure to attain sterilizing immunity in dirty mice even though it is readily achievable in SPF animals.
Figure 3: Reduced immunogenicity and efficacy of live attenuated vaccination in dirty mice.
SPF and dirty mice were untreated or vaccinated with 1000 PFU LAIV i.n. (A) Serum harvested at 30 days post vaccination (dpv) was assessed for anti-PR8 antibodies. (B) 30 days after vaccination mice were challenged with 1000 PFU PR8 and weight loss was monitored. Animals that lost >25% of starting weight were euthanized. (C) Pulmonary virus titer at 3 days post challenge (dpc). (D) Antibodies from (A) were evaluated for neutralization of PR8. Dotted line (C) LOD 37.5, (D) LOD 14. The data (A) are a combination of 2 independent experiments with at least 10 mice per group. The data (B-D) are a combination of 2 independent experiments with at least 6 mice per group. Significance (A) was determined using AUC and one-way ANOVA. Significance (C-D) was determined using one-way ANOVA. Error bars indicate mean ± SEM. *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
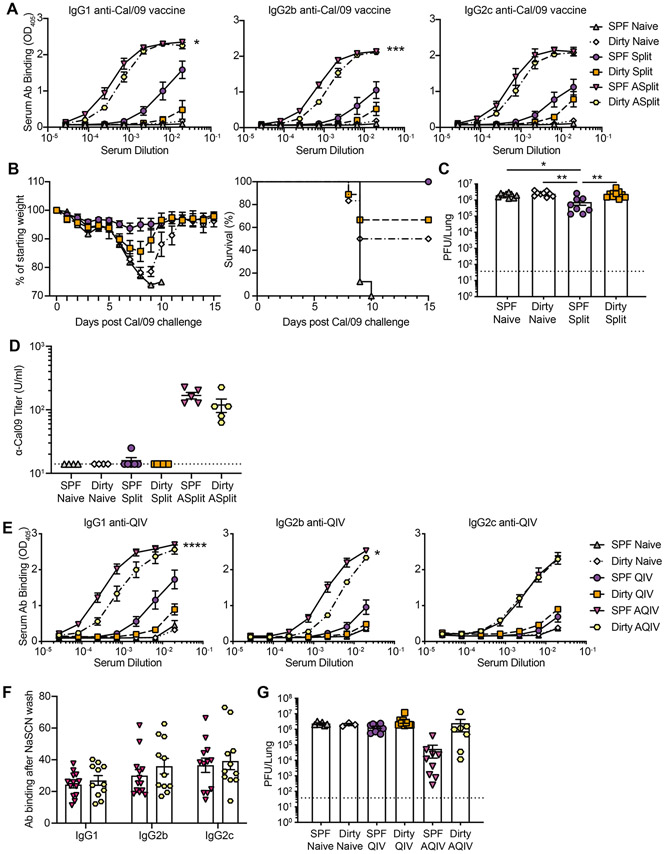
To determine if the defect in sterilizing immunity is specific to vaccination with a live attenuated vaccine, we evaluated killed split vaccines. The adjuvant MF59 enhances immunogenicity in children and the elderly and is critical for inducing protective responses to novel emerging HAs (Tregoning et al., 2018; Wilkins et al., 2017). However, there are significant disparities in the response to adjuvants in small animal models and humans and non-human primates (Chioato et al., 2010; Eisenbarth et al., 2008; Francica et al., 2017; Hornung et al., 2008; Mosca et al., 2008). Therefore, we evaluated the killed split Cal/09 vaccine with and without the adjuvant AddaVax (as a surrogate adjuvant for MF59) in SPF and dirty mice and measured serum antibodies analyzed at 30 days post vaccination. There was a modest reduction in vaccine-specific antibodies in dirty mice (Figure 4A). While both dirty and SPF mice were protected following challenge (Figure 4B), dirty mice had significantly higher viral burden, similar to unvaccinated animals (Figure 4C). We also evaluated the neutralization potential of vaccine specific antibodies demonstrating a slight reduction in dirty mice (Figure 4D). We also evaluated the 2019/2020 seasonal quadrivalent vaccine with and without the adjuvant AddaVax in SPF and dirty mice. Similar to the Cal/09 split vaccine there was a significant defect in the humoral response to vaccine alone in dirty mice that could not be overcome with an adjuvant boost (Figure 4E). To determine if there were differences in anti-vaccine antibody avidity between SPF and dirty mice, we evaluated antibody binding with and without chaotropic agents. These analyses demonstrated no differences in vaccine-specific antibody avidity between SPF and dirty mice (Figure 4F). To determine if the vaccine can control virus replication, we challenged seasonal 2019/2020 vaccinated mice with Cal/09. Because Cal/09 is only partially matched with the 2019/2020 seasonal vaccine this may model seasonal infections with viruses that poorly match the strains chosen for vaccine production. Quadrivalent vaccination failed to reduce viral titers in both SPF and dirty mice (Figure 4G). However, adjuvant boost significantly reduced viral titers in SPF but not dirty mice. Finally, we evaluated the humoral response to the 2019/2020 seasonal vaccine in two analogous models, cohoused BALB/c mice and male C57BL/6 mice housed with the bedding from pet store animals. BALB/c and C57BL/6 mice display differences in Th1/Th2 profiles as well as subclasses of antibodies generated. IgG subclasses can have significant impact on the non-neutralizing functions of antibodies during influenza infection which could impact responses to vaccination (Corti et al., 2011; DiLillo et al., 2016; DiLillo et al., 2014). Both of these approaches drive similar pathogen combinations as cohousing of C57BL/6 mice (Figure S1C). Importantly, both models elicit similar vaccine-specific antibody profiles as cohoused C57BL/6 mice, suggesting that reduced immunogenicity is broadly impacted by diverse immune histories (Figure S3). Together these data suggest that SPF mice have exaggerated humoral immune responses to live, killed split, and adjuvanted influenza vaccines.
Figure 4: Reduced immunogenicity and efficacy of split killed vaccination with and without adjuvant in dirty mice.
(A to D) SPF and dirty mice were untreated or vaccinated with Cal/09 split vaccine with or without AddaVax i.m. (A) Serum harvested at 30 dpv was assessed for anti-Cal/09 vaccine antibodies. (B) 30 dpv mice were challenged with 30,000 PFU Cal/09 and weight loss was monitored. (C) Pulmonary virus titer at 3 dpc. (D) Microneutralization. (E to G) SPF and dirty mice were untreated or vaccinated with 2019-2020 QIV with or without AddaVax i.m. (E) Serum harvested at 30 dpv was assessed for anti-QIV antibodies (F) Antibody avidity from (E) measured after exposure to chaotropic NaSCN. (G) 30 days after vaccination mice were challenged with 75,000 PFU Cal/09. Pulmonary virus titer at 3 dpc. Dotted line (C, and G), LOD 37.5. The data (A) are representative of 2 independent experiments with at least 6 mice per group. The data (B-G) are a combination of 2 independent experiments with at least 6 mice per group. Significance (A and E) was determined using AUC and one-way ANOVA. Significance (C-D and F-G) was determined using one-way ANOVA. Error bars indicate mean ± SEM. *p < 0.05, ** p <0.01, *** p < 0.001, **** p < 0.0001.
Memory responses fail to protect against heterologous challenge
While humoral immunity is the main target of current influenza vaccinations, T cells can potentially provide broad protection and have been the focus of some universal vaccine strategies. Dirty and SPF mice were primed with X31 and challenged with PR8; this classic vaccine strategy exploits conserved internal viral T cell epitopes while avoiding the major neutralizing antibody targets (Effros et al., 1977; Liang et al., 1994; Webster and Askonas, 1980). While vaccination of SPF mice led to survival and reduced viral burden after challenge, protection failed in vaccinated dirty mice (Figure 5A-B). To determine the mechanism driving this disparity, we evaluated memory CD8+ T cell responses before and after challenge. Surprisingly, cohousing did not drive any significant defect in antigen-specific CD8+ T cell numbers, proliferation, capacity to secrete effector molecules IFNγ or TNFα, or markers of residency or activation (CD103 and PD-1) in the lungs, although a larger proportion of antigen-specific T cells were in the lung parenchyma in dirty mice (Figure 5C-F and S4A-H). Additionally, there were no significant alterations to antigen-specific CD4+ T cells numbers or differentiation status after recall (Figure S4I-J). Histological examination demonstrated increased influenza-associated necropurulent bronchiolitis in vaccinated dirty mice after lethal challenge (Figure S5A-H). Together, these data suggest that the increased disease in vaccinated dirty mice after challenge is due to an immunological failure to control the infection and not a result of increased immunopathology. To determine if the increased lethality in dirty mice is due to an antigen-specific failure to protect or an increased susceptibility to a secondary pulmonary infection we primed mice with X31 and challenged with antigenically distinct influenza B virus (IBV). There was no difference in morbidity or mortality after IBV infection suggesting that dirty mice are not more susceptible to sequential pulmonary challenges due to alterations in innate immune responses or failure to repair damage after the primary infection (Figure S5I). Finally, in an effort to determine if CD8+ T cells alone were responsible for the failure in protection we attempted to deplete CD8+ T cells from SPF and dirty mice. However, even with high doses of antibody we were unable to achieve sufficient depletion in dirty mice (Figure S5J). This is likely due to the human-like high levels of basal memory T cells in the tissues in dirty mice and represents an experimental limitation of the cohousing system.
Figure 5: Memory CD8+ T cells fail to protect dirty mice upon influenza virus challenge.
SPF and dirty mice were untreated or vaccinated with 1000 PFU X31 and were challenged after 30 days with 1000 PFU PR8. Animals were evaluated for weight loss (A) and pulmonary virus titers on 3 dpc (B). Dotted line, LOD 37.5. (C) Number of H-2Db-NP366+ CD8+ T cells in the lung at 30 dpv (left) or 3 dpc (right) (D) On 3 dpc, the percentage of H-2Db-NP366+ CD8+ T cells that are Ki67+ from the lung, mediastinal lymph node (mLN) and spleen. (E to F) IFN-γ expression H-2Db-NP366+ CD8+ T cells was determined 31 dpv ex vivo (E), or 3 dpc in vivo (F). (G to I) Whole lungs were harvested and RNA was extracted for RNA-seq from untreated, 31 dpv and 2 dpc SPF and dirty mice. Multidimensional scaling plot demonstrating transcript alterations between groups (G). Heatmap of differentially expressed genes (H). (I) Gene ontology analysis was performed using Panther on genes in each cluster with an adjusted p value <0.01 and LogFC >0.5, FDR <0.01, and FE >20. The data (A-E) are combined from 2-3 independent experiments with at least 6 animals per group. The data (F) are from 1 of 2 representative experiments. Significance (B) was determined using one-way ANOVA. Significance (C to F) was determined using student unpaired two-tailed t-test. Error bars indicate mean ± SEM. *p < 0.05, ** p < 0.01, *** p < 0.001, **** p <0.0001.
To globally evaluate the failure to control challenge in dirty mice, we profiled the transcriptome from whole lung of SPF and dirty mice at baseline, after vaccination, and two days post challenge. Multidimensional scaling demonstrates significant differences between SPF and dirty mice at baseline and post challenge (Figure 5G). SPF and dirty mice have similar transcriptional profiles after vaccination. Interestingly, after vaccination of SPF mice the transcriptional profile is similar to unvaccinated dirty mice at baseline, suggesting that both infection with IAV and natural mouse pathogens from cohousing, drive fundamental changes to the transcriptional response in the lungs after resolution of the infection. To further interrogate the differences between dirty and SPF mice, we evaluated the interaction effect of cohousing on the response to challenge. Semi-supervised clustering demonstrated distinct gene expression patterns impacted by cohousing (Figure 5H). Gene ontology of these clusters identified that cohousing leads to muted B cell, chemotaxis, phagocytic, and other innate immune responses after vaccination and challenge (Figure 5I). These results show that multiple arms of the immune system are leveraged in SPF mice following infection and are not employed to the same degree in dirty mice, potentially leading to reduced capacity to control the infection. While CD8+ T cells have been shown to correlate with protection in humans (Grant et al., 2016), SPF mice exaggerate protection from disease. These data suggest that it may be better to test strategies aimed at exploiting CD8+ T cell immunity in dirty mice rather than SPF mice, as the latter model may fail to translate into humans, while dirty mice may improve translational success.
Discussion
Mice are often used as the first model for the assessment of vaccine immunogenicity and are also used to identify correlates of protection. Unfortunately, studies utilizing traditional SPF mice do not always serve as a faithful predictor of immune responses in humans. Several recent studies have demonstrated that mouse models with complex immune histories better recapitulate human immune responses and can predict responses to immune-based therapies that were successful in SPF mice but failed in humans (Beura et al., 2016; Reese et al., 2016; Rosshart et al., 2019; Rosshart et al., 2017; Takeda et al., 2019). Additionally, a sequential infection strategy to generate mice with diverse infections, similar to dirty mice, demonstrated subdued antibody responses to yellow fever vaccine compared to standard mice, which more closely resembled human responses (Reese et al., 2016). Here we use influenza vaccines as a model to evaluate the differences in vaccine responses between standard SPF mice and dirty mice with complex immune histories. We demonstrate that dirty mice better recapitulate transcriptional signatures observed after human vaccinations. Importantly, we further show that dirty mice have altered humoral responses to influenza vaccines. While SPF mice mount robust responses, dirty mice are more muted and better phenocopy the cellular, humoral, and transcriptional responses observed in humans. These alterations are consistent across two major inbred mouse strains and are independent of the combination of transmitted microbes. We hypothesize that alterations in baseline immune cell activation profiles and composition impact the magnitude of subsequent immune responses preventing robust responses to vaccination. This hypothesis is supported by the observation that vaccines are often less efficacious in individuals from developing countries where the number of previous microbial encounters is increased compared to people in developed areas of the world (Gil et al., 2015; Levine, 2010; Lopman et al., 2012; Parker et al., 2018). The reasons for this shortfall in protection are likely complex, but elevated inflammatory profiles have been noted to detrimentally impact germinal center formation and the development of antibody responses (Matar et al., 2015; Ryg-Cornejo et al., 2016). Whether the persistent elevation in cytokines observed in dirty mice or the numerous changes in immune cell numbers and activation status (Beura et al., 2016; Huggins et al., 2019) is ultimately responsible for altered adaptive immunity is unknown but should be investigated in future work.
Adjuvants can increase the efficacy of influenza vaccines in high-risk populations. Despite this, currently, there is only one approved adjuvant for seasonal influenza vaccines (CDC.gov). It has been well established that adjuvants in SPF mice do not drive the same responses in humans, presenting a major hurdle to testing of novel vaccine adjuvant-antigen combinations (Bucasas et al., 2011; Caproni et al., 2012; Francica et al., 2017; Mosca et al., 2008; Obermoser et al., 2013). Importantly, here we demonstrate that AddaVax in combination with an influenza vaccine drives responses in dirty mice that are more similar to humans than SPF animals, potentially bridging the gap between mice and humans for evaluation of adjuvants. Additionally, we found that vaccine responsive genes from dirty mice are profoundly different from SPF mice and were more enriched in human influenza vaccine data included IFNγ, IFNα, and nitric oxide production, which have been shown to correlate with stronger antibody responses in people after influenza vaccination (Li et al., 2014; Nakaya et al., 2011). These data suggest that the pathways driving antibody responses in environments that severely limit microbial exposure may be fundamentally different from those in dirty mice and humans.
Seasonal influenza vaccination strategies aim to generate robust, ideally sterilizing, humoral immunity. However, sterilizing immunity to seasonal IAV vaccines is not always achieved in humans (Bouvier, 2018). We demonstrated that dirty mice were unable to induce sterilizing immunity in response to LAIV but were protected from morbidity and mortality after lethal challenge. Therefore, additional correlates of protection should be assessed beyond sterilizing immunity when evaluating vaccine candidates. Initial experiments could be performed in dirty mice, instead of more expensive larger animal models or human studies where access to mucosal tissues and reagents for assessment are limited.
Cell-mediated immune responses are the target of some universal influenza vaccine strategies that focus on conserved regions of the virus that do not significantly vary from season to season. Studies in mice have demonstrated CD8+ T cells can provide potent heterosubtypic protection between IAV strains, but the data in humans is less clear (Clemens et al., 2018). Dirty mice responded similarly to SPF mice in the generation and phenotype of IAV-specific CD8+ T cells and did not exhibit lung immunopathology after challenge. But, our cellular protection experiments showed that dirty mice were not protected from viral challenge. Given these data, dirty mice might serve as a better model to evaluate preclinical vaccines targeting cell-mediated immunity.
There are several distinct model systems aimed at providing SPF with natural microbiomes and/or infection histories. One strategy uses laboratory mice reconstituted with wild mice microbiota lacking SPF-banned pathogens (Rosshart et al., 2017). Interestingly infection in this model led to different outcomes during acute influenza A virus infection than we observed here (Rosshart et al., 2017). Previous work from members of our group have demonstrated that at baseline there are similar differences in microbiome between SPF and dirty mice versus wild reconstituted and lab mice with both models seeing increased abundance of Proteobacteria, Bacteroidetes and decreased Verrucomicrobia (Huggins et al., 2019; Rosshart et al., 2017). However, despite the similarities in the microbiome between dirty mice and wild reconstituted mice there are significant differences in basal cytokine levels. This includes proinflammatory cytokines/chemokines TNF, CXCL10, CCL4, and IL-6 which are all increased in dirty compared to SPF and, conversely, are decreased in wild reconstituted lab compared to SPF mice. Together these data demonstrate fundamental differences between these two attempts to normalize microbial experience and suggest that additional microbial communities, including potential pathogens, may be responsible for these differences. Peripheral immune responses can also be dramatically impacted by exposure to environmental acquired microbes (Lin et al., 2020; Yeung et al., 2020). The complex trans-kingdom environment in free living mammals, which has been excluded in traditional SPF laboratory mice, can significantly impact the development and function of the immune system. Together these complementary models provide opportunities to rigorously evaluate how more natural microbial exposure impacts immune responses.
Due to introduction and transmission of natural mouse pathogens this model necessitates isolation from standard SPF animals. We approached this by housing the animals in a BSL3 facility. While exceeding the required biosafety level for the pathogens present this ensured protection for the SPF mouse colonies on campus. This approach has been successfully adopted by several other groups (Choi et al., 2019; Takeda et al., 2019). An alternative strategy to protect SPF animals is to perform cohousing in a facility that is not connected to any other animal husbandry. These different approaches and sources of pet store animals will almost certainly lead to different infection profiles. However, as we demonstrate here the impact of the immune response is independent of any particular pathogen or combination of pathogens. Therefore, the variation in transmitted pathogens across pet stores from different locations will likely not impact subsequent responses to immunologically distinct pathogens or vaccines. Additionally, key findings were recapitulated in another inbred mouse strain and by using contaminated bedding instead if cohousing, further highlighting the plasticity of this model.
While dirty mice better recapitulate key immune signatures observed in humans, they still present some limitations for evaluating vaccines. There are genetic differences between mice and humans that could impact vaccine responses, particularly for adjuvants that target TLR8 (Heil et al., 2004). There are also differences in virus tropism and pathogenesis and some virus strains need to be mouse adapted to achieve infection. Additionally, the short life span of mice makes it difficult to evaluate preexisting influenza immunity and imprinting. Dirty mice would serve as a good model for immunologically naïve children, or adults infected with a novel strain, that are mounting responses against IAV antigens for the first time. Despite these limitations, dirty mice can serve as an alternative to standard SPF mice to increase the translation potential of vaccine candidates. The current global pandemic driven by SARS-CoV-2 highlights the critical importance of having animal models that faithfully recapitulate immunogenicity in humans where rapid translation to the clinic is essential.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ryan A. Langlois (langlois@umn.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
RNA-seq data have been deposited at GEO (GSE182858) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All (serology, flow cytometry, ELISA, titering, morbidity and mortality, ELISA and neutralization) data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD8β antibody (clone lyt3.2) | Bio X Cell | BE0223 RRID: AB_2687706 |
| CD49a (clone Hα31/8) | BD Biosciences | 740262 RRID: AB_2740005 |
| RORγt (clone Q31-378) | BD Biosciences | 562894 RRID: AB_2687545 |
| B220 (clone cRA3-6B2) | Biolegend | 103255 RRID: AB_2563491 |
| CD4 (clone GK1.5) | Biolegend | 100447 RRID: AB_2564586 |
| CD4 (clone GK1.5) BUV395 | BD Biosciences | 563790 RRID: AB_2738426 |
| CD8-β (clone YTS156.7.7) | Biolegend | 126610 RRID: AB_2260149 |
| CD44 (clone IM7) | Tonbo | 65-0441-U100 RRID: AB_2621891 |
| CD44 (clone IM7) | BD Biosciences | 563736 RRID: AB_2738395 |
| CD45 (clone 104) | Tonbo | 60-0451-U100 RRID: AB_2621848 |
| CD45 (clone 30-F11) vF450 | Tonbo | 75-0451-U100 RRID: AB_2621947 |
| CD45 (clone 30-F11) FITC | eBioscience | 11-0451-81 RRID: AB_465049 |
| CD62L (clone MEL-14) | Biolegend | 104440 RRID: AB_2629685 |
| CD69 (clone H1.2F3) | Biolegend | 104512 RRID: AB_493564 |
| CD103 (clone 2E7) | Biolegend | 121414 RRID: AB_1227502 |
| CXCR5 (clone L138D7) | Biolegend | 145517 RRID: AB_2562453 |
| F4/80 (clone BM8) | Biolegend | 123147 RRID: AB_2564588 |
| IFN-γ (clone XMG1.2) | Biolegend | 505826 RRID: AB_2295770 |
| Ki67 (clone 16A8) | Biolegend | 652411 RRID: AB_2562663 |
| KLRG1 (clone 2F1/KLRG1) | Biolegend | 138416 RRID: AB_2561736 |
| PD-1 (clone 29F.1A12) | Biolegend | 25-5982-80 RRID: AB_2573508 |
| T-bet (clone 4B10) | Biolegend | 644814 RRID: AB_10901173 |
| TCR-β (clone 4B10) | eBioscience | 47-5961-82 RRID: AB_1272173 |
| TCR-β (clone H57-597) BV510 | BD Biosciences | 563221 RRID: AB_2738078 |
| TNF-α (clone MP6-XT22) | Biolegend | 506308 RRID: AB_315429 |
| CD45.1 (clone A20) | eBioscience | 47-0453-82 RRID: AB_1582228 |
| Foxp3 (clone FJK-16s) | eBioscience | 53-5773-82 RRID: AB_763537 |
| I-A/I-E (clone M5/114.15.2) | Biolegend | 107639 RRID: AB_2565894 |
| CD8-α (clone 53-6.7) | Tonbo | 35-0081-U100 RRID: AB_2621671 |
| CD8-α (clone 53-6.7) BUV737 | BD Biosciences | 612759 RRID: AB_2870090 |
| HRP-anti-mouse IgG1 | Southern Biotech | 1070-05 RRID: AB_2650509 |
| HRP-anti-mouse IgG2b | Southern Biotech | 1090-05 RRID: AB_2794521 |
| HRP-anti-mouse IgG2c | Southern Biotech | 1079-05 RRID: AB_2794466 |
| HRP-anti-mouse IgG2a | Southern Biotech | 1080-05 RRID: AB_2734756 |
| Polyclonal Anti-Influenza Virus, A/Puerto Rico/8/1934 (H1N1) (antiserum, Rooster) | BEI Resources, NIAID, NIH | V301-511-552, NR-3098 |
| HRP rabbit anti-chicken IgG | Jackson Immuno Research | 303-035-003 RRID: AB_2339290 |
| Bacterial and virus strains | ||
| IAV/PR8 | In house | N/A |
| IAV/Cal/09 | In house (Hai et al., 2010) | N/A |
| LAIV_PR8 | In house (Waring et al., 2018) | N/A |
| IAV/X31 | In house | N/A |
| IBV/Mal/04 | In house (Hamilton et al., 2016) | N/A |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| H-2Db-PA224 (SSLENFRAYV) tetramer | NIH Tetramer Core Facility | N/A |
| H-2Db-NP366 (ASNENMETM) tetramer | NIH Tetramer Core Facility | N/A |
| I-Ab NP311 (QVYSLIRPNENPAHK) tetramer | In House | N/A |
| Influenza A (H1N1) 2009 Monovalent Vaccine | BEI Resources | NR-20347 |
| 2019-2020 quadrivalent influenza vaccine | Sanofi Pasteur | NDC 49281-719-10 |
| AddaVax | InvivoGen | Cat# vac-adx-10 |
| NP366 (ASNENMETM) peptide, no N or C terminal modifications | Biosynn | N/A Custom |
| collagenase type I | Worthington | Cat# LS004196 |
| ABTS Peroxidase Substrate | SeraCare | Cat# 5120-0043 |
| TrueBlue peroxidase substrate | Kirkegard & Perry Laboratories | Cat# 50-674-28 |
| Ghost Dye Red 780 | Tonbo | 13-0865-T100 |
| Cytofix/Cytoperm Fixation/Permeablization Kit | BD Biosciences | Cat# 554714 RRID: AB_2869008 |
| Foxp3 / Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 |
| GolgiPlug Protein Transport Inhibitor | BD Biosciences | Cat# 555029 RRID: AB_2869014 |
| Critical commercial assays | ||
| EZ-spot and PCR Rodent Infections Agent (PRIA) array methods | Charles River Laboratories | |
| AllPrep DNA/RNA kit | QIAGEN | Cat# 80204 |
| RNeasy Micro Plus Kit | QIAGEN | Cat# 74034 |
| Deposited data | ||
| Mouse PBMC RNAseq after (A)QIV, Raw data files | This paper | GEO: GSE182858 |
| Mouse whole lung RNAseq, Raw data files | This paper | GEO: GSE182858 |
| Customized gene set | This paper | c7.custom.immune.datasets.gmt |
| Time series of global gene expression after trivalent influenza vaccination in humans, human expression profiling by array | (Franco et al., 2013) | GEO: GSE48024 |
| Systems biology of immunity to MF59-adjuvanted versus non-adjuvanted trivalent seasonal influenza vaccines in early childhood, human expression profiling by array | (Nakaya et al., 2016) | GEO: GSE74975 |
| Systems biology of vaccination for seasonal influenza in humans, human expression profiling by array | (Nakaya et al., 2011) | GEO: GSE29619 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat# CRL-3216 |
| MDCK (NLB-2) | ATCC | Cat# CCL-34 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: C57BL/6 | Charles River | Strain code: 027 |
| Mouse: BALB/c | Charles River | Strain code: 028 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| FlowJo™ Software For Mac | Becton, Dickinson and Company | Version 9.9.6 |
| GraphPad Prism 9 for macOS | GraphPad Software | Version 9.2.0 (283) |
| R | https://www.r-project.org | Version 4.1.0 |
| Bowtie aligner | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | bowtie2 version 2.3.4.1 |
| Subread software package | http://subread.sourceforge.net | version 1.5.1 |
| bioconductor package edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | version 3.24.3 |
| FactoMineR package | https://CRAN.R-project.org/package=FactoMineR | Version 2.4 |
| Adobe Illustrator | Adobe | Version 25.4.1 |
| stats R package | https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html | Version 4.1.0 |
| ggplot2 R package | https://ggplot2.tidyverse.org | Version 3.3.5 |
| Other | ||
This paper does not report original code.
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice.
Pet store mice were purchased from various Twin Cities area pet stores. Pet store mice were cohoused with 8-week-old female C57BL/6 or BALB/c mice (The Jackson Laboratory or Charles River Laboratories) or dirty bedding from pet store mice was transferred into the cages of male C57BL/6 mice. Cohousing occurred within a BSL-3 facility. C57BL/6 mice were cohoused for 60 days, bled for flow cytometry analysis and screened for infectious agents using EZ-spot and PCR Rodent Infections Agent (PRIA) array methods (Charles River Laboratories). Age-matched mice were maintained in SPF facilities. Males cannot be cohoused as this creates animal welfare concerns due to fighting, aggression, and social defeat. Therefore, we can ethically only use female mice for cohousing experiments. Male mice were used for bedding transfer fomite experiments. Care and use of the animals was in accordance with The Guide for the Care and Use of Laboratory Animals from the National Research Council and the USDA Animal Care Resource Guide. All experimental protocols involving the use of mice were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Viruses.
Viruses (PR8, X31, PR8-LAIV and IBV/Mal/04) were rescued via HEK293T transfection and amplified in embryonated chicken eggs as previously described (Langlois et al., 2012), (Waring et al., 2018), (Hamilton et al., 2016). Eggs were obtained from Charles River Laboratories and were grown at 37°C until 12 days of embryonation. Cal/09 was rescued via HEK293T transfection and amplified in Madin-Darby canine kidney (MDCK) (ATCC) cells as previously described (Hai et al., 2010). Rescued viruses were sequence confirmed and titered on MDCK cells.
Cell lines and maintenance.
HEK293T and MDCK cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS and 1% pen-strep and grown at 37°C. Both HEK293T and MDCK cells derived from female donors. All cell lines were tested for mycoplasma contamination using LookOut Mycoplasma PCR Detection Kit (Sigma-Aldrich, St. Louis, MO, USA) and resulted mycoplasma-free. Cell lines purchased from ATCC as gift were not authenticated since they were purchased from ATCC.
METHOD DETAILS
Infections and vaccinations.
For IAV infections, mice were anesthetized using a weight-based dose of ketamine/xylazine delivered intraperitoneally (i.p.). Mice were infected intranasally (i.n.) with 40 plaque forming units (PFUs) of PR8, or 5,000 PFU of Cal/09. Mice were vaccinated i.n. with 1000 PFU of X31 or 1000 PFU of PR8-LAIV, or vaccinated intramuscularly (i.m.) with 1.5 μg of Cal/09 split vaccine (BEI Resources, NIAID, NIK: Influenza A (H1N1) 2009 Monovalent Vaccine, NR-20347), or 180 μg of 2019-2020 seasonal quadrivalent influenza vaccine (Sanofi Pasteur) with or without 25 μL AddaVax (InvivoGen). For challenge, mice were infected i.n. with 1000 PFU of PR8, 30,000 to 75,000 PFU of Cal/09, or 1000 PFU of IBV/Mal/04. During infection, all mice having weight loss exceeding 25% of their starting weight were sacrificed. For CD8+ T cell depletion experiments, X31 memory mice were injected with 300 μg of anti-CD8β antibody (lyt3.2, Bio X Cell) at days −6, −5, −3 and −2 prior to harvest.
Mouse PBMC RNAseq and GSEA.
Mouse PBMCs were isolated using Ficol®-Paque PREMIUM (Millipore Sigma) and RNA was extracted using RNeasy Micro Plus Kit (Qiagen). The cDNA library was prepared using strand-specific RNA-sequencing protocols. Samples were run on an Illumina NovaSeq (150 bp paired-end). We obtained an average of 35 million read pairs per sample. Sequencing reads were mapped to the mouse genome (GRCm38) using Bowtie aligner (bowtie2 version 2.3.4.1) with local mode, -L 22 and -N 1 parameters (Langmead and Salzberg, 2012). Reads were assigned to Ensembl gene models (Mus_musculus.GRCm38.87.gtf) with featureCounts of the Subread software package (version 1.5.1) (Liao et al., 2014). The reads count matrices were organized corresponding to experimental design and used for subsequent statistical analysis using the bioconductor package edgeR (version 3.24.3) (McCarthy et al., 2012; Robinson et al., 2010). The raw reads count table were normalized by using default method in the package prior to generating statistics. The normalized reads table of mouse blood samples were reformatted to meet the requirements for the subsequent GSEA analysis (Mootha et al., 2003; Subramanian et al., 2005). The gene lists were prepared from human vaccination data and merged into an immune geneset (c7.immune.datasets.gmt) to generate a customized geneset (c7.custom.immune.datasets.gmt) (Mootha et al., 2003; Subramanian et al., 2005). The mouse datasets originated from either SPF or dirty mouse models were computed against the customized geneset by using desktop GSEA analysis engine with default parameters except the enrichment plot generation. Sequencing data were deposited under Gene Expression Omnibus series accession number GSE182858.
Flow cytometry and reagents.
Single cell suspensions were washed with 1 X PBS and stained with a fixable viability dye for 30 min on ice, Ghost Dye™ Red 780 (Tonbo). Cells were washed once with FACS buffer (cold HBSS supplemented with 2% bovine serum), stained with surface Abs, then washed before flow cytometric detection on a BD LSRFortessa (Becton Dickinson). For ex vivo IFN-γ staining, lung single cell suspensions were incubated in complete T cell media with/without 1 μg/mL NP366 peptide for 4 h at 37°C in the presence of GolgiPlug (BD Biosciences). For positive intracellular staining controls, cells were stimulated with eBioscience™ Cell Stimulation Cocktail. Cells were washed 2x with FACS buffer and stained as above. For in vivo IFN-γ staining, mice were challenged with 1000 PFU of PR8 i.n., lungs were harvested (see below) at 3 dpc and single cell suspensions were generated in the presence of GolgiPlug (BD Biosciences). For intracellular staining cells were fixed with BD Cytofix/Cytoperm (BD Biosciences), incubated on ice for 30 min, washed 2x with 1 X BD Perm/Wash buffer, then incubated with Abs for 30 min. Cells were washed 2x with 1 X BD Perm/Wash buffer and resuspended in FACS buffer. Complete T cell media consisted of RPMI 1640 with 10% FBS, 4 mM L-glutamate, 0.1mM nonessential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin and streptomycin, 10 mM HEPES, and 5 mM 2-ME. IAV-specific cells were gated as Live/Dead−, Dump− (F4/80, CD4, B220), CD8+, H-2Db-PA224+ or H-2Db-NP366+.
Isolation of lymphocytes from the lung.
To discriminate parenchymal cells from blood-borne cells, mice were given an intravenous (i.v.) injection of anti-CD8α or anti-CD45 (3 μg diluted in 200 μl PBS) for 3 min, as described (Anderson et al., 2014). Mice were euthanized and spleen, draining parenchymal lymph nodes and lung were harvested. Tissues were minced and washed 2x with harvest buffer (cold RPMI 1640 supplemented with 5% bovine serum, 4 mM L-glutamate and 10 mM HEPES). Lungs were incubated in a solution of RPMI 1640/ 10% bovine serum/ 2 mM MgCl2/ 2mM CaCl2/ 10 mM HEPES/ 4 mM L-glutamate medium containing 100 U/mL of collagenase type I (Worthington) for 45 min at 37°C. Lung pieces were then incubated in a solution of RPMI 1640/10% bovine serum/10 mM HEPES/4 mM L-glutamate medium containing 1.3 mM EDTA (Calbiochem) for 45 min at 37°C. Single cell suspensions of all tissues were generated and stained for flow cytometry as described above.
Histology.
On indicated days post infection/vaccination lungs were harvested and fixed in 4% paraformaldehyde in PBS. 1 week post fixation, samples were transferred to the Comparative Pathology Laboratory (University of Iowa Carver College of Medicine). Tissues were routinely processed, paraffin-embedded, sectioned (~4 μm) onto glass slides and stained with hematoxylin and eosin (H&E). Tissues were evaluated by a boarded veterinary pathologist using a post-examination method of masking to group assignments (Meyerholz and Beck, 2018).
Serum antibody detection by ELISA.
On indicated days post infection/vaccination, mice were bled and serum was isolated. 96 well plates were coated with a 1:25 dilution of UV-killed PR8, a 1:100 dilution of the Influenza A (H1N1) 2009 Monovalent Vaccine (BEI Resources) or a 1:250 dilution of the 2019-2020 quadrivalent influenza vaccine (Sanofi Pasteur) diluted in PBS. All antigen coated plates were blocked with 1% BSA in PBS prior to addition of serum. Serial dilutions were added to coated and blocked plates and bound Ig was detected with HRP-anti-mouse Ig antibodies (IgG1, IgG2b and IgG2c) (Southern Biotech) followed by ABTS Peroxidase Substrate (SeraCare). OD405 was detected by a Synergy H1 plate reader (BioTek). For chaotropic ELISAs, serum bound plates were incubated in 1.5M NaSCN for 15 min. Plates were washed prior to addition of HRP-anti-mouse Ig antibodies and treated as above.
Plaque assay.
Infections of MDCK cells were carried out in infection medium PBS with 10% CaMg, 1% pen-strep, 5% bovine serum albumin) at 37°C for 1 hr. Infection medium was replaced with an agar overlay (MEM, 1 mg/mL tosyl_sulfonyl phenylalanyl chloromethlyl ketone trypsin, 1% DEAE-dextran, 5% NaCO3, 2% agar), and cells were cultured at 37°C for 40 h and then fixed with 4% formaldehyde. Blocking and immunostaining were done for 1 hr at 25°C in 5% milk using the following antibodies: polyclonal anti-IAV PR8/34, 1:5,000 (V301-511-552), and peroxidase rabbit anti-chicken IgG, 1:5,000 (303-035-003; Jackson Immuno Research). TrueBlue peroxidase substrate (Kirkegard & Perry Laboratories) was used as directed for detection of virus plaques.
Microneutralization assay.
MDCK cells were plated in 96 well plates. Serum samples were heat-inactivated at 56°C for 30 min and analyzed in quadruplicate. Serum samples were diluted in virus growth media containing DMEM with 0.5% bovine serum albumin, 1% pen-strep. IAV, either PR8 or Cal/09 was diluted to 2500 x TCID50 in virus growth media and incubated with diluted sera for 1 hr at 37°C. Virus-serum mixture was added to MDCKs in the presence of TPCK for 40 hours at 37°C, fixed with 4% formaldehyde, stained with crystal violet and cytopathic effects (CPE) were assessed. MN antibody titers are expressed as the reciprocal of the highest serum dilution causing protection from virus induced CPE.
Whole Lung RNAseq.
Whole lungs were harvested and RNA was extracted using AllPrep DNA/RNA kit (Qiagen). The cDNA library was prepared using strand-specific RNA-sequencing protocols. Samples were run on an Illumina NovaSeq (50 bp paired-end). We obtained an average of 27 million read pairs per sample. Sequence processing and mapping was performed as described above. Multidimensional scaling was performed with edgeR using the top 500 differentially expressed genes across samples. The regression model in the edgeR package with block design and interaction model was used for statistical analysis to select the corresponding significant genes. Gene ontology analysis was performed using Panther on genes in each cluster with an adjusted p value <0.01 and LogFC >0.5. Sequencing data were deposited under Gene Expression Omnibus series accession number GSE182858.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics.
GraphPad Prism was used to determine statistical significance. Student unpaired two-tailed t-test or one-way ANOVA was used. For ELISAs, area under the curve, ignoring peaks defined by fewer than 2 adjacent points, was determined and one-way ANOVAs were performed to determine significance between groups. A p value of <0.05 was considered statistically significant. To illustrate the similarities among cohoused mice based on past pathogen exposure, a distance matrix was calculated for all cohoused mice using the presence or absence of pathogen exposure based on EZ-spot and PRIA assay results. The distance matrix was plotted in two dimensions using multidimensional scaling with the stats package in R (Team, 2012). Additionally, serology data (n=14 binary categorical variables) were combined with CD44% (continuous variable) and used as input for Factor Analysis of Mixed Data (FAMD) to generate principal components and explore the contribution of individual pathogen exposure and T cell activation to the principal dimensions. FAMD was done in R using the FactoMineR package (Lê et al., 2008). MDS plots were made using the ggplot2 R package (Wickham, 2016). Data for FMD plots were exported from R and graphed using GraphPad Prism.
Supplementary Material
Highlights.
Cohousing SPF and pet store mice led to diverse microbes but stable immune impacts
Vaccine-induced transcriptional signatures are similar between cohoused mice and humans
Cohoused mice have dampened humoral responses to vaccinations compared to SPF animals
Heterologous protection is readily achieved in SPF mice but not cohoused mice or humans
Acknowledgements
This work was supported by NIH R01 AI132962 to RAL and NIH R01 AI150600 to RAL and DM. NIH CIVIC Contract No. 75N93019C00051 to DM and RAL. NIH R01 AI116678 to SEH. JKF, KEB and WEM were supported by T32 HL007741. This project was also funded in part with Federal funds from NIAID, NIH and the department of Health and Human Services, under CEIRS Contract No. HHSN272201400005C. We acknowledge the NIH Tetramer Core Facility for providing H-2Db-PA224 and H-2Db-NP366 tetramers. We thank Dr. Thomas Griffith for I-Ab NP311 tetramer. We also thank the UMN Flow Cytometry Resource Facility, UMN Genomics Center, CFI Dirty Mouse Colony and BSL-3 Program for support. We thank Dr. Steve Jameson for discussions during early stages of the work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors have no competing interests or conflicts.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
References
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, and Masopust D (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9, 209–222. 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, Thompson EA, Fraser KA, Rosato PC, Filali-Mouhim A, et al. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 532, 512–516. 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM (2018). The Future of Influenza Vaccines: A Historical and Clinical Perspective. Vaccines (Basel) 6. 10.3390/vaccines6030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracci L, Canini I, Venditti M, Spada M, Puzelli S, Donatelli I, Belardelli F, and Proietti E (2006). Type I IFN as a vaccine adjuvant for both systemic and mucosal vaccination against influenza virus. Vaccine 24 Suppl 2, S2–56–57. 10.1016/j.vaccine.2005.01.121. [DOI] [PubMed] [Google Scholar]
- Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Nino D, Arden N, Quarles JM, Couch RB, and Belmont JW (2011). Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis 203, 921–929. 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caproni E, Tritto E, Cortese M, Muzzi A, Mosca F, Monaci E, Baudner B, Seubert A, and De Gregorio E (2012). MF59 and Pam3CSK4 boost adaptive responses to influenza subunit vaccine through an IFN type I-independent mechanism of action. J Immunol 188, 3088–3098. 10.4049/jimmunol.1101764. [DOI] [PubMed] [Google Scholar]
- Chioato A, Noseda E, Felix SD, Stevens M, Del Giudice G, Fitoussi S, and Kleinschmidt A, (2010). Influenza and meningococcal vaccinations are effective in healthy subjects treated with the interleukin-1 beta-blocking antibody canakinumab: results of an open-label, parallel group, randomized, single-center study. Clin Vaccine Immunol 17, 1952–1957. 10.1128/cvi.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Kim S, Choi Y, Nielsen TB, Yan J, Lu A, Ruan J, Lee HR, Wu H, Spellberg A, and Jung JU (2019). SERPINBI-mediated checkpoint of inflammatory caspase activation. Nat Immunol 20, 276–287. 10.1038/s41590-018-0303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens EB, van de Sandt C, Wong SS, Wakim LM, and Valkenburg SA (2018). Harnessing the Power of T Cells: The Promising Hope for a Universal Influenza Vaccine. Vaccines (Basel) 6. 10.3390/vaccines6020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CL, Davis HL, Morris ML, Efler SM, Krieg AM, Li Y, Laframboise C, Al Adhami MJ, Khaliq Y, Seguin I, and Cameron DW (2004). Safety and immunogenicity of CPG 7909 injection as an adjuvant to Fluarix influenza vaccine. Vaccine 22, 3136–3143. 10.1016/j.vaccine.2004.01.058. [DOI] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856. 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- Couch RB, Atmar RL, Cate TR, Quarles JM, Keitel WA, Arden NH, Wells J, Niño D, and Wyde PR (2009). Contrasting effects of type I interferon as a mucosal adjuvant for influenza vaccine in mice and humans. Vaccine 27, 5344–5348. 10.1016/j.vaccine.2009.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MM (2008). A prescription for human immunology. Immunity 29, 835–838. 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Palese P, Wilson PC, and Ravetch JV (2016). Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 126, 605–610. 10.1172/jci84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo DJ, Tan GS, Palese P, and Ravetch JV (2014). Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 20, 143–151. 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effros RB, Doherty PC, Gerhard W, and Bennink J (1977). Generation of both cross-reactive and virus-specific T-cell populations after immunization with serologically distinct influenza A viruses. J Exp Med 145, 557–568. 10.1084/jem.145.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, and Flavell RA (2008). Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453, 1122–1126. 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francica JR, Zak DE, Linde C, Siena E, Johnson C, Juraska M, Yates NL, Gunn B, De Gregorio E, Flynn BJ, et al. (2017). Innate transcriptional effects by adjuvants on the magnitude, quality, and durability of HIV envelope responses in NHPs. Blood Adv 1, 2329–2342. 10.1182/bloodadvances.2017011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco LM, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, Arden N, Renwick A, Yu P, Quarles JM, et al. (2013). Integrative genomic analysis of the human immune response to influenza vaccination. Elife 2, e00299. 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A, Kenney LL, Mishra R, Watkin LB, Aslan N, and Selin LK (2015). Vaccination and heterologous immunity: educating the immune system. Trans R Soc Trop Med Hyg 109, 62–69. 10.1093/trstmh/tru198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant EJ, Quiñones-Parra SM, Clemens EB, and Kedzierska K (2016). Human influenza viruses and CD8(+) T cell responses. Curr Opin Virol 16, 132–142. 10.1016/j.coviro.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, Garcia-Sastre A, Tumpey TM, and Palese P (2010). PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J Virol 84, 4442–4450. 10.1128/JVI.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton JR, Sachs D, Lim JK, Langlois RA, Palese P, and Heaton NS (2016). Club cells surviving influenza A virus infection induce temporary nonspecific antiviral immunity. Proc Natl Acad Sci U S A 113, 3861–3866. 10.1073/pnas.1522376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SE, Badovinac VP, Beura LK, Pierson M, Jameson SC, Masopust D, and Griffith TS (2020). New Insights into the Immune System Using Dirty Mice. J Immunol 205, 3–11. 10.4049/jimmunol.2000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, and Bauer S (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303, 1526–1529. 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, and Latz E (2008). Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9, 847–856. 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins MA, Sjaastad FV, Pierson M, Kucaba TA, Swanson W, Staley C, Weingarden AR, Jensen IJ, Danahy DB, Badovinac VP, et al. (2019). Microbial Exposure Enhances Immunity to Pathogens Recognized by TLR2 but Increases Susceptibility to Cytokine Storm through TLR4 Sensitization. Cell Rep 28, 1729–1743.e1725. 10.1016/j.celrep.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson SC, and Masopust D (2018). What Is the Predictive Value of Animal Models for Vaccine Efficacy in Humans? Reevaluating the Potential of Mouse Models for the Human Immune System. Cold Spring Harb Perspect Biol 10. 10.1101/cshperspect.a029132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff WC, Burton DR, Johnson PR, Walker BD, King CR, Nabel GJ, Ahmed R, Bhan MK, and Plotkin SA (2013). Accelerating next-generation vaccine development for global disease prevention. Science 340, 1232910. 10.1126/science.1232910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Varble A, Chua MA, García-Sastre A, and tenOever BR (2012). Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci U S A 109, 12117–12122. 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Josse J, and Husson F (2008). FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software 25, 1–18. [Google Scholar]
- Levine MM (2010). Immunogenicity and efficacy of oral vaccines in developing countries: lessons from a live cholera vaccine. BMC Biol 8, 129. 10.1186/1741-7007-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al. (2014). Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 15, 195–204. 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Mozdzanowska K, Palladino G, and Gerhard W (1994). Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J Immunol 152, 1653–1661. [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin JD, Devlin JC, Yeung F, McCauley C, Leung JM, Chen YH, Cronkite A, Hansen C, Drake-Dunn C, Ruggles KV, et al. (2020). Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe 27, 830–840.e834. 10.1016/j.chom.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman BA, Pitzer VE, Sarkar R, Gladstone B, Patel M, Glasser J, Gambhir M, Atchison C, Grenfell BT, Edmunds WJ, et al. (2012). Understanding reduced rotavirus vaccine efficacy in low socio-economic settings. PLoS One 7, e41720. 10.1371/journal.pone.0041720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoli L, Salanti G, De Vito C, Boccia A, Ioannidis JP, and Villari P (2009). Immunogenicity and adverse events of avian influenza A H5N1 vaccine in healthy adults: multiple-treatments meta-analysis. Lancet Infect Dis 9, 482–492. 10.1016/s1473-3099(09)70153-7. [DOI] [PubMed] [Google Scholar]
- Matar CG, Anthony NR, O'Flaherty BM, Jacobs NT, Priyamvada L, Engwerda CR, Speck SH, and Lamb TJ (2015). Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity. PLoS Pathog 11, e1004858. 10.1371/journal.ppat.1004858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, and Hughes CC (2004). Of mice and not men: differences between mouse and human immunology. J Immunol 172, 2731–2738. 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Meyerholz DK, and Beck AP (2018). Principles and approaches for reproducible scoring of tissue stains in research. Lab Invest 98, 844–855. 10.1038/s41374-018-0057-0. [DOI] [PubMed] [Google Scholar]
- Moldoveanu Z, Love-Homan L, Huang WQ, and Krieg AM (1998). CpG DNA, a novel immune enhancer for systemic and mucosal immunization with influenza virus. Vaccine 16, 1216–1224. 10.1016/s0264-410x(98)80122-9. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C, O'Hagan D, Rappuoli R, and De Gregorio E (2008). Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A 105, 10501–10506. 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE, Patel NB, Zak DE, Aderem A, Dong T, et al. (2016). Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc Natl Acad Sci U S A 113, 1853–1858. 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, et al. (2011). Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12, 786–795. 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, Thompson-Snipes L, Ranganathan R, Zeitner B, Bjork A, et al. (2013). Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 38, 831–844. 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker EP, Ramani S, Lopman BA, Church JA, Iturriza-Gómara M, Prendergast AJ, and Grassly NC (2018). Causes of impaired oral vaccine efficacy in developing countries. Future Microbiol 13, 97–118. 10.2217/fmb-2017-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KJ, and Crooks GM (2007). Immune-cell lineage commitment: translation from mice to humans. Immunity 26, 674–677. 10.1016/j.immuni.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Proietti E, Bracci L, Puzelli S, Di Pucchio T, Sestili P, De Vincenzi E, Venditti M, Capone I, Seif I, De Maeyer E, et al. (2002). Type I IFN as a natural adjuvant for a protective immune response: lessons from the influenza vaccine model. J Immunol 169, 375–383. 10.4049/jimmunol.169.1.375. [DOI] [PubMed] [Google Scholar]
- Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Burger MC, Pulendran B, Sekaly RP, Jameson SC, Masopust D, et al. (2016). Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe 19, 713–719. 10.1016/j.chom.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J (2012). Animal models: Not close enough. Nature 484, S9. 10.1038/nature11102. [DOI] [PubMed] [Google Scholar]
- Rivera J, and Tessarollo L (2008). Genetic background and the dilemma of translating mouse studies to humans. Immunity 28, 1–4. 10.1016/j.immuni.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roestenberg M, Hoogerwerf MA, Ferreira DM, Mordmüller B, and Yazdanbakhsh M (2018). Experimental infection of human volunteers. Lancet Infect Dis 18, e312–e322. 10.1016/s1473-3099(18)30177-4. [DOI] [PubMed] [Google Scholar]
- Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, Leonardi I, et al. (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365. 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, et al. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell 171, 1015–1028.e1013. 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryg-Cornejo V, loannidis LJ, Ly A, Chiu CY, Tellier J, Hill DL, Preston SP, Pellegrini M, Yu D, Nutt SL, et al. (2016). Severe Malaria Infections Impair Germinal Center Responses by Inhibiting T Follicular Helper Cell Differentiation. Cell Rep 14, 68–81. 10.1016/j.celrep.2015.12.006. [DOI] [PubMed] [Google Scholar]
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, et al. (2013). Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 110, 3507–3512. 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda AJ, Maher TJ, Zhang Y, Lanahan SM, Bucklin ML, Compton SR, Tyler PM, Comrie WA, Matsuda M, Olivier KN, et al. (2019). Human PI3Kγ deficiency and its microbiota-dependent mouse model reveal immunodeficiency and tissue immunopathology. Nat Commun 10, 4364. 10.1038/s41467-019-12311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.C. (2012). A language and environment for statistical computing (Foundation for Statistical Computing; ). [Google Scholar]
- Tregoning JS, Russell RF, and Kinnear E (2018). Adjuvanted influenza vaccines. Hum Vaccin Immunother 14, 550–564. 10.1080/21645515.2017.1415684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Herrath MG, and Nepom GT (2005). Lost in translation: barriers to implementing clinical immunotherapeutics for autoimmunity. J Exp Med 202, 1159–1162. 10.1084/jem.20051224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring BM, Sjaastad LE, Fiege JK, Fay EJ, Reyes I, Moriarity B, and Langlois RA (2018). MicroRNA-Based Attenuation of Influenza Virus across Susceptible Hosts. J Virol 92. 10.1128/jvi.01741-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, and Askonas BA (1980). Cross-protection and cross-reactive cytotoxic T cells induced by influenza virus vaccines in mice. EurJ Immunol 10, 396–401. 10.1002/eji.1830100515. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2 : Elegant Graphics for Data Analysis, 2nd Edition (Springer International Publishing : Imprint: Springer,). 10.1007/978-3-319-24277-4. [DOI] [Google Scholar]
- Wilkins AL, Kazmin D, Napolitani G, Clutterbuck EA, Pulendran B, Siegrist CA, and Pollard AJ (2017). AS03- and MF59-Adjuvanted Influenza Vaccines in Children. Front Immunol 8, 1760. 10.3389/fimmu.2017.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, Devlin JC, Hansen C, Cronkite A, Stephens Z, Drake-Dunn C, et al. (2020). Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe 27, 809–822.e806. 10.1016/j.chom.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BE, Sadarangani SP, and Leo YS (2015). The avian influenza vaccine Emerflu. Why did it fail? Expert Rev Vaccines 14, 1125–1134. 10.1586/14760584.2015.1059760. [DOI] [PubMed] [Google Scholar]
- Zhou F, Trieu MC, Davies R, and Cox RJ (2018). Improving influenza vaccines: challenges to effective implementation. Curr Opin Immunol 53, 88–95. 10.1016/j.coi.2018.04.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO (GSE182858) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All (serology, flow cytometry, ELISA, titering, morbidity and mortality, ELISA and neutralization) data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-CD8β antibody (clone lyt3.2) | Bio X Cell | BE0223 RRID: AB_2687706 |
| CD49a (clone Hα31/8) | BD Biosciences | 740262 RRID: AB_2740005 |
| RORγt (clone Q31-378) | BD Biosciences | 562894 RRID: AB_2687545 |
| B220 (clone cRA3-6B2) | Biolegend | 103255 RRID: AB_2563491 |
| CD4 (clone GK1.5) | Biolegend | 100447 RRID: AB_2564586 |
| CD4 (clone GK1.5) BUV395 | BD Biosciences | 563790 RRID: AB_2738426 |
| CD8-β (clone YTS156.7.7) | Biolegend | 126610 RRID: AB_2260149 |
| CD44 (clone IM7) | Tonbo | 65-0441-U100 RRID: AB_2621891 |
| CD44 (clone IM7) | BD Biosciences | 563736 RRID: AB_2738395 |
| CD45 (clone 104) | Tonbo | 60-0451-U100 RRID: AB_2621848 |
| CD45 (clone 30-F11) vF450 | Tonbo | 75-0451-U100 RRID: AB_2621947 |
| CD45 (clone 30-F11) FITC | eBioscience | 11-0451-81 RRID: AB_465049 |
| CD62L (clone MEL-14) | Biolegend | 104440 RRID: AB_2629685 |
| CD69 (clone H1.2F3) | Biolegend | 104512 RRID: AB_493564 |
| CD103 (clone 2E7) | Biolegend | 121414 RRID: AB_1227502 |
| CXCR5 (clone L138D7) | Biolegend | 145517 RRID: AB_2562453 |
| F4/80 (clone BM8) | Biolegend | 123147 RRID: AB_2564588 |
| IFN-γ (clone XMG1.2) | Biolegend | 505826 RRID: AB_2295770 |
| Ki67 (clone 16A8) | Biolegend | 652411 RRID: AB_2562663 |
| KLRG1 (clone 2F1/KLRG1) | Biolegend | 138416 RRID: AB_2561736 |
| PD-1 (clone 29F.1A12) | Biolegend | 25-5982-80 RRID: AB_2573508 |
| T-bet (clone 4B10) | Biolegend | 644814 RRID: AB_10901173 |
| TCR-β (clone 4B10) | eBioscience | 47-5961-82 RRID: AB_1272173 |
| TCR-β (clone H57-597) BV510 | BD Biosciences | 563221 RRID: AB_2738078 |
| TNF-α (clone MP6-XT22) | Biolegend | 506308 RRID: AB_315429 |
| CD45.1 (clone A20) | eBioscience | 47-0453-82 RRID: AB_1582228 |
| Foxp3 (clone FJK-16s) | eBioscience | 53-5773-82 RRID: AB_763537 |
| I-A/I-E (clone M5/114.15.2) | Biolegend | 107639 RRID: AB_2565894 |
| CD8-α (clone 53-6.7) | Tonbo | 35-0081-U100 RRID: AB_2621671 |
| CD8-α (clone 53-6.7) BUV737 | BD Biosciences | 612759 RRID: AB_2870090 |
| HRP-anti-mouse IgG1 | Southern Biotech | 1070-05 RRID: AB_2650509 |
| HRP-anti-mouse IgG2b | Southern Biotech | 1090-05 RRID: AB_2794521 |
| HRP-anti-mouse IgG2c | Southern Biotech | 1079-05 RRID: AB_2794466 |
| HRP-anti-mouse IgG2a | Southern Biotech | 1080-05 RRID: AB_2734756 |
| Polyclonal Anti-Influenza Virus, A/Puerto Rico/8/1934 (H1N1) (antiserum, Rooster) | BEI Resources, NIAID, NIH | V301-511-552, NR-3098 |
| HRP rabbit anti-chicken IgG | Jackson Immuno Research | 303-035-003 RRID: AB_2339290 |
| Bacterial and virus strains | ||
| IAV/PR8 | In house | N/A |
| IAV/Cal/09 | In house (Hai et al., 2010) | N/A |
| LAIV_PR8 | In house (Waring et al., 2018) | N/A |
| IAV/X31 | In house | N/A |
| IBV/Mal/04 | In house (Hamilton et al., 2016) | N/A |
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| H-2Db-PA224 (SSLENFRAYV) tetramer | NIH Tetramer Core Facility | N/A |
| H-2Db-NP366 (ASNENMETM) tetramer | NIH Tetramer Core Facility | N/A |
| I-Ab NP311 (QVYSLIRPNENPAHK) tetramer | In House | N/A |
| Influenza A (H1N1) 2009 Monovalent Vaccine | BEI Resources | NR-20347 |
| 2019-2020 quadrivalent influenza vaccine | Sanofi Pasteur | NDC 49281-719-10 |
| AddaVax | InvivoGen | Cat# vac-adx-10 |
| NP366 (ASNENMETM) peptide, no N or C terminal modifications | Biosynn | N/A Custom |
| collagenase type I | Worthington | Cat# LS004196 |
| ABTS Peroxidase Substrate | SeraCare | Cat# 5120-0043 |
| TrueBlue peroxidase substrate | Kirkegard & Perry Laboratories | Cat# 50-674-28 |
| Ghost Dye Red 780 | Tonbo | 13-0865-T100 |
| Cytofix/Cytoperm Fixation/Permeablization Kit | BD Biosciences | Cat# 554714 RRID: AB_2869008 |
| Foxp3 / Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 |
| GolgiPlug Protein Transport Inhibitor | BD Biosciences | Cat# 555029 RRID: AB_2869014 |
| Critical commercial assays | ||
| EZ-spot and PCR Rodent Infections Agent (PRIA) array methods | Charles River Laboratories | |
| AllPrep DNA/RNA kit | QIAGEN | Cat# 80204 |
| RNeasy Micro Plus Kit | QIAGEN | Cat# 74034 |
| Deposited data | ||
| Mouse PBMC RNAseq after (A)QIV, Raw data files | This paper | GEO: GSE182858 |
| Mouse whole lung RNAseq, Raw data files | This paper | GEO: GSE182858 |
| Customized gene set | This paper | c7.custom.immune.datasets.gmt |
| Time series of global gene expression after trivalent influenza vaccination in humans, human expression profiling by array | (Franco et al., 2013) | GEO: GSE48024 |
| Systems biology of immunity to MF59-adjuvanted versus non-adjuvanted trivalent seasonal influenza vaccines in early childhood, human expression profiling by array | (Nakaya et al., 2016) | GEO: GSE74975 |
| Systems biology of vaccination for seasonal influenza in humans, human expression profiling by array | (Nakaya et al., 2011) | GEO: GSE29619 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat# CRL-3216 |
| MDCK (NLB-2) | ATCC | Cat# CCL-34 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: C57BL/6 | Charles River | Strain code: 027 |
| Mouse: BALB/c | Charles River | Strain code: 028 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| FlowJo™ Software For Mac | Becton, Dickinson and Company | Version 9.9.6 |
| GraphPad Prism 9 for macOS | GraphPad Software | Version 9.2.0 (283) |
| R | https://www.r-project.org | Version 4.1.0 |
| Bowtie aligner | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | bowtie2 version 2.3.4.1 |
| Subread software package | http://subread.sourceforge.net | version 1.5.1 |
| bioconductor package edgeR | https://bioconductor.org/packages/release/bioc/html/edgeR.html | version 3.24.3 |
| FactoMineR package | https://CRAN.R-project.org/package=FactoMineR | Version 2.4 |
| Adobe Illustrator | Adobe | Version 25.4.1 |
| stats R package | https://stat.ethz.ch/R-manual/R-devel/library/stats/html/00Index.html | Version 4.1.0 |
| ggplot2 R package | https://ggplot2.tidyverse.org | Version 3.3.5 |
| Other | ||
This paper does not report original code.
Any additional information required to reanalyze the data reported in the paper is available from the lead contact upon request.