Abstract
The pathogenic cascade of Alzheimer's disease (AD) characterized by amyloid-β protein accumulation is still poorly understood, partially owing to the limitations of relevant models without in vivo neural tissue microenvironment to recapitulate cell–cell interactions. To better mimic neural tissue microenvironment, three-dimensional (3D) core-shell AD model constructs containing human neural progenitor cells (NSCs) with 2% matrigel as core bioink and 2% alginate as shell bioink have been bioprinted by a co-axial bioprinter, with a suitable shell thickness for nutrient exchange and barrier-free cell interaction cores. These constructs exhibit cell self-clustering and -assembling properties and engineered reproducibility with long-term cell viability and self-renewal, and a higher differentiation level compared to 2D and 3D MIX models. The different effects of 3D bioprinted, 2D, and MIX microenvironments on the growth of NSCs are mainly related to biosynthesis of amino acids and glyoxylate and dicarboxylate metabolism on day 2 and ribosome, biosynthesis of amino acids and proteasome on day 14. Particularly, the model constructs demonstrated Aβ aggregation and higher expression of Aβ and tau isoform genes compared to 2D and MIX controls. AD model constructs will provide a promising strategy to facilitate the development of a 3D in vitro AD model for neurodegeneration research.
Keywords: 3D bioprinting, Co-axial, In vitro AD model, Self-assembling
Graphical abstract
Highlights
-
•
3D bioprinted co-axial core-shell in vitro Alzheimer’s disease (AD) model constructs.
-
•
Co-axial constructs provide neural fiber-shaped microenvironment, a suitable shell thickness and a barrier-free core.
-
•
AD models show cell self-clustering and -assembling features and engineered reproducibility with long-term cell viability.
-
•
NSCs with FAD mutations show a higher level of self-renewal and differentiation in AD model than that in 2D and MIX models.
-
•
In vitro AD models present Aβ aggregation and higher expression of Aβ and tau isoform genes compared to 2D and MIX models.
1. Introduction
Alzheimer's disease (AD), a type of age-related neurodegenerative disease, accounts for 50–70% of all dementia cases with mixed brain pathologies [1,2]. It is reported that approximately 35.6 million dementia cases occurred in 2010, particularly among very old people [3]. If therapy for dementia is not improved, it is estimated that there will be 115.4 million people with dementia worldwide by 2050 [4], making dementia a global concern. With the aging of populations, the incidences of dementia and AD are increasing exponentially [5]. Therefore, AD has resulted in serious social and economic burdens worldwide. However, there is still no cure in the pathogenic cascade of AD [6], and recent understanding of the mechanisms underlying AD pathogenesis represents a gap in the recent failures of anti-Aβ therapies. This is because drug therapies based on AD amyloid-β precursor proteins (APP) mouse models are not only expensive and time-consuming but also exhibit no effectiveness in human clinical studies [7,8]. It is an urgent challenge to construct a reliable pathological model for effective drug development to reveal AD pathology and drug screening.
Recent advancements in in vitro brain AD models allow the study of AD pathological hallmarks, including extracellular aggregates of amyloid-β proteins (Aβ), Aβ-induced pathological tau aggregation, and complex neuroinflammatory [[9], [10], [11]]. However, there are still some limitations in the current in vitro three-dimensional (3D) AD models. For instance, a neural stem cell-encapsulated hydrogel mixer in a well was used as the 3D AD culture model and it exhibited two specific proteins, Aβ and tau, for the pathological study of AD [11]; however, cells in this mixer 3D model remained in the originally implanted position, and it was difficult for cell-encapsulated hydrogel mixer 3D model to achieve such high cell density in in vivo brain cortical regions (1––10 × 108 cells mL−1) [12]. Since hydrogel biomaterial was used as supporting functional material in the cell-encapsulated hydrogel mixer 3D model, this biomaterial would take up much space in mixer system. Therefore, a cell-encapsulated hydrogel mixer could not hold such large amounts of stem cells in the certain volumetric unit, and many cells could also often leak out from hydrogel mixer system. Paşca and the colleagues demonstrated that a laminated cerebral cortex–like structure, named human cortical spheroids containing neural cell, were generated by cell proliferation in low-attachment plates for a detailed interrogation of human cortical disease [13]. However, it is challenging to generate homogeneous and reproducible systems.
3D bioprinting is a promising technique for fabricating biomimetic structures for complex composite human-scale living tissues and various human models for biomedical development with the help of recent advancements in 3D printing technology, cell biology, and materials science [[14], [15], [16]]. Co-axial bioprinting based on 3D extrusion bioprinting is used to bioprint core-shell fiber constructs [17,18], exhibiting good cell proliferation [19,20] and significant potential for neural tissue engineering applications [21]. This potential could result from several mechanisms based on our previous study [22,23]: (1) The core part is barrier-free for cells with high density (>108 mL−1), and cell interaction is not limited to developing cell self-assembling and organizing properties to generate complex regeneration tissue structures; (2) the bioprinted engineered biomimetic structures provide fiber-shaped reconstruction of functional tissues that mimic muscle fibers or nerve networks in vivo [18]. Therefore, 3D co-axial bioprinting may be considered an effective and promising strategy for 3D in vitro AD neurodegeneration models.
In this study, 3D co-axial bioprinting was used to bioprint AD model constructs. Our bioprinted constructs were fiber-shaped core-shell structures in which a barrier-free core part could improve human neural progenitor cells (NSCs) on cell self-assembling clusters and cell development to form complex AD neural structures without the limitation of solid hydrogel materials. And biocompatible alginate was used for outer-layer supporting shell to maintain the integrity of fiber-shaped core-shell structures. To construct a bioprinted in vitro AD model, we developed APP-NSCs with familial Alzheimer's disease (FAD) mutation, characterized the core-shell constructs for nutrient exchange, and evaluated the effect of bioprinting processes on cell viability and growth. The cell self-clustering growth and neural differentiation of NSCs underlying the role of different microenvironments were observed and compared. Moreover, we explored the pathway mechanisms of the effects of different microenvironments on the fate of NSCs. Furthermore, this new bioprinted in vitro AD model construct demonstrated Aβ aggregation and higher expression of Aβ and tau isoform genes compared to 2D and 3D MIX control groups (Fig. 1).
Fig. 1.
Schematic of the potential of 3D bioprinted Alzheimer's model. NSCs indicate human neural progenitor cells, APP indicates amyloid-β precursor proteins, Aβ indicates amyloid-β proteins, AD indicates Alzheimer's disease.
2. Materials and methods
2.1. Cell culture
Human ReNcell VM neural progenitor cells (NSCs) were purchased from EMD Millipore (Temecula, CA). The cells were cultured on BD Matrigel (BD Biosciences, San Jose, CA)-coated T25 and T75 cell-culture flasks (BD Biosciences, San Jose, CA) in DMEM/F12 (Gibco) media supplemented with 2 μg mL−1 heparin (StemCell Technologies, Vancouver, Canada), 2% (v/v) B27 neural supplement (Life Technologies, Grand Island, NY, USA), 20 ng mL−1 EGF, 20 ng mL−1 bFGF and 1% (v/v) penicillin/streptomycin solution in a CO2 cell-culture incubator. Cell-culture media were changed every 3 d until cells were confluent.
2.2. Hydrogel material preparation
Sodium alginate was purchased from Aladdin (Aladdin, China). It was used as the outer shell material in bioprinted 3D co-axial core-shell constructs. The bioprinted shell bioink was prepared by dissolving 2% alginate in 0.9% sodium chloride (NaCl) solution (Kelun, China), followed by heat sterilization. Reduced growth factor BD Matrigel purchased from BD Biosciences was added into cell suspension as supplementary and used as the inner core component. Calcium chloride (CaCl2), sodium citrate and EDTA were purchased from Sigma-Aldrich (Sigma-Aldrich, Shanghai, China). For crosslinking of alginate constructs, CaCl2 was dissolved into deionized water at a final concentration of 3% (w/v) and filtered through 0.45 μm syringe filters (Thermo Scientific). For de-crosslinking of alginate, 55 mM sodium citrate and 20 mM EDTA in 0.9% NaCl solution was prepared [24].
2.3. Lentiviral gene vector packaging and infection of NSCs
The mutagenesis determination and packaging of the lentiviral gene vector construct encoding full-length human β-amyloid precursor protein (APP) with the V642I (London) and K595 N/M596L (Swedish) mutations were performed by Genechem Co., Ltd (Genechem, Shanghai, China). To transduce the NSCs with the lentiviral vectors, 50–100 μL viral solution (1 × 108 TU mL−1, MOI = 100) were added to 20–30% confluent proliferating NSCs in 6-well plate, incubated for 18–24 h, and exchanged with fresh medium three times to stop the infection. After the transfected NSCs were developed in fresh medium for 48–72 h, the expression of the infected genes could be confirmed by GFP expression by fluorescence microscopy and western blot analysis. To enrich the transfected NSCs, the cell screening was applied by culturing with using medium containing puromycin.
2.4. 3D co-axial core-shell bioprinting
3D concentric circle core-shell structures were bioprinted with a customized computer-aided co-axial extruding bioprinting device (Livprint, Medprin, China). Firstly, the customized concentric print head with two chambers and nozzles was fabricated. Outer chamber was used for shell structure, and inner chamber for core structure. Then outer chamber was connected to a 10 mL syringe containing 2% sodium alginate (w/v) dissolved in 0.9% NaCl solution (w/v) which were printed to form shell. And the inner chamber was connected to another 10 mL syringe consisted of cell suspension (1 × 108 cells mL−1) bioink with the additive of 2% Matrigel (v/v) which were printed to form core part. Two syringes were independently controlled by different pumps. The extrusion bioprinting speed was set as 15 mL h−1 for shell structure and 5 mL h−1 for core part. During bioprinting, co-axial structures were bioprinted on the receiving crosslinking platform which was a tank containing 3% CaCl2 solution. After printing, core-shell structures were washed with 0.9% NaCl solution for three times to remove remaining calcium ions, and then placed in the fresh medium. The same crosslinking and washing procedures were performed for mixed control groups consisted of 2% sodium alginate (w/v) and cell suspension (1 × 108 cells mL−1) bioink with the additive of 2% Matrigel (v/v).
2.5. Cell viability assay
Cell viability was assessed using a live/dead assay kit (KeyGEN BioTECH, Nanjing, China) according to the manufacturer's instructions. Briefly, 8 μM propidium iodide (PI) and 2 μM Calcein-AM were mixed with 10 mL 0.9% NaCl solution. The bioprinted constructs were incubated in the staining assay solution in dark at room temperature for 15 min. After gentle washing with 0.9% NaCl solution, live and dead cells were imaged using fluorescence microscope (Eclipse Ti2, Nikon, Japan). The number of live cells (green) and dead cells (red) were counted in at least five random sights under 100× magnification, and cell viability (%) was calculated (n = 3).
2.6. 3D cell cultures of NSCs
Before 3D cultures of bioprinted core-shell constructs, shell width was measured by microscope. The bioprinted NSC-laden constructs were incubated for 10–14 days depending on the experiments; proliferation media was changed every 2–3 days. Cell proliferation was evaluated via Alamar Blue Kit (MAIBIO, Shanghai, China) according to the manufacturer's instructions. Briefly, cell-laden core-shell constructs were washed with 0.9% NaCl solution three times and incubated in 6-well plates with 2 mL working solution (Alamar Blue: Fresh medium = 1:10) at 37 °C for 3 h in dark incubator. After incubation, 100 μL solution from each sample were transferred to a 96-well plate, and the optical density (OD) value was measured at wavelength of 570 nm and 630 nm by microplate reader (BioTek ELX800, VT, USA). This procedure was repeated every other day for 9 days and performed on the 2D and mixed control groups as well. The OD of all three groups were normalized to day 1 for plotting and statistics.
2.7. Proteomics mass spectrometry analysis
Proteomics of conditional medium from 3D bioprinted core-shell constructs, and 2D and mixed control groups, were analyzed by label-free quantitative proteomics mass spectrometry. The medium samples from different conditional culture (3D, MIX (non-printed) and 2D environment) at two time points including day 2 and day 14 were collected and sent for the proteomics detection with LC-MS and MaxQuant software (1.6.2.10) analysis (BiotechPack scientific, Beijing, China). To prepare the medium samples for proteomics mass spectrometry analysis, fresh media for cells in 3D, MIX (non-printed) and 2D environment were replaced every 2 days.
2.8. Scanning electron microscopy (SEM) analysis
The surface observation of outer shell and inner core constructs was performed by SEM. After being cultured for 10 days, the bioprinted constructs were washed by PBS and fixed in 2.5% glutaraldehyde for 1 h at room temperature, and dehydrated with a series of ethanol solutions (70%, 80%, 90%, 95%, 100%, 100%) for 20 min in each solution, and then frozen at −80 °C overnight. The samples were then lyophilised using a freeze-drier (LyoQuest-85 PLUS, Telstar, Spain). Hydrogel constructs were cut using a sharp scalpel for various views and sputter-coated with gold, images were acquired from SEM (Phenom, China).
2.9. Paraffin embedding and sectioning of core-shell 3D constructs
For paraffin embedding, core-shell 3D constructs were fixed in 4% paraformaldehyde (PFA) at room temperature. The PFA-fixed 3D constructs were sent for paraffin embedding (Servicebio, Wuhan, China). The paraffin blocks were then cut into 3 μm thick sections via a microtome (SLEE, Germany), mounted on polylysine-coated glass slides (Thermo Scientific), and incubated at 45 °C overnight. The sections were deparaffinized by two changes of xylene for 5 min each and then serially transferred to 100%, 90%, 70% ethanol solution for 3 min each. The sections were then rinsed with distilled water for 5 min three times. The same procedures were performed for mixed control groups.
2.10. Histological and immunohistochemical (IHC) staining
For cell observation, the sections of core-shell 3D constructs were stained with H&E according to routine histology protocols. To analysis the specific protein deposition, the IHC procedures for the sections were also performed according to routine protocols. Briefly, the sections were permeabilized and blocked by using the blocking solution at 4 °C for 12 h. After blocking endogenous peroxidase activities with 3% (v/v) H2O2 solution in TBS for 5 min at room temperature, the core-shell 3D construct sections were washed with TBST three times and then incubated with the blocking solution at room temperature for 2 h. Finally, the core-shell 3D construct sections were incubated with the primary antibody solutions at 4 °C overnight, washed five times with TBST, and incubated with second antibody solutions for 30–60 min, washed five times for 10 min each with TBST and developed by chromogenic substrate. The same procedures were performed for mixed control groups. The primary antibodies used in this study included anti-Aβ 1–16 antibody (803015, Biolegend, China) and anti-p-tau antibody (MN1020, Invitrogen, USA).
2.11. Immunofluorescence analysis
To identify the biomarkers of cells in core-shell 3D constructs, immunofluorescence analysis was performed. The intact bioprinted core-shell 3D constructs were fixed with 4% PFA at room temperature for 1 h. The fixed cells in constructs were then permeabilized by using 0.1% Triton X-100 at room temperature for 4 h, and blocked by incubating with a blocking solution (KeyGEN BioTECH, China) at 4 °C overnight. After being washed with TBST solution (Solarbio, China), the core-shell 3D constructs were incubated with primary antibodies in the diluted solution (KeyGEN BioTECH, China) at 4 °C overnight. After being washed three times with TBST, the core-shell 3D constructs were then incubated AlexaFluor secondary antibodies (Abcam, USA) at room temperature for 4 h. After being washed three times with TBST, cell nucleus was stained by using 4′,6-diamidino-2-phenylindole (DAPI, Solarbio, China) at room temperature for 10 min. To avoid fluorescence quenching, a drop of anti-fade solution (Solarbio, China) was added on the stained the core-shell 3D constructs before imaging. The fluorescence images for expression of cell surface markers was captured with a confocal laser scanning microscope (C2+/C2si+, Nikon, Japan), and the image analysis and 3D reconstitution were performed with using software provided with the microscope. The primary antibodies used in this study included anti-Nestin antibody (ab22035, Abcam); anti-SOX2 antibody (ab93689, Abcam); anti-β-tubulin type III (Tuj1, ab78078, Abcam); anti-MAP2 antibody (ab32454, Abcam); anti-GFAP antibody (ab10062, Abcam); anti-ALDH1L1 antibody (ab190298, Abcam); anti-Synaptophysin antibody (ab32127, Abcam); anti-Aβ 1–42 antibody (D9A3A, Cell Signaling Technology). The secondary antibodies included AlexaFluor 488 anti-mouse and-rabbit secondary antibodies (ab150113 and ab150077, Abcam) and AlexaFluor 568 anti-mouse and-rabbit secondary antibodies (ab175473 and ab175471, Abcam).
2.12. Alginate shell removal
After being cultured for 10 days, to obtain cells in the biprinted constructs, the core-shell 3D constructs were soaked into PBS containing 55 mM sodium citrate and 20 mM EDTA (Sigma Aldrich, Shanghai, China) for 10 min to remove the alginate shell.
2.13. Quantitative real time PCR (qRT-PCR) analysis
To analyze gene expression levels of stemness, differentiation and specific proteins of Alzheimer's disease, qRT-PCR was conducted with several kits according to the manufacturer's instructions. The bioprinted 3D co-axial cells were extracted by dissociation solution as described above. Total RNA was isolated and extracted by total RNA extraction kit (Takara). The quantity and purity of RNA was assessed using a NanoDrop Spectrophotometer (Thermo Scientific). RNA was reversely transcribed into cDNA with reverse transcription kit (Beyotime Biotechnology) according to the protocol. For qRT-PCR, pairing primer sequences were used (Table S1), including Nestin, Tubulin (TUJ1), glial fibrillary acidic protein (GFAP), APP, 3-repeat and 4-repeat tau (3R4RTau), 3-repeat tau (3RTau), 4-repeat tau (4RTau) and β-actin (ACTB). PCR amplification was performed using SYBR Green qPCR SuperMix (C11733046, Invitrogen) with an ABI PRISM® 7500 Sequence Detection System with the procedure of 95 °C 2 min; 95 °C 15 s, 60 °C 30 s, reading plate, 40 cycles. Each sample was repeated 3 times. Relative gene expression was calculated using the 2−ΔΔCt method. The same procedures were performed for 2D and mixed control groups.
2.14. Western blotting analysis
The cells were harvested from the shell-core constructs as described above and lysed on ice with 150 μL RIPA lysate (KeyGEN BioTECH, China). The lysed cells were centrifuged at 10,000×g for 20 min at 4 °C. The concentration of total protein was measured using BCA assay kit (KeyGEN BioTECH, China). 20–30 μg of protein were resolved on 10% SDS-PAGE gels and the proteins were transferred to nylon membranes (Bio-Rad). After being blocked with 5% skim milk solution for 2 h at room temperature, the proteins on nylon membranes were incubated with primary antibodies at 4 °C overnight. After being washed with TBST three times for 10 min, the proteins on nylon membranes were then stained with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Western blot images were visualized by enhanced chemiluminescence (ECL) solution (Beyotime Biotechnology, China) and captured by using BioMax film (GelView 6000 M, BLT PHOTO TECHNOLOGY, China). The protein images were quantitated by Image J software (Rawak Software, Inc., Germany), and the expression of target protein was normalized with the expression of internal reference GAPDH for plotting and statistics. The primary antibodies used in this study included anti-Aβ (6E10, 803014, BioLegend); anti-DDDDK tag (ab205606, Abcam); anti-GAPDH (HC301, TRANS). The secondary antibodies included Goat Anti-mouse IgG (HRP) (ab205719, Abcam) and Goat Anti-Rabbit IgG (HRP) (ab6721, Abcam). The same procedures were performed for 2D and mixed control groups.
2.15. Statistical analysis
Each experiment was repeated at least 3 times and results are presented as mean ± S.D. Comparison between multiple groups was performed using one-way or two-way analysis of variance (ANOVA). Statistical significance was attained at p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), p < 0.0001 (****). Comparison between two groups was performed using the paired t-test. Statistical significance was attained as asterisk (*). The results were analyzed and exported using GraphPad Prism 8.0 or Image J software.
3. Results
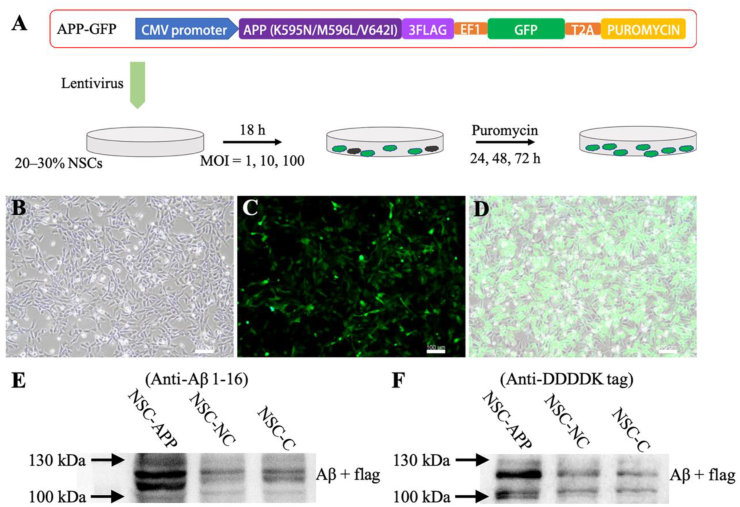
3.1. Generation of NSCs with FAD mutation using lentiviral infection
To produce neural cells that overexpress high levels of neurotoxic Aβ, NSCs derived from a human fetal brain were infected with lentiviral constructs (Fig. 2). These viral vector constructs were designed to overexpress human APP with V642I (London) and K595 N/M596L (Swedish) mutations and transfected into NSCs (Fig. 2A). Cells infected with GFP and puromycin were enriched using the conditional medium containing the suitable puromycin solution and confirmed based on GFP using fluorescence imaging (Fig. 2A–D). Moreover, to test whether the enriched cells could successfully produce Aβ, western blot (WB) analysis demonstrated that high expression of APP fragments in these NSCs with FAD mutations was detected using Anti-Aβ 1–16 and anti-DDDDK tag for fusion protein of APP and flag (Fig. 2E and F). The molecular weight of the fusion protein was approximately 120 kDa, which was greater than that of full-length APP (Fig. 2E and F).
Fig. 2.
Generation of NSCs with FAD mutation. A: Diagrams showing lentiviral vector constructs and infection. B–D: Culture observation and green fluorescence signals confirming the successful viral gene infection and expression. E–F: Images of western blotting of fusion protein of APP and flag showing the successful establishment and expression of the lentiviral vector; NSC-APP indicated the transfected cells with APP lentiviral vector constructs; NSC-NC indicated the transfected cells with negative control lentiviral vector constructs; NSC-C indicated normal NSCs. Scale bar (B–D): 100 μm.
3.2. Bioprinting of 3D cell-laden co-axial core-shell AD constructs
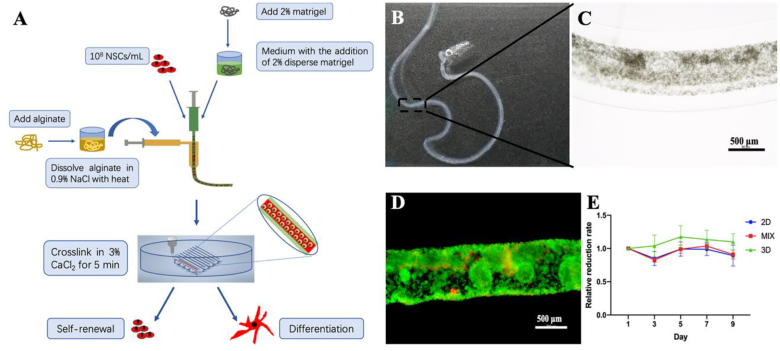
3D cell-laden coaxial core-shell AD constructs were bioprinted using a coaxial 3D bioprinter, with alginate as the shell and a high-density cell suspension (1 × 108 cells mL−1) containing the addition of low Matrigel as the core (Fig. 3A–C). In this study, we set proper extrusion parameters to print core-shell scaffolds with nutrition-permeabilized shell thickness and structural integrity (Fig. 3B and C). When the extrusion speed rates of the alginate shell and cell suspension core were set as 15 mL h−1 and 5 mL h−1, respectively, the outer shell thickness was 253.56 ± 50.75 μm (Fig. S1). The bioprinted constructs were cultured under fresh media for 10–14 days depending on the experiments. And the media was changed every 2–3 days. The same incubation procedures were performed in the MIX control groups which were fabricated by mixing the same number of cells, the same amount of alginate and matrigel as 3D bioprinted constructs with syringe.
Fig. 3.
3D co-axial bioprinting. A: Schematic illustrating the processes of 3D co-axial bioprinting for 3D culture and differentiation. B, C: Images of 3D bioprinted co-axial AD constructs. D: Live & dead staining for cell viability after bioprinting. E: Proliferation evaluation after bioprinting for 9 days compared to cells in 2D and mixed control groups.
Cell viability and proliferation were detected in bioprinted NSCs to evaluate the effect of the bioprinting process on cell survival. Live and dead staining assays were performed immediately after bioprinting, and the results demonstrated that most of the cells in the core part were green and few were red (Fig. 3D), indicating that the cell survival rate was high (90.03 ± 2.97%) and the bioprinting process did not influence the cell viability. Moreover, cell proliferation for 9 days was assessed using the Alamar Blue assay to further explore cell survival. The relative proliferation rate of the 3D co-axial constructs maintained positive growth for a long time compared to the first day, and those of 2D and MIX control groups were lower than that on the first day (Fig. 3E), indicating that 3D co-axial constructs had favorable cell survival for a large number of cells for a long time, and the potential for tissue regeneration required a long-term in vitro culture.
3.3. Different growth morphologies among 3D co-axial AD constructs and control groups
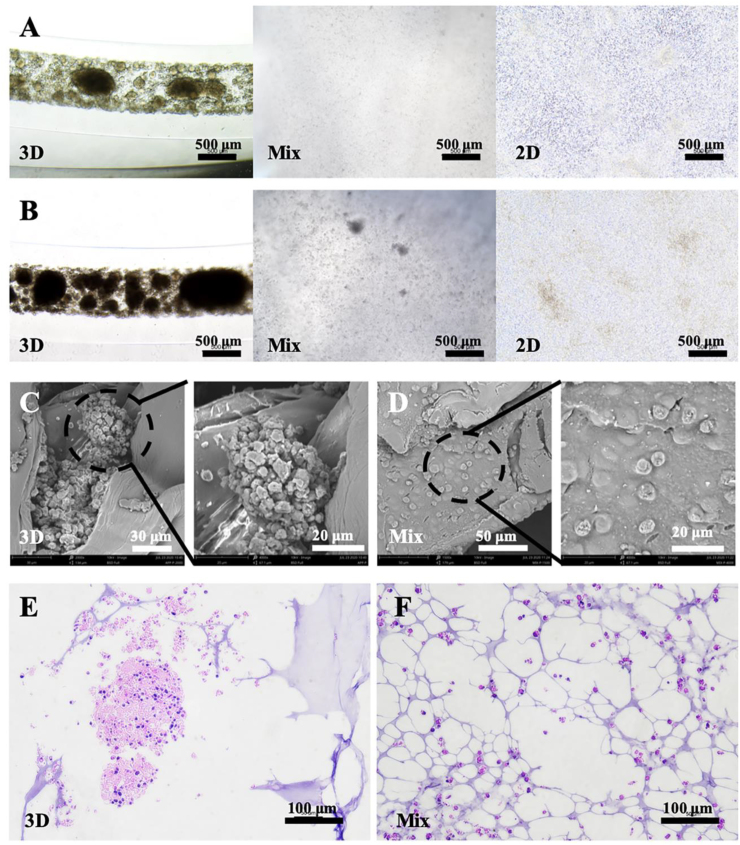
To characterize the growth behavior of NSCs with high cell survival, microscopic observation, scanning electron microscopy (SEM) imaging, and H&E, staining of 3D co-axial AD constructs was performed compared to the control groups. The 2-day culture microscopic observation showed that NSCs in co-axial constructs grew and clustered without space barriers, but NSCs in the MIX group maintained single cells and cells in 2D were crowded and floating because of lack of space (Fig. 4A). The 7-day culture observation further demonstrated that NSCs in co-axial constructs gathered more and larger clusters and the clusters mixed compared to the MIX group with a material barrier, and more cells in 2D were floating than before (Fig. 4B). After a 14-day culture, SEM imaging of 3D co-axial constructs characterized the thin alginate shell, cell clusters, and favorable growth in the core (Fig. 4C). However, SEM imaging of the MIX group revealed that NSCs almost remained single without cell proliferation (Fig. 4D). Simultaneous H&E staining of 3D co-axial constructs and MIX group was consistent with the above results (Fig. 4E and F). These results demonstrate that barrier-free NSC cores in 3D co-axial constructs provided a more favorable 3D microenvironment for NSC growth than the material barrier of the MIX group.
Fig. 4.
Observation of bioprinted constructs. Images of the morphology of NSCs in 3D co-axial constructs, and 2D and mixed control groups, for 2-day (A) and 7-day cultures (B), respectively. SEM images of 3D co-axial constructs (C) and mixed control (D) for a 10-day culture. H&E staining images of 3D co-axial constructs (E) and mixed control (F) for the 10-day culture.
3.4. 3D bioprinted co-axial AD constructs natively induced NSCs proliferation and differentiation
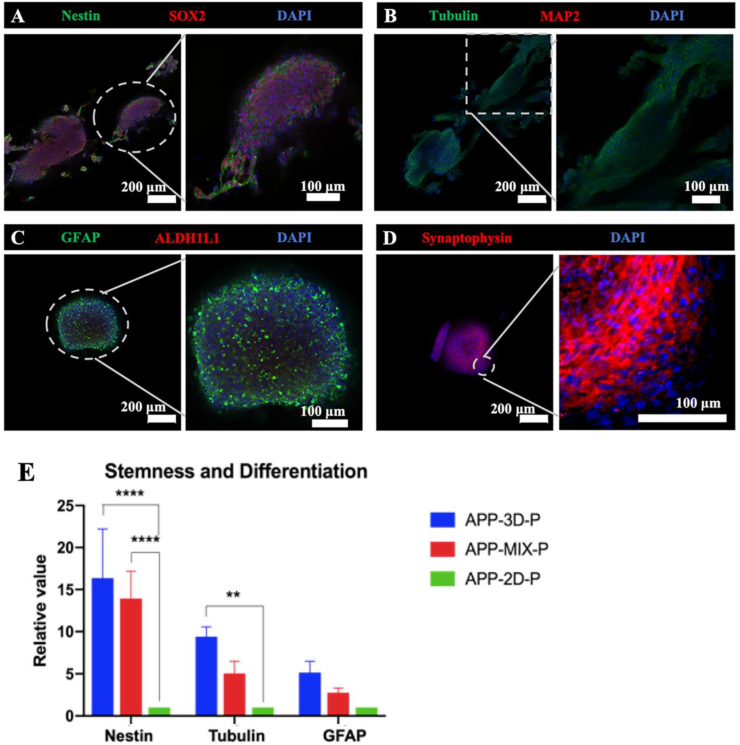
To demonstrate favorable proliferation and differentiation of NSCs in 3D co-axial cultures, in situ cells within 3D co-axial AD constructs were analyzed by immunofluorescence (IF) for the presence of neural stemness as well as neuronal and astrocyte biomarkers. After a 14-day culture in 3D co-axial constructs, in situ NSCs expressed stemness markers of Nestin and SRY-related high-mobility-group (HMG)-box protein-2 (SOX2), indicating self-renewal for proliferation (Fig. 5A), neuronal marker of class III beta-tubulin (Tuj1) (Fig. 5B), glial markers of glial fibrillary acidic protein (GFAP) and negligible aldehyde dehydrogenase 1 family, member L1 (ALDH1L1) (Fig. 5C), and synaptogenesis marker of presynaptic protein synaptophysin (Fig. 5D), showing cell differentiation for a complex neural network compared to NSCs in 2D culture (Figs. S2 and S3).
Fig. 5.
Immunofluorescence analysis and gene expression of FAD NSCs encapsulated within bioprinted core-shell constructs and control groups. The fluorescence images of cells (Within co-axial constructs, 14-day post-printing) stained with DAPI and expressing NSC markers of Nestin and SOX2 (A), neuronal marker of Tubulin (B), astrocyte marker of GFAP and negligible ALDH1L1 (C), and synaptogenesis marker of presynaptic protein synaptophysin (D). (E) Comparative gene expression of cells after removal of the shell among 3D bioprinted NSC and control groups. Relative gene expression was normalized to ACTB and compared to 2D. Mean ± S.D.; n = 3. Two-way ANOVA. **p < 0.01; ****p < 0.0001.
Meanwhile, gene expression of stemness (Nestin), neuronal (Tubulin), and glial (GFAP) markers in 3D co-axial construct NSCs for the 14-day culture were quantified via quantitative real time PCR (RT-PCR) analysis (Fig. 5E). RT-PCR results for Nestin, Tubulin, and GFAP markers revealed that self-renewal proliferation as well as neuronal and astrocyte differentiation of NSCs were promoted in 3D co-axial constructs compared to 2D and MIX control groups. Particularly, gene expression of stemness Nestin and neuronal Tubulin markers was significantly increased by 16.4-fold and 9.4-fold, respectively, in 3D co-axial constructs compared to the 2D control group. Nestin gene expression in the mixed 3D group was significantly different from that in the conventional 2D culture (two-way ANOVA, Fig. 5E). These differences may indicate the potential of a favorable 3D microenvironment for NSCs to improve the proliferation and differentiation (or maturation) of NSCs.
3.5. 3D co-axial microenvironment influenced the fate of NSCs
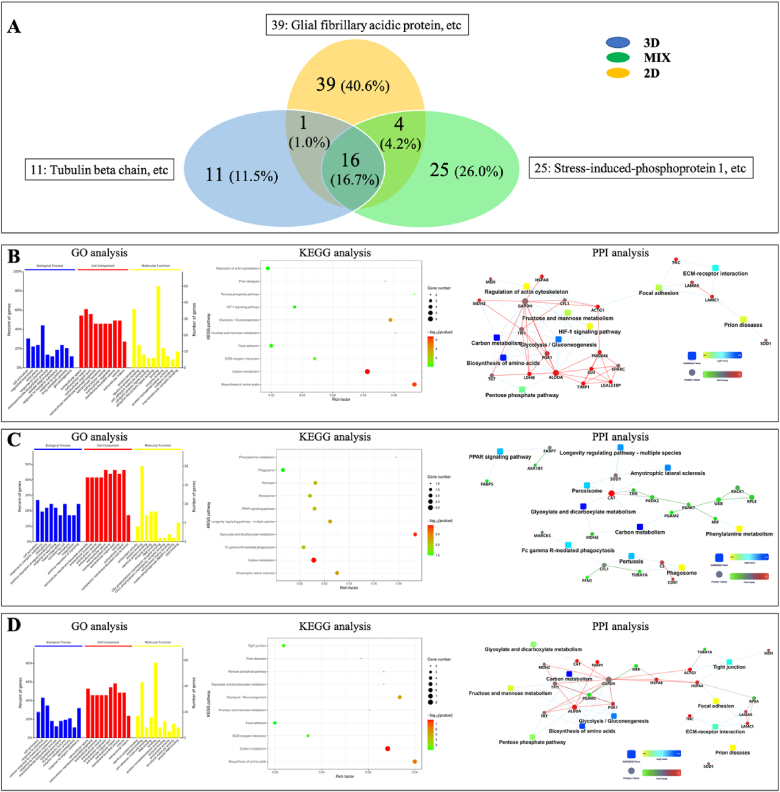
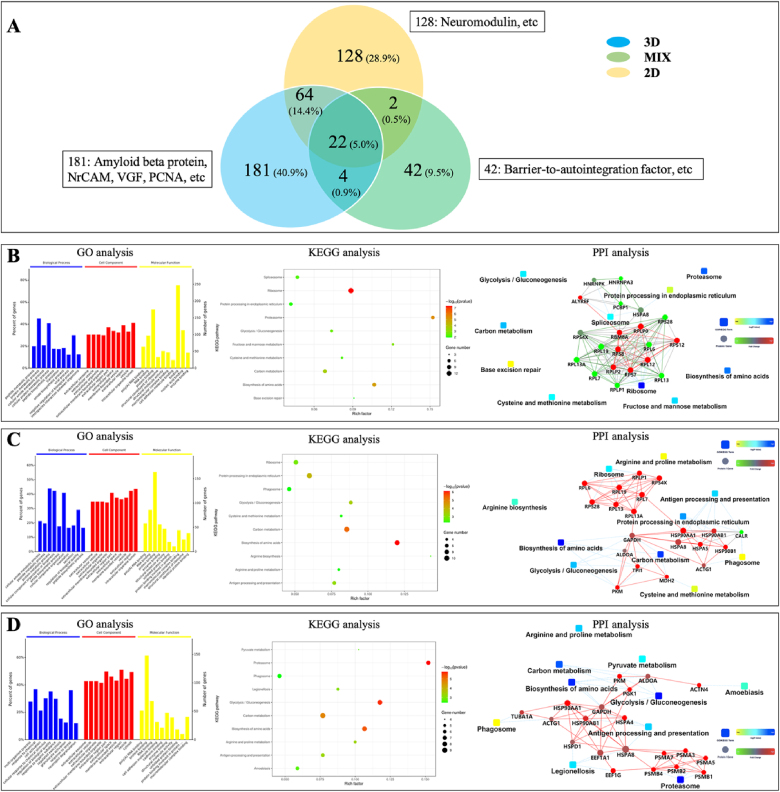
To further explore the effect of the microenvironment on the development of NSCs, secreted protein content by 3D co-axial AD constructs, MIX hydrogel, and 2D culture on day 2 and day 14 was detected using liquid chromatography–mass spectrometry. In our study, the number of total identified proteins on day 14 (443 types) increased dramatically as compared to total enriched proteins on day 2 (96 types). Meanwhile, the metabolic secretory proteins in each 14-day sample from different 3D (271 types), MIX (70 types), and 2D (217 types) cultures were more abundant than those in each 2-day sample from 3D (28 types), MIX (45 types), and 2D (60 types) cultures, respectively (Fig. 6, Fig. 7A, Tables S2 and S3). The dramatically increased proteins in 3D bioprinted microenvironment indicated the validity of our bioprinted constructs for AD model during a long time period and showed 3D co-axial microenvironment had the potential to influence the development of NSCs compared to 2D and MIX control groups [22]. Furthermore, unique and sharing proteins between different samples were compared and analyzed to demonstrate the validity and potential of our bioprinted AD model constructs (Fig. 6, Fig. 7A). The comparison indicated that cells in 3D bioprinted microenvironment represented more complex metabolic development for long time culture due to itself increased abundant unique protein content (from 11 to 181 types) and had well-regulated growth environment through producing lots of the sharing proteins with conventional 2D culture (from 17 to 86 types) from day 2 to day 14 as compared to cells in MIX and 2D cultures. And the relative few unique (from 25 to 42 types) and sharing proteins (from 20 to 28 types) in MIX 3D hydrogel culture reflected the limitation of non-bioprinted 3D microenvironment from day 2 to day 14. Compared to the results of specific proteins of 2-day samples, amyloid beta protein, neuronal cell adhesion molecule (NrCAM), VGF and proliferating cell nuclear antigen (PCNA) were expressed in 3D bioprinted AD model constructs on Day 14, and both tubulin beta chain and glial fibrillary acidic protein were produced in 3D bioprinted and 2D cultures on Day 14 (Fig. 6, Fig. 7A, Tables S2 and S3). These results confirmed the validity and potential of our bioprinted AD model constructs on the fate and development of NSCs.
Fig. 6.
Detection of proteomics in a 2-day medium of 3D bioprinted FAD NSCs and control groups. Venn diagram of proteins found in 3D and control groups (A). Gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG), and protein–protein interaction network (PPI) analysis of the differences of proteins between 2D and 3D (B), 3D and MIX (C), and 2D and MIX (D). GO analysis listed 10 significant enrichment categories in a biological process, cellular component, and molecular function (P-values set to 0.05). KEGG analysis also listed the 10 most significant pathway entries.
Fig. 7.
Detection of proteomics in a 14-day medium of 3D bioprinted FAD NSCs and control groups. Venn diagram of proteins found in 3D and control groups (A). Gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG), and protein–protein interaction network (PPI) analysis of the differences of proteins between 2D and 3D (B), 3D and MIX (C), and 2D and MIX (D). GO analysis listed 10 significant enrichment categories in a biological process, cellular component, and molecular function (P-values set to 0.05). KEGG analysis also listed the 10 most significant pathway entries.
Next, based on the significantly different proteins between different culture methods, we systematically elucidated the related pathways underlying the role of microenvironment affecting the proliferation and development of NSCs by gene ontology (GO), Kyoto encyclopedia of genes and genomes (KEGG), and protein–protein interaction network (PPI) analysis. The day 2 results showed that when metabolic pathways between 3D bioprinted and 3D MIX cultures were compared to those between 3D bioprinted and 2D cultures, there was a distinct difference except similar enrichment categories in the cellular component, indicating that 3D MIX microenvironment provided markedly different stimuli to AD NSCs compared to the 2D environment. Meanwhile, the functional pathway analysis between 3D MIX and 2D culture exhibited high similarity to that between 3D bioprinted and 2D cultures, and there were just some alterations. For example, KEGG analysis revealed that the 10 most significant pathway entries based on the differentially expressed proteins/genes were similar to those between 3D and 2D, except for tight junctions as well as glyoxylate and dicarboxylate metabolism, rather than regulation of actin cytoskeleton and HIF-1 signaling pathway. These results demonstrated that 3D microenvironments between 3D and MIX produced some same induction and stimulus on day 2, but different bio-fabrication strategies between 3D bioprinted and 3D MIX needed further exploration to demonstrate different effect in the cell fate of growth and differentiation. And the detail results were shown in Fig. 6, Table S2. On day 2, by analyzing the characteristics of the differentially expressed proteins between two different culture environments and the relationship of multiple biological pathways involved in NSC growth and development via functional pathway analysis, we found that the pathway of metabolism of amino acid biosynthesis consisting of abundant proteins with a high p-value and rich factor mainly contributed to the different effects on the development of NSCs between 3D and 2D culture environments or between MIX and 2D cultures. This pathway mainly involved aldolase A (ALDOA), reduced glyceraldehyde-phosphate dehydrogenase (GAPDH), cGMP-dependent protein kinase 1 (PKG1), transketolase (TKT), triosephosphate isomerase (TPI1), and phosphoglycerate mutase M isozymes (PGAM2) proteins/genes (Fig. 6B and D). Moreover, the different effects of 3D bioprinted and 3D MIX microenvironments on NSC growth and differentiation were related to glyoxylate and dicarboxylate metabolism involving catalase (CAT) and malate dehydrogenase (MDH2), which represented a high p-value (Red in KEGG analysis) and rich factor (Fig. 6C).
Furthermore, to understand the regulation of our bioprinted AD model constructs on the fate and development of NSCs after a longer time period, the related pathways on day 14 were also analyzed. The results showed the functional metabolic pathway between different two cultures represented distinct difference, such as the regulation of negative regulation of biological process in GO analysis and splicesome KEGG pathway between 3D bioprinted and 2D cultures, the effect of response to stress and cadherin binding process in GO analysis between 3D MIX and 2D cultures (See details in Fig. 7, Table S3). The difference results reflected the different effects of 3D bioprinted, 3D MIX, and 2D environments on the fate and development of NSCs after a longer time period. Moreover, compared to day 2 results, we found that the effects of different environments on metabolic pathways of NSCs resulted from totally different reasons, suggesting complex regulation of environment for 14-day culture. The results revealed that the pathway of metabolism of ribosome involving 60S acidic ribosomal protein (RPLP), 60S ribosomal protein (RPL), and 40S ribosomal protein (RPS) mainly effected the different development of NSCs between 3D and 2D culture environments, because of abundant genes, a high p-value and rich factor in this pathway (Fig. 7B). Similarly, the different effects on NSC growth were mainly regulated by biosynthesis of amino acids involving ALDOA, GAPDH, TPI1, MDH2, and pyruvate kinase (PKM) between 3D bioprinted and 3D MIX microenvironments (Fig. 7C), and the metabolic pathway of proteasome involving proteasome subunit alpha (PSMA) and proteasome subunit beta (PSMB) between MIX and 2D cultures (Fig. 7D).
3.6. Increased Aβ and gene expression of Aβ and tau in the 3D bioprinted co-axial AD model constructs
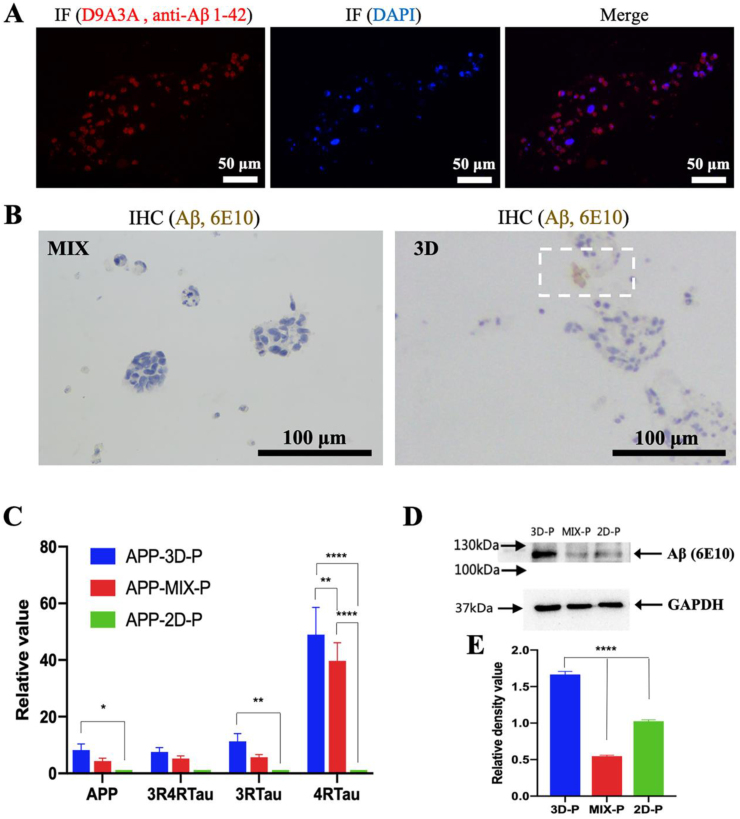
To determine whether our 3D bioprinted co-axial AD model constructs promoted Aβ expression and accelerated Aβ accumulation, a pathological hallmark of AD [6], we compared the level of Aβ in the 3D bioprinted AD culture models with 2D and 3D MIX control groups for the 14-day normal culture by using various molecular analyses. Aβ expression in NSCs in 3D bioprinted AD cultures was evaluated via IF analysis for the presence of Aβ markers. Fixed NSCs robustly expressed 42-amino-acid amyloid-β isoforms (Aβ 1–42) on day 14 (Fig. 8A). To further explore pathology development in 3D bioprinted co-axial AD model constructs, Aβ accumulation plaques by 3D bioprinted AD NSCs, 3D MIX, and 2D AD NSCs were detected via immunohistochemical (IHC) analysis. The presence of Aβ plaques in 3D bioprinted AD constructs was also confirmed by IHC staining using the Aβ 1–16 antibody 6E10, but not in the 3D MIX control group (Fig. 8B).
Fig. 8.
Evaluation of amyloid-β and p-tau levels in 3D bioprinted FAD NSCs. A: Immunofluorescence (IF) analysis of Aβ (red) with a paraffin section. B: Immunohistochemical (IHC) staining of Aβ plaques (brown) with a paraffin section. C: Comparative APP-related gene expression of cells after removal of the shell among 3D bioprinted NSC constructs (APP-3D-P) and control groups (APP-MIX-P and APP-2D-P). Relative gene expression was normalized to ACTB and compared to 2D. Mean ± S.D.; n = 3. Two-way ANOVA. *p < 0.05; **p < 0.01; ****p < 0.0001. D: Different expression of fusion protein of APP and flag was detected via western blotting (WB) analysis among 3D bioprinted NSC constructs (3D-P) and control groups (MIX-P and 2D-P). E: Comparative protein relative expression analysis based on the results of WB among 3D bioprinted NSC constructs (3D-P) and control groups (MIX-P and 2D-P). The relative value was normalized to GAPDH and compared to 2D with ImageJ and Prism 8 software. Mean ± S.D.; n = 3. One-way ANOVA. ****p < 0.0001.
Moreover, Aβ expression was quantified using RT-PCR and WB assays. RT-PCR analysis of APP gene expression revealed that the level in 3D bioprinted AD constructs was higher than that in 3D MIX or 2D AD cultures. The expression level of NSCs in 3D bioprinted AD constructs significantly increased by 8.2-fold in 14-day culture compared to 2D AD control (p < 0.05, Fig. 8C). Our WB analysis also demonstrated the higher expression of Aβ in 3D bioprinted AD constructs compared to 3D MIX and 2D AD control cells (Fig. 8D). Therefore, based on WB results, we found that 3D bioprinted AD constructs exhibited 3.0-fold and 1.6-fold more Aβ expression in extracted cells than in 3D MIX and 2D AD cultures, respectively (Fig. 8E). These results indicated accelerated Aβ accumulation in 3D bioprinted co-axial AD constructs for the 14-day culture, but insufficient secretion in 3D MIX and 2D AD cultures.
Increased tau level alteration was also demonstrated via RT-PCR analysis with elevated p-tau gene expression. Our WB results demonstrated that NSCs in 3D bioprinted co-axial AD constructs exhibited a dramatic increase in mature human 4-repeat tau isoform (4Rtau) expression, which may play an important role in reconstituting tauopathy, compared to the other two control groups. Meanwhile, the level of 3-repeat tau isoforms (3Rtau) also exhibited significant enhancement in 3D bioprinted co-axial AD constructs compared to 2D culture (Fig. 8C). These data suggest that tauopathy is positively related to the excessive accumulation of Aβ in our 3D bioprinted co-axial AD model constructs.
4. Discussion
Favorable in vitro models of human AD brain for the study platforms of AD mechanisms and drug application must have the ability to recapitulate complex brain structures where multiple brain cell types are present, and to generate homogeneous and reproducible disease model systems [25,26]. However, most of the currently relevant in vitro AD disease models, including engineered 3D cell culture [11] and self-assembling brain organoid models [26], do not meet the above requirements simultaneously. To overcome these limitations, we fabricated a bioprinted 3D co-axial model construct that can provide a nerve-fiber-like 3D microenvironment to simultaneously develop cell renewal and differentiation and accelerate Aβ accumulation. Our bioprinted core-shell co-axial in vitro human AD culture model in which the core was barrier-free for cell suspension growth, could utilize the self-assembly and self-organizing properties to provide an engineered platform for complex mechanistic studies and drug screening.
Derived from 3D extrusion bioprinting platforms, which is promising for meeting the requirements of constructing complex composite tissue constructs [16], our bioprinted 3D co-axial constructs depended on two key factors, cell resource and structure design, to achieve the successful establishment of an in vitro AD model. In the native brain of patients with AD, where neurons and astrocytes are present, human neurons have exhibited elevated levels of toxic Aβ species and phosphorylated tau; thus, it is important to construct suitable cell resources. Similar to transgenic mice overexpressing APP with FAD mutations, which have been used as the standard AD model for basic mechanistic studies and drug discovery [27], we successfully generated human AD APP NSCs harboring FAD mutations by lentiviral infection. Moreover, the differentiation potential of NSCs has made it possible to obtain human AD neurons and astrocytes [28,29]. The nutrient transport of media and the mechanical integrity based on the engineered structure of the in vitro AD model should also be considered [30]. In this study, core-shell scaffolds with structural integrity and nutrition-permeabilized shell thickness were bioprinted successfully, with an outer shell thickness of 240.79 ± 28.41 μm. The shell thickness was approximately 200 μm, which is the hydrogel diffusion limit distance; therefore, the integrated bioprinted constructs were theoretically allowed to provide sufficient nutrient transport from the media [31]. Our results on cell viability and proliferation demonstrated the fitness of our bioprinted co-axial constructs for the in vitro AD model.
The bioprinted co-axial constructs exhibited a more physiologically relevant microenvironment for NSCs characterized by self-clustering and -assembling cell interactions. Our results demonstrated that NSCs in co-axial AD constructs gathered and formed several large clusters and the clusters mixed together compared to the MIX group with a material barrier, and several cells in 2D were floating during culturing. We speculated that the barrier-free 3D microenvironment in the core part of the co-axial constructs provided a large space for growth and differentiation, and co-axial fiber constructs mimicked nerve networks in vivo for the reconstruction of fiber-shaped functional tissues [18]. However, 3D MIX hydrogel separated cell–cell interactions and limited nutrient transport into deep inner parts [32]. Our lab's previous research results also showed that neural cells were completely separated by hydrogel in printed cell-encapsulated structures and mixed groups, and tended to maintain stemness states. However, in co-axial core-shell fiber-shaped constructs, cells were connected with neighboring cells in all directions and had a strong differentiation tendency [22]. Meanwhile, our lab previous study found that different cells would form multicellular self-assembled heterogeneous tumor tissue and cell-cell interactions demonstrated by CRE-LOXP switch gene system in co-axial core-shell fiber-shaped constructs [23]. Therefore, favorable constructs for neural tissue needed a favorable 3D microenvironment to improve proliferation and differentiation (or maturation) of NSCs [21,33]. NSCs in our bioprinted co-axial AD model constructs presented not only self-renewal characterized by expression of stemness (Nestin, SOX2) markers and significant gene expression (Nestin) but also stem cell differentiation with expression of neuronal (Tubulin) and astrocyte (GFAP, ALDH1L1) markers as well as higher Tubulin and GFAP gene expression compared to 2D and MIX control groups. These results demonstrate that the 3D co-axial microenvironment provided a barrier-free core part for cell self-assembling growth and differentiation, consistent with previous reports [34]. However, MAP2, another neuronal marker, did not be detected by IF analysis. As known, TuJ1 has been a marker of neurons in the central and peripheral nervous systems from the early stage of neuronal morphologic differentiation [35]. And since MAP2 is largely located in neuronal cell dendrites where it is mainly associated with microtubules and serves as substrates for most of protein kinases and phosphatases present in neurons, MAP2 has been proposed to play important roles in the outgrowth of neuronal processes [36]. Therefore, we indicated that after 14-day culture, neurons differentiated from neural stem cells in our bioprinted constructs might be at the early stage of neural differentiation, and these neurons did not express MAP2 until further development of neuronal processes.
The role of the microenvironment in the self-renewal and differentiation of AD NSCs in an in vitro AD model is not fully understood. To explore the effects of 3D bioprinted, 3D MIX, and conventional 2D culture microenvironments on the related pathways of protein metabolic processes, we analyzed cell-secreted proteomics in conditional media on day 2 and day 14 using label-free quantitative proteomics mass spectrometry. We found that most of enriched proteins were identified as unique proteins existing only one culture environment in not only day 2 samples (78.1%) but also day 14 samples (79.25), exhibiting a significantly different expression robustly and the effects of in vivo-like microenvironment for guiding the differentiation of NSCs [37]. Successful 3D brain disease cell culture models must have the ability to recapitulate various aspects of the human neural physiology in vitro to replicate basic disease processes of AD [38] and this complex interaction in physiology was supported with the dramatical increase of the number of total identified proteins and unique proteins in each sample after 14 days culture as compared to day 2 results. Particularly, NSCs in 3D bioprinted AD model constructs showed a faster increase of unique protein content from day 2 to day 14 than cells in MIX and 2D cultures, suggesting the improvement of our bioprinted AD constructs on the complex interactions of AD in physiology for a long time. This improvement could be demonstrated by the enriched expression of unique proteins of the day 14 sample from 3D bioprinted AD model constructs, including amyloid beta protein, and neural development factors, such as NrCAM [39], VGF [40], and PCNA [41]. Especially amyloid beta protein which was only identified in day 14 sample from our AD model constructs had a distinct role in AD pathogenic cascade [42]. And abundant tubulin beta chain and glial fibrillary acidic protein in 3D bioprinted constructs on day 14 also confirmed our lab previous findings that co-axial core-shell fiber-shaped constructs had an important role in differentiation tendency for neural stem cells [22]. These results reflected the NSC fate of growth and differentiation induced by hydrogel environmental factors [43].
Furthermore, we found that on day 2, the mainly related pathway underlying the role of microenvironment affecting the proliferation and development of NSCs between 3D and 2D culture environments or between MIX and 2D cultures was metabolism of amino acid biosynthesis involving ALDOA, GAPDH, TKT, PGAM2 and PKG1 proteins/genes. ALDOA [44], GAPDH [45], TKT [46] and PGAM2 [47] as functional factors are associated with the regulation of cell proliferation and they are usually overexpressed in tumor cells. PKG1 is present and functionally important in intrinsic primary afferent neurons [48]. Particularly, TPI deficiency has also been shown to lead to neurological symptoms [49]. GAPDH has been shown to interact with neurodegenerative disease-associated proteins, including the amyloid-beta protein precursor [50,51]. These proteins were significantly upregulated in 2D culture, indicating wild proliferation, which may lead to cell death because of the leakage of space and the impairment of cell crowding. However, TPI- and GAPDH-related neurologic symptoms exhibited mild expression levels in 3D bioprinted or 3D MIX microenvironment, indicating that the in vivo-like 3D microenvironment not only promotes cell self-renewal but also induces NSC differentiation into neurons.
Moreover, the different effects on NSC growth and differentiation between 3D bioprinted and 3D MIX microenvironments were mainly related to glyoxylate and dicarboxylate metabolism involving CAT and MDH2. CAT, an enzyme for reactive oxygen species, was upregulated in 3D bioprinted co-axial constructs to increase cell viability and lifespan [52]. However, a novel tumor suppressor gene, MDH2, which is associated with paraganglioma development [53], was downregulated in 3D bioprinted co-axial constructs. The aforementioned results were consistent with the long-term viability and higher level of differentiation into neurons and astrocytes in 3D bioprinted co-axial constructs compared to the MIX culture.
However, due to the complex interactions of neural physiology for AD after 14-day development, the related pathways for the regulation on the fate and development of NSCs were changed between different two cultures. We found the ribosome involving RPLP, RPL and RPS mainly affected the different development of NSCs between 3D and 2D culture environments. The different expression of these ribosomal proteins between 3D and 2D cultures showed different effects of culture environment on cells since the expression of ribosome biogenesis was regulated by the response to environmental conditions to control cell growth [54]. And the mainly related pathway for the different effects on NSC growth between 3D bioprinted and 3D MIX microenvironments was changed into biosynthesis of amino acids and similar to the pathway on day 2 between 3D and 2D culture environments or between MIX and 2D cultures. The above results showed biosynthesis of amino acids was involved in cell proliferation and proliferation, indicating that 3D core-shell constructs could have particularly different effects on the development of NSCs as compared to 2D culture environment. Meanwhile, the metabolic pathway of proteasome involving PSMA and PSMB mainly influenced the different growth of cells between MIX and 2D cultures. Proteasome regulates protein degradation which plays major roles in cell life and death [55] and had higher expression in 2D than that in MIX culture, suggesting that the inactivity of cells in MIX culture [56].
Our results indicate a favorable microenvironment with nutrient exchange and barrier-free cell development in 3D bioprinted co-axial construct culture conditions, and diffusion of Aβ into the media in 2D cultures and 3D in vitro co-axial constructs for the AD model were bioprinted. The amyloid hypothesis of AD supposes that the excessive accumulation of amyloid-β peptide may lead to neurodegeneration with memory deficits in the brain [57]. Our results demonstrated that Aβ deposition was associated with increased Aβ expression and accumulation. The higher level of AD neuron-derived Aβ was quantified using RT-PCR and WB techniques compared to 2D and MIX cultures. Moreover, the gene expression of 4Rtau and 3Rtau proteins was significantly improved. The six tau isoforms in a rat FAD model were found to be related to some aspects of tauopathy [58]. Therefore, our 3D bioprinted in vitro co-axial constructs provided an effective and promising strategy for the establishment of a 3D in vitro AD model.
However, this study has several limitations. Recent advancements in stem cells have made induced pluripotent stem cells (iPSCs) from specific AD patients available to be studied for AD pathology [59]. iPSCs carrying the identical genetic information of patients with AD can directly be used to study the mechanisms of AD and potential drug screening without the overexpression of key genes. iPSC technology allows investigators to identify and validate new pathways of the AD pathogenic cascade in a human-brain-like environment. Further studies should take advantage of patient-derived iPSCs for personalized medicine applications. Another limitation is the lack of exploration of neuroinflammation in the AD brain, which is a key component of neurodegenerative disorders in AD. Neuroinflammation is commonly induced through the activation of microglia and astrocytes [42]. Importantly, the in vitro AD model could not include the study of similar consciousness effects of animal models and the response to external stimuli such as drug discovery [25]. In the future, we plan to use 3D bioprinting for small circular core-shell constructs with injectability, where there are neural stem cells in the core and microglial cells in the shell, and current co-axial core-shell AD model constructs to engineering application.
5. Conclusions
Our 3D bioprinted in vitro AD model constructs based on 3D co-axial bioprinting were successfully established with the advantages of nutrient exchange and barrier-free structure. 3D bioprinted co-axial AD model constructs exhibited improvement in self-clustering and -assembling cells, and they engineered reproducibility via core-shell extrusion bioprinting. Our model could maintain long-term cell viability and self-renewal, and it represents a higher level of differentiation than 2D and 3D MIX (non-printed) models. We revealed the different effects of 3D bioprinted co-axial, 2D, and 3D MIX microenvironments on the growth and development of NSCs by mainly regulating the biosynthesis of amino acids as well as glyoxylate and dicarboxylate metabolism on day 2 and ribosome, biosynthesis of amino acids and proteasome on day 14., indicating that 3D bioprinted co-axial in vivo-like microenvironment not only promotes cell self-renewal but also induces NSC differentiation into neurons for long life spans. Moreover, our 3D bioprinted in vitro co-axial constructs demonstrated increased Aβ expression and Aβ accumulation, and an effective and promising strategy for the establishment of a 3D in vitro AD model for AD neurodegeneration scientific research.
Author contributions
Y.Z. and T.X., outlined the paper, conceived, and designed the experiments. Y.Z. wrote the main paper text, did most experiments and data analysis. HY.C. performed WB and RT-PCR experiments. XY.L. cultured cells. All authors discussed the results and commented on the paper.
CRediT authorship contribution statement
Yi Zhang: Conceptualization, Methodology, Software, Writing – original draft, Writing – review & editing. Haiyan Chen: Investigation, Funding acquisition, Writing – review & editing. Xiaoyan Long: Investigation. Tao Xu: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported from the Key Research and Development Projects of Chinese People's Liberation Army (Grant No. BWS17J036). Natural Science Foundation of China (Grant No. 32000956), China Postdoctoral Science Foundation (Grant No.2020M670294).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.09.023.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schneider J.A., Arvanitakis Z., Bang W., Bennett D.A. Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 2007;69:2197–2204. doi: 10.1212/01.wnl.0000271090.28148.24. [DOI] [PubMed] [Google Scholar]
- 2.Lobo A., Launer L.J., Fratiglioni L., Andersen K., Hofman A. Prevalence of dementia and major subtypes in Europe: a collaborative study of population-based cohorts. Neurology. 2000;54:S4–S9. [PubMed] [Google Scholar]
- 3.Ngandu T., Mangialasche F., Kivipelto M. In: Global Clinical Trials for Alzheimer's Disease. Bairu M., Weiner M.W., editors. Academic Press; San Diego: 2014. Chapter 1-the epidemiology and prevention of Alzheimer's disease and projected burden of disease; pp. 3–20. [Google Scholar]
- 4.Prince M., Bryce R., Albanese E., Wimo A., Ribeiro W., Ferri C.P. The global prevalence of dementia: a systematic review and metaanalysis, Alzheimer's Dementia. 2013;9:63–75. doi: 10.1016/j.jalz.2012.11.007. e2. [DOI] [PubMed] [Google Scholar]
- 5.Wu Y.-T., Beiser A.S., Breteler M.M.B., Fratiglioni L., Helmer C., Hendrie H.C., Honda H., Ikram M.A., Langa K.M., Lobo A., Matthews F.E., Ohara T., Pérès K., Qiu C., Seshadri S., Sjölund B.-M., Skoog I., Brayne C. The changing prevalence and incidence of dementia over time — current evidence, Nat. Rev. Neurol. 2017;13:327–339. doi: 10.1038/nrneurol.2017.63. [DOI] [PubMed] [Google Scholar]
- 6.Tanzi R.E., Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective, Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Oxford A.E., Stewart E.S., Rohn T.T. Clinical trials in Alzheimer's disease: a hurdle in the path of remedy, Int. J. Alzheimer's Dis. 2020:5380346. doi: 10.1155/2020/5380346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings J., Lee G., Ritter A., Sabbagh M., Zhong K. Alzheimer's disease drug development pipeline: 2019. Alzheimer's & Dementia: Transl. Resear. Clin. Intervent. 2019;5:272–293. doi: 10.1016/j.trci.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwak S.S., Washicosky K.J., Brand E., von Maydell D., Aronson J., Kim S., Capen D.E., Cetinbas M., Sadreyev R., Ning S., Bylykbashi E., Xia W., Wagner S.L., Choi S.H., Tanzi R.E., Kim D.Y. Amyloid-β42/40 ratio drives tau pathology in 3D human neural cell culture models of Alzheimer's disease. Nat. Commun. 2020;11:1377. doi: 10.1038/s41467-020-15120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J., Wetzel I., Marriott I., Dréau D., D'Avanzo C., Kim D.Y., Tanzi R.E., Cho H. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer's disease. Nat. Neurosci. 2018;21:941–951. doi: 10.1038/s41593-018-0175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J., Klee J.B., Zhang C., Wainger B.J., Peitz M., Kovacs D.M., Woolf C.J., Wagner S.L., Tanzi R.E., Kim D.Y. A three-dimensional human neural cell culture model of Alzheimer's disease, Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morrison B., Cullen D.K., LaPlaca M. In: Neural Tissue Biomechanics. Bilston L.E., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Vitro models for biomechanical studies of neural tissues; pp. 247–285. [Google Scholar]
- 13.Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.-Y., O'Rourke N.A., Nguyen K.D., Smith S.J., Huguenard J.R., Geschwind D.H., Barres B.A., Paşca S.P. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W.J., Zhong Z., Hu N., Zhou Y.X., Maggio L., Miri A.K., Fragasso A., Jin X.Y., Khademhosseini A., Zhang Y.S. Coaxial extrusion bioprinting of 3D microfibrous constructs with cell-favorable gelatin methacryloyl microenvironments. Biofabrication. 2018;10:10. doi: 10.1088/1758-5090/aa9d44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mota F., Braga L., Rocha L., Cabral B. 3D and 4D bioprinted human model patenting and the future of drug development. Nat. Biotechnol. 2020;38:689–694. doi: 10.1038/s41587-020-0540-1. [DOI] [PubMed] [Google Scholar]
- 16.Kang H.-W., Lee S.J., Ko I.K., Kengla C., Yoo J.J., Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity, Nat. Biotechnol. 2016;34:312–319. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 17.Gong J.X., Schuurmans C.C.L., van Genderen A.M., Cao X., Li W.L., Cheng F., He J.J., Lopez A., Huerta V., Manriquez J., Li R.Q., Li H.B., Delavaux C., Sebastian S., Capendale P.E., Wang H.M., Xie J.W., Yu M.F., Masereeuw R., Vermonden T., Zhang Y.S. Complexation-induced resolution enhancement of 3D-printed hydrogel constructs. Nat. Commun. 2020;11:14. doi: 10.1038/s41467-020-14997-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onoe H., Okitsu T., Itou A., Kato-Negishi M., Gojo R., Kiriya D., Sato K., Miura S., Iwanaga S., Kuribayashi-Shigetomi K., Matsunaga Y.T., Shimoyama Y., Takeuchi S. Metre-long cell-laden microfibres exhibit tissue morphologies and functions. Nat. Mater. 2013;12:584–590. doi: 10.1038/nmat3606. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Kankala R.K., Zhu K., Wang S.-B., Zhang Y.S., Chen A.-Z. Coaxial extrusion of tubular tissue constructs using a gelatin/GelMA blend bioink. ACS Biomater. Sci. Eng. 2019;5:5514–5524. doi: 10.1021/acsbiomaterials.9b00926. [DOI] [PubMed] [Google Scholar]
- 20.Cao X., Ashfaq R., Cheng F., Maharjan S., Li J., Ying G.L., Hassan S., Xiao H.Y., Yue K., Zhang Y.S. A tumor-on-a-chip system with bioprinted blood and lymphatic vessel pair, Adv. Funct. Mater. 2019;29:13. doi: 10.1002/adfm.201807173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato-Negishi M., Onoe H., Ito A., Takeuchi S. Rod-shaped neural units for aligned 3D neural network connection. Adv. Healthc. Mater. 2017;6:1700143. doi: 10.1002/adhm.201700143. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Wang X., Chen H., Jin Z., Dai X., Zhang X., Zhang L., Xu T. A comparative study of the behavior of neural progenitor cells in extrusion-based in vitro hydrogel models, Biomed. Mater. 2019;14 doi: 10.1088/1748-605X/ab3b4b. [DOI] [PubMed] [Google Scholar]
- 23.Dai X., Liu L., Ouyang J., Li X., Zhang X., Lan Q., Xu T. Coaxial 3D bioprinting of self-assembled multicellular heterogeneous tumor fibers. Sci. Rep. 2017;7:1457. doi: 10.1038/s41598-017-01581-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaetani R., Doevendans P.A., Metz C.H.G., Alblas J., Messina E., Giacomello A., Sluijter J.P.G. Cardiac tissue engineering using tissue printing technology and human cardiac progenitor cells. Biomaterials. 2012;33:1782–1790. doi: 10.1016/j.biomaterials.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Quinti L., Park J., Brand E., Tanzi R.E., Kim D.Y. In: Neurotherapeutics in the Era of Translational Medicine. Smith R.A., Kaspar B.K., Svendsen C.N., editors. Academic Press; 2021. Chapter 13-3D Alzheimer's disease in a dish: implications for drug discovery; pp. 311–331. [Google Scholar]
- 26.Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaguri H., Nilsson P., Hashimoto S., Nagata K., Saito T., De Strooper B., Hardy J., Vassar R., Winblad B., Saido T.C. APP mouse models for Alzheimer's disease preclinical studies. EMBO J. 2017;36:2473–2487. doi: 10.15252/embj.201797397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu Q., Tomaskovic-Crook E., Lozano R., Chen Y., Kapsa R.M., Zhou Q., Wallace G.G., Crook J.M. Functional 3D neural mini-tissues from printed gel-based bioink and human neural stem cells. Adv. Healthc. Mater. 2016;5:1429–1438. doi: 10.1002/adhm.201600095. [DOI] [PubMed] [Google Scholar]
- 29.D'Avanzo C., Aronson J., Kim Y.H., Choi S.H., Tanzi R.E., Kim D.Y. Alzheimer's in 3D culture: challenges and perspectives. Bioessays. 2015;37:1139–1148. doi: 10.1002/bies.201500063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drury J.L., Mooney D.J. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials. 2003;24:4337–4351. doi: 10.1016/s0142-9612(03)00340-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y., Yu Y., Chen H., Ozbolat I.T. Characterization of printable cellular micro-fluidic channels for tissue engineering. Biofabrication. 2013;5 doi: 10.1088/1758-5082/5/2/025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giandomenico S.L., Lancaster M.A. Probing human brain evolution and development in organoids. Curr. Opin. Cell Biol. 2017;44:36–43. doi: 10.1016/j.ceb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Sugai K., Nishimura S., Kato-Negishi M., Onoe H., Iwanaga S., Toyama Y., Matsumoto M., Takeuchi S., Okano H., Nakamura M. Neural stem/progenitor cell-laden microfibers promote transplant survival in a mouse transected spinal cord injury model. J. Neurosci. Res. 2015;93:1826–1838. doi: 10.1002/jnr.23636. [DOI] [PubMed] [Google Scholar]
- 34.Li X., Zhou D., Jin Z., Chen H., Wang X., Zhang X., Xu T. A coaxially extruded heterogeneous core–shell fiber with Schwann cells and neural stem cells. Regener. Biomater. 2019:131–139. doi: 10.1093/rb/rbz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katsetos C.D., Legido A., Perentes E., Mörk S.J. Class III β-tubulin isotype: a key cytoskeletal protein at the crossroads of developmental neurobiology and tumor neuropathology. J. Child Neurol. 2003;18:851–866. doi: 10.1177/088307380301801205. [DOI] [PubMed] [Google Scholar]
- 36.Sánchez C., Díaz-Nido J., Avila J. Phosphorylation of microtubule-associated protein 2 (MAP2) and its relevance for the regulation of the neuronal cytoskeleton function. Prog. Neurobiol. 2000;61:133–168. doi: 10.1016/s0301-0082(99)00046-5. [DOI] [PubMed] [Google Scholar]
- 37.Han S., Yang K., Shin Y., Lee J.S., Kamm R.D., Chung S., Cho S.-W. Three-dimensional extracellular matrix-mediated neural stem cell differentiation in a microfluidic device. Lab Chip. 2012;12:2305–2308. doi: 10.1039/c2lc21285d. [DOI] [PubMed] [Google Scholar]
- 38.Jorfi M., D'Avanzo C., Kim D.Y., Irimia D. Three-dimensional models of the human brain development and diseases. Adv. Healthc. Mater. 2018;7:1700723. doi: 10.1002/adhm.201700723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakurai T. The role of NrCAM in neural development and disorders—beyond a simple glue in the brain, Mol. Cell. Neurosci. 2012;49:351–363. doi: 10.1016/j.mcn.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Salton S.R.J., Ferri G.-L., Hahm S., Snyder S.E., Wilson A.J., Possenti R., Levi A. VGF: a novel role for this neuronal and neuroendocrine polypeptide in the regulation of energy balance, front. Neuroendocrinology. 2000;21:199–219. doi: 10.1006/frne.2000.0199. [DOI] [PubMed] [Google Scholar]
- 41.Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Heneka M.T., Carson M.J., Khoury J.E., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M., Herrup K., Frautschy S.A., Finsen B., Brown G.C., Verkhratsky A., Yamanaka K., Koistinaho J., Latz E., Halle A., Petzold G.C., Town T., Morgan D., Shinohara M.L., Perry V.H., Holmes C., Bazan N.G., Brooks D.J., Hunot S., Joseph B., Deigendesch N., Garaschuk O., Boddeke E., Dinarello C.A., Breitner J.C., Cole G.M., Golenbock D.T., Kummer M.P. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee A., Arha M., Choudhary S., Ashton R.S., Bhatia S.R., Schaffer D.V., Kane R.S. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells, Biomaterials. 2009;30:4695–4699. doi: 10.1016/j.biomaterials.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamczur P., Gamian A., Kolodziej J., Dziegiel P., Rakus D. Nuclear localization of aldolase A correlates with cell proliferation. Biochim. Biophys. Acta Mol. Cell Res. 2013;1833:2812–2822. doi: 10.1016/j.bbamcr.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Nicholls C., Li H., Liu J.P. GAPDH: a common enzyme with uncommon functions, Clin. Exp. Pharmacol. Physiol. 2012;39:674–679. doi: 10.1111/j.1440-1681.2011.05599.x. [DOI] [PubMed] [Google Scholar]
- 46.Qin Z.Y., Xiang C., Zhong F., Liu Y., Dong Q.Z., Li K., Shi W.H., Ding C., Qin L.X., He F.C. Transketolase (TKT) activity and nuclear localization promote hepatocellular carcinoma in a metabolic and a non-metabolic manner. J. Exp. Clin. Canc. Res. 2019;38:21. doi: 10.1186/s13046-019-1131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J., Yu L., Fu Q., Gao J., Xie Y., Chen J., Zhang P., Liu Q., Zhao S. Mouse phosphoglycerate mutase M and B isozymes: cDNA cloning, enzyme activity assay and mapping. Gene. 2001;264:273–279. doi: 10.1016/s0378-1119(00)00597-7. [DOI] [PubMed] [Google Scholar]
- 48.Li Z.S., Hung L.Y., Margolis K.G., Ambron R.T., Sung Y.J., Gershon M.D. The alpha isoform of cGMP-dependent protein kinase 1 (PKG1 alpha) is expressed and functionally important in intrinsic primary afferent neurons of the Guinea pig enteric nervous system. Neurogastroenterol. Motil. 2021:17. doi: 10.1111/nmo.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roland B.P., Richards K.R., Hrizo S.L., Eicher S., Barile Z.J., Chang T.-C., Savon G., Bianchi P., Fermo E., Ricerca B.M., Tortorolo L., Vockley J., VanDemark A.P., Palladino M.J. Missense variant in TPI1 (Arg189Gln) causes neurologic deficits through structural changes in the triosephosphate isomerase catalytic site and reduced enzyme levels in vivo. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2019;1865:2257–2266. doi: 10.1016/j.bbadis.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butterfield D.A., Hardas S.S., Lange M.L.B. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and alzheimer's disease: many pathways to neurodegeneration. J. Alzheim. Dis. 2010;20:369–393. doi: 10.3233/JAD-2010-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cumming R.C., Schubert D. Amyloid-beta induces disulfide bonding and aggregation of GAPDH in Alzheimer's disease. Faseb. J. 2005;19 doi: 10.1096/fj.05-4195fje. 2060-+ [DOI] [PubMed] [Google Scholar]
- 52.Schriner S.E., Linford N.J., Martin G.M., Treuting P., Ogburn C.E., Emond M., Coskun P.E., Ladiges W., Wolf N., Van Remmen H., Wallace D.C., Rabinovitch P.S. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 53.Cascon A., Comino-Mendez I., Curras-Freixes M., de Cubas A.A., Contreras L., Richter S., Peitzsch M., Mancikova V., Inglada-Perez L., Perez-Barrios A., Calatayud M., Azriel S., Villar-Vicente R., Aller J., Setien F., Moran S., Garcia J.F., Rio-Machin A., Leton R., Gomez-Grana A., Apellaniz-Ruiz M., Roncador G., Esteller M., Rodriguez-Antona C., Satrustegui J., Eisenhofer G., Urioste M., Robledo M. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. Jnci-J. Nat. Canc. Inst. 2015;107:5. doi: 10.1093/jnci/djv053. [DOI] [PubMed] [Google Scholar]
- 54.Martin D.E., Soulard A., Hall M.N. TOR regulates ribosomal protein gene expression via PKA and the forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 55.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction, Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 56.Xingliang D., Cheng M., Qing L., Tao X. 3D bioprinted glioma stem cells for brain tumor model and applications of drug susceptibility. Biofabrication. 2016;8 doi: 10.1088/1758-5090/8/4/045005. [DOI] [PubMed] [Google Scholar]
- 57.Hardy J., Selkoe D.J. Medicine - the amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 58.Cohen R.M., Rezai-Zadeh K., Weitz T.M., Rentsendorj A., Gate D., Spivak I., Bholat Y., Vasilevko V., Glabe C.G., Breunig J.J., Rakic P., Davtyan H., Agadjanyan M.G., Kepe V., Barrio J.R., Bannykh S., Szekely C.A., Pechnick R.N., Town T. A transgenic Alzheimer rat with plaques, tau pathology, behavioral impairment, oligomeric aβ, and frank neuronal loss, J. Neurosci. 2013;33:6245–6256. doi: 10.1523/JNEUROSCI.3672-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldstein L.S.B., Reyna S., Woodruff G. Probing the secrets of Alzheimer's disease using human-induced pluripotent stem cell technology. Neurotherapeutics. 2015;12:121–125. doi: 10.1007/s13311-014-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.