Abstract
Despite advancements in treatments, oral squamous cell carcinoma (OSCC) has not significantly improved in prognosis or survival rate primarily due to the presence of treatment‐resistant OSCC. The intercellular communication between tumour cells is a molecular mechanism involved in acquiring OSCC treatment resistance. Extracellular vesicles (EVs) and encapsulated miRNAs are important mediators of intercellular communication. Here, we focused on EVs released from clinically relevant radioresistant (CRR) OSCC cells. Additionally, we evaluated the correlation between miRNA expression in the serum samples of patients who showed resistance to radiotherapy and in EVs released from CRR OSCC cells. We found that EVs released from CRR OSCC cells conferred radioresistance to radiosensitive OSCC cells via miR‐503‐3p contained in EVs. This miR‐503‐3p inhibited BAK and impaired the caspase cascade to suppress radiation‐induced apoptosis. Furthermore, OSCC cells with BAK knockdown had increased radioresistance. Additionally, the expression of circulating miR‐503‐3p in patients with OSCC was correlated with a poor treatment response and prognosis of radiotherapy. Our results provide new insights into the relationship between EVs and the radioresistance of OSCC and suggest that the miR‐503‐3p–BAK axis may be a therapeutic target and that circulating miR‐503‐3p is a useful prognostic biomarker in the radiotherapy of OSCC.
Keywords: BAK, extracellular vesicles (EVs), microRNAs, oral squamous cell carcinoma (OSCC), radioresistance
1. INTRODUCTION
Oral squamous cell carcinoma (OSCC) is one of the most common cancers of the oral cavity (Siegel et al., 2017). However, the survival rate has not improved despite advancements in diagnostic modalities and treatments. Thus, the prognosis of advanced OSCC remains poor, with a 5‐year survival rate of approximately 50% (Gupta et al., 2009). This stagnation in the survival rate is mainly attributed to the existence of high‐grade malignant cells that display important hallmarks of cancer, such as resistance to chemotherapy or radiotherapy, abnormal proliferation, and invasion or metastasis (Hanahan & Weinberg, 2011).
Recently, it has been clarified that tumour heterogeneity is closely related to treatment resistance (Naik et al., 2016). Intercellular communication among tumour cells or between tumour cells and stromal cells is deeply involved in acquiring high‐grade malignant traits (Wang et al., 2018). However, the role of intercellular communications in treatment resistance has not been fully elucidated in OSCC. To date, it has become clear that extracellular vesicles (EVs) play an important role as a medium for intercellular communications (Raposo & Stoorvogel, 2013). EVs are classified into large extracellular vesicles (lEVs; also known as microvesicles, approximately 100–1000 nm in diameter) and small extracellular vesicles (sEVs or exosomes, approximately 50–150 nm in diameter) according to their size (Thery et al., 2018). Cancer cells acquire malignant traits by transmitting information to their surrounding cells via EVs and regulating the tumour microenvironment (Becker et al., 2016; Naito et al., 2017). In OSCC, EVs derived from OSCC cells or the surrounding stromal cells are associated with tumour growth, infiltration, and metastasis (Sento et al., 2016; Xie et al., 2019). Research on the relationship between treatment resistance in OSCC, especially radioresistance and EVs, is underway (Khoo et al., 2019).
EVs contain microRNA (miRNA) (Valadi et al., 2007), which are noncoding 20‐ to 25‐nucleotide‐long RNAs that regulate cell death, proliferation, metastasis, and treatment resistance (Garzon et al., 2010). The miRNAs contained in EVs are important mediators of intercellular communications (Kosaka et al., 2013). The usefulness of diagnostic and treatment methods targeting miRNAs contained in EVs has been reported (Kohlhapp et al., 2015; Tominaga et al., 2015). Recent studies have demonstrated that miRNAs are secreted from various cells, including cancer cells, via EVs into body fluids such as the blood, urine, breast milk, and saliva (Fiskaa et al., 2016; Hu et al., 2012). Extracellular miRNAs present in the blood are unstable and may be derived from other cells, such as inflammatory cells and immune cells, in addition to cancer cells, with potentially fundamentally different profiles (Swarup & Rajeswari, 2007). However, miRNA contained in EVs can be stable, even in the blood (Cheng et al., 2014). Recently, EV‐encapsulated miRNAs in the circulation have been proposed as biomarker candidates for disease monitoring and prognosis in the surveillance or monitoring of cancer (Joyce et al., 2016; Pfeffer et al., 2015). Summerer et al. (Summerer et al., 2015) reported that circulating miR‐142, miR‐186, miR‐195, miR‐374b and miR‐574 are prognostic biomarkers for head and neck cancer. However, it is not clear how EV‐derived miRNAs correlate with the therapeutic effect of concurrent chemoradiotherapy (CCRT) and patient prognosis in oral cancer.
In the present study, we focused on EVs released from a clinically relevant radioresistant (CRR) cell line established from an OSCC cell line (Kuwahara et al., 2010) and examined how they affect the radiosensitivity of non‐radiation‐resistant OSCC cells. Then, we analysed the expression of miRNAs contained in EVs, clarified a part of the molecular mechanism of radioresistance, and investigated whether EV‐derived miRNAs could be a new therapeutic strategy and prognostic marker of radiotherapy in OSCC.
2. MATERIALS AND METHODS
2.1. Cell lines and culture
The human OSCC cell lines, SAS, and HSC‐2 were purchased from the Japanese Collection of Research Bioresources Bank of the National Institutes of Biomedical Innovation, Health and Nutrition (Osaka, Japan). The CRR cell line SAS‐R was derived from SAS cells, by exposing them to gradually increasing X‐ray doses (Kuwahara et al., 2010). CRR cells continued to proliferate with daily 2 Gy irradiation for more than 30 days in vitro and were resistant to irradiation. Cells were cultured in DMEM (D6429; Sigma‐Aldrich, Saint Louis, MO, USA) supplemented with 10% FBS (Sigma‐Aldrich) in a humidified atmosphere of 5% CO2 at 37°C.
2.2. Short tandem repeat analysis
Genomic DNA was extracted from SAS and SAS‐R using NucleoSpin Tissue kit (MACHEREY‐NAGEL, Duren, Germany), and short tandem repeat (STR) analysis of the extracted gDNA was performed by Takara Bio (Shiga, Japan). The results of the analysis were checked against the STR analysis results of SAS registered in JCBR (National Institutes of Biomedical Innovation, Health and Nutrition, Osaka, Japan) to confirm the origin of the cells (Table S1).
2.3. Irradiation
Irradiation doses of 2, 4, and 6 Gy were administered with a 150‐KVp X‐ray generator (MBR‐1520R; Hitachi, Tokyo, Japan) with a total filtration of 0.5‐mm aluminium plUS 0.1‐mm copper filter. The dose rate (1.01 Gy·min−1) was measured with a thimble ionization chamber (IC 17A; Far West Technology, Goleta, CA, USA).
2.4. Isolation of EVs from cell cultures
SAS and SAS‐R cells (1 × 106) were seeded in 100 mm tissue culture dishes (AGG Inc., Tokyo, Japan). After 48 h, the conditioned medium (CM) was replaced with 10 ml DMEM containing 10% Exo‐FBS Exosome‐depleted FBS (System Biosciences (SBI), Palo Alto, CA, USA). After a further 24 h, CM was collected and ultrafiltered with a 100‐kDa cut‐off Ultrafiltration filter (Amicon Ultra‐15; MERCK, Tokyo, Japan). The CM (10 ml) was concentrated to approximately 500–700 μl and loaded on a size‐exclusion chromatography (SEC) column (EVSecond L70; GL Sciences, Tokyo, Japan).The fractions were collected according to the manufacturer's protocol. The EV elution fractions were examined by western blotting for CD9, CD81, and ALIX. Moreover, relative quantification of the exosomes was performed using a CD9/CD63 exosome ELISA kit (COSMO BIO, Tokyo, Japan), which could measure exosomes comprising a combination of CD9 and CD63. Furthermore, serum protein concentration was measured using the TaKaRa Bradford Protein Assay Kit (Takara Bio). Subsequently, the eluted samples were pooled. EVs were recovered in phosphate‐buffered saline (PBS) and stored at −80°C. EVs isolated from SAS cells (SAS EVs) and EVs isolated from SAS‐R cells (SAS‐R EVs) were used in subsequent experiments.
2.5. Characterization of EVs
The presence of exosome‐specific protein markers and the absence of non‐EV markers on the isolated vesicles were determined by western blotting. Whole‐cell proteins or EVs (5 μg) were separated by 10%–20% SDS‐PAGE, transferred to nitrocellulose membranes, and blocked for 60 min. The membranes were incubated in primary antibody cocktails (diluted in TBS‐T with 5% BSA) overnight at 4°C, washed thrice for 10 min each in TBS‐T, and incubated in secondary antibody cocktails for 60 min at room temperature. The membranes were washed thrice for 10 min each in TBS‐T and developed using the ECL prime detection kit (GE Healthcare, Chicago, IL, USA). The emitted light was measured using the C‐DiGit blot scanner, and the images were analysed using Image studio for C‐DiGit (LI‐COR Biosciences, Lincoln, NE, USA). The antibodies used were mouse anti‐CD9 antibody (12A12; COSMO BIO), 1/1,000; mouse anti‐CD81 antibody (12C4; COSMO BIO), 1/1,000; mouse anti‐ALIX antibody (ab117600; Abcam, Cambridge, UK), 1/500; mouse anti‐GAPDH antibody (ab8245; Abcam), 1/1,000; rabbit anti‐calnexin antibody (2679; Cell Signalling Technology, Danvers, MA, USA), 1/1,000; anti‐mouse IgG, HRP‐linked antibody (7076; Cell Signalling Technology), 1/2,000; and anti‐rabbit IgG, HRP‐linked antibody (7074; Cell Signalling Technology), 1/2,000.
2.6. Observation of EVs
EVs were visualized with a transmission electron microscope (HT7700; Hitachi, Tokyo, Japan) at 80 kV. EVs samples were processed by the Exosome‐TEM‐easy kit (101 Bio, Palo Alto, CA, USA) according to the manufacturer's instructions.
2.7. Quantification of EVs
EV protein was quantified using the colorimetric BCA protein assay (Thermo Fisher Scientific, Waltham, MA, USA), per the manufacturer's instructions. Nanoparticle tracking analysis (NTA) (NanoSight NS300, Wiltshire, UK) was used to check the size distribution and the concentration of the vesicles. For NTA analysis of the EVs, PBS was used as a diluent; a syringe pump with a constant flow injection was used, and five videos of 60 s each were captured with 1498 frames and a camera level at 15. The videos were recorded and analyzsd with NTA software version 3.3 to determine the size and concentration of the particles.
2.8. Uptake of EVs
SAS EVs or SAS‐R EVs (2 μg) were stained with the red fluorescent dye PKH26 (Sigma‐Aldrich) according to the manufacturer's instructions. The excess dye was removed using a centrifugal extrafiltration filter (Amicon Ultra‐0.5 100K device; MERCK). PKH26‐stained EVs were added to 5000 SAS cells or HSC‐2 cells. After 24 h, the uptake of PKH‐stained EVs into target cells was observed with the fluorescence microscope BZ‐X800 (KEYENCE, Osaka, Japan). Next, whole‐slide images were obtained at the same brightness/contrast with the BZ‐X800 (KEYENCE) utilizing the optical sectioning mode and capturing Z‐stacks of 16–18 μm at recommended step sizes and projected onto a full‐focus image using the BZ‐Analyzer. For all experimental conditions, the exposure times were maintained, and the number of positive cells per 50 cells was counted.
2.9. Transfection of exosome cyto‐tracer into cells
SAS and SAS‐R cells were seeded in 24‐well plates (5 × 104 cells/well). After 24 h, pCT‐CD9‐GFP (pCMV, Exosome/Secretory, CD9 Tetraspanin Tag, Virus; SBI) at a final concentration of 30 virus particles per cell was transfected into the SAS cells using TransDux MAX (SBI) according to the manufacturer's instructions. After 72 h, a cell selection procedure was performed for 7 days using puromycin (5 μg/ml). In the same way, pCT‐CD9‐RFP (SBI) was transfected into SAS‐R cells.
2.10. Cell transfection
The mirVana miRNA mimic negative control #1 (Ambion, Austin, TX, USA), hsa‐miR‐296‐5p mirVana miRNA mimic, hsa‐miR‐503‐3p mirVana miRNA mimic, or hsa‐miR‐6748‐3p mirVana miRNA mimic (# 4464066; Ambion) at a final concentration of 30 nM for the overexpression of each miRNAs were transfected into the target cells using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For decreasing BAK expression, BAK siRNA (10 nM; Stealth siRNA; Invitrogen) were transfected with RNAiMAX into the target cells.
2.11. RNA isolation and real‐time polymerase chain reaction analysis
Total RNA was isolated using the FastGeneTM RNA Basic Kit (NIPPON Genetics, Tokyo, Japan) and then reverse‐transcribed to cDNA using the ReverTra Ace qPCR RT Kit (Toyobo, Osaka, Japan). For total RNA recovery after miRNA transfection, the cultured cells were treated with RNAase (10 μg/ml) at room temperature for 30 min. The cells were then washed twice with PBS, after which they were immediately treated with RNAlater RNA Stabilization Reagent (QIAGEN) to inactivate RNAase before recovery. Real‐time PCR was performed using Thunderbird SYBR qPCR Mix (Toyobo) on a Light Cycler 1.5 (Roche, Basel, Switzerland). Data obtained from RT‐qPCR were analysed using the 2−∆∆Ct method (Livak & Schmittgen, 2001), with glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as a housekeeping gene. Each sample was run in triplicate. The primer sets are listed in Table S2. PCR data were collected from three independent experiments.
2.12. Extraction of total RNA and quantitative real‐time RT‐PCR for microRNAs
Total RNA was extracted from cells and EVs using miRNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. First‐strand cDNA was synthesized using the miScript II RT kit (QIAGEN). Real‐time PCR was run using a miScript SYBR Green PCR kit (QIAGEN) on a LightCycler 1.5 system (Roche, Indianapolis, IN, USA). The primer sets were purchased from QIAGEN (Table S3). Data obtained from RT‐qPCR were analysed using the 2−∆∆Ct method (Livak & Schmittgen, 2001). The exosomal miRNA expression levels were normalized using Cel‐miR‐39. RNU6B was used as a reference gene to evaluate intracellular miRNA expression; conversely, miR‐16‐2‐3p was used as a reference gene to evaluate miRNA expression in serum based on the results of array analysis using patient serum. In addition, we confirmed that the miRNAs were stably expressed in serum to the same extent, and conducted further studies.
2.13. Dual‐luciferase reporter assay
A luciferase reporter containing the full‐length 3′UTR of BAK (wild type and mutant type) was constructed by Active Motif, Inc. (Carlsbad, CA, USA). SAS cells were co‐transfected in a 96‐well white culture plate with 50 ng wild type or mutant type BAK‐3′UTR‐luciferase vectors using FuGENE HD transfection reagent (Promega, Madison, WI, USA) and 30 nM hsa‐miR‐503‐3p mirVana miRNA mimic (# 4464066; Ambion) or mirVana miRNA mimic negative control #1 (Ambion) using Lipofectamine RNAi MAX (Invitrogen). After 48 h, the luciferase activity was measured using the dual‐luciferase reporter assay system (Promega).
2.14. High‐density survival assay
High‐density survival (HDS) assays were performed as described by Kuwahara et al. (Kuwahara et al., 2010; Matsuoka et al., 2016). Exponentially growing cells (5 × 105) were seeded in 60 mm tissue culture dishes (AGG Inc.) and incubated in DMEM supplemented with 10% FBS for 48 h. Cells were exposed to irradiation and then treated with EVs or transfected with miRNA mimic and siRNA. After 72 h, 10% of the cells in each flask were seeded into a new 60‐mm culture dish and incubated for 72 h. Total cell numbers and cell survival in each culture dish were determined using trypan blue exclusion assay.
2.15. Co‐culture assay
After irradiating SAS cells or HSC‐2 cells with 6 Gy, cells (1 × 103) were seeded in an indirect co‐culture plate (NICO‐1; Ginrei Lab, Ishikawa, Japan). Irradiated cells were co‐cultured with SAS cells (1 × 104) or SAS‐R cells (1 × 104) in DMEM containing 10% Exo‐FBS Exosome‐depleted FBS (SBI) for 10 days. Cells were fixed with 99.5% methanol at room temperature and stained with Giemsa solution (Wako, Osaka, Japan). In the image acquisition of the transferred exosomes, whole‐slide images were obtained at the same brightness/contrast using the BZ‐X800 (KEYENCE) via its optical sectioning mode, and Z‐stacks of 16–18 μm at recommended step sizes were captured and projected onto a full‐focus image using the BZ‐Analyzer.
2.16. Cell proliferation assays
For assessing normal proliferation, viable cells (1 × 103 cells/well in 96‐well plates) under EV treatment (0, 10, 100 ng/ml and 1 μg/ml) were quantified every 24 h using the Cell Counting Kit‐8 (Dojindo, Kumamoto, Japan).
2.17. miRNA screening with 3D‐gene microarray
From the advanced OSCC patients who received chemoradiotherapy, serum samples were obtained from three patients who responded to chemoradiotherapy and four who did not (Table S4). Total RNA was extracted from the serum samples, SAS EVs, and SAS‐R EVs using 3D‐Gene RNA extraction reagent (Toray, Kamakura, Japan) according to the manufacturer's instructions. Extracted total RNA was checked by Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and labelled with 3D‐Gene miRNA labelling kit (Toray). Half volumes of labelled RNAs were hybridized onto a 3D‐Gene Human miRNA Oligo chip (Toray). The annotation and oligonucleotide sequences of the probes were conformed to the miRBase miRNA database (http://microrna.sanger.ac.uk/sequences/). After stringent washes, fluorescent signals were scanned with the 3D‐Gene Scanner (Toray) and analysed using 3D‐Gene Extraction software (Toray). The raw data of each spot was normalized by substitution with a mean intensity of the background signal determined by all blank spot signal intensities of 95% confidence intervals. Measurements of spots with the signal intensities greater than two standard deviations (SD) of the background signal intensity were considered valid. The relative expression level of a given miRNA was calculated by comparing the signal intensities of the valid spots throughout the microarray experiments. The normalized data were globally normalized per array, such that the median of the signal intensity was adjusted to 25.
2.18. Clinical serum sample collection and analysis
For the miRNA screening with 3D‐Gene microarray, pretreatment serum samples were obtained from seven patients with locally advanced OSCC who underwent preoperative CRT at the Kumamoto University Hospital between October 2003 and January 2009. Apart from the screening cohort, to elucidate the clinicopathological significance of miR‐503‐3p, pretreatment serum samples were obtained from 55 patients with locally advanced OSCC, who were treated at the Kumamoto University Hospital. We excluded human papillomavirus (HPV)‐positive tumours from the analysis‐based immunostaining results for p16, a surrogate marker for HPV infection. All 55 patients underwent curative surgery following preoperative CRT. The protocol for the preoperative CRT was as previously described (Nomura et al., 2010). Total RNA was extracted from the serum samples of 55 patients using miRNeasy Serum/Plasma Kit (QIAGEN) according to the manufacturer's instructions. The patients were dichotomized into two groups according to their median level of miR‐503‐3p expression. Detailed patient data and miRNA measurements are shown in Table S6. The staging and determination of tumour differentiation were performed according to the seventh AJCC cancer staging manual (Edge et al., 2010). The histological response to CRT was graded using specimens obtained during surgery based on the criteria proposed by Shimosato et al. (Shimosato et al., 1971), as follows: grade I, tumour structures are not destroyed; grade IIa, destruction of the tumour structure is mild (i.e., “viable tumour cells” are frequently observed); grade IIb, destruction of the tumour structure is severe (i.e., “viable tumor cells” are few); grade III, nonviable tumour cells are present; and grade IV, no tumour cells remain.
The Chi‐squared test was performed to determine the associations between the miR‐503‐3p expression status and clinical or pathological variables. The overall survival (OS) and disease‐free survival (DFS) were defined as the time from CRT treatment initiation to the date of death of any cause and the date of recurrence of the cancer or death from any cause, respectively. The Kaplan–Meier method was used to estimate the probability of OS and DFS as a function of time, and the statistical differences in the survival of the subgroups of patients were compared by the log‐rank test. This study was performed with the approval of the Ethics Committee of Kumamoto University (approval number: 174) and in accordance with the Good Clinical Practice and the Declaration of Helsinki guidelines.
2.19. Western blotting
Whole‐cell proteins (Minute Cytoplasmic and Nuclear Extraction Kits; Invent Biotechnologies, Inc., Plymouth, MN, USA) were separated by 10%–20% SDS‐PAGE, transferred to nitrocellulose membranes, and blocked for 2 h. The membranes were incubated in primary antibody cocktails (diluted in TBS‐T containing 5% BSA) overnight at 4°C, washed thrice for 10 min each in TBS‐T, and incubated in secondary antibody cocktail for 60 min at room temperature. The membranes were washed thrice for 10 min each in TBS‐T and developed using the ECL prime detection kit (GE Healthcare, Chicago, IL, USA). The emitted light was measured using the C‐DiGit blot scanner, and the images were analysed using Image studio for C‐DiGit (LI‐COR Biosciences, Lincoln, NE, USA). A list of antibodies used in this study is shown in Table S5. The relative expression of each protein was calculated using the Image J 1.52q software (National Institutes of Health, Bethesda, MD, USA).
2.20. MitoMP assay
The mitochondrial membrane potential (MitoMP) was measured by the JC‐1 MitoMP detection kit (Dojindo). JC‐1 aggregate generated red fluorescence, indicating a normal function for MitoMP. By contrast, the JC‐1 monomer form generated green fluorescence, indicating dysfunction for MitoMP. After EV treatment, miRNA mimic transfection, or siRNA transfection, cells were treated with JC‐1 (4 μM, 37°C, 30 min). Finally, the MitoMP level was analysed by calculating the fluorescence intensity ratio (red: 535 nm (Ex), 595 nm (Em)/green: 485 nm (Ex), 535 nm (Em)) using a fluorescence plate reader (SpectraMax i3x; Molecular Devices, San Jose, CA, USA).
2.21. Apoptosis analysis
SAS cells or HSC‐2 cells were exposed to 6 Gy irradiation and then treated with EVs or transfected with miRNA mimic and siRNA. After 48 h, cells were stained with Apotracker Green (BioLegend, San Diego, CA, USA) and Hoechst 33342 (ThermoFisher Scientific, Waltham, MA, USA). The fluorescence intensity ratio (Apotracker Green: 500 nm (Ex), 520 nm (Em); Hoechst 33342: 350 nm (Ex), 461 nm (Em)) were calculated using a fluorescence plate reader (SpectraMax i3x; Molecular Devices, San Jose, CA, USA).
2.22. Statistical analysis
Differences in mean values between two groups were analysed using Student's t‐tests, while differences in mean values among multiple groups were analysed by one‐way ANOVA with the Bonferroni/Dunn test. We utilized the Kaplan–Meier method to estimate the probability of OS and DFS as a function of time and compared the statistical differences in survival between the patient groups using the log‐rank test. We performed multivariate survival analysis using the Cox regression model to study the effects of pretreatment with miR‐503‐3p on DFS and OS. All p‐values were based on two‐tailed statistical analyses; p‐values < 0.05 were considered statistically significant. All analyses were performed with JMP‐9 software (SAS Institute Inc, Cary, NC, USA).
3. RESULTS
3.1. Characterization of SAS EVs and SAS‐R EVs
Size‐exclusion chromatography was used for EV extraction, and the results of the CD9/CD63 exosome ELISA and Bradford Protein Assay showed that serum proteins and EVs in the peak fraction of the elution were effectively separated. Moreover, the expression of the typical EV markers CD9, CD81, and ALIX was also confirmed by western blotting, in close agreement with the peak fraction of the elution (Figure S1). These preliminary experiments confirmed that high‐purity EVs could be extracted, and we proceeded with subsequent experiments. Structures and properties of EVs released from SAS or SAS‐R cells were validated according to the minimal information for studies of extracellular vesicles proposed by the International Society for Extracellular Vesicles (ISEV) in 2018 (MISEV2018) (Thery et al., 2018). The number of cells at the time of EV recovery was about 6.7 × 106 for SAS cells and about 6.1 × 106 for SAS‐R cells. Both EV types were characterized by the expression of known EV biomarkers (CD9, CD81, ALIX) and the absence of calnexin (Figure 1a). EV protein concentrations, as measured by the BCA protein assay, were 56.8 μg/ml (42.1 to 79.0 μg/ml) for SAS EVs and 57.1 μg/ml (39.1 to 78.4 μg/ml) for SAS‐R EVs collected from 10 ml CM (Figure 1b). NTA demonstrated that the SAS EV particles had a mode diameter of 102.6 ± 5.0 nm and a concentration of 5.62 × 109 ± 5.05 × 108 particles/ml, whereas SAS‐R EVs had a mode diameter of 88.7 ± 3.0 nm and a concentration of 5.43 × 109 ± 4.05 × 108 particles/ml (Figure 1c). EVs from both groups were seen as round‐shaped structures on transmission electron microscopy. The diameter of both EVs was approximately 100 nm; the diameters of SAS EVs were larger than those of SAS‐R EVs (Figure 1d). Collectively, these results indicated no significant differences in the mass, number of particles, or structure of the sEVs released from SAS and SAS‐R cells.
FIGURE 1.
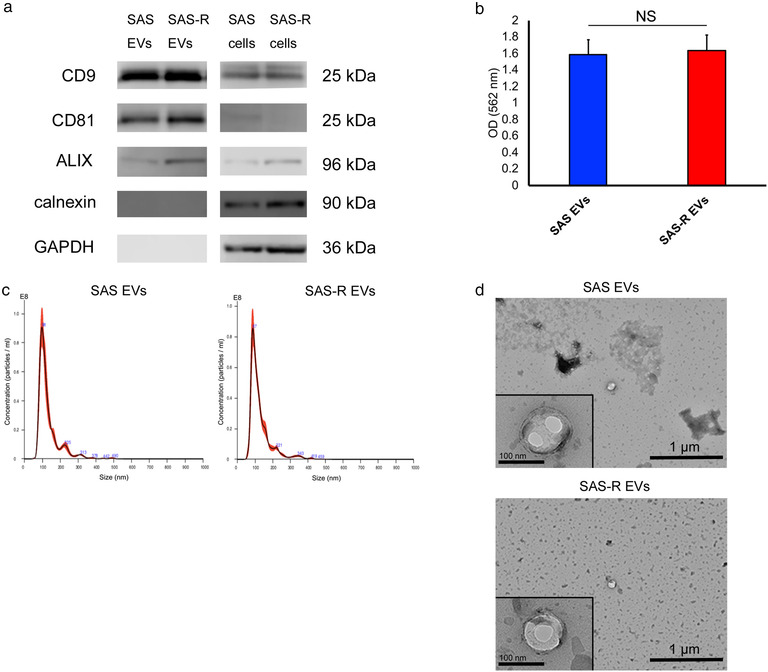
Characterization of OSCC cells derived EVs. (a) Western blotting of EVs and whole‐cell proteins was performed to confirm exosome marker proteins (CD9, CD81, ALIX), calnexin, and GAPDH. (b) The colorimetric BCA protein assay was performed to measure the EV protein. (c) The size distribution and the concentration of the EVs were measured with Nanoparticle Tracking Analysis. (d) Transmission electron micrograph (wide‐field and close‐up) of SAS EVs and SAS‐R EVs. Values are expressed as mean ± standard deviation of triplicate samples. EV, extracellular vesicles; OSCC, oral squamous cell carcinoma; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; NS, no significant differences
3.2. EVs released from radioresistant OSCC cells confer radioresistance
The uptake of EVs into the cells was confirmed by staining EVs with the fluorescent labelling dye PKH26. The uptake of both EV types into SAS cells was confirmed and quantified by fluorescence microscopy (Figure 2a). No significant difference was observed with respect to the uptake of SAS and SAS‐R EVs into SAS cells. To examine whether EVs affect the proliferation of OSCC cells, we assessed the proliferation activity of SAS cells under EV treatment conditions. No significant difference was observed in cell proliferation, even when the dose was changed (Figure 2b), and neither SAS EVs nor SAS‐R EVs affected cell proliferation in SAS cells. Next, we performed the HDS assay to assess the influence of EVs on the radiosensitivity of SAS cells. SAS cells treated with SAS‐R EVs showed significantly increased radioresistance compared with untreated SAS cells and SAS cells treated with SAS EVs (Figure 2c). The results confirmed the uptake of artificially extracted and administered EVs by the cultured cells. However, in order to confirm whether EVs pass between cells in the same manner under normal culture, a co‐culture experiment using the Exosome Cyto‐Tracer that entails fluorescent labelling of the endogenous EVs of SAS and SAS‐R was performed. The SAS cells after irradiation were co‐cultured with SAS or SAS‐R cells using an indirect co‐culture plate (NICO‐1). In co‐culture, an exosome cytotracer was introduced into the EVs to confirm their movement to the other incubator after passage through the filter. Fluorescence microscopy confirmed that each EV passed through the central filter and was taken up by SAS cells in the other incubator (Figure 2d). SAS cells co‐cultured with SAS‐R cells formed more colonies (Figure 2e). These results indicate that SAS‐R EVs confer radioresistance to the surrounding cells.
FIGURE 2.
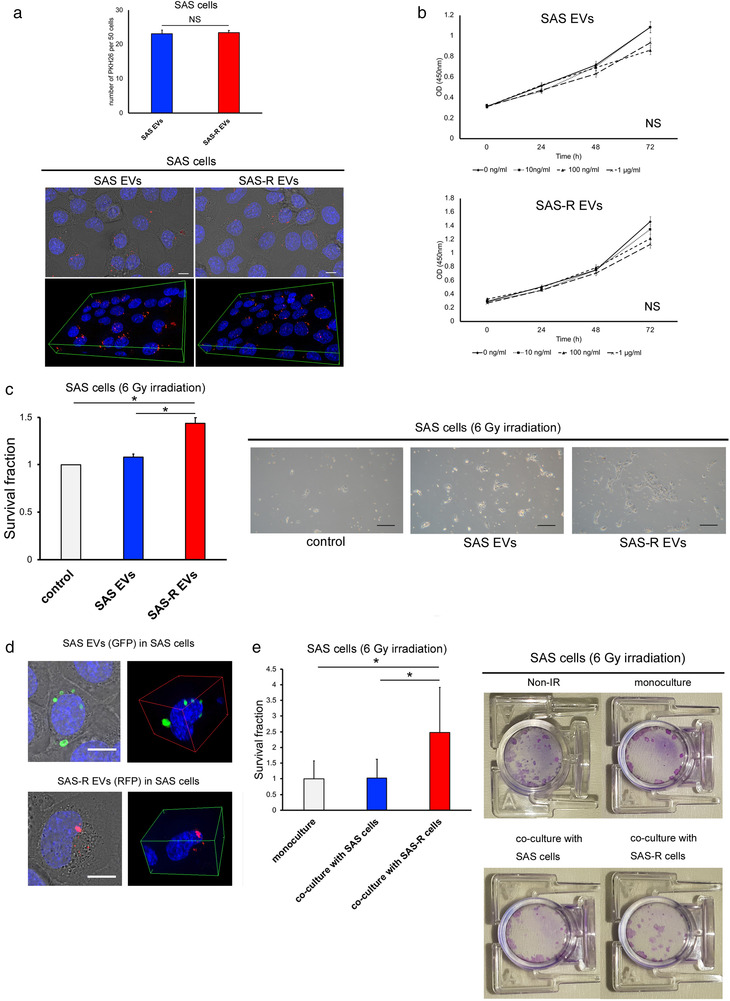
The effect of EVs on OSCC cells. (a) Representative fluorescent microscopic images for PKH26‐labeled EVs uptake after 24 h incubation. EVs are stained red, and nuclei are stained blue with Hoechst 33342. SAS cells containing PKH26‐labeled EVs per 50 cells were quantified. Scale bar, 50 μm. (b) Proliferation of SAS cells cultivated for 3 days with various concentrations of SAS EVs or SAS‐R EVs (0, 10, 100 ng/ml or 1 μg/ml). (c) The survival fraction of SAS cells after exposure to 6 Gy X‐rays was evaluated by high‐density survival assay under SAS EVs or SAS‐R EVs (20 μg/ml). As a control an equal amount of phosphate‐buffered saline without EVs was added to SAS cells. Scale bar, 200 μm. (d) Fluorescent microscopic images of SAS cells incubated for 3 days after docking the two incubators: SAS EVs (green), SAS‐R EVs (red), and nuclei (blue with Hoechst 33342). Fluorescent labelled EVs pass through the central filter and are taken up by SAS cells in the other incubator. (e) Photographs of the cell survival fraction as evaluated by clonogenic assay. After exposure to 6 Gy, SAS cells were co‐cultured with SAS cells or SAS‐R cells in an indirect co‐culture plate. After 10 days, cells were fixed and visualized. Values are expressed as mean ± standard deviation of triplicate samples. *p < 0.05; EV, extracellular vesicles; NS, no significant differences
3.3. Exosomal‐miR‐503‐3p is a candidate miRNA involved in OSCC radioresistance
In this study, based on the results that EV treatment of radioresistant OSCC reduced the radiosensitivity of several OSCC cell lines, we explored the possibility that exosomal miRNAs are involved in the radioresistance regulation mechanism. To extract exosomal miRNAs involved in the regulation of OSCC radioresistance, we first performed a comprehensive analysis of miRNAs in the EVs of radioresistant and parental cell lines. However, the comparison of the exosomal miRNAs of SAS and SAS‐R itself resulted in a large number of candidate miRNAs, and it was difficult to verify whether each miRNA was involved in radioresistance. Therefore, we narrowed down the candidate miRNAs as follows:
For exosomal miRNAs, the global normalization values (FPKM values) of SAS EVs and SAS‐R EVs were compared, and 142 miRNAs that showed increased expression in SAS‐R EVs were selected. Next, the serum miRNAs showed a significant difference (t‐test) between the mean values of global normalization values (FPKM values) of three cases in the CCRT‐responder group and four cases in the CCRT non‐responder group, and 42 miRNAs whose expression was upregulated in the CCRT non‐responders were selected. We compared and verified the miRNAs selected between serum and EVs, and selected three miRNAs whose expression was commonly upregulated in both. (Figure 3a, Table 1). Microarray expression profiling is available on NCBI's GEO (https://www.ncbi.nlm.nih.gov/geo/); data accession number GSE160122. When the expression of the extracted candidate miRNAs was confirmed in SAS and SAS‐R cells, the expression levels of miR‐503‐3p and miR‐6748‐3p were increased in SAS‐R cells. By contrast, miR‐296‐5p had increased expression in SAS cells. When the same verification was performed in EVs, the three miRNAs were more expressed in SAS‐R EVs than in SAS EVs (Figure 3b). The effect of these three miRNAs on the radiosensitivity of SAS cells was examined by transient forced expression. Of the three miRNAs, miR‐503‐3p significantly affected radioresistance compared with the negative control or other miRNAs in SAS cells (Figure 3c and d). Thus, we focused on the involvement of miR‐503‐3p in the radioresistance of OSCC.
FIGURE 3.
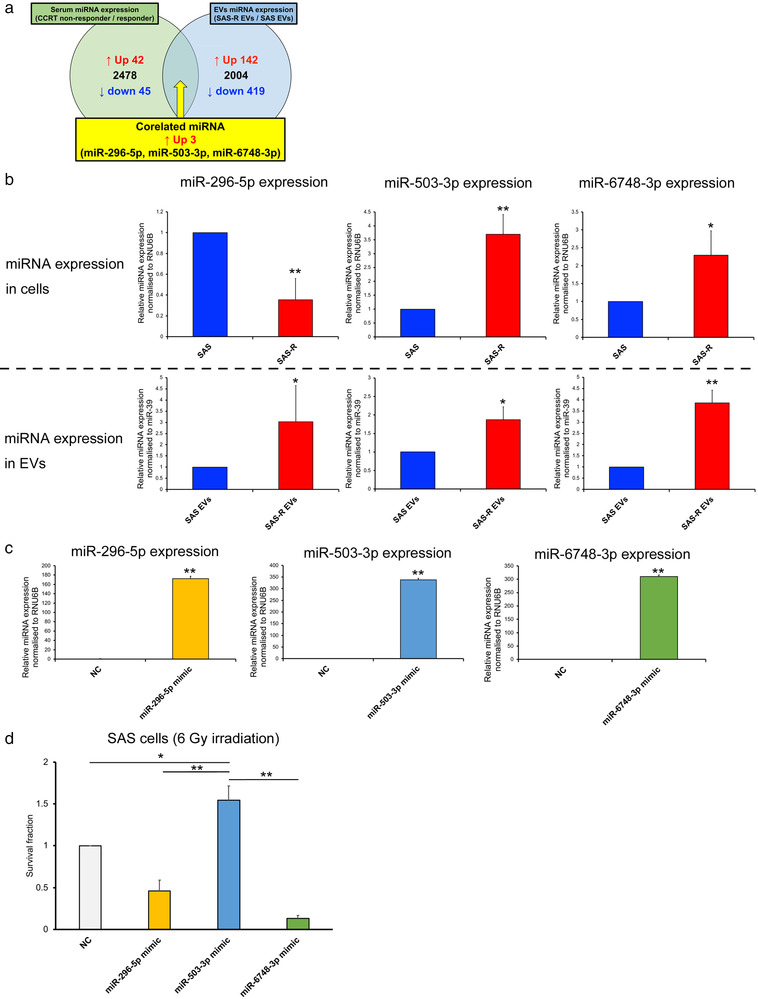
Identification of miRNA involved in radioresistance of OSCC. (a) Venn diagram showing miRNAs that were commonly upregulated in CRT nonresponder serum samples (green) and SAS‐R EVs (blue). (b) Upper row: Comparison of miR‐296‐5p, miR‐503‐3p, and miR‐6748‐3p expression between SAS cells and SAS‐R cells. Lower row: Comparison of miR‐296‐5p, miR‐503‐3p, and miR‐6748‐3p expression between SAS EVs and SAS‐R EVs. The expression of miRNAs was evaluated by RT‐qPCR. (c) Expression of miR‐296‐5p, miR‐503‐3p, and miR‐6748‐3p in SAS cells 48 h after transfection with negative control or miRNA mimic. The efficiency of transfection was evaluated by RT‐qPCR. (d) SAS cells were exposed to 6 Gy of X‐ray and then transfected with negative control or miRNA mimic; the survival fraction was evaluated by high‐density survival assay. Values are expressed as mean ± standard deviation of triplicate samples. *p < 0.05; **p < 0.01. OSCC, oral squamous cell carcinoma; RT‐qPCR, real‐time quantitative polymerase chain reaction
TABLE 1.
Global normalization and relative ratio of corelated 3 miRNAs
| Serum | Cell‐EVs | |||||
|---|---|---|---|---|---|---|
| Global normalization | Global normalization | |||||
| miRNA | CCRT non responder | CCRT responder | Ratio CCRT non responder / CCRT responder | SAS‐R EVs | SAS EVs | Ratio SAS‐R EVs / SAS EVs |
| hsa‐miR‐296‐5p | 117 | 77 | 1.52 | 126 | 122 | 1.1 |
| hsa‐miR‐503‐3p | 16 | 11 | 1.43 | 22 | Not Detected | 25.55 |
| hsa‐miR‐6748‐3p | 11 | 9 | 1.24 | 12 | 5 | 2.69 |
3.4. SAS‐R EVs and miR‐503‐3p suppress the radiation‐induced apoptosis of SAS cells
To elucidate the mechanism underlying radioresistance by SAS‐R EVs and miR‐503‐3p, the apoptotic reaction after irradiation was evaluated. As shown in Figure 4a, SAS cells treated with SAS‐R EVs after irradiation (6 Gy) showed a significant decrease in apoptosis. Similarly, the radiation‐induced apoptosis of SAS cells transfected with miR‐503‐3p was also reduced (Figure 4a). The MitoMP of the irradiated SAS cells treated with SAS‐R EVs was hardly decreased. Similarly, the MitoMP of the irradiated SAS cells transfected with miR‐503‐3p was only very slightly decreased (Figure 4b). We searched for a gene involved in the mitochondrial apoptosis pathway as a target gene for miR‐503‐3p using bioinformatic algorithms (TargetScanHuman, miRDB) and found BAK as a candidate gene. To validate whether BAK mRNA is a direct target of miR‐503‐3p through its cognate site on the 3′UTR of BAK, we performed a luciferase assay. As shown in Figure 4c, miR‐503‐3p significantly reduced the relative luciferase activity of the wild type BAK‐3′UTR reporter gene, but did not reduce the relative luciferase activity of the mutant type BAK‐3′UTR reporter gene. These results indicate that BAK is the target gene for miR‐503‐3p and that miR‐503‐3p specifically binds to the 3′UTR region of BAK mRNA. We also evaluated the expression of BAK in SAS cells after irradiation by western blotting and real‐time RT‐PCR. The irradiated SAS cells had increased BAK expression (Figure S2). However, SAS cells treated with SAS‐R EVs or transfected with miR‐503‐3p had decreased BAK expression after irradiation (Figure 4d).
FIGURE 4.
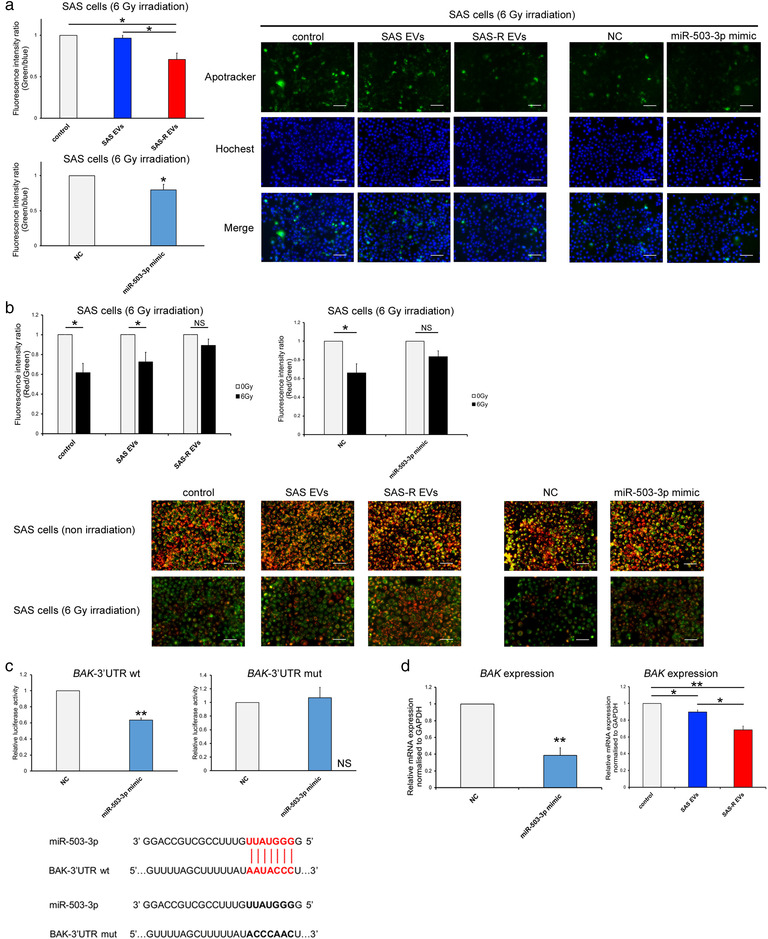
Elucidation of radioresistance acquisition mechanism of OSCC cells. (a) The graph on the left shows the fluorescence intensity ratio of green (Apotracker) and blue (Hochest 33342) in SAS cells 48 h after irradiation with 6 Gy. Each representative immunofluorescence image of Apotracker in SAS cells (treated with phosphate‐buffered saline (control) or SAS EVs or SAS‐R EVs/transfected with negative control (NC) or miR‐503‐3p mimic) is shown on the right, 48 h after irradiation. (b) The change in mitochondrial membrane potential (MitoMP) level of irradiated (6 Gy) SAS cells after EV treatment or miRNA mimic transfection. A representative immunofluorescence image at 48 h after irradiation is shown below the graph. When the MitoMP is high, the cells fluoresce red, and when it is low, they fluoresce green. (c) Luciferase activity of wild type (wt) BAK‐3′UTR reporter gene and mutant type (mut) BAK‐3′UTR reporter gene in SAS cells. Cells were transiently transfected with miR‐503‐3p mimic or negative control. The target region of miR‐503‐3p in the 3′UTR region of BAK is shown on the right of the graph. (d) Expression of BAK in SAS cells (6 Gy irradiated) after EV treatment or miRNA mimic transfection. Values are expressed as mean ± standard deviation of triplicate samples. *p < 0.05; **p < 0.01; OSCC, oral squamous cell carcinoma; NS, no significant differences
3.5. BAK contributes to radiation‐induced apoptosis in OSCC
To confirm whether miR‐503‐3p suppresses radiation‐induced apoptosis and confers radioresistance via silencing BAK expression in irradiated SAS cells, we assessed the effect of BAK knockdown on radiosensitivity by the transfection of BAK siRNA into the irradiated SAS cells. The effects of BAK siRNAs in SAS cells 48 h after transfection on BAK mRNA and BAK protein are shown in Figure S3. To verify the radiosensitivity, SAS cells were irradiated and then transfected with BAK siRNA, and an HDS assay was performed. As shown in Figure 5a, the radioresistance of BAK siRNA‐treated SAS cells was significantly increased. Additionally, the radiation‐induced apoptosis of BAK siRNA‐treated SAS cells was significantly decreased (Figure 5b). MitoMP of BAK siRNA‐treated SAS cells did not decrease (Figure 5c). Confirming our previous experimental results (Figure 4d and Figure S3), BAK mRNA expression in SAS cells significantly decreased due to SAS‐R EVs, miR‐503‐3p, and siBAK treatments (Figure S4). Additionally, although the results of the quantitative analysis varied (Figure S5), the release of cytochrome C and the downstream expressions of Apaf‐1, caspase 9, caspase 3, and cleaved caspase 3 decreased in the irradiated SAS cells treated with SAS‐R EVs or transfected with miR‐503‐3p and BAK siRNA‐treated SAS cells (Figure 5d, Figure S4).
FIGURE 5.
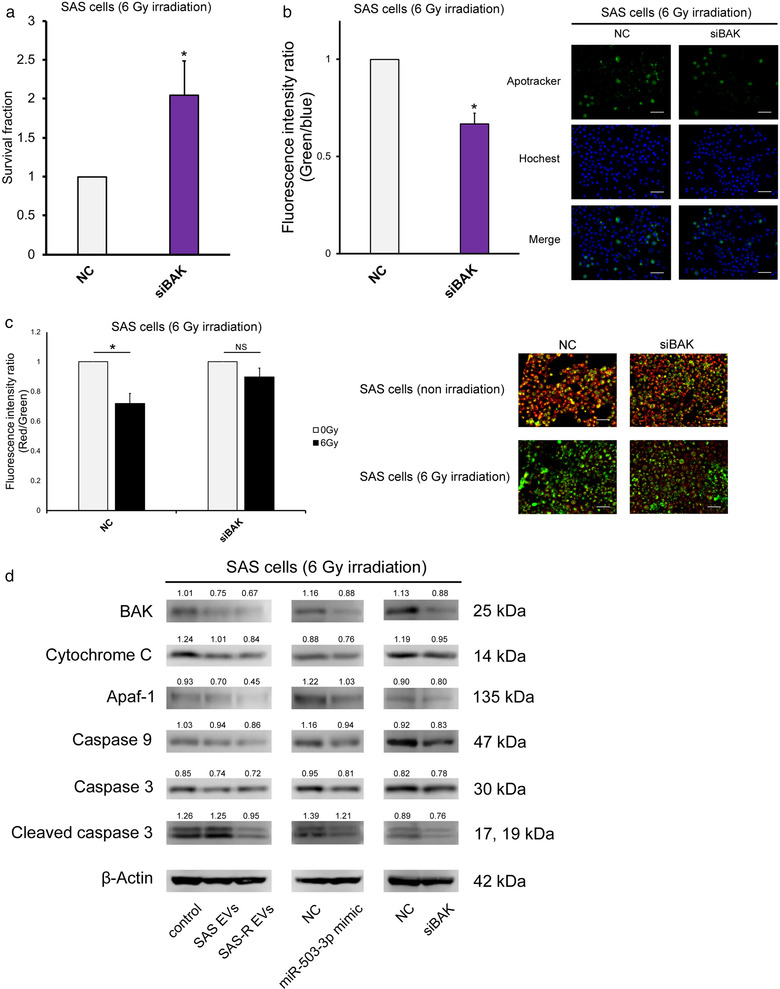
BAK silencing mimics the biological effects of miR‐503‐3p overexpression in SAS cells. (a) SAS cells were exposed to 6 Gy and then transfected with negative control or siBAK; the survival fraction was evaluated by high‐density survival assay. (b) The graph on the left shows the fluorescence intensity ratio of green (Apotracker) and blue (Hochest 33342) 48 h after irradiation with 6 Gy. Each representative immunofluorescence image of Apotracker in SAS cells (transfected with negative control or siBAK) is shown on the right. These images are in the state 48 h after irradiation. (c) Change in mitochondrial membrane potential (MitoMP) level of irradiated (6 Gy) SAS cells after siBAK transfection. A representative immunofluorescence image is shown right the graph (48 h after irradiation). When the MitoMP is high, the cells fluoresce red, and when it is low, they fluoresce green. (d) Western blots of apoptosis molecular component (BAK, cytochrome C, Apaf‐1, caspase 9, caspase3, cleaved caspase 3) in SAS cells 48 h after 6 Gy irradiation. Mean values were obtained using an image analyser (Figure S5) from at least three independent experiments are shown at the top of each band. Values are expressed as mean ± standard deviation of triplicate samples. *p < 0.05; NS, no significant differences
3.6. SAS‐R EVs also confer radiation resistance to other OSCC cell lines via the miR‐503‐3p‐BAK axis
In order to confirm whether the results obtained in the previous experiments can be generalized to OSCC, a verification experiment was conducted using HSC‐2 cells. We confirmed the uptake of SAS EVs and SAS‐R EVs into HSC‐2 cells by fluorescence microscopy (Figure S6). In the HDS assay, the radioresistance of HSC‐2 cells treated with SAS‐R EVs or transfected with miR‐503‐3p mimic or BAK siRNA was significantly increased, similar to SAS cells (Figure 6A). HSC‐2 cells co‐cultured with SAS‐R cells formed more colonies compared to cells co‐cultured with SAS cells (Figure S7). Furthermore, MitoMP of HSC‐2 cells treated with SAS‐R EVs or transfected with miR‐503‐3p mimic or BAK siRNA showed no significant changes compared with each control (Figure 6c). In the case of SAS cells, BAK mRNA expression in HSC‐2 cells significantly decreased due to SAS‐R EVs, miR‐503‐3p, and siBAK treatments (Figure S8). In addition, although the results of the quantitative analysis varied (Figures S8 and S9), the radiation‐induced apoptosis of HSC‐2 cells treated with SAS‐R EVs or transfected with miR‐503‐3p mimic or BAK siRNA also decreased (Figure 6d). These results suggest that EVs secreted from radiation‐resistant OSCCs contribute to the acquisition of radiation resistance to some extent.
FIGURE 6.
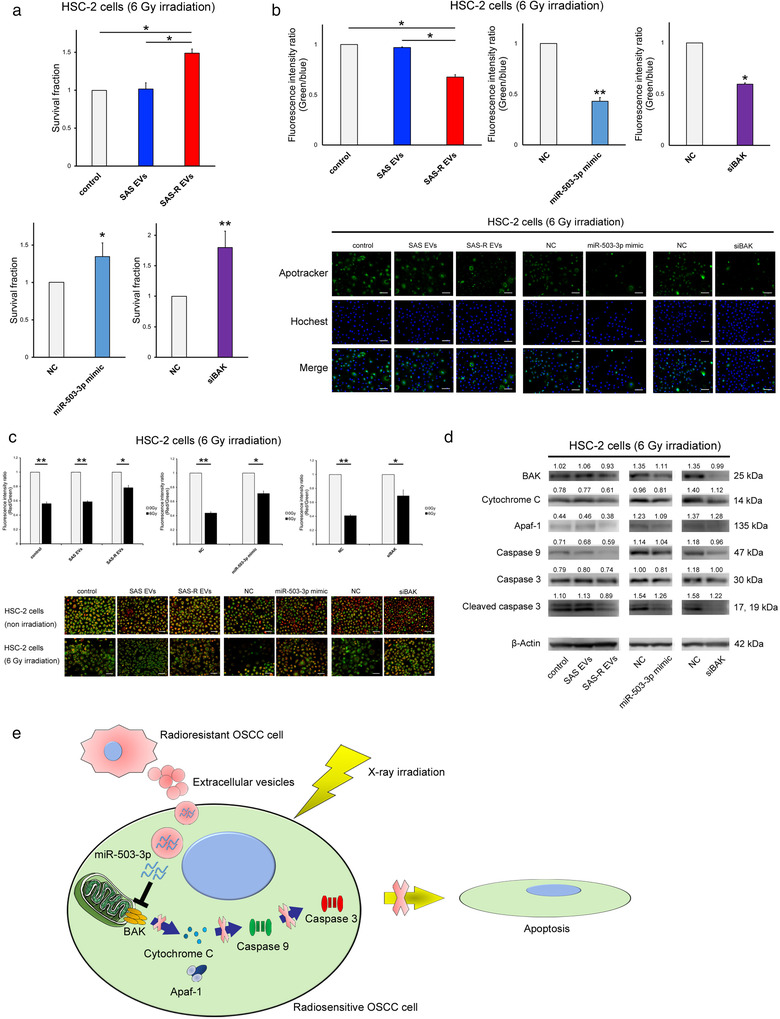
Reproducibility experiment using HSC‐2 cells. (a) HSC‐2 cells were exposed to 6 Gy and then treated with phosphate‐buffered saline (control) or SAS‐R EVs (10 μg) / transfected with negative control (NC) or miR‐503‐3p mimic/transfected with NC or siBAK; the survival fraction was evaluated by HDS assay. (b) Graph showing the fluorescence intensity ratio of green (Apotracker) and blue (Hochest 33342) 48 h after irradiation with 6 Gy. Each representative immunofluorescence image of Apotracker in HSC‐2 cells is shown below the graph. These images are captured 48 h after irradiation. (c) Change in mitochondrial membrane potential (MitoMP) level of irradiated (6 Gy) HSC‐2 cells. A representative immunofluorescence image is shown below the graph (48 h after irradiation). When the MitoMP is high, the cells emit red fluorescence, and when it is low, the cells emit green fluorescence. (d) Western blots of apoptosis molecular component (BAK, cytochrome C, Apaf‐1, caspase 9, caspase 3, cleaved caspase 3) in HSC‐2 cells at 48 h after 6 Gy irradiation. (e) Schematic illustration of the EVs released from radioresistant OSCC cells‐mediated radioresistance model. When radiosensitive or radioresistant OSCC cells were exposed to X‐ray irradiation, EVs released from radioresistant OSCC cells were taken up by the radiosensitive OSCC cells. Subsequently, there was suppression of BAK by miR‐503‐3p contained in EVs, and finally inhibition of the apoptotic pathway controlled by downstream molecules of BAK, and radiosensitive cells acquire radioresistance. Mean values obtained using an image analyser (Figure S9) from at least three independent experiments are shown at the top of each band. Values are expressed as mean ± standard deviation of triplicate samples. *p < 0.05; **p < 0.01
3.7. Clinical significance of circulating miR‐503‐3p in patients with OSCC who underwent CCRT
To confirm the results of in vitro data in clinical OSCC, the clinical significance of circulating miR‐503‐3p expression in OSCC patients who underwent preoperative CRT was examined. A total of 55 OSCC patients were dichotomized into two groups according to their median level of miR‐503‐3p expression. Distribution of the clinical background characteristics of the patients is shown in Table 2 and Table S6. The frequency of miR‐503‐3p‐High patients was significantly higher among cases that showed lymph node metastasis (p = 0.043) and a poor pathological response to preoperative CRT (p = 0.003). No significant difference was found between miR‐503‐3p expression in variables, such as age, sex, primary tumour site, T‐category, lymphovascular invasion (LI), and extracapsular extension (ECE). Additionally, a higher circulating miR‐503‐3p expression level was associated with poorer survival; in the miR‐503‐3p‐High and miR‐503‐3p‐Low groups, the median OS was 29.0 months (95% CI = 31.0–70.5) and 48.0 months (95% CI = 65.8–96.0; p = 0.027; Figure 7a), respectively, and the median DFS was 21.4 months (95% CI = 31.1–68.3) and 50.3 months (95% CI = 58.7–92.9; p = 0.026; Figure 7b), respectively. Furthermore, in the Cox proportional hazards regression model, after adjusting for age, sex, primary site, T‐category, N‐category, differentiation, LI, ECE, and pathological response, the influence of miR‐503‐3p expression on DFS (hazard ratio, 3.008; 95% CI 1.086–9.207; p = 0.033) (Table 3) remained.
TABLE 2.
Correlation between the miR‐503‐3p expression and clinicopathological factors in 55 patients with OSCC
| miR‐503‐3p expression | ||||
|---|---|---|---|---|
| High | Low | |||
| Characteristic | Total | n (%) | n (%) | p‐value |
| Age (years) | ||||
| Median | 67.2 | 66.7 | 67.5 | |
| Range | 39‐85 | 39‐85 | 41‐85 | |
| ≤ 65 | 21 | 11(52.3) | 10(47.7) | 0.864 |
| > 65 | 34 | 17(50) | 17(50) | |
| Sex | ||||
| Male | 32 | 18(56.2) | 14(43.8) | 0.350 |
| Female | 23 | 10(43.5) | 13(56.5) | |
| Primary site | ||||
| Tongue | 16 | 8(50) | 8(50) | 0.881 |
| Mandible | 20 | 9(45) | 11(55) | |
| Maxilla | 6 | 3(50) | 3(50) | |
| Oral floor | 4 | 2(50) | 2(50) | |
| Buccal mucosa | 9 | 6(66.7) | 3(33.3) | |
| T‐category | ||||
| T2 | 18 | 8(44.4) | 10(55.6) | 0.800 |
| T3, T4 | 37 | 20(54.1) | 17(45.9) | |
| N‐category | ||||
| N0 | 23 | 8(34.8) | 15(65.2) | 0.043* |
| ≥ N1 | 32 | 20(62.5) | 12(37.5) | |
| Differentiation | ||||
| Well | 43 | 22(51.1) | 21(48.9) | 0.943 |
| Moderate, poor | 12 | 6(50) | 6(50) | |
| Lymphovascular invasion | ||||
| Yes | 3 | 1(33.3) | 2(66.7) | 0.531 |
| No | 52 | 27(51.9) | 25(48.1) | |
| Extracapsular extension | ||||
| Yes | 5 | 3(60) | 2(40) | 0.899 |
| No | 27 | 17(62.3) | 10(37.7) | |
| Pathological response | ||||
| Grade I, IIa (poor response) | 12 | 8(66.7) | 4(33.3) | 0.003** |
| Grade IIb (partical response) | 13 | 6(46.1) | 7(53.9) | |
| Grade ≥ III (complete response) | 30 | 14(46.7) | 16(53.3) | |
| Recurrence | ||||
| Yes | 12 | 9(75) | 3(25) | 0.059 |
| No | 43 | 19(44.2) | 24(55.8) | |
The chi‐square test was used to examine the relationships between miR‐503‐3p expression and clinicopathologic factors. *p < 0.05 and **p < 0.01.
Abbreviation: OSCC, oral squamous cell carcinoma.
FIGURE 7.
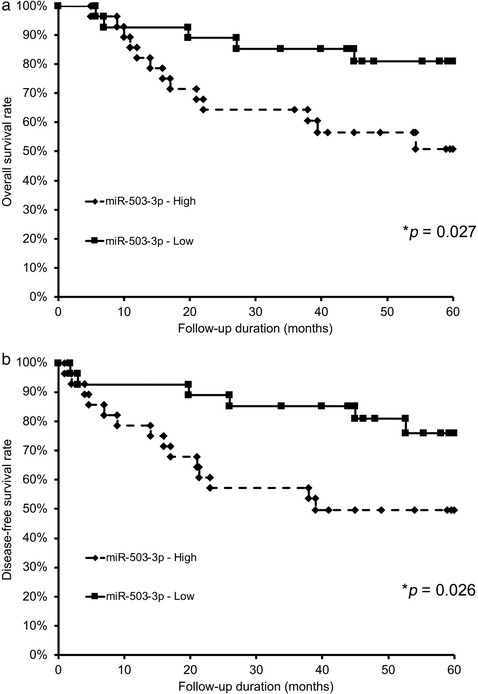
Relationships between circulating miR‐503‐3p expression and survival in patients with OSCC. For the Kaplan–Meier survival analysis, the patients were divided into two groups based on high or low miR‐503‐3p expression. (a) Overall survival of the 55 OSCC patients based on miR‐503‐3p expression. (b) Disease‐free survival of 55 OSCC patients based on miR‐503‐3p expression. OSCC, oral squamous cell carcinoma
4. DISCUSSION
In the research on EVs, the EV extraction process and quality checks are extremely important (Thery et al., 2018). Therefore, we isolated EVs from OSCC cells according to the MISEV2018 (Thery et al., 2018) and evaluated their quality. The vesicles obtained by the isolation procedures in our study would be appropriately classified as sEVs, according to MISEV2018 (Thery et al., 2018). There were no significant differences between SAS EVs and SAS‐R EVs in their composition, size, or amount of release (Figure 1). In past studies, highly malignant cells, such as metastatic cells, released more EVs than primary cells, and the size of EVs became larger (Ono et al., 2018). In contrast to previous study, since SAS‐R cells were established by irradiating the SAS cells with more than 60 Gy, there was assumed to be no difference in the essence of both cells (Livak & Schmittgen, 2001). In quality confirmation using molecular markers, the expressions of ALIX and GAPDH varied among EVs, and the expression patterns differed from those of the derived cells. In general, proteins that are not expressed in cells are not included in EVs. On the other hand, Yoshioka et al. reported that the expression patterns of intracellular proteins and proteins in EVs were not always positively correlated in the verification using various cell lines (Yoshioka et al., 2013). Some proteins are possibly selectively loaded into EVs, whereas some proteins are not. Regarding ALIX, the expression of the cell‐extracted protein differed between SAS and SAS‐R (Figure 1a), which is considered to reflect this phenomenon. On the other hand, GAPDH was probably not loaded into EVs for some reason. In any case, research on EVs molecular markers in the field of oral cancer is still underway, and research on EVs‐specific molecular markers derived from oral cancer is expected in the future (Yap et al., 2020). There are multiple methods for EVs isolation, each with advantages and disadvantages (Thery et al., 2018). Therefore, a quality check is indispensable for any isolation methods, and the EVSecond L70 SEC column is a method that can isolate EVs with a quality that can be sufficiently used for experiments, at least in OSCC cells.
At present, there is no consensus on the effect of EVs on proliferation in cancer cells (Jella et al., 2014; Pace et al., 2019; Qu et al., 2009; Raimondo et al., 2015). In the present study, there was no change in the proliferation of SAS cells treated with SAS EVs or SAS‐R EVs. However, the significant increase of radioresistance in SAS cells and EV uptake was confirmed in co‐culture and EV tracking system (Figure 2b, c). These results demonstrate that SAS‐R EVs are involved explicitly in conferring radioresistance to radiosensitive cells regardless of their proliferative activity. Recently, Mutschelknaus et al. (Mutschelknaus et al., 2017; Mutschelknaus et al., 2016) reported that in head and neck cancer, radiation increases the uptake of EVs from cells and that the administration of EVs after irradiation increased radioresistance. Although no significant differences were found in our preliminary experiments, the uptake of EVs and radioresistance in SAS cells tended to increase after irradiation (Figure S10). Moreover, EVs had no significant effect on radiosensitivity in SAS cells treated with EVs before irradiation (Figure S11). These results indicate that EVs exert their effects by acting specifically on the site where cells are irradiated and some kind of damage has occurred. On the contrary, in contrast to the reports by Mutschelknaus et al. (Mutschelknaus et al., 2017; Mutschelknaus et al., 2016), EVs from irradiated SAS cells did not affect the radioresistance of SAS cells (data not shown). Collectively, the present data indicate that SAS‐R EVs potentially have a higher ability for radiosensitivity regulation compared to SAS EVs.
The regulation of malignant phenotypes by miRNA contained in EVs has been well reported (Kulkarni et al., 2019). However, although there are several reports of EV‐miRNA profiles in head and neck cancer (Langevin et al., 2017; Li et al., 2016), no studies have focused on radioresistance. When considering that there is no difference in the amount of release between SAS EVs and SAS‐R EVs, the difference in components, such as miRNA contained in EVs, may have a great influence on EVs conferring radioresistance to SAS cells. Therefore, we focused on specific miRNA profiles contained in EVs derived from CRR cells and blood samples of patients whose response to CCRT was poor. We found miR‐503‐3p as the candidate miRNA involved in EV‐mediated radioresistance in OSCC (Figure 3). Recently, it was discovered that microRNAs can be actively sorted into exosomes in complex with proteins, such as Argonaute, then released into the circulation (Bell & Taylor, 2016). Indeed, in several malignancies, positive correlations were found between tumour‐specific miRNA in the tumour and exosomes in circulation (Rabinowits et al., 2009; Taylor & Taylor, 2008; Tsukamoto et al., 2017). We also confirmed the positive correlation between miR‐503‐3p in cancer cells and EVs (Figure 3b). Therefore, the present data suggest that CRR cells carry and release radioresistance‐related miRNAs in the tumour microenvironment via exosomes. To date, there have been no studies in OSCC that have screened candidate miRNAs using cultured cells and patient fluid samples. Therefore, the results of the present study could likely be applied to clinical practice, especially as a liquid biopsy. Additionally, although it has been reported that miR‐503‐3p promotes the apoptosis of lung cancer cells and promotes the epithelial‐mesenchymal transition of breast cancer (Sun et al., 2017; Zhao et al., 2016), the present study is the first to report the relationship between miR‐503‐3p and radioresistance in a malignant tumour.
In the present study, the forced expression of miR‐503‐3p clearly showed the same results in EVs uptake experiments. These results strongly support the hypothesis that SAS‐R EVs are taken up into radiosensitive cells by radiation stimulation and that miR‐503‐3p contained in SAS‐R EVs confer radioresistance. Generally, it has been regarded that irradiation induces various cell death pathways, such as apoptosis, mitotic catastrophe, necrosis, and autophagy, in cancer cells and exerts a therapeutic effect by suppressing cell proliferation (Baskar et al., 2012). Because apoptosis is a typical example of radiation‐induced cell death (Dewey et al., 1995; Rupnow & Knox, 1999), we focused on apoptosis‐related genes and selected BAK as a target gene related to radiation‐induced apoptosis by in silico analysis. BAK is a member of the BCL‐2 family of proteins that permeates mitochondrial membranes, activates the caspase cascade, and promotes apoptosis (Edlich, 2018; Eskes et al., 1998). There are several reports on the BAK‐related apoptotic response after irradiation. Przemeck et al. (Przemeck et al., 2007) reported that the radiation‐induced apoptosis of gastric epithelial cells is regulated by BAK. Choi et al. (Choi et al., 2006) reported that activation of the p38 MAPK pathway in response to radiation signals conformational changes in BAK, resulting in mitochondrial activation‐mediated apoptotic cell death in human non‐small cell lung cancer cells. Under either EV‐treatment or miR‐503‐3p‐treatment conditions, the present data were in agreement with these reports (Figure 4). Moreover, in our BAK knockdown experiments, the decrease of radiosensitivity was also observed in SAS cells as with EV treatment and miR‐503‐3p treatment. In addition, it was suggested that SAS‐R EVs confer radiation resistance in HSC‐2 cells as well as in SAS cells using the miR‐503‐3p‐BAK axis (Figure 6). Taken together, the findings indicate the involvement of the miR‐503‐3p–BAK axis in the EV‐mediated acquisition of radioresistance by regulating apoptosis in OSCC (Figure 6e). On the contrary, miRNAs as well as proteins and lipids are involved in the regulation of cancer malignancy by EVs (Raposo & Stoorvogel, 2013). Interestingly, Fitzgerald et al. (Fitzgerald et al., 2018) reported the existence of a novel cell–cell communication mechanism through cytokines encapsulated in EVs. As not all results in the present study can be explained by miR‐503 encapsulated in EVs, the regulatory mechanism for radioresistance conferred by cytokines contained in EVs should be investigated in future studies.
Several studies are associated with cancer treatment resistance and miRNA; in pancreatic ductal adenocarcinoma, high expression of miR‐155‐5p in serum and tumour correlates with resistance to chemotherapy and poor prognosis (Greither et al., 2010; Mikamori et al., 2017). Pang et al. (Pang et al., 2016) reported a low expression of miR‐497‐5p in the plasma of patients with osteosarcoma resistant to chemotherapy. We propose miR‐503‐3p as a candidate biomarker based on the in vitro results and clinical samples herein. As a result of the verification using patient serum, high expression of circulating miR‐503‐3p was correlated with a poor pathological response to CRT (Table 2). Additionally, high expression of circulating miR‐503‐3p was also associated with the shortening of OS and DFS (Figure 7). Furthermore, circulating miR‐503‐3p status is one of the independent prognostic factors in the present cohort (Table 3). However, our preliminary experiments showed that the induced expression of miR503‐3p has no significant effect on the sensitivity of 5‐FU treatment in vitro (data not shown). Therefore, these results suggest that serum miR‐503‐3p is a useful biomarker for predicting the therapeutic effects of radiotherapy on and the prognosis of patients with OSCC. In addition, in our preliminary experiments, we found that in some cases EVs contained very small amounts or only about half of the miRNAs in serum (data not shown), which is consistent with several recent reports (Arroyo et al., 2011; Endzelins et al., 2017; Otahal et al., 2021). Especially, it is now recognized that not all miRNAs in the blood are derived from EVs, and in some cases there may be a certain amount of miRNAs present in the Ago2 ribonucleoprotein complex or in other states (Arroyo et al., 2011). Therefore, it is important to consider these possibilities when applying the results of this study to actual clinical practice.
TABLE 3.
Multivariate analysis of prognostic factors by the Cox proportional hazards regression models
| OS | DFS | ||||
|---|---|---|---|---|---|
| Variables | n | Hazard ratio (95% CI) | p‐value | Hazard ratio (95% CI) | p‐value |
| T‐category | |||||
| T3, T4 | 37 | 3.842 (1.202–17.289) | 0.021* | 3.145 (0.947–14.456) | 0.062 |
| T2 | 18 | ||||
| N‐category | |||||
| ≧ N1 | 32 | 1.334 (0.485–3.985) | 0.579 | 2.224 (0.735–8.204) | 0.161 |
| N0 | 23 | ||||
| Differentiation | |||||
| Moderate, poor | 12 | 3.121 (1.145–8.168) | 0.027* | 2.697 (0.916–7.501) | 2.697 |
| Well | 43 | ||||
| Lymphovascular invasion | |||||
| Yes | 3 | 0.248 (0.012–1.507) | 0.145 | 3.117 (0.496–61.798) | 0.254 |
| No | 52 | ||||
| Extracapsular extension | |||||
| Yes | 5 | 3.348 (0.819–11.647) | 0.088 | 2.685 (0.642–9.616) | 0.164 |
| No | 27 | ||||
| Pathological response | |||||
| Grade I, II | 25 | 2.907 (1.097–8.532) | 0.031* | 2.204 (0.774–6.739) | 0.138 |
| Grade ≧ III | 30 | ||||
| miR‐503‐3p expression | |||||
| High | 28 | 3.006 (1.086–9.207) | 0.033* | 2.803 (0.948–9.421) | 0.062 |
| Low | 27 | ||||
Progressive advances in the development of nucleic acid drugs using miRNA have been made in recent years, and these drugs have been used in the treatment of patients with liver diseases (Janssen et al., 2013; van der Ree et al., 2017). Our results may lead to drug discovery and development of new therapies targeting miR‐503‐3p or EVs derived from radiation‐resistant OSCC. However, our findings are based on in vitro experiments, and especially the experiments of introducing miRNA into cells have a strong artificial aspect. Therefore, future studies are required to verify the results of this study in vivo.
There are some limitations to the present study. It is necessary to examine how CRR cell‐derived EVs and miR‐503‐3p affect stromal cells, such as fibroblasts and macrophages. Additionally, as the present clinicopathological analysis targeted patients who received 5‐FU‐based preoperative chemoradiotherapy in our previous Phase 2 study (Nomura et al., 2010), it is necessary to verify the clinical significance of miR‐503‐3p in patients with OSCC treated with CDDP‐based chemoradiotherapy, which is the standard treatment for head and neck cancer. In addition, the significant role of miR‐503‐3p in regulating radiation‐induced apoptosis is unclear. Furthermore, the components that control DNA double‐strand break repair are possibly also involved in the acquisition of radiation resistance of cells. Furthermore, the present study also demonstrates that miR‐503‐3p is possibly involved, not only in the radioresistance of OSCC but possibly in the control of other malignant phenotypes as well, based on its correlation with clinicopathological factors (Table 2) and the results of the miR‐503‐3p forced expression experiments (Figures 2 and 3). Finally, in this study, we focused on miRNAs that are altered between radioresistant and sensitive OSCC cells and those that confer radioresistance. However, the approach of extracting miRNAs with radiosensitizing effects using methods such as high‐throughput screening to elucidate the radiotolerance mechanism may also be useful, and further research is warranted. Despite these limitations, the present study is the first to elucidate that EVs released from CRR cells confer radioresistance to surrounding cells. Furthermore, we first demonstrated that the miR‐503‐3p–BAK axis regulates radiation‐induced apoptosis and contributes to acquiring radiation resistance in OSCC. Furthermore, we discovered that circulating miR‐503‐3p in the serum of patients with OSCC was a potential biomarker. These findings will provide new insights into the study of the relationship between EVs and treatment resistance, especially radiation resistance, in OSCC.
CONFLICT OF INTERESTS
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
ETHICAL APPROVAL
This study was performed with the approval of the Ethics Committee of Kumamoto University (approval number: 174, 1427) and in accordance with the Good Clinical Practice and the Declaration of Helsinki guidelines.
Supporting information
Supporting Information
ACKNOWLEDGEMENTS
The authors would like to thank Enago (www.enago.jp) for the English language review. The author(s) declare the receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by a Grant‐in‐Aid for Scientific Research (C) (18K09771) from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Yamana, K. , Inoue, J. , Yoshida, R. , Sakata, J. , Nakashima, H. , Arita, H. , Kawaguchi, S. , Gohara, S. , Nagao, Y. , Takeshita, H. , Maeshiro, M. , Liu, R. , Matsuoka, Y. , Hirayama, M. , Kawahara, K. , Nagata, M. , Hirosue, A. , Toya, R. , Murakami, R. , … Nakayama, H. (2021). Extracellular vesicles derived from radioresistant oral squamous cell carcinoma cells contribute to the acquisition of radioresistance via the miR‐503‐3p‐BAK axis. Journal of Extracellular Vesicles, 10, e12169. 10.1002/jev2.12169
Keisuke Yamana and Junki Inoue contributed equally.
Ryoji Yoshida and Hideki Nakayama are co‐corresponding authors.
REFERENCES
- Arroyo, J. D. , Chevillet, J. R. , Kroh, E. M. , Ruf, I. K. , Pritchard, C. C. , Gibson, D. F. , Mitchell, P. S. , Bennett, C. F. , Pogosova‐Agadjanyan, E. L. , Stirewalt, D. L. , Tait, J. F. , & Tewari, M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. PNAS, 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar, R. , Lee, K. A. , Yeo, R. , & Yeoh, K. W. (2012). Cancer and radiation therapy: Current advances and future directions. International Journal of Medical Sciences, 9(3), 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, A. , Thakur, B. K. , Weiss, J. M. , Kim, H. S. , Peinado, H. , & Lyden, D. (2016). Extracellular vesicles in cancer: Cell‐to‐cell mediators of metastasis. Cancer Cell, 30: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, E. , & Taylor, M. A. (2016). Functional roles for exosomal microRNAs in the tumour microenvironment. Computational and Structural Biotechnology Journal, 15: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L. , Sharples, R. A. , Scicluna, B. J. , & Hill, A. F. (2014). Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell‐free blood. Journal of Extracellular Vesicles, 3: 23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S. Y. , Kim, M. J. , Kang, C. M. , Bae, S. , Cho, C. K. , Soh, J. W. , Kim, J. H. , Kang, S. , Chung, H. Y. , Lee, Y. S. , & Lee, S. J. (2006). Activation of Bak and Bax through c‐abl‐protein kinase Cdelta‐p38 MAPK signaling in response to ionizing radiation in human non‐small cell lung cancer cells. Journal of Biological Chemistry, 281(11), 7049–7059. [DOI] [PubMed] [Google Scholar]
- Dewey, W. C. , Ling, C. C. , & Meyn, R. E. (1995). Radiation‐induced apoptosis: Relevance to radiotherapy. International Journal of Radiation and Oncology in Biology and Physics, 33(4), 781–796. [DOI] [PubMed] [Google Scholar]
- Edge, S. , Byrd, D. R. , & Compton, C. C. (2010). AJCC cancer staging manual (7th ed.). New York: Springer. [Google Scholar]
- Edlich, F. (2018). BCL‐2 proteins and apoptosis: Recent insights and unknowns. Biochemical and Biophysical Research Communications, 500(1), 26–34. [DOI] [PubMed] [Google Scholar]
- Endzelins, E. , Berger, A. , Melne, V. , Bajo‐Santos, C. , Sobolevska, K. , Abols, A. , Rodriguez, M. , Santare, D. , Rudnickiha, A. , Lietuvietis, V. , Llorente, A. , & Line, A. (2017). Detection of circulating miRNAs: Comparative analysis of extracellular vesicle‐incorporated miRNAs and cell‐free miRNAs in whole plasma of prostate cancer patients. BMC Cancer [Electronic Resource], 17(1), 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes, R. , Antonsson, B. , Osen‐Sand, A. , Montessuit, S. , Richter, C. , Sadoul, R. , Mazzei, G. , Nichols, A. , & Martinou, J. C. (1998). Bax‐induced cytochrome C release from mitochondria is independent of the permeability transition pore but highly dependent on Mg2+ ions. Journal of Cell Biology, 143: 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskaa, T. , Knutsen, E. , Nikolaisen, M. A. , Jorgensen, T. E. , Johansen, S. D. , Perander, M. , & Seternes, O. M. (2016). Distinct small RNA signatures in extracellular vesicles derived from breast cancer cell lines. Plos One, 11: E0161824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, W. , Freeman, M. L. , Lederman, M. M. , Vasilieva, E. , Romero, R. , & Margolis, L. (2018). A system of cytokines encapsulated in extracellular vesicles. Scientific Reports, 8(1), 8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon, R. , Marcucci, G. , & Croce, C. M. (2010). Targeting microRNAs in cancer: Rationale, strategies and challenges. Nature Reviews Drug Discovery, 9(10), 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greither, T. , Grochola, L. F. , Udelnow, A. , Lautenschlager, C. , Wurl, P. , & Taubert, H. (2010). Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. International Journal of Cancer, 126: 73–80. [DOI] [PubMed] [Google Scholar]
- Gupta, S. , Kong, W. , Peng, Y. , Miao, Q. , & Mackillop, W. J. (2009). Temporal trends in the incidence and survival of cancers of the upper aerodigestive tract in Ontario and the United States. International Journal of Cancer, 125(9), 2159–2165. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. , & Weinberg, R. A. (2011). Hallmarks of cancer: The next generation. Cell, 144(5), 646–674. [DOI] [PubMed] [Google Scholar]
- Hu, G. , Drescher, K. M. , & Chen, X. M. (2012). Exosomal miRNAs: Biological properties and therapeutic potential. Frontiers in Genetics, 3: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, H. L. , Reesink, H. W. , Lawitz, E. J. , Zeuzem, S. , Rodriguez‐Torres, M. , Patel, K. , van der Meer, A. J. , Patrick, A. K. , Chen, A. , Zhou, Y. , Persson, R. , King, B. D. , Kauppinen, S. , Levin, A. A. , & Hodges, M. R. (2013). Treatment of HCV infection by targeting microRNA. New England Journal of Medicine, 368(18), 1685–1694. [DOI] [PubMed] [Google Scholar]
- Jella, K. K. , Rani, S. , O'Driscoll, L. , McClean, B. , Byrne, H. J. , & Lyng, F. M. (2014). Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiation Research, 181(2), 138–145. [DOI] [PubMed] [Google Scholar]
- Joyce, D. P. , Kerin, M. J. , & Dwyer, R. M. (2016). Exosome‐encapsulated microRNAs as circulating biomarkers for breast cancer. International Journal of Cancer, 139(7): 1443–1448. [DOI] [PubMed] [Google Scholar]
- Khoo, X. H. , Paterson, I. C. , Goh, B. H. , & Lee, W. L. (2019). Cisplatin‐ resistance in oral squamous cell carcinoma: Regulation by tumor cell‐derived extracellular vesicles. Cancers, 11(8), 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhapp, F. J. , Mitra, A. K. , Lengyel, E. , & Peter, M. E. (2015). MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene, 34(48), 5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka, N. , Yoshioka, Y. , Hagiwara, K. , Tominaga, N. , Katsuda, T. , & Ochiya, T. (2013). Trash or treasure: Extracellular microRNAs and cell‐to‐cell communication. Frontiers in Genetics, 4:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, B. , Kirave, P. , Gondaliya, P. , Jash, K. , Jain, A. , Tekade, R. K. , & Kalia, K. (2019). Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discovery Today, 24(10), 2058–2067. [DOI] [PubMed] [Google Scholar]
- Kuwahara, Y. , Mori, M. , Oikawa, T. , Shimura, T. , Ohtake, Y. , Mori, S. , Ohkubo, Y. , & Fukumoto, M. (2010). The modified high‐density survival assay is the useful tool to predict the effectiveness of fractionated radiation exposure. Journal of Radiation Research, 51(3), 297–302. [DOI] [PubMed] [Google Scholar]
- Langevin, S. , Kuhnell, D. , Parry, T. , Biesiada, J. , Huang, S. , Wise‐Draper, T. , Casper, K. , Zhang, X. , Medvedovic, M. , & Kasper, S. (2017). Comprehensive microRNA‐sequencing of exosomes derived from head and neck carcinoma cells in vitro reveals common secretion profiles and potential utility as salivary biomarkers. Oncotarget, 8(47), 82459–82474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Li, C. , Wang, S. , Wang, Z. , Jiang, J. , Wang, W. , Li, X. , Chen, J. , Liu, K. , Li, C. , & Zhu, G. (2016). Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR‐21 to normoxic cells to elicit a prometastatic phenotype. Cancer Research, 76(7), 1770–1780. [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2–ΔΔCT method. Methods (San Diego, Calif.), 25(4), 402–408. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y. , Nakayama, H. , Yoshida, R. , Hirosue, A. , Nagata, M. , Tanaka, T. , Kawahara, K. , Sakata, J. , Arita, H. , Nakashima, H. , Shinriki, S. , Fukuma, D. , Ogi, H. , Hiraki, A. , Shinohara, M. , Toya, R. , & Murakami, R. (2016). IL‐6 controls resistance to radiation by suppressing oxidative stress via the Nrf2‐antioxidant pathway in oral squamous cell carcinoma. British Journal of Cancer, 115(10), 1234–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikamori, M. , Yamada, D. , Eguchi, H. , Hasegawa, S. , Kishimoto, T. , Tomimaru, Y. , Asaoka, T. , Noda, T. , Wada, H. , Kawamoto, K. , Gotoh, K. , Takeda, Y. , Tanemura, M. , Mori, M. , & Doki, Y. (2017). MicroRNA‐155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Scientific Reports, 7: 42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschelknaus, L. , Azimzadeh, O. , Heider, T. , Winkler, K. , Vetter, M. , Kell, R. , Tapio, S. , Merl‐Pham, J. , Huber, S. M. , Edalat, L. , Radulovic, V. , Anastasov, N. , & Atkinson, M. J. (2017). Moertl S. Radiation alters the cargo of exosomes released from squamous head and neck cancer cells to promote migration of recipient cells. Scientific Reports, 7(1), 12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschelknaus, L. , Peters, C. , Winkler, K. , Yentrapalli, R. , Heider, T. , Atkinson, M. J. , & Moertl, S. (2016). Exosomes derived from squamous head and neck cancer promote cell survival after ionizing radiation. Plos One, 11(3), e0152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, P. P. , Das, D. N. , Panda, P. K. , Mukhopadhyay, S. , Sinha, N. , Praharaj, P. P. , Agarwal, R. , & Bhutia, S. K. (2016). Implications of cancer stem cells in developing therapeutic resistance in oral cancer. Oral Oncology, 62: 122–135. [DOI] [PubMed] [Google Scholar]
- Naito, Y. , Yoshioka, Y. , Yamamoto, Y. , & Ochiya, T. (2017). How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cellular and Molecular Life Sciences, 74: 697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, T. , Murakami, R. , Toya, R. , Teshima, K. , Nakahara, A. , Hirai, T. , Hiraki, A. , Nakayama, H. , Yoshitake, Y. , Ota, K. , Obayashi, T. , Yamashita, Y. , Oya, N. , & Shinohara, M. (2010). Phase II study of preoperative concurrent chemoradiation therapy with S‐1 in patients with T4 oral squamous cell carcinoma. International Journal of Radiation and Oncology in Biology and Physics; 76(5), 1347–1352. [DOI] [PubMed] [Google Scholar]
- Ono, K. , Eguchi, T. , Sogawa, C. , Calderwood, S. K. , Futagawa, J. , Kasai, T. , Seno, M. , Okamoto, K. , Sasaki, A. , & Kozaki, K. (2018). HSP‐enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. Journal of Cellular Biochemistry, 119(9), 7350–7362. [DOI] [PubMed] [Google Scholar]
- Otahal, A. , Kuten‐Pella, O. , Kramer, K. , Neubauer, M. , Lacza, Z. , Nehrer, S. , & De Luna, A. (2021). Functional repertoire of EV‐associated miRNA profiles after lipoprotein depletion via ultracentrifugation and size exclusion chromatography from autologous blood products. Scientific Reports, 11: 5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, K. R. , Dutt, R. , & Galileo, D. S. (2019). Exosomal L1CAM stimulates glioblastoma cell motility, proliferation, and invasiveness. International Journal of Molecular Sciences, 20(16), 3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P. C. , Shi, X. Y. , Huang, W. L. , & Sun, K. (2016). miR‐497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. European Review for Medical and Pharmacological Sciences, 20: 3765–3769. [PubMed] [Google Scholar]
- Pfeffer, S. R. , Grossmann, K. F. , Cassidy, P. B. , Yang, C. H. , Fan, M. , Kopelovich, L. , Leachman, S. A. , & Pfeffer, L. M. (2015). Detection of exosomal miRNAs in the plasma of melanoma patients. Journal of Clinical Medicine, 4: 2012–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck, S. M. , Duckworth, C. A. , & Pritchard, D. M. (2007). Radiation‐induced gastric epithelial apoptosis occurs in the proliferative zone and is regulated by p53, bak, bax, and bcl‐2. American Journal of Physiology. Gastrointestinal and Liver Physiology, 292(2), 620–627 [DOI] [PubMed] [Google Scholar]
- Qu, J.‐L. , Qu, X.‐J. , Zhao, M.‐F. , Teng, Y.‐E. , Zhang, Y. , Hou, K.‐Z. , Jiang, Y.‐H. , Yang, X.‐H. , & Liu, Y.‐P. (2009). Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Digestive and Liver Disease, 41: 875–880. [DOI] [PubMed] [Google Scholar]
- Rabinowits, G. , Taylor, C. G. , Day, J. M. , Taylor, D. D. , & Kloecker, G. H. (2009). Exosomal microRNA: A diagnostic marker for lung cancer. Clinical Lung Cancer, 10(1), 42–46. [DOI] [PubMed] [Google Scholar]
- Raimondo, S. , Saieva, L. , Corrado, C. , Fontana, S. , Flugy, A. , Rizzo, A. , Leo, G. D. , & Alessandro, R. (2015). Chronic myeloid leukemia‐derived exosomes promote tumor growth through an autocrine mechanism. Cell Communication and Signaling, 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo, G. , & Stoorvogel, W. (2013). Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology, 200(4): 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnow, B. A. , & Knox, S. J. (1999). The role of radiation‐induced apoptosis as a determinant of tumor responses to radiation therapy. Apoptosis, 4: 115–143. [DOI] [PubMed] [Google Scholar]
- Sento, S. , Sasabe, E. , & Yamamoto, T. (2016). Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell‐derived exosomes. Plos One, 11: E0148454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimosato, Y. , Oboshi, S. , & Baba, K. (1971). Histological evaluation of effects of radiotherapy and chemotherapy for carcinomas. Japanese Journal of Clinical Oncology, 1: 19–35. [Google Scholar]
- Siegel, R. L. , Miller, K. D. , & Jemal, A. (2017). Cancer statistics, 2017. CA: A Cancer Journal for Clinicians, 67(1), 7–30. [DOI] [PubMed] [Google Scholar]
- Summerer, I. , Unger, K. , Braselmann, H. , Schuettrumpf, L. , Maihoefer, C. , Baumeister, P. , Kirchner, T. , Niyazi, M. , Sage, E. , Specht, H. M. , Multhoff, G. , Moertl, S. , Belka, C. , & Zitzelsberger, H. (2015). Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. British Journal of Cancer, 113(1), 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Li, L. , Xing, S. , Pan, Y. , Shi, Y. , Zhang, L. , & Shen, Q. (2017). miR‐503‐3p induces apoptosis of lung cancer cells by regulating p21 and CDK4 expression. Cancer Biomarkers, 20: 597–608. [DOI] [PubMed] [Google Scholar]
- Swarup, V. , & Rajeswari, M. R. (2007). Circulating (cell‐free) nucleic acids–a promising, non‐invasive tool for early detection of several human diseases. Febs Letters; 581: 795–799. [DOI] [PubMed] [Google Scholar]
- Taylor, D. D. , & Taylor, C. G. (2008). MicroRNA signatures of tumor‐derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology, 110(1), 13–21. [DOI] [PubMed] [Google Scholar]
- Thery, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga, N. , Kosaka, N. , Ono, M. , Katsuda, T. , Yoshioka, Y. , Tamura, K. , Lotvall, J. , Nakagama, H. , & Ochiya, T. (2015). Brain metastatic cancer cells release microRNA‐181c‐containing extracellular vesicles capable of destructing blood‐brain barrier. Nature Communications, 6: 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto, M. , Iinuma, H. , Yagi, T. , Matsuda, K. , & Hashiguchi, Y. (2017). Circulating exosomal microRNA‐21 as a biomarker in each tumor stage of colorectal cancer. Oncology, 92(6), 360–370. [DOI] [PubMed] [Google Scholar]
- Valadi, H. , Ekstrom, K. , Bossios, A. , Sjostrand, M. , Lee, J. L. , & Lotvall, J. O. (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 9(6), 654–659. [DOI] [PubMed] [Google Scholar]
- van der Ree, M. H. , de Vree, J.M. , Stelma, F. , Willemse, S. , van der Valk, M. , Rietdijk, S. , Molenkamp, R. , Schinkel, J. , van Nuenen, A. C. , Beuers, U. , Hadi, S. , Harbers, M. , van der Veer, E. , Liu, K. , Grundy, J. , Patick, A. K. , Pavlicek, A. , Blem, J. , Huang, M. , … Reesink, H. W. (2017). Safety, tolerability, and antiviral effect of RG‐101 in patients with chronic hepatitis C: A phase 1B, double‐blind, randomized controlled trial. Lancet, 389(10070): 709–717. [DOI] [PubMed] [Google Scholar]
- Wang, J. J. , Lei, K. F. , & Han, F. (2018). Tumor microenvironment: recent advances in various cancer treatments. European Review for Medical and Pharmacological Sciences, 22(12), 3855–3864. [DOI] [PubMed] [Google Scholar]
- Xie, C. , Du, L. Y. , Guo, F. , Li, X. , & Cheng, B. (2019). Exosomes derived from microRNA‐101‐3p‐overexpressing human bone marrow mesenchymal stem cells suppress oral cancer cell proliferation, invasion, and migration. Molecular and Cellular Biochemistry, 458(1‐2), 11–26 [DOI] [PubMed] [Google Scholar]
- Yap, T. , Pruthi, N. , Seers, C. , Belobrov, S. , McCullough, M. , & Celentano, A. (2020). Extracellular vesicles in oral squamous cell carcinoma and oral potentially malignant disorders: A systematic review. International Journal of Molecular Sciences; 21(4), 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka, Y. , Konishi, Y. , Kosaka, N. , Katsuda, T. , Kato, T. , & Ochiya, T. (2013). Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of Extracellular Vesicles, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. , Fan, X. , Jiang, L. , Xu, Z. , Xue, L. , Zhan, Q. , & Song, Y. (2016). miR‐503‐3p promotes epithelial‐mesenchymal transition in breast cancer by directly targeting SMAD2 and E‐cadherin. Journal of Genetics and Genome, 44: 75–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


