Abstract
This review is focused on the subset of antibacterial agents whose action involves one-on-one targeting of infecting bacteria. These agents target individual bacteria and their efficacy is based on particle numbers in contrast to chemical agents such as antibiotics, whose efficacy is based on minimal inhibitory concentrations. Four extant members of this class are predatory bacteria, functional (plaque-forming) phages, and engineered particulate systems, phagemids (plasmids that contain a phage packaging signal) and antibacterial drones (ABDs) that package chromosomal island DNA carrying antibacterial genes. We differentiate the natural predators, phages and predatory bacteria, from the engineered delivery vehicles, phagemids and ABDs, because the latter are much more versatile and can largely bypass the historical warfare that informs the predator-prey interactions.
Introduction
In the throes of the antimicrobial resistance crisis, considerable funding has engendered a large array of research projects, ranging from the search for scarcer and scarcer new antibiotics, through the development of antibacterial peptides, bacteriocins, antisense RNAs, to novel small molecules targeting essential bacterial functions [1]. In this review, we focus on an additional and somewhat distinct class of anti-bacterials, which target individual bacteria rather than depending on the achievement of a body-wide MIC, and we single them out for general evaluation and comparison. Included are predatory bacteria, functional (plaque-forming) phages, and two types of engineered particulate systems, phagemids (plasmids that contain a phage packaging signal) and antibacterial drones (ABDs) that package chromosomal island DNA carrying antibacterial genes. They have important advantages over antibacterial chemicals and have several common and distinguishing features: Their efficacy is measured by their efficiency of killing or disabling target bacteria rather than by minimal inhibitory concentrations; they cause no dysbiosis of the microbiome, there are probably few if any pre-existing allergies, they have no known toxic side effects and are not themselves toxic and they can, in principle, be engineered to avoid resistance. But they are nowhere near the medical quick-fix of antibiotics in their heyday. And it is ironic that one of their great advantages is their narrow host range, which is actually too narrow since the ideal would be to target all strains of at least one species – clearly the most critical objective for this class of anti-bacterials.
Predatory bacteria
Predatory bacteria, of which the prototype is Bdellovibrio bacteriovorus, were discovered nearly 60 years ago [2], giving rise to a broad field of inquiry in which a large and diverse group of organisms was discovered and many individual species characterized [3]. These fall into three main groups on the basis of their mode of predation: endobiotic, where the predatory organism invades and multiplies within its prey, epibiotic, where the predatory organism attaches to and kills its prey from the outside, and transbiotic or group predators, where the predator secretes lytic proteins and feeds on the released cellular contents of its prey [4]. The endobiotic prototype is Bdellovibrio bacteriovorus, whose biology has been analyzed in great detail [5] and which has been tested in a variety of animal models. From the clinical point of view, the epibiotic prototype is Micavibrio aeruginosavorus, which has also been studied in considerable detail [6]. Transbiotic predators are not of clinical interest per se, although some of their products are of major importance. B. bacteriovorus and M. aeruginosavorus are the only two that have thus far been considered for clinical utility.
Endobiotic predation: B. bacteriovorus
Predation.
B. bacteriovorus is a small Gram-negative motile rod with a polar flagellum responsible for motility. It is an obligatory endoparasite of most Gram-negative bacteria including many major pathogens, (see fig. 1B). Its predatory activity involves an attack phase (AP) in which the free-living organism locates its prey, responds to a specific signal in the cell wall, and enters a growth phase (GP). It attaches to the cell wall, apparently by means of a type IV pilus, and in response to a second cellular signal, secretes hydrolytic enzymes that open a channel in the cell wall through which the organism enters the periplasm [7]. It then causes the cell to form a round ‘bdelloplast’ and grows into a multi-cell filament by metabolizing the prey cell’s contents, then septates and lyses the cell.
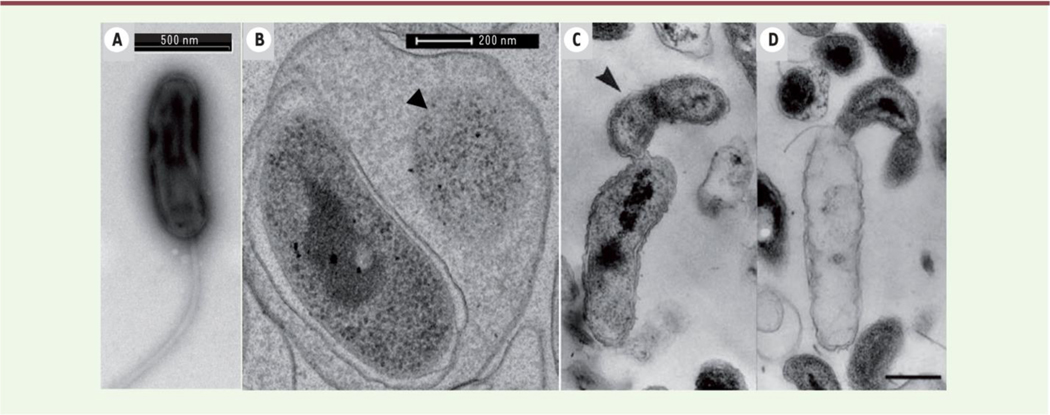
Fig. 1. Transmission electron microscopic observation of endobiotic predation by B. bacteriovorus H100 (B) and epibiotic predation by B. exovorus (C & D.
B. bacteriovorus is shown within the periplasm of an E. coli cell (B) and B. exovorus attached externally to the organism (C & D). One can observe the reduced cytoplasm of the prey organism (B arrow), the extracellular growth and division of the epibiotic predator (C arrow), and the emptied body of the prey organism (D). Reproduced from [3] with permission
Safety; side effects.
B. bacteriovorus is non-toxic for mammals and is cleared from a mouse in about 3 weeks following parenteral administration [8]. It can survive in macrophages, apparently without visible ill effects on the cell [6]. It stimulates modest production of cytokines, especially IL-13 [9] and has no significant effect on the colonic microflora or intestinal histology in rats [8, 9].
In vitro antibacterial activity.
B. bacteriovorus was recognized early as a potential therapeutic antibacterial, and its activity analyzed extensively in vitro. In a study with Klebsiella pneumoniae, it was found to reduce the bacterial population by about 106-fold, but the surviving bacteria became phenotypically resistant and shortly regrew [10]. A possible solution for this appeared serendipitously when B. bacteriovorus was isolated from wastewater in association with a lytic bacteriophage. This association survived filtration and dilution and presumably represented physical attachment of the phage to the B. bacteriovorus cell, perhaps similarly to the recently observed attachment of phage particles to Bacillus subtilis flagellae [11]. Treatment with this lytic phage along with B. bacteriovorus [12], resulted in eradication of the prey organism. In other studies, B. bacteriovorus was observed to penetrate and disrupt biofims [13], and to be active against biofilm-forming oral pathogens [14]. Interestingly, a residual population of prey organisms persists in a biofilm despite prolonged incubation and the continued presence of a greater number of B. bacteriovorus [13]. In this latter study B. bacteriovorus reduced the level of the same prey organisms in the planktonic state to undetectable. The overall view of in vitro predation studies with B. bacteriovorus is that under many conditions a considerable number of prey organisms survive B. bacteriovorus predation, possibly owing to interfering substances such as cyanide and indole produced by the prey organism [15] but that under some conditions it can completely eliminate its prey – an unappealing prospect for an obligatory parasite.
In vivo antibacterial activity.
In one series of experiments with a Klebsiella pneumoniae lung infection in rats, B. bacteriovorus reduced the intrapulmonary bacterial titer by about 103-fold [10]. However, it did not reduce the bacterial burden in various organs in rats infected IV with K. pneumoniae; inexplicably, the B. bacteriovorus-treated rats seemed healthy compared to the untreated controls, which become moribund after 36 h [16]. In other in vivo studies, B. bacteriovorus attenuated Salmonella infections in chicks [17], cured K. pneumoniae infections by cooperative action with immune cells in zebrafish larvae, and rescued mice infected by aerosol with Yersinia pestis [18].
Epibiotic predation: B. bacteriovorus and B. exovorus
The closely related B. exovorus is an epibiotic predator as shown in Fig. 1 C & D, and, somewhat surprisingly, B. bacteriovorus can attack S. aureus epibiotically, both in planktonic suspensions [19, 20] and in biofilms which it can disrupt [21]. Electron microscopy shows B. bacteriovorus attached externally to partially disrupted S. aureus, suggesting that it destroys the cell from without and extracts the contents in the typical epibiotic manner.
Micavibrio aeruginosovorus.
M. aeruginosovorus is an epibiotic predator whose predatory behavior has been studied extensively in comparison with that of the endobiotic B. bacteriovorus, both for its interesting biology as an epibiotic prototype and for its clinical potential [22]. It is very effective against biofilms in vitro [23] and has been tested for safety in animals [8], where it induces a modest cytokine response but has no detectable toxic effects.
Given all of the results described above, and many more in the literature, plus the frequent isolation of B. bacteriovorus from human stools, predatory bacteria have been very widely touted as potential therapeutics for human infections with Gram-negative bacteria. See, for example [24], among many other publications. It would seem that an obvious place to start would be with external or intestinal infections. Nevertheless, it does not appear ever to have been used as such, perhaps because the idea of eating, smearing, or injecting live bacteria to chase other live bacteria in or on the body may simply seem too outlandish.
Bacteriophages
The great expectations for phage therapy that followed the discovery of phages were not met, owing to a profound lack of understanding of phage biology, so that phage therapy (PT) was largely sidelined by the discovery of antibiotics in the late ‘30’s. It has limped along, especially in Eastern Europe, where, largely owing to ease and inexpensiveness of production, it continues to be a cottage industry, largely discredited in the West owing to a chronic lack of rigorous supporting clinical data. If an individual with an infection recovers after anecdotal treatment with a cocktail of 15 or 20 different phages, that seems to be sufficient support for the cocktail strategy, and if the individual does not recover, trop mal (or even, tant pis). In the era of antimicrobial resistance (AMR), phage therapy has had a re-awakening in the West, but rather than simply adopting the rather unsatisfying Eastern European cocktail strategy, research in the West has proceeded by detailed investigations of the biological principles that would have to inform any rational therapeutics. Many of these biological principles were well-established, inasmuch as the study of phages has been a cornerstone of modern molecular biology since the 1950’s. Pre-clinically, simple studies in which animals infected with laboratory strains are treated with phages to which they are known to be susceptible, always result in dramatic cures, serving as ample proof of principle and showing that phages are non-toxic. But in the extrapolation to naturally-occurring strains, a major feature of phage biology reared its ugly head - Phages had been at war with their bacterial hosts since both appeared on the planet, and the bacteria have developed a vast armamentarium of resistances starting with blockage of adsorption, DNA destruction by restriction enzymes and CRISPRs, and extending to every aspect of the phage life cycle with which the host could theoretically interfere [25]. So bacteria, unlike the US capitol, were well prepared for the onslaught of their enemies. Consequently, the problem of limited phage host range has thus far been surmounted only in individual cases in which, with a bacterial diagnosis in hand, an extensive search of phage collections would uncover one to which the infecting organism was sensitive; and in a few cases, anecdotal compassionate use of such phages has saved patients dying from untreatable AMR bacteria [26] [27]. Bacteria recovered from such patients, after successful phage treatment, are, however, generally resistant to the phage [26]. Though highly satisfying, this paradigm hardly represents a general therapeutic platform and is not likely to lead to one. Nor is it likely that the problems of ancient natural or recently acquired phage resistance will easily be surmounted. Instead, the Eastern European cocktail concept has been adopted, after all, but more rationally. For a detailed and thoughtful review see [28]. Well-developed examples include: combinations of several broad-host range phages that can collectively surmount most resistance mechanisms or selection for phage mutants or variants resistant to host defense mechanisms [29] [30] [31]. These strategies can potentially cover 90% or more of strains of a given species; specific bacterial diagnosis, however, will probably always be necessary (though Chan, et al. [28] envision the possibility that carefully constructed cocktails could be used presumptively). A more direct means of broadening specificity and enhancing effectiveness involves engineering of individual phages using the recently described “rebooting” method involving transfection of bacterial L-forms with intact phage genomic DNA [32]. This method can be used to develop individual phages with any combination of features [33] such as modified tails to broaden adsorption specificity [34], resistance to restriction enzymes by incorporating methylases and to CRISPR by incorporating anti-CRISPR genes. An additional strategy, for which a clinical trial is currently underway by Locus Pharmaceuticals, is engineering phages to deliver bactericidal CRISPRs [35]. The advantage of this over phages is not obvious. Ongoing theoretical problems with PT are the potential mobilization of virulence or resistance genes in the target population and release of toxic bacterial components by phage-induced lysis. PT is likely to have an important role in bacterial infection control in the AMR era, but that role will have to be evaluated by double-blind, placebo-controlled trials, which are only just beginning [36] [37].
Phagemids and antibacterial drones (ABDs)
The promising idea has recently emerged of packaging antibacterial genes in phage-like capsids. This avoids some of the consequences of predator-prey warfare and enables precise engineering and limitless versatility of antibacterial cargos, with two different incarnations being phagemids and ABDs.
Phagemids
Phagemids are hybrid plasmids containing a phage packaging module plus added genes of interest, which are packaged in phage particles by the cognate helper phage [38] [39] [40]. In the first published paper to report the potential therapeutic use of a phagemid [38], the intergenic region of filamentous phage f1, which directs f1 packaging, was cloned to a plasmid containing gef or chpBK, each encoding the toxin component of a TA system. The plasmid was transferred into a host cell containing the packaging-defective f1 helper phage, R408. This cell packages and extrudes large numbers of f1-like particles containing the modified plasmid, and only a few ordinary f1 particles. After a demonstration of the ability of these particles to kill F+ bacteria in vitro, they were administered to mice that had been infected with F+ E. coli and they reduced the number of infecting bacteria by about 1000-fold, demonstrating the therapeutic potential of the phagemid. In a second report, Bikard et al [40] cloned the ΦNM1 terminase and rinA genes to the rolling circle staphylococcal plasmid pC194, followed by a CRISPR/cas9 module with a spacer targeting the aphA (kanamycin resistance) gene. In this scheme, phage lysates contained only about 3% phagemid particles (~3×105/ml) and the phagemid was structurally unstable, as is typically the case for rolling circle plasmids with large inserts. These particles were shown to eliminate a kanamycin resistance (KmR) plasmid and to kill cells in which the aphA (KmR) gene was chromosomal, in both cases via Cas9-induced DNA cleavage of the aphA protospacer. This preparation was used to treat a mouse staphylococcal skin infection, causing a decrease (but only a five-fold decrease) in the number of infecting KmR staphylococci, as measured with GFP-tagged cells. In a third phagemid paper, Krom et al. [39] used the same f1/m13 system as Westwater et al [38], but developed a much more elaborate and sophisticated set of constructs to accomplish several different outcomes, successfully targeting F+ E. coli cells in vitro and in vivo.
Although the bactericidal effectiveness of phagemids based on coliphage f1/m15 has been convincingly demonstrated, the host range of these phagemids among natural E. coli populations must be extremely narrow, as only F+ or HFR strains are susceptible and F is a rare, mating-derepressed plasmid; the only possible way to extend this range has been to modify the phage to recognize mating pili of other plasmids [41], which, incidentally, are normally mating-repressed and do not express their mating pili. A much better phagemid strategy is that reported by Bikard, et al. [40], in which the helper phage is a typical staphylococcal siphovirus. Although its host range would be limited by restriction enzymes etc., as is the case for the ABDs, effective remedies for both can be envisioned. In the configuration described by Bikard et al., much improvement could be achieved by eliminating phage maturation and improving plasmid stability and particle yield.
ABDs
The ABDs, developed in and for S. aureus, are based upon the highly mobile, phage-inducible pathogenicity islands known as SaPIs [42] [43], which are ideal for conversion to antibacterial agents. SaPIs are ~15 kb chromosomal elements, long ago derived from prophages; while retaining several key prophage functions, including an integrase, a master repressor, a replicon, and a SaPI-specific terminase small subunit (TerS), they have evolved into distinct genetic elements with a unique lifestyle, including repressors that are not SOS induced, pathogenic cargos, and an important role for their bacterial cells. They are induced to excise and replicate by helper phage-encoded anti-repressors [44] [45], are packaged in small phage-like particles assembled from phage virion proteins [46], and released in very high numbers (~109/ml) [47], disseminating their cargo of superantigen and other virulence genes. Their recent conversion from disease-causing pathogenicity islands to disease curing ABDs involved i) deletion of virulence genes, ii) expansion of cargo capacity by deletion of genes responsible for small capsid formation, and iii) incorporation of antibacterial cargos. In addition, the ABD helper prophage was modified to prevent phage production by deletion of its specific TerS gene. The helper prophage replicates upon induction with mitomycin C but its DNA is not packaged. Its anti-repressor induces the resident ABD, which is packaged in full-sized phage particles, using its specific TerS, with some 30 kb of cloning space available for cargo genes. Initial studies have used ABDs containing tetM or cadA for selection and scoring, and CRISPR/cas9, armed with spacers targeting highly conserved but non-essential genes such as agrA or fnbA. Upon entry into a susceptible cell, Cas9::spacer is produced, binds to and induces a double strand cut in the agrA or fnbA protospacer, which is lethal owing to a lack of non-homologous end joining capability [48] [42]. A similar system using CRISPR/cas3a, which targets RNA nonspecifically, has recently been developed by Kiga, et al. [49].
Either ABD (Fig. 2) caused a 104-fold decrease in viability, with survivors being CRISPR-resistant mutants. An ABD containing the non-nucleolytic CRISPR/dcas9 armed with a spacer targeting the P2P3 regulatory region of the virulence-controlling agr locus (Fig. 3) totally blocked agr expression. These two ABDs were tested in murine subcutaneous abscess [50] and peritoneal lethality [51] models, sharply attenuating the abscesses and rescuing mice infected intraperitoneally with a lethal dose of staphylococci [52]. These results were obtained with strains susceptible to the ABD, including the clinically important methicillin-resistant (MRSA) strain USA300. Some other strains, which were resistant to both the ABD helper phage (80α) and to the broad host range phage K, were nevertheless susceptible to ABD lethality [52]. This was assumed to reflect the much simpler requirement for ABD cargo expression than for phage sensitivity, which requires execution of the entire phage lytic cycle. Nevertheless, many clinical S. aureus isolates are only weakly susceptible to the ABDs. This remains a major problem facing the clinical applicability of the ABD system, as is also the case for any other phage particle-based antibacterial strategy. In the case of the ABDs, since adsorption/injection is largely non-specific in staphylococci, and CRISPRs are very rare, the most serious impediment is almost certainly restriction enzyme degradation; possible solutions are under development in the author’s laboratory.
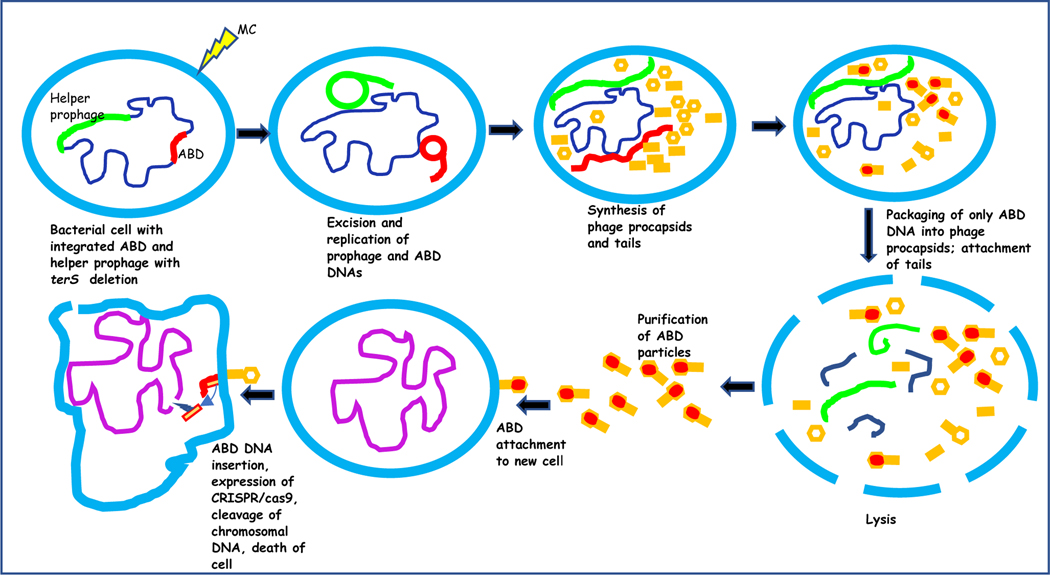
Fig. 2. Production and activity of ABD2003, carrying CRISPR/cas9:: fnbA.
Integrated ABD (red) and helper prophage (green) are induced by mitomycin C (MC) to excise and replicate, followed by procapsid production and packaging of ABD DNA (only), followed by lysis, release and purification of ABD particles. These attach to and infect target cells. ABD DNA is injected, cas9 is expressed and cleaves the agrA protospacer,
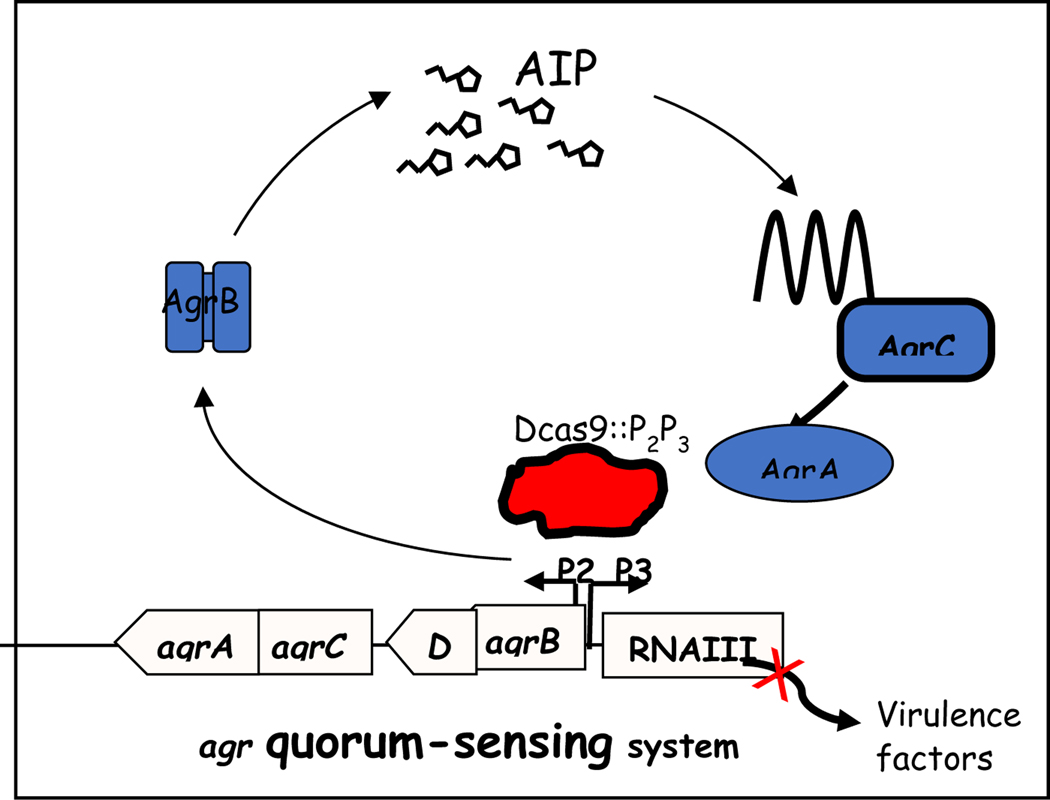
Fig. 3. Blockage of agr expression by ABD2006 containing CRISPR/dcas9::agrP2P3.
Agr is an autocatalytic 2-component signal transduction system consisting of a transmembrane receptor, AgrC, which senses and is activated by a cyclic thiolactone peptide, the AIP, processed from a precursor, AgrD by AgrB, which is also required for its secretion. Activated AgrC transphosphorylates AgrA, which activates the two regulatory promoters, P2 and P3, thus autoactivating the P2 operon as well as the P3 transcript, a regulatory RNA known as RNAIII, which regulates the virulon. dCas9::P2P3 binds to the promoter region, shutting down the regulon
Conclusion
The discovery of antibiotics was followed by the tremendously exciting and extremely rapid development of antimicrobial therapeutics – so much so that the medical world was lulled into a state of complacency – indeed “spoiled” by the power and efficacy of these miracle drugs. The predicted AMR crisis, born in <20 years, has steadily worsened until it has finally occasioned an intense effort to remedy it by any means possible. This effort has generated a wide field of endeavor that has been increasing for at least 40 years but has yet to come up with anything remotely as effective as the antibiotic quick-fix for infections. In this review, we have addressed a specific sub-field in this endeavor, the development of four single-unit systems that attack individual bacteria, one-by-one, in contrast to soluble chemicals, and report that each of them has great promise but is as yet very far from ameliorating the AMR crisis. Three of these use phage particles for delivery of antibacterial agents – phages themselves, phagemids, and ABDs. Each is plagued by pre-existing resistances and host range limitations, the overcoming of which is the key to clinical utility but is highly challenging, to say the least. The use of intact phages has progressed the furthest but remains limited to the rescue of patients fatally infected by pan-resistant bacteria, using pre-tested personalized phages. The fourth, using live predatory bacteria, is highly promising, especially in particular situations, but seems plagued by the distasteful prospect of using live bacteria to chase and attack other live bacteria within or on the human body. On the whole, each of these one-on-one targeting systems has a potential place in antibacterial bacterial infection control in the AMR era, but each has a long way to go.
Acknowledgements
Work from the author’s lab was supported by NIH grant R01-AI139613
References
- 1. Ghosh C, et al. , Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol, 2019. 27(4): p. 323–338. * Thoughtful and comprehensive review - well-worth reading for an overview of possible answers to the AMR problem
- 2.Stolp H. and Starr MP, Bdellovibrio Bacteriovorus Gen. Et Sp. N., a Predatory, Ectoparasitic, and Bacteriolytic Microorganism. Antonie Van Leeuwenhoek, 1963. 29: p. 217–48. [DOI] [PubMed] [Google Scholar]
- 3. Jurkevitch E. and Jacquet S, [Bdellovibrio and like organisms: outstanding predators!]. Med Sci (Paris), 2017. 33(5): p. 519–527. * Report showing both endo- and epibiotic predation by Bdellovibrio. Source of Fig. 1 in the present MS.
- 4.Seccareccia I, Kost C, and Nett M, Quantitative Analysis of Lysobacter Predation. Appl Environ Microbiol, 2015. 81(20): p. 7098–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laloux G, Shedding Light on the Cell Biology of the Predatory Bacterium Bdellovibrio bacteriovorus. Front Microbiol, 2019. 10: p. 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Z, Kadouri DE, and Wu M, Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics, 2011. 12: p. 453. * Description of group or “wolfpack” predation (referred to as “transbiotic” in the text)
- 7. Kuru E, et al. , Fluorescent D-amino-acids reveal bi-cellular cell wall modifications important for Bdellovibrio bacteriovorus predation. Nat Microbiol, 2017. 2(12): p. 1648–1657. Detailed analysis of endobiotic predation by Bdellovibrio
- 8.Shatzkes K, et al. , Examining the safety of respiratory and intravenous inoculation of Bdellovibrio bacteriovorus and Micavibrio aeruginosavorus in a mouse model. Sci Rep, 2015. 5: p. 12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shatzkes K, et al. , Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci Rep, 2017. 7: p. 43483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shatzkes K, et al. , Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio, 2016. 7(6). * Carefully presented report of in vivo antibacterial activity of Bdellovibrio
- 11. Yu Zhuodong Zhu C.S. Liang, Chen Linlin, Shen Yun, Yu Pingfeng, Hitchhiking Behavior in Bacteriophages Facilitates Phage Infection and Enhances Carrier Bacteria Colonization. Environ. Sci. Technol, 2021. 55(4): p. 2462–2472. * Describes penetration of biofilm by B. subtilis with phage particles adherent to the bacterial flagellae, thus enabling phage to penetrate the biofilm along with the bacteria
- 12. Hobley L, et al. , Dual Predation by Bacteriophage and Bdellovibrio bacteriovorus Can Eradicate Escherichia coli Prey in Situations where Single Predation Cannot. J Bacteriol, 2020. 202(6). ** Report of synergistic antibacterial activity of Bdellovibrio and bacteriophage, suggesting tha Bdellovibrio and phage are pysically attached in nature.
- 13.Kadouri D. and O’Toole GA, Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol, 2005. 71(7): p. 4044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dashiff A. and Kadouri DE, Predation of oral pathogens by Bdellovibrio bacteriovorus 109J. Mol Oral Microbiol, 2011. 26(1): p. 19–34. [DOI] [PubMed] [Google Scholar]
- 15.Mun W, et al. , Cyanide Production by Chromobacterium piscinae Shields It from Bdellovibrio bacteriovorus HD100 Predation. mBio, 2017. 8(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shatzkes K, et al. , Examining the efficacy of intravenous administration of predatory bacteria in rats. Sci Rep, 2017. 7(1): p. 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atterbury RJ, et al. , Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl Environ Microbiol, 2011. 77(16): p. 5794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Findlay JS, et al. , Predatory bacteria can protect SKH-1 mice from a lethal plague challenge. Sci Rep, 2019. 9(1): p. 7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pantanella F, et al. , Behaviour of Bdellovibrio bacteriovorus in the presence of Gram-positive Staphylococcus aureus. New Microbiol, 2018. 41(2): p. 145–152. * Demonstration of epibiotic predation of S. aureus by Bdellovibrio
- 20.Iebba V, et al. , Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus Cystic fibrosis isolates. Front Microbiol, 2014. 5: p. 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monnappa AK, et al. , Bdellovibrio bacteriovorus inhibits Staphylococcus aureus biofilm formation and invasion into human epithelial cells. Sci Rep, 2014. 4: p. 3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dashiff A, et al. , Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol, 2011. 110(2): p. 431–44. * Detailed description of epibiotic predation by Micavibrio compared to endobiotic predation by Bdellovibrio
- 23.Kadouri D, Venzon NC, and O’Toole GA, Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol, 2007. 73(2): p. 605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perez J, et al. , The antibiotic crisis: How bacterial predators can help. Comput Struct Biotechnol J, 2020. 18: p. 2547–2555. * Detailed and thoughtful review of the biology and clinical potential of predatory bacteria.
- 25. Moller AG, et al. , Genes Influencing Phage Host Range in Staphylococcus aureus on a Species-Wide Scale. mSphere, 2021. 6(1). *Comprehensive bioinformatic identification of a much wider variety of bacterial genes that could impact phage resistance than heretofore imagined.
- 26. Schooley RT, et al. , Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob Agents Chemother, 2017. 61(10). ** Riveting report of the compassionate use of a personalized phage to rescue a patient dying of a disseminated pan-resistant Acinetobacter baumani infection
- 27. Dedrick RM, et al. , Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med, 2019. 25(5): p. 730–733. * Report of the compassionate use of a customized phage to rescue a cystic fibrosis patient with a fatal mycobacterial infection
- 28. Chan BK, Abedon ST, and Loc-Carrillo C, Phage cocktails and the future of phage therapy. Future Microbiol, 2013. 8(6): p. 769–83. ** Extremely well-presented and thoughtfull analysis of the composition, potential role, and economics of phage cocktais from a contemporary Western point of view.
- 29. Kelly D, et al. , Development of a broad-host-range phage cocktail for biocontrol. Bioeng Bugs, 2011. 2(1): p. 31–7. ** Isolation and testing of 6 restriction-resistant variants of staphylococcal phage K, which, when combined into a cocktail, were active with 97% of clinical strains in a local databank
- 30.Botka T, et al. , Lytic and genomic properties of spontaneous host-range Kayvirus mutants prove their suitability for upgrading phage therapeutics against staphylococci. Sci Rep, 2019. 9(1): p. 5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehman SM, et al. , Design and Preclinical Development of a Phage Product for the Treatment of Antibiotic-Resistant Staphylococcus aureus Infections. Viruses, 2019. 11(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kilcher S, et al. , Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc Natl Acad Sci U S A, 2018. 115(3): p. 567–572. ** Description of an elegant and novel system in which synthetic phage genomes could be transformed into Listeria L-forms and redovered as functional phage particles - a wide-open platfrom for limitless phage engineering
- 33.Brown R, Lengeling A, Wang B, Phage engineering: how advances in molecular biology and synthetic biology are being utilized to enhance the therapeutic potential of bacteriophages. Quantitative Biology, 2017. 5(1): p. 42–54. [Google Scholar]
- 34. Ando H, et al. , Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst, 2015. 1(3): p. 187–196. * Demonstration of enhancement of phage host range by modifying tail fibers
- 35. Park JY, et al. , Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci Rep, 2017. 7: p. 44929. * Phage as delivery system for CRISPR/cas9. Uses astronomical multiplicities that would surely cause lysis from without - see unpublished information from Locus Pharmaceuticals.
- 36. Wright A, et al. , A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol, 2009. 34(4): p. 349–57. * First published clinical trial of phage therapy - not very impressive
- 37. Furfaro LL, Payne MS, and Chang BJ, Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front Cell Infect Microbiol, 2018. 8: p. 376. * Review of SaPI biology for the uninitiated
- 38.Westwater C, et al. , Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother, 2003. 47(4): p. 1301–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Krom RJ, et al. , Engineered Phagemids for Nonlytic, Targeted Antibacterial Therapies. Nano Lett, 2015. 15(7): p. 4808–13. * A superbly sophistocated account of what can be done by engineering a filamentous phage-based vehicle to defeat infections by “male” strains of E.coli.
- 40. Bikard D, et al. , Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol, 2014. 32(11): p. 1146–50. ** The development of a phagemid that sets the stage for widespread application of the phagemid principle, and develops CRISPR/cas9 as an ideal antibacterial cargo
- 41. Marzari R, et al. , Extending filamentous phage host range by the grafting of a heterologous receptor binding domain. Gene, 1997. 185(1): p. 27–33. * Demonstration of the possibility of extending the host range of filamentous phages such as M13 by modifying the adsorption mechanism to recognize conjugative pili encoded by plasmids other than F.
- 42.Novick RP, Christie GE, and Penadés JR, The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol, 2010. 8: p. 541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsay JA, et al. , The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol, 1998. 29: p. 527–543. [DOI] [PubMed] [Google Scholar]
- 44.Ubeda C, et al. , Characterization of mutations defining SaPI functions and enabling autonomous replication in the absence of helper phage. Mol Microbiol, 2008. 67:493–503. [DOI] [PubMed] [Google Scholar]
- 45. Tormo-Mas MA, et al. , Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature, 2010. 465(7299): p. 779–82. * Reports the remarkable identification of a meta.bolic enzyme, dUTPase, that doubles as the helper phage encoded SaPI antirepressor
- 46.Damle PK, et al. , The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology, 2012. 432(2): p. 277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ruzin A, Lindsay J, and Novick RP, Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol Microbiol, 2001. 41(2): p. 365–77. * Description of the first molecular analysis of the SaPIs, which are responsible for the spread of superantigen toxin genes and are the first pathogenicity islands shown to be mobile
- 48. Gomaa AA, et al. , Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. mBio, 2014. 5(1): p. e00928–13. *First demonstration of the lethality of CRISPR/cas9-induced double strand scissions of bacterial DNA
- 49.Kiga K, et al. , Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat Commun, 2020. 11(1): p. 2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barg N, et al. , Murine Model of Cutaneous Infection with Gram-Positive Cocci. Infect. and Immun, 1992. 60: p. 2636–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brouillette E, et al. , DNA immunization against the clumping factor A (ClfA) of Staphylococcus aureus. Vaccine, 2002. 20(17–18): p. 2348–57. [DOI] [PubMed] [Google Scholar]
- 52. Ram G, et al. , Conversion of staphylococcal pathogenicity islands to CRISPR-carrying antibacterial agents that cure infections in mice. Nat Biotechnol, 2018. 36(10): p. 971–976. **Reports the conversion of the highly mobile staphylococcal pathogenicity islands (SaPIs) to potent bactericidal or bacteriostatic agents (ABDs) using CRISPR/cas9 to cure murine infections.