Abstract
Cryptococcus neoformans MATα and MATa pheromones were amplified by direct PCR. Nucleotide sequence analyses revealed unique restriction enzyme sites. Sixty strains were used to devise a restriction fragment length polymorphism typing scheme that yielded three variety-specific patterns. Additionally, pheromone-specific PCR allowed easier identification of diploid C. neoformans strains than flow cytometry.
The encapsulated yeast Cryptococcus neoformans causes cryptococcal meningoencephalitis in healthy and immunocompromised individuals, especially AIDS patients. There are two mating types (α and a), three varieties (Cryptococcus neoformans var. neoformans, var. gattii, and var. grubii), and five serotypes (A, B, C, D, and A/D). Most strains are haploid, although diploid strains have occasionally been reported. Many tests available to distinguish C. neoformans are labor and time intensive or are dependent upon commercial reagents that are not readily available (1, 2, 6, 7, 9). C. neoformans var. gattii was recently distinguished by two α-mating-type-specific genes in a study wherein absence of an α-specific amplicon was considered evidence of the a mating type (3). We aimed to develop a simple test based upon the C. neoformans MATα pheromone described earlier and the MATa pheromone recently characterized in one of our laboratories (8; C. M. McClelland, G. L. Woodlee, T. S. Seymour, and B. L. Wickes, unpublished data).
A PCR protocol was developed for direct amplification of DNA from boiled C. neoformans cells, avoiding the elaborate DNA extraction methods described previously (1). The details of strains used are provided in Table 1. Cultures were grown in yeast extract-peptone-dextrose broth (Difco) at 30°C and 180 rpm, to an optical density at 600 nm of 3 to 4. A 1.5-ml aliquot was washed and resuspended in 700 μl of deionized water and kept in a boiling water bath for 5 min, followed by storage on ice. Five microliters of cell suspension was used as the PCR template. The PCR mixture included 5.0 μl of PCR buffer containing 15 mM MgCl2, 2.5 μl each of both primers (10 mM stock), 1.0 μl of deoxynucleoside triphosphate mix (10 mM each), and 1.8 U of Taq DNA polymerase (Perkin-Elmer, Foster City, Calif.). The α-mating-type-specific 5′ oligonucleotide primer was 5′-CTTCACTGCCATCTTCACCA-3′ and the 3′ oligonucleotide primer was 5′-GACACAAAGGGTCATGCCA-3′. The a-mating-type-specific 5′ oligonucleotide primer was 5′-CGCCTTCACTGCTACCTTCT-3′ and the 3′ oligonucleotide primer was 5′-AACGCAAGAGTAAGTCGGGC-3′. Initial denaturation was done at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 57.5°C for 1 min, amplification at 72°C for 1 min, and final extension at 72°C for 7 min in a GeneAmp PCR System 9600 (Perkin-Elmer). The PCR amplicons and restriction digests were electrophoresed on 3.5% MetaPhor agarose (FMC BioProducts, Rockland, Maine) in Tris-borate-EDTA buffer. All restriction enzymes were used in accordance with the manufacturer's instructions (New England Biolabs, Beverly, Mass.). The amplicons were sequenced on an ABI Prism 377 sequencer with the Big Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer).
TABLE 1.
C. neoformans pheromone sequences from diverse clinical and environmental isolates
| C. neoformans variety | No. of strains showing:
|
GenBank accession no. | ||
|---|---|---|---|---|
| MATα | MATa | MATα/a | ||
| grubii | 1 | AF 226880a | ||
| 6 | AF 226881–226886b | |||
| 2 | AF 226887–226888c | |||
| 1 | AF 226889–226890d | |||
| neoformans | 2 | 2 | AF 226891–226894e | |
| 14 | 1 | AF 226895–226909f | ||
| 2 | AF 226910–226911b | |||
| 2 | AF 226912–226913c | |||
| gattii | 1 | AF 226914e | ||
| 19 | 2 | AF 226915–226935b | ||
| 4 | AF 226936–226939c | |||
| 1 | AF 226940–226941d | |||
New York State Herbarium, Albany, N.Y.
R. Larsen, Los Angeles, Calif.
T. Sorrel, Sydney, Australia.
V. Chaturvedi, Albany, N.Y.
American Type Culture Collection, Manassas, Va.
F. Dromer, Paris, France.
Direct PCR of boiled cell suspensions allowed rapid and reproducible amplification of C. neoformans pheromones. No difference was seen between results obtained with purified C. neoformans genomic DNA and those obtained with a boiled-cell suspension (data not shown). This result was consistent with a report of similar direct PCR amplifications with yeast (5). Initially, four type strains, including C. neoformans var. neoformans (ATCC 28957 and ATCC 28958), C. neoformans var. grubii (NYSD 1649), and C. neoformans var. gattii (ATCC 32609), were used for standardization. A 101-bp MATα fragment and a 117-bp MATa fragment were amplified from type strains. Sequence analyses revealed unique restriction fragment length polymorphism (RFLP), which was confirmed with restriction digests yielding patterns characteristic of known mating types and varieties (Fig. 1). The scheme was further evaluated with 56 geographically diverse strains, which were characterized in our laboratory with standard biochemical and serological tests (Table 1). All strains had the predicted RFLP patterns that define the three varieties. A MATa amplicon was amplified from 2 strains, while 53 strains tested positive for MATα, which is reported to be predominant among clinical isolates (1, 7).
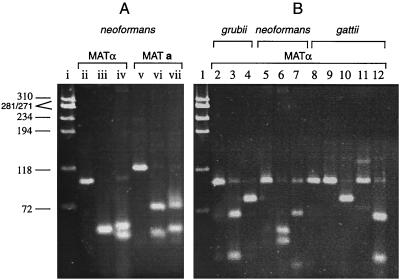
FIG. 1.
C. neoformans pheromone fragments analyzed by PCR-RFLP to determine α and a mating types (A) and three varieties (B). (A) A φX174 DNA size marker was digested with HaeIII (lane i). Strain ATCC 28957 α mating type 101-bp amplicon (lane ii) was digested with EarI to reveal 50- and 51-bp bands migrating together (lane iii) or was digested with AluI to get 53- and 48-bp bands (lane iv); the 117-bp a mating type amplicon of strain ATCC 28958 (lane v) was digested with EarI to yield 71- and 46-bp fragments (lane vi) or was digested with HgaI to reveal 73- and 44-bp bands (lane vii). (B) A φX174 DNA size marker was digested with HaeIII (lane 1). All other lanes have the 101-bp α mating type amplicon. The gel shows the following: a strain NYSD 1649 amplicon (lane 2), an NYSD 1649 amplicon digested with Tsp45I to reveal 67- and 34-bp bands (lane 3) and digested with HpaII, yielding a visible 78-bp band and an invisible 23-bp band (lane 4); a strain ATCC 28957 amplicon (lane 5), an AluI digest with 53- and 48-bp bands (lane 6), an AciI digest yielding a 68-bp band and a faint 33-bp band (lane 7), and a strain ATCC 32609 amplicon (lane 8), no restriction with Tsp45I (lane 9), an HpaII digest with a 78-bp fragment and a 23-bp invisible fragment (lane 10), no restriction with AluI (lane 11), and an AciI digest with 65- and 36-bp bands (lane 12).
Five strains tested positive for both MATα and MATa, which was strongly indicative of diploidy or aneuploidy (7). Flow cytometry was performed on these strains to confirm ploidy as described in an earlier publication (10). Briefly, C. neoformans cultures were grown on YRG (1% yeast extract, 1% peptone, 2% glucose) agar slants for 2 days and then inoculated in YRG broth at an optical density at 600 nm of 0.1 and incubated for 16 h at 30°C on a gyratory shaker (180 rpm). The cells were harvested by centrifugation, washed twice with distilled water, and fixed with 70% ethanol overnight at 4°C. The next day, cells were again washed twice with distilled water and once with NS buffer (9), suspended in a staining solution (NS buffer with 0.5 mg of heat-inactivated RNase/ml and 5 μg of propidium iodide/ml [Sigma Chemical Company]) at a concentration of 107 cells/ml, and incubated at 37°C for 150 min.
The flow cytometry assay was performed with the FASCan flow cytometer (Becton Dickinson) to measure the total DNA content of cells. Data acquisition and analysis were done with Cell Quest software. The instrument settings were as follows: forward scatter, 2.30 linear gain; side scatter, 176 volts log; fluorescence, 457 volts log; and threshold value, 60. More than 20,000 cells were used to measure fluorescence intensity at each data point. The data were displayed as histograms, in which the abscissa represents the channel numbers in proportion to the intensities of the fluorescence and the ordinate shows the number of cells (Fig. 2A). MATα/a strains had twice the DNA content of the standard haploid strains, as was evident from the characteristic doubling of fluorescence intensity, thus confirming diploidy. The same strains were positive for MATα and MATa by PCR, thereby establishing a good correlation between these two methods for determination of C. neoformans ploidy (Fig. 2B).
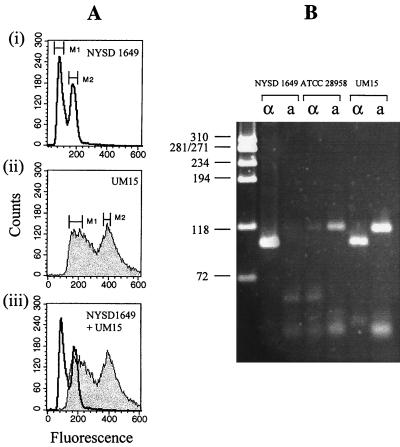
FIG. 2.
Ploidy of three C. neoformans strains analyzed by flow cytometry (A) and PCR (B) for MATα/MATa. (A) Haploid strain NYSD 1649 with nuclear DNA fluorescence intensities distributed from channels 50 to 240, the first peak being near channel 85 and the second peak near channel 164 (graph i); diploid strain UM15 with fluorescence intensities distributed from channels 100 to 440 with two peaks near channels 160 and 388 (graph ii). Graph iii shows that the second peak of NYSD 1649 coincided with the first peak of UM15, thereby indicating a doubling of cellular DNA in the latter strain. (B) Control strains NYSD 1649 (MATα) and ATCC 28958 (MATa) are positive for one of the two pheromone PCR amplicons that correlate with haploid DNA; the diploid strain UM15 was positive for both MATα and MATa amplicons, which confirmed its diploidy as seen with flow cytometry.
The PCR-RFLP typing scheme could prove valuable for the characterization of clinical and environmental isolates obtained in epidemiological investigations. It could enable routine delineation of ploidy, varieties, and mating types relatively easily, based on two PCR amplicons. A second application of this typing scheme is in molecular pathogenesis studies, wherein mating with a congenic pair of C. neoformans tester strains is recommended, for example, to remove extraneous mutations from gene knockout mutants (4). These mating analyses could be rapidly done with PCR-RFLP in conjunction with the technically demanding selection of individual basidiospores followed by mating with tester pairs. Finally, it is expected that future studies of the ploidy of C. neoformans strains and a search for diploids will be greatly facilitated by this scheme, as already has been reported for Saccharomyces cerevisiae (5).
Acknowledgments
The study was financially supported in part by NIH grants R29-AI-41968 (V.C.) and R29-AI-43522 (B.L.W.), the Pfizer Educational Award (V.C.), and the Burroughs Wellcome Young Investigator Award in Medical Mycology (B.L.W.).
Nucleotide sequencing and flow cytometry were performed at the Molecular Genetics and Cellular Immunology cores, respectively, at Wadsworth Center. We thank the colleagues listed in Table 1 for contributing strains used in this study.
REFERENCES
- 1.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: American Society for Microbiology; 1998. [Google Scholar]
- 2.Franzot S P, Salkin I F, Casadevall A. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halliday C L, Bui T, Krockenberger M, Malik R, Ellis D H, Carter D A. Presence of α and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol. 1999;37:2920–2926. doi: 10.1128/jcm.37.9.2920-2926.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heitman J, Allen B, Alspaugh J A, Kwon-Chung K J. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol. 1999;28:1–5. doi: 10.1006/fgbi.1999.1155. [DOI] [PubMed] [Google Scholar]
- 5.Huxley C, Green E D, Dunham I. Rapid assessment of Saccharomyces cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- 6.Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J Clin Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwon-Chung K J, Bennett J A. Medical mycology. Philadelphia, Pa: Lea and Febiger; 1992. [Google Scholar]
- 8.Moore T D E, Edman J C. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki T, Nishibayashi S, Kuroiwa T, Kanbe T, Tanaka K. Variance of ploidy in Candida albicans. J Bacteriol. 1982;152:893–896. doi: 10.1128/jb.152.2.893-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka R, Taguchi H, Takeo K, Miyaji M, Nishimura K. Determination of ploidy in Cryptococcus neoformans by flow cytometry. J Med Vet Mycol. 1996;34:299–301. [PubMed] [Google Scholar]