Abstract
Introduction
Drug resistance remains a major challenge in malaria treatment, especially after the emergence of resistance to artemisinin-based combined therapies. Plasmodium falciparum Kelch13 gene mutations are implicated in conferring artemisinin resistance. Thus, this study was aimed at determining the occurrence of Kelch13 (K13) propeller resistance gene polymorphism mutations in Bushenyi district, Uganda.
Methods
Participants suspected to have malaria were recruited. P. falciparum was confirmed using antigen histidine-rich protein 2 (HRP2) (Pf) (Access Bio, Inc, USA) and microscopy. Malaria-positive patients were treated with artemeter-lumefantrine (AL). Blood was withdrawn from participants who tested positive for parasites after day 3 and kept in blood filter papers (ET31CHR; Whatman Limited, Kent, UK). DNA was extracted using chelex-suspension method. Nested polymerase chain reaction (PCR) was conducted and the second-round products sequenced using Sanger’s method. Sequenced products were analyzed using DNAsp 5.10.01 software and then blasted on to the NCBI for K13-propeller gene sequence identity using the Basic Local Alignment Search Tool (BLAST).
Results
Out of 283 enrolled participants, 194 completed the follow-up schedule. A total of 134 (69%) had no parasites on day 3, while 60 (31%) had parasites on that day. Out of the 60 samples, 40 (62%) were positively amplified as P. falciparum, with polymorphisms in the K13-propeller gene detected in 3 (7.5%) out of the 40 amplicons. Polymorphisms at codon 1929, 1788 and 1801 were detected separately in one sample each. Sequences have been deposited in NCBI with accession numbers PRJNA720348 and PRJNA720800.
Conclusion
Polymorphisms in the K13-propeller gene previously reported to be associated with artemisinin resistance were not detected in the P. falciparum isolates from Bushenyi district, Uganda. More studies need to be conducted on the new mutations detected so as to understand their association, if any, with ACT resistance.
Keywords: artemisinin, resistance, Kelch13, polymorphism
Introduction
Malaria remains the major vector-borne disease globally, with Plasmodium falciparum (P. falciparum) accounting for the majority of clinical cases. The recommended method for clearing malaria infection is through chemotherapy. Artemisinin-based combined therapies (ACTs) were first introduced in Uganda in 2006.1 In Uganda, artemether-lumefantrine has been the primary drug of choice for treating uncomplicated malaria, though some reports from other parts of the world have indicated poor responses or partial resistance.2–4 Drug resistance is majorly associated with single-nucleotide polymorphisms occurring at different genetic loci of resistant genes.5 Previous studies have reported that mutations are generally associated with fitness costs, but are selected as they allow the parasite to survive in the presence of the drug. Once the pressure is removed, the mutation no longer has a selective advantage.6
Currently, P. falciparum K13-propeller single-nucleotide polymorphism arising because of mutations has been implicated in conferring resistance to ACTs.7 This has been supported by previous reports that have indicated that the artemisinin resistance is caused by mutations in a kelch protein encoded by PF3D7_1343700 on P. falciparum chromosome 13 (kelch13).8 This gene is presently monitored to track artemisinin resistance spread in relation to the World Health Organization (WHO) recommendations and at least 200 non-synonymous mutations in the K13 gene have been reported in different parts of the world.9
Resistance to ACTs has been confirmed in the Great Mekong sub-regions of Southeast Asia and some parts of African countries.10 However, artemisinin resistance in Uganda is still under-detected, with only a few studies indicating the possibility of ACT resistance emergence.11 A previous study conducted in northern Uganda, which is a high malaria transmission area, indicated a possibility of increasing resistance to ACTs.12 The study confirmed the presence of polymorphism in the K13 gene mutation at codon 533, previously reported in Cambodia, that is responsible for conferring resistance to ACTs. Other single-nucleotide polymorphisms in the K13 gene have also been reported in other African countries. However, the study in northern Uganda did not completely confirm the presence of resistance in the study area due to insufficient distribution of resistance genes, thus regular surveillance of resistance in the entire country is recommended to monitor the efficacy of ACTs.
Most recently, studies conducted in northern Uganda13 and eastern Uganda14 have indicated that ACT resistance has currently evolved in Uganda. These studies concluded that there was an independent emergence and local spread of clinically artemisinin-resistant P. falciparum. If this scenario replicates in other parts of the country, then efforts made in trying to eradicate malaria may be futile, leading to an increase in the mortality rate. The findings from these studies envisaged a potential risk of resistance spread all over Uganda, hence there is a call for large-scale therapeutic efficacy surveys.
Current assessment of circulating resistant P. falciparum parasites to ACTs is not well defined in the country and in the Bushenyi district of western Uganda, in particular, which is a moderate-high malaria transmission region. As artemisinin resistance continues to evolve, a robust approach is required to identify artemisinin drug resistance patterns earlier. According to WHO, Global Plan for Artemisinin Resistance goal number 2, the world should increase monitoring and surveillance to evaluate the threat of artemisinin resistance. WHO recommends surveillance every 24 months to assess the therapeutic efficacy of malaria drugs. Because of this scenario, in vivo clinical therapeutic efficacy tests (TES) and in vitro tests coupled with molecular determination for precise mutations in the Pfkelch13 gene are the recommended methods for surveillance. But, in vitro and in vivo methods have some limitations since they have sophisticated protocols and the follow-up duration for patients is long. Thus, molecular screening for mutations to detect potential resistance markers is a possible alternative. Therefore, the main aim of this study was to determine the prevalence and molecular diversity of K13-propeller molecular markers, a baseline study in context of artemisinin pressure in the south western region of Uganda. Molecular marker surveillance will serve as an epidemiological tool to assess baseline drug efficacy, which can be used as an indicator of future antimalarial failure and early detection of the same, hence containing its spread.
Materials and Methods
Study Design
This was a therapeutic efficacy health point study done among patients presenting with symptoms of malaria. This study was conducted at four selected health centers in Bushenyi district, Uganda. A cohort of human participants who had been confirmed to have malaria was followed for 28 days after treatment with artemether-lumefatrine (AL) drug. The follow-up was done at fixed checkup visits, ie, days 0, 1, 2, 3, 7, 14 and 28. On day 1 and day 2 the participants were given the first dose and the second dose of the antimalaria drug by following the direct observation treatment (DOT) procedures. On day 3, P. falciparum parasite detection was done and the participants were given the third dose of the AL drug. The first dose was given at the health facilities while the second and third doses were given at the places of residence of the participants. P. falciparum detection was done by performing a thin and thick blood smear. The participants were followed on days 7, 14 and 28, whereby the occurrences of P. falciparum and malaria clinical symptoms were evaluated. Consequently, clinical examinations such as physical examination, body weight and body temperature, and adverse drug effects were conducted by a qualified clinician on all follow-up days. The development of fever was used as a malaria clinical indicator in the subsequent follow-up days. In case of development of severe or complicated malaria during follow-up, participants were referred to the health center for parenteral artesunate administration and discontinued from the study. Patients without parasites after day 3 of follow-up were not considered for genotypic analysis.
Study Area
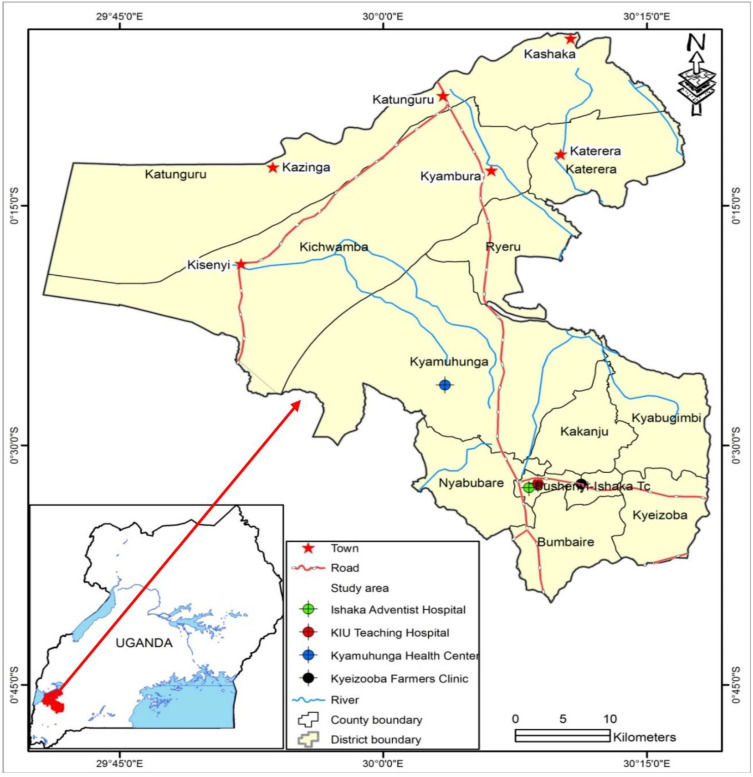
The study was conducted at four health facilities: the Kampala International University-Teaching Hospital (KIU-TH), Ishaka Adventist Hospital, Kyeizooba Health Center III and Kyamuhunga Health Center III in Bushenyi district of south western Uganda. The four hospitals are more than 327 km from the capital city of Kampala. Bushenyi district lies on latitude: 0° 28ʹ 22.8” (0.473°) South, longitude: 30° 16ʹ 8.4” (30.269°) East. The average elevation is about 1649 meters (5410 feet). The current drug of choice for the treatment of uncomplicated malaria is artemether-lumefantrine. Molecular analysis was conducted at Makerere University, College of Health Sciences, Department of Molecular Biology and Immunology. Figure 1 shows a detailed location of the study hospitals.
Figure 1.
Map showing locations of the sampling areas (hospitals).
Study Population and Study Setting
The study recruited participants who had resided in Bushenyi district for at least the last six months before the start of the study, and excluded those who refused to consent to the study and the nomadic participants. In addition, participants with febrile clinical conditions caused by other microorganisms other than malaria were excluded from the study. All adult patients signed the informed consent form and parents or guardians signed the informed consent form on behalf of patients below 18 years. Physical examination, complete medical history, demographic information and contact information were obtained from the participants.
Blood Sample Collection
Blood samples were collected by obtaining 1 mL of venous blood for participants older than 2 years. In the case of children below 2 years of age, 100 μL finger-pricked blood samples were collected. This procedure was repeated during the subsequent follow-up visits. The blood spot was made on chromatography filter paper (ET31CHR; Whatman Limited, Kent, UK), labeled well with the participant identification number and kept in a dust-free lock and key cabinet.
Malaria Diagnosis
All patients who had been suspected to have malaria infection by having fever (≥38°C) or having a history of fever in the past 24 hours were confirmed for the presence of P. falciparum using microscopy as a confirmatory test and rapid diagnostic test as a preliminary screening test. Rapid Diagnostic Test (RDT) analysis was conducted according to a method described by Mutagonda et al,15 using HRP-II (HRP2 (Pf) (Access Bio, Inc, USA). Five (5) µL of blood was added to the test kit well. Three (3) to five (5) drops of assay diluent were added to the well and results read after 30 min. Blood samples were considered positive in cases when both a test and a control line appeared and negative RDT if only a control line appeared in the result window. RDT-positive samples were then subjected to microscopy studies for P. falciparum confirmation. For microscopy, thick and thin blood smears were stained with 2% Giemsa for 30 minutes. A smear was considered negative if no parasites were observed after examination under 100 high-powered fields. For Giemsa-stained thin smears, they were fixed in methanol solution before examination.
DNA Extraction
The malaria-positive samples on day 3 after treatment with ACTs were genotyped for mutations on K13-propeller genes. This was done on blood drawn on day 3 for the parasite recurrent participants and stored as dried blood spots (DBS) on filter papers. DNA was extracted from blood-spotted filter papers using Chelex suspension method,16 with slight modifications. Briefly, the blood-spotted filter papers were soaked for 24 hours in 1-mL of 0.5% saponin-1 phosphate buffered saline. The mixture was incubated at room temperature for 10 min, then centrifuged at 14,000 rpm for 2 min and supernatant discarded. The mixture was washed two (2) times in 1-mL PBS and boiled for 8 min in 100 mL PCR grade water, followed by the addition of 50-µL 20% Chelex. To elute the extracted DNA, 150-µL buffer AE was added to each well using a multi-channel pipette and incubated for 1 min at room temperature. This set up was centrifuged at 3000 rpm for 8 min to recover the DNA as supernatant and stored at −30°C. To test for any cross-contamination, each batch of 10 samples contained 1 negative control, which consisted of a section of plain filter paper cut in the same way as the DBS sample.
PCR Amplification
K13 genes were amplified by nested PCR using a protocol described by Boussaroque et al,17 using the primers listed in Table 1. For the first round PCR, 0.5-mL DNA was amplified with 10-mL PCR mix (1.25 U/mL, Taq DNA polymerase, 0.4 mMdNTP mixture, PCR buffer, and 4 mM Mg2Cl), 0.5 mL forward primer (10 mM), 0.5 mL reverse primer (10 mM), and sterile ultrapure water added to a final volume of 20 mL. For the second round PCR, 0.5 mL primary PCR products were amplified with 40 mL reaction system, including 20 mL PCR mix, 1.0 mL forward primer (10 mM), 1.0 mL reverse primer (10 mM), and H2O. The amplification conditions were maintained at 95°C for 3 min; followed by 30 cycles (95°C for 30 s, 55°C for 30 s, 72°C for 30 s); 72°C for 5 min; then stored at −20°C. All PCR products were analyzed using 1% (w/v) agarose gel (Fisher Scientific, USA), stained with 0.05-μg/mL ethidium bromide (Sigma Aldrich, USA). The expected amplicon size was approximately 800 bp. PCR-grade water was used as a negative control.
Table 1.
Primers and Gene Fragment Used for Evaluating Artemisinin Resistant P. falciparum Markers
| Gene Fragment | PCR Primer Sequence | Assessed Codons |
|---|---|---|
| K13-propeller (1st round PCR) | 5ʹ_CGGAGTGACCAAATCTGGGA-3 | T474I, M476I, A481V, Y493H, T508N, P527T, G533S, N537I, R539T, I543T, P553L, R561H, V568G, P574L, A578S, and C580Y |
| 5ʹ_GGGAATCTGGTGGTAACAGC-3 | ||
| K13-propeller (2nd round PCR) | 5ʹ_TCAACAATGCTGGCGTATGTG-3 | |
| 5ʹ_TGATTAAG GTAATTAAAAGCTGCTCC-3 |
Sequencing of K13-Propeller Gene
The sequencing protocol was adopted from Maiga et al,18 with slight modifications. Briefly, the second round of PCR products were purified using a jet quick PCR product purification kit (Sigma Aldrich, USA). Consequently, 5-µL of the purified second round of PCR products was run on 1.0% (w/v) agarose gel stained with 0.05-μg/mL ethidium bromide (Sigma Aldrich, USA) to counter-check for the presence and concentration of the PCR products. This was followed by bi-directional cycle sequencing of K13 using the second round K133 forward and K132 reverse PCR primers (Eurofins Genomics, Germany), using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The cycle sequencing PCR was performed in a total reaction volume of 20 μL with 6.0 μL of the BigDye Terminator v3.1 5X sequencing buffer (Applied Biosystems, USA). This was accomplished by mixing the above total reaction volume with 2.0 μL BigDye Terminator v3.1 (Applied Biosystems, USA), 1.0 µL of 10 ng/μL (K133 forward or K132 reverse) primers and 6.0-µL nuclease-free water. Finally, 5.0 ng/μL of the purified second round of PCR products was added to make up the total volume. The sequencing was run under a thermal cycler using the following conditions: one cycle of polymerase activation 96°C for 60 s followed by 35 cycles of denaturation at 96°C for 10s, annealing at 53°C for 30 s, extension at 60°C for 300 s (Gene Amp 9700 PCR system, USA). The amplified products were then stored at 4°C until the next step of extension. The extension PCR products were followed by purification using DyeEx2.0 spin kit (QIAGEN™, Maryland, USA). Consequently, 5.0 μL of the purified cycle sequencing PCR products was mixed with 5.0-µL de-ionized formamide (Sigma Aldrich, USA) and then loaded in the 310 genetic analyzer (Applied Biosystems, USA). Finally, the products were bi-sequenced with POP-7™ (Applied Biosystems, USA) as a sequencing polymer.
Sequence Data Analysis
The DNA sequence data were base-called using the sequence analysis software -v5 (soft-ware.broadinstitute.org) and then blasted on to the NCBI sequence data base to confirm the K13-propeller gene sequence’s identity by using the Basic Local Alignment Search Tool (BLAST) at http://blast.ncbi.nlm.nih.gov/Blast.cgi. All sequences were then assembled. The kelch DNA and protein sequences were aligned with ClustalW software using Bio-edit. PF3D7_1343700K13-propeller gene sequences in NCBI database were used as the reference sequence.
Results
Baseline Characteristics and Study Profile
Out of 283 enrolled participants, 194 (68.5%) completed the follow-up schedule;; 31.5% discontinued the study due to various reasons; 134 (69%) had no parasites on day 3, while 60 (31%) had parasites on that day and hence were processed for genotypic analysis. The participants had a mean weight (kg) of 54.1 with standard deviation of 16.8. The female participants were the majority at 32 (53.3%). The median age for females was 27.3 and median age for males was 22.5. Body temperature substantially reduced on day 3 by recording a mean temperature of 36.7°C. The initial parasitemia on day 0 was 12,403 µL (Table 2).
Table 2.
Baseline Characteristics of the Study Participants
| Variable | Microscopy Corrected (Day 3 Follow-Up), n = 60 |
|---|---|
| Weight (kg), mean (SD) | 54.1 (16.8) |
| Gender (male), n (%) | 28 (46.6%) |
| Gender (female), n (%) | 32 (53.3%) |
| Body temperature on day 0, °C, mean (SD) | 37.3 °C S.D. ± 1.3 |
| Body temperature on day 3, °C, mean (SD) | 36.7 °C S.D. ± 0.7 |
| Parasitaemia (µL) on day 0* (95% CI) | 12,403 (8242–13,565) |
| Median age in years (males) | 22.5 |
| Age range in years (males) | (3–82) |
| Median age in years (females) | 27.3 |
| Age range in years (females) | (5–74) |
Notes: °C: degree Celsius; parasitemia*: geometric mean parasite density (asexual parasites/µL); *p < 0.001, the mean was significantly different.
Abbreviations: n, number of patients; SD, standard deviation; 95% CI, 95% confidence interval.
PCR Amplification
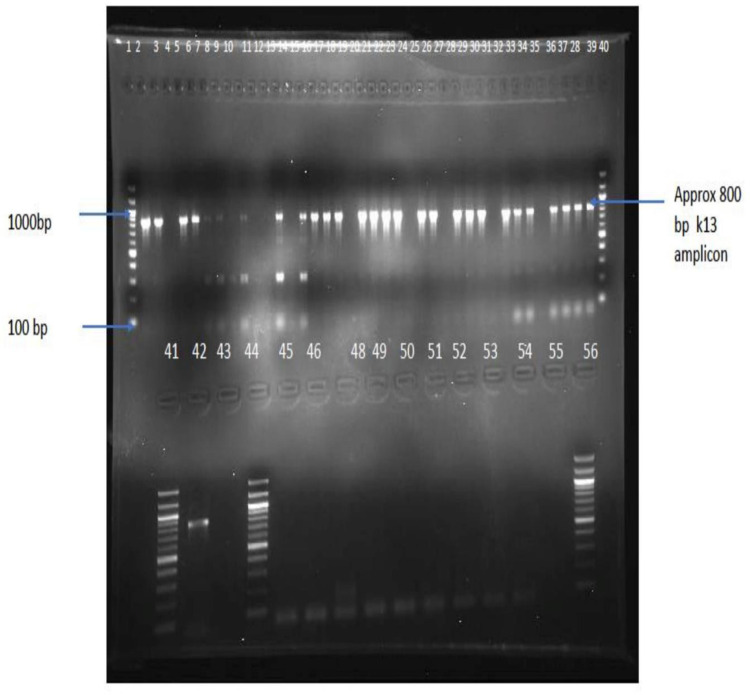
A total of 60 blood samples was processed for DNA isolation and PCR amplifications. For 40 (62%) samples, parasite DNA was successfully amplified, as shown in Figure 2 and Table 3, using nested PCR reaction for the K13-propeller gene, and hence were sequenced.
Figure 2.
Positive PCR products detected using agarose gel electrophoresis. Wells 1, 40, 41 and 56 contain 100bp DNA ladder; wells 42 and 43 contain positive control (P. falciparum 3d7). The rest of the wells contain samples.
Table 3.
Number of Cases Nested PCR+ve, Pfkelch13 Amplified Isolated from Health Centers
| Health Center | No. of Samples (n (%)) | PCR+ve Pf (n (%)) | PfK13 Amplified (n/%) |
|---|---|---|---|
| KIU-TH | 26 (43.3) | 19 (73.07) | 0 (0) |
| Ishaka Adventist Hospital (IAH) | 12 (20) | 6 (50) | 1 (16.66) |
| Kyeizooba Health Center III | 13 (21.6) | 6 (46.15) | 1 (16.66) |
| Kyamuhunga Health Center III | 9 (15) | 9 (100) | 1 (11.1) |
| Total | 60 (100) | 40 (66.6) | 3 (7.5) |
Note: The table provides the P.falciparum PCR positive and the frequencies of the samples that harbored the SNPs of Pfkelch13.
Prevalence of K13-Propeller Mutations
K-13-propeller single-nucleotide polymorphisms were evaluated compared with the 3D7 reference strain (PF3D7-1343700). K13-propeller non-synonymous polymorphism recorded in the current study includes 1929, 1788, and 1801. However, mutation on K13-propeller gene was not detected at positions 580, 578, 574, 568, 561, 553, 543, 539, 537, 533, 527, 508, 493, 481,476, and 474, respectively. Two (2) samples were not analyzed because of poor sample quality of the dried blood spots or inadequate extracted DNA. A detailed analysis of the samples is shown in Table 4.
Table 4.
Polymorphisms Observed in the Kelch13 Fragments and Amino Acid Substitutions
| Sample ID | No. of Codon | Wild Type A.A | Mutant Type A.A | Type of Mutation |
|---|---|---|---|---|
| KIU 45 | 1929 | Glycine | Asparagine | G1929A non-synonymous |
| KYA30 | 1788 | Asparagine | Tyrosine | A1788T non-synonymous |
| IAH 31 | 1801 | Cysteine | Glycine | C1801G non-synonymous |
Notes: The table describes various polymorphisms identified in PfK13 for the 3 resistance profiled samples. Capital letter in the codon sequence indicates the mutated nucleotide.
Abbreviations: AA, amino acid; Gly (G), Glycine; Cys (C), Cysteine; Asp (A), Asparagine; Try (T), Tyrosine.
Discussion
The problem of antimalarial drug resistance has posed major health challenges in under-developed countries, especially in Southeast Asia, South America and Africa,19 thus aggravating the impending problems with drug policies in Uganda and the world as a whole.8 Artemisinin-based combination therapy (ACT) is currently recommended as the first-line drug for the treatment of uncomplicated malaria.1 Surprisingly, there is no substitute for ACTs used for treating uncomplicated malaria because resistance to all available antimalarial drugs has developed, leading to poor efficacy. Furthermore, previously reported artemisinin resistance in Southeast Asian countries and poor responses of the malaria parasites in some parts of Africa and South America remain major threats in efforts to combat malaria in the world.20 With the possibility of malaria resistance spreading worldwide by following the previously recorded patterns of chroloquine and sulphodoxine-pyrimethamine resistance,21 we should develop standard surveillance methods to identify the possible emergence of resistance to artemisinin combined therapies. Thus, this study was aimed at identifying the presence of K13 P. falciparum resistance markers.
Molecular methods use molecular markers to detect resistance genes. They indicate mutations encoding biological resistance to antimalarial drugs by using DNA isolated from parasites. The nested PCR amplifications used here, however, have shown that only 40 of the 60 samples amplified the kelch13 gene despite being positive according to RDT and microscopy. This might be attributable to low parasite infections below 20 parasites/µL of blood, coupled with long storage of blood samples. The blood samples were subjected to PCR amplification after a period of one year since the time of sample collection. However, a previous study, by Kézia et al,22 reported that PCR amplification was more sensitive for DNA isolated from DBS compared with whole preserved blood. The findings from this study, which have indicated for the presence of parasites after day 3 follow-up, may have been as a result of increased gametocyte carriage and thus increased transmissibility of drug-resistant phenotype.
Previous studies have reported twelve (12) polymorphisms in the K13-propeller gene considered as the cause of artemisinin resistance in P. falciparum parasites.23 This study hereby reports the absence of commonly identified artemisinin resistance-associated SNPs reported in other parts of the world, such as Cambodia and Myanmar, which have confirmed resistance. Our study is in contrast to a recent study in Northern Uganda by Miotto et al,24 which reported non-synonymous polymorphism at codon 533 previously detected in Cambodia, an area with confirmed artemisinin resistance. Moreover, a previous study conducted in Eastern Uganda also detected non-synonymous polymorphism at codon 578 in low frequencies in patients who showed prolonged clearance after treatment.25 Our findings are in agreement with a previous molecular epidemiological study, which reported absence of artemisinin resistance mutations among P. falciparum parasites in Zaire and Angola.26 The absence of artemisinin associated molecular markers in the study area are also in agreement with other in vivo, in vitro and molecular therapeutic efficacy studies conducted previously in Africa, which concluded that artemisinin-based combination therapies remain effective in treating malaria caused by P. falciparum. A study conducted in south eastern Gabon from 2011 to 2014 reported limited polymorphism of A578S (0.4%) and two new mutations at A494V (0. %4) and V504A (0.9%).27 Surprisingly, a study conducted by immigrants from Africa to China reported 23 different genotypes of the K13-propeller that were observed in 24 (4.97%) of the samples; R539T and P574L, which have also been associated with ART resistance, were reported in two samples from Angola and Equatorial Guinea. No mutations were detected in 11 samples from North Africa. Epidemiologically, the frequency of the K13-propeller was 6.50% (8/123) in Central Africa, followed by East Africa (1/19, 5.26%), West Africa (9/198, 4.55%), and South Africa (6/132, 4.55%).28 The findings from this study are also supported by a previous study conducted in Uganda, which reported a lack of ACT resistance markers from patients with uncomplicated malaria.29
A recent longitudinal study conducted in Busia and Tororo districts of eastern Uganda during 2016–2019 reported on the presence of K13 mutations at C469T, Ala675, and A578S alleles. These mutations have been reported previously in Southeast Asia and in northern Uganda, where evidence of ACT resistance has been indicated. However, substitution Ala578Ser, which is the PfK13-propeller domain mutation most commonly reported in African isolates, does not mediate delayed clearance of parasites.14
A new study conducted in Gullu district of northern Uganda, a high malaria transmission area, has shown that resistance has evolved to the drug of choice for the treatment of uncomplicated malaria. The study, conducted for 4 years (2015–2019), reported K13 mutations in 20% of blood samples. After genotyping the blood samples, the study reported the presence of mutations in the A675V and C469Y allele in the K13 gene. These mutations have been detected previously in SEA and they have been listed by WHO as the candidate markers for ACT resistance. It was evident from this study that the prevalence of K13 mutations increased from 3.9% in 2015 to 19.8% in 2019. However, the genetic data indicated that the mutations detected from the study emerged independently.13 Meanwhile, an increase in the prevalence of two K13-propeller domain mutations of Cys469Tyr and Ala675Val has been reported in northern Uganda and eastern Uganda.30,31 The occurrences of K13 mutations circulating within northern and eastern Uganda may be as a result of high malaria prevalence and the misuse of antimalarial drugs, hence creating drug resistance.
Mutations at codon positions 1929, 1789 and 1801 found in our study have not been previously reported in other molecular therapeutic studies conducted in Uganda and other parts of the world. However, the role they may play in conferring resistance needs to be further evaluated. This will help track the emergence and spread of artemisinin resistance in parts of the world other than Southeast Asia. Our study is in agreement with previous studies conducted in Uganda,32,33 which also recorded new mutations not previously reported in Southeast Asia. A more recent study34 conducted in Central Africa Republic, a country having close proximity to Uganda, has also reported a non‑synonymous mutation at locus Y653N with a prevalence of 0.5%, a mutation that has not been associated before with artemisinin resistance in Southeast Asia. In addition, other new non-synonymous mutations (M472I; Y558C; K563R; P570L; P615S) have been reported in other places but have not shown any role in conferring resistance.
Surprisingly, previous studies have reported new non-validated mutations, which were present in patients who had poor recovery after treatment with ACTs.35 This raises concern whether they have some role to play in conferring resistance, and hence may be considered in future as molecular markers of ACT resistance. A mutation at codon A675Vwas reported in Rwanda36 and Uganda.37 The same mutation at codon A675V had also previously been reported in SEA,16 an epicenter of emergence and spread of ACT resistance. Unfortunately, our study was unable to conclude if the K13-propeller gene mutations recorded in the Plasmodium falciparum parasite among the isolates from Bushenyi district of Uganda were of local emergence or emerged from other parts of the world.
According to the WWARN K13 Genotype-Phenotype Study Group,10 molecular results need to be compared with either in vivo or in vitro therapeutic efficacy studies (TES). Although in vitro studies were not conducted from samples isolated from this study site, a previous in vivo study38 from the same study area reported reoccurrence of P. falciparum, 3 days and other subsequent days after treatment with ACTs, thus recommending further molecular surveillance for resistance genes circulating in the area. Moreover, other in vivo studies reported in Uganda showed occurrence of malaria parasites despite the absence of resistant molecular markers.39,40 The recurrence of parasites after day 3 of treatment might have been influenced by both intrinsic and extrinsic factors. The factors include, but are not limited to, high parasitemia before treatment, poor host immune responses to malaria parasites and poor diagnosis leading to improper treatment. Limitations associated with the research put forth include that our study did not conduct in vitro studies that would have enriched findings on the susceptibility patterns of this latest identified mutation detected in the study area to ACTs. The second limitation of our study was that it was conducted only in four sentinel sites of Bushenyi district. It is unclear whether such a molecular epidemiological pattern is observed in other parts of the country.
Conclusion
Despite the administration of ACTs as the first-line drug for treating Plasmodium falciparum parasites, this study showed single-nucleotide polymorphism, which is different from the results deduced from the studies reported in Southeast Asia, which is an epicenter for evolution and transmission of ACT resistance. Non-synonymous K13 mutations currently remain uncommon in the study area. The study concludes that artemisinin resistance is yet to be reported and confirmed in Bushenyi district of western Uganda. Moreover, the new single-nucleotide polymorphism mutations detected in our study need to be characterized further to ascertain their role in conferring ACT resistance. In addition, molecular surveillance of drug resistance needs to be scaled up in Uganda as a whole to have regular updates on the possibilities of emergence and spread of ACT resistance for future malaria containment. From previous evidence, this is the first study in western Uganda that investigates the efficacy of ACT resistance by using Pfkelch13 gene propeller polymorphism.
Acknowledgments
We hereby thank the entire clinical staff of Kampala International University-Teaching Hospital (KIU-TH), Ishaka Adventist Hospital, Kyeizooba Health Center and Kyamuhunga Health Center III for their technical assistance. We consequently thank the entire adult participants and the parents or guardians who kindly provided consent for this study. We also thank the village health officers who helped our study team during the follow-up of the participants. This study was funded solely by the authors.
Data Sharing Statement
Data in tables used to support the findings of this study are included within the article. The sequences have been deposited in NCBI with accession numbers PRJNA720348 and PRJNA720800, respectively.
Ethical Clearance
Informed consent was obtained from the participants before inclusion in the study. The study was conducted in accordance with the Declaration of Helsinki on human experimentation. The ethical approval of the study was sought from Mbarara University of Science and Technology (MUST) Institutional Research and Ethics Committee (IREC) on Human Research (Approval no. 06/01-17) and Uganda National Council for Science and Technology (Approval no. HS2241).
Author Contributions
This work was conducted in collaboration between all authors. All authors have made substantial contributions to conception and design, acquisition of data, analysis and interpretation of data; took part in drafting the article and revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Disclosure
All authors declare that there are no conflicts of interest existing in regard to this study and publication of the findings.
References
- 1.National Malaria Control Division. National malaria annual report 2017–2018. Kampala, Uganda: Ministry of Health; 2019. [Google Scholar]
- 2.World malaria report 2020. Geneva: World Health Organization; 2020. [Google Scholar]
- 3.Kamau E, Campino S, Amenga EL. K13-propeller polymorphisms in Plasmodium falciparum parasites from Sub-Saharan Africa. J Infect Dis. 2015;8:1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conrad MD, Rosenthal PJ. Antimalarial drug resistance in Africa: the calm before the storm. Lancet Infect Dis. 2019;9:1371–1380. [DOI] [PubMed] [Google Scholar]
- 5.Miotto O, Sekihara M, Tachibana S-I, et al. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;6:123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui FA, Boonhok R, Cabrera M, et al. Role of Plasmodium falciparum Kelch 13 protein mutations in P. falciparum populations from northeastern Myanmar in mediating artemisinin resistance. MBio. 2020;11:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbengue A, Bhattacharjee S, Pandharkar T. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019). Geneva: World Health Organization; 2020. [Google Scholar]
- 9.World Health Organization. World Malaria Report, 2020 summary; 2015. Available from: https://www.who.int/publications/i/item/9789240015791. Assessed 30 March, 2021.
- 10.WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments—A WWARN individual patient data meta-analysis. BMC Med. 2019;17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad MD, Bigira V, Kapisi J. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One. 2014;9:1371–1380. doi: 10.1371/journal.pone.0105690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ocan F, Bwanga F, Okeng A. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis. 2016;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balikagala B, Naoyuki F, Ikeda M, Katuro O, Tachibana S, Yamauchi M. Evidence of Artemisinin-Resistant Malaria in Africa. N Engl J Med. 2021;385:1163–1171. [DOI] [PubMed] [Google Scholar]
- 14.Tumwebaze P, Katairo T, Okitwi M, et al. Drug susceptibility of Plasmodium falciparum in eastern Uganda: a longitudinal phenotypic and genotypic study. Lancet Microbe. 2021;2:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mutagonda RF, Appolinary AR, Omary MS. Malaria prevalence, severity and treatment outcome in relation to day 7 lumefantrine plasma concentration in pregnant women in Tanzania. Nat Rev Neurol. 2016;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SM, Parobek CM, DeConti DK. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis. 2015;5:680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussaroque A, Fall M, Madamet M. Emergence of mutations in the K13 propeller gene of Plasmodium falciparum isolates from Dakar, Senegal, in 2013–2014. Antimicrob Agents Chemother. 2016;15:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maiga AW, Fofana B, Sagara I. No evidence of delayed parasite clearance after oral artesunate treatment of uncomplicated falciparum malaria in Mali. Am J Trop Med Hygiene. 2012;5:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F, Culleton R, Zhang M. Emergence of Indigenous Artemisinin-Resistant Plasmodium falciparum in Africa. Engl J Med. 2017;376:991–993. doi: 10.1056/NEJMc1612765 [DOI] [PubMed] [Google Scholar]
- 20.Nzoumbou R, Gildas CP, Wockama P. Molecular assessment of kelch13 non‑synonymous mutations in Plasmodium falciparum isolates from Central African Republic (2017–2019). BMC Inf Dis. 2020;6:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashley EA, Dhorda M, Fairhurst RM. Tracking spread of artemisinin resistance in Plasmodium falciparum malaria. Engl J Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kézia KG, Cor JF, Álvaro C, Maria F, Érika M. Low sensitivity of nested PCR using Plasmodium DNA extracted from stained thick blood smears: an epidemiological retrospective study among subjects with low parasitaemia in an endemic area of the Brazilian Amazon region. Malar J. 2004;3:7–16. doi: 10.1186/1475-2875-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talundzic E, Okoth SA, Congpuong K. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog. 2015;11:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miotto O, Sekihara M, Tachibana S-I, et al. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;12:109–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ocan M, Bwanga F, Okeng A. Prevalence of K13-propeller gene polymorphisms among Plasmodium falciparum parasites isolated from adult symptomatic patients in northern Uganda. BMC Infect Dis. 2016;16:1–9. doi: 10.1186/s12879-016-1777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawkes MK, Conroy AL, Opoka RO. Slow clearance of Plasmodium falciparum in severe pediatric Malaria, Uganda, 2011–2013. Emerg Infect. 2015;21:1237–1239. doi: 10.3201/eid2107.150213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fatima D, Voumbo M, Kouna C, et al. Prevalence of Plasmodium falciparum antimalarial drug resistance genes in Southeastern Gabon from 2011 to 2014. Infect Drug Resist. 2018;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C, Zhang H, Zhou R. Polymorphisms of Plasmodium falciparum K13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis. 2017;17. doi: 10.1186/s12879-017-2634-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper A, Conrad MD, Watson QD. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother. 2015;59:5061–5064. doi: 10.1128/AAC.00921-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asua V, Vinden J, Conrad MD, et al. Changing molecular markers of antimalarial drug sensitivity across Uganda. Antimicrob Agents Chemother. 2019;3:1818–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asua V, Conrad MD, Aydemir O, et al. Changing prevalence of potential mediators of aminoquinoline, antifolate, and artemisinin resistance across Uganda. J Infect Dis. 2021;223:985–994. doi: 10.1093/infdis/jiaa687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeka A, Dorsey G, Kamya MR. Artemether-lumefantrine versus dihydro artemisinin-piperaquine for treating uncomplicated malaria: a randomized trial to guide policy in Uganda. PLoS One. 2008;3:184–195. doi: 10.1371/journal.pone.0002390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorsey G, Staedke S, Clark TD. Combination therapy for uncomplicated falciparum malaria in Ugandan children: a randomized trial. J Malaria. 2007;3:5–12. [DOI] [PubMed] [Google Scholar]
- 34.Nzoumbou R, Gildas CB, Wockama P. Molecular assessment of Kelch13 non‑synonymous mutations in Plasmodium falciparum isolates from Central African Republic (2017–2019). BMC Inf. 2020;4:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.silva M, Ferreira PE, Otienoburu SD, et al. Plasmodium falciparum K13 expression associated with parasite clearance during artemisinin-based combination therapy. J Antimicrob Chemother. 2019;18:90–93. [DOI] [PubMed] [Google Scholar]
- 36.Tacoli C, Gai PP, Bayingana C. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg. 2016;5:1090–1093. doi: 10.4269/ajtmh.16-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda M, Kaneko M, Tachibana SI. Artemisinin-resistant Plasmodium falciparum with high survival rates, Uganda, 2014–2016. Emerg Infect Dis. 2018;24:718–726. doi: 10.3201/eid2404.170141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniga JN, Aliero AA, Ibrahim N, Okech MA, Mugasa CM. Plasmodium falciparum malaria clinical and parasitological outcomes after in-vivo Artemether- Lumefantrine (AL) treatment at Bushenyi District Uganda. Int J Trop Dis Health. 2018;8:10–18. [Google Scholar]
- 39.Rasmussen SA, Ceja FG, Conrad MD, et al. Changing antimalarial drug sensitivities in Uganda. Antimicrob Agents Chemother. 2017;3:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeka A, Wallender E, Mulebeke R, et al. Comparative efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for the treatment of uncomplicated malaria in Ugandan children. J Infect Dis. 2019;219:1112–1120. doi: 10.1093/infdis/jiy637 [DOI] [PMC free article] [PubMed] [Google Scholar]