Abstract
The revolution in genetics has rapidly increased our knowledge of human and mouse genes that are critical for the formation of dental enamel and helps us understand how enamel evolved. In this graphical review we focus on the roles of 41 genes that are essential for the secretory stage of amelogenesis when characteristic enamel mineral ribbons initiate on dentin and elongate to expand the enamel layer to the future surface of the tooth. Based upon ultrastructural analyses of genetically modified mice, we propose a molecular model explaining how a cell attachment apparatus including collagen 17, α6ß4 and αvß6 integrins, laminin 332, and secreted enamel proteins could attach to individual enamel mineral ribbons and mold their cross-sectional dimensions as they simultaneously elongate and orient them in the direction of the retrograde movement of the ameloblast membrane.
Keywords: biomineralization, amelogenesis, evolution, basement membrane, SLC13A5, ACP4
1. Introduction
The gar is a fish that has enamel on the surface of its scales and teeth (Kawasaki et al., 2021; Sire, 1995). Enamel was formed by the common ancestor of coelacanth and gar (>450 mya) and appeared first on scales that pre-date the development of enamel on teeth (Braasch et al., 2016). Teeth and scales arise from odontodes that evolved from a common founder odontode and consequently their developmental processes rely upon an overlapping repertoire of genetic elements (Chen et al., 2020). A striking demonstration of this relationship is the significant loss of both scales and teeth in a teleost (Medaka) caused by a splice junction mutation in Edar (Atukorala et al., 2010). In humans, EDAR mutations cause hypohidrotic ectodermal dysplasia showing tooth agenesis and abnormal morphogenesis of teeth, hair, and sweat glands. The founder odontode for teeth apparently depended upon a series of reciprocal epithelial-mesenchymal interactions that were co-opted for the induction of multiple epithelial organs (Thesleff, 2006). This common evolutionary history is a basis for genetic mutations in single genes resulting in the ectodermal dysplasias and other syndromes that combine tooth agenesis and/or dental malformations with aplasia of lacrimal and salivary glands, reductions in tongue fungiform and filiform papillae, hyperkeratosis of the skin, skin fragility, etc (Adaimy et al., 2007).
Because of its evolutionary history, dental enamel formation (amelogenesis) relies upon a variety of genetic elements that are critical for the attachment of skin to its underlying basement membrane and mesenchymal tissues. Because of this, mutations in COL17A1, LAMA3, LAMB3, LAMC2, PLEC, ITGB4 and ITGA6 cause epidermolysis bullosa with skin fragility and enamel malformations (Chung and Uitto, 2010; Fine, 2010). It is curious that the skin attachment apparatus is conserved in ameloblasts, because ameloblasts degrade their basement membrane prior to the onset of amelogenesis, then abruptly lose contact with a mesenchymal matrix once enamel starts to form. While the genes noted above are associated with hemidesmosomes in skin, hemidesmosomes are not observed in ameloblasts before the maturation stage (Sahlberg et al., 1998). Nevertheless, ameloblasts detach from underlying mineral specifically at the onset of enamel formation in Lama3−/− mice, confirming that laminin 332 (comprised of LAMA3, LAMB3, and LAMC2) strengthens the anchorage of secretory ameloblasts to the underlying enamel matrix (Ryan et al., 1999; Sahlberg et al., 1998). Laminin 332 likely binds to secreted enamel proteins that bind directly to the enamel mineral ribbons.
Secreted enamel proteins are encoded by genes belonging to the secretory calcium-binding phosphoprotein (SCPP) gene family (Kawasaki and Weiss, 2003), which was spawned by duplication of SPARC-like 1 (SPARCL1), itself duplicated from secreted protein acidic cysteine-rich (SPARC), a gene that encodes a basement membrane protein (Jayadev and Sherwood, 2017). There are 24 SCPP genes in humans (Kawasaki and Amemiya, 2014). The enamel-associated SCPP genes are amelogenin (AMEL), enamelin (ENAM), ameloblastin (AMBN), amelotin (AMTN), odontogenic, ameloblast associated (ODAM), secretory calcium-binding phosphoprotein proline-glutamine rich 1 (SCPPPQ1) and dentin sialophosphoprotein (DSPP). DSPP is a dentin protein that is transiently expressed by pre-ameloblasts, immediately prior to the onset of enamel mineralization. Three SCPP genes are expressed by secretory ameloblasts: AMEL, ENAM, and AMBN (Fincham et al., 1999).
Strong concepts of how dental enamel forms were formulated long before the genetics revolution and centered attention on amelogenin, the most abundant secreted enamel protein (Simmer and Fincham, 1995; Termine et al., 1980). Amelogenin was the first enamel protein to have its cDNA cloned and to be expressed as a recombinant protein. In vitro studies of recombinant amelogenin under different conditions spawned competing theories of amelogenin’s role in amelogenesis (Bai et al., 2020; Fang et al., 2011; Fincham et al., 1994) and encouraged hope for the production of synthetic enamel as a dental restorative. The genetics of human conditions associated with enamel malformations, however, suggests that amelogenesis is more complex than previously realized.
A search in 2015 of Online Mendelian Inheritance in Man (OMIM) for hereditary conditions that include an enamel phenotype revealed 91 such conditions, 71 with a known molecular etiology or linked genetic loci (Wright et al., 2015). The most common enamel phenotype is enamel hypoplasia, or thin enamel, which is caused by deficiencies affecting the secretory stage of amelogenesis (when the enamel layer achieves its final thickness). A table of 41 human genes that encode proteins necessary for normal formation of the full thickness of the enamel layer is provided (Table S1). Ten of these genes regulate gene expression or cease expression prior to the onset of enamel mineralization and are not discussed further.
2. Ultrastructure of Developmental Changes Along the Ameloblast Distal Membrane
The fundamental feature of forming dental enamel is the growth of characteristic thin mineral ribbons that start on the surface of dentin (Smith et al., 2016) and extend to a mineralization front 250–300Å from the ameloblast plasma membrane (Ronnholm, 1962b) where the mineral ribbons elongate, apparently by the addition of ions or mineral to their tips. The mineralization front is only slightly more distant from the ameloblast membrane as the basement membrane was prior to its earlier degradation and reabsorption (Ronnholm, 1962a).
Enamel formation is a biological process that is accomplished by the ameloblast distal membrane as it progresses sequentially through a series of modifications that can be readily observed at the ultrastructural level, as shown in Fig. 1. All vertebrates yet characterized that form “ancestral” or “true” enamel (Kawasaki et al., 2021) do so by ameloblast-like cells depositing characteristically thin mineral ribbons on mineralized collagen and extending those ribbons in close proximity to the ameloblast membrane. In humans, the cross-sectional dimension of the ribbons at the mineralization front is about 15Å x 150Å (Kerebel et al., 1979). Near the mineralization front where the ribbons elongate, they are comprised of amorphous calcium phosphate, but crystalize with depth where they produce diffraction patterns characteristic of apatite (Beniash et al., 2009). The morphology and organization of the mineral in enamel is determined prior to its crystallization, suggesting that the consistent dimensions of the cross-sections at the mineralization front is accomplished by molding, since non-crystalline minerals such as amorphous calcium phosphate that don’t have crystalline faces cannot be shaped by proteins binding to and selectively inhibiting the growth of specific crystal faces. The smooth increase in cross-sectional dimensions of the crystallites with depth (Nylen et al., 1963; Warshawsky, 1989) is accomplished by a relatively slow, but steady rate of ions depositing onto the sides of the ribbons. Many studies support the concept that secreted enamel proteins, particularly the abundant amelogenins, can bind to the sides of the crystallites and inhibit mineral deposition and regulate the conversion of ACP to apatite (Bartlett et al., 2021; Hu et al., 2016; Margolis et al., 2014; Shaw et al., 2020; Yamazaki et al., 2019). Proteolytic cleavages and influxes of calcium, phosphate, and citrate ions potentially regulate the attachment of proteins to mineral surfaces.
Fig. 1.
Visualization of enamel formation by focused ion beam-backscattered scanning electron microscopy (FIB-bSEM) of a continuously growing 7-week mouse mandibular incisor. A) Ameloblasts are separated from the unmineralized predentin matrix by a well-defined basement membrane (BM). The predentin matrix contains odontoblastic processes, matrix vesicles released by odontoblasts, and numerous collagen fibrils/fibers that are oriented toward the ameloblasts where they appear to attach to the BM. B) The BM thins as the ameloblast membrane becomes less linear. C) Ameloblast finger-like processes penetrate the BM and extend into predentin between the collagen fibrils, as remnants of the BM accumulate along the ameloblast membrane that is reabsorbing it. D) Mineral nuclei appear in predentin some distance from the ameloblast membrane (arrowheads). E) Dentin mineral coalesces into a continuous mineral layer that extends to near the ameloblast membrane. F) Enamel ribbons initiate in patches on dentin mineral associated with the ends of mineralized collagen fibers. G) A continuous segment of mouse incisor completing the formation of initial enamel (above the G) and showing the progressive formation of Tomes processes as the segment continues in a second row below the G. The ends of Tomes’ processes form by the rapid extension of “prongs” of mineral ribbons at interrod growth sites along the peripheral part of the distal ameloblast membrane near the cell junctions. H) High magnification of initial enamel ribbons forming on the ends of collagen fibers and extending at different angles back to the ameloblast membrane, following the finger-like process as it retreated into the cell membrane. I) Rod and interrod enamel after formation of the Tomes process [asterisk indicates a space of Weber (Bartlett et al., 2021)]. J) Tomes’ process showing the positions of rod (r) and interrod (ir) growth sites in the mineralization front. K) Maturation stage enamel has a basal lamina comprised of SCPP proteins bound to the surface enamel. Key: Am, ameloblast; d, dentin, e, enamel, ir, interrod growth site; pd, predentin; r, rod growth site; tp, Tomes’ process; white scale bars (under figure letters) = 500 nm.
Enamel mineral ribbons are oriented with respect to the ameloblast membrane extending them. The ribbons are typically oriented perpendicular to the membrane and in the direction of the membrane’s retrograde movement that provides space for ribbon elongation. After forming a thin initial enamel layer on dentin mineral with essentially all of the mineral ribbons oriented in a parallel array, mammalian ameloblasts develop a characteristic distal membrane specialization called a “Tomes process” by temporarily extending enamel ribbons only from “interrod growth sites” along the periphery of the ameloblast distal membrane near its borders with adjacent ameloblasts. This generates “prongs” of enamel ribbons that cause the central part of the distal membrane to protrude into the forming enamel (Nanci and Warshawsky, 1984). “Rod growth sites” located on the protruding part of the Tomes extend enamel ribbons at a deeper level than the interrod growth sites (Skobe, 2006) and generate enamel rods. Parallel arrays of characteristic enamel mineral ribbons in rods extend at different orientations than the parallel arrays of identical ribbons extending at interrod growth sites. Reproducible variations in the patterns of rod and interrod enamel between different types of mammals and even at different depths within a single tooth provide strong evidence for ameloblast membrane control over the oriented elongation of enamel mineral ribbons (Boyde, 1969). The hierarchical organization of enamel mineral ribbons in mammals appears to be unexplainable by models that form mineral in the presence of amelogenin in vitro without cell participation.
3. Critical Functions of Ameloblasts
Secretory stage ameloblasts are tall, cylindrical, densely-packed cells forming a sheet that delineates the protected space covering the surface of dentin and forming enamel (Smith, 1979). Ameloblasts absorb ions and nutrients while expelling waste products along their proximal membrane. They secrete and reabsorb ions and proteins along their distal membrane, while attaching to and elongating the enamel mineral ribbons. Groups of ameloblasts within the sheet move relative to other groups and retreat from the enamel surface as the enamel layer expands. Many of the critical genes and proteins that carry out these activities have been identified by their contributions to the etiology of inherited enamel defects (Fig. 2; Table S1). Identifying critical genes allows genetically modified mice to be generated and characterized to learn more about the function of each gene and its protein products. The primary objective is to determine the mechanism of enamel mineral ribbon formation, which requires ultrastructural characterization of the mineral generated in the modified mice.
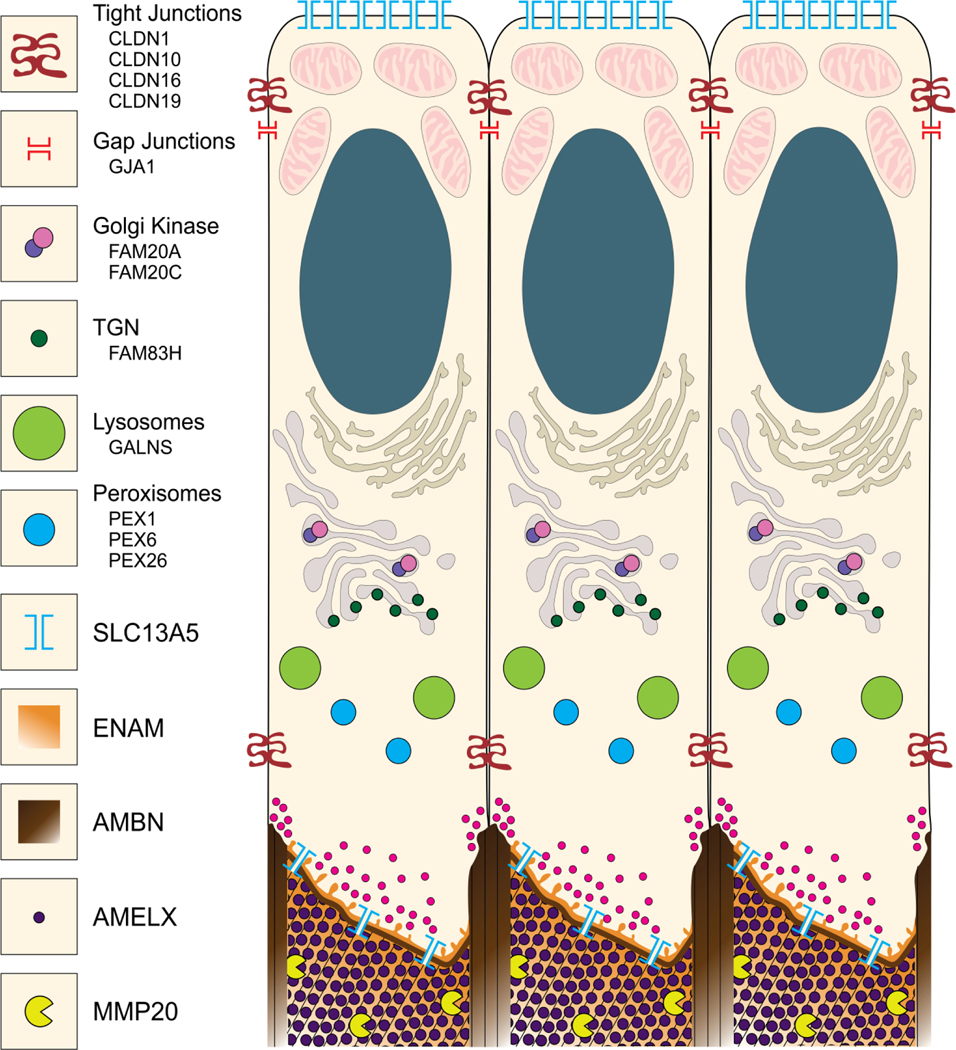
Fig. 2.
Graphical Illustration of ameloblasts showing proteins critical for the secretory stage. CLDN1, CLDN10, CLDN16, and CLDN19 encode tight junction proteins restricting entry of intercellular ions and molecules into the enamel space and also function in cell motility. GJA1 encodes a gap junction protein important for intercellular communication with adjacent ameloblasts. FAM20A and FAM20C encode components of the Golgi casein kinase complex that phosphorylates secreted enamel proteins. FAM83H encodes a cytosolic protein associated with the trans-Golgi network (TGN) and the keratin cytoskeleton. GALNS encodes a lysosomal hydrolase necessary for the degradation of glycosaminoglycans. PEX1, PEX6, and PEX26 are necessary for peroxisome biogenesis. SLC13A5 encodes a membrane citrate channel providing citrate influx proximally and efflux (into the enamel matrix) distally. Small secretory vesicles (magenta) in the proximal and distal parts of the Tomes’ process (distally) secrete AMELX, AMBN, ENAM, and MMP20 at the rod and interrod growth sites. Thin lines in the extracellular matrix (bottom) represent the general direction of enamel mineral ribbons. Not shown: CACNA1C encodes an L-type (voltage-dependent calcium channel) thought to provide regulated Ca2+ influx. SLC10A7 encodes a transmembrane transporter of unknown specificity that causes severe enamel defects when mutated. Graphic by Shelly Zhang.
We have yet to explain the roles of RELT, LTBP3, SLC13A5, ACP4, PLEC, LAMA3, LAMB3, COL17A1, ITGA6, ITGB4, ITGAV, and ITGB6. RELT is a tumor necrosis factor receptor expressed by secretory stage ameloblasts specifically, but its role in amelogenesis is unknown (Kim et al., 2019). LTBP3 is co-secreted with TGFß, functions early in tooth formation, and is associated with tooth agenesis. Plectin (PLEC), Laminin 332 (LAMA3, LAMB3, LAMC2), COL17A1, and α6ß4 (ITGA6, ITGB4) integrin combine to form an attachment complex in skin (Chung and Uitto, 2010) that is also required for attachment of ameloblasts to the underlying mineral (Ryan et al., 1999; Sahlberg et al., 1998). Integrin αvß6 (ITGAV, ITGB6) also contributes to cell attachment (Mohazab et al., 2013). The α6ß4 and αvß6 integrin attachment complexes provide intracellular feedback concerning conditions outside the cell at the mineralization front that significantly affect the expression levels of critical enamel proteins. Itgb6−/− ameloblasts increased Amelx expression 21-fold and Enam expression 7.6-fold (Mohazab et al., 2013).
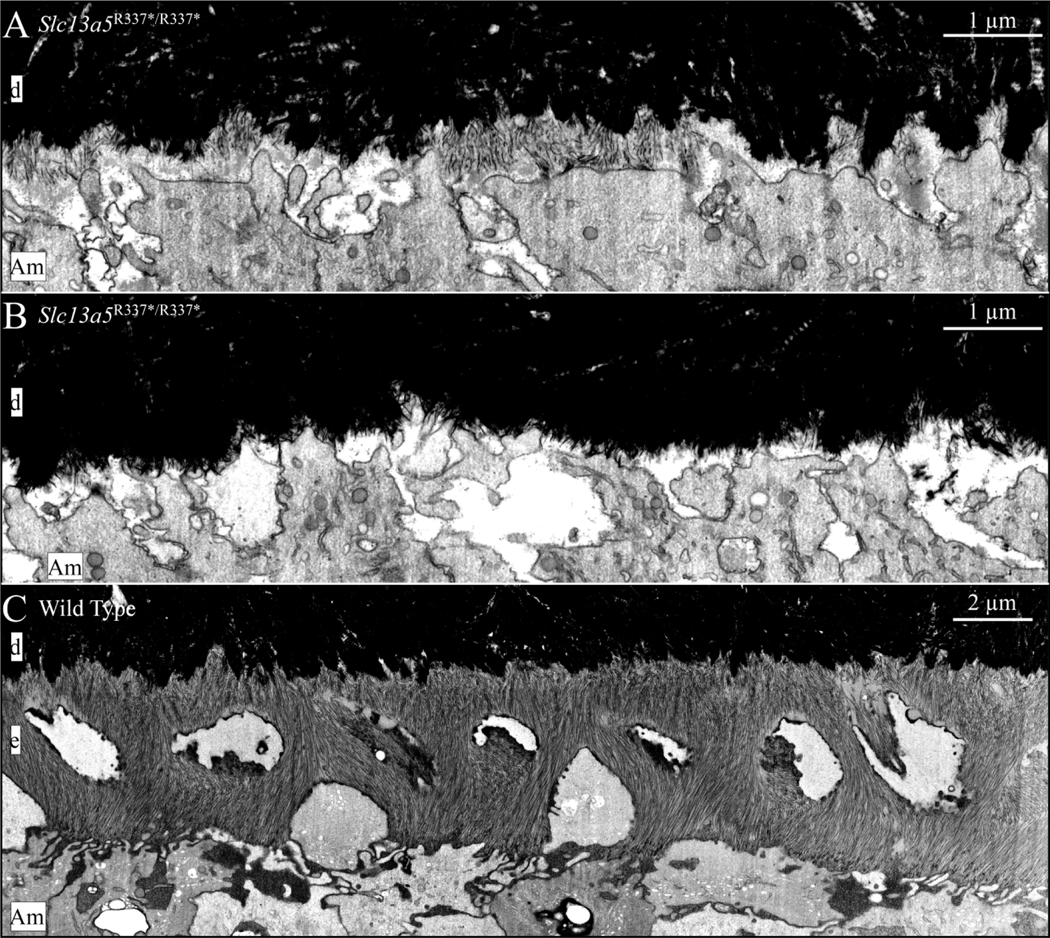
The ultrastructure of the initial enamel formed in wild-type, Amelx−/−, Enam−/−, Ambn−/−, Amelx−/−Ambn−/−, Mmp20−/−, Acp4R110C/R110C, and Slc13a5R337*/R337*mice is shown in Fig. 3. While amelogenesis is seriously disrupted in all of these mice, oriented initial enamel ribbons form in Amelx−/−, Mmp20−/−, and Acp4R110C/R110C mice, whereas none form at all in Enam−/− and Ambn−/− mice. Only isolated patches of short, poorly organized enamel ribbons form in Slc13a5R337*/R337* mice and appear to dissolve before they can extend further (Fig. 4).
Fig. 3.
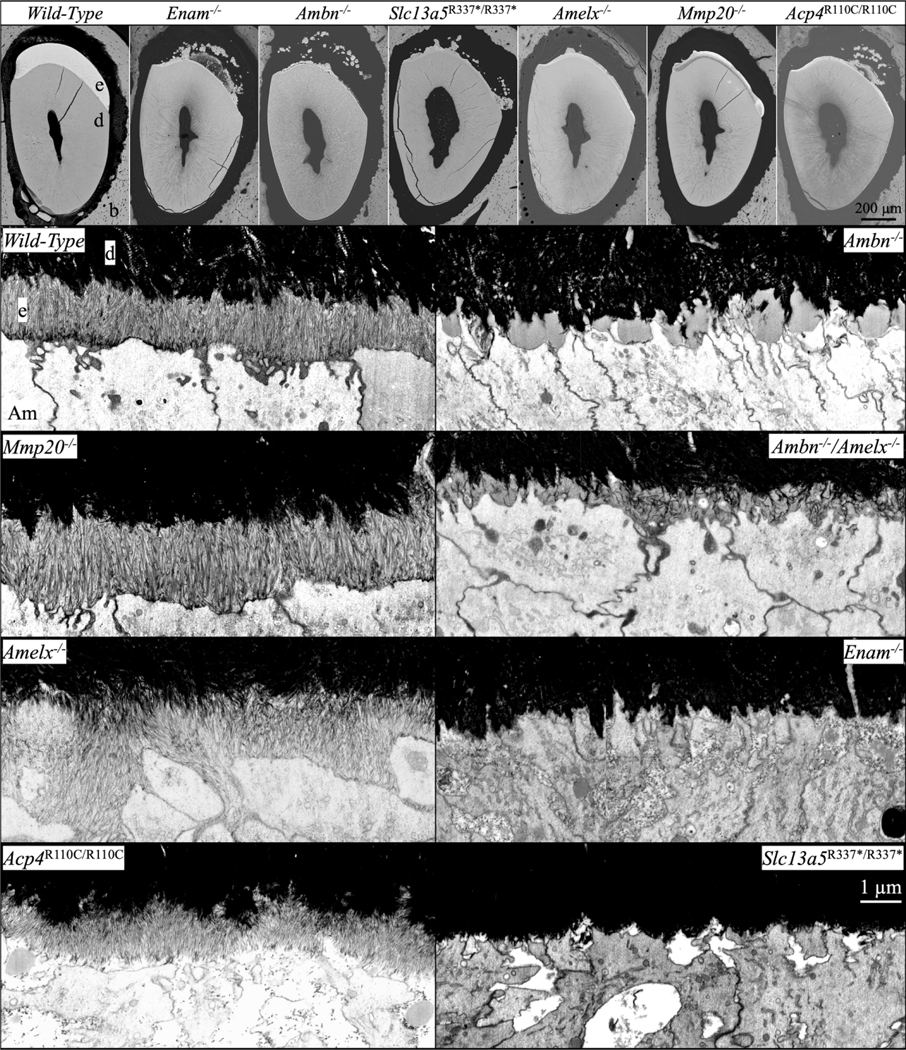
bSEM and FIB-bSEM visualization of initial enamel forming in 7-week mouse mandibular incisors in Amelx−/−, Enam−/−, Ambn−/−, Amelx−/−Ambn−/−, Mmp20−/−, Acp4R110C/R110C, and Slc13a5R337*/R337* mice. Top: bSEM sections of incisor cross-sections at the level of the labial alveolar crest (near the point of eruption). All of the knockouts showed severe enamel hypoplasia, many with ectopic mineral deposition within the overlying enamel organ. Below: FIB-bSEM images of where enamel should be forming in 8 different mouse incisors all at the same magnification. Characteristic initial enamel mineral ribbons form in Mmp20/-, Amelx−/−, and Acp4R110C/R110C mice, while no enamel ribbons form in Enam−/− and Ambn−/− mice. Ambn−/− nulls show accumulations of organic material comprised mainly of amelogenin that are greatly reduced in the Amelx−/−Ambn−/− double null. Key: Am, ameloblast; b, bone; d, dentin; e, enamel.
Fig. 4.
FIB-bSEM visualization of initial enamel forming in 7-week mouse mandibular incisors in in Slc13a5R337*/R337* and wild-type mice. A) Although delayed, patches of enamel ribbons form on the surface of mineralized dentin in Slc13a5R337*/R337* mice. These patches do not appear to be stable and were not observed further incisally (shown in B). This phenotype suggests that tribasic citrate (C6H5O73-) is required to stabilize the thin mineral ribbons. After the initial ribbons disappear or fail to form, the mineral surface on dentin appears “spiky”. These spikes are possibly mineral associated with the frayed ends of collagen fibrils. In some places the ends of these spikes are associated with the ameloblast membrane, suggesting they possibly elongate as enamel mineral ribbons in wild type mice. A spiky dentin surface is also observed in Enam−/−, Ambn−/−, and Ambn−/−Amelx−/− mice after initial enamel deposition fails (Liang et al., 2019). C) Enamel ribbon formation in a wild type incisor at a similar position as the one shown in A above. Note that at this level of development, enamel has already progressed beyond initial enamel formation and is forming rod and interrod enamel. Also note that the wild type enamel mineral ribbons show flexibility and bend without breaking. This flexibility of enamel mineral ribbons may be imparted by their association with enamel proteins and/or citrate. Key: Am, ameloblast; d, dentin; e, enamel.
The formation, organization, and orientation of enamel mineral ribbons are fundamental characteristics that have so far proved to be inseparable genetically. Long, characteristically-shaped enamel mineral ribbons that are oriented haphazardly have never been observed in knockout mice.
4. Enamel Ribbon Attachment, Elongation and Orientation
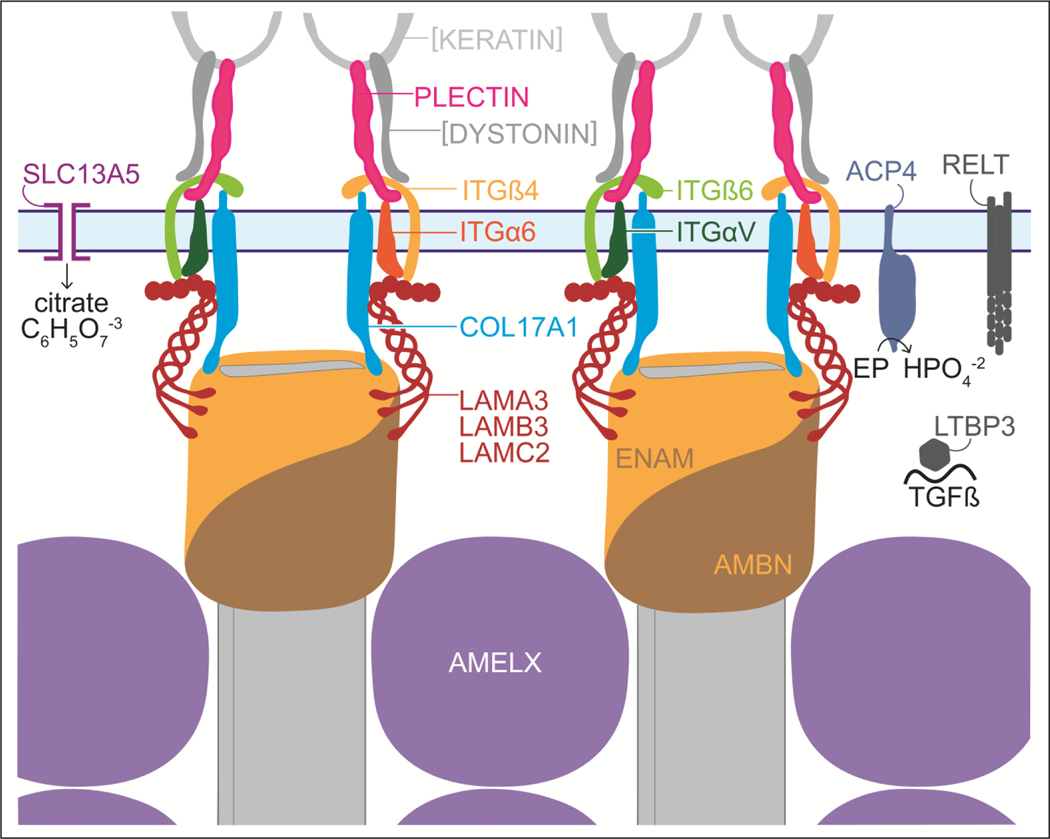
A molecular model of enamel mineral ribbon formation is proposed in Fig. 5. We hypothesize that enamel ribbons are shaped by a molding process that simultaneously lengthens and orients the mineral ribbons by the incremental addition of ions or mineral to the tips of existing enamel ribbons in the direction of the retrograde movement of the ameloblast membrane. We propose that the molds are comprised of secreted enamel proteins, most likely enamelin and ameloblastin, connected to integrin complexes (α6ß4 and/or αvß6 integrin) via laminin 332 and COL17A1. Maintaining a portion of the mold bound to hardened mineral secures the attachment of ameloblasts to the ribbons. Extending the molds proximally allows for daily increments of appositional growth, while degrading the molds distally allows for the slow growth of the enamel ribbons in width and thickness. This model suggests that each ribbon is associated with a specific membrane apparatus throughout the secretory stage, that the membrane assemblies can rotate to allow for the random orientation of enamel ribbons with respect to their A and B axes and can slip past each other in the membrane so that enamel ribbons associated at the dentino-enamel junction (DEJ) are not fixed in their relationship to adjacent ribbons as they elongate.
Fig. 5.
Molecular model for an ameloblast attachment apparatus that simultaneously shapes, elongates, and orients enamel mineral ribbons. The specifics of the model are hypothetical and are based upon a model for skin attachment (Chung and Uitto, 2010). This model proposes that enamel ribbon shape is determined by a molding process that determines the cross-sectional shape of the tips of the enamel ribbons as increments of mineral are added to extend the ribbons in the direction of the retrograde movement of the ameloblast membrane (in light blue and spanned by the SLC13A5 citrate transporter on the left), thereby orienting the ribbons as they elongate. The ENAM/AMBN mold is thought to be extended proximally and disassembled distally, presumably assisted by MMP20 cleavages. Intact enamelin (containing its C-terminus) is only detected at the mineralization front (Hu et al., 1997). Amelogenin is scant at the mineralization front (Nanci et al., 1998; Uchida et al., 1989) and not critical for shaping or orienting enamel ribbons, but is more important in separating the ribbons and guiding their transition from amorphous calcium phosphate into apatite. The connection of the mineral-associated enamel proteins to integrins α6ß4, αVß6 and COL17A1 is mediated by laminin 332. Inside the cell the integrins bind to keratin filaments via plectin. SLC13A5 channels tribasic citrate ions into the enamel matrix. ACP4 is a membrane-bound acid phosphatase associated with the ameloblast Tomes’ process that may function during endocytosis and dephosphorylates enamel proteins (EP), perhaps prior to their reabsorption, providing phosphate in the matrix for mineralization. RELT is a tumor necrosis factor receptor and LTBP3 is co-secreted with TGFß that may mediate matrix-cell interactions. Graphic by Shelly Zhang.
This model explains how the mammalian adaptation of the Tomes’ process (which establishes partially distinct rod and interrod growth sites with retrograde movements in different orientations), can establish rod and interrod organizations of identical crystals oriented in different directions (Nanci and Warshawsky, 1984). In rodent incisors the movement of groups of ameloblasts relative to other groups generates a decussation (X-shaped) pattern of rods in the enamel. Our model predicts that to maintain the cellular connection to each ribbon elongating at the mineralization front, the interrod enamel ribbons (which extend near the cell junctions) must contribute to multiple interrod structures as the ameloblast progresses from the DEJ to the final surface of enamel (Nanci and Warshawsky, 1984).
Future genetic advances will likely lengthen the list of genes critical for the secretory stage of amelogenesis. We haven’t identified the proteins that transport calcium and phosphate ions into the secretory stage enamel matrix, which would be a welcome addition to future amelogenesis models.
5. Conclusion
The model proposes that each mineral ribbon is attached to a COL17A1 and integrin based membrane complex via secreted proteins (laminin 332, ENAM, and AMBN) that mold the cross-sectional dimensions of mineral added to the tip of each ribbon. The density of membrane complexes establishes the average distance separating the ribbons, which determines the crystallites’ final thicknesses during enamel maturation when they interlock with adjacent crystals. ACP4 may dephosphorylate enamel proteins prior to their reabsorption into ameloblasts, and release their phosphate for mineralization. SLC13A5 transports citrate into the enamel matrix, which stabilizes the thin mineral ribbons. AMEL and its MMP20 cleavage products bind, separate, and support the mineral ribbons, and guide their transition into apatite.
Supplementary Material
Highlights.
A Genetic Model for the Secretory Stage of Dental Enamel Formation
Forty one genes are known to cause defects during the secretory stage of amelogenesis. We propose a model for enamel mineral ribbon formation inclusive of these 41 genes. Enamel ribbon formation is an evolutionary adaptation of the basement membrane. Integrins α4/ß6; αVß6, COL17A1, Laminin 332, SLC13A5, ACP4, Enamelin, Ameloblastin.
Acknowledgements
This study was supported by NIDCR/NIH research grants R01DE027675 (JPS), R56DE015846 (JH), UG3DE028849 (JH), and R01DE028297 (JDB), R01DE026769 (Y-HPC), the Ministry of Science and Technology in Taiwan (grant 108-2314-B-002-038-MY3), National Taiwan University Hospital (grant 109-N4534), and by National Research Foundation of Korea (NRF) grant funded by the Korea government NRF2020R1A2C2100543.
Footnotes
Declaration of Competing Interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Credit Author Statement
Graphical Structural Biology Review
VSI: Annual collections - Biomineralization.
James P. Simmer: Writing-Original draft preparation. Jan C-C. Hu: Project administration, Writing- Reviewing and Editing. Yuanyuan Hu: Visualization FIB-bSEM Figures (#1, #3, #4). Shelly Zhang: Graphical Figures (#2 and #5). Tian Liang: Writing- Reviewing and Editing. Shih-Kai Wang: Writing- Reviewing and Editing. Jung-Wook Kim: Writing- Reviewing and Editing. Yasuo Yamakoshi: Writing- Reviewing and Editing. Yong-Hee Chun: Writing- Reviewing and Editing. John D. Bartlett: Writing- Reviewing and Editing. Charles E. Smith: Visualization FIB-bSEM Figures (#1, #3, #4), Writing- Reviewing and Editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adaimy L, Chouery E, Megarbane H, Mroueh S, Delague V, Nicolas E, Belguith H, de Mazancourt P, Megarbane A, 2007. Mutation in WNT10A is associated with an autosomal recessive ectodermal dysplasia: the odonto-onycho-dermal dysplasia. Am J Hum Genet 81, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atukorala AD, Inohaya K, Baba O, Tabata MJ, Ratnayake RA, Abduweli D, Kasugai S, Mitani H, Takano Y, 2010. Scale and tooth phenotypes in medaka with a mutated ectodysplasin-A receptor: implications for the evolutionary origin of oral and pharyngeal teeth. Arch Histol Cytol 73, 139–148. [DOI] [PubMed] [Google Scholar]
- Bai Y, Yu Z, Ackerman L, Zhang Y, Bonde J, Li W, Cheng Y, Habelitz S, 2020. Protein nanoribbons template enamel mineralization. Proc Natl Acad Sci U S A 117, 19201–19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JD, Smith CE, Hu Y, Ikeda A, Strauss M, Liang T, Hsu YH, Trout AH, McComb DW, Freeman RC, Simmer JP, Hu JC, 2021. MMP20-generated amelogenin cleavage products prevent formation of fan-shaped enamel malformations. Sci Rep 11, 10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniash E, Metzler RA, Lam RS, Gilbert PU, 2009. Transient amorphous calcium phosphate in forming enamel. J. Struct. Biol 166, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyde A, 1969. Electron microscopic observations relating to the nature and development of prism decussation in mammalian dental enamel. Bull Group Int Rech Sci Stomatol 12, 151–207. [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, Berlin AM, Campbell MS, Barrell D, Martin KJ, Mulley JF, Ravi V, Lee AP, Nakamura T, Chalopin D, Fan S, Wcisel D, Canestro C, Sydes J, Beaudry FE, Sun Y, Hertel J, Beam MJ, Fasold M, Ishiyama M, Johnson J, Kehr S, Lara M, Letaw JH, Litman GW, Litman RT, Mikami M, Ota T, Saha NR, Williams L, Stadler PF, Wang H, Taylor JS, Fontenot Q, Ferrara A, Searle SM, Aken B, Yandell M, Schneider I, Yoder JA, Volff JN, Meyer A, Amemiya CT, Venkatesh B, Holland PW, Guiguen Y, Bobe J, Shubin NH, Di Palma F, Alfoldi J, Lindblad-Toh K, Postlethwait JH, 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet 48, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Blom H, Sanchez S, Tafforeau P, Märss T, Ahlberg PE, 2020. The developmental relationship between teeth and dermal odontodes in the most primitive bony fish Lophosteus. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Uitto J, 2010. Epidermolysis bullosa with pyloric atresia. Dermatol Clin 28, 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang PA, Conway JF, Margolis HC, Simmer JP, Beniash E, 2011. Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc Natl Acad Sci U S A 108, 14097–14102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP, 1999. The structural biology of the developing dental enamel matrix. J. Struct. Biol 126, 270–299. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC, 1994. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol 112, 103–109. [DOI] [PubMed] [Google Scholar]
- Fine JD, 2010. Inherited epidermolysis bullosa. Orphanet J Rare Dis. 5:12., 10.1186/1750-1172-1185-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP, 1997. Cloning and characterization of porcine enamelin mRNAs. J. Dent. Res 76, 1720–1729. [DOI] [PubMed] [Google Scholar]
- Hu Y, Smith CE, Cai Z, Donnelly LA, Yang J, Hu JC, Simmer JP, 2016. Enamel ribbons, surface nodules, and octacalcium phosphate in C57BL/6 Amelx−/− mice and Amelx+/− lyonization. Mol. Genet. Genomic Med 4, 641–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev R, Sherwood DR, 2017. Basement membranes. Curr Biol 27, R207–r211. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Weiss KM, 2003. Mineralized tissue and vertebrate evolution: the secretory calcium-binding phosphoprotein gene cluster. Proc. Natl. Acad. Sci. U.S.A 100, 4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Amemiya CT, 2014. SCPP genes in the coelacanth: tissue mineralization genes shared by sarcopterygians. J. Exp. Zool. B Mol. Dev. Evol 322, 390–402. [DOI] [PubMed] [Google Scholar]
- Kawasaki K, Keating J, Nakatomi M, Welten M, Mikami M, Sasagawa I, Puttick M, Donoghue P, Ishiyama M, 2021. Coevolution of enamel, ganoin, enameloid, and their matrix SCPP genes in osteichthyans. iScience 24, 102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerebel B, Daculsi G, Kerebel LM, 1979. Ultrastructural studies of enamel crystallites. J. Dent. Res 58, 844–851. [DOI] [PubMed] [Google Scholar]
- Kim JW, Zhang H, Seymen F, Koruyucu M, Hu Y, Kang J, Kim YJ, Ikeda A, Kasimoglu Y, Bayram M, Zhang C, Kawasaki K, Bartlett JD, Saunders TL, Simmer JP, Hu JC, 2019. Mutations in RELT cause autosomal recessive amelogenesis imperfecta. Clin. Genet 95, 375–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Hu Y, Smith CE, Richardson AS, Zhang H, Yang J, Lin B, Wang SK, Kim JW, Chun YH, Simmer JP, Hu JC, 2019. AMBN mutations causing hypoplastic amelogenesis imperfecta and Ambn knockout-NLS-lacZ knockin mice exhibiting failed amelogenesis and Ambn tissue-specificity. Mol. Genet. Genomic Med 7, e929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis HC, Kwak SY, Yamazaki H, 2014. Role of mineralization inhibitors in the regulation of hard tissue biomineralization: relevance to initial enamel formation and maturation. Front Physiol. 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohazab L, Koivisto L, Jiang G, Kytomaki L, Haapasalo M, Owen GR, Wiebe C, Xie Y, Heikinheimo K, Yoshida T, Smith CE, Heino J, Hakkinen L, McKee MD, Larjava H, 2013. Critical role for alphavbeta6 integrin in enamel biomineralization. J Cell Sci 126, 732–744. [DOI] [PubMed] [Google Scholar]
- Nanci A, Warshawsky H, 1984. Characterization of putative secretory sites on ameloblasts of the rat incisor. Am. J. Anat 171, 163–189. [DOI] [PubMed] [Google Scholar]
- Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen W, Krebsbach PH, Yamada Y, Hammarström L, Simmer JP, Fincham AG, Snead ML, Smith CE, 1998. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem 46, 911–934. [DOI] [PubMed] [Google Scholar]
- Nylen MU, Eanes ED, Omnell KA, 1963. Crystal growth in rat enamel. J Cell Biol 18, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnholm E, 1962a. The amelogenesis of human teeth as revealed by electron mircoscopy I. The fine structure of the ameloblasts. J. Ultrastructure Res 6, 229–248. [DOI] [PubMed] [Google Scholar]
- Ronnholm E, 1962b. The amelogenesis of human teeth as revealed by electron mircoscopy II. The development of the enamel crystallites. J. Ultrastructure Res 6, 249–303. [DOI] [PubMed] [Google Scholar]
- Ryan MC, Lee K, Miyashita Y, Carter WG, 1999. Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J Cell Biol. 145, 1309–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlberg C, Hormia M, Airenne T, Thesleff I, 1998. Laminin gamma2 expression is developmentally regulated during murine tooth morphogenesis and is intense in ameloblasts. J Dent Res 77, 1589–1596. [DOI] [PubMed] [Google Scholar]
- Shaw WJ, Tarasevich BJ, Buchko GW, Arachchige RMJ, Burton SD, 2020. Controls of nature: Secondary, tertiary, and quaternary structure of the enamel protein amelogenin in solution and on hydroxyapatite. J Struct Biol 212, 107630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Fincham AG, 1995. Molecular mechanisms of dental enamel formation. Crit. Rev. Oral Biol. Med 6, 84–108. [DOI] [PubMed] [Google Scholar]
- Sire JY, 1995. Ganoine formation in the scales of primitive actinopterygian fishes, lepisosteids and polypterids. Connect. Tissue Res 33, 213–222. [DOI] [PubMed] [Google Scholar]
- Skobe Z, 2006. SEM evidence that one ameloblast secretes one keyhole-shaped enamel rod in monkey teeth. Eur J Oral Sci 114 Suppl 1, 338–342. [DOI] [PubMed] [Google Scholar]
- Smith CE, 1979. Ameloblasts: secretory and resorptive functions. J Dent Res. 58, 695–707. doi: 10.1177/002203457905800221011. [DOI] [PubMed] [Google Scholar]
- Smith CE, Hu Y, Hu JC, Simmer JP, 2016. Ultrastructure of early amelogenesis in wild-type, Amelx−/−, and Enam−/− mice: enamel ribbon initiation on dentin mineral and ribbon orientation by ameloblasts. Mol. Genet. Genomic Med 4, 662–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU, 1980. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem 255, 9760–9768. [PubMed] [Google Scholar]
- Thesleff I, 2006. The genetic basis of tooth development and dental defects. Am J Med Genet A. 140, 2530–2535. [DOI] [PubMed] [Google Scholar]
- Uchida T, Tanabe T, Fukae M, 1989. Immunocytochemical localization of amelogenins in the deciduous tooth germs of the human fetus. Arch Histol Cytol 52, 543–552. [DOI] [PubMed] [Google Scholar]
- Warshawsky H, 1989. Organization of crystals in enamel. Anat Rec 224, 242–262. [DOI] [PubMed] [Google Scholar]
- Wright JT, Carrion IA, Morris C, 2015. The molecular basis of hereditary enamel defects in humans. J. Dent. Res 94, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tran B, Beniash E, Kwak SY, Margolis HC, 2019. Proteolysis by MMP20 Prevents Aberrant Mineralization in Secretory Enamel. J. Dent. Res 98, 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.